Abstract
Lung cancer is a heterogeneous disease encompassing a wide array of genetic abnormalities. The MET receptor tyrosine kinase is altered in many lung cancers, especially non-small cell lung cancer (NSCLC), and clinical trials of MET inhibitors that are underway are documenting cases of acquired resistance. Based on evidence that the RON tyrosine kinase receptor can also be overexpressed in NSCLC, we evaluated the potent MET/RON dual kinase inhibitor LY2801653 in this setting. LY2801653 was more efficacious than the MET/ALK/RON/ROS inhibitor crizotinib with a distinct pattern of downstream signaling effects. Using the PamGene platform, we found that inhibition of MET and RON was associated with decreased phosphorylation of CBL, PI3K and STAT3. In classical and orthotopic mouse xenograft models of lung cancer, LY2801653 decreased tumor growth, dramatically inhibiting mitotic events and angiogenesis. Taken together, our results argued that specific targeting of the MET/RON kinases could provide robust inhibition of cell proliferation and tumor outgrowth in multiple in vitro and in vivo models of NSCLC. These findings offer a robust preclinical proof of concept for MET/RON targeting by LY2801653 as a promising small molecule modality to treat NSCLC.
Keywords: non-small cell lung cancer, MET/RON receptor tyrosine kinase, LY2801653 kinase inhibitor, signal transduction, orthotopic mouse model
Introduction
Lung cancer is a global health problem, and can be divided into NSCLC and small cell lung cancer (SCLC). NSCLC can be further subdivided into various histologies such as adenocarcinoma, squamous cell carcinoma (SCC), and large cell carcinoma. As has been shown recently, there are various molecular subtypes of NSCLC, such as EGFR mutation (1, 2), ALK translocation (3), and ROS1 translocation (4). We have previously shown MET receptor tyrosine can be a good target for NSCLC (5, 6).
MET RTK can be activated through a number of mechanisms, especially with ligand hepatocyte growth factor (HGF) stimulation. In lung cancer, MET can be overexpressed along with HGF. There also can be gain-of-function mutations within the semaphorin and juxtamembrane domain (5, 7, 8). In a subset of NSCLC, MET is amplified (9–11). RON is a RTK similar to MET, however, its activation is dependent on a separate ligand, macrophage-stimulating protein (MSP). RON has been shown to be overexpressed in NSCLC, and there are a number of isoforms in lung cancer (12). Particularly, delta 165 and 155 isoforms are known to be potent oncogenes (13, 14). It appears there is strong interaction of MET with its family member RON (15, 16). Recent data indicate an increasingly important role for receptor cross-talk in the activation of MET and RON. MET/RON complexes are present on the cell surface prior to ligand-induced dimerization, and ligand-stimulated MET activation results in direct transphosphorylation of RON (15). Catenacci DV, et al. have shown that RON and MET co-stimulation led to enhanced malignant phenotypes over stimulation of either receptor alone using gastroesophageal adenocarcinoma cell lines. They have shown that growth inhibition as evidenced by viability and apoptosis assays was optimal using novel blocking monoclonal antibodies to both RON and MET, versus either alone. They have also shown that combined siRNA knockdown of RON and MET inhibited signaling feedback loops and led to optimal anti-tumorigenic effect (16). Since only a subset of MET expressing tumors respond to anti-MET therapeutics, we hypothesized that MET and RON could be better therapeutic targets in NSCLC. There are various strategies to inhibit MET and RON—such as specific antibodies or SMIs (17). The compound LY2801653 is SMI that has shown in vitro activity against both MET and RON receptors at nanomolar concentrations.
In this study, we sought to determine the specific inhibition of NSCLC cell lines with LY2801653. Compared to crizotinib, we found LY2801653 to be more efficacious in A549, H1703 and H1993 cell lines. Concurrently, we were able to show dramatic effects on mouse xenograft models. Interestingly, we show that there are specific signal transduction pathways that are affected which help in understanding the systems biology for MET/RON pathways. These data support potential clinical development of LY2801653 for treatment of NSCLC.
Materials and Methods
Cell Lines
We used one human bronchial epithelial cell line (BEAS-2B) and 10 NSCLC cell lines, A549, H358, H520, H522, H661, H1703, H1993, H2170, H2228, SKMES-1 that were obtained from the American Type Culture Collection (Manassas, VA, USA), and cultured accordingly. A TPR-MET expressing murine pre-B cell line, BaF3 TPR-MET, was generated by transfection of an expression vector containing TPR-MET cDNA as previously described (18). BaF3 was grown in RPMI-1640 containing 10% FBS and 10% WEHI-conditioned medium as a source of murine interleukin-3. A luciferase tagged A549 cell line, A549-luc-C8, was purchased from Caliper Life Sciences (Hopkinton, MA, USA) and grown in RPMI 1640 with 10% FBS.
MET/RON SMI LY2801653
The MET/RON SMI was provided by Eli Lilly and Company (Indianapolis, IN, USA). The drug was received in powder form and dissolved in DMSO according to company instructions and used at the indicated concentration.
Specific Knockdown of MET/RON by siRNA
Targeted siRNA to MET and RON were used on A549 cells individually and in combination for dual knockdown (Santa Cruz; Met siRNA, sc-29197, Ron siRNA, sc-36434, Control siRNA-D, sc-44232) using the Lipofectamine RNAiMAX Transfection Reagent kit (Invitrogen #13778-075). Lysates from each siRNA treatment were immunoblotted to verify gene knockdown and cell viability assessed to determine the effect of gene knockdown relative to siRNA control cells. Detailed methods for this assay are described in Supplementary Materials and Methods.
Cell viability assay
Cell lines’ sensitivity to MET/RON inhibitor LY2801653 or siRNA was determined by performing cell proliferation assay using standard MTT. Detailed methods for this assay are described in Supplementary Materials and Methods. The IC50 was generated for all cell lines. Similarly, the efficacy of crizotinib was determined. Crizotinib was purchased from LC Laboratories (Woburn, MA, USA).
Immunoblot analysis and antibodies
NSCLC cells treated with drug or siRNA were immunoblotted to evaluate the biochemical effects of MET/RON inhibition. Untreated cells or control siRNA were used as controls. Immunoblotting on whole cell lysates was performed following routine protocols (19, 20). All antibodies used for immunoblot analysis are listed in Supplementary Materials and Methods.
PamGene tyrosine kinase array and run analyses
PamGene technology using the PamChip® Tyrosine Kinase Array allows the kinetic detection of the phosphorylation of peptides spotted onto a 3-dimensional porous well of a 4-array chip produced and commercialized by PamGene (‘s-Hertogenbosch, The Netherlands). This technology can be used to measure activity of purified kinases and the effects of kinase inhibitors on them. Phosphorylation on these arrays is measured as described elsewhere (21). Detailed methods for PamGene analyses are provided in Supplementary Materials and Methods. The lysates of H1993 cells treated with LY2801653 (1, 3 and 10 nM) for 4 hours similar to assay conducted for immunoblot analysis were hybridized in four technical replicates. The data generated were analyzed with BioNavigator (Version 0.4.2.82; PamGene).
Pathway and network analyses
Peptides found to be significantly down- or up-regulated in the statistical analysis (p<0.05) in PamGene were used for pathway and network analyses using the bioinformatics analysis software MetaCore™ (GeneGo, Thomson Reuters, St. Joseph, MI, USA) as described (22, 23). Several MetaCore™ tools were used to analyze data and are listed in Supplementary Materials and Methods. MetaCore™’s enrichment analysis tool uses hypergeometric modeling to determine statistical significance of enrichment. ‘GeneGo Map Folders’ (MetaCore™ tool) are organized to collect all GeneGo maps that describe different canonical pathways involved in the same process. The significant process networks were noted, and relevant signaling networks were assembled based on statistically significant data generated by PamGene fold-change calculation.
