Abstract
Skeletal muscle possesses a remarkable capacity to regenerate when injured, but when confronted with major traumatic injury resulting in volumetric muscle loss (VML), the regenerative process consistently fails. The loss of muscle tissue and function from VML injury has prompted development of a suite of therapeutic approaches but these strategies have proceeded without a comprehensive understanding of the molecular landscape that drives the injury response. Herein, we administered a VML injury in an established rodent model and monitored the evolution of the healing phenomenology over multiple time points using muscle function testing, histology, and expression profiling by RNA sequencing. The injury response was then compared to a regenerative medicine treatment using orthotopic transplantation of autologous minced muscle grafts (~1 mm3 tissue fragments). A chronic inflammatory and fibrotic response was observed at all time points following VML. These results suggest that the pathological response to VML injury during the acute stage of the healing response overwhelms endogenous and therapeutic regenerative processes. Overall, the data presented delineate key molecular characteristics of the pathobiological response to VML injury that are critical effectors of effective regenerative treatment paradigms.
Introduction
Skeletal muscle comprises over 40% of body mass in lean individuals, is primarily responsible for coordinating voluntary movements and can readily adapt to its environment. After injury, skeletal muscle is capable of repairing and regenerating through a pool of resident stem cells (satellite cells) that activate, proliferate, differentiate, and fuse to form new or repair existing multinucleated myofibers1. However, after volumetric muscle loss (VML) injury2, which is a type of severe trauma that ablates resident cells and structures primarily responsible for regeneration, the intrinsic muscle regenerative process fails3. Instead, gross compartmental tissue fibrosis manifests4,5 and chronic functional deficits result. VML is a common clinical outcome after open or closed (e.g., crush injury requiring fasciotomy and tissue evacuation) extremity trauma, for which there are currently no regenerative standards of care.
The magnitude and location of VML injuries result in considerable heterogeneity and necessitate a cadre of therapies. For instance, a small VML defect isolated to a peripheral portion of a muscle unit may be best compensated by synergist hypertrophy mediated through physical therapy. Whereas, massive VML defects have been shown to not respond to conventional physical therapy and may require regenerative medicine augmentation. Regenerative treatments for VML have utilized various strategies6–14 but require significant further advancement to be of therapeutic benefit to patients presenting acute or chronic VML injury.
The fairly extensive number of regenerative therapies that have been tested for VML repair have proceeded to date without a comprehensive understanding of the pathophysiology of the injury response. Animal models of VML injury have described15,16 a dysregulated immune response that coincides with aberrant or muted muscle fiber regeneration and fibrosis, although the temporal coordination of these events and drivers of this response trajectory have not been elucidated. Without determination of a clearly effective therapy and lack of knowledge of the molecular phenomenology-mediating injury repair, further advancement of the field depends on understanding and mitigation of the host pathobiological response. Thus, a key step to advance translational medicine in this domain is to establish a comprehensive reference map of the various molecular patterns that drive the diverse sub-processes through time and identify critical factors that influence and modulate gene expression amplitude and dynamics. The formulated unbiased views of the pathophysiology can then serve as a guide to assess and enhance new therapeutic modalities for VML.
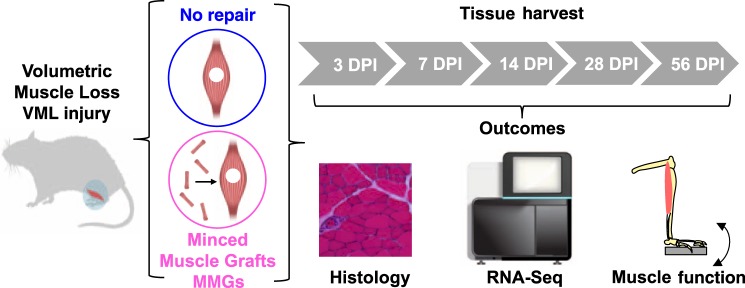
Herein, we administer a VML injury to a rodent model and track the molecular phenomenology and etilogy through multiple time points using muscle function testing, histology, and expression profiling by RNA sequencing (RNA-Seq) (Fig. 1). Next, we evaluate the effect of a regenerative therapy (orthotopic transplantation of autologous minced muscle grafts (MMGs)) and compare readouts of tissue and molecular function. Employing this approach, we were able to quantitatively understand the time course of tissue-level changes induced by a regenerative therapy and establish metrics for assessing progression through discrete healing state transitions. This schema provides a resource to understand the physiologic response to VML as well as to enable sensitive evaluation of regenerative therapies after severe trauma.
Fig. 1. Schematic of experimental approach to define the molecular response to volumetric muscle loss (VML) injury and how transplantation of autologous minced muscle graft (MMGs) impacts regenerative trajectory.
Injured and uninjured muscles were extracted at multiple days post injury (DPI) and characterized using histology, high-throughput sequencing of RNA (RNA-Seq), and muscle function
Results
VML induces fibrotic scarring and poor muscle regeneration
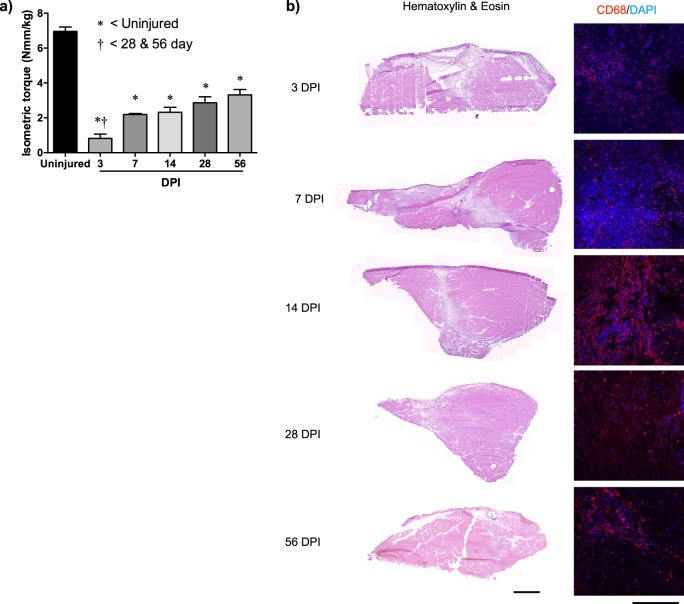
VML injury was surgically created unilaterally in the rat tibialis anterior (TA) muscle and assessed for in vivo neural-evoked strength deficits intermittently post injury (Fig. 2a). At 3, 7, 14, 28, and 56 days post injury, ~88, 69, 67, 59, and 52% of strength was lost, respectively. Histologically, VML injury resulted in a clear muscle defect devoid of muscle fibers, and an associated influx of inflammatory cells over the ensuing weeks (Fig. 2b). Macrophages (CD68+ cells) persisted in the defect for weeks to months after injury (Fig. 2b). Consistent with previous studies demonstrating gross fibrosis within the VML wound bed3,16,17, evidence of muscle fiber regeneration within the defect was limited out to 56 days post injury.
