Abstract
Habitat degradation can affect trophic ecology by differentially affecting specialist and generalist species, and the number and type of interspecific relationships. However, the effects of habitat degradation on the trophic ecology of coral reefs have received limited attention. We compared the trophic structure and food chain length between two shallow Caribbean coral reefs similar in size and close to each other: one dominated by live coral and the other by macroalgae (i.e., degraded). We subjected samples of basal carbon sources (particulate organic matter and algae) and the same 48 species of consumers (invertebrates and fishes) from both reefs to stable isotope analyses, and determined the trophic position of consumers and relative importance of various carbon sources for herbivores, omnivores, and carnivores. We found that both reefs had similar food chain length and trophic structure, but different trophic pathways. On the coral-dominated reef, turf algae and epiphytes were the most important carbon source for all consumer categories, whereas on the degraded reef, particulate organic matter was a major carbon source for carnivores. Our results suggest that the trophic structure of the communities associated with these reefs is robust enough to adjust to conditions of degradation.
Introduction
One of most evident effects of habitat loss and degradation in terrestrial and aquatic ecosystems is a decline in the diversity of ecological communities via changes on species abundance and richness1,2. However, habitat degradation can modify the number of species interactions, potentially altering the trophic ecology3,4. For example, in a forest subjected to selective logging in Borneo, species of ground-feeding and understorey-feeding birds had significantly higher trophic positions than they had in a non-logged forest5. In Moorea, the stable isotopic signatures of marine carbon sources and consumers differed significantly between two bays as a result of different levels of anthropogenic activities causing differences in mean annual river flow to each bay6.
Life history traits may also determine the response of species to habitat degradation, with specialist species generally being more affected than generalist species7,8. For example, butterfly species with a narrow feeding niche and low levels of mobility and reproduction were most strongly affected by habitat loss across a wide range of habitats in America and Europe9. Also, among bird species, long-lived, large, non-migratory, forest specialists were less likely to occur and less abundant in more intensively man-used habitats than were short-lived, small, migratory, habitat generalists10.
Coral reefs are the most biologically diverse ecosystems in tropical waters and provide important ecosystem services to millions of people around the world11. However, coral reefs are being widely affected by a combination of global and local stressors, including climate change-induced coral bleaching, diseases, overfishing, and eutrophication12. Habitat degradation on coral reefs is mainly manifested as declines in the abundance of reef-building corals and their replacement by macroalgae or other organisms13–15. Coral reef degradation is already affecting community structure by changing diversity and abundance of species16,17 as well as ecosystem functioning and services18,19. The removal of particular species (e.g., by overfishing) and the addition or increase in abundance of others may fundamentally change ecological feedbacks, resulting in a transition of the ecosystem to an alternate state4,20. As occurs with terrestrial species, coral reef specialists are expected to be more affected than generalists by reef degradation21–23, further altering the food webs16.
Food-chain length is an important descriptor of community structure and ecosystem functioning24,25. Ecosystem size and disturbance have been examined as factors potentially determining food chain length in some aquatic and terrestrial ecosystems, with results pointing to ecosystem size as having a greater influence than disturbance26,27. Yet, at the global scale, food chain length showed weak or no relationships with ecosystem size28, probably because environmental variables interact in complex ways to structure a community and may affect metrics of food web structure other than food chain length29.
Stable isotopes provide information on the flow of energy or nutrients through food webs30 and a measure of food-chain length that integrates the assimilation of energy or mass flow through all the trophic pathways leading to top predators (i.e., the trophic structure)25,26. Carbon isotope ratios are used as a tracer of food carbon source, whereas nitrogen isotope ratios are indicative of consumer trophic position31. The use of stable isotope analyses and descriptors (e.g., trophic niche size) to examine the trophic structure and functioning of coral reef organisms and communities has been increasing in the last few years32–34. For example, some studies have examined the stable isotope composition of reef organic matter sources and consumers along environmental gradients or in different seasons35–38. Others have examined the potential effect of habitat degradation on coral reef food webs via stable isotope analyses by focusing on species of higher order consumers34.
Here, we analyse the effect of habitat degradation on food webs by comparing the trophic structure and food chain length between two shallow Caribbean coral reefs known as “Limones” and “Bonanza” that are similar in size and subjected to the same environmental conditions, but have contrasting levels of degradation (Fig. 1). Limones is known for its abundance of the reef-building coral Acropora palmata39. Contrarily, Bonanza, which previously held abundant colonies of A. palmata, has sustained a substantial decline in live coral cover and increase in macroalgal cover from values estimated in 1985 (33% and 4%, respectively)40. We first compared the architectural complexity (rugosity index) and percent cover of live coral and different types of functional groups of algae between reefs. We then explored whether the trophic structure and food chain length of associated reef communities differed between Limones and Bonanza by comparing the stable isotopes (δ15N and δ13C) of several basal carbon sources and the same 48 reef-associated consumer species on both reefs as well as the trophic position of the latter. Using bi-plots of δ13C and δ15N values we compared the isotopic niche widths of the different trophic categories of consumers (herbivores, omnivores, carnivores) between reefs by means of several metrics of trophic structure41,42. We hypothesized that, as the coral-dominated Limones provides more habitats and trophic niches than Bonanza, then it will also exhibit a more complex trophic web with a higher trophic position of predators than the degraded reef.
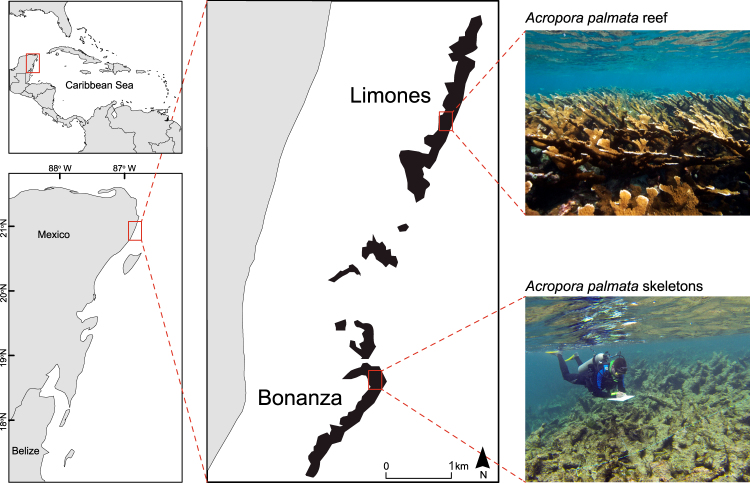
Figure 1.
General location and study sites. (upper left panel) Caribbean region, (bottom left panel) Mesoamerican Reef, (central and right panels), Limones and Bonanza reefs on the Caribbean coast of Mexico. Map produced in QGIS 2.18 (www.qgis.org) using the following data sources: National Geospatial-Intelligence Agency (base map, World Vector Shoreline Plus, 2004. http://shoreline.noaa.gov/data/datasheets/wvs.html). The location of survey sites was obtained from the present study. Data sources are open access under the Creative Commons License (CC BY 4.0).
Results
Characterization of reef and benthic composition
As anticipated, the ecological condition of both reefs differed broadly (Fig. 1). Live coral cover was significantly higher on Limones (50.1 ± 15.1%, mean ± 95% CI) than on Bonanza (7.3 ± 4.6%) (t14 = 5.46, p = 0.0003), as was reef rugosity (Limones: 1.92 ± 0.31; Bonanza: 1.42 ± 0.09; t14 = 3.0, p = 0.025); whereas cover of brown and red fleshy, and green and red calcareous macroalgae was significantly greater on Bonanza than on Limones (Fig. 2). The average cover of all macroalgae (defined as erect fleshy or calcareous algae larger than 2 cm, i.e., excepting algal turf15) was greater on Bonanza (69.9%) than on Limones (26.6%).
