Abstract
The thalamic reticular nucleus (TRN) is a unique brain structure at the interface between the thalamus and the cortex. Because the TRN receives bottom-up sensory input and top-down cortical input, it could serve as an integration hub for sensory and cognitive signals. Functional evidence supports broad roles for the TRN in arousal, attention, and sensory selection. How specific circuits connecting the TRN with sensory thalamic structures implement these functions is not known. The structural organization and function of the TRN is particularly interesting in the context of highly organized sensory systems, such as the primate visual system, where neurons in the retina and dorsal lateral geniculate nucleus of the thalamus (dLGN) are morphologically and physiologically distinct and also specialized for processing particular features of the visual environment. To gain insight into the functional relationship between the visual sector of the TRN and the dLGN, we reconstructed a large number of TRN neurons that were retrogradely labeled following injections of rabies virus expressing enhanced green fluorescent protein (EGFP) into the dLGN. An independent cluster analysis, based on 10 morphological metrics measured for each reconstructed neuron, revealed three clusters of TRN neurons that differed in cell body shape and size, dendritic arborization patterns, and medial-lateral position within the TRN. TRN dendritic and axonal morphologies are inconsistent with visual stream-specific projections to the dLGN. Instead, TRN neuronal organization could facilitate transmission of global arousal and/or cognitive signals to the dLGN with retinotopic precision that preserves specialized processing of foveal versus peripheral visual information.
INDEXING TERMS: TRN, LGN, anatomy, independent cluster analysis, RRID: SCR_001775, RRID: SCR_001622
The thalamic reticular nucleus (TRN) is a thin, shell-like structure wrapping around the thalamus and forming an interface between the thalamus and the cortex that is present in all mammals (Pinault, 2004). Neurons in the TRN receive collateral input from thalamic relay neurons and corticothalamic neurons (Jones, 1985). All TRN neurons are GABAergic and make inhibitory connections with thalamic relay neurons (Jones, 1985; Ohara and Lieberman, 1985; Sanches-Vives and McCormick, 1997; Pinault and Deschenes, 1998; Uhlrich et al., 2003). Based on the organization of thalamic and cortical inputs, the TRN is organized into different sectors, many of which are associated with first-order sensory thalamic nuclei (Jones, 1985). Some sectors of the TRN receive inputs from additional brain areas including the pulvinar (Conley and Diamond, 1990; Baldauf, 2010), the brainstem and basal forebrain (Guillery and Harting, 2003), and the prefrontal cortex (Zikopoulos and Barbas, 2006). Accordingly, the TRN could serve as an integration hub for bottom-up sensory and top-down cognitive signals (Guillery and Harting, 2003). Along these lines, Crick (1984) proposed that the TRN controls the flow of perceptual signals from the thalamus to the cortex. Subsequent studies have provided evidence that TRN neurons are modulated by attention and facilitate interactions between sensory and cognitive signals (McAlonan et al., 2006, 2008; Halassa et al., 2014; Wimmer et al., 2015). Recent studies have even outlined specific genetic contributions to the TRN’s role in sensory selection and attention (Ahrens et al., 2015; Wells et al., 2016).
While it is clear that the TRN is important for a number of broadly defined functions such as sensory selection, attention, and arousal, how circuits connecting TRN neurons with their thalamic targets mediate these functions remains unknown. The specific organization and function of TRN neurons is especially intriguing in the context of highly specialized sensory systems, such as the visual system of the primate. Neurons in the primate retina and dorsal lateral geniculate nucleus of the thalamus (dLGN) are optimized for acuity and color vision—unique specializations among mammals—and also form distinct parallel processing streams to encode a rich representation of the visual world. Primate retinal and dLGN neurons in the magnocellular (M), parvocellular (P), and koniocellular (K) streams are morphologically and physiologically distinct in order to convey information about visual motion, form/acuity, and color in parallel (Kaplan, 2004). M, P, and K neurons are physically segregated into separate layers in the dLGN and synapse in specific laminar compartments within the visual cortex. Given the strict segregation of feed-forward visual signals into parallel streams, it remains an open question whether TRN inputs to the dLGN in the primate maintain this stream-specific segregation or provide a more global input that is not stream-specific. More is known about the morphology, physiology, and organization of TRN neurons in nonprimate species; these findings provide clues about possible primate TRN-dLGN connectivity schemes, discussed below.
Neurons in the visual sector of the TRN receive inputs from and project axons to the dLGN (Sherman and Guillery, 2006). These afferent inputs and efferent projections are retinotopically organized, consistent with the topographic organization of afferent and efferent connections between TRN sectors and their associated sensory thalamic nuclei (Montero et al., 1977; Crabtree and Killackey, 1989; Conley and Diamond, 1990; Uhlrich et al., 2003; Fitzgibbon et al., 2007). Evidence from carnivores, rodents, and Galagos suggests that sectors of the TRN, including the visual sector, are organized into adjacent stripes or tiers including “inner” and “outer” tiers that receive input from higher-order and first-order thalamic nuclei, respectively (Conley and Diamond, 1990; Sherman and Guillery, 2006; Baldauf, 2010; Lam and Sherman, 2011). It is not clear whether the visual sector of the TRN in macaque monkeys is organized into tiers since macaques may have separate TRN sectors devoted to higher-order thalamic nuclei such as the pulvinar (Jones, 1985; Lyon and Rabideau, 2012).
Given the specificity of inputs to TRN sectors/tiers, it is tempting to predict that neurons within distinct TRN sectors/tiers are unique in some respect. Anatomical studies of individual neurons throughout the TRN from a variety of species, including rodents, cats, rabbits, marmosets, and Galagos, have yielded broadly similar findings: TRN neurons are mostly nonspiny, with dendrites that form a polarized or “discoid” arborization pattern such that TRN neurons are oriented in parallel, i.e., horizontal, relative to the borders of the TRN (Scheibel and Scheibel, 1972; Spreafico et al., 1991; Lubke, 1993; Ohara and Havton, 1996; Uhlrich et al., 2003; Fitzgibbon et al., 2007). Variations have been observed in cell body shape—fusiform versus more rounded cell bodies (Spreafico et al., 1991; Pinault, 2004)—and in the extent to which TRN dendrites are polarized or circular. Often these differences relate to medial-lateral position within the TRN (e.g., sector) and/or position in thinner versus thicker portions of the TRN (Scheibel and Scheibel, 1972; Spreafico et al., 1991; but see Lubke, 1993; Pinault, 2004).
Results of both in vitro and in vivo neurophysiological recordings from TRN neurons provide conflicting evidence in favor of homogeneous or heterogeneous populations of TRN neurons. While some studies suggest that TRN neurons display similar spiking characteristics (Spreafico et al., 1988; Pinault, 2004), others provide evidence for distinct TRN types based on physiological properties such as burst firing (Kimura et al., 2012; Lee et al., 2007). Additional evidence in favor of heterogeneous TRN subpopulations comes from studies of local connectivity within the TRN. Multiple groups have demonstrated that TRN neurons are locally interconnected via electrical connections (Landisman et al., 2002; Deleuze and Huguenard, 2006; Lam et al., 2006), although local connectivity may be more limited in the macaque TRN (Williamson et al., 1994). Interestingly, networks of gap-junction connected neurons in the rodent somatosensory sector of the TRN form two different aggregate shapes: discoid/planar networks that are restricted to a single tier of the TRN and more distributed networks that are not restricted to a tier within the TRN (Lee et al., 2014). Taken together, connectivity (afferent, efferent, and local) patterns support a functional segregation of TRN neurons into tiers. Subtle variations in neuronal morphology and physiology also correlate with TRN sector/tier. Thus, observations from a variety of species lend support to a scheme whereby neurons in the visual sector of the primate TRN connect to the dLGN through parallel channels that preserve the segregation of M, P, and K stream visual information.
Our aim was to test three alternative organization and connectivity schemes for the macaque monkey visual TRN: 1) TRN input to the dLGN is organized in parallel streams and originates from morphologically heterogeneous TRN neurons that are separated into tiers or clustered in order to preserve retinotopy; 2) TRN input to the dLGN is global, i.e., not stream-specific, and originates from a single, homogeneous population of TRN neurons; or 3) TRN input to the dLGN is global (not stream-specific), but is organized along an alternative dimension to preserve a unique visual specialization, such as acuity. To test these TRN organization schemes, we reconstructed a large population of dLGN-projecting neurons in the visual sector of the TRN of the macaque monkey that were retrogradely labeled following injections of rabies virus expressing enhanced green fluorescent protein (EGFP) into the dLGN (Briggs et al., 2016). To our knowledge, the dendritic morphology of neurons in the visual sector of the macaque TRN has not been described previously. Additionally, few prior studies have completed a comprehensive morphological survey of TRN neurons projecting to the same thalamic target. We observed morphologically heterogeneous subpopulations of TRN neurons aligned with the medial-lateral axis of the TRN, but no evidence of neuronal differences across tiers in the dorsal-ventral axis. Furthermore, dendritic and axonal morphologies of TRN neurons are inconsistent with a parallel stream organization of TRN inputs to the dLGN. Instead, morphological data are consistent with the third organization scheme proposed above, whereby neurons in the middle of the TRN are specialized for transmitting global (e.g., arousal/cognitive) signals to foveal visual field representations in the dLGN, while neurons toward the medial and lateral aspects of the TRN are specialized for transmitting global signals to peripheral visual field representations in the dLGN.