Xenograft tumor model for LY2801653
Female homozygous athymic nude mice aged 5–6 weeks were purchased from Harlan Laboratories (Indianapolis, IN, USA) and allowed to acclimate for 1 week. Animal care was in accordance with institutional animal care guidelines.
Approximately 2.5×106 A549 or 5×106 H1993 NSCLC cells were inoculated subcutaneously in the right flank of each mouse. Tumor growth was measured with calipers and volume (mm3) calculated as (L × W × H)/2. When the volume reached a mean of 200~400 mm3, mice were randomized into two groups of 10 and 11 and treatment initiated. LY2801653 was administered twice a day for 4 weeks by oral gavage at 20 mg/kg to one group while the vehicle 10% Acacia was delivered to the second group at same duration, route and dose. Body weight and tumor volume were recorded every 3 days until study was terminated and animals euthanized. Tumor tissues were excised and fixed in 10% buffered formalin and embedded in paraffin.
Histology and immunohistochemistry
Paraffin embedded blocks of all tumors were sectioned at 5 μm. Each sample was stained with Hematoxylin and Eosin (H&E) for histopathological analysis. Immunostaining was done using antibodies against cleaved caspase-3 for apoptosis, Ki-67 for cell proliferation, CD31 for angiogenesis, and total MET, phospho-MET, RON-beta, phospho-RON with antibodies described in detail in Supplementary Materials and Methods. All samples from both treatment and vehicle groups (n=10 or 11) were analyzed. In addition to scoring staining intensity, three random fields in each slide were evaluated for mitotic figures and blood vessels (n=30 or 33). Positively and negatively stained cell counts were divided by the total number of cells counted to generate the percentage of positive cells in a field; the area of necrosis was determined and excluded from evaluation of other factors.
Orthotopic lung cancer tumor model and imaging using Xenogen IVIS®
Six-week-old female SCID mice (Harlan Laboratories) were used to establish orthotopic tumors in lung. Tumors were initiated by intrathoracic injection of 5×104 A549-luc-C8 cells in 25 μl of PBS into the right lung percutaneously as described (24). Mice were randomly divided into control and treatment groups (n=9 per group), observed daily and body weight measured twice weekly. One week after inoculation, mice were given LY2801653 or 10% Acacia as vehicle control by oral gavage at 20 mg/kg daily. To detect tumor, mice were imaged with Luciferin-D using the Xenogen IVIS® Imaging System 200 Series (Xenogen Corporation, Alameda, CA, USA) on Days 0, 7, 14, and 21 as described (24, 25). Upon termination of study, mice were euthanized and imaged with open chest. Lungs and other organs were formalin fixed and paraffin embedded. Sections were cut at several depths to evaluate presence or absence of tumor in chest cavity. Bioluminescence data collected from IVIS® was used to generate graphs comparing relative radiance in each mouse.
Statistical analysis
For cell viability assay, data were expressed as mean ± standard error of mean (SEM) and analyzed using GraphPad Prism version 5.0d. For PamGene analyses, log intensity values were analyzed using paired-two-tailed Student’s t test comparing untreated controls versus treated. The statistically “significantly modulated” genes were defined as those that had a p-value of less than 0.05. This statistical analysis was performed using BioNavigator (PamGene). For xenograft tumor model, p-value between mean tumor volumes of vehicle and LY2801653 treated was calculated by two-tailed Student’s t test using GraphPad Prism.
Results
Robust MET and RON expression in NSCLC cell lines
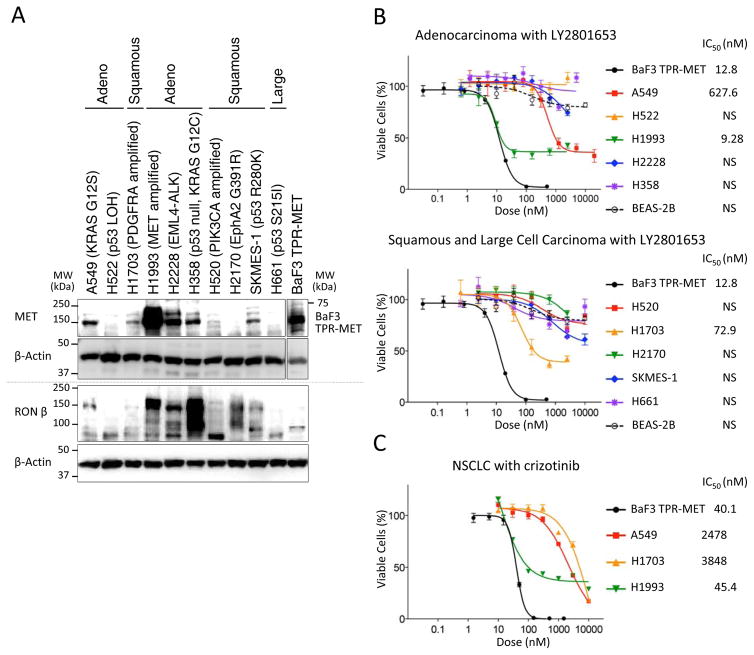
We first examined expression of MET and RON in a panel of NSCLC cell lines of various histologies – adenocarcinoma (A549, H522, H1993, H2228, H358), SCC (H520, H1703, H2170, SKMES-1) and large cell carcinoma (H661). Seven of the 10 NSCLC cell lines (except H522, H2170 and H661) co-expressed both MET and RON, with the highest level of MET being expressed by H1993 (Fig. 1A). MET expression was detected at 145 kDa and RON-beta was detected at 150 kDa. BaF3 TPR-MET, serving as the positive control, showed high level of MET expression recognized at 65kDa.
Figure 1. MET and RON protein expression and effect of LY2801653 on NSCLC cell lines.
A. Immunoblots for protein expression from lysates of the NSCLC cell lines using antibodies against MET, RON. MET expression is detected at 145 kDa. RON expression is detected at 150 kDa. B. Effect of LY2801653 on growth inhibition of NSCLC cells in vitro. Percentage of viable cells was determined relative to the untreated control, which had same dose of DMSO. Each data point represents the mean value and SEM of 6 replicate wells. A549, H1703 and H1993 are sensitive cell lines against LY2801653. C. Effect of crizotinib on growth inhibition of NSCLC cells in vitro. A549, H1703 and H1993 are sensitive cell lines against LY2801653, but IC50 of crizotinib was higher than IC50 of LY2801653 in each cell line.
Inhibitory effect of LY2801653 on growth of NSCLC cell lines
To determine the effect of LY2801653 on cell proliferation, we treated NSCLC cell lines with LY2801653 for 72 hours in a dose range of 1 nM-10 μM. BaF3 TPR-MET was used as a positive control. Of the ten NSCLC lines tested, A549, H1703 and H1993 were found to be sensitive to LY2801653 at IC50 627.6 nM, 72.9 nM and 9.28 nM, respectively (Fig. 1B). Similarly, cells were treated with crizotinib. A549, H1703 and H1993 were found to be sensitive to crizotinib at IC50 2478 nM, 3848 nM and 45.4 nM, respectively (Fig. 1C). BaF3 TPR-MET cells were also found to be sensitive to crizotinib at IC50 40.1 nM. For these cells, the IC50 of crizotinib was higher than the IC50 of LY2801653 (A549; 3.9-fold IC50, H1703; 53-fold IC50, H1993; 4.9-fold IC50, and BaF3 TPR-MET; 3.1-fold IC50). BEAS-2B normal bronchial epithelial cells, which do express low levels of MET (26), did not respond to MET/RON inhibition.