Figure 2.
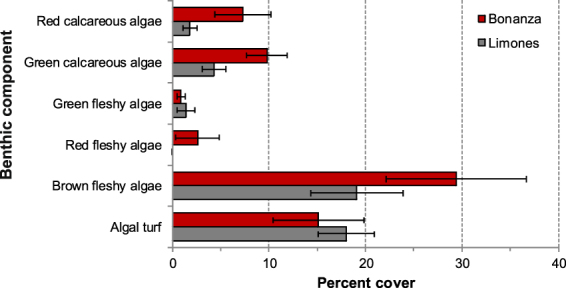
Macroalgal abundance on each reef. Percent cover of different functional groups of macroalgae on Limones (grey columns) and Bonanza (red columns) reefs (N = 8 transects per reef; error bars denote 95% confidence intervals).
Trophic structure and food chain length
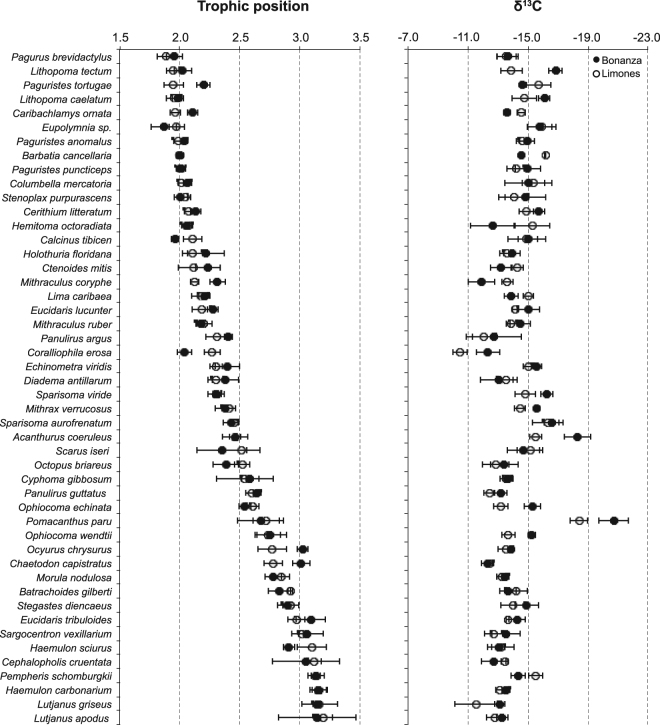
Despite the differences in cover of benthic components, the stable isotope analyses showed that the trophic structure of Limones and Bonanza was generally similar (Fig. 3). However, whereas the overall range of δ15N values was very similar on both reefs, the range in δ13C values was wider on Bonanza than on Limones (Fig. 3, Supplementary Table S1). On both reefs, basal carbon sources had a similar range of δ15N values (0.52 to 2.85‰) (Supplementary Table S1). Among consumers, the lowest δ15N values were obtained for herbivorous and omnivorous invertebrates such as the terebellid polychaete Eupolymnia sp., the hermit crab Pagurus brevidactylus, and the star snails Lithopoma tectum and L. caelatum, whereas the highest δ15N values were obtained for four carnivorous fishes: the schoolmaster snapper Lutjanus apodus, the grey snapper L. griseus, the Caesar grunt Haemulon carbonarium, and the glassy sweeper Pempheris schomburgkii (Supplementary Table S1). Estimates of trophic positions (TP) generally matched the dietary information of these species reported in the literature (Supplementary Table S1). In only eight of the 48 consumer species the TP varied significantly with reef. Five of these species had a higher TP on Bonanza (the scallop Caribachlamys ornata, the hermit crab Paguristes tortugae, the nodose clinging crab Mithraculus coryphe, the yellow-tail snapper Ocyurus chrysurus, and the four-eye butterflyfish Chaetodon capistratus), whereas three had a higher TP on Limones (the orangeclaw hermit Calcinus tibicen, the muricid snail Coralliophila erosa, and the bluestriped grunt Haemulon sciurus). Regardless, there was a high correlation between the TP values of individual consumers from Limones and Bonanza (r = 0.97) (Fig. 4). Moreover, the TP values of the five species of carnivores with the highest TP did not differ significantly between reefs (Fig. 4, Supplementary Table S1), indicating a similar food chain length.
Figure 3.
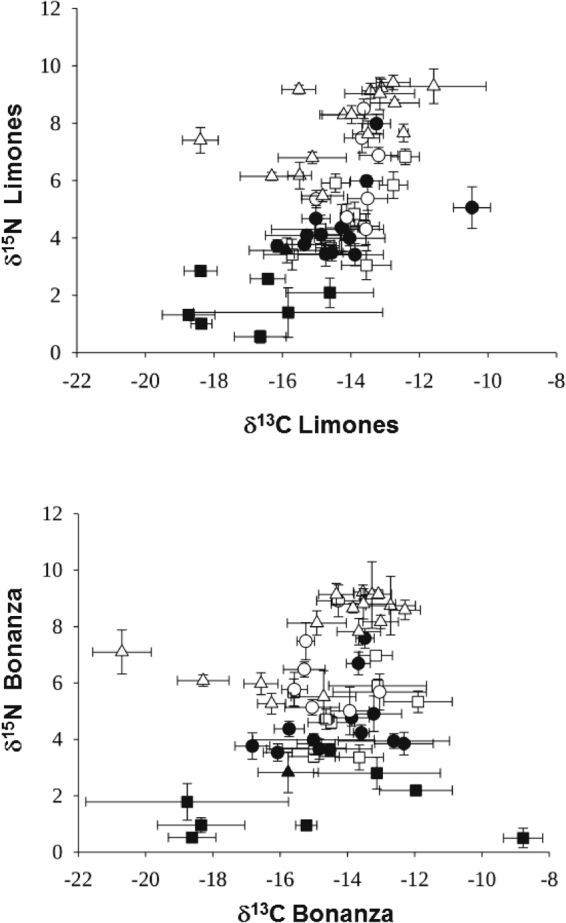
Trophic space for studied reefs. Biplot of mean ± SD δ13C and δ15N values for basal carbon sources (filled squares), annelids (filled triangles), crustaceans (open squares), mollusks (filled circles), echinoderms (open circles), and fishes (open triangles) on Limones (upper panel) and Bonanza (lower panel) reefs.
Figure 4.
Food wed model for Limones and Bonanza reefs. Mean ± 95% confidence interval for trophic position (left panel) and δ13C values (right panel) of 48 species of consumers on Limones (open circles) and Bonanza (filled circles) reefs.
The overall range in δ13C values on the degraded reef (Bonanza) relative to Limones reflects the broader range in δ13C values of basal carbon sources, which ranged from −18.76 ± 3.01 (mean ± SD) for turf algae to −8.78 ± 0.58 for the red coralline algae Amphiroa tribulus (Supplementary Table S1). The δ13C values of carbon sources were generally more enriched on Bonanza, except for algal turf, the green fleshy alga Caulerpa racemosa, and the green calcareous alga Halimeda tuna (Supplementary Table S2). On both reefs, the more depleted δ13C values among consumers were those for the French angelfish Pomacanthus paru, the blue tang Acanthurus coeruleus, the star snail L. tectum, and the redband parrotfish Sparisoma aurofrenatum, and the more enriched for C. erosa and M. coryphe (Supplementary Table S1). Significantly more enriched δ13C values were found for 10 species (nine invertebrates and one fish) on Limones and for eight species (seven invertebrates and one fish) on Bonanza (Supplementary Table S2). Therefore, the correlation between the δ13C values of consumers between both reefs was lower (r = 0.69) than the correlation between the corresponding TP values (Fig. 4).