Fig. 2. VML injury in the rat TA muscle induces chronic strength deficits and prolonged tissue damage.
a TA muscle maximal isometric torque was elicited using common peroneal nerve stimulation in uninjured and VML injured (non-repair) muscle at the specified days post injury (DPI). Values are mean ± sem. *, †: p < 0.05. b Representative TA muscle hematoxylin and eosin (H&E) and macrophage (CD68) probed sections are presented. Scale bars for H&E and CD68 images are 1000 and 150 µm, respectively. TA tibialis anterior, VML volumetric muscle loss
Temporal transcriptomes of VML-injured tissues display pathways associated with heightened and sustained inflammation in combination with excessive fibrogenesis
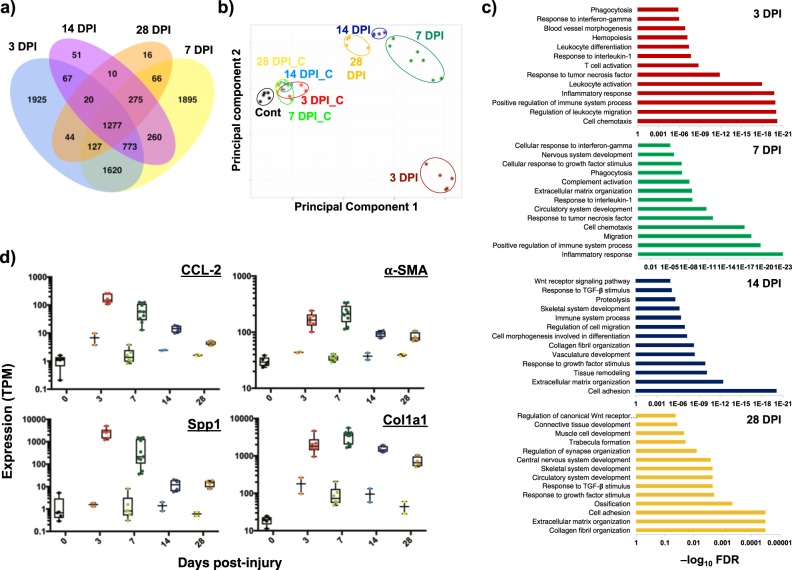
To measure the molecular mechanisms contributing to poor muscle regeneration induced by VML, we performed expression profiling by RNA-Seq for unoperated controls, VML-affected muscles and unaffected contralateral muscle for multiple time points in the acute response phase (3, 7, 14, 28 days post injury). Biological and technical replicates for all three sample types demonstrated excellent correlations and reproducibility across all time points. Samples that did not meet adequate thresholds were discarded. In total, 16,020 mRNAs were detected (TPM > 1) for at least one time point; and 4,897 genes were observed to be dynamic (fold change > 2) at one or more time points (Fig. 3a). Principal component analysis (PCA) of the data sets revealed the contralateral uninjured tissues clustered together for all time points and in close proximity to control tissues (Fig. 3b). PCA also revealed the tissues extracted 3 days post injury displayed significant separation from other time points and the tissues progressed back toward the contralateral uninjured tissues as time progressed. However, for tissues extracted at 28 days post injury, a distinct separation was observed between the injured and uninjured tissues, indicative the tissue did not fully heal as measured by tissue transcriptome.
Fig. 3. Characterization of molecular response to VML injury.
a Venn diagram showing number of unique and overlapping differentially expressed genes for muscle tissues administered VML injury and extracted at different days post injury (DPI). Days 3 and 7 show the most unique differentially expressed genes. b Principal component analysis (PCA) of RNA-Seq data sets from control (cont), injured (example: 3 DPI), and uninjured (example: 3 DPI_C) data sets from different time points harvested show distinct separation between injured and uninjured samples. c Enriched pathways associated with differentially expressed genes from each time point. Enrichment scores are plotted as −log10(FDR), where FDR is the false discovery rate. d Individual graphs of gene expression plotted in TPM (transcripts per million reads) where uninjured samples are plotted first and injured samples are plotted second for each time point. CCL2: C-C Motif chemokine ligand 2, SPP1: secreted phosphoprotein 1, α-SMA: alpha smooth muscle actin, Col1a1: collagen type I alpha chain. VML volumetric muscle loss
Pathway analysis of the differentially expressed genes for 3 days post injury revealed a large number of gene sets associated with chemotaxis, inflammation, and immune cell infiltration. Previous muscle regenerative studies18,19 have shown inflammation -related programs subsided after several days, but many of these genes remained upregulated over the entire time course (Fig. 3c–d). Detection of these transcripts was also consistent with immuno-staining of the injured muscle, whereby invasion of CD68+ cells (monocytes and macrophages) into the injured site was observed 14 and 28 days post injury (Fig. 2b). At 7 days post injury, many of the pathways associated with the inflammatory response and immune system remained upregulated, such as the Complement pathway. Previously, sustained activation of the Complement system in regenerating muscle was shown to stimulate Wnt signaling20 and promote fibrosis by attenuating satellite cell proliferation21,22 and increasing fibroblast production of collagen23. Consistent with this observation, strong increases in expression of a family of genes-associated connective tissue cells (Col1a1, α-SMA/Acta2, S100a4) along with extracellular matrix (ECM) deposition and remodeling such as structural components (collagen and elastin) were detected. This result was also in agreement with histological observations of fibrotic supplantation of muscle tissue (Fig. 2b) at this time period. At 14 and 28 days post injury, fibrogenic pathways such as Wnt, transforming growth factor beta (TGF-β), and other ECM pathways remained enriched and indicated that the regenerative process began to fail and been replaced by fibrosis. Taken together, these results suggest VML induces strong and persistent overexpression of inflammatory transcripts, which in turn provokes dysregulated ECM deposition and may inhibit satellite cell repair.
