Abstract
Background
Clinical and epidemiological findings point to an association between type 2 diabetes (T2D) and low birth weight. However, the nature of the relationship is largely unknown. The aim of this study was to identify novel single nucleotide polymorphisms (SNPs) in T2D and birth weight, and their pleiotropic loci.
Methods
A pleiotropy-informed conditional false discovery rate (cFDR) method was applied to two independent genome-wide association studies (GWAS) summary statistics of T2D (n = 149 821) and birth weight (n = 26 836).
Results
A conditional Q–Q plot showed strong enrichment of genetic variants in T2D conditioned on different levels of association with birth weight. 133 T2D-associated SNPs, including 120 novel SNPs, were identified with a significance threshold of cFDR < 0.05; 13 significant birth weight-associated SNPs, including 12 novel SNPs (cFDR < 0.05) were identified. Conjunctional cFDR (ccFDR) analysis identified nine pleiotropic loci, including seven novel loci, shared by both T2D and birth weight (ccFDR < 0.05). Two novel SNPs located at the CDK5 regulatory subunit-associated protein 1-like 1 (CDKAL1; rs1012635; cFDR < 0.05) and adenylate cyclase 5 (ADCY5; rs4677887; cFDR < 0.05) genes are of note. These two genes increase the risk of T2D and low birth weight through the pathway of the “fetal insulin hypothesis.”
Conclusion
Several pleiotropic loci were identified between T2D and birth weight by leveraging GWAS results. The results make it possible to explain a greater proportion of trait heritability and improve our understanding of the shared pathophysiology between T2D and birth weight.
Keywords: birth weight, genome-wide association study, pleiotropy, type 2 diabetes
Introduction
Diabetes is a group of systemic metabolic disorders characterized by long-term hyperglycemia due to scarcity of insulin secretion and/or deficiency of insulin action.1 Diabetes is an increasingly serious global public health issue. Type 2 diabetes (T2D), the most common form of diabetes, begins with a condition of insulin resistance in which target cells fail to respond to insulin properly.2 Currently, more than 400 million people throughout the world suffer from diabetes.3 Over 90% of cases are T2D,4 which caused 4.9 million deaths in 2014.5 Type 2 diabetes has become one of leading causes of death, disability, and increased health care costs.
There is accumulating evidence showing that early life experiences, such as birth weight, have continuous effects on adult health.6 Numerous studies have demonstrated that low birth weight can lead to increased risk of developing T2D. For example, a prospective study including three large cohorts (n = 149 794) found that participants with low birth weight (2.00–2.75 kg) had significantly higher susceptibility to T2D than those with reference birth weight (3.25–3.75 kg).7 A recent meta-analysis of two studies analyzed 3627 T2D cases and 12 974 control participants of European ancestry.8 A genetic risk score was created on the basis of five low birth weight-related single nucleotide polymorphisms (SNPs). The analysis showed that for each 1 point increment in the genetic risk score, the risk of developing T2D increased by 6%.8 Using Mendelian randomization, it was further demonstrated that the low birth weight was actually causing the excess risk in T2D.8
Previous genome-wide association studies (GWAS) have been successful in identifying potential genetic risk factors for T2D that are associated with birth weight.9 Despite a number of SNPs found to be reproducibly associated with both traits, these SNPs explain a small proportion of heritability for T2D (<5%) and birth weight (~25%),10,11 and new information on the nature of the genetic component of these phenotypes needs to be investigated. In order to explain a greater proportion of genetic mechanisms underlying these highly correlated phenotypes in the pathogenesis of T2D and birth weight, further innovative analytical methods are required to uncover additional novel genes or variants, especially novel shared variants associated with both T2D and birth weight. As a novel analytical approach, the pleiotropy-informed conditional false discovery rate (cFDR) method,12 which requires only summary statistics from GWAS, could provide increased power to detect SNPs in GWAS results and elucidate mechanistic relationships between genetically related phenotypes. Using this approach, Andreassen et al.12 reported genetic overlap between a number of diseases and phenotypes, and successfully identified common variants associated with schizophrenia and bipolar disorder.
In the present study we applied the cFDR method to two large and independent T2D and birth weight GWAS datasets13,14 to identify additional and novel genetic loci of these two traits and to determine whether T2D shares susceptibility loci with birth weight, with the aim of gaining insights into the common pathophysiology between T2D and birth weight.
Methods
Genome-wide association studies datasets
The GWAS summary statistics were obtained from two publicly available datasets. The Diabetes Genetics Replication and Meta-analysis (DIAGRAM) consortium dataset (http://diagram-consortium.org/downloads.html, accessed 1 January 2015) contains the results for association with T2D from 34 840 cases and 114 981 controls. To our knowledge, it is the largest meta-analysis data of the reported T2D GWAS studies at present. The Early Growth Genetics Consortium dataset (http://egg-consortium.org/birth-weight.html, accessed 1 January 2015) contains the results for association with birth weight from a meta-analysis of up to 18 population-based European studies including 26 836 subjects. The two datasets contain summary statistics, providing P-values, size, and direction of effects at over 2 million directly genotyped or imputed SNPs from the HapMap project (release 27; ftp://ftp.ncbi.nlm.nih.gov/hapmap/00README.releasenotes_rel27, accessed 10 February 2015). There were no overlapping subjects between the T2D and birth weight GWAS datasets. The detailed inclusion criteria and phenotype characteristics from different GWAS are described in the original publications.13,14
Data processing
Two combined GWAS meta-analysis summary statistics for the 95 861 common SNPs were annotated, and then pairs of SNPs with large correlations were removed using the linkage disequilibrium (LD)-based pruning method. The LD pruning method begins with a window of 50 SNPs where the LD between each pair of SNPs is calculated, and if pairs have an R2 > 0.2, one of that pair of SNPs (the SNP with the smaller minor allele frequency) is removed. Following this initial removal of SNPs, the window shifts five SNPs forward and the procedure is repeated until there is no pair of SNPs in high LD. The HapMap 3 genotypes (http://www.sanger.ac.uk/resources/downloads/human/hapmap3.html, accessed 15 August 2015) were used to prune the dataset. At the end of the pruning procedure, there were 32 132 variants remaining to be used in the analysis. Genomic control is often needed to adjust GWAS results to ensure that the variance estimates for each SNP are not inflated due to population structure. Genomic control was previously applied by the original authors in the two datasets,13,14 so there was no need to reapply this adjustment in the present study.