Importance of basal carbon sources to consumers
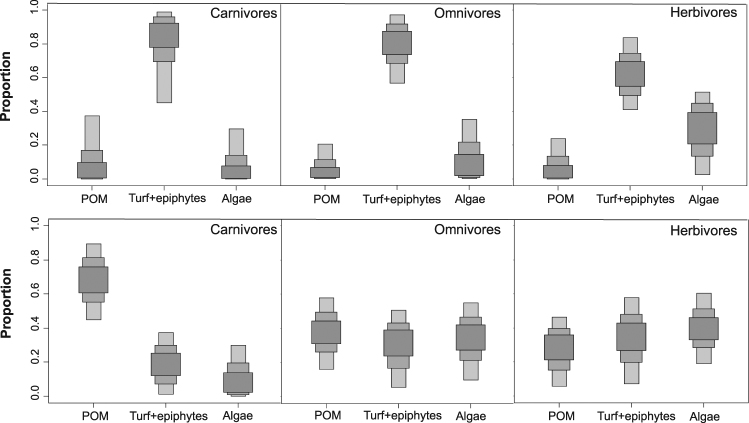
The relative importance of basal carbon sources supporting the food webs on each reef was explored with a Bayesian mixing model. This analysis revealed that the main carbon source for all three consumer categories on Limones was algal turf + epiphytes, which on average contributed approximately 60%, 80%, and 85% of organic carbon to the isotopic signature of herbivores, omnivores, and carnivores, respectively. Macroalgae were the second most important carbon source for herbivores (~29%), whereas POM contributed much smaller percentages to the isotopic signature of all consumer trophic categories on Limones (Fig. 5A). In contrast, at Bonanza all basal carbon sources contributed more evenly to the diet of omnivores and herbivores, but POM emerged as a more important source for carnivores, contributing ~70% to their isotopic signature versus ~20% from turf + epiphytes and ~10% from macroalgae (Fig. 5B).
Figure 5.
Importance of carbon sources for consumers. The relative importance of basal carbon sources for carnivores, omnivores, and herbivores in Limones (upper panel) and Bonanza (lower panel) reefs. Boxes in different gray shading denote 95% (light), 75% (darker) and 50% (darkest) credibility intervals.
Trophic niche of consumers
The total area of the trophic niche for each consumer category was compared between reefs. The difference was much greater for herbivores than for omnivores and carnivores (Fig. 6). Herbivores from Limones had a relatively narrow δ13C range (from −16.31‰ for S. aurofrenatum to −13.51‰ for the long-spined sea urchin Diadema antillarum) and a smaller standard ellipse area (SEAc) (Table 1). In contrast, herbivores from Bonanza had a broader δ13C range (from −18.28‰ for A. coeruleus to −11.90‰ for M. coryphe), resulting in a greater niche size, as indicated by the convex hull area (TA) and SEAc (Table 1), with much less overlap (47%) between their SEAc and the SEAc of herbivores from Limones than the other way around (88%) (Table 1, Fig. 6). In omnivores, the magnitude of the TAs was large because P. paru was relatively depleted in δ13C on both reefs, but more so in Bonanza, constituting an outlier and resulting in a relatively large convex hull in which much of the contained niche space was unoccupied. In this case, the SEAc provides a better characterization of the niche width30. The SEAc of omnivores from Limones and Bonanza had a great overlap (100% and 84%, respectively) (Table 1, Fig. 6). TAs were much smaller for carnivores than omnivores, and SEAc of carnivores showed a substantial overlap between reefs (69% for Limones and 76% for Bonanza) (Table 1, Fig. 6). Overall, these results strongly suggest that Bonanza has a greater range of benthic basal carbon sources than Limones, which is consistent with the differential percent cover of macroalgae between reefs (see Fig. 2).
Figure 6.
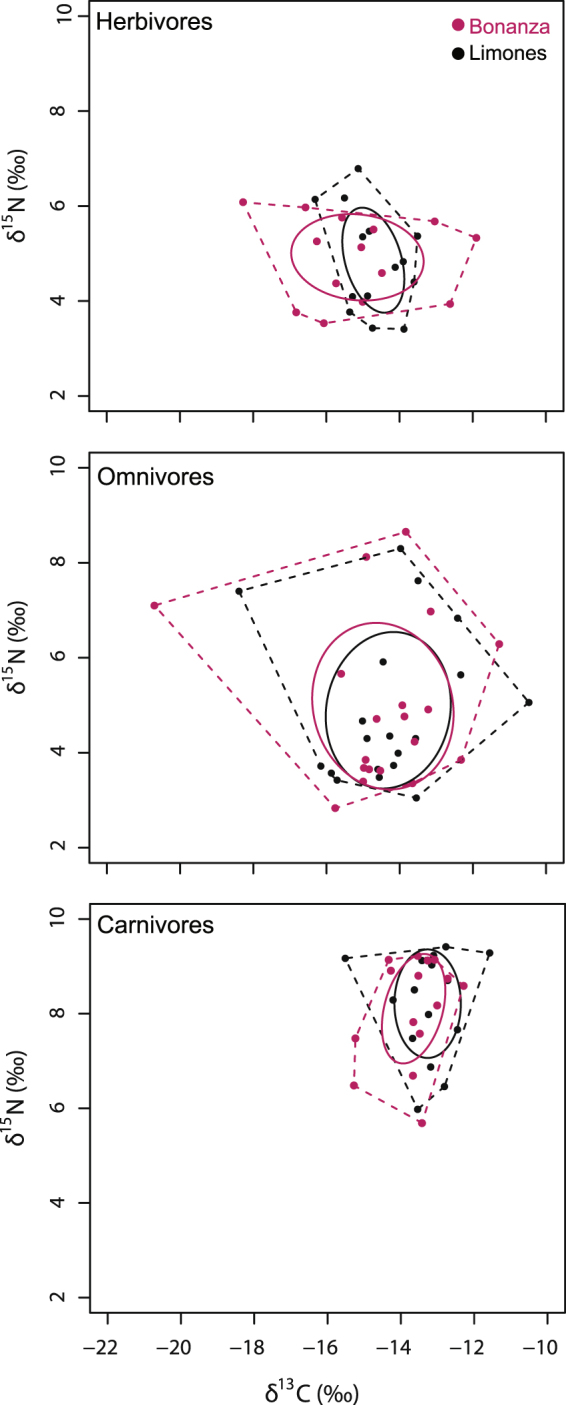
Isotopic niche by trophic category of consumers. Mean δ13C and δ15N values of herbivores (top panel), omnivores (center panel) and carnivores (bottom panel) in Limones (black) and Bonanza (red) reefs. Dotted lines: convex hull area (TA). Solid lines: standard ellipse area corrected for small sample sizes (SEAc).
Table 1.
