Abstract
The rise of advanced technologies for characterizing human populations at the molecular level, from sequence to function, is shifting disease prevention paradigms toward personalized strategies. Because minimization of adverse outcomes is a key driver for treatment decisions for diseased populations, developing personalized therapy strategies represent an important dimension of both precision medicine and personalized prevention. In this commentary, we highlight recently developed enabling technologies in the field of DNA damage, DNA repair, and mutagenesis. We propose that omics approaches and functional assays can be integrated into population studies that fuse basic, translational and clinical research with commercial expertise in order to accelerate personalized prevention and treatment of cancer and other diseases linked to aberrant responses to DNA damage. This collaborative approach is generally applicable to efforts to develop data-driven, individualized prevention and treatment strategies for other diseases. We also recommend strategies for maximizing the use of biological samples for epidemiological studies, and for applying emerging technologies to clinical applications.
Keywords: DNA damage, Comet, H2AX, Host cell reactivation, DNA repair, DNA damage response, Precision medicine
1. Introduction
The National Institute of Environmental Health Sciences (NIEHS) sponsored a workshop in June 2015 entitled “Workshop on New Approaches for Detecting DNA Damage and Mutation in Population Studies”. This commentary emerged from a consensus-building discussion that followed technology-focused presentations by attendees, including several of the authors. Attendees broadly agreed that the field of DNA damage, repair, and mutagenesis is uniquely positioned to take a leading role in developing strategies for personalized disease prevention. The purpose of this publication, therefore, is to propose a framework for promoting personalized prevention through collaborative population-based studies that engage cutting-edge technologies.
In the quarter century since the human genome project was launched it has become apparent that the molecular basis for inter-individual differences includes much more than just the DNA sequence. Environmental exposures and stochastic phenomena produce enormous complexity in biological response at the level of epigenetics, transcriptional and translational regulation, and posttranslational modifications of proteins. Furthermore, every individual possesses a mosaic of heterogeneous cells. This staggering variability leads to a unique set of risks and vulnerabilities for each individual and calls into question the standard approach that has dominated preventive medicine since its inception.
An increasing focus on “Precision Medicine” [1] at the national level reflects the growing recognition that, because no two individuals are exactly alike, a tailored approach to treatment based on either germline genetics and/or tumor-specific genetics is likely to provide the largest benefit to patients and to best uphold the principle of primum non nocere. Here, we discuss the role that DNA repair phenotype assays and new DNA sequencing approaches can play in improving precision medicine and cancer prevention. As precision medicine methods apply to tertiary prevention, we extend these principles to secondary (screening) and primary prevention under a general framework of “Precision prevention” and its importance in exposure biology. The same principles of targeted therapy apply, at likely much higher benefit given the lower cost of treatment to patients when interventions are made further upstream. Thus, rather than waiting for a potentially incurable disease to manifest, one can instead address the specific needs of individuals through disease-preventing interventions, or detection and treatment at the earliest possible stage. Precision prevention focuses on being able to predict who is at high risk for a given disease and thereby target screening frequency and onset as well as primary prevention interventions earlier in life to alter disease susceptibility. Individualized prediction is derived from the integrated impact of individual inherent factors (the individual’s genome and epigenome), individual physiological factors (e.g., inflammation and comorbidities) as well as individual biomarkers and response to environmental factors (e.g., individual responses to exposure to air, water, soil, and food). For example, while almost everybody may be exposed to certain pollutants in the environment, such as polycyclic aromatic hydrocarbons (PAH), some individuals may be more susceptible to their health effects based on having deficient DNA repair capacity (DRC). Thus, in this example, measuring DRC in combination with measures of individual PAH metabolites can help in terms of risk stratification and risk assessment. In general, information about inter-individual differences in the ability to respond to environmental exposures and physiological stress are potentially useful for personalized prevention of any disease for which risk is governed by gene-environment interactions.
Precision prevention requires screening tools that enable stratification to identify groups that would most benefit from interventions. Furthermore, fine-tuned tools are needed to prevent ineffective focus on individuals who would not benefit greatly from primary and secondary prevention interventions. Precision prevention promises to identify at-risk individuals, empowering educated decisions on prevention. Importantly, the concept of precision prevention applies not only to the identification of risk-prone individuals, but is also relevant to the evaluation of risk-associated exposures. For example, with the advent of robust analytical tools, we are now poised to break down complex mixtures so that effort(s) can be made toward mitigating the effects of key harmful constituents. Importantly, precision prevention will certainly reduce health care costs over time, because small advances in disease prevention among many add up to a significant reduction in the socioeconomic burden of disease.
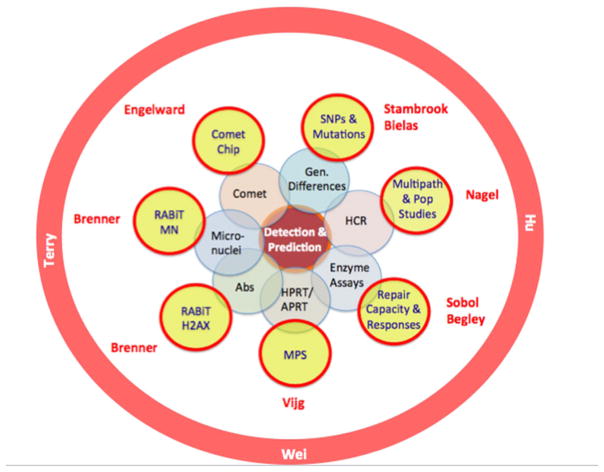
This review focuses on emerging methods for developing better predictors for disease risk in populations exposed to known or unknown agents that can induce DNA damage or alter the ability to repair DNA damage. DNA damage can lead to mutations and cell death; inefficient DNA repair is associated with cancer, neurological disease, immune dysfunction, and developmental disorders, making the damage response of central importance in precision prevention. For decades, researchers have sought to understand what makes some people more prone to disease than others. Prior to discussing today’s cutting-edge approaches, it is helpful to look back at past high impact discoveries. Importantly, for many of these key discoveries, there is now a ‘modernized’ version. Fig. 1 shows how several current technologies are connected to prior advances in our understanding of the DNA damage response, DNA repair, and mutagenesis.
Fig. 1.
From traditional assays (center) to today’s tools for population studies (outer circle). The impact of new technologies emerges through their integration into population studies (orange circle).
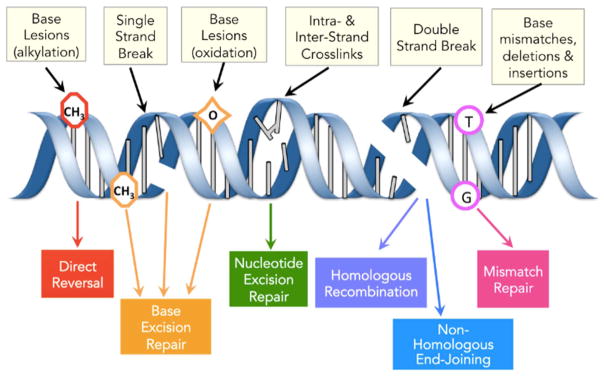
The field of DNA damage and repair began with studies of mutagenesis. Even before the structure of the double helix was known, scientists were working to understand how our environment impacts our genes. Hermann Muller first showed that radiation can lead to mutations [2], and Charlotte Auerbach founded the field of chemical and environmental mutagenesis when she demonstrated that mustard gas, like radiation, can induce mutations [2]. Other early key contributions included those of Alexander Hollaender, who showed that UV wavelength correlates with the spectrum of nucleic acids absorption and with mutagenesis, arguing that nucleic acids were the genetic material at a time when dogma had proteins playing that role [3]. He subsequently demonstrated that under reduced oxygen tension, the mutagenic effects of radiation and of certain chemicals was reduced [4]. Also key were the studies of J. Weigle [5], showing that DNA can be ‘reactivated’ in the cell through a process that we now know is DNA repair. Propelled by these advances, our understanding of environmentally-induced mutations leapt forward in the 1990s with the discovery of translesion DNA polymerases [6]. There is now significant literature, spanning from molecular to population-based studies, that uncovers both chemical and biochemical processes that cause DNA damage and a subsequent increase in mutations, associated with disease. As expected, varied types of environmental exposure will give rise to different DNA lesions, and these lesions can be repaired by one or more of six major DNA repair pathways (Fig. 2). Though the bulk of research findings are based on cellular and animal models, we suggest that fundamental concepts drawn from laboratory findings and some relevant population studies can now be applied to a precision prevention strategy. Using datasets derived from well-examined gene-environmental disease entities provides an illustration of how precision prevention can provide insights into disease origins and mechanisms, as well as needed screening assays and early biomarkers that will underlie preventive/intervention strategies.
Fig. 2.
Major classes of DNA damage and major DNA repair pathways.
1.1. Towards precision prevention: the example of breast cancer
As with any disease, precision prevention of breast cancer first requires prediction of individuals at high risk. Historically, high-risk individuals were identified based on their family history, but as the incidence of breast cancer has been increasing in women under 40 years in the U.S. and women under 50 around the world [7,8], it is increasingly inaccurate to define high risk purely based on family history. Identifying high-risk individuals more accurately is essential as the most effective primary prevention options include chemoprevention (e.g., tamoxifen) and risk reducing surgeries. These options are not without side effects and thus require accurate targeting. Greater accuracy is also needed for secondary prevention as MRI screening is more sensitive than mammography in young women and more frequent screenings may be needed. For example, if DNA repair phenotypic markers can help to improve the accuracy of the estimate for a given woman’s underlying risk, then clinical decision-making can be improved. Precision prevention will mean that some “high-risk” women based on their family history may actually be at much lower risk if they have relatively robust DRC compared to the population average, and therefore those individuals should not undergo risk-reducing surgeries including mastectomies and oophorectomies, which obviously have major harmful effects on overall mortality for average risk women. Similarly, precision prevention should also improve that identification of truly high-risk women, irrespective of family history, who may benefit from early initiation of chemoprevention, early and frequent MRI screening, and/or risk-reducing surgeries at an earlier age.
Improved risk stratification is also needed to better understand how the environment modifies underlying genetic susceptibility. Environmental factors are likely to have their impact temporally over one’s lifespan, such as in the well-described relationship between exposure to diethylstilbestrol and disease [9,10]. Given the influence of exposures on cancer risk across life course, improved delineation of risk is essential to inform prevention efforts. Thus, although new, non-invasive approaches are in development [11], we focus here on the precision prediction of breast cancer as an example.
Whilst a few high-penetrance allelic variants are strongly associated with high individual risk of breast cancer, most women, perhaps as many as 90%, who develop breast cancer are not carriers of BRCA1 or BRCA2. Rather, underlying genetic susceptibility to breast cancer is likely driven by interactions between multiple alleles, rather than by a single or a few major variants. In other words, for many women individual susceptibility to breast cancer is driven by inherited combinations of multiple low penetrance alleles [12]. This observation leads to two potential approaches to individualized risk prediction. One is to identify these multiple low-penetrance alleles and develop tests for them – a genotypic approach that has been somewhat successful for those in the most extreme risk category. Alternatively, a phenotypic approach can be taken; for example, inducing DNA damage in a tissue sample (e.g., blood), and then measuring the rate of repair or the frequency of unrepaired damage. Because not all phenotypic variation can be predicted from sequence, and because not all samples are amenable to phenotypic assays, the two approaches are complementary. In this review we will address both genotypic and phenotypic approaches.