Statistical analysis
Pleiotropic enrichment estimation
In GWAS studies, quantile–quantile (Q–Q) plots are commonly used to show the observed association (y-axis) across SNPs compared with the expected distribution of association test statistics (x-axis) under the null hypothesis. Any deviation from the identity line implies either incorrect assumed distribution or a true association. In the present study, to estimate the pleiotropic enrichment of association compared with that expected under the null hypothesis, conditional Q–Q plots were constructed by successively conditioning the principal trait on the SNPs with varying strengths of association in the conditional trait as per Andreassen et al.12 The Q–Q curves were plotted for quantiles of nominal −log10(P) values for association of the subset of variants below each significance threshold in the conditional trait. Specifically, nominal P-values (−log10(P)) were plotted on the y-axis and empirical quantiles (−log10(q)) were plotted on the x-axis for T2D and birth weight, respectively. Pleiotropy enrichment can be assessed from the degree of leftward shift from the expected identity line as the principal phenotype is successively conditioned on more stringent significance criteria in the conditional phenotype. Greater spacing between conditional Q–Q curves intuitively indicates a stronger trend of pleiotropic enrichment shared between the principal and conditional traits.
Calculation of the cFDR
In order to identify novel loci associated with T2D and birth weight, the cFDR was computed, an extension of the standard FDR framework that incorporates GWAS summary statistics from the pruned dataset to demonstrate the probability that a random SNP is null for association with the principal phenotype given that the observed P-values for the principal and conditional phenotypes are both smaller than two predefined disease-specific significance thresholds.15 In the present study, the cFDR was calculated for each SNP where T2D was the principal phenotype conditioned on the strength of association with birth weight (T2D|birth weight) and vice versa (birth weight|T2D). To assess whether the cFDR method leads to enrichment of specific loci, we successively confined the subset of SNPs being tested based on the level of significance for the association of each variant with the conditional trait using the following criteria: P < 1 (all SNPs), P < 0.1, P < 0.01, and P < 0.001. A cFDR of 0.05 was used to distinguish whether an SNP is significantly associated with the principal phenotype. The procedures used are described in detail by Andreassen et al.12 To visualize the localization of significant loci associated with T2D given their association with birth weight and vice versa, a cFDR Manhattan plot was constructed, which marks the significance of various SNPs and their chromosomal locations.
Calculation of conjunctional cFDR
To determine pleiotropic loci, the conjunctional cFDR (ccFDR) was calculated, which refers to the possibility that a given SNP has a false positive association with both the principal and conditional traits. The ccFDR was computed as the maximum cFDR values (i.e. T2D| birth weight and birth weight|T2D) of the two traits. A ccFDR of 0.05 was used to identify whether an SNP is a pleiotropic locus. To visualize the localization of significant pleiotropic loci, ccFDR Manhattan plots were constructed on the basis of the ranking of ccFDR.
Function annotation of pleiotropic SNPs
To explore whether any of the identified pleiotropic SNPs may play a functional role in T2D and birth weight, each pleiotropic SNP was annotated to corresponding DNA features or regulatory elements in non-coding regions using HaploReg (http://www.broadinstitute.org/mammals/haploreg/haploreg.php, acc essed 1 March 2016) and RegulomeDB (http://www.regulomedb.org/, accessed 15 March 2016) tools. HaploReg retrieves the ENCODE annotation for the SNP of interest as well as other SNPs in LD; the user can configure values (such as the LD threshold and the reference population used) from 1000 genomes datasets. RegulomeDB retrieves the ENCODE annotation and calculates a score for the regulatory potential of this region. It also has a database of predicted functional SNPs, by disease or trait and by SNP. Then, the program GOEAST (http://omicslab.genetics.ac.cn/GOEAST/, accessed 10 May 2016) was used to identify significantly enriched gene ontology (GO) terms among the list of genes associated with pleiotropic SNPs. The P-values were calculated by hypergeometric tests and adjusted for multiple comparisons by stringent Yekutieli (FDR under dependency) adjustment.16 In order to partially explore and characterize the functional relationship of the T2D genes identified, the corresponding protein association networks were constructed using the STRING 10.0 database (http://string-db.org/, accessed 1 May 2016).