Isotopic niche metrics for consumers on Limones and Bonanza reefs. NR (δ15N range), CR (δ13C range), convex hull total area (TA), Bayesian standard ellipse area (SEA), Bayesian-corrected estimate of the standard ellipse area (SEAc), overlap in SEAc between reefs for each category, and percentage of overlap with SEAc of the same trophic category from the other reef.
| Trophic category | Reef | NR | CR | TA units² | SEA units² | SEAc units² | SEAc overlap units² (%) |
|---|---|---|---|---|---|---|---|
| Herbivores | Limones | 4.88 | 2.80 | 6.06 | 2.54 | 2.75 | 2.42 (88%) |
| Bonanza | 4.59 | 6.58 | 11.00 | 4.75 | 5.15 | 2.42 (47%) | |
| Omnivores | Limones | 6.12 | 6.31 | 24.56 | 8.33 | 8.82 | 8.78 (100%) |
| Bonanza | 6.01 | 7.55 | 32.02 | 10.01 | 10.60 | 8.78 (84%) | |
| Carnivores | Limones | 4.36 | 3.75 | 7.45 | 3.03 | 3.26 | 2.25 (69%) |
| Bonanza | 5.38 | 2.39 | 6.93 | 2.76 | 2.97 | 2.25 (76%) |
Discussion
Habitat degradation is thought to be a major driver of change in the species composition and trophic structure of ecosystems2,5,9. In coral reefs, recent studies have shown that alterations to habitat structure may result in the shortening of food chains through a decrease in the size of reef fishes16, eutrophication33,43, or changes in prey availability for mesopredators34. Here, we show that the food chain length and the trophic space of 48 reef-associated species were essentially unaltered when comparing a coral-dominated reef (Limones) and a degraded, macroalgal-dominated reef (Bonanza). We chose two reefs of a similar size to minimize the potential effect of ecosystem size on food chain length24,27. Our results also do not imply similarities in community structure because we intentionally sampled the same species on both reefs. Yet, the structure of the coral reef food web and the trophic position of high-order consumers (mesopredators) were similar between both reefs despite the difference in degradation, although consumers showed greater enrichment of δ13C on the degraded reef.
On coral reefs, baseline carbon sources may include benthic macroalgae, reef-associated detritus, particulate organic matter (POM), coral tissue, and phytoplankton38,44. We focused our samples of baseline carbon sources on erect macroalgae and turf, which cover a large percentage of reef area on both reefs (see Fig. 2), and on POM, which is an important carbon source in many coral reefs45,46. In a coral reef of New Caledonia, Le Bourg et al.46 compared food chains based on POM between two sites distant from each other by less than 10 km, a reef lagoon and the outer slope of the reef. They found that δ15N did not differ between zones, whereas δ13C values were significantly higher in the lagoon than on the outer slope, suggesting that the two food chains were based on different primary sources of carbon46. We obtained a similar result, as values of δ15N and the derived trophic positions of species did not differ between Limones and Bonanza but the contribution of different carbon sources varied substantially between the two food chains, even though we compared the same zone (back reef) of two separate reefs distant from each other by ~2 km.
Although results from mixing models should be taken with caution because they are based on fixed trophic enrichment factors47, our results strongly suggest that on the coral-dominated reef, turf algae and epiphytes constitute a key source of carbon for consumers of the different trophic categories, whereas macroalgae and POM are considerably less important. Turf is an important component of coral reef food webs because many more herbivorous fishes feed on turf algae than on erect macroalgae48,49, which they often find unpalatable. In addition, turf traps vast amounts of detritus, and some fishes that are considered as herbivorous actually feed on the detritus trapped in the turf50. Also importantly, algal turf constitutes one of the most productive habitats for small reef mesograzer invertebrates, in particular small crustaceans such as amphipods, isopods, ostracods, and tanaids51,52, which can be consumed by fishes53,54. Therefore, turf-feeding herbivores constitute an important trophic link between benthic primary production and secondary consumers55,56, especially since the use of stable isotopes has revealed that predators on coral reefs consume more herbivorous prey (both fishes and invertebrates) than previously reported57. This appears to be the case in Limones, where the isotopic signature of consumers in all trophic categories was consistent with a more benthic-derived carbon pathway. Similarly, benthic primary production was found to be an important source for consumer production, including apex predators, in some coral-dominated coral reefs of Australia and Hawaii32,35,38.
In contrast to Limones, the distribution of the carbon signal on Bonanza was broader and originated from different sources, potentially increasing trophic diversity (indicated by a broader range in δ13C) at the base of the food web41. Among consumers, herbivores in particular exhibited a broader isotopic niche along the carbon axis on Bonanza compared with Limones. On Bonanza, macroalgae, algal turf, and POM were identified as being more or less equally important as carbon sources for herbivores and omnivores, but not for carnivores. Degraded reefs are characterized by increases in algal cover that can benefit some herbivorous fishes but only in the short term, as some large erect macroalgae (such as species in the genera Dictyota, Lobophora, Laurencia, and Sargassum) are often unpalatable to fishes58,59. These macroalgae are readily consumed by small mesograzers and omnivores, such as sea hares, some snails, amphipods, and majoid crabs60,61, but the ecological effects of these small animals tend to be more localized than those of herbivorous fishes or urchins62. Therefore, macroalgal-feeding herbivores may not represent a major food source for carnivores on Bonanza, for which POM was identified as the main carbon source. On coral reefs, POM includes phytoplankton as well as other edible particles such as phytobenthos debris, mucus, and faeces45. Small pelagic and benthic fishes associated with coral reefs, such as silversides (Atherinidae) and basslets (Grammatidae), feed on POM and zooplankton, as do juveniles of surgeonfishes (Acanthuridae), grunts (Haemulidae), jacks (Carangidae), and yellowtail snappers (Ocyurus chrysurus)63. Other fishes that are mainly herbivorous can change feeding modes depending on the ecological context58,64. These fishes are prey for many reef mesopredators, which may further use resources differentially depending on the seascape configuration. For instance, the blackspot snapper Lutjanus ehrenbergii shifted from a benthic macroalgal food web on shelf reefs to a phytoplankton-based food web on oceanic reefs in the Red Sea44. Planktonic-derived production was also identified as important for sustaining key predatory species in reef fisheries in Australia45,65. On Bonanza, reef degradation appears to be causing a temporal shift from a benthic algal-based food web to a phytoplankton-based food web.
Omnivory is common in marine food webs66 and many of the species that we sampled on both reefs were omnivores with broader isotopic niches than those of herbivores or carnivores, as would be expected. On both reefs, the wider niche breadth of omnivores was mainly caused by the extreme position in the niche space of the carbon-depleted French angelfish, P. paru. Marine benthic consumers that rely upon phytoplankton for sustenance will generally display low (depleted) δ13C values67. French angelfish can consume algae and invertebrates, but feed mostly on sponges63, which are filter feeders and hence display low values of δ13C. For example, an average δ13C value of −17.2‰ was recorded for coral reef sponges in Florida68. Nevertheless, even after accounting for outliers such as P. paru, omnivores from Bonanza had the largest overall isotopic niche as revealed by the size of the SEAc. Omnivores play an important role in dampening potential trophic cascades69 and would be expected to have a greater potential of short-term adaptive response to changes in habitat degradation.
Phase shifts are becoming increasingly common in coral reefs with different levels of habitat degradation70,71. The current paradigm is that habitat loss and degradation are the main drivers of biodiversity loss, but our results cannot be discussed in terms of diversity or abundance because that was not the aim of our study. We found that coral cover was much lower and macroalgal cover much higher in Bonanza than in Limones. Yet, despite the differences in habitat degradation, food chain length was similar on both reefs, suggesting that the trophic structure of the communities associated with these reefs is robust enough to adjust to conditions of degradation. However, we do not interpret our finding as indicative of a lack of effect of habitat degradation on the trophic ecology of these reefs, because the range in δ13C values for carbon sources and herbivorous species was wider in Bonanza than in Limones, and the carbon pathways appear to differ between both reefs despite their proximity. Future research should include sampling on more coral- and algal-dominated reefs to examine the generality of our findings, and exploring the presence of potential sublethal effects (e.g. lower nutritional condition59,72) of habitat degradation on high-order consumers.