MATERIALS AND METHODS
The tissue examined in this study was prepared as a part of a separate study (Briggs et al., 2016). Therefore, all of the experimental methods involving the use of animals have been described in detail in the Experimental Methods section of Briggs et al. (2016). All animal procedures conducted in the prior study were approved by the Institutional Animal Care and Use Committees. Briefly, the experimental procedures of the prior study involved surgical injection of a modified rabies virus carrying the gene encoding EGFP, SADΔG-EGFP (Wickersham et al., 2007), into the dLGN in two adult male macaque monkeys (Macacca mulatta). SADΔG-EGFP lacks the gene encoding an essential glycoprotein and cannot move trans-synaptically. The modified rabies virus then acts like a retrograde tracer because it is taken up by axon terminals at the injection site and moves in a retrograde direction to the cell body, where it replicates and makes EGFP. The benefit of virus-mediated retrograde labeling over conventional retrograde tracers is infected/labeled neurons express EGFP throughout their arbors creating Golgi-like neuronal fills and enabling complete reconstruction of neuronal dendritic morphology. Additionally, the modified rabies virus is quite effective at infecting and causing EGFP expression in large numbers of retrograde-labeled neurons, enabling reconstruction of hundreds of neurons per animal (Wickersham et al., 2007; Osakada et al., 2011; Briggs et al., 2016).
In sterile recovery surgery performed while animals were fully anesthetized, total volumes of 5–30 μl of SADΔG-EGFP were injected at 4–6 separate injection sites over 2–3 pipette penetrations such that virus injections spanned the six layers of the dLGN at different retinotopic locations (see Fig. 1Aii,Bii). Depths of dLGN layers were identified by neurophysiological recording of light responses prior to injections. At each injection site, 1–5 μl of virus was injected over 5–15 minutes with 2–5-minute rest periods before repositioning injection pipettes. Animals recovered for 7 days following virus injection and were then euthanized and perfused. Frozen tissue blocks containing the thalamus were sectioned coronally at a thickness of 50 μm. Sections were stained for cytochrome oxidase activity and against GFP using a biotinylated secondary antibody to facilitate permanent staining of labeled neurons via DAB/peroxidase reactivity. Sections were mounted on glass slides, defatted, and coverslipped.
Figure 1.
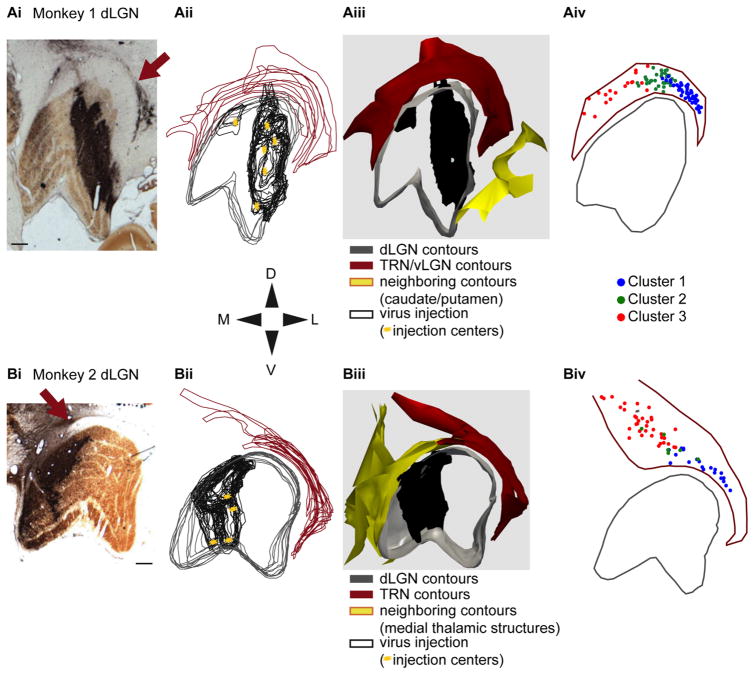
Virus injections restricted to the dLGN and map of reconstructed TRN neurons. Ai,Bi: Photographs of coronal sections through the dorsal lateral geniculate nucleus (dLGN) of Monkeys 1 and 2 stained for cytochrome oxidase and against EGFP such that the dLGN layers and injection site are visualized, respectively. Arrows indicate regions of dense retrogradely labeled thalamic reticular nucleus (TRN) neurons. Section orientation follows the dorsal-ventral/medial-lateral (DV/ML) compass below and scale bars represent 500 μm for all parts of A,B. Aii,Bii: Contour outlines for the dLGN (gray) and the visual sector of the TRN (maroon) for all sections containing injected virus. Black contours outline regions with injected virus in each section and yellow stars indicate the centers of each injection (six injections in Monkey 1, four injections in Monkey 2). Section orientation and scale bars as in Ai,Bi. Aiii,Biii: 3D renderings of the contours and injected virus for each animal with colors, orientation, and scale bars as in Aii,Bii. Aiv,Biv: Maps of the locations of each reconstructed TRN cell body (99 neurons from Monkey 1 [Aiv], 61 neurons from Monkey 2 [Biv], 160 neurons in total), color-coded according to cluster assignment (see legend), within a single aggregate TRN contour (maroon). Orientations and scale bars as in Ai,Bi.
Reconstructing neurons in the visual sector of the TRN
The experimental aim of the current study was to reconstruct a large number of individual labeled neurons in the visual sector of the TRN, in order to characterize the morphology of visual TRN neurons in the macaque monkey. Importantly, we verified in both animals that virus injections were entirely restricted to within the dLGN and did not encroach into any neighboring structures such as the TRN or the ventral LGN (Fig. 1). We observed no labeled neurons outside of layer 6 in the visual cortex, indicating that virus was not injected into the ventral LGN or pulvinar nucleus (Lund et al., 1975; Conley and Friederich-Ecsy, 1993; Rockland, 1994; Briggs et al., 2016). We also verified that virus was not injected into the TRN by observing a lack of labeled neurons in the basal forebrain, a structure that projects to the TRN and not the LGN (Jones, 2002; Guillery and Harting, 2003; Briggs et al., 2016).
Having confirmed that all labeled neurons in the TRN resulted from retrograde virus infection following virus injection into the dLGN in both animals, we reconstructed a total of 160 neurons in the visual sector of the TRN (99 neurons from Monkey 1 and 61 neurons from Monkey 2). Neurons were selected for reconstruction if they were reasonably isolated from neighboring labeled neurons in order to ensure that all reconstructed processes belonged to the same neuron. We also attempted to reconstruct neurons from all regions of the visual sector of the TRN in order to obtain a comprehensive dataset (Fig. 1Aiv,Biv); however, some areas were difficult to sample, given dense retrograde labeling. A small number of the most rostral and medial neurons reconstructed from Monkey 1 (see Fig. 1Aiv) may be located within the ventral LGN, a structure that is adjacent to and contiguous with the visual sector of the TRN surrounding the rostral portion of the dLGN (Harrington, 1997). Reconstructions of structural contours and injection site contours as well as neuronal reconstructions were made using a Neuroludica system (MicroBrightField, Williston, VT; RRID: SCR_001775) with an Optronics camera attached to a Nikon E800M microscope (Nikon Instruments, Melville, NY). Contours for the dLGN, TRN, and injection site were traced for every section containing a reconstructed neuron. Contours for all sections containing injection site label were overlaid and 3D renderings of these contours were created in order to visualize the structure of the TRN, dLGN, and injection sites in both animals (Fig. 1Aii–iii,-Bii–iii). For each neuronal reconstruction, the distance from the cell body to the center of the TRN in the medial-lateral (ML) axis was calculated. Distances from the cell body to the dorsal border of the TRN and the dorsal border of the dLGN were also calculated. Additionally, the dorsal-ventral (DV) thickness of the TRN, measured at the ML position of the cell body, was determined for each contour containing a reconstructed neuron in order to calculate neuronal position within the TRN as a percentage of DV TRN thickness.
We preferentially reconstructed neurons whose cell bodies were located within a single section in order to unambiguously trace the largest extent of each cell body, taking into account cell body orientation relative to the plane of section, and accurately estimate its area and roundness (see below for treatment of neurons whose cell bodies spanned multiple sections). We then reconstructed the dendritic processes of each neuron, following dendrites through at least three adjacent sections (one on each side of the “home” section containing the cell body) and up to a maximum of eight adjacent sections. A node was placed at each branch point along the dendrite.
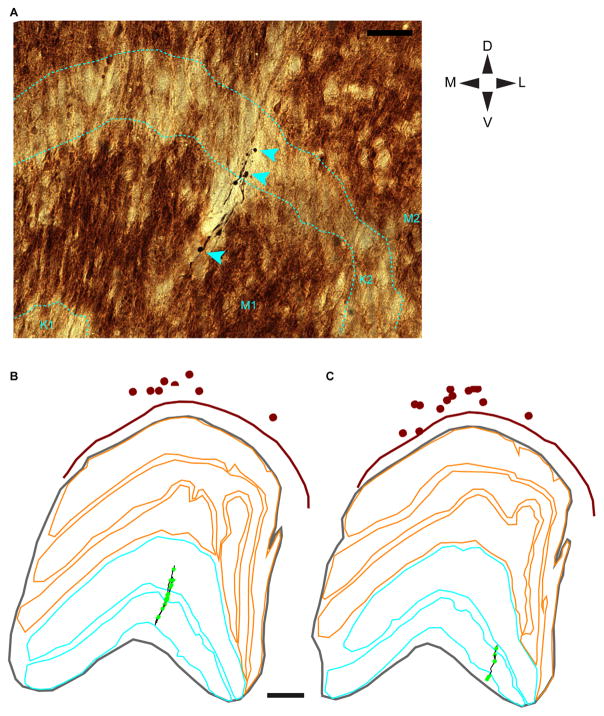
Axons are typically not reliably labeled following rabies virus injection (see Briggs et al., 2016) and we were unable to trace local axons within the TRN, as these were not clearly labeled in either animal. However, we were able to reconstruct parts of three separate putative TRN axons, including terminal boutons, within the dLGN for Monkey 2. For these axonal reconstructions, we traced the contours of each dLGN layer and we placed markers at the centers of cell bodies for all TRN neurons within the same section. With these data, we were able to estimate the ML position of labeled putative TRN axons relative to labeled TRN cell bodies and determine the dLGN laminar location of putative TRN axonal boutons (see Fig. 7).
