Knockdown of microRNA166 increases drought resistance in rice via decreased stem water conductivity and reduced leaf transpiration rates.
Abstract
MicroRNAs are 19- to 22-nucleotide small noncoding RNAs that have been implicated in abiotic stress responses. In this study, we found that knockdown of microRNA166, using the Short Tandem Target Mimic (STTM) system, resulted in morphological changes that confer drought resistance in rice (Oryza sativa). From a large-scale screen for miRNA knockdown lines in rice, we identified miR166 knockdown lines (STTM166); these plants exhibit a rolled-leaf phenotype, which is normally displayed by rice plants under drought stress. The leaves of STTM166 rice plants had smaller bulliform cells and abnormal sclerenchymatous cells, likely causing the rolled-leaf phenotype. The STTM166 plants had reduced stomatal conductance and showed decreased transpiration rates. The STTM166 lines also exhibited altered stem xylem and decreased hydraulic conductivity, likely due to the reduced diameter of the xylem vessels. Molecular analyses identified rice HOMEODOMAIN CONTAINING PROTEIN4 (OsHB4), a member of HD-Zip III gene family, as a major target of miR166; moreover, rice plants overexpressing a miR166-resistant form of OsHB4 resembled the STTM166 plants, including leaf rolling and higher drought resistance. The genes downstream of miR166-OsHB4 consisted of polysaccharide synthesis-related genes that may contribute to cell wall formation and vascular development. Our results suggest that drought resistance in rice can be increased by manipulating miRNAs, which leads to developmental changes, such as leaf rolling and reduced diameter of the xylem, that mimic plants’ natural responses to water-deficit stress.
Leaf architecture, an agriculturally important trait contributing to rice (Oryza sativa) productivity, modulates photosynthesis and thus regulates plant growth and development. The growth of leaves initiates at the peripheral zone of the shoot apical meristem and then proceeds in a three-dimensional manner, moving from proximal to distal, adaxial to -abaxial, and medial to lateral (Moon and Hake, 2011). Leaves often change their architecture to cope with changes in the environment such as light intensity, moisture, temperature, etc., and moderate leaf rolling can enhance rice yield in multiple ways (Sakamoto et al., 2006; Lang et al., 2003; Eshed et al., 2001; Wu, 2009; Yan et al., 2012a). Rolled leaves maintain their upright posture, thus minimizing shadowing among leaves and improving overall photosynthetic efficiency of the plants. Leaf rolling also decreases stomatal conductance and reduces water loss, especially under drought-stress conditions. Moreover, leaf rolling delays the onset of senescence, thus increasing source-to-sink translocation of photoassimilates. Leaf rolling is a spontaneous response of plants to environmental conditions for maximizing productivity, and identification of the genetic component that modulates leaf rolling may provide tools to enhance rice yield under stress conditions.
Several genetic regulators of leaf rolling have been identified in rice, and some of the recently identified ones include ROLLED LEAF9/SHALLOT-LIKE1 (Zhang et al., 2009), SHALLOT-LIKE2 (Zhang et al., 2015), ROLLED AND ERECT LEAF1 (Chen et al., 2015), ROLLED AND ERECT LEAF2 (Yang et al., 2016), SEMI-ROLLED LEAF1 (Xiang et al., 2012), SEMI-ROLLED LEAF2 (Liu et al., 2016), ROLLING LEAF14 (Fang et al., 2012), and LATERAL ORGAN BOUNDARIES DOMAIN GENE (Li et al., 2016b). Mutants of these genes show either adaxially (inward) or abaxially (outward) rolled-leaf phenotypes, which is usually associated with altered development of bulliform cells. The bulliform cells are a group of large and highly vacuolated cells that mainly exist in monocots and play a crucial role in modulating the status of leaf rolling, as bulliform cells lose turgor under water-deficit conditions, which results in leaf rolling. When water deficit is relieved, bulliform cells absorb water and swell up, thus flattening the leaves (Price et al., 1997).