Methods
Study site
The study was conducted in the Puerto Morelos Reef National Park, located on the Mexican portion of the Mesoamerican Reef System (see Fig. 1). The Puerto Morelos reef system encompasses a series of shallow reef units and patches constituting an extended fringing reef system separated from the shoreline by a shallow lagoon. There is more coral cover on the crest and back-reef zones (down to approximately 5 m in depth) than on the fore-reef zone, which is mostly of low relief40. Habitat degradation varies among reef units and patches23. We selected two reef units exhibiting visibly contrasting levels of degradation: “Limones” and “Bonanza”. Limones (centred at 20°59.1′N, 86°47.9′W) is considered an exceptional site for Acropora palmata within the Mesoamerican Reef System due to its large populations of this branching coral species that cover nearly 40% of the reef substrata39 (Fig. 1). In contrast, Bonanza (centred at 20°57.6′N, 86°48.9′W), which in 1985 had 33% cover of live coral and 4% cover of algae40, currently exhibits extensive areas of dead Acropora skeletons (Fig. 1) and a predominance of erect macroalgae. Fishing activities are banned on both reefs since 1996. Given the high ecological value of Limones, tourist activities are not allowed in this reef since 201439, whereas Bonanza is open to visitation. The southern limit of Limones is separated from the northern limit of Bonanza by a distance of ~1900 m. The two reefs are similar in size (~1500 m in length), depth range (1–5 m), and distance from the shoreline. Samples for stable isotope analyses (water, plants, invertebrates, and fishes) were collected on the back reef to crest zones within an area of ~500 m2 near the centre of each reef.
Characterisation of reef and benthic composition
To compare the current status of Limones and Bonanza, benthic habitat composition and architectural complexity were described for each reef using eight randomly selected 10-m transects within the same area in which samples were collected on each reef. At each transect, coral cover was measured by means of the line-point counts method. A surveyor recorded the benthic component (live coral in general and different functional groups of algae) intercepting the line every 10 cm (i.e., 100 points per transect), and cover of each component was estimated as a percentage of the number of points overlaying the transect. Also, at each transect, reef complexity was measured using the rugosity index, which is the ratio of a length of a chain moulded to the reef surface to the linear distance between its start and end point18. A perfectly flat surface would have a rugosity index of one, with larger numbers indicating more complex surfaces.
Sample collection and preparation
The same species of consumers and types of basal carbon sources were sampled on both reefs by SCUBA diving and transported to the laboratory at UASA-UNAM (Puerto Morelos, Mexico) to be processed for stable isotope analyses. Consumers included 48 species, of which 32 were of invertebrates (including annelids (1 species), bivalves (4), gastropods (8), chitons (1), octopuses (1), decapod crustaceans (10), sea urchins (4), brittle stars (2), and sea cucumbers (1 species)) and 16 were of fishes (see Supplementary Table S1 for the full list of species). Our main criterion for selecting consumer species was that they were reef-associated species, preferentially with limited movement ranges, both to reduce the possibility of organisms shifting between reefs and because this type of species can be presumably more susceptible to reef habitat degradation. Basal carbon sources included particulate organic matter (POM), four species of macroalgae (Amphiroa tribulus, Caulerpa racemosa, Dictyota cervicornis, Halimeda tuna), epiphytes, and turf algae. For all consumer species and basal carbon sources, at least three to five replicates were sampled from each reef, yielding 514 samples in total. To minimize potential seasonal effects on the isotopic composition of organisms, all samples were obtained between late September 2015 and early February 2016, and samples from both reefs were interspersed throughout this period.
POM was obtained from seawater collected with a dark 5-L carboy. Then, 1 L of seawater was gently vacuumed using Whatman GF/F (0.7 µm) filters previously incinerated at 450 °C for 4 h. Samples of algal turf were taken from the top of dead coral using scalpel and forceps under a magnifying glass to avoid the presence of contaminants (e.g., endolithic animals)73. Macroalgae were collected by hand. Epiphytes were carefully removed from macroalgal fronds using a glass slide. Invertebrates were collected by hand or with the help of tweezers or nets, and transported live to the laboratory, where they were kept in containers with filtered seawater for 24–48 h with the aim of emptying their gut contents, which can affect the isotopic composition74. Individuals were then frozen and preserved at −20 °C. Prior to analyses, the calcareous shells or exoskeletons of invertebrates were removed. A sample of muscle was taken from the abdomen of crustaceans, the foot of chitons, gastropods and bivalves, the Aristotle’s lantern of urchins and brittle stars, the body wall of sea cucumbers, and the tentacles of octopuses. Fishes (16 species) were identified in situ and caught with Hawaiian spears. A sample of muscle was taken from the dorsal region and immediately frozen until processing75. All samples were acidified with HCl (1 N) to remove inorganic carbon76 and then rinsed with Milli-Q water. Samples were dried at 60 °C for 48 h in aluminium foil trays, then ground to a fine powder and homogenized with agate mortar and pestle. Powdered sub-samples were weighed and sent for isotopic analysis in ultra-pure tin capsules.
Stable isotope analyses
Elemental and stable isotope analyses were carried out in a Finnigan Delta V Plus mass spectrometer (Thermo Scientific) interfaced with an elemental analyser (Elemental Combustion System, Costech model 4010) at the Mass Spectrometry Laboratory of Centro Interdisciplinario de Ciencias Marinas, Instituto Politécnico Nacional, La Paz, Mexico. The average precision across runs was 0.1‰ for δ15N and 0.02‰ for δ13C. Carbon and nitrogen ratios were expressed in delta (δ) notation, defined as parts per mil (‰) of difference relative to an international standard:
| 1 |
where X is N (nitrogen) or C (carbon), and R is the ratio of the heavier, rarer isotopes (13C and 15N) to the lighter, more common isotopes (12C and 14N, respectively). Delta values are reported relative to the international standards of Vienna Pee-Dee Belemnite (VPDB) carbon and atmospheric nitrogen26.
Data analyses
Student’s t tests were used to test the null hypotheses of no significant differences in the mean percent coral cover, rugosity index, and δ15N and δ13C composition of each basal resource and consumer species between Limones and Bonanza. Bi-plots of δ13C and δ15N values of basal carbon sources and consumers were used to visualize the food web structure at each reef and the differences in isotope values of basal carbon sources and consumers among reefs.
The relative trophic position (TP) of all species of consumers from each reef was calculated using the following equation from the meta-analysis performed by Hussey et al.77:
| 2 |
where TPbase was set at a trophic level 2 baseline using the mean δ15N of the epifaunal reef clam Barbatia domingensis (=B. cancellaria), which we chose as the baseline organism given that filter-feeding bivalves are good integrators of the isotopic variation at the base of pelagic and benthic food webs26,35; δ15Nlim is the saturating isotope limit as TP increases; δ15Nbase is the isotope value for a known baseline consumer in the food web (in this case, B. domingensis), and k is the rate at which δ15NTP approaches δ15Nlim per TP step. Estimates of k and δ15Nlim are given by:
| 3 |
| 4 |
with values for the intercept β0 = 5.924 and the slope β1 = −0.271. These values characterize the change in δ15N as dietary δ15N values increase77.