Results
Pleiotropic enrichment of T2D SNPs conditional on association with birth weight and vice versa
Conditional Q–Q plots are a common method to graphically assess the pleiotropic enrichment of genetic loci. Interestingly, we observed a strong enrichment of T2D-associated SNPs, with the proportion of true effects in T2D varying considerably depending on different levels of association for birth weight (Fig. 1a) because there appears to be a greater amount of separation between the different curves. As shown in Fig. 1b, there is a less robust enrichment pattern for birth weight conditioned on T2D compared with the pattern for T2D conditioned on birth weight. The presence of an earlier leftward shift indicates a greater proportion of true associations for the principal trait given the nominal P-value of the conditional trait.
Figure 1.
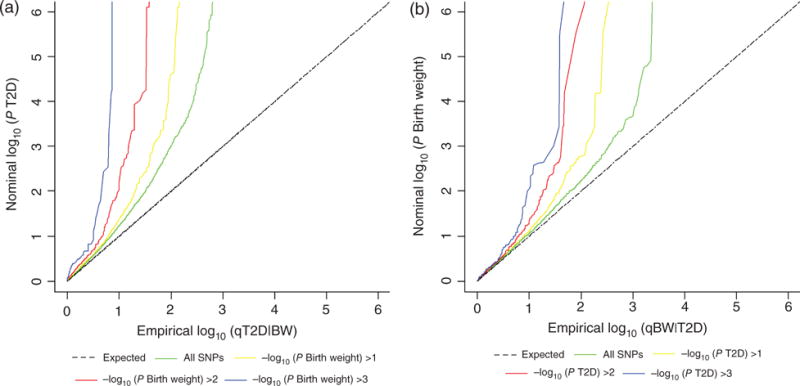
Conditional Q–Q plot. Stratified Q-Q plot of enrichment versus nominal −log10 P-values (corrected for inflation) in (a) type 2 diabetes (T2D) as a function of significance of the association with birth weight (T2D|birth weight) and (b) birth weight as a function of significance of the association with T2D (birth weight|T2D) below the standard genome-wide association study threshold of P < 5 × 10−8 at the level of −log10(P) > 0, −log10(P) > 1, −log10(P) > 2, and −log10(P) > 3 corresponding to P < 1, P < 0.1, P < 0.01, and P < 0.001, respectively. Dashed lines indicate the null-hypothesis.
Type 2 diabetes loci identified with cFDR
As shown in the cFDR Manhattan plot for T2D conditioned on birth weight (see Fig. S1, available as Supplementary Material to this paper), 133 significant SNPs were identified with a significance threshold of cFDR < 0.05 on 21 different chromosomes (Table S1). Interestingly, 13 of these significant SNPs reached genome-wide significance at 5 × 10−8 in the original meta-analysis for T2D,13 including two loci (rs2881654 and rs2283228) also reported in previous T2D GWAS.17,18 More importantly, 120 novel loci were identified that had been overlooked in the original meta-analysis.13 Using the more conservative threshold of cFDR < 0.01, 62 significant loci remained (Table S1). Interestingly, there were five significant SNPs, including three novel SNPs rs163177 (cFDR = 1.50 × 10−8), rs234857 (cFDR = 3.47 × 10−3) and rs3852527 (cFDR = 4.73 × 10−3) located at the potassium voltage-gated channel subfamily Q member 1 (KCNQ1) gene (11p15.4), that were associated with an increased risk of T2D susceptibility in previous studies.18,19 To explore the functional association among identified T2D target genes and networks involved in the biological function of T2D, the genes that include the 133 significant SNPs were uploaded into the STRING 10.0 database. Interestingly, the network consisted of positive regulation of cellular process genes and negative regulation of transcription from RNA polymerase II promoter genes, showing a strong protein– protein interaction among proteins corresponding to the T2D target genes (Fig. 2).
Figure 2.
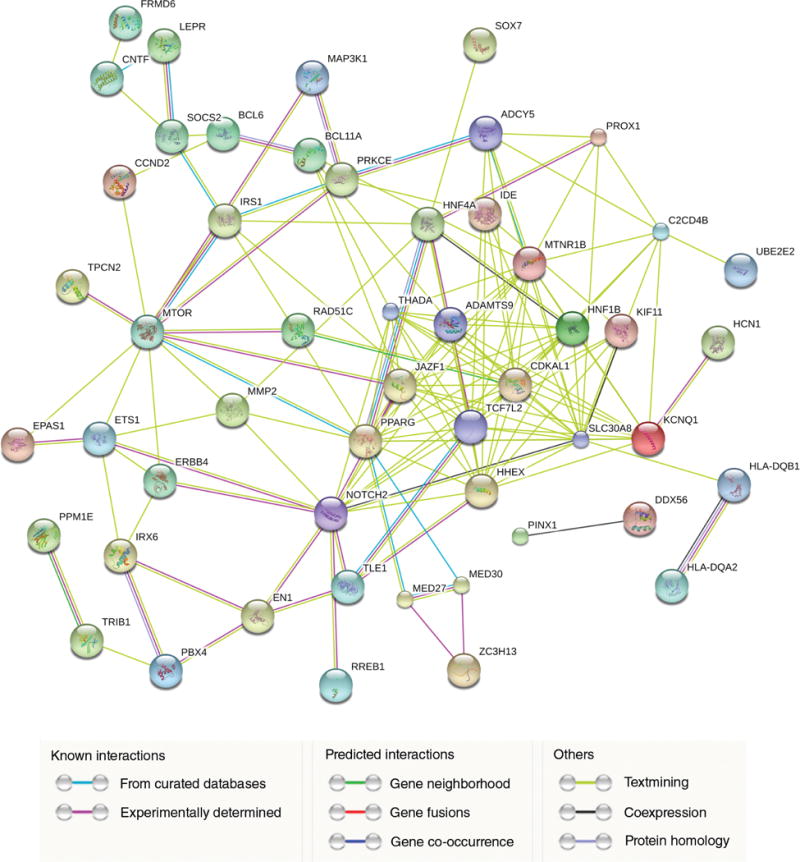
Functional protein association network analysis for type 2 diabetes (T2D) susceptibility genes. Connections are based on coexpression and experimental evidence with a STRING 10.0 (http://string-db.org/, accessed 1 May 2016) summary score above 0.4. The network that related to positive regulation of cellular process and negative regulation of transcription from RNA polymerase II promoter showed significant enrichment for T2D susceptibility genes. Each filled node denotes a gene; edges between nodes indicate protein–protein interactions between protein products of the corresponding genes. Different edge colors represent the types of evidence for the association. Definitions for all protein symbols are given in Table S2.