At the molecular level, leaf shape in monocots, either flat or rolled, is dependent upon the antagonistic expression patterns of transcription factors and small RNAs targeting these transcription factors at adaxial (top) and abaxial (bottom) sides that are functionally specialized for light capture and gas exchange, respectively (Moon and Hake, 2011; Merelo et al., 2016). Homeodomain Leu zipper III transcription factors (HD-ZIP III, a group of homeobox transcription factors) such as PHABULOSA, PHAVOLUTA (McConnell et al., 2001), REVOLUTA (Otsuga et al., 2001), and ROLLED1 (Nelson et al., 2002) determine the adaxial polarity, while members of KANAD I family, such as MILK-WEED POD1 (Candela et al., 2008) and YABBY (YAB) family such as YAB2 and YAB3 (Eshed et al., 2004), determine the development of abaxial cells. Two small RNAs, the transacting short interfering RNAs TAS3 and the microRNA miR165/166, exhibit opposite polar distribution and facilitate the establishment of adaxial-abaxial leaf polarity, including proper development of leaf vasculature (Nogueira et al., 2007). The miR165/166 family members regulate leaf development by targeting HD-ZIP III genes (Fouracre and Poethig, 2016). The HD-Zip III family in rice consists of five genes: Oshox10, Oshox9, Oshox33, Oshox32, and Oshox29, which are also named OSHB1 to OSHB5, respectively (Agalou et al., 2008).
The plant vascular system is composed of xylem and phloem, which are specialized for the transport of water and photoassimilates, respectively. In leaves, xylem is present at the adaxial side and phloem is positioned abaxially in the vascular bundles. The rolled leaf phenotype is directly related to water transport through the xylem. Xylem vessels/conduits mainly consist of tracheary elements, which undergo complete clearance of cellular components through programmed cell death during cell differentiation, forming an apoplastic route to transport water and solutes. In addition, development of a cellulose-rich thick secondary cell wall provides mechanical support to the plants (De Rybel et al., 2016; Heo et al., 2017).
Since xylem conduits are connected through dead cell wall skeletons, the driving force for water/solute transport is mainly mediated by transpiration. Differences in the diameter of xylem vessels are regarded as some of the most important parameters in plant-water relations. According to the Hagen-Poiseuille equation, the flow rate of water along a tube is proportional to the fourth power of its radius. Thus, a small change in vessel diameter can result in a considerable change in flow rate or conductivity. Xylem area-specific conductivity (Ks) can be considered as the proxy for transport efficiency and can strongly impact plant responses to freezing-induced embolism or drought-induced cavitation (McCulloh et al., 2010; Hacke et al., 2017). Owing to its direct relationship with transpiration, vascular transport is also dependent on stomatal density and the status of stomatal opening/closing.
Recently, a large-scale knockdown of different miRNA families in rice was attempted using the Short Tandem Target Mimic (STTM) technology, and transgenic plants were phenotyped under natural paddy field conditions (Zhang et al., 2017). In this study, we characterized the miR166 knockdown plants (STTM166), which exhibit rolled leaves, smaller bulliform cells in the leaves, and altered xylem vessels in the stem. The STTM166 plants displayed an elevated drought resistance likely due to a reduced transpiration rate. MiR166 was localized in the xylem vessels. Overexpression of a miR166-resistant version of OsHB4 resulted in phenotypes resembling those of the STTM166 plants, which supports the idea that OsHB4 is one of the major targets of miR166 in rice. RNA-seq-based transcriptomic analyses revealed that the expression of cell wall biosynthesis-related genes was altered in STTM166 plants. Our study provided genetic evidence supporting the role of miR166-OsHB4 as a regulatory module in xylem development, leaf rolling, and drought resistance in rice.