To quantify the relative importance of basal carbon sources supporting the food webs on each reef, we applied a Bayesian mixing model using SIAR (Stable Isotope Analysis in R) version 3.0.278,79. Mixing models require the use of trophic enrichment factors and for these purposes we used the mean (±SD) values proposed by Post26 for δ13C (0.40 ± 1.30‰) and δ15N (3.40 ± 1.00‰). The estimated values of the dietary proportion were obtained via a Markov-Chain Monte Carlo (MCMC) simulation78 on stable isotope data from consumers. Each consumer species was previously categorized as herbivore, omnivore, or carnivore based on literature reports on its type of diet (see Supplementary Table S1). For each trophic category on each reef, we calculated NR (δ15N range) and CR (δ13C range), measures that provide information on the food chain length and maximum trophic position within the community, and the diversity of basal carbon sources, respectively. We also inferred the total isotopic niche width using the total area (TA) index, which is a measure of the area of a polygon drawn through the most extreme data points of the population in the isotopic niche space (i.e., the convex hull)41, and the standard ellipse area (SEA), which measures the isotopic niche width of the mean core community and contains approximately 40% of the data42. To compare the total trophic niche area for each consumer trophic category (i.e., herbivores, omnivores, carnivores) between reefs, we used the Bayesian standard ellipse area corrected for sample size (SEAc), estimated and plotted using the SIBER routine for the SIAR package in R42. For each trophic category, distinction between the two SEAc for each reef was inferred by posterior probabilities derived by Bayesian inference based on 100,000 MCMC draws, and the trophic niche overlap was calculated as the proportion of SEAc overlapping between the two reefs.
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).
Electronic supplementary material
Acknowledgements
We thank Fernando Negrete-Soto and Cecilia Barradas-Ortiz for their invaluable technical assistance throughout the development of this research. We also thank Iris Segura-García for her advice and training on sampling for stable isotope analyses; Ángel H. Ruvalcaba-Díaz for his technical assistance in the stable isotope analyses; Roberto González-Gómez, Alí Espinosa-Magaña, Leslie Cid-González, and Martha Oropesa-Ríos for their assistance in field and laboratory activities, and Laura Celis-Gutiérrez for her help obtaining the literature for species identification. This study received funding from Universidad Nacional Autónoma de México (Program UNAM-DGAPA-PAPIIT, project IN-205614), granted to L.A.-E. UNAM-DGAPA also provided two years of financial support to P.S.M.-V through a postdoctoral fellowship. Open access was partially paid by Instituto de Ciencias del Mar y Limnología,UNAM.
Author Contributions
E.L.-A., L.A.-F., and P.B.-F. designed the research; P.S.M.-V. performed the research and collected data; P.S.M.-V., E.L.-A., P.B.-F., L.A.-F., S.A.-G., and A.S.-G. analysed data; E.L.-A., P.S.M.-V., P.B.-F., L.A.-F. wrote the paper. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-22463-x.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Duffy JE. Biodiversity loss, trophic skew and ecosystem functioning. Ecol. Lett. 2003;6:680–687. doi: 10.1046/j.1461-0248.2003.00494.x. [DOI] [Google Scholar]
- 2.Dobson A, et al. Habitat loss, trophic collapse, and the decline of ecosystem services. Ecology. 2006;87:1915–1924. doi: 10.1890/0012-9658(2006)87[1915:HLTCAT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 3.Winemiller, K. O. & Polis, G. A. Food webs: what can they tell us about the world? In Food webs: Integration of patterns and dynamics (eds Polis, G. A. & Winemiller, K. O.) 1–22 (Kluwer/Academic,1996).
- 4.Estes JA, et al. Trophic downgrading of planet Earth. Science. 2011;333:301–306. doi: 10.1126/science.1205106. [DOI] [PubMed] [Google Scholar]
- 5.Hamer KC, et al. Impacts of selective logging on insectivorous birds in Borneo: the importance of trophic position, body size and foraging height. Biol. Conserv. 2015;188:82–88. doi: 10.1016/j.biocon.2014.09.026. [DOI] [Google Scholar]
- 6.Letourneur Y, et al. Identifying carbon sources and trophic position of coral reef fishes using diet and stable isotope (δ15N and δ13C) analyses in two contrasted bays in Moorea, French Polynesia. Coral Reefs. 2013;32:1091–1102. doi: 10.1007/s00338-013-1073-6. [DOI] [Google Scholar]
- 7.Le Viol I, et al. More and more generalists: two decades of changes in the European avifauna. Biol. Lett. 2012;8:780–782. doi: 10.1098/rsbl.2012.0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Álvarez-Filip L, Paddack MJ, Collen B, Robertson DR, Côté IM. Simplification of Caribbean reef-fish assemblages over decades of coral reef degradation. PLoS One. 2015;10:e0126004. doi: 10.1371/journal.pone.0126004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Öckinger E, et al. Life-history traits predict species responses to habitat area and isolation: a cross-continental synthesis. Ecol. Lett. 2010;13:969–979. doi: 10.1111/j.1461-0248.2010.01487.x. [DOI] [PubMed] [Google Scholar]
- 10.Newbold T, et al. Ecological traits affect the response of tropical forest bird species to land-use intensity. Proc. R. Soc. Lond. Ser. B: Biol. Sci. 2013;280:2012–2131. doi: 10.1098/rspb.2012.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moberg F, Folke C. Ecological goods and services of coral reef ecosystems. Ecol. Econ. 1999;29:215–233. doi: 10.1016/S0921-8009(99)00009-9. [DOI] [Google Scholar]
- 12.Hughes TP, et al. Climate change, human impacts, and the resilience of coral reefs. Science. 2003;301:929–933. doi: 10.1126/science.1085046. [DOI] [PubMed] [Google Scholar]
- 13.Gardner TA, Côté IM, Gill JA, Grant A, Watkinson AR. Long-term region-wide declines in Caribbean corals. Science. 2003;301:958–960. doi: 10.1126/science.1086050. [DOI] [PubMed] [Google Scholar]
- 14.Bruno JF, Selig EZ. Regional decline of coral cover in the Indo-Pacific: timing, extent, and subregional comparisons. PLoS One. 2007;2:e711. doi: 10.1371/journal.pone.0000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson, J. B. C., Donovan, M. K., Cramer, K. L. & Lam, V. V. (Editors). Status and Trends of Caribbean Coral Reefs: 1970–2012, 304 pp. (Global Coral Reef Monitoring Network, International Union for the Conservation of Nature, 2014).