In attempting to determine the potential power of these approaches, a number of models have been developed to predict individualized breast cancer risk based on epidemiological and clinical risk factors, but with limited success [13]. In studies in which these factors were augmented with information from two phenotypic DNA repair assays, however, the predictive power for breast cancer was markedly increased [14,15]. Importantly, these studies showed that genotype was not equivalent to phenotype and that the phenotypic markers of DNA repair were much stronger in predicting why one sister was diagnosed with breast cancer when the other sister was not [14,15]. To date such phenotypic approaches have been laborious to perform, with low throughput and thus remain impractical for large-scale use. However, improvements in existing assays, as well as development of novel ones, are emerging that will accelerate precision prevention. These assays provide valuable insights into multiple cellular pathways simultaneously, are high throughput, and/or require only limited tissue or blood volumes. Validation is ongoing, wherein the pre-requisites for such assays are characterization of reproducibility, specificity, and, critically, the association with eventual disease or other known biomarkers.
1.2. Moving from classical measures of DNA damage, repair and mutagenesis to contemporary methodologies
Given the myriad significant health problems that can result from DNA damage, great effort has been spent developing technologies to monitor DNA damage and to measure DNA repair. Here, we show how some of our most fundamental assays for assessing DNA damage and repair (Fig. 1, inner circles) are now being eclipsed by high-throughput, highly sensitive platforms (Fig. 1, outer circles). Herein, we discuss how these new technologies might impact the field. Importantly, we call attention to exciting opportunities to work synergistically, so that multiple platforms can be incorporated into population studies.
2. Tools and technologies in DNA damage, repair and mutagenesis for personalized prevention
Functional assays and sequencing technology have taken major steps forward in the last decade. Together, these technologies constitute a diverse toolbox of complementary methods that can potentially be used to develop individualized disease risk assessments and prevention programs.
2.1. Emerging technologies to quantify DNA damage, DNA damage response and DNA repair capacity for personalized risk and exposure assessments
In this section we describe several cutting-edge and emerging technologies that address the question of “what does it take” to start using the technology among basic scientists, clinical researchers, and population study investigators. The first five approaches, CometChip, RABiT (Rapid Automated Biodosimetry Technology) γ-H2AX, FM-HCR (Fluorescence-based multiplexed host cell reactivation), DNA Repair Beacons, and ECL-DDR (Electrochemiluminescence-based DNA Damage Response) are phenotypic in concept, while others (e.g., CypherSeq, discussed in the next section) are genotypically oriented. A key consideration will be practicality and throughput. If these approaches are to be of use for precision prevention, the assays need to be simple to perform, cost-effective, and high-throughput. Some of the assays described here already meet this goal, in that they are high-throughput modifications of existing assays, while others would need additional development to reach this goal.
2.1.1. CometChip
The CometChip is based upon the traditional Comet assay, wherein the extent of DNA damage is quantified based upon the extent to which DNA migrates away from the nucleus when electrophoresed. As an example, a normal healthy cell has highly supercoiled intact DNA that does not migrate during electrophoresis. However, if a cell is exposed to ionizing radiation or other DNA damaging agents, the DNA can become nicked and fragmented, and is thus able to migrate away from the nucleus when electrophoresed. The assay was originally developed in the 1980s by Ostling and Johansen and Singh [16–18], and it has been used in thousands of studies. Nevertheless, it is often affected by low reproducibility and relatively low throughput. To overcome these limitations, the CometChip was developed, and this approach has been shown to increase throughput by more than ~100X together with increased sensitivity and reproducibility. The approach is described below.
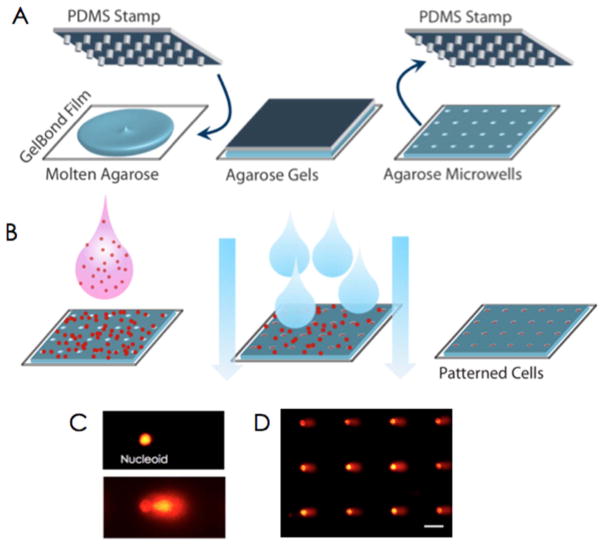
The CometChip (Fig. 3) works by taking advantage of photolithography to create a stamp with pegs that are approximately the diameter of a single mammalian cell. The mold is then pressed into molten agarose, the temperature is dropped, and the mold is removed to reveal an array of microwells. A cell suspension is placed on top of the microwells and mammalian cells are loaded by gravity. The arrayed cells can then be manipulated (e.g., exposed to DNA damage and allowed to repair for different lengths of time), and ultimately processed the same way as for a traditional comet assay (namely, lysis, incubation in high pH buffer, and electrophoresis). Using this approach, each well of a 96-well plate can contain ~300 microwells at the base, enabling 96 sample conditions to be assayed in parallel. Sensitivity is also increased in part due to reduced comet-to-comet variation. Using the CometChip, the repair kinetics (multiple time points) of 24 human cell lines were recently analyzed in parallel [19]. Approximately 1000 samples were analyzed in a single experiment, something that could not be done using the traditional comet assay due to experimental noise.
Fig. 3.
CometChip uses photolithography to create a mold with micrometer scale pegs. A) The mold creates an array of microwells. B) Cells are loaded into the wells by gravity and excess cells are removed by shear force. C) Comet data from an undamaged cell (top) and a heavily damaged cell (bottom). D) An array of comets resulting from the CometChip.
2.1.1.1. Readiness/Needs for potential application to precision prevention
Variation in DRC has been clearly linked with risk for several types of cancer. The CometChip technique is therefore ready for epidemiological studies of risk prediction.
2.1.2. RABiT- γH2AX and global DNA repair capacity
The γ-H2AX assay has been widely used to quantify the yield of DNA double strand breaks after radiation or other genotoxic exposures [20]. The RABiT (Rapid Automated Biodosimetry Technology) approach for (among other endpoints) high-throughput γ-H2AX measurements was initially developed as an ultra-high-throughput technology for biodosi-metric reconstruction of past radiation exposures, measuring micro-nucleus or γ-H2AX yields in a fingerstick of blood, with a throughput of ~30,000 samples per day [21]. This fully-automated high-throughput methodology has been adapted to assay global DRC through automated quantification of the time-dependent kinetics of the disappearance of γ-H2AX foci after a radiation challenge [22].
The current automated methodology, using an automated robotic workstation, involves acquiring fresh fingerstick blood samples, which are then centrifuged, irradiated and dispensed into a 96 well plate format. A key aspect here is the use of a small, inexpensive automated capillary fingerstick irradiator [22]. The multi-well plates containing the cells are maintained in the RABiT incubator, and the post-irradiation, time-dependent γ-H2AX yield measurements are based on sequential automated samplings from these lymphocytes at five post-irradiation times from 0.5 to 24 h. A typical result from a study of 94 individuals is shown in Fig. 4 [21], and we note here that the technique requires a small number of cells per sample – typically 25,000 lymphocytes for the 5 samplings (i.e., 5000 lymphocytes per time point). As illustrated in Fig. 4, the key quantities characterizing global DRC are the characteristic decay constant of the γ-H2AX (DSB) yield (Kdec in Fig. 4), and the yield of long-term unrepaired breaks (Fres in Fig. 4).
Fig. 4.

γ-H2AX yields and DNA repair kinetics as measured with the high-throughput fully-automated RABiT system [21,22], from a study of finger stick blood samples from 94 healthy individuals [23]. Experimental data and model fit (see [23]) pooled from 94 donors exposed ex vivo to 4 Gy gamma radiation, assayed at 0.5, 2, 4, 7 and 24 h post irradiation.
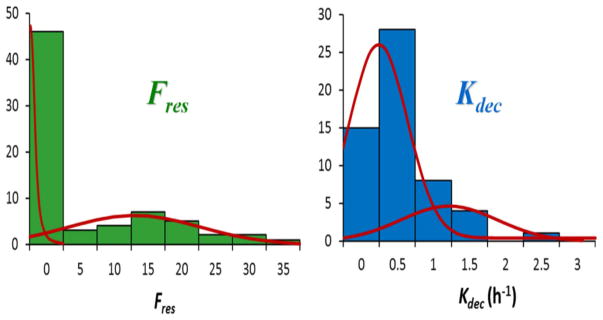
Fig. 5 shows the person-to-person distributions of the parameters Fres (residual DSBs) and Kdec (characteristic DSB decay time) derived from the recent study of 94 healthy individuals [23] using the RABiT system. The red curves show fits to biphasic normal distributions, which might be interpreted as relating to normal and radiation-sensitive sub-populations; interestingly, there is a statistically significant separation of the two distributions for the yield of long-term breaks (Fres).
Fig. 5.
Distributions of the DNA repair parameters Fres (residual DSBs) and Kdec (characteristic DSB decay time) derived from a recent study [23] of 94 healthy individuals using the high throughputfully-automated RABiT system [21,22]. The red curves show fits to biphasic normal distributions, showing evidence for distinct subpopulations with different DNA repair capacities.
The RABiT methodology to date has been developed on a dedicated purpose-built robotic workstation. In the past few years, however, commercial robotically-based, high-throughput cell handling workstations have become very common. The RABiT protocols are being adopted for use on commercial high-content, high-throughput cellular screening systems [24].
2.1.2.1. Readiness/Needs for potential application to precision prevention
Several studies have suggested that DRC is a major risk factor for development of many cancers including lung, breast, and bladder. Thus, the high-throughput RABiT γ-H2AX approach is potentially ready for population studies to measure the predictive power of global DNA repair in a variety of cancers.
2.1.3. FM-HCR
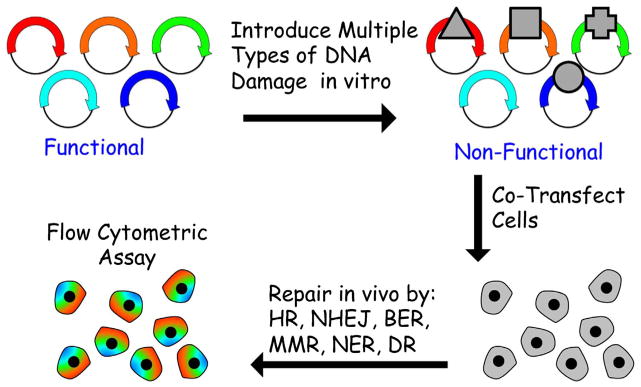
Historically, measuring DRC has been a laborious, time-consuming activity requiring extensive training and unique methods to analyze each of the repair pathways. This reality has contributed to DNA repair experts working in silos defined by single repair pathways, and has represented a major barrier to including functional repair assays in epidemiological studies [25]. Fluorescence-based multiplexed host cell reactivation (FM-HCR) assays measure the ability of cells to repair lesions which altogether are substrates for all 6 of the major DNA repair pathways illustrated in Fig. 1, using transiently transfected episomal fluorescent reporter vectors (Fig. 6) [26]. Multiple repair pathways can be monitored simultaneously because each pathway is reported by a different colored fluorescent reporter protein. Detection is carried out by flow cytometry, but the assays are also amenable to high-throughput imaging analysis. Key strengths of FM-HCR include the ability to measure repair capacity in multiple pathways simultaneously in live cells, the use of a single approach with a quantitative readout for all pathways (transfection and fluorescence measurements), and the flexibility to measure repair capacity in any transfectable tissue with a turnaround time of less than 24 h.