Biochemical effects of inhibition of MET and RON
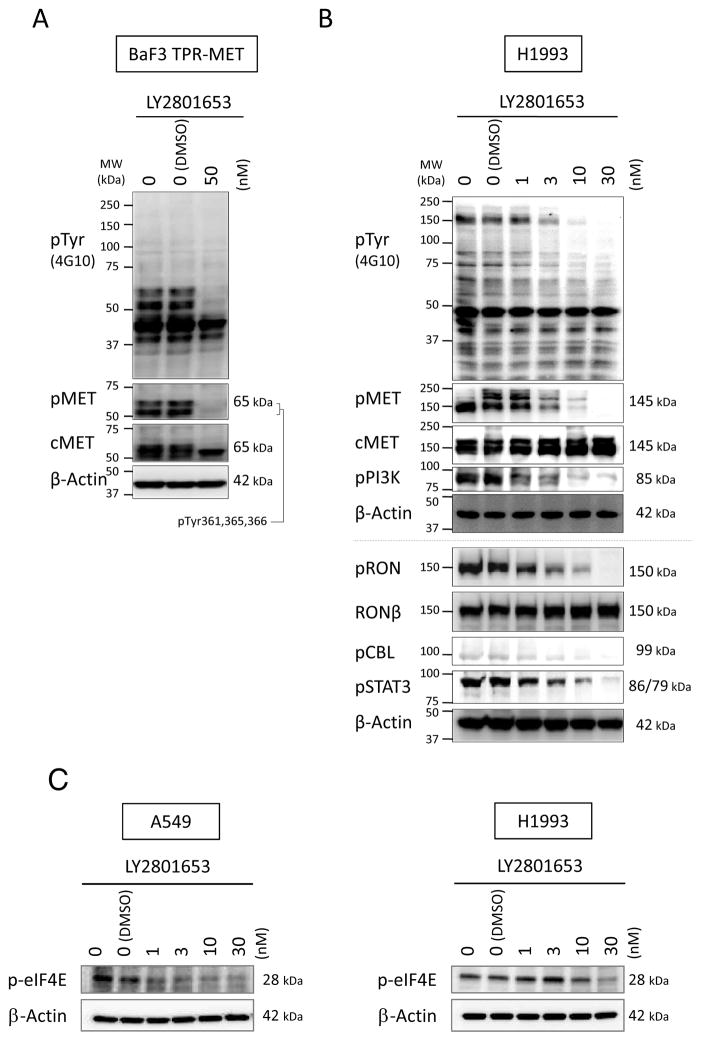
To determine the biochemical consequences of MET and RON kinase inhibition by LY2801653, we first used BaF3 TPR-MET cells. Changes in tyrosine phosphorylation of cellular proteins were evaluated using phospho-specific antibodies against tyrosine phosphorylation sites in MET. We found that 50 nM LY2801653 inhibited phosphorylation at Tyr361/365/366 autophosphorylation sites on TPR-MET (Fig. 2A). To determine whether LY2801653 also inhibited activation of MET and RON in lung cancer cells, we used H1993 cells. Treatment of H1993 cells with LY2801653 at doses 1, 3, 10 and 30 nM reduced global tyrosine phosphorylation of cellular proteins in a dose-dependent manner, as well as tyrosine phosphorylation of MET at Tyr1230/1234/1235 (homologous to Tyr361/365/366 in TPR-MET) and tyrosine phosphorylation of RON at Tyr1238/1239 (Fig. 2B).
Figure 2. Biochemical effects of inhibition of MET and RON.
A. Immunoblotting of BaF3 TPR-MET cells treated with LY2801653 (MET/RON small-molecule inhibitor) at 50 nM for 4 hours. B. Immunoblotting of H1993 cells treated with LY2801653 (1, 3, 10 and 30 nM) for 4 hours. Phosphorylation of cellular proteins was determined by immunoblotting whole cell lysates using anti-phosphotyrosine antibody (4G10), anti-pY1230/1234/1235-MET antibody (recognizing the corresponding pY361/365/366 sites in TPR-MET (A)), anti-pY458/pY199-PI3K p85/p55 antibody (B), anti-pY1238/1239-RON antibody (B), anti-pY674-CBL antibody (B), and anti pY705-STAT3 antibody (B). C. Immunoblotting to detect level of p-eIF4E in A549 and H1993 cells treated with LY2801653 at increasing doses for 4 hours.
Next, tyrosine phosphorylation of CBL (Tyr674), PI3K p85/p55 (Tyr458/199) and STAT3 (Tyr705) was examined. We have previously studied the relationship between CBL and NSCLC (27, 28), PI3K and MET in SCLC (29), and STAT3 and MET/RON in gastroesophageal adenocarcinoma (16). Because of our interest in these pathways, we picked CBL, PI3K and STAT3 from the PamGene and GeneGo analyses of significant genes associated with H1993 treatment with 3 nM LY2801653. Our data shows that treating H1993 cells with LY2801653 reduced the phosphorylation of these proteins in a dose-dependent manner (Fig. 2B). We utilized Tyr674 for CBL as both Tyr674 and Tyr700 are involved in cell motility.
One of the pathways LY2801653 may potentially be exerting its effect is via inhibition of MKNK1/2, as reported by Yan SB et al. (30). MKNK1/2 activities are measured by the phosphorylation of its substrate Eukaryotic Initiation Factor 4E (eIF4E) at Ser209. We analyzed the phosphorylated eIF4E (ser209) level of A549 and H1993 cells treated with LY2801693 (Fig. 2C). Our data show that there was incremental loss of p-eIF4E protein expression with increasing dose of the drug in both the cell lines, indicating that the MKNK1/2 pathway may be one of the mechanisms by which LY2801653 inhibition works. This provides a potential mechanism of action for LY2801653.
Identification of 13 key peptides whose phosphorylation was modulated by treatment with LY2801653 (3 nM) in H1993 cells
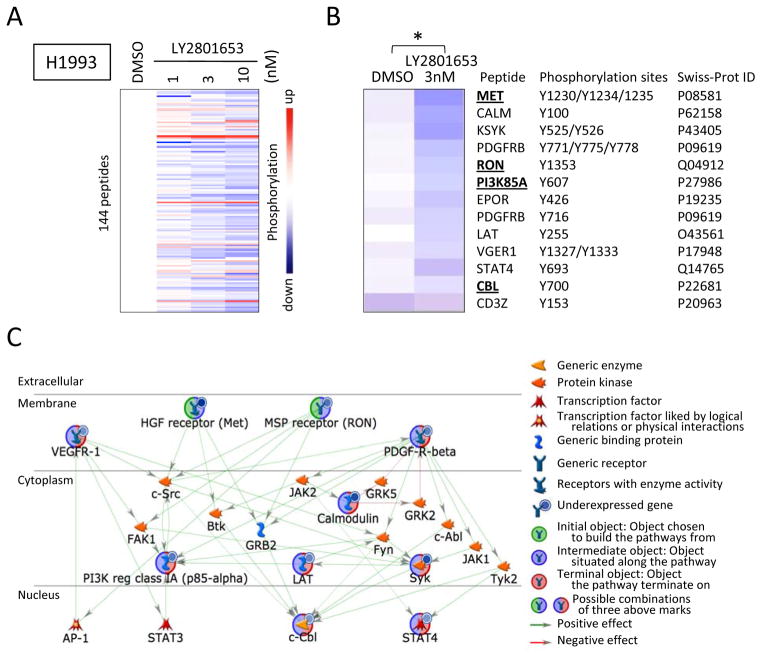
Using the PamGene kinase array platform, we demonstrate that treating H1993 cells with LY2801653 has significant effect on the phosphorylation of many peptides (Fig. 3A and B; Supplementary Fig. S1). The tyrosine kinase activity responsible for most peptides was down regulated in a concentration–dependent manner (Fig. 3A).