RESULTS
STTM166 Plants Exhibit a Rolled-Leaf Phenotype
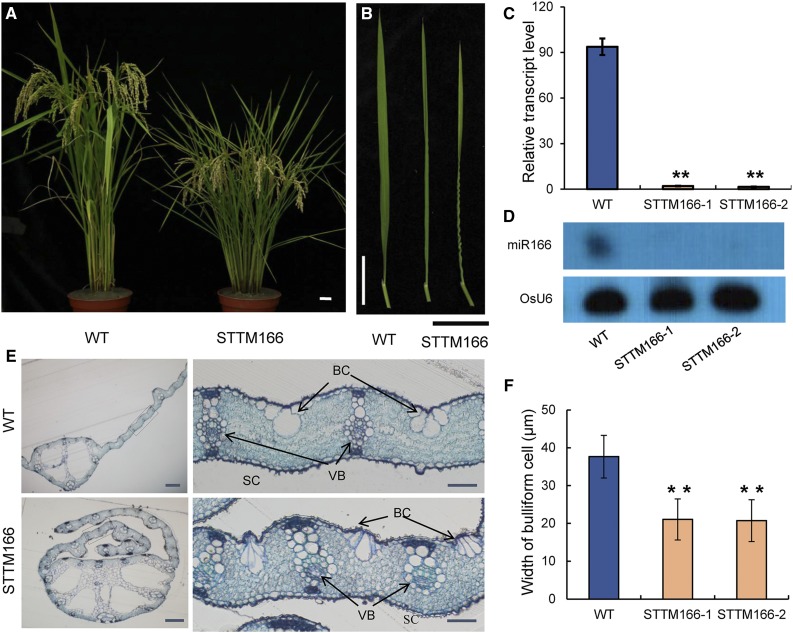
To identify miRNAs that modulate plant morphology and other agronomic traits, we selected 37 different miRNA families in rice and generated knockdown lines using the STTM technology (Tang et al., 2012; Yan et al., 2012b). Phenotyping of the STTM plants grown under natural paddy field conditions revealed that miR166 knockdown plants (STTM166 lines) exhibited multiple phenotypic alterations, including alterations in plant height, leaf morphology, and seed size. At maturity, plant height was reduced by 33.9% in STTM166 plants compared with that in the wild type (Fig. 1A; Supplemental Fig. S1A). The reduced height in STTM166 was associated with decreases in the length of the first, second, and third internodes by 36.5%, 43.4%, and 49.5%, respectively, but the length of the fourth internode did not show a significant difference between STTM166 and wild-type plants (Supplemental Fig. S1C). Although the seed size and weight were changed in STTM166 plants (Supplemental Fig. S1, D–G), the rates of seed setting were comparable between STTM166 and the wild type, which suggests that miR166 knockdown did not affect sexual reproduction. The grain yield per plant was reduced by 16.5% and 23.2%, respectively, in STTM166-1 and STTM166-2 when compared with the wild type under normal growth conditions (Supplemental Fig. S2A). Another prominent feature of STTM166 plants is adaxially rolled leaves (Fig. 1B), which is a typical phenotype of the class III HD-Zip mutants (Emery et al., 2003; Juarez et al., 2004). The expression level of miR166 was quantified in two independent lines of STTM166 plants using stem-loop quantitative reverse-transcript PCR (Fig. 1C) and small RNA northern blot analysis (Fig. 1D), and both methods detected a significant reduction in the mature form of miR166 in two independent STTM166 lines. These results indicate that the observed phenotypes of STTM166 plants is attributed to the down-regulation of miR166.
Figure 1.
Morphological and growth phenotypes of STTM166 plants. A, Wild-type and STTM166 plants at maturity. Scale bar, 2 cm. B, The rolled-leaf phenotype of STTM166 plants at maturity. C, Expression of mature miR166 in wild-type and STTM166 plants assayed by using quantitative stem-loop real-time PCR. D, The expression levels of mature miR166 in wild-type and STTM166 plants detected using RNA gel blotting. U6 gene served as a loading control. E, The cross section of leaf in wild-type and STTM166 plants at maturity. BC, Bulliform cell; VB, vascular bundle; SC, sclerenchymatous cell. Scale bar represents 200 μm in leaf and 50 μm in blade. F, The width of bulliform cells in leaf adaxial epidermis. Data are presented as means ± sd (n = 3 in E and F). **P < 0.01, two-tailed, two-sample Student’s t test.