Fig. 6.
Schematic of FM-HCR. Fluorescence based multiplex host cell reactivatio (FM-HCR) assays use unique fluorescent reporter plasmids to measure repair capacity in multiple DNA repair pathways in parallel in live cells (Nagel et al. (2014) PNAS 111(18), E1823–32).
FM-HCR involves in vitro preparation of reporter plasmids containing specific types of DNA damage that alters either the efficiency or the fidelity of transcription after transfection into cells. The earliest HCR assays were based upon the ability of UV-induced DNA damage to block replication of viral DNA; viral transduction efficiency was proportional to the ability of the host cell to repair and subsequently replicate the damaged viral DNA. Since the advent of recombinant DNA, HCR assays have made use of transiently transfected plasmid vectors that express reporter proteins in human cells. Some types of DNA damage, such as strand breaks, UV-induced photoproducts, and DNA cross-links, block transcription unless they are repaired. Thus, expression of the plasmid encoded reporter protein is proportional to repair capacity. FM-HCR has recently extended this paradigm to include DNA lesions that do not block transcription, such as O6-methylguanine or 8-oxoguanine. These DNA lesions are bypassed by the RNA polymerase, however they induce transcriptional errors via a process that has been termed transcriptional mutagenesis [27]. FM-HCR uses plasmids containing site-specific DNA damage to measure repair by transducing lesion-induced transcriptional errors into measurable fluorescent signals that are proportional to repair capacity for the lesion of interest. Because cytotoxic and mutagenic DNA lesions often either block transcription or cause transcriptional errors, FM-HCR is broadly applicable to measuring repair efficacy in all of the major DNA repair pathways.
FM-HCR works with transfection and analysis in a 96-well format, allowing for automated flow cytometric analysis and batch processing of samples. This allows for analysis of repair capacity in 4 pathways for 48 samples in approximately 2 h of active laboratory time.
2.1.3.1. Readiness/Requirements for potential application to precision prevention
Pioneering work published over 20 years ago has already demonstrated that HCR assays are ready for applications in precision prevention [28,29], and the new FM-HCR assays can now build upon this paradigm by way of population studies in cells isolated from normal human tissues. In particular, measuring multiple DNA repair pathways using relatively accessible primary blood cells and epithelial cells can provide estimates of inter-individual variation, tissue-specific variation, and will provide key information about the association between DRC and disease risk.
2.1.4. DNA repair beacons
DNA repair pathways maintain the integrity of the genome and thereby help prevent the onset of cancer, disease and aging phenotypes [30]. As such, the critical requirement for DNA repair proteins and pathways in response to radiation and genotoxic chemotherapeutics implicates DNA repair proteins as prime targets for improving response to currently available anti-cancer regimens [31]. In this vein, inhibitors to the DNA repair proteins PARP1, ATM, APE1, WRN and BLM (among others) have been developed and are either undergoing clinical testing or are being considered for such [32–38]. Although defects in critical DNA repair pathways or proteins can predispose to cancer onset [39,40], such cancer-specific DNA repair defects offer novel approaches for tumor-selective therapy [41]. These repair defects may manifest as genomic loss (LOH or mutations), suppression of mRNA expression via promoter methylation, defects in mRNA stability by aberrant miRNA expression, or loss of protein expression. Many of these cancer-specific DNA repair defects [42] can be detected using current omics technologies. However, there are many defects that can only be detected from an analysis of either pathway- or protein-specific DRC. The Sobol lab has developed a DNA Repairomics platform as an essential tool to address this need. This platform offers a high degree of flexibility, may be utilized with standard laboratory equipment, will be critical in biomarker analysis, and will have immediate application in screening and structure-activity relationship (SAR) analysis for DNA repair protein inhibitors using purified proteins.
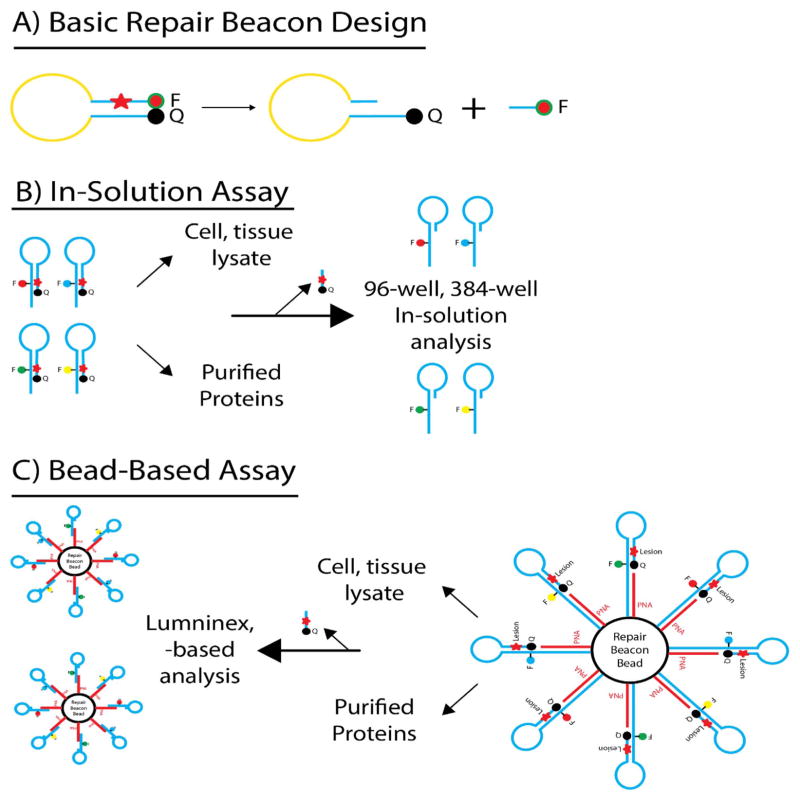
The overall structure of a DNA Repair Beacon, as recently described by Sobol and colleagues [43], is shown in Fig. 7A. The DNA Repair Beacon consists of a deoxyoligonucleotide containing a single base lesion with a 6-Carboxyfluorescein (6-FAM) moiety conjugated to the 5′ end and a Dabcyl moiety conjugated to the 3′ end of the oligonucleo-tide. The base excision repair (BER) molecular beacon is 43 bases in length and the sequence is designed to promote the formation of a stem-loop structure with 13 nucleotides in the loop and 15 base pairs in the stem [32,44]. When folded in this configuration the 6-FAM moiety is quenched by Dabcyl in a non-fluorescent manner via Förster Resonance Energy Transfer (FRET) [45,46]. The lesion is positioned such that following base lesion removal and strand scission the remaining 5 base oligonucleotide containing the 6-FAM moiety is released from the stem. The DNA repair beacons are incubated with cell extracts or purified proteins to facilitate lesion removal and DNA strand cleavage. The subsequent release and detachment of the 6-FAM containing DNA from the quencher (Dabcyl) results in an increase of fluorescence that is proportionate to the level of DNA repair. By collecting multiple reads of the fluorescence values, real-time assessment of repair activity is possible. Using standard quantitative real-time PCR instruments allows for the simultaneous analysis of numerous samples. To provide multiplexing capacity, the beacons are being optimized for multiple sets of fluor/quencher pairs that will allow the assay to be used in 96- or 384-well platforms for high-throughput application. To complement the beacon ‘In Solution’ assay (96-well plate), the platform has been modified using microspheres or bead-based Beacons (Fig. 7B and C). These include an extended 5′ arm containing biotin to allow the use of optically encoded microspheres (beads). Bead-based tethering provides a high-degree of multiplexing as well as side-by-side analysis of DNA repair protein levels with additional Luminex™-based endpoints from the same lysate sample.
Fig. 7.
DNA Repair Molecular Beacons – (A) Overall design of the DNA repair molecular beacons – a deoxyoligonucleotide containing a single base lesion with a 6-Carboxyfluorescein (6-FAM) moiety conjugated to the 5′ end and a Dabcyl moiety conjugated to the 3′ end of the oligonucleotide. (B) Schematic representation of utility of the DNA repair molecular beacon assay in 96- or 384-well plates for analysis of cell and tissue lysates or purtified proteins. (C) Modification of the DNA repair molecular beacon platform – microspheres or bead-based Beacons for increased multiplexing capacity.
2.1.4.1. Readiness/Requirements for potential application to precision prevention
DNA Repair Beacons represent a novel approach, utilizing state-of-the-art nucleic acid-based technologies for enzymatic activity profiling useful in biomarker analysis and in the development of specific DNA repair inhibitors. Since the ‘In-Solution’ assay is a real-time, quantitative assay that measures fluorescence, the assessment of activity is achieved with standard quantitative real-time PCR instruments, allowing the simultaneous analysis of numerous samples. The bead-based assay has the advantage of being able to be combined with other bead-based analysis tools simultaneously. Overall, this platform is amenable to kinetic analyses, DNA Repair quantification and inhibitor validation and is adaptable for quantification of DRC with purified proteins, with tissue and tumor cell lysates and with application for functional biomarker measurements [43]. The use of beads, the design of the unbiased discovery platform, and the adaptability of the DNA Repair Beacon to many DNA repair protein substrates that can be modified to provide specificity for damage-specific nucleases, structure-specific nucleases, helicases, and topoisomerase all contribute to the development of a complete DNA Repairomics platform that can be applied in future studies.
2.1.5. ECL-DDR
Quantitative measures of all DNA Damage Response (DDR) and DNA repair proteins in a cell or tissue sample have the potential to pinpoint defects in key response pathways that can identify individuals susceptible to environmentally-induced disease or can be exploited for development of cancer therapeutics. The γ-H2AX assay, described above, is a single protein analytic that reflects activation of the DDR and can be considered a surrogate measure of DNA strand breaks. To activate one arm of the DDR, Ataxia telangiectasia mutated (ATM) autophosphorylates in response to DNA strand breaks [47–49]. Activated ATM then phosphorylates histone protein H2A leading to the formation of γ-H2AX foci at sites of DNA damage, [50] and promotes the phosphorylation and activation of hundreds of downstream proteins including checkpoint kinase 2 (CHEK2) and tumor protein p53 (p53) [48,51]. Activation of the DDR regulates the activity of > 100 DNA repair and cell cycle proteins vital to genome stability [52]. Severe defects in the DNA damage response can predispose individuals to cancer and neurological diseases [53]. In addition, individuals heterozygous for defective DDR and DNA repair genes have increased rates of cancer incidence [54], which further highlights the importance of measuring DRC. The link between cancer incidence and defects in DDR highlights the need for proper activation of damage-responses that is essential for preventing disease.