Figure 3. The protein tyrosine kinase activity profile on PamChip® microarrays and their network analyses using MetaCore™ (GeneGo).
A. The protein tyrosine kinase activity profile obtained by using H1993 lysates treated with LY2801653 (3 nM) as tested on PamChip® microarrays. The color-coded response signature is shown as a heatmap where treatment-related upregulation of kinase activity is reflected by red and a decrease by blue. The protein tyrosine kinase activity is down regulated in a concentration-dependent manner for most peptides. B. Thirteen significant tyrosine kinase substrate modulation by LY2801653 (3 nM) treatment as tested on PamChip® microarrays is shown (*: p-value <0.05, paired-two-tailed Student’s t test). C. Network between the PamGene significant genes associated with H1993 treatment with LY2801653 (3 nM) using MetaCore™ (GeneGo). Objects highlighted with green and blue represents ‘Initial and Intermediate object’ which means object chosen to build the pathways from and situated along the pathway. Object highlighted with blue and red represents ‘Intermediate and Terminal object’ which means object situated along the pathway and chosen to build the termination of pathways.
Analysis shows that thirteen substrate sites on 12 genes were significantly modulated by LY2801653 (p<0.05). The kinases that were inhibited are: MET (Tyr1230/1234/1235), calmodulin (CALM) (Tyr100), tyrosine-protein kinase SYK (KYSK) (Tyr525/526), beta-type platelet-derived growth factor receptor precursor (PDGFRB) (Tyr771/775/778, Tyr716), RON (Tyr1353), PI3K regulatory subunit alpha (PI3K85A) (Tyr607), erythropoietin receptor precursor (EPOR) (Tyr426), linker for activation of T-cells family member 1 (LAT) (Tyr255), vascular endothelial growth factor receptor 1 precursor (VEGFR1) (Tyr1327/1333), STAT4 (Tyr693), and E3 ubiquitin-protein ligase CBL (Tyr700) (p<0.05). Interestingly, the T-cell surface glycoprotein CD3 zeta chain precursor (CD3Z) (Tyr153) was the only site, which was found to be significantly upregulated (p<0.05) (Fig. 3B). A network between these 12 modulated genes was created utilizing the ‘Shortest Paths Algorithm, Build Network’ of GeneGo (Fig. 3C). EPOR and CD3Z, were excluded from the network to simplify visualization because they are not related to NSCLC directly. 47 tyrosine kinase substrate sites on 40 genes were significantly modulated when cells were treated with 10 nM LY2801653 (p<0.05) (Supplementary Fig. S1).
STAT3 is one of the key transcription factors involved with the genes significantly modulated by LY2801653 in H1993 cells
To determine key transcription factors involved with genes most significantly affected by MET/RON inhibition with LY2801653 in the PamGene kinase array, we analyzed statistically significant data generated by fold-change calculations (12 genes for 3 nM; 40 genes for 10 nM) by Pathway Analysis Software GeneGo’s robust and manually-curated MetaCore™ pathway analysis solution. STAT3 and SP1 were found to be the key transcription factors affected by LY2801653 (3 nM) treatment in H1993 (p=2.15E-43). The top six transcription factors involved in the 12 genes significantly modulated are derived using the query ‘What are the key transcription factors and target genes in my data? Most Popular Questions’ of GeneGo and detailed in Table 1. The table shows that the top six transcription factors were related to 8–11 of the total 12 significantly altered genes. For 10 nM treatment, SP1 was the first key transcription factor (p=9.49E-100) and STAT3 was the second key transcription factor (p=1.86E-71) affected (data not shown).
Table 1. The Key Transcription Factors of H1993 treatment with LY2801653 (3 nM).
Metacore™ (GeneGo) analysis based on identified PamGene 12 siginificant genes (Fig. 3B) which are modulated by LY2801653 treatment (3 nM). Note: ‘Total Nodes’ indicate the total number of genes analyzed whereas ‘Seed Nodes’ indicate the number of genes out of the Total Nodes that were involved with that particular transcription factor.
| No | Gene | GO processes | Total Nodes | Seed Nodes | p-Value |
|---|---|---|---|---|---|
| 1 | STAT3 | enzyme linked receptor protein signaling pathway (75.0%), cell surface receptor linked signaling pathway (100.0%), transmembrane receptor protein tyrosine kinase signaling pathway (66.7%), protein phosphorylation (66.7%), signal transduction (100.0%) | 12 | 11 | 2.15E-43 |
| 2 | SP1 | enzyme linked receptor protein signaling pathway (66.7%), cell surface receptor linked signaling pathway (91.7%), transmembrane receptor protein tyrosine kinase signaling pathway (58.3%), protein phosphorylation (58.3%), signal transduction (91.7%) | 12 | 11 | 2.15E-43 |
| 3 | p53 | enzyme linked receptor protein signaling pathway (72.7%), cell surface receptor linked signaling pathway (100.0%), protein phosphorylation (63.6%), signal transduction (100.0%), transmembrane receptor protein tyrosine kinase signaling pathway (54.5%) | 11 | 10 | 5.75E-39 |
| 4 | c-Myc | cell surface receptor linked signaling pathway (100.0%), enzyme linked receptor protein signaling pathway (66.7%), protein phosphorylation (66.7%), interspecies interaction between organisms (55.6%), positive regulation of transport (55.6%) | 9 | 8 | 1.33E-30 |
| 5 | c-Jun | cell surface receptor linked signaling pathway (100.0%), enzyme linked receptor protein signaling pathway (66.7%), interspecies interaction between organisms (55.6%), signal transduction (100.0%), signaling (100.0%) | 9 | 8 | 1.33E-30 |
| 6 | STAT1 | cell surface receptor linked signaling pathway (100.0%), enzyme linked receptor protein signaling pathway (66.7%), positive regulation of transport (55.6%), signal transduction (100.0%), transmembrane receptor protein tyrosine kinase signaling pathway (55.6%) | 9 | 8 | 1.33E-30 |
Gene-Specific Dual Inhibition of MET/RON
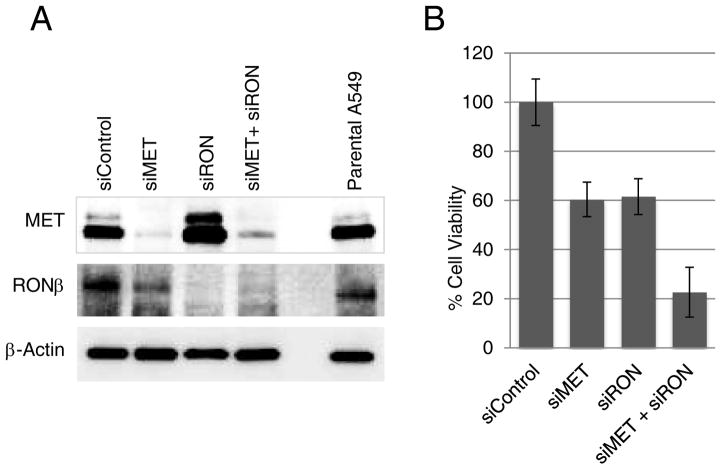
In order to provide evidence that dual inhibition of MET and RON by LY2801653 may be more beneficial than either MET or RON inhibition alone, we conducted gene-specific inhibition studies. siRNA was used to silence either MET, RON or both MET and RON in A549 cells by transient transfection. Gene silencing was confirmed by immunoblot analysis showing a marked reduction in MET and RON protein expression compared to control siRNA (Fig. 4A). Parental A549 is shown to express high levels of MET and RON, which remain unchanged with control siRNA. siMET eliminated MET protein expression almost completely with no change in RON expression. siRON eliminated RON expression almost completely. With MET/RON dual silencing, both proteins were reduced. 96 hours after transfection, the viability of cells silenced for MET or RON or dual MET and RON was measured and compared against control siRNA. We show that while cell viability was reduced by 60% in either MET or RON silenced cells, the dual inhibition resulted in a reduced viability to 22% (Fig. 4B). These results not only show that specific inhibition of MET and RON serve to reduce cell viability in A549 cells but also that dual MET/RON inhibition has a synergistic effect, as depicted via LY2801653 treatment.