Figure 7.
Putative TRN axons within the dLGN. A: Photograph of a putative TRN axon in a single dLGN section illustrating boutons (cyan arrowheads) in different dLGN layers. Layers of the dLGN are outlined with dashed cyan lines and labeled at the bottom of the photograph. Orientation is according to the DV/ML compass and the scale bar (top right) = 50 μm. B,C: Reconstructions of two partial putative TRN axons within the dLGN. Axonal arbors in B include those illustrated in A. Axonal arbors are illustrated in black and boutons are represented by bright green dots. TRN contours are illustrated in maroon, dLGN contours in gray, M dLGN layers in cyan, and P dLGN layers in orange. K dLGN layers are the zones in between the M and P layers. TRN cell bodies are represented by maroon dots. Scale bar = 500 μm in B,C.
Analysis of morphological data
We exported data from each neuronal reconstruction to define morphological metrics within four categories. In the first category, to quantify the position of individual neurons in the TRN, three metrics were extracted: 1) ML position relative to the center of the TRN (in microns); 2) DV position within the TRN as a percentage of the DV thickness in the home section; and 3) distance to the dorsal border of the dLGN in proportion to DV position within the TRN.
In the second category, to quantify the size and shape of cell bodies, two metrics were extracted: 1) cell body area; and 2) cell body roundness, a value between 0 and 1 where 0 is completely planar and 1 is perfectly circular. Importantly, in order to avoid a potential confound whereby cell body area and roundness could be biased due to orientation relative to the plane of section, especially when cell bodies were not restricted to the home section, we included cell body area and roundness measurements for cell body reconstructions through serial sections for 29 TRN neurons whose cell bodies spanned more than one section.
In the third category, to quantify the number and length of dendrites, five metrics were extracted: 1) the number of dendritic trees, where each individual tree originates at the cell body; 2) the number of dendritic nodes or branch points; 3) the number of dendritic endings; 4) the total length of dendrite; and 5) the length of each type of dendrite, termed 1st order if they originate at the cell body, 2nd order if they originate at the first node from the cell body, etc. The following metrics were then calculated from the dendritic data: 1) average dendritic distance to nodes per neuron; 2) average dendritic distance to endings per neuron; 3) average length of 1st order dendrite per neuron; 4) average length of 2nd order dendrite per neuron; and 5) average length of 3rd and higher-order dendrite per neuron. All TRN neurons in the dataset had 1st and 2nd order dendrites and most (all but nine) TRN neurons had 3rd order dendrites. Because not all TRN neurons in the dataset had dendrites of higher than 3rd order, we averaged the length of all 3rd and higher-order dendrites.
In the fourth category, to quantify the shape of each dendritic arborization, two metrics were extracted: 1) the angle of each dendrite, measured as the angle of the dendrite exiting the cell body or node (where 0 is always upward in the dorsal direction); and 2) the length of dendrite per 30° phase angle bin (e.g., the amount of dendrite in each wedge of a pie centered at the cell body with 12 equal-sized wedges). The following metrics were then calculated from the dendritic angle data: 1) the average angle of 1st, 2nd, and 3rd and higher-order dendrites in degrees; 2) the average angle of all dendrites for each neuron, calculated by transforming angular data into unit vectors and averaging these to generate an average angle per neuron in radians (Berens, 2009); 3) the circular variance of the total dendritic arborization, calculated as 1 – R where R is the absolute value of the average angle corrected for phase angle bin size (Berens, 2009); and 4) the angular deviation of the total dendritic arborization, calculated as √(2*circular variance), which provides a measurement analogous to the standard deviation of dendritic angles per neuron (Berens, 2009). For each TRN neuron, the dendrite angle metrics provide information about the angle at which dendrites exit the cell body (e.g., average 1st order dendrite), the orientation of the dendritic arborization relative to dorsal (average dendritic angle), and the circularity of the dendritic arborization (circular variance and angular deviation). For example, low angular deviation values indicate consistently rounded dendritic arbors, while high angular deviation values indicate more polarized dendrites.
In order to determine whether the TRN neurons in our dataset were homogeneous or heterogeneous in their morphology, we performed an independent cluster analysis using 10 morphological metrics measured from each of our 160 neuronal reconstructions as the 10 input parameters for each neuron. The clustering algorithm assumes that each parameter used for clustering is independent (Thorndike, 1953; Cauli et al., 2000). Therefore, we removed any parameters that were dependent on one another. The 10 parameters that we used for the cluster analysis were: cell body area, cell body roundness, ML position relative to the center of the TRN, DV position within the TRN, number of dendritic trees, average dendritic distance to nodes, average length of 3rd and higher-order dendrites, average angle of 1st order dendrites, average phase angle of the total dendritic arborization, and angular deviation of the total dendritic arborization (Table 1). In our cluster analysis, each of the 10 parameters had equal weight. We used the “pdist” function in MatLab (MathWorks, Natick, MA; RRID: SCR_001622) to calculate the Euclidean distance between each neuron, defined as a point in a 10-dimensional space. We then used the “linkage” function in MatLab, specifying Ward’s method to define clusters by the inner squared distance between neurons. The “dendrogram” function in MatLab was then used to visualize the linkage tree showing the relationship between clusters. Removing or changing one parameter did not significantly redistribute neurons among clusters.
TABLE 1.
Statistical Comparisons for All 10 Morphological Metrics Across Clusters 1, 2, and 3
| Cell body area | Cell body roundness | Cell body position relative to top TRN | Cell body position relative to ML center | Number of dendritic trees | Average distance to nodes | Average length of 3rd + order dendrites | Average angle of 1st order dendrite | Average angle (radians) | Angular deviation (from circular) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Mean ± SEM Cluster 1 | 235 ± 11 μm2 | 0.6 ± 0.01 | 45 ± 4% DV depth | 1274 ± 60 μm Lateral | 3.8 ± 0.2 | 105 ± 9 μm | 143 ± 11 μm | 181 ± 4 ° | 0.4 ± 0.2 radians | 1.34 ± 0.004 |
| Number neurons = 68 | ||||||||||
| Mean ± SEM Cluster 2 | 231 ± 16 μm2 | 0.5 ± 0.02 | 54 ± 4% DV depth | 358 ± 20 μm Lateral | 3.6 ± 0.2 | 127 ± 14 μm | 172 ± 16 μm | 187 ± 7 ° | 1.1 ± 0.3 radians | 1.32 ± 0.006 |
| Number neurons = 39 | ||||||||||
| Mean ± SEM Cluster 3 | 310 ± 15 μm2 | 0.5 ± 0.02 | 46 ± 3% DV depth | 754 ± 87 μm Medial | 3.8 ± 0.2 | 132 ± 8 μm | 122 ± 12 μm | 162 ± 5 ° | 0.4 ± 0.3 radians | 1.34 ± 0.006 |
| Number neurons = 53 | ||||||||||
| P value | 6.2 × 10−5 | 0.002 | 0.19 | 7.6 × 10−31 | 0.87 | 0.025 | 0.015 | 0.001 | 0.08 | 0.013 |
| Cluster differences | 3 >1 & 2 | 1 >2 & 3 | all different | 3 >1 | 2 >3 | 1 & 2 >3 | (2 >1, p= 0.02) | 1 & 3 >2 |
Independent of the cluster analysis, we examined the relationships between morphological metrics for all TRN neurons by comparing two metrics and measuring linear regression fits to the resultant scatterplots. We estimated the goodness of each fit with R2 values. We next examined the statistical relationships between neurons in the three clusters defined by the cluster analysis using nonparametric multiple-comparisons tests (Kruskal–Wallis one-way analysis of variance). We also used two-sample comparisons tests (paired t-test or Wilcoxon rank-sum test) to determine whether the number of neurons per cluster differed and to assess whether values for a single metric differed significantly across neurons in two clusters. The results of statistical tests for all 10 morphological metrics included in the cluster analysis are listed in Table 1. All other comparisons are described in the Results.