Anatomical analysis of leaves was performed to analyze the changes in STTM166 plants at the cellular and tissue levels. The adaxial rolling was clearly evidenced by the circularly shaped cross section of a leaf in the STTM166 plant (Fig. 1E, left). In contrast to the wild type, some of the small veins in the laterally rolled region did not form normal sclerenchymatous cells at the abaxial side (Fig. 1E, right). However, the midrib region and most of the secondary veins in STTM166 leaves showed a cellular organization similar to that in the wild type. The bulliform cells at adaxial side in STTM166 plants were deflated (Fig. 1E, right). Since the size and number of bulliform cells vary depending on their locations in the leaf, we selected the bulliform cells adjacent to the midrib and measured the width of the bulliform cells. The average width of bulliform cells was reduced by 43.9% in STTM166 leaves as compared with that in the wild type (Fig. 1F). Thus, both abnormality of sclerenchyma cells and deflated bulliform cells likely contribute to the adaxially rolled leaf phenotype of STTM166 plants.
STTM166 Plants Display Reduced Transpiration and Enhanced Drought Resistance
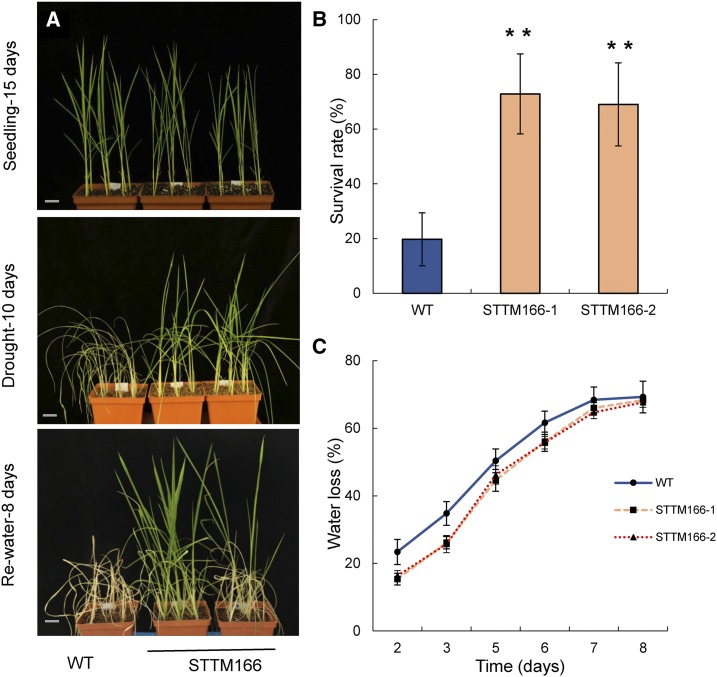
Since leaf rolling is considered as an adaptive mechanism to conserve water (O’Toole and Cruz, 1980; Zhang et al., 2009), we asked whether STTM166 plants may be more resistant to drought stress. Drought resistance assays were performed using pot-grown plants in a phytotron growth chamber. Seedlings were established under normal growth conditions for 15 d, and drought was then initiated by withholding water. In wild-type plants, leaf rolling appeared on the fifth day after withholding water, and the leaves became wilted on the eighth day, whereas STTM166 leaves did not wilt during the 10-d drought treatment (Fig. 2A; Supplemental Fig. S3). On the 10th day after the drought-stress treatment, recovery of plants was attempted by rewatering, and the survival rate was recorded on the eighth day after rewatering. Plants exhibiting growth of new leaves were considered as surviving plants. The survival rate was increased by 53.2% and 49.3% in STTM166-1 and STTM166-2, respectively, as compared with that in wild type (Fig. 2B). In the paddy field, while the spikelet fertility, a main indicator for drought resistance, of STTM lines was unchanged when compared with the wild type under normal growth conditions, the STTM 166 lines displayed significantly higher spikelet fertility than that of wild-type plants under drought-stress conditions (Supplemental Fig. S2, B and C). During the drought-stress treatment, water loss was also monitored, and the result shows that STTM166 plants lost water more slowly than the wild type (Fig. 2C).