Figure 4. Effect of specific inhibition on MET, RON and dual MET/RON on protein expression and cell viability.
A. A549 cells were transfected with siRNA against MET, RON or MET+RON simultaneously. The figure shows immunoblot analysis 72 hours after transfection. B. The viability of cells silenced for MET or RON or dual MET and RON was measured and compared against control siRNA.
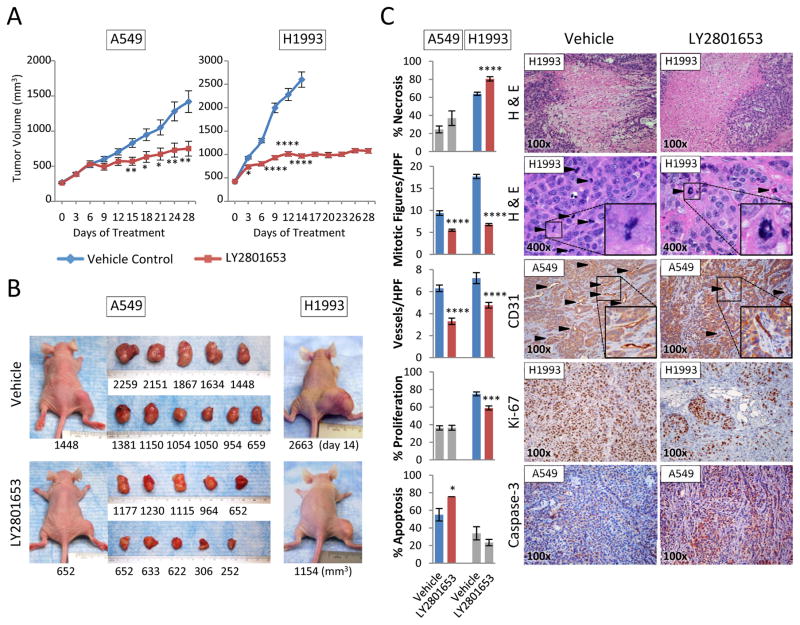
LY2801653 inhibits growth of A549 and H1993 tumor xenografts in nude mice
To further investigate the effect of MET/RON inhibition by LY2801653 in tumor proliferation, we next used the mouse flank xenograft model. Two NSCLC cell lines, A549 and H1993, were chosen based on their MET/RON expression profile and in vivo tumorigenicity. Treatment with LY2801653 at 20 mg/kg/day reduced A549 and H1993 tumor growth significantly relative to vehicle control (p<0.05) (Fig. 5A and B) with unchanging body weight. Due to the rapid increase in tumor volume in untreated vehicle control mice, all mice in the vehicle groups had to be terminated on Day 14 of the study.
Figure 5. LY2801653 inhibits growth of tumor xenografts in nude mice.
A. Results from tumor xenograft experiments testing the efficacy of LY2801653 in inhibiting the growth of A549 or H1993 cell line tumors in nude mice. Oral gavage treatment with LY2801653 at 20 mg/kg/day reduced A549 and H1993 tumor growth significantly relative to vehicle control (p<0.05). B. Representative images of mice and tumors treated with LY2801653 and control. C. Immunohistochemical staining and evaluation. H&E histologic staining for necrosis and mitotic figures, CD31 for angiogenesis, Ki-67 antibody staining for cell proliferation, and caspase-3 for apoptosis were evaluated via immunohistochemistry. Bar graphs indicate quantification of 10 or 11 mice samples (n=10 or 11). Three random fields in each mouse sample were evaluated for mitotic figures and vessels (n=30 or 33). Error bars represent SEM. P-value between mean tumor volumes, the percentage of positive cells, or the numbers of mitotic figures and vessels of vehicle control and LY2801653 by two-tailed Student’s t test (*: p-value <0.05, **: p-value <0.01, ***: p-value <0.001, ****: p-value <0.0001).
Consistent with the in vitro experiments of GeneGo analyses, mitotic figures (H&E staining) and vessels (CD31 staining) of tumor specimens were significantly decreased in LY2801653-treated mice in both A549 and H1993 studies. Moreover, in the treated arms of H1993, necrosis was increased and cell proliferation (Ki-67) was decreased. In the treated arms of A549, apoptosis (Caspase-3 staining) in tumor cells was increased (Fig. 5C). Tumor specimen from the LY2801653- and vehicle-treated mice were immunostained for phosphorylated MET and RON. Phosphorylated MET was decreased modestly in LY2801653-treated mice in both A549 and H1993 studies. Moreover, in the treated arms of H1993, phosphorylated RON was decreased modestly in LY2801653-treated mice (Supplementary Fig. S3).
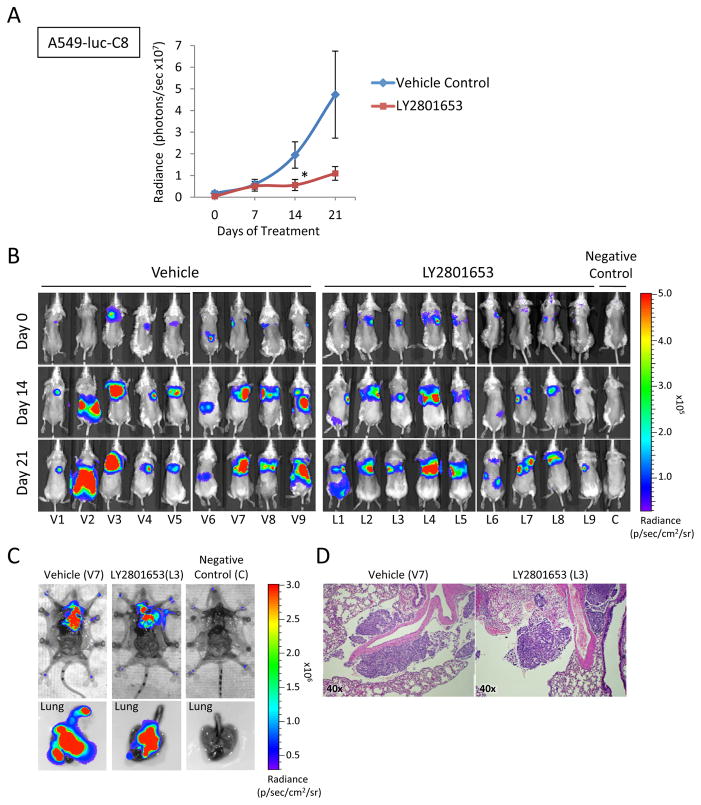
LY2801653 inhibits growth of A549-luc-C8 lung orthotopic tumor xenografts in SCID mice
To examine the effects of LY2801653 in an orthotopic tumor model, we established orthotopic lung tumors using A549-luc-C8 cells in the right lung of SCID mice. Tumor progression was monitored weekly by detecting bioluminescence using IVIS® imaging as described in materials and methods. Treatment with LY2801653 at 20 mg/kg/day reduced A549-luc-C8 tumor growth significantly relative to vehicle control on day 14 (p<0.05) (Fig. 6A and B) with unchanging body weight. Vehicle and treatment groups were euthanized and imaged with open chest and lung (Fig. 6C). Serial sections were cut of each right and left lung and stained with H&E to evaluate presence or absence of tumor in primary lung, contra-lateral lung and chest cavity. We were able to confirm the presence of tumors in the lung of all these mice (Fig. 6D).
