Abstract
Cancer cells possessing “stemness,” or stem-cell properties, are referred to as cancer stem cells (CSC) or cancer-initiating cells. The concept that these cells rest at the apex of the cancer hierarchy is an evolving theme in cancer research. These cells are by definition primarily responsible for initiation and propagation of tumors as well as relapse after therapy, and they are therefore of major scientific interest. Several studies indicate that hepatocellular carcinomas (HCC) that harbor phenotypic features of stem cells and progenitor cells constitute a subclass of therapeutically challenging cancers that are associated with a particularly poor prognosis. We have recently demonstrated tha any cell type in the mouse hepatic lineage can undergo oncogenic reprogramming into a CSC by activating different cell type–specific pathways (1). Identification of common and cell of origin–specific phenotypic and genetic changes could provide new therapeutic targets for liver cancer.
Keywords: liver, cancer, stemness, cancer stem cells
Introduction
The biological similarity of cancer and stem cells has long been recognized, and consequently the concept of cancer as a disease of abnormal stem cells is not new (2). Furthermore, the extensive confirmation of the cancer stem cell (CSC) concept in multiple tumor types, has decisively established the importance of the CSC paradigm in both experimental and human neoplasia (3). In this context it is worth emphasizing the importance of the discovery of Takahashi and Yamanaka demonstrating that introduction and expression of only four genes (e.g., Oct-3/4, Sox2, c-Myc, and KLF4) into adult mouse fibroblasts, and later other cell types such as hepatocytes, could reprogram these cells into functional embryonic stem cells entitled induced pluripotent stem cells (iPSCs) for the cancer field (4). It is noteworthy that most of the effective reprogramming factors have an established role in oncogenesis (e.g., c-Myc, Oct-4, Sox-2, Nanog, Lin28, and p53). These discoveries strongly indicate that differentiated cells (e.g. hepatocytes) could be the source of CSC, and capable of supporting both inter- and intratumoral phenotypic and genetic heterogeneity seen in most human cancers (5).
The evolution of CSCs from cells at different stages of differentiation may contribute, at least in part, to the phenotypic and genetic heterogeneity seen in liver cancer. However, whether a lineage stage may be a factor in acquisition of stemness properties at the cellular and molecular levels is not yet understood. We have recently addressed whether the differentiation stage of distinct hepatic lineage cells: (a) dictates the acquisition of CSC properties and (b) contributes to the phenotypic and genetic heterogeneity of liver cancer (1). These data will be summarized and discussed here.
Results and Discussion
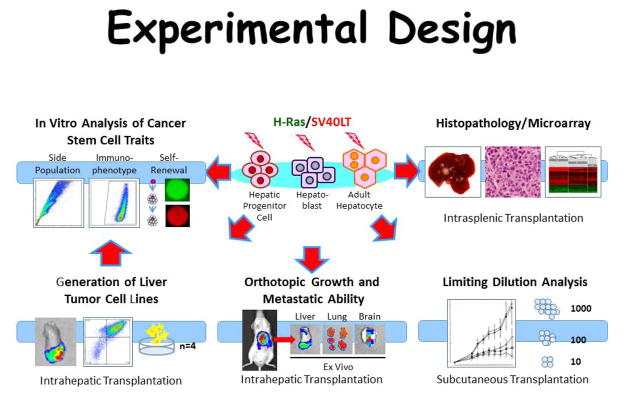
Three cell types from mouse liver were used (i.e. adult hepatocytes (AH) hepatoblasts (HB) and hepatic progenitor cells (HPC) to perform a sytematic side by side comparision of tumorigenesis after oncogeneic transformation. The cell types were isolated at high purity and subjected to lentiviral transduction with vectors encoding oncogenic H-ras and SV40LT (experimental proceedures summarized in Fig. 1). All three lineages gave rise to tumors with varying HCC and cholangiocarcinoma (CCA) content i.e. AH- tumors showed mostly HCC, HB-tumors mostly CCA, and HPC-tumors mostly anaplastic histopathology, indicating a epithelial to mesenchymal transition. Since HCCs, CCAs, and anaplastic tumors were generated in the model and produced by the same initial oncogenetic drivers, multible side-by-side comarisons were possible independent of confounding factors caused by different oncogenic drivers. The key observations included: (1) a 590-gene signature with significant common dysregulation in all 3 tumor types (2) hierarchical clustering of common genes separated the tumors in accordance with the originally transformed cell type; (3) the identified gene signature was able to separate human HCCs from CCAs and combined hepatocellular CCAs; (4) AH-derived induced pluripotent stem cell gene signature was enriched in AH-derived but not in HB- and HPC-derived tumors; and (5) c-myc was strongly induced only in AH-derived tumors, and RNA-mediated knockdown re-affirmed the central role of c-myc in reprogramming and oncogenic transformation of AH (6).