Measuring the levels and activity of the > 100 proteins and associated damage-induced post-translational modifications (PTMs) participating in DDR and DNA repair is technically challenging. The associated technology platform requires (1) a large dynamic range measure low-, medium- and highly- expressed proteins and PTMs, (2) high-content and high-throughput capabilities, and (3) ease-of-use and robustness to effectively transition into practice in clinical or laboratory settings. A commercially available high-throughput and high-content capable electrochemiluminescence (ECL)-based platform has been adapted for use in measuring DDR proteins and damage-induced PTMs. Available from MesoScale Discoveries, the ECL platform uses an electrode-lined well coated with a specific antibody to bind a target protein, a second target specific antibody labeled with a light emitting tag, an electrical-to-chemical signal initiation, and an amplification cycle to generate a luminescence signal that quantitates target proteins or PTMs over a 6-log dynamic range (www.mesoscale.com) [55–58]. Further, the 96-well and 384-well plate designs, coupled with 4- to 10-addressable spots in each well, endow the ECL-platform with a high-content capability that could measure thousands of DDR and DNA repair proteins and associated PTM’s simultaneously. Five antibody pairs specific to protein and phosphorylated DDR components were identified and optimized for use in the ECL-platform, as demonstrated by cell line- and tumor-specific responses to ionizing radiation and other classic DNA damaging agents (Hseih et al., in preparation). The technology platform and validated assays were also assessed using clinical samples. A study design that utilized patient blood draws, pre-and post-diagnostic CT-scans, was utilized. Diagnostic CT-scans that expose patients to ~30 mSv of radiation during each medical procedure have been shown to activate the DDR. ECL-based analysis of patient-matched leukocytes using the DDR-specific assays successfully identified activation of the DDR in the post-CT scan sample for most patients (Hseih et al., in preparation). Further, the study demonstrated that the ECL technology platform is amenable for use with clinical samples. Validation of new measures of DDR or DNA repair components in clinical samples is needed to promote wide-spread use of next generation measures of DRC. Clinical samples derived from patients undergoing radiation-associated medical procedures represent a well-controlled study of patients with an environmental exposure to DNA damaging agents, and this study design can be used to validate other platforms or assays.
2.1.5.1. Readiness/Requirements for potential application to precision prevention
The ECL-platform is commercialized and ready for deployment in pre-clinical and population-based studies. It has been shown to be technically reproducible in lab and clinical studies, with radiation-associated medical procedures identified as an optimal exposure for technology validation. Thousands of commercial antibodies specific to DDR and DNA repair proteins are available for testing in the ECL-platform. Near complete coverage of the hundreds to thousands of protein participating in the DDR and DNA repair pathways requires robust analysis of existing antibodies and/or development of antibody pairs to each target. Further, the use of good manufacturing practices (GMP) to produce high-coverage and technically-reproducible ECL-plates is needed so that researchers can assess DDR and DNA repair and associated PTM-protein levels in patient studies with a 1000 or more participants.
2.2. Emerging technologies to quantify mutation frequencies for personalized risk and exposure assessments
Environmental mutagens can cause disease in somatic cells by inducing mutations in critical genes. If, for example, the genes are drivers of malignant transformation, the resulting outcome/disease is cancer. Thus, monitoring the frequency of mutation in humans has the potential to provide early markers of exposure and increased risk of developing disease. Methods to measure induced mutations have focused on (1) housekeeping genes that, when mutated, offer a selective growth advantage, (2) plasmid reporter genes, and (3) mice with reporter genes integrated into their chromosomes [59–61]. Data produced by these methodologies have been used to estimate human mutagenesis, but a major concern in genetic toxicology is the ability to extrapolate the genotoxicity observed with high-dose mutagen exposure in model animals or human cells in culture to that which occurs at low, environmentally-relevant doses in humans [62]. Human studies have been limited to those involving cells in culture, at mutational targets subject to selection (i.e., the hprt gene), and therefore are subject to biases and lack sufficient sensitivity to detect mutation resulting from low-dose exposure to environmental chemicals. Thus, overly conservative risk assessments are typically made based on assays that monitor DNA damage rather than mutation, and maximum tolerated doses (MTDs) are calculated based on extrapolation of these high-dose experiments (usually in model organisms or cell culture) to low-dose, environmentally relevant exposure in humans. In the case of DNA damaging agents, these MTDs ignore the efficiency of DNA repair in removing the lesions, as well as the vast range in mutagenic potential of various DNA adducts. However, with the advent of next-generation sequencing technologies, we are now in a position to establish the unbiased frequency and spectrum of spontaneous mutations throughout the human genome, and to identify regions of the genome that are the most tractable for use as robust biomarkers of mutagenic exposure.
Massively-parallel sequencing is revolutionizing biomedical research, enabling high-throughput and low-cost sequencing of hundreds of billions of bases in about one day [63,64]. However, these technologies are limited by error rates of 0.05% to 1% [65,66] resulting in millions of sequencing errors per experiment. This level of inaccuracy hinders our ability to apply these promising sequencing technologies to the detection and quantification of somatic mutations, an application that requires very high sensitivity and specificity [67–70]. One way to circumvent this problem and quantify mutational heterogeneity within tissues is via whole genome sequencing of a representative number of single cells [71]. However, the level of accuracy in single-cell genome sequencing may not be sufficient for detecting differences in exposure-induced mutation load between individuals. Accurate measurement of all types of genetic changes accumulated over a lifetime in single-cell lineages recently performed with the fibroblast clones from healthy human donors provided proof-of-principle that the load of base substitutions can serve as an overall dosimeter of environmentally-(UV-) induced mutagenesis [72]. While such technologies are extremely powerful for investigating the fundamental mechanisms of mutagenesis, they are limited in throughput and, thus, in their utility to monitor mutagenic exposure in human populations. While several groups have devised methods to improve the accuracy of bulk massively parallel sequencing, these methods do not overcome the three main technological barriers that have precluded the application of next generation sequencing- (NGS-) based assays for monitoring the extremely low frequency of somatic mutations in populations, namely: (1) the intrinsic error frequency of high-throughput sequencing, (2) the number of reads a sequencing platform can produce, and (3) the amount of input DNA available.
Many groups have worked to improve the error rate of NGS with both computational [73] and molecular approaches [74–76]. The CAPP-Seq system uses statistical models to parse error from real variants, which permits a mutation to be detected among a background of 5000 nucleotides or 2 × 10−4 substitutions errors per nucleotide [73]. To date, the most accurate molecular approaches for error correction are based on DNA barcoding technologies in which each read is assigned a unique identifier and amplified. Multiple copies of each read are then sequenced, and a consensus is created. Utilizing 12–14 base pair single stranded barcodes, the Safe-Sequencing System improves mutation detection down to roughly 10−5 mutations per base pair [76]. Several other groups have also described similar molecular barcode-based error-reducing methodologies [75,77]. One notable example is the Duplex Sequencing method in which each double-stranded template molecule is tagged with a double-stranded barcode [75]. The use of double-stranded barcodes permitted the detection of 1 mutation in 4 × 105 wild-type base pairs, though, theoretically, double-stranded barcoding should permit the resolution of < 1 mutant base among 109 wild-type nucleotides [75].
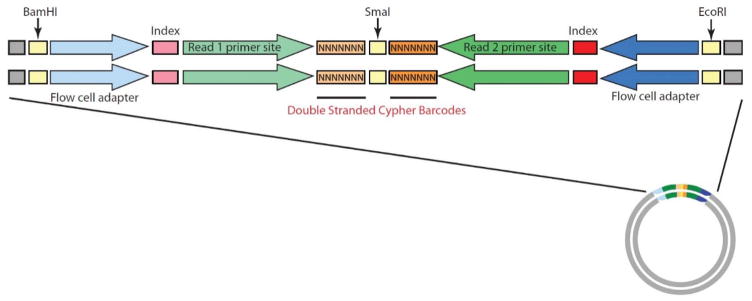
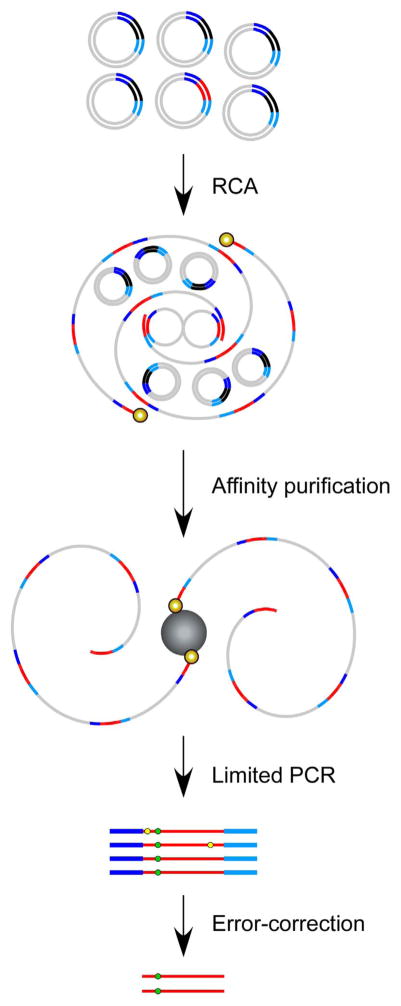
To overcome these barriers to rare variant detection, Bielas and colleagues have designed and established a novel mutation detection method, termed CypherSeq [78]; a circular, double-stranded, dual-barcoded sequencing methodology that combines barcoding, targeted rolling circle amplification (RCA), bead-based enrichment, and massively parallel sequencing into a single assay. The circular nature of CypherSeq libraries (Fig. 8) offers several distinct advantages over other technologies. Circular DNA is inherently more stable than linear DNA, and can be further preserved via transformation into E. coli and preparation of glycerol stocks. Transforming CypherSeq libraries into bacteria allows users to titrate the number of barcodes used in a sequencing run. Additionally, CypherSeq libraries can be proliferated in bacteria as a pre-amplification step prior to sequencing, which, thanks to the repair pathways of E. coli, offers much greater fidelity than PCR and reduces data loss during correction. Furthermore, the circular nature of the plasmid-based sequencing library permits enrichment for specific targets using RCA, which serves to reduce off-target reads and maximize read depth (Fig. 9).
Fig. 8.
Schematic representation of the double-stranded CypherSeq construct. The CypherSeq construct includes all of the components necessary for sequencing on Illumina platforms, plus two 7-nucleotide double-stranded, randomly generated barcodes (flanking a blunt-SmaI restriction site). Sheared genomic DNA is ligated into the vector at the SmaI site. The library is then amplified (via E. coli transformation or PCR) and deep sequenced. The resulting sequence reads are filtered, computationally de-convoluted, and error-corrected via the double-stranded CypherSeq barcodes.
Fig. 9.
Overview of rolling circle amplification (RCA) enrichment from CypherSeq libraries. A CypherSeq vector library is amplified by extension of biotinylated, target-specific primers using the strand displacement synthesis-proficient polymerase. Two primers, one targeting each of the complementary strands, must be used to achieve double-strand molecular barcoded error correction. Template CypherSeq vectors containing non- target sequences remain unamplified while templates containing the target sequence are amplified via RCA into long single-stranded products containing redundant copies of the target sequence and sequencing cassette. The RCA products are purified using magnetic streptavidin-coated beads, subjected to limited PCR with the library preparation primers, and sequenced. Reads are computationally compiled by barcode and a consensus is made for each barcode family independently. Substitutions occurring in < 90% of the reads within a family are rejected as artifacts, while substitutions present in all or nearly all (> 90%) of a family are accepted as true mutations.
The CypherSeq approach corrects the errors inherent to NGS sequencing, allowing detection of mutations at frequencies as low as 2.4 × 10−7 per base pair [78]. However, by increasing the number of base pairs sequenced, the sensitivity of the CypherSeq methodology can be increased, as double-stranded barcoding-based error correction can theoretically permit the resolution of mutation frequencies as low as 10−9 to 10−10 per nucleotide [75]. As such, the CypherSeq methodology allows for exact determination of mutation frequencies in high-throughput screens that interrogate millions of base pairs simultaneously, and can permit the first high-resolution estimate of the rate of somatic mutation throughout the human genome. These data would provide the first available measurements of random mutations throughout the genome in humans, and will permit us to delineate the impact that genomic architecture, sequence context, replication timing and transcription have on genome-wide mutation frequency and spectrum. Moreover, this technology allows human mutagen exposure to be monitored via DNA biomarkers, through the direct assessment of mutation at neutral (free of selection pressure) target sites enriched via RCA. This data would serve as a historical record of environmental mutagenic exposure, and potentially provide the ideal biomarker for human risk assessment, as the measured endpoint (somatic mutation) is a driver of carcinogenesis.