Figure 6. LY2801653 inhibits growth of orthotopic tumor xenografts in SCID mice.
A. Quantitative analysis of bioluminescence photon counts as a measure of A549-luc-C8 cell line tumor growth in SCID. Oral gavage treatment with LY2801653 at 20 mg/kg/day reduced A549-luc-C8 tumor growth significantly relative to vehicle control (p<0.05). B. Representative images of the whole body. C. Representative fluorescence images of whole body with open chest and lung. D. H&E histologic staining to evaluate presence or absence of tumor in chest cavity. Error bars represent SEM. P-value between mean radiance of vehicle control and LY2801653 by two-tailed Student’s t test (*: p-value <0.05).
Discussion
In this report, we describe a novel ATP-competitive, potent and highly selective dual SMI of MET/RON, LY2801653 and its effect on NSCLC. MET/RON are important pathways in NSCLC. In vitro and in vivo, there was considerable cell death with LY2801653 treatment.
Over the past decade, there has been considerable data generated on MET biology and therapy for lung cancer. In lung cancer, MET and phosphorylated MET (especially juxtamembrane domain site pY1003 and autocatalytic site pY1234/1235) are overexpressed in 40% of tumor tissues (31), and are prognostic biomarkers (32). Like MET, RON and phosphorylated RON also are overexpressed in lung cancer (12), but unlike MET, which can be mutated in lung cancer, RON is not mutated. However, RON can have differential transcripts in various tumors (33, 34) and it is not known which transcripts exist in lung cancer. This would be important to determine since oncogenic functionality of epithelial cells in the lung may be dependent on different RON transcripts.
As has been shown, MET can be overexpressed or amplified in tumors resistant to cytotoxic agents such as cisplatin (35) or SMIs such as gefitinib (36–38). Recently, MET resistance was observed utilizing anti-MET antibody (MetMAb) (39). The mechanism by which MET acquires resistance is not known. MET can also synergize with other targets such as EGFR in NSCLC (40). A number of MET inhibitors are being tested with EGFR inhibitors in lung cancers (41–43). However, in the most recent trial utilizing tivantinib with erlotinib versus erlotinib alone (Phase III trial NCT01244191) (44), no difference in overall survival was observed (45). Our data shows that MET/RON may be important to co-target in NSCLC. In order to better treat tumors, a strategy that employs MET and RON dual inhibition would be more desirable than inhibiting one pathway alone.
We found LY2801653 to be more efficacious than crizotinib in A549 (KRAS G12S), H1703 (PDGFRA amplified), and H1993 (MET amplified) NSCLC cell lines. We also showed that LY2801653 was effective in in vivo models. Due to the focused studies on LY2801653, it would be useful for the future to compare all the small molecule inhibitors versus antibody inhibition of MET versus MET and RON. Interestingly, LY2801653 may also be effective against tumors via inhibition of MKNK1/2 (30). Thus, for the future, it would be interesting to determine the therapeutic inhibition of MET/RON in lung cancer patients harboring various mutations. We have determined the sensitivities of the LY2801653 in various NSCLC cell lines harboring different “driver” mutations. Since NSCLC is a heterogeneous group of diseases, it is important to note that not all NSCLC will respond to the same strategy of inhibition. LY2801653 inhibited A549, H1703 and H1993 cells but not others tested. It is possible that this is related to the various other abnormalities that exist within these cells. As well, these are in vitro experiments, and more in vivo experiments should be done for the future to determine if tumor-stromal interactions play important roles in the inhibition of the various cells. For the future, it will also be prudent to have isogenic cell lines with the various oncogenic and tumor suppressor abnormalities that exist or co-exist in NSCLC.
Kinase assays on PamChip® peptide microarrays provide rapid identification of substrates for the kinase under investigation. PamGene assay removes many limitations to assay development because 144 peptides can be tested simultaneously. Computational pathway analysis (MetaCore™, GeneGo) provides detailed knowledge of the highest-ranked common network and help validated the implicated disease pathways. Using PamGene and MetaCore™ tools, we found that several downstream signal transduction molecules were affected with MET/RON inhibition, including CBL, PI3K and STAT3 (Fig. 2B, Fig. 3, Table 1).
CBL is an E3 ubiquitin ligase that regulates RTK activity in a number of different receptors, including MET, EGFR, PDGFR, and VEGFR. In our initial study, we evaluated expression and mutational status of CBL in archival NSCLC tumor tissue collected from African-Americans, Caucasians and East Asians (27). Interestingly, we found that CBL was expressed at relatively low levels and was mutated occasionally. The overall mutation rate of CBL in NSCLC was 8.4% (10/119). Preliminary functional studies of these mutants revealed defects in their ability to ubiquitinate MET, and no discernable effects on the ubiquitination of EGFR. Moreover, forced expression of these mutants in an NSCLC cell line enhanced cell viability in vitro (28). From these data and our data, this pathway may play an important role in lung cancer carcinogenesis.
The PI3K pathway is fundamental for cell development, growth, and survival. Its deregulation and contribution to carcinogenesis has been well documented and reviewed in the past, including in carcinomas of the lung. We have reported that PI3K pathway is crucially targeted via MET stimulation in SCLC (29). Now, this study finds that inhibition of the MET by LY2801653 targets the PI3K pathway. Hence, PI3K also may be an important target in NSCLC.
STAT3, one of the seven members of the STAT family, has been the most strongly implicated member in tumorigenesis (46, 47). We have previously shown in a gastric cancer cell line that STAT3 is activated upon RON and/or MET stimulation (16). As shown here in the MetaCore™ Analysis Data, STAT3 is the key transcription factor affected by LY2801653 treatment of H1993 cells. Further, tyrosine phosphorylation of STAT3 was also reduced by LY2801653. The HGF/MET signaling pathway is responsible for stimulation of cell motility and invasion (20, 48), and STAT3 is responsible for at least part of the MET signal.