Birth weight loci identified with cFDR
As shown in the cFDR Manhattan plot for birth weight (Fig. S2), 13 significant SNPs were identified with a significance threshold of cFDR < 0.05 for birth weight variation on their association with T2D; these were mapped to nine different chromosomes (Table 1). Using the more conservative threshold of conditional cFDR < 0.01, three significant loci remained. Of interest, the current pleiotropy-informed cFDR method validated a locus (rs1042725) that was identified in a previous birth weight GWAS study.14 Twelve new loci were discovered for the 13 significant SNPs at cFDR <0.05, including a novel SNP rs1012635 observed at the previously identified T2D-associated gene CDK5 regulatory subunit-associated protein 1-like 1 (CDKAL1).20
Table 1.
Conditional false discovery rate: birth weight loci given type 2 diabetes (birth weight|T2D)
| SNP | SNP chromosome | SNP position | Birth weight P-value | cFDR (birth weight|T2D) | Nearby gene |
|---|---|---|---|---|---|
| rs1012635 | 6 | 20783274 | 4.10 × 10−3 | 1.91 × 10−2 | CDK5 regulatory subunit-associated protein 1-like 1 (CDKAL1) |
| rs1042725 | 12 | 64644614 | 2.80 × 10−9 | 2.21 × 10−7 | High mobility group AT-hook 2 (HMGA2) |
| rs10882028 | 10 | 93933880 | 2.90 × 10−6 | 1.07 × 10−4 | Cytoplasmic polyadenylation element binding protein 3 (CPEB3) |
| rs12610185 | 19 | 19582722 | 7.70 × 10−3 | 4.62 × 10−2 | PBX homeobox 4 (PBX4) |
| rs231354 | 11 | 2662927 | 2.30 × 10−3 | 2.45 × 10−2 | Potassium voltage-gated channel subfamily Q member 1 (KCNQ1) |
| rs2782980 | 10 | 115771517 | 6.40 × 10−5 | 8.30 × 10−3 | Adrenoceptor beta 1 (ADRB1) |
| rs4677887 | 3 | 124582913 | 3.60 × 10−4 | 1.67 × 10−2 | Adenylate cyclase 5 (ADCY5) |
| rs4753073 | 11 | 92357123 | 2.70 × 10−3 | 2.23 × 10−2 | Melatonin receptor 1B (MTNR1B)||solute carrier family 36 member 4 (SLC36A4) |
| rs916419 | 22 | 48555425 | 3.50 × 10−4 | 4.05 × 10−2 | Bromodomain containing 1 (BRD1) |
| rs871961 | 3 | 150061208 | 6.70 × 10−5 | 4.26 × 10−2 | Carboxypeptidase B1 (CPB1)||carboxypeptidase A3 (CPA3) |
| rs1258191 | 10 | 50663125 | 1.20 × 10−5 | 2.71 × 10−2 | Mitogen-activated protein kinase 6 pseudogene 6 (MAPK6PS6)|| LOC727726 |
| rs6016373 | 20 | 38587509 | 1.60 × 10−5 | 4.06 × 10−2 | Heat shock protein family E (Hsp10) member 1 pseudogene 1 (HSPE1P1)||MAF bZIP transcription factor B (MAFB) |
| rs222857 | 17 | 7105287 | 1.70 × 10−5 | 4.02 × 10−2 | Claudin 7 (CLDN7) |
The chromosome position in the GRCh37.p5 sequence of Genome Build 37.3 (ftp://ftp.ncbi.nlm.nih.gov/genomes/Homo_sapiens/ARCHIVE/BUILD.37.3, accessed 1 May 2015).
SNP, single nucleotide polymorphism; cFDR, conditional false discovery rate.
Pleiotropic loci in T2D and birth weight identified with ccFDR
To investigate whether any of the SNPs associated with T2D conditioned on birth weight were also significantly associated with birth weight conditioned on T2D, ccFDR was calculated and a ccFDR Manhattan plot was constructed (Fig. 3). Nine independent pleiotropic SNPs on a total of seven chromosomes reached a significance level of ccFDR < 0.05 (Table 2). Of the nine independent pleiotropic SNPs, rs231354 has previously been associated with T2D21 and rs1042725 has been associated with both T2D and birth weight.14,22 The present analysis reports seven novel pleiotropic SNPs not previously detected. These seven loci annotated at eight different genes, of which six (cytoplasmic polyadenylation element binding protein 3 [CPEB3], PBX homeobox 4 [PBX4], adrenoceptor beta 1 (ADRB1), melatonin receptor 1B [MTNR1B], solute carrier family 36 member 4 [SLC36A4], and bromodomain containing 1 [BRD1]) are novel genes and another two genes (CDKAL1 and adenylate cyclase 5 [ADCY5]) were reported in previous T2D and birth weight GWAS.9
Figure 3.