Figure 2.
The drought resistance phenotype of STTM166 plants. A, Drought resistance assay in wild-type and STTM166 plants. Scale bar, 2 cm. B, Survival rate after drought and rewatering treatments. C, The water loss during drought stress. Data are presented as means ± sd (n = 36 in B and C). **P < 0.01, two-tailed, two-sample Student’s t test.
To corroborate the water loss data, we examined leaf surface temperature using infra-red imaging. Under normal growth conditions in a phytotron growth chamber, the surface temperature of STTM166 leaves was higher than that of wild type (Fig. 3A), suggesting reduced transpiration from leaves in STTM166 plants. Moreover, in the paddy field conditions, the transpiration rate was consistently lower (P < 0.001) in STTM166 plants than in wild type at the three time points tested during the day (Fig. 3B). Since transpiration is tightly associated with stomata, we quantified the density and size of stomata. The stomatal density was increased by 23.4% and 16.8% (Supplemental Fig. S4, A and B), while stomata size was decreased by 49.3% and 50.2% (Supplemental Fig. S5, A and B) in STTM166-1 and STTM166-2, respectively, as compared with those in wild-type plants. Under normal growth conditions, the stomatal conductance was significantly reduced (Fig. 3C), while leaf water potential remained unchanged in these two STTM lines when compared with the wild type (Fig. 3D). However, the STTM166 lines exhibited significantly higher leaf water potential than wild type under drought-stress conditions (Fig. 3D). In both wild-type and STTM166 plants, there was no difference in stomatal density and size between adaxial and abaxial surface of leaf blade (Supplemental Fig. S4, A and C). Measurements of ABA contents indicated that STTM166 and wild-type plants had comparable ABA levels under both normal and drought-stress conditions (Supplemental Fig. S6, A and B). These results suggest that increased leaf water potential and decreased transpiration rate in STTM166 plants were not mediated by changes in ABA content but were more likely due to the morphological changes in leaves and stems of STTM166 plants.
Figure 3.
Leaf transpiration rate and leaf water potential in STTM166 plants. A, The surface temperature of wild-type and STTM166 leaves. Scale bar, 2 cm. B, The transpiration rates of wild-type and STTM166 plants grown in a paddy field. C, The stomatal conductance of STTM166 and wild-type plants under normal growth conditions in paddy field. D, The leaf water potential of STTM166 and wild-type plants under normal and drought-stress conditions. Data are presented as means ± sd (n = 5 in B–D). **P < 0.01, two-tailed, two-sample Student’s t test.