RESULTS
Our aim was to test three alternative hypotheses regarding the organization and connectivity between the visual sector of the TRN and the dLGN in the macaque monkey. Under the first organizational scheme, TRN-to-dLGN connectivity is organized into parallel streams (M, P, K) and originates from heterogeneous TRN neurons that are separated into tiers or organized into clusters in order to preserve the retinotopy of TRN-dLGN connections. Under the second organizational scheme, TRN-to-dLGN connectivity is global (i.e., not parallel stream-specific) and originates from a homogeneous population of TRN neurons. Under the third organizational scheme, TRN-to-dLGN connectivity is not stream-specific, but originates from a heterogeneous population of TRN neurons organized along an alternative dimension in order to accommodate a primate-specific visual specialization such as acuity. To test these alternative organization schemes, we conducted a comprehensive survey of the morphology of dLGN-projecting neurons within the visual sector of the TRN in the macaque monkey. We utilized a virus-mediated retrograde tracing method that produces Golgi-like complete fills of the dendritic arbors of infected neurons and enables observation of hundreds of infected/labeled neurons including rarer neuronal types (Briggs et al., 2016). A modified rabies virus containing the EGFP gene was injected in the dLGN of two monkeys resulting in retrograde infection and labeling of neurons in the visual sector of the TRN in both animals (Fig. 1). The tissue examined in this study was prepared as a part of a prior study (Briggs et al., 2016). As documented in Briggs et al. (2016), virus injections in both animals were entirely restricted to the dLGN and did not leak into neighboring structures such as the TRN, ventral LGN, or pulvinar nucleus (Fig. 1Ai–iii,Bi–iii) because labeled neurons were never observed outside of layer 6 in the visual cortex nor were they observed in the basal forebrain, a structure that projects to the TRN but not the LGN (Lund et al., 1975; Conley and Friederich-Ecsy, 1993; Rockland, 1994; Jones, 2002; Guillery and Harting, 2003; Briggs et al., 2016). Accordingly, all of the TRN neurons observed in this study shared a common trait, namely, axonal projections to the dLGN.
Having confirmed that virus injections were entirely restricted to the dLGN in both animals, we then reconstructed the complete dendritic arbors of 160 neurons in the visual sector of the TRN (99 neurons from Monkey 1 and 61 neurons from Monkey 2). In order to generate a comprehensive dataset, we reconstructed neurons throughout the ML axis of the TRN in both animals (Fig. 1Aiv,Biv). Virus injections were in different regions of the dLGN in the two animals—the injections in Monkey 1 were more rostral in the dLGN and the injections in Monkey 2 were more caudal in the dLGN. As a result of the more rostral injection in Monkey 1, it is possible that we reconstructed a small number of neurons in the ventral LGN, a structure that is adjacent to and contiguous with the TRN surrounding the rostral portion of the dLGN and is known to contain neurons that project to the dLGN (Harrington, 1997). As described below, possible inclusion of a small number of ventral LGN neurons did not bias the clustering results as all clusters contained equal distributions of neurons from both animals.
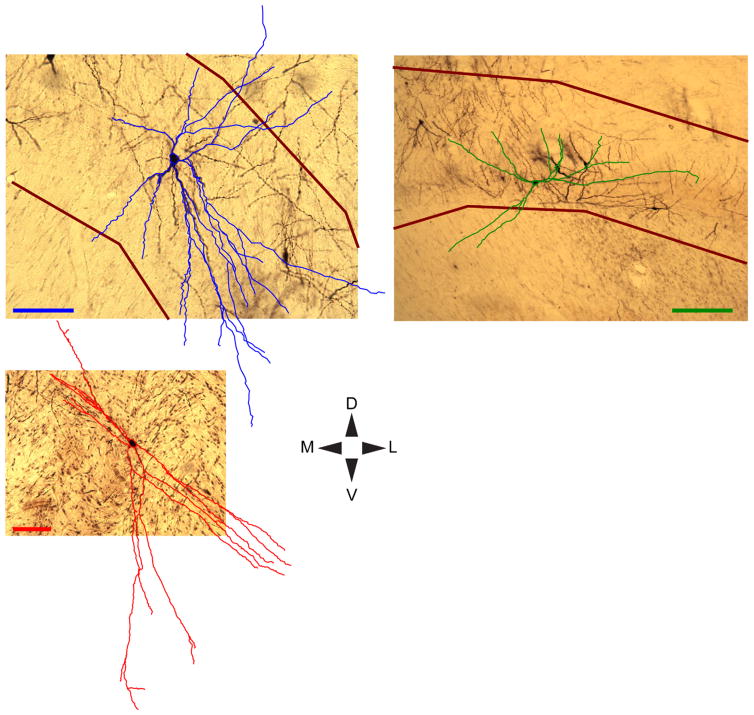
Neurons in the visual sector of the TRN lacked spines (Fig. 2), consistent with prior work that identified TRN neurons as nonspiny and GABAergic (Jones, 1985; Ohara and Lieberman, 1985; Spreafico et al., 1991; Lubke, 1993; Sanches-Vives and McCormick, 1997; Pinault and Deschenes, 1998). TRN neurons had predominantly circular or elongated cell bodies (Fig. 2Aii–vi,Bii–vii) and their dendritic trees all originated at the cell body, i.e., there were no TRN neurons with apical dendrites like those observed in cortical pyramidal neurons (Figs. (2 and 3)). Qualitative observation of the pattern of dendritic arborization suggested that TRN neurons in the more medial (warm colors in Fig. 2Ai,Bi and Fig. 3, bottom left) and lateral (cool colors in Fig. 2Ai,Bi and Fig. 3, top left) regions of the TRN were more polarized or “discoid” (Sherman and Guillery, 2006) while TRN neurons closer to the ML midline (greenish colors in Fig. 2Ai,Bi and Fig. 3, top right) had more circularly arranged dendrites.
Figure 2.
Representative TRN neurons in each animal. Ai: Aggregate contours of the TRN (black) and dLGN (gray) with five representative reconstructed TRN neurons colored warm to cool according to their relative ML position within the TRN of Monkey 1. Reconstructions are flattened for visualization (compressed in the z-axis). Orientations are according to the DV/ML compass and scale bars = 500 μm. Aii–vi: Photographs of the same five TRN neurons with color-matched scale bars = 100 μm. Bi: Aggregate contours of the TRN and dLGN with six representative reconstructed TRN neurons from Monkey 2, conventions as in Ai. Bii–vii: Photographs of the same six TRN neurons with color-matched scale bars = 100 μm.
Figure 3.
Detailed reconstructions of representative TRN neurons. Reconstructed dendrites of representative TRN neurons (blue, green, red) are overlaid onto photographs of each neuron in its home section. Darkly stained cell bodies are visible through cell body contours. Reconstructions are flattened for visualization (compressed in the z-axis corresponding to the rostral-caudal axis). Maroon lines indicate TRN borders. TRN borders are outside the image frame for the lower left neuron, but are ventral and dorsal to the neuron. Orientations are according to the DV/ML compass and scale bars each = 100 μm (note differences in scale). The upper left TRN neuron (blue) is grouped with Cluster 1, the upper right TRN neuron (green) is grouped with Cluster 2, and the lower left TRN neuron (red) is grouped with Cluster 3.
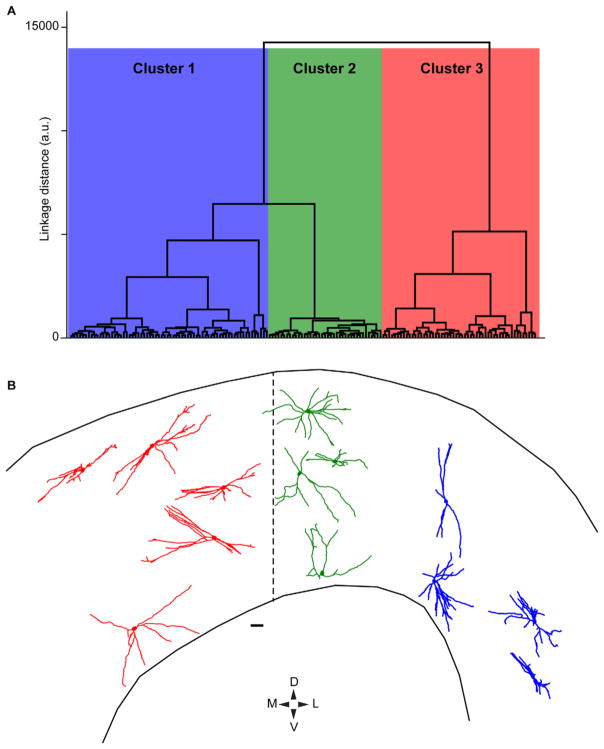
In order to test whether the visual sector TRN neurons are morphologically homogeneous or heterogeneous, we conducted an independent cluster analysis using 10 morphological metrics calculated for each of the 160 TRN neurons in our dataset. The 10 metrics included in the analysis are described in the Materials and Methods section and are listed in Table 1. Importantly, each of the 10 metrics included in the cluster analysis were independent from one another. The cluster analysis revealed three different clusters of TRN neurons from our dataset (Fig. 4A). Cluster 1 contained 68 TRN neurons (48 from Monkey 1, 20 from Monkey 2), Cluster 2 contained 39 TRN neurons (33 from Monkey 1, 6 from Monkey 2), and Cluster 3 contained 53 TRN neurons (18 from Monkey 1, 35 from Monkey 2). The numbers of neurons per cluster from the two animals were not significantly different (P = 0.2, paired t-test) and any unevenness in the distributions of neurons per cluster was likely due to nonuniform sampling across the TRN in each animal because neurons within dense regions of retrograde label could not be accurately reconstructed. Importantly, the fact that TRN neurons from both animals were distributed throughout the three clusters suggests that the same morphological neuron types were reconstructed in both animals in spite of different virus injection locations in more rostral or caudal regions of the dLGN in Monkey 1 and Monkey 2, respectively. This is further indicated by Figure 4B, which illustrates 13 TRN neurons, seven from Monkey 1 and six from Monkey 2, that are now color-coded according to their cluster assignment (Cluster 1 in blue, Cluster 2 in green, Cluster 3 in red). TRN neurons in Cluster 1 are positioned laterally relative to the midline of the TRN in the ML axis; Cluster 2 neurons are positioned close to the midline; and Cluster 3 neurons are positioned medially relative to the midline. Note also that the dendritic arborization patterns of TRN neurons in Cluster 2 appear qualitatively more circular compared to those of Clusters 1 and 3.