2.3. Importance of animal models in understanding gene and environment effects on DNA damage response and disease
Population science is an invaluable tool for identifying biological markers that associate with environmental exposure and disease. These associations, however, require validation. To facilitate implementation of preventative measures and identification of possible therapeutic targets requires a basic understanding of target organ biology. Animal models, therefore, play an important role in the validation process and in deciphering perturbed mechanisms. Such models allow testing for effects of environmental exposures or interacting genes on a disease process and have the potential of uncovering associations that may not be obvious from human epidemiology studies. In the case of cancer, the effect may be to accelerate or retard the incidence of disease or rate of tumor progression.
An example of the latter is the role of low penetrance CHEK2 alleles in breast cancer. The CHEK2*1100delC allele has a C deletion at position 1100 to produce a truncated protein lacking a kinase domain. It clearly impacts breast cancer risk since first degree relatives of patients with bilateral disease who are heterozygous for CHEK2*1100delC are at three times greater risk for breast cancer than first degree relatives of patients with bilateral disease who are CHEK2 wild-type. Risk increases eight-fold compared with women in the general population [79]. In the mouse, some of the biological consequences of homozygosity at Chk2*1100delC are predictable whereas others are unexpected. The Chk2 kinase participates in cell cycle and checkpoint regulation by phosphorylating Cdc25A and promoting its proteasome-mediated degradation. Following DNA damage, it perturbs the integrity of the G1/S checkpoint, producing genomic instability [80]. The mouse model, in fact, displays constitutive DNA damage, an altered cell cycle profile, and an elevated level of polyploidy and multinucleated cells [81]. The mice also develop tumors, not restricted to the mammary gland, and expire only after 12 months of age [82]. Strikingly, only female mice develop tumors above the level of control mice [82], suggestive of a hormonal contribution. In humans, male breast cancer is very rare [83] but has been associated with obesity and with hormonal imbalances [84]. It has also been associated with gynecomastia (enlarged breasts in men) and excessive levels of estrogen [85]. It should be instructive to ascertain whether or not the sexual dimorphism observed in mice homozygous at Chk2*1100delC is hormonally related and whether such imbalance may apply to the human condition.
Whether environmental exposure or genetic interactions affect the time of onset or severity of the disease in women who harbor the CHEK2*1100delC allele is not known but has been addressed in the mouse. Exposure to the carcinogen 7,12-dimethylbenz(a)anthracene (DMBA) causes tumor formation in mice, which is accelerated in similarly treated mice homozygous for Chk2*1100delC [82]. Likewise, mice that overexpress the oncogenic receptor tyrosine kinase RON in the mammary gland form tumors in about 40 weeks. In contrast, the time of tumor formation in mice that overexpress RON and that are also homozygous for Chk2*1100delC is reduced to 34 weeks [86]. Thus, both genetic constitution and environmental exposure interact with the Chk2*1100delC allele to produce a more severe phenotype.
While rodent models have been invaluable for monitoring responses to environmental challenges and genetic interactions, they are limited by cost, by their generally long response time, and by the fact that their responses may differ from those of humans. Response time and cost for assessing exposure and for modeling human disease can be overcome by model systems that use lower organisms such as Drosophila [87,88] or C. elegans [89]. Although informative, these models, however, do not necessarily mimic the human condition or effects at target organs. In principle, most of these concerns can now be overcome by the use of human induced pluripotent stem (iPS) cells [90,91] that have been coaxed to differentiate into a lineage representative of the human target cell type [92]. While this approach represents an advance over current commonly used approaches, it still assumes that responses to exposure are cell autonomous. The use of iPS cell-derived organoids that mimic the complexity of a target tissue could circumvent this last concern, and its advantages for studying cellular responses to drug exposure have been discussed [93]. Further possibilities for establishing mechanisms underlying responses to environmental exposure or drug administration may be provided by patient-derived iPS cells [94] or by precision editing of iPS cells [95] prior to their differentiation into specific lineages and into organoids. One can thus envision a personalized approach to prevention or reduction of exposure to environmental toxicants.
2.4. Integration across methods, fields, and disciplines to achieve predictive power
Significant advances in predictive power should be realized as an integrated landscape emerges based on multiple assays that address similar aspects of the same question. Thus, researchers in assay development need to collaborate to perform complementary studies. By sharing the same sample set, we will learn about the strengths and weaknesses of different approaches, affording new opportunities for methods refinement. Therefore, it is of particular importance for basic researchers not only to team up with clinical researchers and population scientists, but additionally to join forces with other methods development teams so that data can be collected from a shared sample source. Ideally, if subpopulations of susceptible individuals are identified, these could be cross-examined using complementary methods to explore assay integration. As an example, DNA double strand breaks can now be measured in more than one way. Ideally, multiple different approaches (for example, the comet and γ-H2AX assays) should be brought to bear on a shared sample set to learn more about the strengths and weaknesses of each approach and the ability of data collected using divergent methods to combine in order to strengthen data analysis. Ultimately, many of the methods described here will need to be used in parallel both to validate the differing approaches, and to pave the way for more robust analysis tools. Moreover, for individuals with high risk, whole-genome or whole-exome sequencing of single-cells, cell clones or micro-biopsies can be used to validate extreme risk prediction and to support prevention recommendations.
3. Overcoming barriers to personalized prevention
3.1. Considerations from the epidemiology perspective: practical considerations for collecting and preserving biological samples for phenotypic assays
In epidemiological studies, phenotypic assays complement genomic approaches that cannot directly assess pathway function. While molecular epidemiology studies using genetic biomarkers, such as germline mutations or single nucleotide polymorphisms, have advanced rapidly, leading to genome-wide association studies [96] and whole genome sequencing efforts, studies with phenotypic assays are limited. This is partly because viable cells are often needed for cellular functional testing. In such cases, time course and dose escalation are vital for assessing cellular functions, and the use of live cells to reffect the individual’s biological response to exposures and the need to keep the cells proliferating normally are required to simulate in vivo biology. Therefore, to address the requirement for viable lymphocytes in performing phenotypic assays for biomarker studies, both fresh and cryopreserved lymphocytes have been used for cell culture-based functional assays. The use of fresh lymphocytes does not allow most assays to be conducted in batches, while cryopreservation of isolated lymphocytes results in a considerable loss of viable cells. Both fresh and cryopreserved lymphocytes have been used to assess the extent of DNA damage and the kinetics of repair as phenotypic assays. This includes testing for DRC using cells that are exposed to a known carcinogen in cell culture assays, such as Comet assay [97–99], or γ-H2AX foci formation [100] or to a substrate harboring DNA damage, such as the host-cell reactivation (HCR) assay.
Although freshly isolated lymphocytes and cryopreserved lymphocytes have their respective advantages and drawbacks, both lymphocyte sources appear to produce valid data regarding repair capacity. When the feasibility of using cryopreserved whole blood as a source of viable lymphocytes in molecular epidemiology studies was compared with the use of whole blood cryopreserved by traditional methods, using HCR and mutagen sensitivity assays, the outcome was similar with a correlation 0.77 (P < 0.001) for paired blood samples [101]. The Wei lab has also shown that the baseline of γ-radiation-induced chromatid breaks, as measured by the mutagen sensitivity assay were not significantly different between lymphocytes from either frozen blood or fresh blood. Although the correlation between the numbers of chromatid breaks in the paired blood samples was statistically significant, the lymphocytes from frozen whole blood were more sensitive to γ-radiation, with a higher mean level of chromatid breaks than that in fresh blood. Overall, these data suggest that within the limits of the parameters investigated, cryopreserved whole blood is a good source of viable lymphocytes for biomarker assays in molecular epidemiological studies.
While either T- or B-lymphocytes may be used in biomarker-based molecular epidemiology studies, in practice, T-lymphocytes are more easily maintained through PHA stimulation in cell culture. In contrast, B-lymphocytes have to be obtained by transformation with EB virus, a procedure that has limited success. Whereas T-lymphocytes play a central role in cell-mediated immunity and B-lymphocytes are responsible for the humoral immunity of the adaptive immune system, both types of cells are responsive to antigens through their specific receptors in different ways [102]. More importantly, T-lymphocytes, once stimulated, have all the biologic features that B-lymphocytes do, including DRC and apoptotic response when the damage overwhelms their DRC. Based on a series of studies of cancer susceptibility with DRC measured by the HCR assay using T-lymphocytes from the peripheral blood, the DRC of lymphocytes was shown to accurately reflect the repair capacity of the donor [103]. Given the limited availability of viable biopsies, studies integrating genetic analyses with functional assays in multiple tissues are needed to establish whether genetic analyses can predict function, and whether function in blood cells is representative of function in other tissues.
3.2. Importance of approaches for enhancing estimates of disease risk, treatment effects, and susceptible populations
The role of DRC in cancer susceptibility has been demonstrated in several environmentally induced cancers using blood samples. For example, reduced DRC in T-lymphocytes from blood, based on the HCR assay, is associated with sunlight-induced skin cancers, including non-melanoma (i.e., basal cell and squamous cell carcinomas) [104] and cutaneous melanoma [105]. In these studies, an engineered expression vector harboring well-defined DNA damage, such as thymidine-thymidine induced by UV light exposure, can be transfected into cells and used to monitor the host cells’ nucleotide excision repair pathway. An inherited low level of DRC may explain why some of the exposed people contracted skin cancers but others did not. A similar approach also demonstrated an association between lower DRC and the risk of prostate and breast cancers [106,107]. This DRC assay also detected low-level DRC that is responsible for elevated risk of tobacco-induced lung cancer [108] and head and neck cancer [109]. In these experiments, a tobacco carcinogen, benzo[a]pyrene diol epoxide, was used to form DNA adducts on the expression vector that was transfected into the host cells. A recent genome-wide association study of lung cancer has shown that the DRC-associated risk of lung cancer has a genetic basis that could be detected by genetic variants in blood samples [110]. It should be noted that while these studies provide a foundation for personalized prevention based on inter-individual differences, the studies reach their conclusions by measuring significantly different average repair capacity in case versus control study groups. While the existence of such measurable differences is necessary, for personalized prevention strategies to be feasible, it is not sufficient. In practice, precision prevention demands assays that are sufficiently reproducible to provide information that can be used to reliably assign individual people to risk groups and guide clinical decisions. This higher bar may require improvements to the reproducibility of assays to enable measuring differences between large populations.
3.3. Limitations and barriers to applying DNA repair functional assays in population studies
Blood-based biomarker studies in molecular epidemiology would be ideally performed in population-based case-control studies, particularly in well-defined cohort studies, for example in a nested case-control study. However, for practical reasons, including the timing of blood sample collection, field transportation, sample storage, cell culture, and experimental batch effects in performing the phenotypic assay (including DRC assays that require viable cells), hospital-based case-control studies are preferred. These types of studies allow for consistency in obtaining experimental data that are comparable across batches, particularly for repeated experiments that ideally are performed on the same day under the same experimental conditions, to minimize variation due to laboratory conditions, reagent batch order, and cell culture medium preparation.