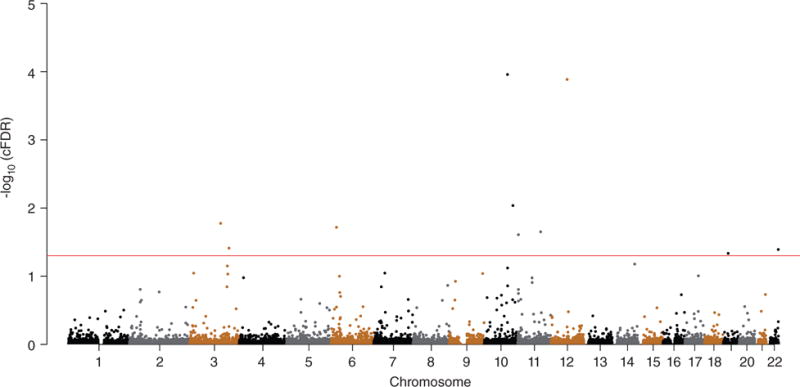
“Conjunctional Manhattan plot” of conjunctional −log10 (conditional false discovery rate [cFDR]) values for type 2 diabetes (T2D) and birth weight. Single nucleotide polymorphisms with conjunctional −log10 cFDR >1.3 (i.e. cFDR < 0.05) are shown above the red line. The figure marks the chromosomal locations of significant loci. Details for all significant loci are given in Table 2.
Table 2.
Conjunctional conditional false discovery rate: pleiotropic loci in type 2 diabetes and birth weight
| SNP | SNP chromosome | SNP position* | cFDR
|
ccFDR | Nearby gene | |
|---|---|---|---|---|---|---|
| T2D|birth weight | Birth weight|T2D | |||||
| rs1012635 | 6 | 20783274 | 3.36 × 10−8 | 1.91 × 10−2 | 1.91 × 10−2 | CDK5 regulatory subunit-associated protein 1-like 1 (CDKAL1) |
| rs1042725 | 12 | 64644614 | 1.29 × 10−4 | 2.21 × 10−7 | 1.29 × 10−4 | High mobility group AT-hook 2 (HMGA2) |
| rs10882028 | 10 | 93933880 | 1.10 × 10−4 | 1.07 × 10−4 | 1.10 × 10−4 | Cytoplasmic polyadenylation element binding protein 3 (CPEB3) |
| rs12610185 | 19 | 19582722 | 2.35 × 10−5 | 4.62 × 10−2 | 4.62 × 10−2 | PBX homeobox 4 (PBX4) |
| rs231354 | 11 | 2662927 | 2.94 × 10−5 | 2.45 × 10−2 | 2.45 × 10−2 | Potassium voltage-gated channel subfamily Q member 1 (KCNQ1) |
| rs2782980 | 10 | 115771517 | 9.18 × 10−3 | 8.30 × 10−3 | 9.18 × 10−3 | Adrenoceptor beta 1 (ADRB1) |
| rs4677887 | 3 | 124582913 | 3.68 × 10−3 | 1.67 × 10−2 | 1.67 × 10−2 | Adenylate cyclase 5 (ADCY5) |
| rs4753073 | 11 | 92357123 | 2.67 × 10−5 | 2.23 × 10−2 | 2.23 × 10−2 | Melatonin receptor 1B (MTNR1B)||solute carrier family 36 member 4 (SLC36A4) |
| rs916419 | 22 | 48555425 | 2.11 × 10−2 | 4.05 × 10−2 | 4.05 × 10−2 | Bromodomain containing 1 (BRD1) |
The chromosome position in the GRCh37.p5 sequence of Genome Build 37.3 (ftp://ftp.ncbi.nlm.nih.gov/genomes/Homo_sapiens/ARCHIVE/BUILD.37.3, accessed 1 May 2015).
SNP, single nucleotide polymorphism; cFDR, conditional false discovery rate; ccFDR, conjunctional cFDR; T2D, type 2 diabetes.
Functional and pathway analysis for pleiotropic loci
A series of bioinformatics analyses was conducted to explore the potential regulatory functions for nine pleiotropic SNPs. As annotated using HaploReg and RegulomeDB databases, rs1012635 (CDKAL1), rs1042725 (high mobility group AT-hook 2 [HMGA2]), rs12610185 (PBX4), rs231354 (KCNQ1), rs4753073 (MTNR1B) and rs2782980 (ADRB1) overlapped with open chromatin in a number of ENCODE cell lines, such as hepatocyte, fibroblast and osteoblast cell lines, which potentially related to T2D or birth weight phenotypes. Furthermore, we identified six pleiotropic SNPs, namely rs12610185 (PBX4), rs231354 (KCNQ1OT1), rs2782980 (ADRB1), rs4677887 (ADCY5), rs4753073 (MTNR1B) and rs916419 (BRD1), that fell into the enhancer regions of the corresponding genes annotated to these SNPs in a number of organs, such as the liver, pancreas, colonic mucosa and small intestine, confirming that these pleiotropic SNPs are highly enriched within regions of active chromatin state. To systematically investigate whether the observed pleiotropic SNPs were T2D and birth weight specific, GO analysis was conducted (Table 3), revealing a variety of biological processes, (e.g. adenosine receptor signaling pathway [P = 4.29 × 10−6], G-protein-coupled purinergic receptor signaling pathway [P = 4.29 × 10−6] and adenylate cyclase-activating dopamine receptor signaling pathway [P = 9.79 × 10−6]) that are closely related to T2D and birth weight.