Figure 4.
Cluster dendrogram of 160 TRN neurons and representative reconstructions of neurons in each cluster. A: Dendrogram illustrating linkage distances between 160 reconstructed TRN neurons based on 10 independent morphological metrics (see Materials and Methods and Table 1). Three main clusters are illustrated in blue, green, and red. B: Representative reconstructions of 13 TRN neurons color-coded according to their cluster (reconstructions are flattened in the z-axis). ML positions within the TRN are relative to the dashed line representing the center of the visual sector of the TRN, orientation is according to the DV/ML compass below and scale bar = 100 μm. Note the DV thickness of the TRN is not to scale in order to enhance display clarity.
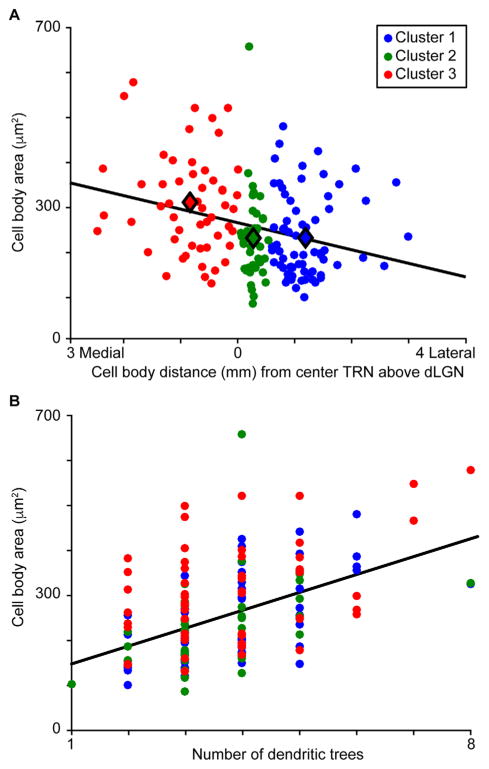
Independent of the cluster analysis, we compared morphological metrics across our dataset of reconstructed TRN neurons to determine whether there were any general relationships between morphological metrics among TRN neurons. We observed three general relationships across all 160 TRN neurons. First, there was a relationship between cell body area and ML position within the TRN such that more medial TRN neurons had larger cell bodies, while more lateral TRN neurons had smaller cell bodies (Fig. 5A). Furthermore, there were significant differences across clusters in both cell body area and ML position (Table 1 and Figs. 5A, 6A). Second, there was a positive relationship between cell body area and the number of dendritic trees (Fig. 5B), even though there were no significant differences in the number of dendritic trees across clusters (Table 1). Third, there was a relationship between cell body roundness and ML position (R2 = 0.1, data not shown). Additionally, TRN neurons in Cluster 1, positioned laterally, had significantly more rounded cell bodies compared to neurons in Clusters 2 and 3 (Table 1, Fig. 6B). The cell body roundness measurement could be confounded by variations in cell body orientation relative to the plane of section. The majority of TRN neurons in the dataset (131 out of 160) had cell bodies within a single 50-μm section and contours were reconstructed through their largest girth, taking orientation into account. For 29 TRN neurons, cell bodies spanned two adjacent sections, so serial cell body reconstructions were made for these neurons (see Materials and Methods). Only three Cluster 2 neurons had cell bodies spanning adjacent sections, likely because neurons in this cluster had the smallest cell bodies (Table 1, Fig. 6A). Consistent with the results of the overall cluster analysis, Cluster 3 neurons with cell bodies in adjacent sections had significantly larger cell bodies compared to Cluster 1 neurons with cell bodies in adjacent sections (P = 0.02; Cluster 1 cell body area = 194 ± 18 μm2, n = 13; Cluster 3 cell body area = 271 ± 26 μm2, n = 13). Similarly, Cluster 1 neurons with cell bodies in adjacent sections had significantly more rounded cell bodies compared to Cluster 3 neurons with cell bodies in adjacent sections (P = 0.008; Cluster 1 cell body roundness = 0.56 ± 0.03; Cluster 3 cell body roundness = 0.46 ± 0.02). Because the results for neurons with cell bodies in adjacent sections match those for the whole TRN dataset, cell body orientation relative to the plane of section was not a confound in the overall cluster analysis.
Figure 5.
Position and size of TRN neurons. A: Scatterplot illustrating the negative relationship between cell body area and ML position of the cell body within the TRN such that more medial TRN neurons had larger cell bodies (R2 = 0.08). Data from all 160 TRN neurons are illustrated. Cluster assignment illustrated by color according to the legend (for number of neurons per cluster, refer to Table 1). Black line illustrates linear regression fit to the data. Filled black diamonds illustrate average cell body area and ML position for each cluster. B: Scatterplot illustrating the positive relationship between number of dendritic trees and cell body area for all 160 TRN neurons regardless of cluster assignment (R2 = 0.22). Conventions as in A.
Figure 6.
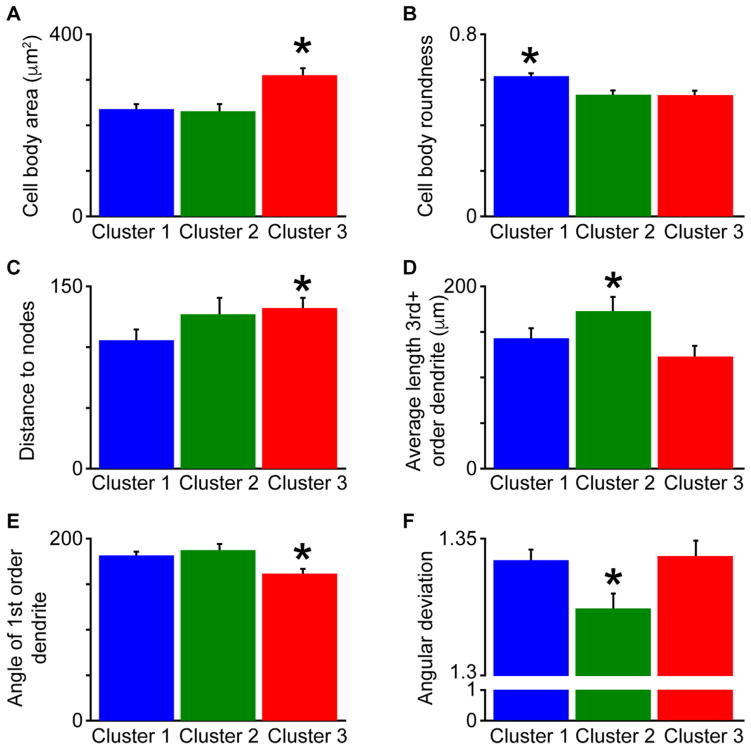
Cluster-specific differences in morphological metrics for all 160 TRN neurons. A: Cluster 3 neurons had significantly larger cell body areas compared to Cluster 1 and Cluster 2 neurons, indicated by the asterisk (*P < 6.2 × 10−5). For numbers of neurons per cluster, refer to Table 1. B: Cluster 1 neurons had significantly more rounded cell bodies compared to Cluster 2 and Cluster 3 neurons (*P < 0.002). C: Cluster 3 neurons had significantly longer dendritic distances to nodes compared to Cluster 1 neurons (*P < 0.025). D: Cluster 2 neurons had significantly more 3rd and higher-order dendrites compared to Cluster 3 neurons (*P < 0.015). E: Cluster 3 neurons had significantly lower 1st order dendrite angles compared to Cluster 1 and Cluster 2 neurons (*P < 0.001). F: Cluster 2 neurons were significantly more circular as defined by lower angular deviation values compared to Cluster 1 and Cluster 3 neurons (*P < 0.013).
For 8 out of the 10 morphological metrics included in the cluster analysis, there were significant differences across the three clusters (Table 1). Only two metrics were not significantly different across clusters and these included the dorsal-ventral (DV) position within the TRN and the number of dendritic trees. It is possible that the visual sector of the TRN is too thin, on average 538 ± 25 μm thick in our dataset, to observe significant differences in DV position within such a small range. However, the lack of differences in DV position across visual sector TRN neurons suggests a lack of morphological segregation by tier (see Discussion). The fact that TRN neurons across all three clusters had the same number of dendritic trees suggests that differences across clusters were related to the orientation and complexity of dendritic arborization patterns rather than the total number of dendritic trees.