Orthotopic transplantation of autologous MMGs stimulates muscle repair and regeneration
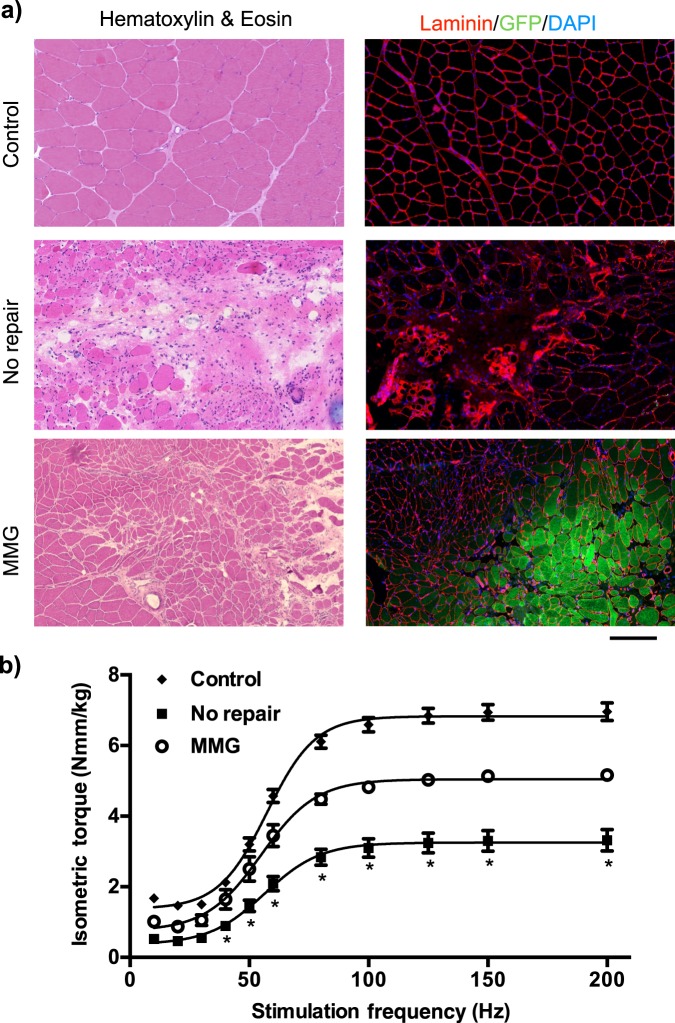
Autologous MMGs have previously been shown to orchestrate de novo muscle fiber regeneration11,24. MMGs derived from syngeneic green fluorescent protein (GFP) Lewis rats were surgically implanted in a collagen gel in the VML defect site and progenitor cells derived from the minced grafts were observed to contribute to de novo muscle fiber regeneration, evidenced by the presence of GFP+ fibers 56 days post injury (Fig. 4a). Compared to non-repaired muscles, minced graft repair of VML injury significantly improved TA muscle strength across a range of peroneal nerve stimulation frequencies by 56–95% by 56 days post injury; however, minced graft repaired muscles still presented an ~26% deficit of maximal strength (Fig. 4b).
Fig. 4. Autologous MMGs promote partial muscle fiber regeneration and functional recovery.
a TA muscle sections stained with hematoxylin and eosin or probed for laminin, GFP, nuclei from control (uninjured), VML injured no repair and minced graft repaired muscles 56 days post injury are presented. Scale bar is 200 µm. b Maximal TA muscle isometric torque was measured as a function of stimulation frequency in each group at 56 days post injury. Values are mean ± sem. *p < 0.05. MMG minced muscle graft, TA tibialis anterior
Gene expression programs after regenerative treatment display similar response trajectories to non-repaired tissues
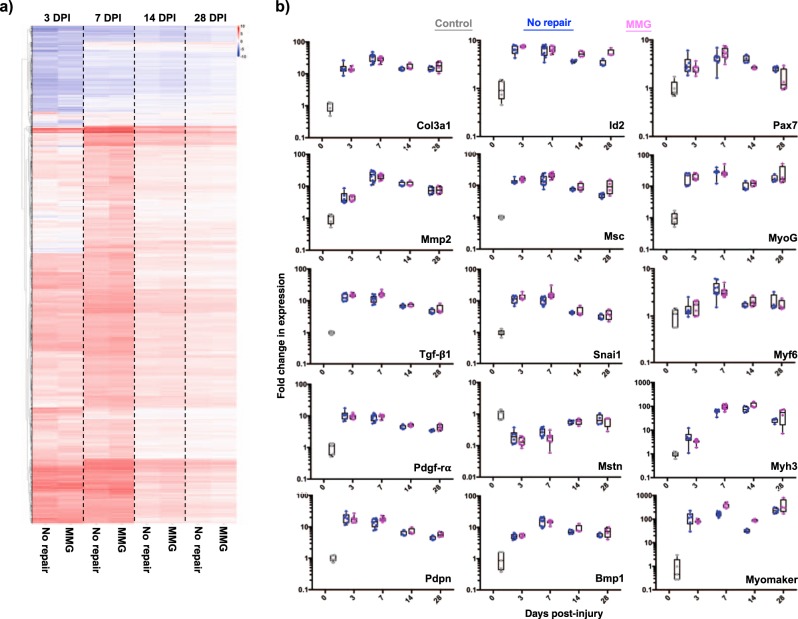
To quantitatively understand how a regenerative medicine therapy converges to positively influence endogenous gene expression programs after VML, whole-tissue RNA-Seq was performed for each time point and condition. Differential expression (DE) analysis and hierarchical clustering of the data sets through time revealed highly similar patterns for the non-repaired samples with those that were treated with MMGs (Fig. 5a). This observation is clearly illustrated by a family of genes nominally associated with fibrosis (collagen 3-Col3a1, matrix-metalloproteinase 2-MMP2, TGF-β1, platelet-derived growth factor receptor α-PDGFRα, podoplanin-PDPN), where the expression pattern appeared invariant with treatment (Fig. 5b). This result was also viewed for multiple genes associated with negative regulation of myogenesis (inhibitor of differentiation 2-Id2, musculin-MSC, snail family transcriptional repressor 1-Snai1, myostatin-MSTN, bone morphogenetic protein 1-Bmp1). Comparing the genes associated with positive myogenic regeneration (paired box protein 7-Pax7, myogenin-MyoG, myogenic factor 6-Myf6, embryonic myosin heavy chain-Myh3, Tmem8c-Myomaker) from the unrepaired samples with those treated with MMGs, increases in expression were observed over the time course for both types of samples but did not vary considerably with treatment. This result suggests that the acute VML response results in a robust tissue transcriptome characterized by a fibrotic pathology that suppresses relative expression of canonical myogenic pathways, regardless of treatment.