Table 3.
Top five most significant gene ontology terms enriched for genes associated with pleiotropic SNPs
| GO ID | GO terms | log (OR) | P-value |
|---|---|---|---|
| GO:0001973 | Adenosine receptor signaling pathway | 9.66 | 4.29 × 10−6 |
| GO:0035588 | G-Protein-coupled purinergic receptor signaling pathway | 9.66 | 4.29 × 10−6 |
| GO:0007195 | Adenylate cyclase-inhibiting dopamine receptor signaling pathway | 9.14 | 9.79 × 10−6 |
| GO:0035587 | Purinergic receptor signaling pathway | 8.66 | 2.43 × 10−5 |
| GO:0007191 | Adenylate cyclase-activating dopamine receptor signaling pathway | 8.47 | 3.42 × 10−5 |
Gene ontology (GO) enrichment analysis was performed using the GOEAST program (http://omicslab.genetics.ac.cn/GOEAST/, accessed 10 May 2016), which gave “P > 0” when the obtained P-value was less than the minimum float value (1.17549435082229 × 10−38). OR, odds ratio.
Discussion
By applying the stratified cFDR method to the summary statistics of T2D and birth weight GWAS, 133 T2D susceptibility loci were identified, including 120 novel loci that were missed in the original GWAS meta-analysis for T2D.13 Furthermore, 13 significant birth weight-associated SNPs conditioned on T2D (cFDR < 0.05) were identified that were mapped to nine different chromosomes. Importantly, nine pleiotropic SNPs were identified suggesting a shared genetic mechanism among them. The results demonstrate that GWAS from birth weight may improve discovery of T2D susceptibility loci and enhance our understanding of the effect of common genetic variants on both traits.
In the present study, the most significant novel T2D susceptibility SNP identified by the cFDR method was rs163177. This SNP is located at the T2D risk gene KCNQ1 (11p15.4). The KCNQ1 gene, encoding the voltage-gated K+ channel KvLQT1 subunit,23 regulates insulin secretion function through the voltage-gated K+ channel, which drives an electrical signal in pancreatic β-cells to promote glucose-stimulated insulin release.24 In addition, Torekov et al. reported that KCNQ1 long QT syndrome patients are susceptible to symptomatic hypoglycemia and hyperinsulinemia, because KCNQ1 gene mutations can delay repolarization of β-cells and result in increased insulin secretion.25 Another interesting SNP, rs4848526 (2q14.2), is located in the intron of the engrailed homeobox 1 (EN1) gene, which plays an important role in WNT signaling activation.26 The WNT signaling pathway is associated with many physiological and pathophysiological activities, including T2D.27 It is involved in the pathogenesis of T2D by regulating β-cell genesis and proliferation,28 modulating lipid metabolism and insulin secretion,29 and mediating the production of the incretin hormone glucagon-like peptide-1 (GLP-1).30 Based on known and predicted protein–protein interactions, network analysis of the T2D target genes further illustrated the interactions of molecules that regulate metabolism of T2D and clustered these genes into functional categories. Interestingly, the network clusters that involved positive regulation of cellular process, negative regulation of transcription from RNA polymerase II promoter, positive regulation of lipid metabolic process, hepatoblast differentiation, and hepatocyte differentiation may be the functionally regulated modules for pathophysiology of T2D.
Furthermore, the current pleiotropy cFDR method identified 13 significant SNPs for birth weight conditioned on T2D (cFDR < 0.05), including SNP rs1042725, which was reported to be associated with birth weight in a previous GWAS.14 Twelve new loci were discovered. For example, the novel significant SNP rs1012635 was annotated at gene CDKAL1, which showed a strong association with birth weight in a previous study.20 The CDKAL1 gene is located on chromosome 6p22.3, is 698 kbp long and encodes a member of the methylthiotransferase family. Previous studies suggested that CDKAL1 was expressed in human pancreatic islets and may be associated with insulin secretion by pancreatic β-cells by interacting with cyclin dependent kinase 5 (CDK5).31 However, the exact function of CDKAL1 remains unclear.
Importantly, nine pleiotropic variants were identified and the SNP rs1042725 in HMGA2 (12q14.3) has been associated with both T2D and birth weight.14,22 As an important genetic determinant of birth weight and human height,14,32 HMGA2 was also considered a T2D risk gene.33 In white adipose tissue, high expression of HMGA2 can drive cellular senescence to increase susceptibility to T2D.33 More importantly, seven novel pleiotropic loci were identified (rs1012635 in CDKAL1, rs10882028 in CPEB3, rs12610185 in PBX4, rs2782980 in ADRB1, rs4677887 in ADCY5, rs4753073 in MTNR1B [SLC36A4] and rs916419 in BRD1) that have not been reported in previous studies. Both CDKAL1 and ADCY5 were confirmed to increase risk of T2D and low birth weight through the “fetal insulin hypothesis,”9 suggesting that gene variation related to pancreatic β-cell function or insulin secretion during embryonic development can lead to decreased birth weight as well as subsequent development of T2D.9 The GO analysis revealed an enrichment of biological processes that are closely related to T2D and birth weight, including the adenosine receptor signaling pathway and the G-protein-coupled purinergic receptor signaling pathway, among others. A previous study reported that the adenosine A2B receptor signaling pathway modulates glucose, lipid homeostasis and chronic inflammation, and regulates the activity of resident macrophages in adipose tissue in T2D.34 Interestingly, changes in cardiac glucose metabolism are associated with low birth weight,35 suggesting a potential role for the adenosine receptor in low birth weight.