In some studies, measurements can be made with minimal sample volume. However, in most other studies the amount of blood available may severely restrict the number of measurements per patient. One question often asked is whether, when considering sample size for such biomarker studies, more subjects should be recruited or more experiments or measurements be made. These considerations should take into account both the financial constraints and the correlation between measurements. For the study design, one approach is to balance the variance and the financial constraints by minimizing the variance to maximize the power, because the power to detect a difference under the alternative is inversely related to the variance [111]. This calls for well-established, reliable phenotypic assays that can be performed consistently with a minimal co-efficient of variation. Another issue is whether DRC in lymphocytes can reflect DRC in target tissues, such as tumors. Considering the influence of tumor microenvironment as well as somatic mutations that may impact DRC in tumors, it is not likely DRC in lymphocytes will reflect DRC in tumors. However, the current application of DRC in lymphocytes has been validated as a biomarker for genetic predisposition to cancer [110].
4. Concluding thoughts
Current and emerging technologies for measuring inter-individual differences in DNA damage and repair, the DNA damage response, and mutagenesis have the potential to make Precision Prevention a reality for many diseases and many people. To realize this potential, the performance factors for the assays used in this effort must include reproducibility and ease-of-use; compatibility must be adequately addressed to allow the transition of DNA repair measures from research laboratories to clinical testing. Many of the assays described above, for example, originated in academic research labs with limited resources or expertise to identify factors that affect technical variance in assay results. These factors can include instrument calibration, environmental fluctuations, human errors, and quality control, among others. Engineering an assay for ease of use, as well as reproducibility, are additional factors that limit the broad use of DNA damage and repair assays. As a practical matter, converting an assay into an easy-to-use design is not supported through most research funding mechanisms, although small business (SBIR) grants do provide a potential path toward this goal. However, SBIR grants require a biotech or commercial partnership, with a small business with the resources to bring such a product to market. Notably, the ability for DNA damage and DRC assays to make large impacts in disease prevention efforts will require widespread testing in a clinical setting, thus necessitating a “kit” and a highly reproducible assay. Once performance factors are validated, broader assay implementation factors including, e.g., manufacturability, shelf-life, commercialization, and health insurance mandates must be addressed, or at least discussed, before measures of DNA damage and repair can impact Precision Prevention and Precision Medicine decisions for specific diseases.
A potential strategy for dealing with issues relating to large-scale assay performance would be for the NIH to engage the Federal Drug Administration (FDA) and industries with proven expertise in developing and using high quality and manufacturer-ready diagnostic assays to help guide assay development. The FDA has years of regulatory experience that could be of great benefit to laboratories with little experience in transitioning a laboratory test into a clinical diagnostic. For applications outside the purview of the FDA, industry partnerships could promote assay development. The engagement of industry early in the assay development pipeline could help to speed up the transition of a laboratory assay to a clinical assay and to provide economic insight into making DNA damage and repair measures mainstream diagnostic tools. Industries that could help support assay transition, as well as benefit from the final product, could include pharmaceutical and biotechnology companies that have expert knowledge in high throughput assay development, Good Laboratory Practice- (GLP-) based expertise that will be needed for a marketed diagnostic, as well as potential products that could be coupled with diagnostic assays. A main conclusion that should be acted upon is that scientists developing useful DNA damage and DRC assays need to engage with experts that have business and manufacturing expertise to transition their assay to a clinically useful product. Similarly, the DDR community must work together with federal entities to continue to show the importance of measuring these factors to highlight their value in Precision Prevention.
The confluence of new high throughput phenotypic assays, advances in DNA sequencing technology, and methods for analyzing large data sets have already begun to revolutionize the practice of medicine. The field of DNA damage, repair and mutagenesis has played a leading role in these advances. The field has also been deeply involved in developing current strategies for predicting disease susceptibility and establishing relationships between environmental exposure and risk at the level of populations, particularly in the context of preventing carcinogenic exposures. We are now poised to refine these strategies to assess individual susceptibility and take key steps toward the practice of precision prevention.
Acknowledgments
Funding
The following authors have funding from the National Institutes of Health: ES019319 to J.H.B.; ES02116 to B.P.E.; ES019492 to T.J.B.; ES019494 to D.J.B.; CA148629 and ES025139 to R.W.S. ES022576 and CA092584 to Z.D.N.; ES011038 to P.J.S. and ES011740 to Q.W. M-B.T. is funded by the Breast Cancer Research Foundation.
Footnotes
Conflict interests
J.H.B. has consultancy, stock ownership, and royalties with Grail Inc. B.P.E. is a co-inventor on a patent for CometChip that has been licensed to Trevigen, Inc. R.W.S. is a scientific consultant for Trevigen, Inc.
References
- 1.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auerbach C. Chemical Mutagenesis. Biol Rev Camb Philos Soc. 1949;24:355–391. doi: 10.1111/j.1469-185x.1949.tb00580.x. [DOI] [PubMed] [Google Scholar]
- 3.HoIlaender A, Emmons CW. Wavelength dependence of mutation production in the ultraviolet with special emphasis on fungi. Cold Spring Harb Symp Quant Biol. 1941:179–186. [Google Scholar]
- 4.Hollaender A, Baker WK, Anderson EH. Effect of oxygen tension and certain chemicals on the x-ray sensitivity of mutation production and survival. Cold Spring Harb Symp Quant Biol. 1951;16:315–326. doi: 10.1101/sqb.1951.016.01.023. [DOI] [PubMed] [Google Scholar]
- 5.Weigle JJ, Bertani G. Multiplicity reactivation of bacteriophage inactivated by ionizing radiations. Virology. 1956;2:344–355. doi: 10.1016/0042-6822(56)90029-0. [DOI] [PubMed] [Google Scholar]
- 6.Ghosal G, Chen J. DNA damage tolerance: a double-edged sword guarding the genome. Transl Cancer Res. 2013;2:107–129. doi: 10.3978/j.issn.2218-676X.2013.04.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson RH, Chien FL, Bleyer A. Incidence of breast cancer with distant involvement among women in the United States, 1976–2009. JAMA. 2013;309:800–805. doi: 10.1001/jama.2013.776. [DOI] [PubMed] [Google Scholar]
- 8.Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends – an update. Cancer Epidemiol Biomark Prev. 2016;25:16–27. doi: 10.1158/1055-9965.EPI-15-0578. [DOI] [PubMed] [Google Scholar]
- 9.Macon MB, Fenton SE. Endocrine disruptors and the breast: early life effects and later life disease. J Mammary Gland Biol Neoplasia. 2013;18:43–61. doi: 10.1007/s10911-013-9275-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoover RN, Hyer M, Pfeiffer RM, Adam E, Bond B, Cheville AL, Colton T, Hartge P, Hatch EE, Herbst AL, Karlan BY, Kaufman R, Noller KL, Palmer JR, Robboy SJ, Saal RC, Strohsnitter W, Titus-Ernstoff L, Troisi R. Adverse health outcomes in women exposed in utero to diethylstilbestrol. N Engl J Med. 2011;365:1304–1314. doi: 10.1056/NEJMoa1013961. [DOI] [PubMed] [Google Scholar]
- 11.Brenner DJ, Shuryak I, Russo S, Sachs RK. Reducing second breast cancers: a potential role for prophylactic mammary irradiation. J Clin Oncol. 2007;25:4868–4872. doi: 10.1200/JCO.2007.11.0379. [DOI] [PubMed] [Google Scholar]
- 12.Pharoah PD, Antoniou AC, Easton DF, Ponder BA. Polygenes, risk prediction, and targeted prevention of breast cancer. N Engl J Med. 2008;358:2796–2803. doi: 10.1056/NEJMsa0708739. [DOI] [PubMed] [Google Scholar]
- 13.Meads C, Ahmed I, Riley RD. A systematic review of breast cancer incidence risk prediction models with meta-analysis of their performance. Breast Cancer Res Treat. 2012;132:365–377. doi: 10.1007/s10549-011-1818-2. [DOI] [PubMed] [Google Scholar]
- 14.Kennedy DO, Agrawal M, Shen J, Terry MB, Zhang FF, Senie RT, Motykiewicz G, Santella RM. DNA repair capacity of lymphoblastoid cell lines from sisters discordant for breast cancer. J Natl Cancer Inst. 2005;97:127–132. doi: 10.1093/jnci/dji013. [DOI] [PubMed] [Google Scholar]
- 15.Machella N, Terry MB, Zipprich J, Gurvich I, Liao Y, Senie RT, Kennedy DO, Santella RM. Double-strand breaks repair in lymphoblastoid cell lines from sisters discordant for breast cancer from the New York site of the BCFR. Carcinogenesis. 2008;29:1367–1372. doi: 10.1093/carcin/bgn140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ostling O, Johanson KJ. Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochem Biophys Res Commun. 1984;123:291–298. doi: 10.1016/0006-291x(84)90411-x. [DOI] [PubMed] [Google Scholar]
- 17.Ostling O, Johanson KJ. Bleomycin, in contrast to gamma irradiation, induces extreme variation of DNA strand breakage from cell to cell. Int J Radiat Biol Relat Stud Phys Chem Med. 1987;52:683–691. doi: 10.1080/09553008714552201. [DOI] [PubMed] [Google Scholar]