Fig. 5. Transcriptional response of regenerative (MMGs) treatment for VML show marginal effects compared to non-repaired tissue at time points sampled.
a Heatmap of differentially expressed genes (injured vs. uninjured) for each treatment and time point. Data are plotted as union of biological replicates. b Individual box plots of gene expression plotted as fold change (injured vs. uninjured) for four different time points (Sham-gray, No Repair-blue, MMG-purple). DPI days post-injury, MMG minced muscle graft, VML volumetric muscle loss
Discussion
Currently, very little is known about the global molecular response after VML and why this injury exceeds skeletal muscle’s endogenous regenerative capabilities through satellite cells. Recently, we administered a VML injury to a swine model16 and observed upregulation of inflammatory and fibrotic transcriptional programs at 3 months post injury. Understanding the molecular signals that induce the response of skeletal muscle to adopt a non-regenerative phenotype and when these signals are presented is paramount in advancing our understanding of VML pathogenesis and therapeutic efficacy. In this study, we performed multi-dimensional profiling of VML-injured tissues during the healing process using muscle function, histology, and transcriptomics and compared the response to treatment with autologous MMGs. Profiling of unfractionated muscle tissue from the VML injured site enabled detection of transcripts from multiple cell types and insights into tissue-level responses.
For muscles that were non-repaired after VML, in all time points the tissue appeared to be overwhelmed by persistent inflammatory and fibrogenic signals. Histological observation of the non-repaired muscles highlighted inadequate muscle fiber regeneration and a dense population of macrophages in the defect area. This result is consistent with sustained upregulation of multiple components of the Complement system, which has previously been shown to impair muscle regeneration20 and promote fibrotic deposition. In contrast to muscle regenerative studies, whereby the cellular infiltrate transitions25 from a pro-inflammatory population to a primarily anti-inflammatory phenotype after several days, many of the inflammatory and immune-related signaling pathways remained upregulated over the entire time course. Persistent inflammation and inadequate debris clearance during muscle regeneration have been shown to influence fibrosis by inducing fibro-adipogenic progenitors (FAPs)26,27 to proliferate and differentiate into fibroblasts or adipocytes. We viewed sustained transcriptional overexpression of PDGFRα, a marker for FAPs, in parallel to sustained increases in expression of TGF-β1. Increases in TGF-β1 signaling have been shown to block FAP apoptosis and induce fibrogenic differentiation28, which our current findings suggest. Moreover, elevated TGF-β1 has been shown to suppress the regenerative functions of satellite cells29. The collective integration of these gene expression programs for non-repaired tissues suggests a model of continuous infiltration of immune cells and perpetual inflammation that in turn regulates mesenchymal progenitors and cells to produce excess matrix30. A similar model of chronic inflammation that contains both pro-inflammatory and anti-inflammatory immune cells has been shown for Duchenne muscular dystrophy, which also uniquely exhibits fibrosis and FAP infiltration and differentiation31.
The similar gene expression programs generated from VML-injured tissues without and treated with MMGs suggests unrelenting immune and fibrotic responses following VML injury. These results support the hypothesis that persistent inflammation and strong fibrosis overwhelm and dampen myogenic repair. Previously11,32,33, MMGs were found to promote muscle regeneration after extended post-surgical times (8–16 weeks), but in the early time periods sampled here (3–28 days) myogenic regenerative expression patterns did not vary considerably compared to non-repaired samples. These early time periods were deterministically sampled, as immediate interventions have been shown to favorably promote long-term outcomes for VML. However, multiple factors may be contributing to prevention of regenerative pathways from these treatments during the early time periods sampled such as the strong upregulation of negative regulators of myogenic differentiation such as TGF-β, SMAD2/3, Snai, musculin (MSC), myostatin (MSTN), and inhibitor of differentiation (Id) proteins (Id2 and Id3). Since muscle repair and regeneration canonically utilizes the sequential activation of specific basic helix-loop-helix transcription factors34 (MyoD, MyoG, and Mef2), potent overexpression of negative regulators may block these factors from appropriately activating myogenic genes. Thus, even if satellite cells or differentiated progenitors migrate or are transplanted into the defect site, their differentiation is initially obstructed by overexpression of negative regulators.
Another factor that may be contributing to suboptimal regenerative outcomes and inhibiting treatment efficacy at the time periods sampled is the aberrant deposition and remodeling of ECM in the defect area. Multiple ECM transcripts were strongly upregulated after VML such as fibrillar collagens, lysyl oxidase, and proteoglycans in comparison to proteases that breakdown matrix. The resulting dense composition of the deposited ECM may confer differential integrin signaling35 for satellite cells or myogenic progenitors in the defect site, which in turn would augment their differentiation and fusion. Loss of adhesive cues and increases in integrins36 (which were also detected) has previously been shown to alter muscle stem cell dynamics in aged tissues37 and produce weakened healing responses. Thus, the coupled action of negative regulators and dysregulated ECM produce an environment that is unfavorable for resident or transplanted myogenic cells to mediate optimal myogenic outcomes.