The major strengths of the present study are that by leveraging of GWAS results from T2D and birth weight phenotypes, we successfully improved the detection of uncovered T2D associated loci without additional large datasets and identified several novel pleiotropic SNPs using the cFDR method. These findings present novel insights for exploring common underlying molecular mechanisms and offer promising clues for further experimental studies. There may be some limitations to the present study. First, we were unable to associate the genetic findings with clinical outcomes due to our inability to access raw clinical data. However, the primary purpose of the study was to improve the identification of disease-associated genes and explore the overlapping biological mechanisms between T2D and birth weight. In addition, we did not identify all the previously implicated genes in T2D and birth weight because the present study analyzed only a subset of available GWAS to efficiently reveal the missing heritability in the two traits. Therefore, clinical replications and further biological experiments are necessary to validate our findings.
In conclusion, the present study demonstrated high efficiency of the cFDR method in improving the identification of novel genetic variants of both T2D and birth weight. The findings offer novel insights into potential shared genetic mechanisms in T2D and birth weight, which may form a basis for further biological experiments and clinical replication.
Supplementary Material
Figure S1. Conditional Manhattan plot of conditional −log10(False discovery rate) values for type 2 diabetes (T2D) given birth weight.
Figure S2. Conditional Manhattan plot of conditional −log10(False discovery rate) values birthweight given T2D.
Table S1. Conditional false discovery rate (cFDR): T2D loci given birth weight (T2D|birth weight (cFDR < 0.05.
Table S2. Gene symbol and definitions.
Highlights.
This study demonstrates high efficiency of the conditional false discovery rate (cFDR) method in improving the identification of novel genetic variants of both type 2 diabetes (T2D) and birth weight.
The findings offer novel insights into potential shared genetic mechanisms in T2D and birth weight, which may form a basis for further biological experiments and clinical replication.
The study identified two novel pleiotropic loci that may be related to the processes that affect T2D metabolism and may therefore contribute to the genetic susceptibility to T2D.
Acknowledgments
HWD was supported, in part, by grants from the National Institutes of Health (R01AR059781, R01AR050496, R01AR057049), and the Edward G. Schlieder Endowment fund from Tulane University. YCC was supported, in part, by the Foundation from China Scholarship Council (20150322). The authors thank the DIAGRAM Consortium and the Early Growth Genetics Consortium for the summary statistics of genome-wide association studies data.
Footnotes
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Disclosure
The authors declare that they have no conflicts of interest.
References
- 1.American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2009;32(Suppl 1):S62–7. doi: 10.2337/dc09-S062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ozcan U, Cao Q, Yilmaz E, et al. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–61. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 3.NCD Risk Factor Collaboration. Worldwide trends in diabetes since 1980: A pooled analysis of 751 population-based studies with 4.4 million participants. Lancet. 2016;387:1513–30. doi: 10.1016/S0140-6736(16)00618-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: Estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27:1047–53. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 5.Saddik B, Al-Dulaijan N. Diabetic patients’ willingness to use tele-technology to manage their disease: A descriptive study. Online J Public Health Inform. 2015;7:e214. doi: 10.5210/ojphi.v7i2.6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gluckman PD, Hanson MA, Bateson P, et al. Towards a new developmental synthesis: Adaptive developmental plasticity and human disease. Lancet. 2009;373:1654–7. doi: 10.1016/S0140-6736(09)60234-8. [DOI] [PubMed] [Google Scholar]
- 7.Zimmermann E, Gamborg M, Sorensen TI, Baker JL. Sex differences in the association between birth weight and adult type 2 diabetes. Diabetes. 2015;64:4220–5. doi: 10.2337/db15-0494. [DOI] [PubMed] [Google Scholar]
- 8.Wang T, Huang T, Li Y, et al. Low birthweight and risk of type 2 diabetes: A Mendelian randomisation study. Diabetologia. 2016;59:1920–7. doi: 10.1007/s00125-016-4019-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersson EA, Pilgaard K, Pisinger C, et al. Type 2 diabetes risk alleles near ADCY5, CDKAL1 and HHEX-IDE are associated with reduced birthweight. Diabetologia. 2010;53:1908–16. doi: 10.1007/s00125-010-1790-0. [DOI] [PubMed] [Google Scholar]