As stated above, TRN neurons in each cluster differed in ML position, cell body area, and cell body roundness and these morphological features were correlated across the visual sector TRN population (Figs. 5A,B, 6A,B, Table 1). There were also cluster-specific differences in the average dendritic distances to nodes and the amount of 3rd and higher-order dendrites (Fig. 6C,D, Table 1). Specifically, nodes of Cluster 3 neurons were further from the cell body compared to Cluster 1 neurons, while Cluster 2 neurons had significantly more higher-order dendritic branches compared to Cluster 3 neurons. Cluster 3 neurons also had more polarized dendritic arborization patterns as the angles of 1st order dendrites, those connected to the cell body, were significantly less than those for Cluster 1 and 2 neurons (Fig. 6E, Table 1) and the dendritic arborization patterns of Cluster 3 neurons were significantly less circular compared to those of Cluster 2 neurons (Fig. 6F, Table 1). The dendritic arborization patterns of Cluster 1 neurons were also less circular compared to those of Cluster 2 neurons (Fig. 6F, Table 1). Finally, the mean phase angles of dendritic trees for Cluster 2 neurons were significantly greater than those for Cluster 1 neurons and also greater than those for Cluster 3 neurons (Table 1), suggesting that the dendritic arbors of neurons in Clusters 1 and 3 were oriented in parallel with the TRN and those of Cluster 2 neurons were not. Together these results support the qualitative observation (Fig. 4) that Cluster 2 neurons had dendritic trees that were spaced more evenly around the cell body, forming a more circular dendritic arborization pattern compared to neurons in Clusters 1 and 3, making them less likely to be orientated in parallel with the DV boundaries of the TRN.
To summarize the dendritic data, Cluster 1 neurons were located laterally within the TRN, had small and rounder cell bodies with nodes closer to cell bodies, and more polarized dendritic trees that were orientated in parallel with the DV TRN boundaries. Cluster 2 neurons were located centrally within the TRN, had the smallest cell bodies and dendritic trees with more higher-order branches that were more evenly spaced around the cell body, giving these neurons a circular dendritic arborization pattern such that they were not oriented in any specific direction relative to the TRN boundaries. Cluster 3 neurons were located medially within the TRN, had large, less rounded cell bodies with nodes further from cell bodies, less dendritic branching, and more polarized dendritic trees that were oriented in parallel with the DV TRN boundaries.
Together, the dendritic data suggest that TRN neurons are morphologically heterogeneous and neuronal types are separated along the ML axis, not into tiers. These findings are consistent with the third organizational scheme whereby TRN-to-dLGN connectivity is not parallel stream-specific, but is organized along an alternative dimension. To provide additional confirmation of this organizational scheme, we examined the morphology of three partial putative TRN axonal arbors within separate sections of the dLGN. We presume the axons we observed in the dLGN originated from the TRN based on striking homology to TRN axons described previously (Uhlrich et al., 2003) and a lack of homology to retinogeniculate or corticogeniculate axons, which are mostly restricted to single eye-specific layers within the dLGN, or pretectal axons, which are not restricted to columns within the dLGN (Uhlrich and Manning, 1995). However, since we cannot be certain about the origin of the observed axons, we refer to them as putative TRN axons. Putative TRN axons were oriented along the DV axis and appeared restricted to a single retinotopic column within the dLGN (Fig. 7A–C), suggesting that TRN-to-dLGN connections are precisely retinotopic. Interestingly, terminal boutons from the same axons were located in multiple dLGN layers (Fig. 7A–C). The first axon, illustrated in the photograph in Figure 7A and the partial reconstruction in Figure 7B, had 17 total boutons that were evenly distributed across the M1, K2, and M2 layers (four boutons in M1; six boutons in K2; seven boutons in M2). Similarly, the second axon, illustrated in the partial reconstruction in Figure 7C, had three boutons in K1, three boutons in M1, and one bouton in K2; and the third axon had 28 boutons in K1, five boutons in M1, and two boutons in K2. All three axonal arbors had boutons in a mixture of M and K layers within a retinotopic column of the dLGN. Additionally, all three axonal arbors had boutons in a combination of ipsilateral and contralateral eye layers within the dLGN. Together these findings further support the hypothesis that TRN-to-dLGN connections convey parallel stream-mixed and ipsilateral/contralateral eye-mixed signals to the dLGN in a precisely retinotopically organized manner (Uhlrich et al., 2003).
DISCUSSION
Neurons in the TRN are uniquely positioned to influence the feedforward flow of sensory signals traveling from the thalamus to the cortex (Jones, 1985; Pinault, 2004). The TRN could serve as an integration hub for bottom-up and top-down inputs from structures throughout the brain (Guillery and Harting, 2003). Accordingly, theoretical and functional studies suggest that the TRN controls the flow of sensory information from thalamus to cortex and that global cognitive signals, like attention and arousal, modulate the activity of TRN neurons (Crick, 1984; Fuentealba and Steriade, 2005; McAlonan et al., 2008; Halassa et al., 2014; Kimura, 2014; Wimmer et al., 2015). Neurons throughout the TRN in a variety of mammals display broadly similar morphological and neurophysiological characteristics (Scheibel and Scheibel, 1972; Spreafico et al., 1988; Spreafico et al., 1991; Lubke, 1993; Ohara and Havton, 1996; Uhlrich et al., 2003; Fitzgibbon et al., 2007), perhaps supporting a more generalized function of the TRN, such as gating the feedforward flow of sensory signals. However, the organization of inputs to the TRN into separate sectors and/or tiers and the topographic organization of many TRN sectors (Montero et al., 1977; Jones, 1985; Crabtree and Killackey, 1989; Conley and Diamond, 1990; Uhlrich et al., 2003; Sherman and Guillery, 2006; Fitzgibbon et al., 2007; Baldauf, 2010; Lam and Sherman, 2011) also suggest that neurons in different compartments within the TRN each play separate roles in regulating thalamic activity. A generalized functional role of the TRN—e.g., in gating arousal/attention—does not preclude additional, more specialized functions for specific TRN sectors. For example, in highly organized sensory systems such as the early visual system of the primate, the visual sector of the TRN could serve as a general gate for arousal state while also maintaining retinotopically organized connections with the dLGN to preserve specializations for acuity and/or color processing (along the lines of the third organizational scheme proposed above). Alternatively, the visual sector of the TRN in primates could be organized into parallel M, P, and K streams, in line with the first organizational scheme proposed above. Along these lines, a prior study of retrogradely labeled neurons following injections of two distinct tracers into different layers and different rostral/caudal regions of macaque dLGN demonstrated nonoverlapping populations of labeled TRN neurons (Bickford et al., 2000), supporting the notion that macaque TRN is retinotopically organized and perhaps lending support to the hypothesis that TRN neurons are organized into parallel streams.
We aimed to specifically test whether TRN neurons in the visual sector of the TRN in the macaque monkey are organized to provide M, P, K parallel stream-specific connections to the dLGN or organized along an alternative dimension to support transmission of global signals. We performed a comprehensive morphological characterization of neurons within the visual sector of the TRN that all shared the same postsynaptic target, the dLGN. Independent clustering of 10 morphological metrics measured from 160 TRN neurons yielded three clusters of visual sector TRN neurons in the macaque (Fig. 4). Neurons in the three clusters differed significantly in 8 of the 10 morphological metrics included in the analysis (Table 1). Neurons in each cluster displayed unique characteristics: Cluster 1 neurons, located laterally, had small, rounded cell bodies and polarized dendritic arbors with nodes close to the cell body; Cluster 2 neurons, located near the midline, had the smallest cell bodies and more complex dendritic arbors that were circularly organized; Cluster 3 neurons, located medially, had large cell bodies and polarized dendritic arbors that were less complex, with nodes further from the cell body. Strikingly, neurons in the three clusters were organized into different compartments in the ML axis of the visual sector of the TRN, but there was no evidence for segregation of morphologically distinct TRN neurons into tiers (Fig. 1Aiv,Biv). Additionally, we observed broader trends across our dataset including a relationship between cell body size and ML position within the TRN and also between cell body size and the number of dendritic trees per neuron (Fig. 5). Finally, putative TRN axons within the dLGN were strictly oriented along the DV axis and had terminals in multiple dLGN layers (Fig. 7). The axonal termination patterns we observed are consistent with those of TRN axons in Galagos (Uhlrich et al., 2003) and suggest that individual TRN axons target a mixture of M, P, and K stream neurons in both ipsilateral and contralateral eye layers of the dLGN.
Together, the dendritic and axonal morphology of visual sector TRN neurons in the macaque, combined with morphological evidence from studies of TRN neurons in other species, support a connectivity scheme whereby the TRN transmits a global signal (e.g., mixed across the parallel visual streams and mixed across eye-specific layers) to the dLGN that is precisely retinotopically organized. It is well established that the visual sector of the TRN has a retinotopic map such that TRN neurons in adjacent stripes or columns, running perpendicular to the TRN borders (along the DV axis), respond to neighboring regions in visual space (Sherman and Guillery, 2006; Fitzgibbon et al., 2007; Vaingankar et al., 2012). The retinotopic map of the dLGN is similarly organized into stripes or columns that run perpendicular to the dLGN layers along the DV axis (Sherman and Guillery, 2006). If separate clusters of TRN neurons projected to M, P, and K dLGN neurons in a manner that preserved retinotopy in the two structures, neurons in each cluster would either be organized into adjacent tiers or clustered within each TRN stripe. We did not observe this organization for neurons in the three clusters; instead, neurons in the clusters were organized into compartments spanning the ML axis of the TRN. Interestingly, the lack of stream-specific organization of TRN neurons in this dataset contrasts with the stream-specific organization of corticogeniculate neurons in the same animals, labeled following the same virus injections in the dLGN and the same data analysis methods (Briggs et al., 2016).