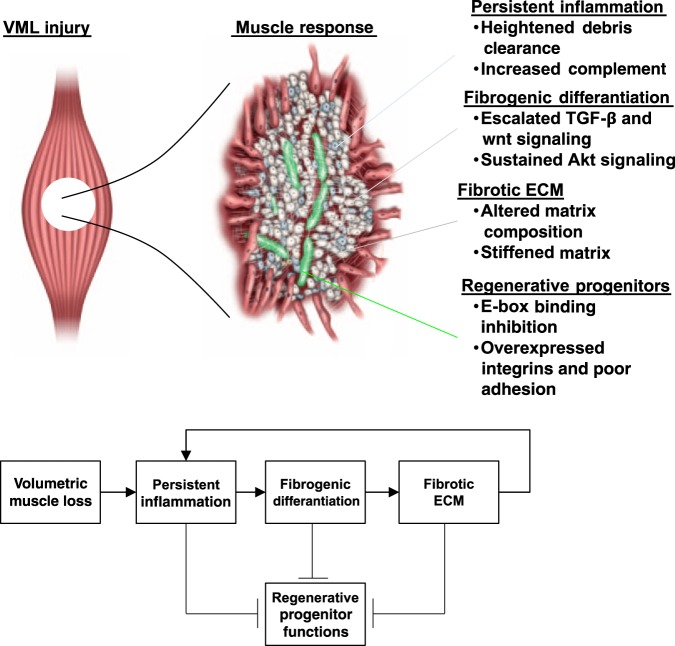
Integrating these results implies that VML injury drives formation of an inhibitive feedback loop38 that effectively limits therapies such as autologous grafts during these early periods (Fig. 6). Thus, in order to treat the response that results from a severe VML insult, treatment strategies need to be directed toward amelioration of this feedback loop. Since the immune response is the first reaction after VML injury, modulating early inflammatory effects may be effective for subsequent treatments (such as with progenitors). Indeed, recent approaches have taken aim at modulating early immune-related responses in the remaining musculature after VML injury to improve muscle regeneration10,39. Previously, it was shown that the behavior of mesenchymal progenitors40 and their differentiation into matrix-producing cells was affected by pro-inflammatory and sustained TGF-β signaling. Thus, immuno-modulatory drugs and technologies may be beneficial therapies but require consideration of their influence on the FAP population and subsequent differentiation. Another avenue of therapies requiring further investigation that may play an effective role when combined with immuno-modulation are anti-fibrotic agents41, which have been shown to limit pro-fibrotic TGF-β effects and persistent protein kinase B (Akt) signaling that perpetuate the feedback loop. Anti-fibrotic agents can induce apoptosis of FAPs and differentiated fibrogenic cells and when combined with inhibition of pro-inflammatory signals may permit a more favorable environment for myogenic regenerative cells to “break” the feedback loop. Combinatory treatment of inflammatory and fibrotic signals has previously been shown to attenuate muscular dystrophy pathologies42,43 and requires further investigation for treatment of VML. The generation of a more favorable microenvironment would then provide invading or transplanted myogenic progenitors to properly differentiate and regenerate tissue. In addition to encountering a more favorable environment, it was shown myogenic progenitors in fast twitch muscles protect against fibrosis via emission of exosomes containing microRNA-206 (miR-206)44, which represses ribosome-binding protein 1, a regulator of collagen biosynthesis. Thus, restoring the lack of or reduced capacity of satellite cells in the defect area may simultaneously attenuate fibrosis45 and promote regeneration.
Fig. 6. VML induces formation of an inhibitive feedback response loop that modulates the regenerative actions of myogenic progenitors.
The abrupt tissue removal engenders a sustained inflammatory response that invokes fibrogenic cells to differentiate and produce excess matrix. The modified matrix and milieu in turn modifies the regenerative functions of myogenic progenitors and prevents healing. ecm extracellular matrix, VML
In summary, our work provides definitive evidence of the molecular networks that underpin the VML response and progressive degeneration and fibrosis. We used a large-scale data approach to understand how these networks are adjusted when treated with a regenerative treatment strategy. The results of a common response behavior for both treated and unrepaired samples resemble muscular dystrophies, non-dystrophic myopathies, and amyotrophic lateral sclerosis, where continuous cycles of myofiber degeneration and incessant inflammation drive fibrosis. Accordingly, the mechanisms that ultimately drive impaired tissue regeneration after VML suggest this injury may be effectively treated with therapies for dystrophic muscle or other chronic pathologies.
Materials and methods
Animals
A total of 76 Lewis rats were used in this study. All animal procedures were approved by the Institutional Animal Care and Use Committee and were conducted in compliance with the Animal Welfare Act and in accordance with the principles of the Guide for the Care and Use of Laboratory Animals at the United States Institute of Surgical Research. Inbred male Lewis rats (350–400 g; ~4 months of age) were purchased from Harlan Laboratories (Envigo) and a subset of Lewis GFP rats (n = 4) were purchased from the Rat Resource Center, all rats were housed in a specific pathogen-free animal facility and given food and water ad libitium. All rats received a pre-surgical (~30 min prior) administration of buprenorphine-SR (1.2 mg/kg; s.c.) for pain management. No adverse events were observed.
VML injury and repair
Rats underwent surgical creation of a VML injury in the left TA muscle using aseptic technique. Briefly, the lateral incision was made along the lower leg and the skin and fascia overlying the TA muscle were reflected. A spatula tool was placed under the middle third of the TA muscle and a 6-mm biopsy punch was used partially ablate muscle tissue from the region of the muscle. Approximately, 1-mm width of tissue remained on either side of the defect, which provided continuity of proximal and distal regions of the remaining muscle mass. The wet weight of the excised tissue was measured (~86 mg). A subset of rats underwent VML injury repair with autologous MMGs (~1 mm3 pieces of muscle tissue). The minced grafts were prepared from the excised tissue using Vanna scissors. The fascia was closed using 6-0 Vicryl and the skin was closed using surgical staples.