miR166 Regulates Stem Vasculature and Hydraulic Conductivity
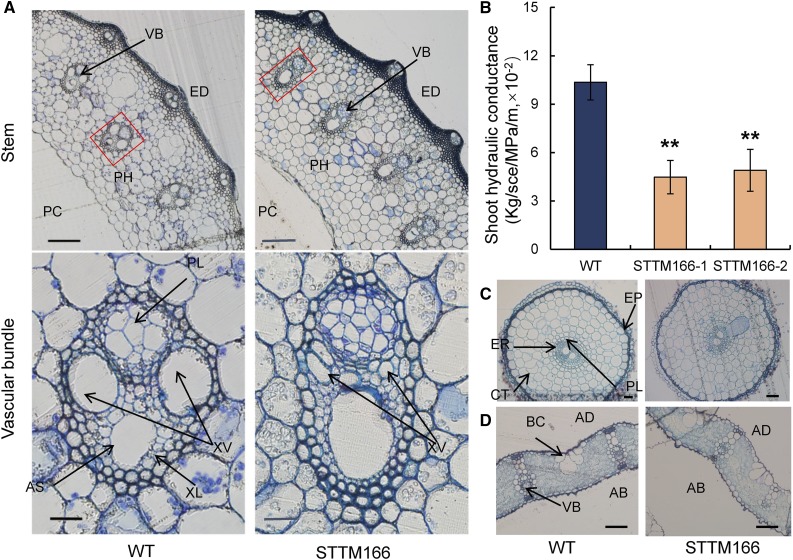
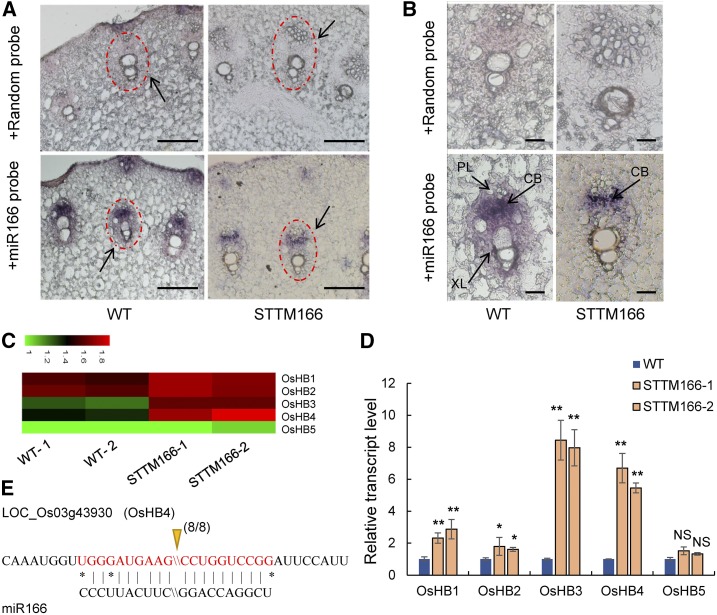
Our anatomical analyses also revealed structural changes in the vascular bundle of stems of STTM166 plants. The diameter of xylem vessels was significantly decreased in STTM166 stems as compared with those in the wild type (Fig. 4A). The decreased xylem area in STTM166 was expected to result in changes in shoot hydraulic conductivity (Kshoot). Indeed, measurements of Kshoot, which were normalized by the stem-supporting leaf area (Brodribb and Feild, 2000), by using a high-pressure flow meter (HPFM) revealed that, under both phytotron growth chamber and paddy field conditions, Kshoot was decreased by 56.8% and 52.7% in STTM166-1 and STTM166-2, respectively, as compared to those in the wild type (Fig. 4B). Interestingly, as a part of the interconnected vascular system, the vascular bundles of root (Fig. 4C) and leaf (Fig. 4D) did not show significant morphological differences between STTM166 and wild-type plants under normal growth conditions. These results suggest that miR166 plays an important role in the vascular development in the stem. This notion was further supported by the study of miR166 localization. By in situ hybridization using an LNA-miR166 antisense probe, miR166 was detected in the stem vascular bundle in wild-type plants, including the phloem and cambium (Fig. 5A), and cambium cells displayed the highest level of miR166, which might be associated with the origin of miR166 precursor formation. The formation of miRNA precursor in meristematic cells and mobility of mature miRNA have been demonstrated earlier (Tretter et al., 2008; Chitwood et al., 2009). In STTM166 plants, only a weak miR166 signal was detected in the cambium (Fig. 5, A and B). Furthermore, the expression of five selected aquaporin genes in the root was analyzed, and no significant difference was observed between STTM166 and wild-type plants (Supplemental Fig. S7). This suggests that the decreased transpiration rate was unlikely due to differences in water uptake in roots of STTM166 plants.
Figure 4.