- 18.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 19.Ge J, Prasongtanakij S, Wood DK, Weingeist DM, Fessler J, Navasummrit P, Ruchirawat M, Engelward BP. CometChip: a high-throughput 96-well platform for measuring DNA damage in microarrayed human cells. J Vis Exp. 2014:e50607. doi: 10.3791/50607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Redon CE, Dickey JS, Bonner WM, Sedelnikova OA. gamma-H2AX as a biomarker of DNA damage induced by ionizing radiation in human peripheral blood lymphocytes and artificial skin. Adv Space Res. 2009;43:1171–1178. doi: 10.1016/j.asr.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garty G, Chen Y, Salerno A, Turner H, Zhang J, Lyulko O, Bertucci A, Xu Y, Wang H, Simaan N, Randers-Pehrson G, Yao YL, Amundson SA, Brenner DJ. The RABIT: a rapid automated biodosimetry tool for radiological triage. Health Phys. 2010;98:209–217. doi: 10.1097/HP.0b013e3181ab3cb6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turner HC, Sharma P, Perrier JR, Bertucci A, Smilenov L, Johnson G, Taveras M, Brenner DJ, Garty G. The RABiT: high-throughput technology for assessing global DSB repair. Radiat Environ Biophys. 2014;53:265–272. doi: 10.1007/s00411-014-0514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma PM, Ponnaiya B, Taveras M, Shuryak I, Turner H, Brenner DJ. High throughput measurement of gammaH2AX DSB repair kinetics in a healthy human population. PLoS One. 2015;10:e0121083. doi: 10.1371/journal.pone.0121083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Repin M, Turner HC, Garty G, Brenner DJ. Next generation platforms for high-throughput biodosimetry. Radiat Prot Dosim. 2014;159:105–110. doi: 10.1093/rpd/ncu161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagel ZD, Chaim IA, Samson LD. Inter-individual variation in DNA repair capacity: a need for multi-pathway functional assays to promote translational DNA repair research. DNA Repair. 2014;19:199–213. doi: 10.1016/j.dnarep.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagel ZD, Margulies CM, Chaim IA, McRee SK, Mazzucato P, Ahmad A, Abo RP, Butty VL, Forget AL, Samson LD. Multiplexed DNA repair assays for multiple lesions and multiple doses via transcription inhibition and transcriptional mutagenesis. Proc Natl Acad Sci U S A. 2014;111:E1823–1832. doi: 10.1073/pnas.1401182111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doetsch PW. Translesion synthesis by RNA polymerases: occurrence and biological implications for transcriptional mutagenesis. Mutat Res-Fundam Mol Mech Mutagen. 2002;510:131–140. doi: 10.1016/s0027-5107(02)00258-0. [DOI] [PubMed] [Google Scholar]
- 28.Athas WF, Hedayati MA, Matanoski GM, Farmer ER, Grossman L. Development and field-test validation of an assay for DNA-repair in circulating human lymphocytes. Cancer Res. 1991;51:5786–5793. [PubMed] [Google Scholar]
- 29.Li C, Wang LE, Wei Q. DNA repair phenotype and cancer susceptibility – a mini review. Int J Cancer. 2009;124:999–1007. doi: 10.1002/ijc.24126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2. ASM Press; Washington, D.C: 2006. [Google Scholar]
- 31.Ljungman M. Targeting the DNA damage response in cancer. Chem Rev. 2009;109:2929–2950. doi: 10.1021/cr900047g. [DOI] [PubMed] [Google Scholar]
- 32.Tang JB, Svilar D, Trivedi RN, Wang XH, Goellner EM, Moore B, Hamilton RL, Banze LA, Brown AR, Sobol RW. N-methylpurine DNA glycosylase and DNA polymerase beta modulate BER inhibitor potentiation of glioma cells to temozolomide. Neuro Oncol. 2011;13:471–486. doi: 10.1093/neuonc/nor011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rai G, Vyjayanti VN, Dorjsuren D, Simeonov A, Jadhav A, Wilson DM, 3rd, Maloney DJ. Synthesis, biological evaluation, and structure-activity relationships of a novel class of apurinic/apyrimidinic endonuclease 1 inhibitors. J Med Chem. 2012;55:3101–3112. doi: 10.1021/jm201537d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srinivasan A, Wang L, Cline CJ, Xie Z, Sobol RW, Xie XQ, Gold B. Identification and characterization of human apurinic/apyrimidinic endonuclease-1 inhibitors. Biochemistry. 2012;51:6246–6259. doi: 10.1021/bi300490r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beltran H, Eng K, Mosquera JM, Sigaras A, Romanel A, Rennert H, Kossai M, Pauli C, Faltas B, Fontugne J, Park K, Banfelder J, Prandi D, Madhukar N, Zhang T, Padilla J, Greco N, McNary TJ, Herrscher E, Wilkes D, MacDonald TY, Xue H, Vacic V, Emde AK, Oschwald D, Tan AY, Chen Z, Collins C, Gleave ME, Wang Y, Chakravarty D, Schiffman M, Kim R, Campagne F, Robinson BD, Nanus DM, Tagawa ST, Xiang JZ, Smogorzewska A, Demichelis F, Rickman DS, Sboner A, Elemento O, Rubin MA. Whole-exome sequencing of metastatic cancer and biomarkers of treatment response. JAMA Oncol. 2015;1:466–474. doi: 10.1001/jamaoncol.2015.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi S, Gamper AM, White JS, Bakkenist CJ. Inhibition of ATM kinase activity does not phenocopy ATM protein disruption: implications for the clinical utility of ATM kinase inhibitors. ABBV Cell Cycle. 2010;9:4052–4057. doi: 10.4161/cc.9.20.13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen GH, Dexheimer TS, Rosenthal AS, Chu WK, Singh DK, Mosedale G, Bachrati CZ, Schultz L, Sakurai M, Savitsky P, Abu M, McHugh PJ, Bohr VA, Harris CC, Jadhav A, Gileadi O, Maloney DJ, Simeonov A, Hickson ID. A small molecule inhibitor of the BLM helicase modulates chromosome stability in human cells. Chem Biol. 2013;20:55–62. doi: 10.1016/j.chembiol.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aggarwal M, Sommers JA, Shoemaker RH, Brosh RM., Jr Inhibition of helicase activity by a small molecule impairs Werner syndrome helicase (WRN) function in the cellular response to DNA damage or replication stress. Proc Natl Acad Sci U S A. 2011;108:1525–1530. doi: 10.1073/pnas.1006423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sobol RW. Genome instability caused by a germline mutation in the human DNA repair gene POLB. PLoS Genet. 2012;8:e1003086. doi: 10.1371/journal.pgen.1003086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keimling M, Deniz M, Varga D, Stahl A, Schrezenmeier H, Kreienberg R, Hoffmann I, Konig J, Wiesmuller L. The power of DNA double-strand break (DSB) repair testing to predict breast cancer susceptibility. FASEB J. 2012;26:2094–2104. doi: 10.1096/fj.11-200790. [DOI] [PubMed] [Google Scholar]
- 41.Peralta-Leal A, Rodriguez MI, Oliver FJ. Poly(ADP-ribose)polymerase-1 (PARP-1) in carcinogenesis: potential role of PARP inhibitors in cancer treatment. Clin Transl Oncol. 2008;10:318–323. doi: 10.1007/s12094-008-0207-8. [DOI] [PubMed] [Google Scholar]
- 42.Alberts B. Redefining cancer research. Science. 2009;325:1319. doi: 10.1126/science.1181224. [DOI] [PubMed] [Google Scholar]
- 43.Svilar D, Vens C, Sobol RW. Quantitative, real-time analysis of base excision repair activity in cell lysates utilizing lesion-specific molecular beacons. J Vis Exp. 2012:e4168. doi: 10.3791/4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mutamba JT, Svilar D, Prasongtanakij S, Wang XH, Lin YC, Dedon PC, Sobol RW, Engelward BP. XRCC1 and base excision repair balance in response to nitric oxide. DNA Repair (Amst) 2011;10:1282–1293. doi: 10.1016/j.dnarep.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clegg RM. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 1992;211:353–388. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
- 46.Yaron A, Carmel A, Katchalski-Katzir E. Intramolecularly quenched fluorogenic substrates for hydrolytic enzymes. Anal Biochem. 1979;95:228–235. doi: 10.1016/0003-2697(79)90210-0. [DOI] [PubMed] [Google Scholar]
- 47.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 48.Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 49.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 50.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 51.Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge SJ. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci U S A. 2000;97:10389–10394. doi: 10.1073/pnas.190030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 53.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2. ASM Press; 2005. [Google Scholar]
- 54.Seemanova E, Jarolim P, Seeman P, Varon R, Digweed M, Swift M, Sperling K. Cancer risk of heterozygotes with the NBN founder mutation. J Natl Cancer Inst. 2007;99:1875–1880. doi: 10.1093/jnci/djm251. [DOI] [PubMed] [Google Scholar]
- 55.Gowan SM, Hardcastle A, Hallsworth AE, Valenti MR, Hunter LJ, de Haven Brandon AK, Garrett MD, Raynaud F, Workman P, Aherne W, Eccles SA. Application of meso scale technology for the measurement of phosphoproteins in human tumor xenografts. Assay Drug Dev Technol. 2007;5:391–401. doi: 10.1089/adt.2006.044. [DOI] [PubMed] [Google Scholar]
- 56.Liang M, Klakamp SL, Funelas C, Lu H, Lam B, Herl C, Umble A, Drake AW, Pak M, Ageyeva N, Pasumarthi R, Roskos LK. Detection of high- and low-affinity antibodies against a human monoclonal antibody using various technology platforms. Assay Drug Dev Technol. 2007;5:655–662. doi: 10.1089/adt.2007.089. [DOI] [PubMed] [Google Scholar]
- 57.Cao L, Yu Y, Darko I, Currier D, Mayeenuddin LH, Wan X, Khanna C, Helman LJ. Addiction to elevated insulin-like growth factor I receptor and initial modulation of the AKT pathway define the responsiveness of rhabdomyosarcoma to the targeting antibody. Cancer Res. 2008;68:8039–8048. doi: 10.1158/0008-5472.CAN-08-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fichorova RN, Richardson-Harman N, Alfano M, Belec L, Carbonneil C, Chen S, Cosentino L, Curtis K, Dezzutti CS, Donoval B, Doncel GF, Donaghay M, Grivel JC, Guzman E, Hayes M, Herold B, Hillier S, Lackman-Smith C, Landay A, Margolis L, Mayer KH, Pasicznyk JM, Pallansch-Cokonis M, Poli G, Reichelderfer P, Roberts P, Rodriguez I, Saidi H, Sassi RR, Shattock R, Cummins JE., Jr Biological and technical variables affecting immunoassay recovery of cytokines from human serum and simulated vaginal fluid: a multi-center study. Anal Chem. 2008;80:4741–4751. doi: 10.1021/ac702628q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swiger RR, Cosentino L, Shima N, Bielas JH, Cruz-Munoz W, Heddle JA. The cII locus in the MutaMouse system. Environ Mol Mutagen. 1999;34:201–207. [PubMed] [Google Scholar]
- 60.Monroe JJ, Kort KL, Miller JE, Marino DR, Skopek TR. A comparative study of in vivo mutation assays: analysis of hprt, lacI, cII/cI and as mutational targets for N-nitroso-N-methylurea and benzo[a]pyrene in Big Blue mice. Mutat Res. 1998;421:121–136. doi: 10.1016/s0027-5107(98)00171-7. [DOI] [PubMed] [Google Scholar]
- 61.Shima N, Swiger RR, Heddle JA. Dietary restriction during murine development provides protection against MNU-induced mutations. Mutat Res. 2000;470:189–200. doi: 10.1016/s1383-5718(00)00104-2. [DOI] [PubMed] [Google Scholar]
- 62.Eder Budiawan E. Cancer risk assessment for the environmental mutagen and carcinogen crotonaldehyde on the basis of TD(50) and comparison with 1,N(2)-propanodeoxyguanosine adduct levels. Cancer Epidemiol Biomark Prev. 2001;10:883–888. [PubMed] [Google Scholar]
- 63.Miller NA, Farrow EG, Gibson M, Willig LK, Twist G, Yoo B, Marrs T, Corder S, Krivohlavek L, Walter A, Petrikin JE, Saunders CJ, Thiffault I, Soden SE, Smith LD, Dinwiddie DL, Herd S, Cakici JA, Catreux S, Ruehle M, Kingsmore SF. A 26-hour system of highly sensitive whole genome sequencing for emergency management of genetic diseases. Genome Med. 2015;7:100. doi: 10.1186/s13073-015-0221-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 65.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, Chodon T, Nelson SF, McArthur G, Sosman JA, Ribas A, Lo RS. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center’s improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA. Human cancers express a mutator phenotype. Proc Natl Acad Sci U S A. 2006;103:18238–18242. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bielas JH, Loeb LA. Quantification of random genomic mutations. Nat Methods. 2005;2:285–290. doi: 10.1038/nmeth751. [DOI] [PubMed] [Google Scholar]