- 16.Álvarez-Filip L, Gill JA, Dulvy NK. Complex reef architecture supports more small-bodied fishes and longer food chains on Caribbean reefs. Ecosphere. 2011;2(10):art118. doi: 10.1890/ES11-00185.1. [DOI] [Google Scholar]
- 17.Álvarez-Filip L, Carricart-Ganivet JP, Horta-Puga G, Iglesias-Prieto R. Shifts in coral assemblage composition do not ensure persistence of reef functionality. Sci. Rep. 2013;3:1–5. doi: 10.1038/srep03486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Álvarez-Filip L, Dulvy NK, Gill JA, Côté IM, Watkinson AR. Flattening of Caribbean coral reefs: region-wide declines in architectural complexity. Proc. R. Soc. Lond., Ser. B: Biol. Sci. 2009;276:3019–3025. doi: 10.1098/rspb.2009.0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferrario F, et al. The effectiveness of coral reefs for coastal hazard risk reduction and adaptation. Nat. Commun. 2014;5:3794. doi: 10.1038/ncomms4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandin SA, McNamara DE. Spatial dynamics of benthic competition on coral reefs. Oecologia. 2012;168:1079–1090. doi: 10.1007/s00442-011-2156-0. [DOI] [PubMed] [Google Scholar]
- 21.Munday PL. Habitat loss, resource specialisation, and extinction on coral reefs. Global Change Biol. 2004;10:1642–1647. doi: 10.1111/j.1365-2486.2004.00839.x. [DOI] [Google Scholar]
- 22.Graham NAJ, et al. Extinction vulnerability of coral reef fishes. Ecol. Lett. 2011;14:341–348. doi: 10.1111/j.1461-0248.2011.01592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lozano-Álvarez E, et al. Does reef architectural complexity influence resource availability for a large reef-dwelling invertebrate? J. Sea Res. 2017;128:84–91. doi: 10.1016/j.seares.2017.08.010. [DOI] [Google Scholar]
- 24.Post DM, Pace ML, Hairston NGJ. Ecosystem size determines food-chain length in lakes. Nature. 2000;405:1047–1049. doi: 10.1038/35016565. [DOI] [PubMed] [Google Scholar]
- 25.Post DM. The long and short of food-chain length. Trends Ecol. Evol. 2002;17:269–277. doi: 10.1016/S0169-5347(02)02455-2. [DOI] [Google Scholar]
- 26.Post DM. Using stable isotopes to estimate trophic position: models, methods, and assumptions. Ecology. 2002;8:703–710. doi: 10.1890/0012-9658(2002)083[0703:USITET]2.0.CO;2. [DOI] [Google Scholar]
- 27.Takimoto G, Spiller DA, Post DM. Ecosystem size, but not disturbance, determines food-chain length on islands of the Bahamas. Ecology. 2008;89:3001–3007. doi: 10.1890/07-1990.1. [DOI] [PubMed] [Google Scholar]
- 28.Vander Zanden MJ, Fetzer WW. Global patterns of aquatic food chain length. Oikos. 2007;116:1378–1388. doi: 10.1111/j.0030-1299.2007.16036.x. [DOI] [Google Scholar]
- 29.Schriver TA. Food webs in relation to variation in the environment and species assemblage: a multivariate approach. PLoS One. 2015;10:e0122719. doi: 10.1371/journal.pone.0122719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Layman CA, et al. Applying stable isotopes to examine food web structure: an overview of analytical tools. Biol. Rev. 2012;87:545–562. doi: 10.1111/j.1469-185X.2011.00208.x. [DOI] [PubMed] [Google Scholar]
- 31.Vander Zanden MJ, Rasmussen JB. Variation in δ15N and δ13C trophic fractionation: implications for aquatic food web studies. Limnol. Oceanogr. 2001;46:2061–2066. doi: 10.4319/lo.2001.46.8.2061. [DOI] [Google Scholar]
- 32.Hilting AK, Currin CA, Kosaki RK. Evidence for benthic primary production support of an apex predator–dominated coral reef food web. Mar. Biol. 2013;160:1681–1695. doi: 10.1007/s00227-013-2220-x. [DOI] [Google Scholar]
- 33.Kolasinski J, et al. Stable isotopes reveal spatial variability in the trophic structure of a macro-benthic invertebrate community in a tropical coral reef. Rapid Commun. Mass Spectrom. 2016;30:433–446. doi: 10.1002/rcm.7443. [DOI] [PubMed] [Google Scholar]
- 34.Hempson TN, et al. Coral reef mesopredators switch prey, shortening food chains, in response to habitat degradation. Ecol. Evol. 2017;7:2626–2635. doi: 10.1002/ece3.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wyatt ASJ, Waite AM, Humphries S. Stable isotope analysis reveals community-level variation in fish trophodynamics across a fringing coral reef. Coral Reefs. 2012;31:1029–1044. doi: 10.1007/s00338-012-0923-y. [DOI] [Google Scholar]
- 36.Kürten B, et al. Influence of environmental gradients on C and N stable isotope ratios in coral reef biota of the Red Sea, Saudi Arabia. J. Sea Res. 2014;85:379–394. doi: 10.1016/j.seares.2013.07.008. [DOI] [Google Scholar]
- 37.Briand MJ, Bonnet X, Goiran C, Guillou G, Letourneur Y. Major sources of organic matter in a complex coral reef lagoon: identification from isotopic signatures (δ13C and δ15N) PLoS One. 2015;10:e0131555. doi: 10.1371/journal.pone.0131555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Briand MJ, Bonnet X, Guillou G, Letourneur Y. Complex food webs in highly diversified coral reefs: Insights from δ13C and δ15N stable isotopes. Food Webs. 2016;8:12–22. doi: 10.1016/j.fooweb.2016.07.002. [DOI] [Google Scholar]
- 39.Rodríguez-Martínez RE, Banaszak AT, McField MD, Beltrán-Torres AU, Álvarez-Filip L. Assessment of Acropora palmata in the Mesoamerican Reef System. PLoS One. 2014;9:e96140. doi: 10.1371/journal.pone.0096140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jordán-Dahlgren, E. Atlas de los Arrecifes Coralinos del Caribe Mexicano, 110 pp. (Centro de Investigaciones de Quintana Roo, 1993).
- 41.Layman CA, Arrington DA, Montaña CG, Post DM. Can stable isotope ratios provide for community-wide measures of trophic structure? Ecology. 2007;88:42–48. doi: 10.1890/0012-9658(2007)88[42:CSIRPF]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 42.Jackson AL, Inger R, Parnell AC, Bearhop S. Comparing isotopic niche widths among and within communities: SIBER–Stable Isotope Bayesian Ellipses in R. J. Anim. Ecol. 2011;80:595–602. doi: 10.1111/j.1365-2656.2011.01806.x. [DOI] [PubMed] [Google Scholar]
- 43.Kolasinski J, Rogers K, Cuet P, Barry B, Froui P. Sources of particulate organic matter at the ecosystem scale: a stable isotope and trace element study in a tropical coral reef. Mar. Ecol. Prog. Ser. 2011;443:77–93. doi: 10.3354/meps09416. [DOI] [Google Scholar]
- 44.McMahon KW, Thorrold SR, Houghton LA, Berumen ML. Tracing carbon flow through coral reef food webs using a compound-specific stable isotope approach. Oecologia. 2016;180:809–821. doi: 10.1007/s00442-015-3475-3. [DOI] [PubMed] [Google Scholar]
- 45.Wyatt ASJ, Lowe RJ, Humphries S, Waite AM. Particulate nutrient fluxes over a fringing coral reef: source-sink dynamics inferred from carbon to nitrogen ratios and stable isotopes. Limnol. Oceanogr. 2013;58:409–427. doi: 10.4319/lo.2013.58.1.0409. [DOI] [Google Scholar]
- 46.Le Bourg, B. et al. The same but different: stable isotopes reveal two distinguishable, yet similar, neighbouring food chains in a coral reef. J. Mar. Biol. Assoc. U.K. Preprint at 10.1017/S0025315417001370 (2017).