Cross sections of stem, root, and leaf and hydraulic conductivity through the shoot of wild-type and STTM166 plants. A, The cross sections of stem and the vascular bundle in wild-type and STTM166-1 plants. Scale bar represents 200 μm in stem and 20 μm in vascular bundle. VB, Vascular bundle; ED, epidermis; PH, pith (parenchyma); PC, pulp cavity; XV, xylem vessel; AS, air space; PL, phloem; XL, xylem. B, Water conductivity through the shoot of wild-type and STTM166 plants. “kg/s/MPa/m” represents Kshoot, the shoot hydraulic conductivity per unit leaf area. C, Cross sections of roots of wild-type and STTM166 plants. Scale bar, 50 μm. PL, Pericycle; CT, cortex; ER, endodermis; EP, epidermis. D, Cross sections of leaves of wild-type and STTM166 plants. Scale bar, 50 μm. BC, bulliform cell; AD, adaxial surface; AB, abaxial surface; VB, vascular bundle. Data are presented as means ± sd (n = 5 in B). **P < 0.01, two-tailed, two-sample Student’s t test.
Figure 5.
In situ hybridization of miR166 and the expression profiles of OsHB genes. A, In situ hybridization of miR166 in stem of STTM166-1 and wild-type plants. Scale bar, 200 μm. Black arrow indicates vascular bundle. B, Magnified images of vascular bundles after in situ hybridization. Scale bar, 20 μm. XL, Xylem; PL, phloem; CB, cambium. C, The heat map of expression patterns of OsHB genes from the RNA-seq data. D, The expression profiles of OsHB genes in STTM166 and wild-type plants. E, The cleavage site of OsHB4 mRNAs mediated by miR166. Solid arrow indicates the frequency of RACE clones corresponding to the site. Data are presented as means ± sd (n = 3 in D). **P < 0.01, two-tailed, two-sample Student’s t test.
A Subset of HD-Zip III Genes Are Regulated by miR166
In Arabidopsis (Arabidopsis thaliana), miR165/166 is predicted to target the transcripts of HD-Zip III genes (Floyd and Bowman, 2004; Mallory et al., 2004). The sequences of miR165 and miR166 in Arabidopsis have only one nucleotide difference, and both miRNAs are thought to target the same genes (Rhoades et al., 2002). miR166, but not miR165, has been reported in rice, and all five OsHB genes were found to contain the miRNA166 binding sequence and thus could be the targets of miR166 (Nagasaki et al., 2007; Luo et al., 2013). However, experimental evidence for miR166-mediated cleavage of OsHB genes is still lacking.
Gene expression analysis revealed that all OsHB genes were highly expressed in leaf and OsHB1–4, but not OsHB5, were also abundantly expressed in stem (Supplemental Fig. S8A). OsHB1 and OsHB4 were induced, while OsHB2, OsHB3, and OsHB5 were suppressed by drought-stress treatment (Supplemental Fig. S8B). To identify which OsHB gene is mainly responsible for the phenotypes of STTM166 plants, we examined the expression levels of the five OsHB genes from the RNA-seq data in STTM166 and wild-type plants. The expression levels of OsHB3 and OsHB4 were significantly higher, while the expression levels of OsHB1, OsHB2, and OsHB5 were either unchanged or only moderately increased in STTM166 leaves as compared with those in the wild type (Fig. 5C). The RNA-seq data were further validated by quantitative real-time PCR (Fig. 5D). These results suggest that OsHB3 and OsHB4 could be the main targets of miR166 in rice. This is consistent with the finding that OsHB3 and OsHB4 expression levels are comparatively higher in stem than in any other tissues (Itoh et al., 2008). The modified RLM-RACE method was used to detect cleavage in all five OsHB genes, and cleavage in only OsHB4 transcripts was confirmed (Fig. 5E). The mapped cleavage site was found to be identical to those reported in Arabidopsis (Emery et al., 2003; Tang et al., 2003). Therefore, OsHB4 could be a direct target of miR166 and regulate vascular development in the stem of rice.