In this study, the antitumor efficacy of LY2801653 was evaluated in tumor models that (i) express MET/RON (ii) harbor MET gene amplification or (iii) harbor KRAS mutation. We have confirmed the effect of LY2801653 against NSCLC cell lines using both heterotopic and orthotopic human xenograft mouse models. In the heterotopic mouse model, LY2801653 inhibited the proliferation of tumors and decreased mitotic cells and vessels significantly. Modest but not notable differences in the expression of phosphorylated MET and RON was detected in the treated versus untreated tumor tissues although we had seen considerable effects on tumor growth. It is unclear if there is a potential feed-back loop that is cross-activating the phosphorylation and needs further exploration. In the orthotopic mouse model, LY2801653 also inhibited tumor proliferation.
The tumor microenvironment plays a major role in promoting tumor growth (49). In xenografts models, tumor location is important in relation to sensitivity to chemotherapy (50) and is a critical factor in targeted therapies. Taking these points into consideration, orthotopic models have advantages over heterotopic models. These advantages include use of the relevant site for tumor-host interaction, the ability to study site-specific dependence of therapy, organ-specific expression of genes and replication of clinical scenarios. For these reasons, we used the heterotopic models with two NSCLC cell lines, and only the luciferase tagged A549-luc-C8 for the orthotopic modeling. A luciferase tagged H1993 cell was not obtainable.
Finally, others and we have previously shown gain of function mutations of MET in lung cancers (5, 8, 20). There also seems to be differential response to the MET SMI SU11274 in E168D versus N375S MET mutant lung cancer cells (8). Although MET and RON synergize in function, in terms of mutated MET, it is not known if the interaction between a mutated MET and RON remain the same. It would now be interesting to test the various MET mutations in combination with RON for efficacy of the LY2801653 inhibitor. As further therapeutics are developed, such as antibodies specific to MET or RON, these would be important to test in combination with the SMIs. Several inhibitors of MET and RON are currently being investigated in clinical trials (17, 43). LY2801653 is also being investigated in patients with advanced cancer (adenocarcinoma of the colon or rectum, head and neck squamous cell carcinoma, and cholangiocarcinoma) (Phase I trial I3O-MC-JSBA, NCT01285037). Because of its potent efficacy at lower concentration and against multi-targeted kinase especially MET/RON, we believe that LY2801653 has a promising therapeutic potential in treating NSCLC.
In summary, LY2801653 is a novel, orally available multi-targeted kinase inhibitor, which inhibits MET and RON signaling. It has significant in vitro and in vivo antitumor activities against MET- and RON-driven tumors. The results described in this report provide justification for advancing LY2801653 to clinical development.
Supplementary Material
Acknowledgments
Supported in part by: NIH/NCI (5R01CA129501-05 and 5R01CA100750-09), Lilly (RS) and ASCO Translational Award (EEV).
The authors thank Sau-chi Betty Yan and Ling Liu (Lilly Research Laboratories, Eli Lilly and Company, Indianapolis, IN) for providing LY2801653.
Footnotes
Disclosure of Potential Conflict of Interest: No potential conflicts of interest were disclosed.
References
- 1.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 4.Bergethon K, Shaw AT, Ou SH, Katayama R, Lovly CM, McDonald NT, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30:863–70. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sattler M, Reddy MM, Hasina R, Gangadhar T, Salgia R. The role of the c-Met pathway in lung cancer and the potential for targeted therapy. Ther Adv Med Oncol. 2011;3:171–84. doi: 10.1177/1758834011408636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sadiq AA, Geynisman DM, Salgia R. Inhibition of MET receptor tyrosine kinase and its ligand hepatocyte growth factor. J Thorac Oncol. 2011;6:S1810–1. doi: 10.1097/01.JTO.0000407568.45147.43. [DOI] [PubMed] [Google Scholar]
- 7.Sattler M, Salgia R. The MET axis as a therapeutic target. Update on cancer therapeutics. 2009;3:109–18. doi: 10.1016/j.uct.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krishnaswamy S, Kanteti R, Duke-Cohan JS, Loganathan S, Liu W, Ma PC, et al. Ethnic differences and functional analysis of MET mutations in lung cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:5714–23. doi: 10.1158/1078-0432.CCR-09-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okuda K, Sasaki H, Yukiue H, Yano M, Fujii Y. Met gene copy number predicts the prognosis for completely resected non-small cell lung cancer. Cancer science. 2008;99:2280–5. doi: 10.1111/j.1349-7006.2008.00916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cappuzzo F, Marchetti A, Skokan M, Rossi E, Gajapathy S, Felicioni L, et al. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J Clin Oncol. 2009;27:1667–74. doi: 10.1200/JCO.2008.19.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Go H, Jeon YK, Park HJ, Sung SW, Seo JW, Chung DH. High MET gene copy number leads to shorter survival in patients with non-small cell lung cancer. J Thorac Oncol. 2010;5:305–13. doi: 10.1097/JTO.0b013e3181ce3d1d. [DOI] [PubMed] [Google Scholar]
- 12.Kanteti R, Krishnaswamy S, Catenacci D, Tan YH, El-Hashani E, Cervantes G, et al. Differential expression of RON in small and non-small cell lung cancers. Genes, chromosomes & cancer. 2012;51:841–51. doi: 10.1002/gcc.21968. [DOI] [PubMed] [Google Scholar]
- 13.Zhou YQ, He C, Chen YQ, Wang D, Wang MH. Altered expression of the RON receptor tyrosine kinase in primary human colorectal adenocarcinomas: generation of different splicing RON variants and their oncogenic potential. Oncogene. 2003;22:186–97. doi: 10.1038/sj.onc.1206075. [DOI] [PubMed] [Google Scholar]
- 14.Zhou D, Pan G, Zheng C, Zheng J, Yian L, Teng X. Expression of the RON receptor tyrosine kinase and its association with gastric carcinoma versus normal gastric tissues. BMC Cancer. 2008;8:353. doi: 10.1186/1471-2407-8-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Follenzi A, Bakovic S, Gual P, Stella MC, Longati P, Comoglio PM. Cross-talk between the proto-oncogenes Met and Ron. Oncogene. 2000;19:3041–9. doi: 10.1038/sj.onc.1203620. [DOI] [PubMed] [Google Scholar]
- 16.Catenacci DV, Cervantes G, Yala S, Nelson EA, El-Hashani E, Kanteti R, et al. RON (MST1R) is a novel prognostic marker and therapeutic target for gastroesophageal adenocarcinoma. Cancer biology & therapy. 2011;12:9–46. doi: 10.4161/cbt.12.1.15747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang MH, Padhye SS, Guin S, Ma Q, Zhou YQ. Potential therapeutics specific to c-MET/RON receptor tyrosine kinases for molecular targeting in cancer therapy. Acta Pharmacol Sin. 2010;31:1181–8. doi: 10.1038/aps.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sattler M, Pride YB, Ma P, Gramlich JL, Chu SC, Quinnan LA, et al. A novel small molecule met inhibitor induces apoptosis in cells transformed by the oncogenic TPR-MET tyrosine kinase. Cancer research. 2003;63:5462–9. [PubMed] [Google Scholar]
- 19.Maulik G, Kijima T, Ma PC, Ghosh SK, Lin J, Shapiro GI, et al. Modulation of the c-Met/hepatocyte growth factor pathway in small cell lung cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2002;8:620–7. [PubMed] [Google Scholar]
- 20.Ma PC, Kijima T, Maulik G, Fox EA, Sattler M, Griffin JD, et al. c-MET mutational analysis in small cell lung cancer: novel juxtamembrane domain mutations regulating cytoskeletal functions. Cancer research. 2003;63:6272–81. [PubMed] [Google Scholar]