Muscle histology
TA muscle tissue isolation and freezing were performed as described previously11. The muscle was frozen in melting isopentane and stored at −80 °C until further analysis. Sections of 8-μm thickness were stained with hematoxylin and eosin (H&E). Composite brightfield images of the complete TA muscle were acquired using Axio Scan.Z1 microscope and ZEN imaging software (Carl Zeiss Microscopy). Additional labeling of muscle sections was completed by combinations of antibodies and probes including DAPI (1 µg/ml; Molecular Probes D21490), monoclonal anti-mouse CD68 (1 µg/ml; Biorad MCA341R), monoclonal anti-rat laminin (2.5 µg/ml; Abcam ab11576), and polyclonal anti-goat GFP (1 µg/ml; Abcam ab6673). Appropriate isotype-specific-conjugated secondary antibodies (1:200; Molecular Probes A21125, A11055, or A21202) were then applied. Fluorescent imaging of the TA muscle was conducted with an Olympus FluoView FV1000 laser scanning confocal microscope (Olympus America Inc., Melville, NY) mounted on an inverted Olympus IX81 microscope and equipped with Multi-line Argon (458, 488, and 515 nm), HeNeG (543 nm), diode (405 nm), and diode (635 nm) lasers using an Olympus UPLSAPO 20×/0.85 N.A. oil immersion lens. All images were acquired as 12-bit multi-TIFF files.
Muscle functional assessment
In vivo functional testing of TA muscles was performed using methodology previously described11. Briefly, TA muscle in vivo physiological properties were measured in anesthetized rats (isoflurane 1.5–2.0%) using a dual-mode muscle lever system (Aurora Scientific, Inc., Mod. 305b). Subcutaneous needle electrodes were inserted in the posterior compartment of the lower limb on each side of the common peroneal nerve. Optimal voltage (2–5 V) was set with a series of tetanic contractions (5–10 contractions; 150 Hz, 0.1 ms pulse width, 400 ms train). Then, a skin incision was made at the antero-lateral aspect of the ankle and the distal tendons of the extensor digitorum longus and extensor hallicus longus muscles was isolated and severed above the retinaculum11. TA muscle maximal isometric tetanic torque was measured as a function of stimulation frequency (10–200 Hz) with the ankle at a right angle. The servomotor input and force and displacement transducer outputs are controlled and acquired, respectively, using a PC equipped with a data acquisition board (National Instruments) and custom designed Lab View (National Instruments)-based software program.
RNA extraction, quantification, preparation, and sequencing
TA muscles were harvested from euthanized rats, immersed in Trizol (Thermo Fisher) and snap frozen on liquid nitrogen and stored at −80 °C for subsequent use. The tissues were then thawed, homogenized (Tissue Ruptor, Qiagen) for 30 s at room temperature and total RNA was isolated from the slurry using the miRNeasy Mini Kit (Qiagen) as per the manufacturer’s instructions. RNA concentration and integrity were measured with a Nanodrop spectrophotometer (Nanodrop 2000c) and Bioanalyzer (Agilent 2100), respectively. Samples that did not pass quality metrics (RIN > 7) were precluded from further processing. 500 ng of isolated total RNA was used to produce cDNA libraries using the Truseq protocol (Illumina), as per the manufacturer’s instructions. Individual libraries were pooled and sequenced using 76 base-pair paired-end reads on a Hi-Seq 2500 in high output mode to an average depth of 35 M reads per library.
RNA-Seq data processing
High-throughput RNA sequencing reads were pseudo-aligned to the Genbank-annotated Rattus norvegicus (Rat) genome (build 6.0), transcript abundances were quantified and gene counts were generated using Kallisto46 with default parameters. To determine sample quality for downstream analysis, per sample read depth was measured, and outliers were detected by conducting PCA on the log2-transformed gene counts. Samples that contained poor read depth (less than 5 million reads) or were determined to be outliers by PCA were removed from downstream analysis.
DE of gene counts was calculated using the R/Bioconductor implementation of DESeq247. Analysis of expression data, such as set analysis based on treatment-specific expression profiles over time, PCA of treatment by time gene profiles, and correlation of inter-treatment gene expression were performed with the R base package. DE of genes in treated samples relative to uninjured controls was determined using a log2 fold-change threshold greater than 1 or less than −1 and significance was measured using an adjusted p-value less than 0.05 (Benjamini–Hochberg correction for multiple testing). PCA plots from significant DE genes were generated using base R libraries and ggplot2 (version 2.1.0).
Pathway analysis of significant gene sets was performed using the GAGE R/Bioconductor package48. Pathway activation/repression was determined by gene set enrichment analysis performed by GAGE with pathway annotations from Gene Ontology Consortium49, KEGG and Reactome50 databases. Significance was measured using an adjusted p-value less than 0.05 (Benjamini–Hochberg correction for multiple testing).
Statistical analyses
Data were analyzed using Prism 6 for Mac OSX (Graphpad Inc.; La Jolla, CA). All dependent variables were analyzed separately using repeated measures ANVOAs, when appropriate Tukey HSD analysis was performed. Significance was accepted at the α < 0.05 level and data are reported as mean ± SE.