- 10.Ali O. Genetics of type 2 diabetes. World J Diabetes. 2013;4:114–23. doi: 10.4239/wjd.v4.i4.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magnus P, Gjessing HK, Skrondal A, Skjaerven R. Paternal contribution to birth weight. J Epidemiol Community Health. 2001;55:873–7. doi: 10.1136/jech.55.12.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andreassen OA, Thompson WK, Schork AJ, et al. Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLoS Genet. 2013;9:e1003455. doi: 10.1371/journal.pgen.1003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris AP, Voight BF, Teslovich TM, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–90. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horikoshi M, Yaghootkar H, Mook-Kanamori DO, et al. New loci associated with birth weight identify genetic links between intrauterine growth and adult height and metabolism. Nat Genet. 2013;45:76–82. doi: 10.1038/ng.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liley J, Wallace C. A pleiotropy-informed Bayesian false discovery rate adapted to a shared control design finds new disease associations from GWAS summary statistics. PLoS Genet. 2015;11:e1004926. doi: 10.1371/journal.pgen.1004926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yekutieli D, Benjamini Y. Resampling-based false discovery rate controlling multiple test procedures for correlated test statistics. J Stat Plann Inference. 1999;82:171–96. [Google Scholar]
- 17.Elbein SC, Gamazon ER, Das SK, Rasouli N, Kern PA, Cox NJ. Genetic risk factors for type 2 diabetes: A trans-regulatory genetic architecture? Am J Hum Genet. 2012;91:466–77. doi: 10.1016/j.ajhg.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Unoki H, Takahashi A, Kawaguchi T, et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet. 2008;40:1098–102. doi: 10.1038/ng.208. [DOI] [PubMed] [Google Scholar]
- 19.Hanson RL, Guo T, Muller YL, et al. Strong parent-of-origin effects in the association of KCNQ1 variants with type 2 diabetes in American Indians. Diabetes. 2013;62:2984–91. doi: 10.2337/db12-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao J, Li M, Bradfield JP, et al. Examination of type 2 diabetes loci implicates CDKAL1 as a birth weight gene. Diabetes. 2009;58:2414–8. doi: 10.2337/db09-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Travers ME, Mackay DJ, Dekker Nitert M, et al. Insights into the molecular mechanism for type 2 diabetes susceptibility at the KCNQ1 locus from temporal changes in imprinting status in human islets. Diabetes. 2013;62:987–92. doi: 10.2337/db12-0819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saxena R, Elbers CC, Guo Y, et al. Large-scale genecentric meta-analysis across 39 studies identifies type 2 diabetes loci. Am J Hum Genet. 2012;90:410–25. doi: 10.1016/j.ajhg.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanguinetti MC, Curran ME, Zou A, et al. Coassembly of K(V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature. 1996;384:80–3. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- 24.Ashcroft FM. ATP-sensitive potassium channelopathies: Focus on insulin secretion. J Clin Invest. 2005;115:2047–58. doi: 10.1172/JCI25495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torekov SS, Iepsen E, Christiansen M, et al. KCNQ1 long QT syndrome patients have hyperinsulinemia and symptomatic hypoglycemia. Diabetes. 2014;63:1315–25. doi: 10.2337/db13-1454. [DOI] [PubMed] [Google Scholar]
- 26.Adamska M, MacDonald BT, Sarmast ZH, Oliver ER, Meisler MH. En1 and Wnt7a interact with Dkk1 during limb development in the mouse. Dev Biol. 2004;272:134–44. doi: 10.1016/j.ydbio.2004.04.026. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Shan J, Chang W, et al. Chemical and genetic evidence for the involvement of Wnt antagonist Dickkopf2 in regulation of glucose metabolism. Proc Natl Acad Sci U S A. 2012;109:11402–7. doi: 10.1073/pnas.1205015109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rulifson IC, Karnik SK, Heiser PW, et al. Wnt signaling regulates pancreatic beta cell proliferation. Proc Natl Acad Sci U S A. 2007;104:6247–52. doi: 10.1073/pnas.0701509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin T. The Wnt signalling pathway and diabetes mellitus. Diabetologia. 2008;51:1771–80. doi: 10.1007/s00125-008-1084-y. [DOI] [PubMed] [Google Scholar]
- 30.Yi F, Sun J, Lim GE, Fantus IG, Brubaker PL, Jin T. Cross talk between the insulin and Wnt signaling pathways: Evidence from intestinal endocrine l cells. Endocrinology. 2008;149:2341–51. doi: 10.1210/en.2007-1142. [DOI] [PubMed] [Google Scholar]
- 31.Steinthorsdottir V, Thorleifsson G, Reynisdottir I, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39:770–5. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 32.Buysse K, Reardon W, Mehta L, et al. The 12q14 microdeletion syndrome: Additional patients and further evidence that HMGA2 is an important genetic determinant for human height. Eur J Med Genet. 2009;52:101–7. doi: 10.1016/j.ejmg.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 33.Markowski DN, Thies HW, Gottlieb A, et al. HMGA2 expression in white adipose tissue linking cellular senescence with diabetes. Genes Nutr. 2013;8:449–56. doi: 10.1007/s12263-013-0354-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnston-Cox H, Koupenova M, Yang D, et al. The A2B adenosine receptor modulates glucose homeostasis and obesity. PLoS One. 2012;7:e40584. doi: 10.1371/journal.pone.0040584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang KC, Lim CH, McMillen IC, Duffield JA, Brooks DA, Morrison JL. Alteration of cardiac glucose metabolism in association to low birth weight: Experimental evidence in lambs with left ventricular hypertrophy. Metabolism. 2013;62:1662–72. doi: 10.1016/j.metabol.2013.06.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Conditional Manhattan plot of conditional −log10(False discovery rate) values for type 2 diabetes (T2D) given birth weight.
Figure S2. Conditional Manhattan plot of conditional −log10(False discovery rate) values birthweight given T2D.
Table S1. Conditional false discovery rate (cFDR): T2D loci given birth weight (T2D|birth weight (cFDR < 0.05.
Table S2. Gene symbol and definitions.