- 47.Viana IG, Valiela I, Martinetto P, Monteiro-Pierce R, Fox SE. Isotopic studies in Pacific Panama mangrove estuaries reveal lack of effect of watershed deforestation on food webs. Mar. Environ. Res. 2015;103:95–102. doi: 10.1016/j.marenvres.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 48.Bellwood DR, Hughes TP, Hoey AS. Sleeping functional group drives coral reef recovery. Curr. Biol. 2006;16:2434–2439. doi: 10.1016/j.cub.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 49.Ledlie MH, et al. Phase shifts and the role of herbivory in the resilience of coral reefs. Coral Reefs. 2007;26:641–653. doi: 10.1007/s00338-007-0230-1. [DOI] [Google Scholar]
- 50.Wilson SK, Bellwood DR, Choat JH, Furnas MJ. Detritus in the epilithic algal matrix and its use by coral reef fishes. Oceanogr. Mar. Biol. Annu. Rev. 2003;41:279–309. [Google Scholar]
- 51.Cowles A, Hewitt JE, Taylor RB. Density, biomass and productivity of small mobile invertebrates in a wide range of coastal habitats. Mar. Ecol. Prog. Ser. 2009;384:175–185. doi: 10.3354/meps08038. [DOI] [Google Scholar]
- 52.Berthelsen AK, Taylor RB. Arthropod mesograzers reduce epiphytic overgrowth of subtidal coralline turf. Mar. Ecol. Prog. Ser. 2014;515:123–132. doi: 10.3354/meps11025. [DOI] [Google Scholar]
- 53.Kramer MJ, Bellwood O, Bellwood DR. The trophic importance of algal turfs for coral reef fishes: the crustacean link. Coral Reefs. 2013;32:575–583. doi: 10.1007/s00338-013-1009-1. [DOI] [Google Scholar]
- 54.Dromard CR, Bouchon-Navaro Y, Harmelin-Vivien M, Bouchon C. Diversity of trophic niches among herbivorous fishes on a Caribbean reef (Guadeloupe, Lesser Antilles), evidenced by stable isotope and gut content analyses. J. Sea Res. 2015;95:124–131. doi: 10.1016/j.seares.2014.07.014. [DOI] [Google Scholar]
- 55.Choat JH, Clements KD, Robbins WD. The trophic status of herbivorous fishes on coral reefs. I. Dietary analysis. Mar. Biol. 2002;140:613–623. doi: 10.1007/s00227-001-0715-3. [DOI] [Google Scholar]
- 56.Kramer MJ, Bellwood O, Fulton CJ, Bellwood DR. Refining the invertivore: diversity and specialisation in fish predation on coral reef crustaceans. Mar. Biol. 2015;162:1779–1786. doi: 10.1007/s00227-015-2710-0. [DOI] [Google Scholar]
- 57.Page HM, et al. Stable isotopes reveal trophic relationships and diet of consumers in temperate kelp forest and coral reef ecosystems. Oceanography. 2013;26(3):180–189. doi: 10.5670/oceanog.2013.61. [DOI] [Google Scholar]
- 58.McClanahan TR, Hendrick V, Rodrigues MJ, Polunin NVC. Varying responses of herbivorous and invertebrate-feeding fishes to macroalgal reduction on a coral reef. Coral Reefs. 1999;18:195–203. doi: 10.1007/s003380050181. [DOI] [Google Scholar]
- 59.Pratchett MS, et al. Effects of climate-induced coral bleaching on coral-reef-fishes—ecological and economic consequences. Oceanogr. Mar. Biol. Annu. Rev. 2008;46:251–296. [Google Scholar]
- 60.Coen LD. Herbivory by crabs and the control of algal epibionts on Caribbean host corals. Oecologia. 1988;75:198–203. doi: 10.1007/BF00378597. [DOI] [PubMed] [Google Scholar]
- 61.Stachowicz JJ, Hay ME. Mutualism and coral persistence: the role of herbivore resistance to algal chemical defense. Ecology. 1999;80:2085–2101. doi: 10.1890/0012-9658(1999)080[2085:MACPTR]2.0.CO;2. [DOI] [Google Scholar]
- 62.Hay ME. The ecology and evolution of seaweed-herbivore interactions on coral reefs. Coral Reefs. 1997;16(Suppl):S67–S76. doi: 10.1007/s003380050243. [DOI] [Google Scholar]
- 63.Randall JE. Food habits of reef fishes of the West Indies. Stud. Trop. Oceanogr. 1967;5:655–847. [Google Scholar]
- 64.Ferreira CEL, Gonçalves JEA. J. Fish Biol. 2006. Community structure and diet of roving herbivorous reef fishes in the Abrolhos Archipelago, south-western Atlantic; pp. 1533–1551. [Google Scholar]
- 65.Frisch AJ, Ireland M, Baker R. Trophic ecology of large predatory reef fishes: energy pathways, trophic level, and implications for fisheries in a changing climate. Mar. Biol. 2014;161:61–73. doi: 10.1007/s00227-013-2315-4. [DOI] [Google Scholar]
- 66.Thompson RM, Hemberg M, Starsomski BM, Shurin JB. Trophic levels and trophic tangles: the prevalence of omnivory in real food webs. Ecology. 2007;88:612–617. doi: 10.1890/05-1454. [DOI] [PubMed] [Google Scholar]
- 67.France RL. Carbon-13 enrichment in benthic compared to planktonic algae: foodweb implications. Mar. Ecol. Prog. Ser. 1995;124:307–312. doi: 10.3354/meps124307. [DOI] [Google Scholar]
- 68.Lamb K, Swart PK, Altabet MA. Nitrogen and carbon isotopic systematics of the Florida reef tract. Bull. Mar. Sci. 2012;88:119–146. doi: 10.5343/bms.2010.1105. [DOI] [Google Scholar]
- 69.Bruno JF, O’Connor MI. Cascading effects of predator diversity and omnivory in a marine food web. Ecol. Lett. 2005;8:1048–1056. doi: 10.1111/j.1461-0248.2005.00808.x. [DOI] [Google Scholar]
- 70.Hughes TP, Graham NAJ, Jackson JBC, Mumby PJ, Steneck RS. Rising to the challenge of sustaining coral reef resilience. Trends Ecol. Evol. 2010;25:633–642. doi: 10.1016/j.tree.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 71.Bruno JF, Precht WF, Vroom PS, Aronson RD. Coral reef baselines: How much macroalgae is natural? Mar. Pollut. Bull. 2014;80:24–29. doi: 10.1016/j.marpolbul.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 72.Hempson, T. N., Graham, N. A. J., McNeil, M. A., Bodin, N. & Wilson, S. K. Regime shifts shorten food chains for mesopredators with potential sublethal effects. Funct. Ecol. Preprint at 10.1111/1365-2435.13012 (2018).
- 73.Titlyanov EA, Kiyashko SI, Titlyanova TV, Kalita TL, Raven JA. δ13C and δ15N values in reef corals Porites lutea and P. cylindrica and in their epilithic and endolithic algae. Mar. Biol. 2008;155:353–361. doi: 10.1007/s00227-008-1025-9. [DOI] [Google Scholar]
- 74.Newman Gearing, J. The study of diet and trophic relationships through natural abundance 13C in Carbon IsotopeTechniques (eds. Coleman, D. C. & Fry, B.) 201–218 (Academic Press, 1991).
- 75.Stallings CD, et al. Effects of preservation methods of muscle tissue from upper-trophic level reef fishes on stable isotope values (δ13C and δ15N) PeerJ. 2015;3:e874. doi: 10.7717/peerj.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boutton, T. W. Stable carbon isotope ratios of natural materials: I. Sample preparation and mass spectrometric analyses in Carbon isotope techniques (eds. Coleman, D. C. & Fry, B.) 155–171 (Academic Press, 1991).
- 77.Hussey NE, et al. Rescaling the trophic structure of marine food webs. Ecol. Lett. 2014;17:239–250. doi: 10.1111/ele.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Parnell AC, Inger R, Bearhop S, Jackson AL. Source partitioning using stable isotopes: coping with too much variation. PLoS One. 2010;5:e9672. doi: 10.1371/journal.pone.0009672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/ (2015).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).