Figure 1.
Schematic represenation of the experimental designs.
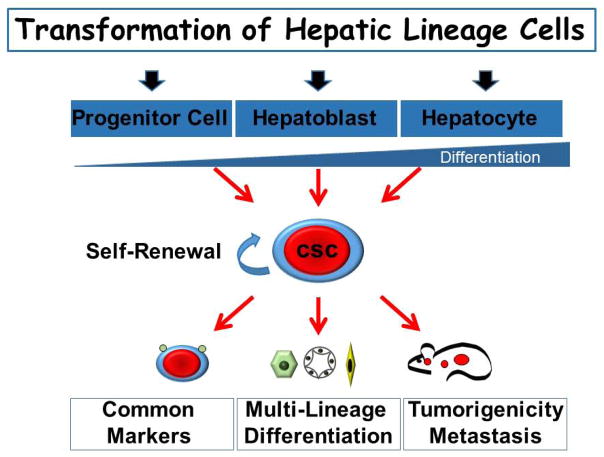
In conclusion, this study provides the first comprehensive and systematic comparison of hepatocarcinogenesis initiated by controlled oncogenic transformation of cells at specific stages of hepatic lineage. Differentiated hepatocytes, hepatoblasts, and adult hepatic progenitor cells were isolated at high purity and efficiently transduced with the same combination of H-Ras and SV40LT oncogenes. This permitted a unique and direct side-by-side comparison of cellular and molecular characteristics of transformed cells both in vitro and in vivo. The study formally showed that any hepatic lineage cell can be reprogrammed into CSC by activating diverse cell type-–specific pathways (Fig. 2). It also described common and cell of origin–specific phenotypic and genetic changes that correctly identified the murine tumors according to their cellular origin, providing an important means to phenotypically classify morphologically diverse human primary hepatocellular cancers. Accordingly, identification of both cells that are susceptible to oncogenic transformation and relevant molecular pathways is essential for a deeper understanding of the origin of liver cancer and development of more effective therapeutic strategies.
Figure 2.
Schematic illustration of the transformation of hepatic lineage cells.
References
- 1.Holczbauer A, Factor VM, Andersen JB, Marquardt JU, Kleiner DE, Raggi C, Kitade M, Seo D, Akita H, Durkin ME, Thorgeirsson SS. Modeling pathogenesis of primary liver cancer in lineage-specific mouse cell types. Gastroenterology. 2013 Jul;145(1):221–31. doi: 10.1053/j.gastro.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sell S. Stem cell origin of cancer and differentiation therapy. Crit Rev Oncol Hematol. 2004;51:1–28. doi: 10.1016/j.critrevonc.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell. 2015;16:225–238. doi: 10.1016/j.stem.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.((insert blockade))
- 4.Campos-Sanchez E, Cobaleda C. Tumoral reprogramming: plasticity takes a walk on the wild site. Biochim Biophys Acta. 2015;1849:436–447. doi: 10.1016/j.bbagrm.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Marquardt JU, Andersen JB, Thorgeirsson SS. Functional and genetic deconstruction of the cellular origin in liver cancer. Nat Rev Cancer. 2015;15:653–667. doi: 10.1038/nrc4017. [DOI] [PubMed] [Google Scholar]
- 6.Kaposi-Novak P, et al. Central role of c-Myc during malignant conversion in human hepatocarcinogenesis. Cancer Res. 2009;69:2775–2782. doi: 10.1158/0008-5472.CAN-08-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]