Furthermore, the heterogeneity of TRN dendritic morphology across the ML axis and lack of segregation of morphologically distinct TRN neurons into tiers suggest that subpopulations of TRN neurons could be specialized to process signals from different visual field representations. One of the unique adaptations among primates is acuity vision and the primate visual system evolved a number of specializations to accommodate high acuity, including an expansion of the number of neurons at the fovea in the retina and at foveal and parafoveal representations in the dLGN and visual cortex. Neurons with foveal and parafoveal receptive fields are located in the middle of the dLGN with the most foveal neurons at the caudal pole, while lower and upper peripheral visual fields are represented by neurons in the more medial and lateral portions of the dLGN (Malpeli and Baker, 1975). Neurons clustered near the middle of the TRN (Cluster 2 neurons) have the smallest cell bodies, the most circularly oriented arbors, and the most complex dendritic branches—possible morphological specializations for integrating a large number of inputs from dLGN neurons with overlapping foveal receptive fields. Given the precise retinotopic organization of connections between the dLGN and TRN, it is possible that this subpopulation of neurons in the middle of the TRN connect to dLGN neurons with foveal and parafoveal receptive fields. Accordingly, the organization of TRN neurons across the ML axis combined with the retinotopy of TRN axons could reflect morphological specializations for the transmission of information at foveal versus peripheral visual field representations. From an ethological perspective, a visual field-specific organization for a global arousal signal makes sense. In primates, where foveal and peripheral visual signals are processed quite differently, it seems appropriate that arousal or attention signals originating at the center of gaze may also be processed differently from arousal/attention signals originating in the periphery.
The morphological differences between TRN neurons in the three clusters are subtle. In this sense, visual sector TRN neurons in the macaque monkey are similar to TRN neurons in a variety of mammals in that they have generally similar morphologies (Lubke, 1993; Ohara and Havton, 1996; Fitzgibbon et al., 2007) with some subtle variations across TRN regions (Scheibel and Scheibel, 1972; Spreafico et al., 1991; Pinault, 2004). Some have proposed that morphological differences observed across TRN neurons reflect anatomical constraints due to the shape of the TRN (Scheibel and Scheibel, 1972; Spreafico et al., 1991) rather than differences in inputs, outputs, or neuronal computations. Along these lines, the morphological differences we observe in the macaque TRN could reflect a continuum of variation across neurons. Importantly, neither a continuum of morphological types nor distinct neuronal subpopulations organized along the ML axis of the TRN are consistent with stream-specific organization of TRN neurons into tiers or clusters. Thus, whether morphological differences across the visual sector of the TRN reflect a continuum or discrete subpopulations, the functional impact of the TRN on the dLGN remains the same. In both cases, the retinotopic organization of connections, paired with the lack of parallel stream specificity, suggest that TRN neurons transmit a global (arousal/attention) signal to the dLGN that is retinotopically organized.
Our findings lead to a number of intriguing predictions about the functional organization of the TRN and its influence on visual information processing through the dLGN. First, our data suggest that TRN inputs to the dLGN are global, and not specific to the M, P, and K parallel processing streams. Since dLGN neurons maintain their M, P, or K stream identity and monocular responses regardless of arousal or attentive state, the influence of the TRN on the dLGN must be subtle and/or involve inhibitory modulation that does not alter the receptive field properties of dLGN neurons. Optogenetic approaches, such as those utilized in rodents (e.g., Wimmer et al., 2015), may help resolve the nature of TRN influence on dLGN visual responses.
A second possible prediction of our findings is that the visual sector of TRN in the macaque may not have tiers. Rather than tiers, the TRN in the macaque may be organized into separate visual sectors: one connected to the dLGN and another connected to the visual portion of the pulvinar. In carnivores and some primates (Galagos, marmosets), the visual sector of the TRN has two tiers that receive input from the visual portion of the pulvinar and the dLGN (Conley and Diamond, 1990; Sherman and Guillery, 2006; Fitzgibbon et al., 2007; Baldauf, 2010). In these species, the visual portion of the pulvinar is located within the same coronal plane and in close proximity to the dLGN. Thus, a single “visual sector” of the TRN in carnivores and some primates may contain tiers receiving inputs from the pulvinar and dLGN. In macaques, the visual portion of the pulvinar (the ventrolateral and inferior pulvinar) is located caudal and medial to the dLGN (Kaas and Lyon, 2007). The visual pulvinar in macaques is connected to a portion of the TRN that is caudal, dorsal, and medial to the dLGN-projecting sector of the TRN (Lyon and Rabideau, 2012) (specifically, compare their fig. 5 to our Fig. 1Aiv,Biv to view pulvinar-projecting TRN neurons that are caudal, dorsal, and medial to the TRN neurons in this study). It is therefore possible that in the macaque there is a “visual pulvinar” sector of the TRN that is caudal, dorsal, and medial to a separate visual dLGN-projecting sector of the TRN, removing the need for tiers within each of these sectors.
Broadly speaking, our findings support the notion that there is diversity in neuronal structure and function within a single sector of the TRN and among neurons projecting to a common postsynaptic target. While the TRN remains a complex and poorly understood brain structure, our findings suggest that it may also be highly flexible, such that neurons in a given sector of the TRN can evolve and/or adapt to accommodate species-specific specializations such as acuity vision. Methodologies like virus-mediated circuit tracing and optogenetics that enable selective labeling and targeted manipulation of neurons will undoubtedly increase our understanding of the organization and function of this important part of the brain.
Acknowledgments
We thank Drs. Ed Callaway and Marty Usrey for allowing us to use the tissue prepared as a part of a prior study.
Footnotes
CONFLICT OF INTEREST
None of the authors on this study have any real or potential conflict of interest that could inappropriately influence this study.
ROLE OF AUTHORS
All of the authors had full access to all of the data in this study and take responsibility for the integrity of the data and the accuracy of the data analyses. F.B., E.M.B., E.A.F., and S.L. conceived of and designed the study. E.M.B., E.A.F., and S.L. completed the neuronal reconstructions. E.M.B. and F.B. analyzed the data and wrote the article.
LITERATURE CITED
- Ahrens S, Jaramillo S, Yu K, Ghosh S, Hwang G-R, Paik R, Lai C, He M, Huang ZJ, Li B. ErbB4 regulation of a thalamic reticular nucleus circuit for sensory selection. Nat Neurosci. 2015;18:104–111. doi: 10.1038/nn.3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf ZB. Dual chemoarchitectonic lamination of the visual sector of the thalamic reticular nucleus. Neuroscience. 2010;165:801–818. doi: 10.1016/j.neuroscience.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berens P. CircStat: A Matlab toolbox for circular statistics. J Stat Softw. 2009;31:1–21. [Google Scholar]
- Bickford ME, Ramcharan E, Godwin DW, Erisir A, Gnadt J, Sherman SM. Neurotransmitters contained in the subcortical extraretinal inputs to the monkey lateral geniculate nucleus. J Comp Neurol. 2000;424:701–717. doi: 10.1002/1096-9861(20000904)424:4<701::aid-cne11>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Briggs F, Kiley CW, Callaway EM, Usrey WM. Morphological substrates for parallel streams of corticogeniculate feedback originating in both V1 and V2 of the macaque monkey. Neuron. 2016;90:388–399. doi: 10.1016/j.neuron.2016.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauli B, Porter JT, Tsuzuki K, Lambolez B, Rossier J, Quenet B, Audinat E. Classification of fusiform neocortical interneurons based on unsupervised clustering. Pros Natl Acac Sci U S A. 2000;97:6144–6149. doi: 10.1073/pnas.97.11.6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley M, Diamond IT. Organization of the visual sector of the thalamic reticular nucleus in galago. Eur J Neurosci. 1990;2:211–226. doi: 10.1111/j.1460-9568.1990.tb00414.x. [DOI] [PubMed] [Google Scholar]
- Conley M, Friederich-Ecsy B. Functional organization of the ventral lateral geniculate complex of the tree shrew (Tupaia belangeri): II. Connections with the cortex, thalamus, and brainstem. J Comp Neurol. 1993;328:21–42. doi: 10.1002/cne.903280103. [DOI] [PubMed] [Google Scholar]
- Crabtree JW, Killackey HP. The topographical organization of the axis of projection within the visual sector of the rabbit’s thalamic reticular nucleus. Eur J Neurosci. 1989;1:94–109. doi: 10.1111/j.1460-9568.1989.tb00777.x. [DOI] [PubMed] [Google Scholar]
- Crick F. Function of the thalamic reticular complex: the searchlight hypothesis. Neurobiology. 1984;81:4586–4590. doi: 10.1073/pnas.81.14.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleuze C, Huguenard JR. Distinct electrical and chemical connectivity maps in the thalamic reticular nucleus: potential roles in synchronization and sensation. J Neurosci. 2006;26:8633–8645. doi: 10.1523/JNEUROSCI.2333-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgibbon T, Szmajda BA, Martin PR. First order connections of the visual sector of the thalamic reticular nucleus in marmoset monkeys (Callithrix jacchus) Vis Neurosc. 2007;24:857–874. doi: 10.1017/S0952523807070770. [DOI] [PubMed] [Google Scholar]