- 69.Taylor SD, Ericson NG, Burton JN, Prolla TA, Silber JR, Shendure J, Bielas JH. Targeted enrichment and high-resolution digital profiling of mitochondrial DNA deletions in human brain. Aging Cell. 2014;13:29–38. doi: 10.1111/acel.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vermulst M, Bielas JH, Kujoth GC, Ladiges WC, Rabinovitch PS, Prolla TA, Loeb LA. Mitochondrial point mutations do not limit the natural lifespan of mice. Nat Genet. 2007;39:540–543. doi: 10.1038/ng1988. [DOI] [PubMed] [Google Scholar]
- 71.Gravina S, Ganapathi S, Vijg J. Single-cell, locus-specific bisulfite sequencing (SLBS) for direct detection of epimutations in DNA methylation patterns. Nucleic Acids Res. 2015;43:e93. doi: 10.1093/nar/gkv366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saini N, Roberts SA, Klimczak LJ, Chan K, Grimm SA, Dai S, Fargo DC, Boyer JC, Kaufmann WK, Taylor JA, Lee E, Cortes-Ciriano I, Park PJ, Schurman SH, Malc EP, Mieczkowski PA, Gordenin DA. The impact of environmental and endogenous damage on somatic mutation load in human skin fibroblasts. PLoS Genet. 2016;12:e1006385. doi: 10.1371/journal.pgen.1006385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE, Shrager JB, Loo BW, Jr, Alizadeh AA, Diehn M. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lou DI, Hussmann JA, McBee RM, Acevedo A, Andino R, Press WH, Sawyer SL. High-throughput DNA sequencing errors are reduced by orders of magnitude using circle sequencing. Proc Natl Acad Sci U S A. 2013;110:19872–19877. doi: 10.1073/pnas.1319590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schmitt MW, Kennedy SR, Salk JJ, Fox EJ, Hiatt JB, Loeb LA. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A. 2012;109:14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Casbon JA, Osborne RJ, Brenner S, Lichtenstein CP. A method for counting PCR template molecules with application to next-generation sequencing. Nucleic Acids Res. 2011;39:e81. doi: 10.1093/nar/gkr217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gregory MT, Bertout JA, Ericson NG, Taylor SD, Mukherjee R, Robins HS, Drescher CW, Bielas JH. Targeted single molecule mutation detection with massively parallel sequencing. Nucleic Acids Res. 2016;44:e22. doi: 10.1093/nar/gkv915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Johnson N, Fletcher O, Naceur-Lombardelli C, dos Santos Silva I, Ashworth A, Peto J. Interaction between CHEK2*1100delC and other low-penetrance breast-cancer susceptibility genes: a familial study. Lancet. 2005;366:1554–1557. doi: 10.1016/S0140-6736(05)67627-1. [DOI] [PubMed] [Google Scholar]
- 80.Falck J, Mailand N, Syljuasen RG, Bartek J, Lukas J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature. 2001;410:842–847. doi: 10.1038/35071124. [DOI] [PubMed] [Google Scholar]
- 81.Bahassi el M, Penner CG, Robbins SB, Tichy E, Feliciano E, Yin M, Liang L, Deng L, Tischfield JA, Stambrook PJ. The breast cancer susceptibility allele CHEK2*1100delC promotes genomic instability in a knock-in mouse model. Mutat Res. 2007;616:201–209. doi: 10.1016/j.mrfmmm.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 82.Bahassi el M, Robbins SB, Yin M, Boivin GP, Kuiper R, van Steeg H, Stambrook PJ. Mice with the CHEK2*1100delC SNP are predisposed to cancer with a strong gender bias. Proc Natl Acad Sci U S A. 2009;106:17111–17116. doi: 10.1073/pnas.0909237106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Humphries MP, Jordan VC, Speirs V. Obesity and male breast cancer: provocative parallels? BMC Med. 2015;13:134. doi: 10.1186/s12916-015-0380-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brinton LA, Cook MB, McCormack V, Johnson KC, Olsson H, Casagrande JT, Cooke R, Falk RT, Gapstur SM, Gaudet MM, Gaziano JM, Gkiokas G, Guenel P, Henderson BE, Hollenbeck A, Hsing AW, Kolonel LN, Isaacs C, Lubin JH, Michels KB, Negri E, Parisi D, Petridou ET, Pike MC, Riboli E, Sesso HD, Snyder K, Swerdlow AJ, Trichopoulos D, Ursin G, van den Brandt PA, Van Den Eeden SK, Weiderpass E, Willett WC, Ewertz M, Thomas DB European Rare Cancer Study G. Anthropometric and hormonal risk factors for male breast cancer: male breast cancer pooling project results. J Natl Cancer Inst. 2014;106:djt465. doi: 10.1093/jnci/djt465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brinton LA, Key TJ, Kolonel LN, Michels KB, Sesso HD, Ursin G, Van Den Eeden SK, Wood SN, Falk RT, Parisi D, Guillemette C, Caron P, Turcotte V, Habel LA, Isaacs CJ, Riboli E, Weiderpass E, Cook MB. Prediagnostic sex steroid hormones in relation to male breast cancer risk. J Clin Oncol. 2015;33:2041–2050. doi: 10.1200/JCO.2014.59.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Meyer SE, Peace BE, Bahassi el M, Kavanaugh GM, Wagh PK, Robbins SB, Yin M, Wells SI, Zinser GM, Stambrook PJ, Waltz SE. Chk2*1100delC acts in synergy with the Ron receptor tyrosine kinase to accelerate mammary tumorigenesis in mice. Cancer Lett. 2010;296:186–193. doi: 10.1016/j.canlet.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Varga SJ, Qi C, Podolsky E, Lee D. A new Drosophila model to study the interaction between genetic and environmental factors in Parkinson’s disease. Brain Res. 2014;1583:277–286. doi: 10.1016/j.brainres.2014.08.021. [DOI] [PubMed] [Google Scholar]
- 88.Na J, Musselman LP, Pendse J, Baranski TJ, Bodmer R, Ocorr K, Cagan R. A Drosophila model of high sugar diet-Induced cardiomyopathy. PLoS Genet. 2013;9:e1003175. doi: 10.1371/journal.pgen.1003175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Leung MCK, Williams PL, Benedetto A, Au C, Helmcke KJ, Aschner M, Meyer JN. Caenorhabditis elegans: an emerging model in biomedical and environmental toxicology. Toxicol Sci. 2008;106:5–28. doi: 10.1093/toxsci/kfn121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 91.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 92.Zhang R, Takebe T, Sekine K, Koike H, Zheng Y, Taniguchi H. Identification of proliferating human hepatic cells from human induced pluripotent stem cells. Transplant Proc. 2014;46:1201–1204. doi: 10.1016/j.transproceed.2013.12.021. [DOI] [PubMed] [Google Scholar]
- 93.Takebe T, Taniguchi H. Human iPSC-Derived miniature organs: a tool for drug studies. Clin Pharmacol Ther. 2014;96:310–313. doi: 10.1038/clpt.2014.110. [DOI] [PubMed] [Google Scholar]
- 94.Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Charpentier E, Doudna JA. Biotechnology: rewriting a genome. Nature. 2013;495:50–51. doi: 10.1038/495050a. [DOI] [PubMed] [Google Scholar]
- 96.Ku CS, Loy EY, Pawitan Y, Chia KS. The pursuit of genome-wide association studies: where are we now? J Hum Genet. 2010;55:195–206. doi: 10.1038/jhg.2010.19. [DOI] [PubMed] [Google Scholar]
- 97.Azqueta A, Slyskova J, Langie SA, O’Neill Gaivao I, Collins A. Comet assay to measure DNA repair: approach and applications. Front Genet. 2014;5:288. doi: 10.3389/fgene.2014.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lockett KL, Hall MC, Clark PE, Chuang SC, Robinson B, Lin HY, Su LJ, Hu JJ. DNA damage levels in prostate cancer cases and controls. Carcinogenesis. 2006;27:1187–1193. doi: 10.1093/carcin/bgi288. [DOI] [PubMed] [Google Scholar]
- 99.Smith TR, Miller MS, Lohman KK, Case LD, Hu JJ. DNA damage and breast cancer risk. Carcinogenesis. 2003;24:883–889. doi: 10.1093/carcin/bgg037. [DOI] [PubMed] [Google Scholar]
- 100.Valdiglesias V, Giunta S, Fenech M, Neri M, Bonassi S. γH2AX as a marker of DNA double strand breaks and genomic instability in human population studies. Mutat Res/Rev Mutat Res. 2013;753:24–40. doi: 10.1016/j.mrrev.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 101.Cheng L, Wang LE, Spitz MR, Wei Q. Cryopreserving whole blood for functional assays using viable lymphocytes in molecular epidemiology studies. Cancer Lett. 2001;166:155–163. doi: 10.1016/s0304-3835(01)00400-1. [DOI] [PubMed] [Google Scholar]
- 102.Miller JF, Sadelain M. The journey from discoveries in fundamental immunology to cancer immunotherapy. Cancer Cell. 2015;27:439–449. doi: 10.1016/j.ccell.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 103.Li C, Wang LE, Wei Q. DNA repair phenotype and cancer susceptibility – a mini review. Int J Cancer. 2009;124:999–1007. doi: 10.1002/ijc.24126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang LE, Li C, Strom SS, Goldberg LH, Brewster A, Guo Z, Qiao Y, Clayman GL, Lee JJ, El-Naggar AK, Prieto VG, Duvic M, Lippman SM, Weber RS, Kripke ML, Wei Q. Repair capacity for UV light induced DNA damage associated with risk of nonmelanoma skin cancer and tumor progression. Clin Cancer Res. 2007;13:6532–6539. doi: 10.1158/1078-0432.CCR-07-0969. [DOI] [PubMed] [Google Scholar]
- 105.Wei Q, Lee JE, Gershenwald JE, Ross MI, Mansfield PF, Strom SS, Wang LE, Guo Z, Qiao Y, Amos CI, Spitz MR, Duvic M. Repair of UV light-induced DNA damage and risk of cutaneous malignant melanoma. J Natl Cancer Inst. 2003;95:308–315. doi: 10.1093/jnci/95.4.308. [DOI] [PubMed] [Google Scholar]
- 106.Hu JJ, Hall MC, Grossman L, Hedayati M, McCullough DL, Lohman K, Case LD. Deficient nucleotide excision repair capacity enhances human prostate cancer risk. Cancer Res. 2004;64:1197–1201. doi: 10.1158/0008-5472.can-03-2670. [DOI] [PubMed] [Google Scholar]
- 107.Matta J, Echenique M, Negron E, Morales L, Vargas W, Gaetan FS, Lizardi ER, Torres A, Rosado JO, Bolanos G, Cruz JG, Laboy J, Barnes R, Medina SS, Romero A, Martinez R, Dutil J, Suarez E, Alvarez-Garriga C, Bayona M. The association of DNA Repair with breast cancer risk in women. A comparative observational study. BMC Cancer. 2012;12:490. doi: 10.1186/1471-2407-12-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wei Q, Cheng L, Amos CI, Wang LE, Guo Z, Hong WK, Spitz MR. Repair of tobacco carcinogen-induced DNA adducts and lung cancer risk: a molecular epidemiologic study. J Natl Cancer Inst. 2000;92:1764–1772. doi: 10.1093/jnci/92.21.1764. [DOI] [PubMed] [Google Scholar]
- 109.Wang LE, Hu Z, Sturgis EM, Spitz MR, Strom SS, Amos CI, Guo Z, Qiao Y, Gillenwater AM, Myers JN, Clayman GL, Weber RS, El-Naggar AK, Mao L, Lippman SM, Hong WK, Wei Q. Reduced DNA repair capacity for removing tobacco carcinogen-induced DNA adducts contributes to risk of head and neck cancer but not tumor characteristics. Clin Cancer Res. 2010;16:764–774. doi: 10.1158/1078-0432.CCR-09-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang LE, Gorlova OY, Ying J, Qiao Y, Weng SF, Lee AT, Gregersen PK, Spitz MR, Amos CI, Wei Q. Genome-wide association study reveals novel genetic determinants of DNA repair capacity in lung cancer. Cancer Res. 2013;73:256–264. doi: 10.1158/0008-5472.CAN-12-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lai D, King TM, Moye LA, Wei Q. Sample size for biomarker studies: more subjects or more measurements per subject? Ann Epidemiol. 2003;13:204–208. doi: 10.1016/s1047-2797(02)00261-2. [DOI] [PubMed] [Google Scholar]