Acknowledgements
The authors gratefully acknowledge Mrs. Monica Jalomo, Mrs. Shayna Levine, and Ms. Monica Farley for technical support during these experiments, Tom Sheehan for assistance with artwork, Tamara Mason, Tyler Miselis, Fontina Kelley, and the Broad Institute Genomics Platform for sequencing and technical assistance, and Darrell O. Ricke for insightful discussions. This material is based on work supported under Air Force Contract No. FA8721-05-C-0002 and/or FA8702-15-D-0001. This work was also supported by the Neuromusculoskeletal Injuries Research Award, Clinical and Rehabilitative Medicine Research Program, United States Army Medical Research, and Materiel Command (MR140099). Opinions, interpretations, recommendations, and conclusions are those of the authors and are not necessarily endorsed by the United States Government. Any opinions, findings, conclusions, or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the U.S. Air Force GSM3150345, GSM3150346, GSM3150347, GSM3150348, GSM3150349, GSM3150350, GSM3150351, GSM3150352, GSM3150353, GSM3150354, GSM3150355, GSM3150356, GSM3150357, GSM3150358, GSM3150359, GSM3150360, GSM3150361, GSM3150362, GSM3150363, GSM3150364, GSM3150365, GSM3150366, GSM3150367, GSM3150368, GSM3150369, GSM3150370, GSM3150371, GSM3150372, GSM3150373, GSM3150374, GSM3150375, GSM3150376, GSM3150377, GSM3150378, GSM3150379, GSM3150380, GSM3150381, GSM3150382, GSM3150383, GSM3150384, GSM3150385, GSM3150386, GSM3150387, GSM3150388, GSM3150389, GSM3150390, GSM3150391, GSM3150392, GSM3150393, GSM3150394, GSM3150395, GSM3150396, GSM3150397, GSM3150398, GSM3150399, GSM3150400, GSM3150401, GSM3150402, GSM3150403, GSM3150404, GSM3150405, GSM3150406, GSM3150407, GSM3150408, GSM3150409, GSM3150410, GSM3150411, GSM3150412, GSM3150413, GSM3150414, GSM3150415, GSM3150416, GSM3150417, GSM3150418, GSM3150419, GSM3150420, GSM3150421, GSM3150422, GSM3150423, GSM3150424, GSM3150425, GSM3150426, GSM3150427, GSM3150428, GSM3150429, GSM3150430, GSM3150431, GSM3150432, GSM3150433, GSM3150434, GSM3150435, GSM3150436, GSM3150437, GSM3150438, GSM3150439, GSM3150440, GSM3150441, GSM3150442, GSM3150443, GSM3150444, GSM3150445, GSM3150446, GSM3150447, GSM3150448, GSM3150449, GSM3150450, GSM3150451, GSM3150452, GSM3150453, GSM3150454, GSM3150455, GSM3150456, GSM3150457, GSM3150458, GSM3150459, GSM3150460, GSM3150461, GSM3150462, GSM3150463, GSM3150464, GSM3150465, GSM3150466, GSM3150467, GSM3150468, GSM3150469, GSM3150470, GSM3150471, GSM3150472, GSM3150473, GSM3150474, GSM3150475, GSM3150476, GSM3150477, GSM3150478, GSM3150479, GSM3150480, GSM3150481, GSM3150482.
Author contributions
C.A.A. and B.T.C. designed the study, C.A.A., S.M.G., S.M.G., C.P., C.Z., and B.T.C. performed experiments and collected data, C.A.A., A.W., and J.L. conducted analysis, C.A.A., S.M.G., and B.T.C. wrote the manuscript.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Edited by A. Rufini.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
7/23/2018
Correction to: Cell Death and Discovery (2018) 4, 33; 10.1038/s41420-018-0027-8; published online 20 February 2018.
References
- 1.Scharner J, Zammit PS. The muscle satellite cell at 50: the formative years. Skelet. Muscle. 2011;1:28. doi: 10.1186/2044-5040-1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grogan BF, Hsu JR. Volumetric muscle loss. J. Am. Acad. Ortho. Surg. 2011;19:S35–S37. doi: 10.5435/00124635-201102001-00007. [DOI] [PubMed] [Google Scholar]
- 3.Garg K, et al. Volumetric muscle loss: persistent functional deficits beyond frank loss of tissue. J. Ortho. Res. 2015;33:40–46. doi: 10.1002/jor.22730. [DOI] [PubMed] [Google Scholar]
- 4.Corona BT, Wenke JC, Ward CL. Pathophysiology of volumetric muscle loss injury. Cells Tissues Organs. 2015;202:180–188. doi: 10.1159/000443925. [DOI] [PubMed] [Google Scholar]
- 5.Corona BT, et al. The promotion of a functional fibrosis in skeletal muscle with volumetric muscle loss injury following the transplantation of muscle-ECM. Biomaterials. 2013;34:3324. doi: 10.1016/j.biomaterials.2013.01.061. [DOI] [PubMed] [Google Scholar]
- 6.Dziki JL, Sicari BM, Wolf MT, Cramer MC, Badylak SF. Immunomodulation and mobilization of progenitor cells by extracellular matrix bioscaffolds for volumetric muscle loss treatment. Tissue Eng. Part A. 2016;22:1129–1139. doi: 10.1089/ten.tea.2016.0340. [DOI] [PubMed] [Google Scholar]
- 7.Grasman JM, Zayas MJ, Page RL, Pins GD. Biomimetic scaffolds for regeneration of volumetric muscle loss in skeletal muscle injuries. Acta Biomater. 2015;25:2–15. doi: 10.1016/j.actbio.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aurora A, Roe JL, Corona BT, Walters TJ. An acellular biological scaffold does not regenerate appreciable de novo muscle tissue in rat models of volumetric muscle loss injury. Biomaterials. 2015;67:393–407. doi: 10.1016/j.biomaterials.2015.07.040. [DOI] [PubMed] [Google Scholar]
- 9.Sicari BM, et al. An acellular biological scaffold promotes skeletal muscle formation in mice and humans with volumetric muscle loss. Sci. Transl. Med. 2014;6:234ra58. doi: 10.1126/scitranslmed.3008085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadtler K, et al. Developing a pro-regenerative biomaterial scaffold microenvironment requires T helper 2 cells. Science. 2016;352:366–370. doi: 10.1126/science.aad9272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corona BT, et al. Autologous minced muscle grafts: a tissue engineering therapy for the volumetric muscle loss of skeletal muscle. Am. J. Physiol. Cell Physiol. 2014;305:C761–C775. doi: 10.1152/ajpcell.00189.2013. [DOI] [PubMed] [Google Scholar]
- 12.Aurora A, Garg K, Corona BT, Walters TJ. Physical rehabilitation improves muscle function following volumetric loss injury. BMC Sports Sci. Med. Rehab. 2014;6:41–51. doi: 10.1186/2052-1847-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quarta M, et al. Bioengineered constructs combined with exercise enhance stem cell-mediated treatment of volumetric muscle loss. Nat. Commun. 2017;8:15613. doi: 10.1038/ncomms15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machingal MA, et al. A tissue-engineered muscle repair construct for functional restoration of an irrecoverable muscle injury in a murine model. Tissue Eng. Part A. 2011;17:2291–2303. doi: 10.1089/ten.tea.2010.0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurtgen BJ, et al. Autologous minced muscle grafts improve endogenous fracture healing and muscle strength after musculoskeletal trauma. Physiol. Rep. 2017;5:e13362. doi: 10.14814/phy2.13362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greising SM, et al. Unwavering pathobiology of volumetric muscle loss injury. Sci. Rep. 2017;7:13179. doi: 10.1038/s41598-017-13306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garg K, Corona BT, Walters TJ. Losartan administration reduces fibrosis but hinders functional recovery after volumetric muscle loss injury. J. Appl. Phys. 2014;117:1120–1131. doi: 10.1152/japplphysiol.00689.2014. [DOI] [PubMed] [Google Scholar]