- Fuentealba P, Steriade M. The reticular nucleus revisited: intrinsic and network properties of a thalamic pacemaker. Prog Neurobiol. 2005;75:125–141. doi: 10.1016/j.pneurobio.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Guillery RW, Harting JK. Structure and connections of the thalamic reticular nucleus: advancing views over half a century. J Comp Neurol. 2003;463:360–371. doi: 10.1002/cne.10738. [DOI] [PubMed] [Google Scholar]
- Halassa MM, Chen Z, Wimmer RD, Brunetti PM, Zhao S, Zikopoulos B, Wang F, Brown EN, Wilson MA. State-dependent architecture of thalamic reticular subnetworks. Cell. 2014;158:808–821. doi: 10.1016/j.cell.2014.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington ME. The ventral lateral geniculate nucleus and the intergeniculate leaflet: interrelated structures in the visual and circadian systems. Neurosci Behav Rev. 1997;21:705–727. doi: 10.1016/s0149-7634(96)00019-x. [DOI] [PubMed] [Google Scholar]
- Jones EG. The thalamus. New YorkPlenum Press; 1985. [Google Scholar]
- Jones EG. Thalamic organization and function after Cajal. Prog Brain Res. 2002;136:333–357. doi: 10.1016/s0079-6123(02)36029-1. [DOI] [PubMed] [Google Scholar]
- Kaas JH, Lyon DC. Pulvinar contributions to the dorsal and ventral streams of visual processing in primates. Brain Res Rev. 2007;55:285–296. doi: 10.1016/j.brainresrev.2007.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan E. The M, P, and K pathways of the primate visual system. In: Chalupa L, Werner J, editors. The visual neurosciences. Cambridge, MA: MIT Press; 2004. pp. 481–493. [Google Scholar]
- Kimura A. Diverse subthreshold cross-modal sensory interactions in the thalamic reticular nucleus: implications for new pathways of cross-modal attentional gating function. Eur J Neurosci. 2014;39:1405–1418. doi: 10.1111/ejn.12545. [DOI] [PubMed] [Google Scholar]
- Kimura A, Yokoi I, Imbe H, Donishi T, Kaneoke Y. Distinctions in burst spiking between thalamic reticular nucleus cells projecting to the dorasl lateral geniculate and lateral posterior nuclei in the anesthetized rat. Neuroscience. 2012;226:208–226. doi: 10.1016/j.neuroscience.2012.09.016. [DOI] [PubMed] [Google Scholar]
- Lam Y-W, Sherman SM. Functional organization of the thalamic input to the thalamic reticular nucleus. J Neurosci. 2011;31:6791–6799. doi: 10.1523/JNEUROSCI.3073-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam Y-W, Nelson CS, Sherman SM. Mapping of the functional interconnections between thalamic reticula neurons using photostimulation. J Neurophysiol. 2006;96:2593–2600. doi: 10.1152/jn.00555.2006. [DOI] [PubMed] [Google Scholar]
- Landisman CE, Long MA, Beierlein M, Deans MR, Paul DL, Connors BW. Electrical synapses in the thalamic reticular nucleus. J Neurosci. 2002;22:1002–1009. doi: 10.1523/JNEUROSCI.22-03-01002.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-H, Govindaiah G, Cox CL. Heterogeneity of firing properties among rat thalamic reticular nucleus neurons. J Physiol. 2007;582:195–208. doi: 10.1113/jphysiol.2007.134254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-C, Patrick SL, Richardson KA, Connors BW. Two functionally distinct networks of gap junction-coupled inhibitory neurons in the thalamic reticular nucleus. J Neurosci. 2014;34:13170–13182. doi: 10.1523/JNEUROSCI.0562-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubke J. Morphology of neurons in the thalamic reticular nucleus (TRN) of mammals as revealed by intracellular injections into fixed brain slices. J Comp Neurol. 1993;329:458–471. doi: 10.1002/cne.903290404. [DOI] [PubMed] [Google Scholar]
- Lund JS, Lund RD, Hendrickson AE, Bunt AH, Fuchs AF. The origin of efferent pathways from the primary visual cortex, area 17, of the macaque monkey as shown by retrograde transport of horseradish peroxidase. J Comp Neurol. 1975;164:287, 303. doi: 10.1002/cne.901640303. [DOI] [PubMed] [Google Scholar]
- Lyon DC, Rabideau C. Lack of robust LGN label following transneuronal rabies virus injections into macaque area V4. J Comp Neurol. 2012;520:2500–2511. doi: 10.1002/cne.23050. [DOI] [PubMed] [Google Scholar]
- Malpeli JG, Baker FH. The representation of the visual field in the lateral geniculate nucleus of macaca mulatta. J Comp Neurol. 1975;161:569–594. doi: 10.1002/cne.901610407. [DOI] [PubMed] [Google Scholar]
- McAlonan K, Cavanaugh JR, Wurtz RH. Attentional modulation of thalamic reticular neurons. J Neurosci. 2006;26:4444–4450. doi: 10.1523/JNEUROSCI.5602-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlonan K, Cavanaugh JR, Wurtz RH. Guarding the gateway to cortex with attention in visual thalamus. Nature. 2008;456:391–394. doi: 10.1038/nature07382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero VM, Guillery RW, Woolsey CN. Retinotopic organization within the thalamic reticular nucleus demonstrated by a double label autoradiographic technique. Brain Res. 1977;138:407–421. doi: 10.1016/0006-8993(77)90681-3. [DOI] [PubMed] [Google Scholar]
- Ohara PT, Havton LA. Dendritic arbors of neurons from different regions of the rat thalamic reticular nucleus share a similar orientation. Brain Res. 1996;731:236–240. doi: 10.1016/0006-8993(96)00706-8. [DOI] [PubMed] [Google Scholar]
- Ohara PT, Lieberman AR. The thalamic reticular nucleus of the adult rat: experimental anatomical studies. J Neurocytol. 1985;14:365–411. doi: 10.1007/BF01217752. [DOI] [PubMed] [Google Scholar]
- Osakada F, Mori T, Cetin A, Marshel JH, Virgen B, Callaway EM. New rabies virus variants for monitoring and manipulating activity and gene expression in defined neural circuits. Neuron. 2011;71:617–631. doi: 10.1016/j.neuron.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinault D. The thalamic reticular nucleus: structure, function and concept. Brain Res Rev. 2004;46:1–31. doi: 10.1016/j.brainresrev.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Pinault D, Deschenes M. Projection and innervation patterns of individual thalamic reticular axons in the thalamus of the adult rat: a three-dimensional, graphic, and morphometric analysis. J Comp Neurol. 1998;391:180–203. doi: 10.1002/(sici)1096-9861(19980209)391:2<180::aid-cne3>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Rockland KS. Further evidence for two types of cortico-pulvinar neurons. NeuroReports. 1994;5:1865, 1868. doi: 10.1097/00001756-199410000-00006. [DOI] [PubMed] [Google Scholar]
- Sanches-Vives MV, McCormick DA. Functional properties of perigeniculate inhibition of dorsal lateral geniculate nucleus thalamocortical neurons in vitro. J Neurosci. 1997;17:8880–8893. doi: 10.1523/JNEUROSCI.17-22-08880.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibel ME, Scheibel AB. Specialized organizational patterns within the nucleus reticularis thalami of the cat. Exp Neurol. 1972;34:316–322. doi: 10.1016/0014-4886(72)90177-x. [DOI] [PubMed] [Google Scholar]
- Sherman SM, Guillery RW. Exploring the thalamus and its role in cortical function. 2. Boston: MIT Press; 2006. [Google Scholar]
- Spreafico R, De Curtis M, Frassoni C, Avanzini G. Electrophysiological characteristics of morphologically identified reticular thalamic neurons from rat slices. Neuroscience. 1988;27:629–638. doi: 10.1016/0306-4522(88)90294-1. [DOI] [PubMed] [Google Scholar]
- Spreafico R, Battaglia G, Frassoni C. The reticular thalamic nucleus (RTN) of the rat: cytoarchitectural, Golgi, immunocytochemical, and horseradish peroxidase study. J Comp Neurol. 1991;304:478–490. doi: 10.1002/cne.903040311. [DOI] [PubMed] [Google Scholar]
- Thorndike RL. Who belongs in the family? Psychometrika. 1953;18:267–276. [Google Scholar]
- Uhlrich DJ, Manning KA. Projection of individual axons from the pretectum to the dorsal lateral geniculate complex in the cat. J Comp Neurol. 1995;363:147–159. doi: 10.1002/cne.903630112. [DOI] [PubMed] [Google Scholar]
- Uhlrich DJ, Manning KA. Laminar and cellular targets of individual thalamic reticular nucleus axons in the lateral geniculate nucleus in the prosimian primate galago. J Comp Neurol. 2003;458:128–143. doi: 10.1002/cne.10568. [DOI] [PubMed] [Google Scholar]
- Vaingankar V, Soto-Sanchez C, Wang X, Sommer FT, Hirsch JA. Neurons in the thalamic reticular nucleus are selective for diverse and complex visual features. Front Integr Neurosci. 2012;6:1–17. doi: 10.3389/fnint.2012.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells MF, Wimmer RD, Schmitt LI, Feng G, Halassa MM. Thalamic reticular impairment underlies attention deficit in Ptchd1(Y/−) mice. Nature. 2016;532:58–63. doi: 10.1038/nature17427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Finke S, Conzelmann KK, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nature Methods. 2007;4:47–49. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson AM, Ohara PT, Ralston DD, Milroy AM, Ralston HJ. Analysis of gamma-aminobutyric acidergic synaptic contacts in the thalamic reticular nucleus of the monkey. J Comp Neurol. 1994;349:182–192. doi: 10.1002/cne.903490203. [DOI] [PubMed] [Google Scholar]
- Wimmer RD, Schmitt LI, Davidson TJ, Nakajima M, Deisseroth K, Halassa MM. Thalamic control of sensory selection in divided attention. Nature. 2015 doi: 10.1038/nature15398. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zikopoulos B, Barbas H. Prefrontal projections to the thalamic reticular nucleus form a unique circuit for attentional mechanisms. J Neurosci. 2006;26:7348–7361. doi: 10.1523/JNEUROSCI.5511-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]