- 21.Versele M, Talloen W, Rockx C, Geerts T, Janssen B, Lavrijssen T, et al. Response prediction to a multitargeted kinase inhibitor in cancer cell lines and xenograft tumors using high-content tyrosine peptide arrays with a kinetic readout. Molecular cancer therapeutics. 2009;8:1846–55. doi: 10.1158/1535-7163.MCT-08-1029. [DOI] [PubMed] [Google Scholar]
- 22.Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, et al. Molecular definition of breast tumor heterogeneity. Cancer cell. 2007;11:259–73. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 23.Butovsky O, Siddiqui S, Gabriely G, Lanser AJ, Dake B, Murugaiyan G, et al. Modulating inflammatory monocytes with a unique microRNA gene signature ameliorates murine ALS. The Journal of clinical investigation. 2012;122:3063–87. doi: 10.1172/JCI62636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mordant P, Loriot Y, Lahon B, Castier Y, Leseche G, Soria JC, et al. Bioluminescent orthotopic mouse models of human localized non-small cell lung cancer: feasibility and identification of circulating tumour cells. PloS one. 2011;6:e26073. doi: 10.1371/journal.pone.0026073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertino P, Piccardi F, Porta C, Favoni R, Cilli M, Mutti L, et al. Imatinib mesylate enhances therapeutic effects of gemcitabine in human malignant mesothelioma xenografts. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:541–8. doi: 10.1158/1078-0432.CCR-07-1388. [DOI] [PubMed] [Google Scholar]
- 26.Jagadeeswaran R, Surawska H, Krishnaswamy S, Janamanchi V, Mackinnon AC, Seiwert TY, et al. Paxillin is a target for somatic mutations in lung cancer: implications for cell growth and invasion. Cancer research. 2008;68:132–42. doi: 10.1158/0008-5472.CAN-07-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan YH, Krishnaswamy S, Nandi S, Kanteti R, Vora S, Onel K, et al. CBL is frequently altered in lung cancers: its relationship to mutations in MET and EGFR tyrosine kinases. PloS one. 2010;5:e8972. doi: 10.1371/journal.pone.0008972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lo FY, Tan YH, Cheng HC, Salgia R, Wang YC. An E3 ubiquitin ligase: c-Cbl: a new therapeutic target of lung cancer. Cancer. 2011;117:5344–50. doi: 10.1002/cncr.26153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maulik G, Madhiwala P, Brooks S, Ma PC, Kijima T, Tibaldi EV, et al. Activated c-Met signals through PI3K with dramatic effects on cytoskeletal functions in small cell lung cancer. Journal of cellular and molecular medicine. 2002;6:539–53. doi: 10.1111/j.1582-4934.2002.tb00453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan SB, Peek VL, Ajamie R, Buchanan SG, Graff JR, Heidler SA, et al. LY2801653 is an orally bioavailable multi-kinase inhibitor with potent activity against MET, MST1R, and other oncoproteins, and displays anti-tumor activities in mouse xenograft models. Investigational new drugs. 2012 doi: 10.1007/s10637-012-9912-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma PC, Tretiakova MS, MacKinnon AC, Ramnath N, Johnson C, Dietrich S, et al. Expression and mutational analysis of MET in human solid cancers. Genes, chromosomes & cancer. 2008;47:1025–37. doi: 10.1002/gcc.20604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tretiakova M, Salama AK, Karrison T, Ferguson MK, Husain AN, Vokes EE, et al. MET and phosphorylated MET as potential biomarkers in lung cancer. Journal of environmental pathology, toxicology and oncology : official organ of the International Society for Environmental Toxicology and Cancer. 2011;30:341–54. doi: 10.1615/jenvironpatholtoxicoloncol.v30.i4.70. [DOI] [PubMed] [Google Scholar]
- 33.Eckerich C, Schulte A, Martens T, Zapf S, Westphal M, Lamszus K. RON receptor tyrosine kinase in human gliomas: expression, function, and identification of a novel soluble splice variant. Journal of neurochemistry. 2009;109:969–80. doi: 10.1111/j.1471-4159.2009.06027.x. [DOI] [PubMed] [Google Scholar]
- 34.Ma Q, Zhang K, Yao HP, Zhou YQ, Padhye S, Wang MH. Inhibition of MSP-RON signaling pathway in cancer cells by a novel soluble form of RON comprising the entire sema sequence. Int J Oncol. 2010;36:1551–61. doi: 10.3892/ijo_00000642. [DOI] [PubMed] [Google Scholar]
- 35.Akervall J, Guo X, Qian CN, Schoumans J, Leeser B, Kort E, et al. Genetic and expression profiles of squamous cell carcinoma of the head and neck correlate with cisplatin sensitivity and resistance in cell lines and patients. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10:8204–13. doi: 10.1158/1078-0432.CCR-04-0722. [DOI] [PubMed] [Google Scholar]
- 36.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 37.Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suda K, Murakami I, Katayama T, Tomizawa K, Osada H, Sekido Y, et al. Reciprocal and complementary role of MET amplification and EGFR T790M mutation in acquired resistance to kinase inhibitors in lung cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16:5489–98. doi: 10.1158/1078-0432.CCR-10-1371. [DOI] [PubMed] [Google Scholar]
- 39.Catenacci DV, Henderson L, Xiao SY, Patel P, Yauch RL, Hegde P, et al. Durable complete response of metastatic gastric cancer with anti-Met therapy followed by resistance at recurrence. Cancer discovery. 2011;1:573–9. doi: 10.1158/2159-8290.CD-11-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puri N, Salgia R. Synergism of EGFR and c-Met pathways, cross-talk and inhibition, in non-small cell lung cancer. Journal of carcinogenesis. 2008;7:9. doi: 10.4103/1477-3163.44372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bagai R, Ma PC. Combined treatment with MET inhibitors and other therapies in lung cancer. Transl Lung Cancer Res. 2012;1:214–8. doi: 10.3978/j.issn.2218-6751.2012.09.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Padda S, Neal JW, Wakelee HA. MET inhibitors in combination with other therapies in non-small cell lung cancer. Transl Lung Cancer Res. 2012;1:238–53. doi: 10.3978/j.issn.2218-6751.2012.10.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menis J, Giaj Levra M, Novello S. MET inhibition in lung cancer. Transl Lung Cancer Res. 2013;2:23–39. doi: 10.3978/j.issn.2218-6751.2012.12.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scagliotti GV, Novello S, Schiller JH, Hirsh V, Sequist LV, Soria JC, et al. Rationale and design of MARQUEE: a phase III, randomized, double-blind study of tivantinib plus erlotinib versus placebo plus erlotinib in previously treated patients with locally advanced or metastatic, nonsquamous, non-small-cell lung cancer. Clinical lung cancer. 2012;13:391–5. doi: 10.1016/j.cllc.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 45.ArQule II. Woburn, MA: ArQule, Inc; [cited 2012 Oct 2]. Available from: http://investors.arqule.com/releasedetail.cfm?ReleaseID=710618. [Google Scholar]
- 46.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 47.Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–88. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 48.Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G. Targeting MET in cancer: rationale and progress. Nature reviews Cancer. 2012;12:89–103. doi: 10.1038/nrc3205. [DOI] [PubMed] [Google Scholar]
- 49.Whiteside TL. The tumor microenvironment and its role in promoting tumor growth. Oncogene. 2008;27:5904–12. doi: 10.1038/onc.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teicher BA. Tumor models for efficacy determination. Molecular cancer therapeutics. 2006;5:2435–43. doi: 10.1158/1535-7163.MCT-06-0391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.