- 18.Arnold L, et al. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J. Exp. Med. 2007;204:1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aguilar CA, et al. In vivo monitoring of transcriptional dynamics after lower-limb muscle injury enables quantitative classification of healing. Sci. Rep. 2015;5:13885. doi: 10.1038/srep13885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naito AT, et al. Complement C1q activates canonical Wnt signaling and promotes aging-related phenotypes. Cell. 2012;149:1298–1313. doi: 10.1016/j.cell.2012.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serrano AL, et al. Cellular and molecular mechanisms regulating fibrosis in skeletal muscle repair and disease. Curr. Top. Dev. Biol. 2011;96:167–201. doi: 10.1016/B978-0-12-385940-2.00007-3. [DOI] [PubMed] [Google Scholar]
- 22.Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Develop. 2011;138:3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brack AS, et al. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- 24.Carlson BM. The regeneration of minced muscles. Monogr. Dev. Biol. 1972;4:1–128. [PubMed] [Google Scholar]
- 25.Tidball JG, Villalta SA. Regulatory interactions between muscle and the immune system during muscle regeneration. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010;298:R1173–R1187. doi: 10.1152/ajpregu.00735.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joe AWB, et al. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat. Cell Biol. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uezumi A, Fukada S, Yamamoto N, Takeda S, Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat. Cell Biol. 2010;12:143–152. doi: 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- 28.Lemos DR, et al. Nilotinib reduces muscle fibrosis in chronic muscle injury by promoting TNF-mediated apoptosis of fibro/adipogenic progenitors. Nat. Med. 2015;21:786–794. doi: 10.1038/nm.3869. [DOI] [PubMed] [Google Scholar]
- 29.Carlson ME, et al. Relative roles of TGF-ß1 and Wnt in the systemic regulation and aging of satellite cell responses. Aging Cell. 2009;8:676–689. doi: 10.1111/j.1474-9726.2009.00517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mann CJ, et al. Aberrant repair and fibrosis development in skeletal muscle. Skelet. Muscle. 2011;1:21. doi: 10.1186/2044-5040-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Villalta SA, Nguyen HX, Deng B, Gotoh T, Tidball JG. Shifts in macrophage phenotypes and macrophage competition for arginine metabolism affects the severity of muscle pathology in muscular dystrophy. Hum. Mol. Genet. 2011;18:482–496. doi: 10.1093/hmg/ddn376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corona BT, Henderson BE, Ward CL, Greising SM. Contribution of minced muscle graft progenitor cells to muscle fiber formation after volumetric muscle loss injury in wild-type and immune-deficient mice. Physiol. Rep. 2017;5:e13249. doi: 10.14814/phy2.13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward CL, Li J, Corona BT. An autologous muscle tissue expansion approach for the treatment of volumetric muscle loss. Biores. Open Acc. 2015;4:198–208. doi: 10.1089/biores.2015.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aguilar CA, et al. Transcriptional and Chromatin Dynamics of Muscle Regeneration after Severe Trauma. Stem Cell Rep. 2016;7:1–15. doi: 10.1016/j.stemcr.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moreno-Layseca P, Streuli CH. Signaling pathways linking integrins with cell cycle progression. Matrix Biol. 2014;34:144–153. doi: 10.1016/j.matbio.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 36.Lukjanenko L, et al. Loss of fibronectin from the aged stem cell niche affects the regenerative capacity of skeletal muscle in mice. Nat. Med. 2016;22:897–905. doi: 10.1038/nm.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conboy IM, et al. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 2005;433:760–764. doi: 10.1038/nature03260. [DOI] [PubMed] [Google Scholar]
- 38.Parker MW, et al. Fibrotic extracellular matrix activates a profibrotic positive feedback loop. J. Clin. Invest. 2014;124:1622–1635. doi: 10.1172/JCI71386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.San Emeterio C, Olingy CE, Chu Y, Botchwey EA. Selective recruitment of non-classical monocytes promotes skeletal muscle repair. Biomaterials. 2017;117:32–43. doi: 10.1016/j.biomaterials.2016.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer GA, Lieber RL. Skeletal muscle fibrosis develops in response to desmin deletion. Am. J. Physiol. Cell Physiol. 2012;302:C1609–C1620. doi: 10.1152/ajpcell.00441.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palumbo-Zerr K, et al. Orphan nuclear receptor NR4A1 regulates transforming growth factor-β signaling and fibrosis. Nat. Med. 2015;21:150–158. doi: 10.1038/nm.3777. [DOI] [PubMed] [Google Scholar]
- 42.Huang P, Zhao XS, Fields M, Ransohoff RM, Zhou L. Imatinib attenuates skeletal muscle dystrophy in mdx mice. FASEB J. 2009;23:2539–2548. doi: 10.1096/fj.09-129833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dadgar SZ, et al. Asynchronous remodeling is a driver of failed regeneration in Duchenne muscular dystrophy. J. Cell Biol. 2014;207:139–158. doi: 10.1083/jcb.201402079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fry CS, Kirby TJ, Kosmac K, McCarthy J, Peterson CA. Myogenic progenitor cells control extracellular matrix production by fibroblasts during skeletal muscle hypertrophy. Cell Stem Cell. 2017;20:56–69. doi: 10.1016/j.stem.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fry CS, et al. Inducible depletion of satellite cells in adult, sedentary mice impairs muscle regeneration capacity without affecting sarcopenia. Nat. Med. 2015;21:76–80. doi: 10.1038/nm.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotech. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 47.Love M, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinform. 2009;10:161–178. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ashburner M, et al. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fabregat A, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2016;44:D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]