Overexpression of miR166-Resistant OsHB4 Phenocopies STTM166 Plants
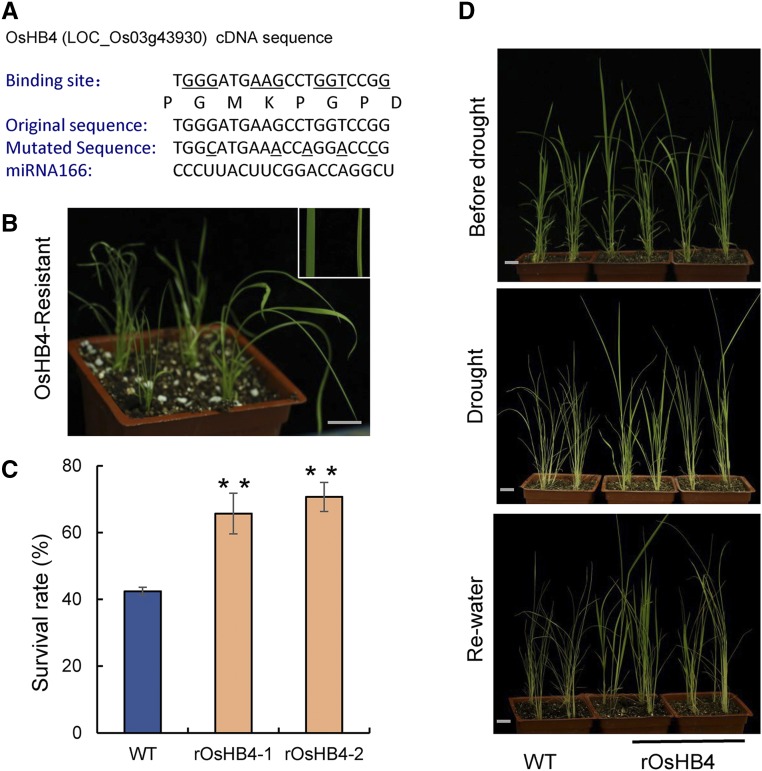
The miR166-mediated morphological changes and drought resistance through the regulation of OsHB4 were further studied by constructing and overexpressing a miR166-resistant version of OsHB4 (rOsHB4) in rice (Fig. 6A). At the seedling stage, several independent transgenic lines displayed an adaxially rolled leaf phenotype (Fig. 6B). Additionally, rOsHB4 overexpression also led to improved drought resistance in rice (Fig. 6D). The survival rates after 10 d of drought treatment increased by 22.3% and 28.3% in two independent rOsHB4 overexpression lines, rOsHB4-1 and rOsHB4-2, respectively, as compared with that in the wild type (Fig. 6C).
Figure 6.
The phenotypes of rOsHB4 overexpression plants. A, The mutations generated at the miR166 binding site of OsHB4 gene to create miR166-resistant OsHB4 (rOsHB4). B, The rolled-leaf phenotype of the rOsHB4-overexpression transgenic seedlings. C, Survival rate after drought and rewatering treatments. D, Drought resistance phenotype of the rOsHB4 transgenic lines. Scale bar, 2 cm. Data are presented as means ± sd (n = 36 in C). **P < 0.01, two-tailed, two-sample Student’s t test.
A mutant with a single-nucleotide deletion in the OsHB4 gene generated using CRISPR/Cas9 and plants with overexpression of miR166 (OE-MIR166) using the miR166 precursor sequence were also generated and phenotyped. However, neither the Oshb4 mutant nor the OE-MIR166 plants showed significant differences in leaf and vasculature morphology and development when compared with the wild type (Supplemental Fig. S9).
Transcriptomic Analysis Identifies miR166-Regulated Genes
MiR166 is known to regulate the class III HD-Zip genes encoding a family of transcription factors that mediate downstream functional genes participating in organ and vascular development or meristem maintenance (Ariel et al., 2007). To identify genes with altered expression levels in the STTM166 plants compared with those in the wild type, RNA sequencing and transcriptomic analysis were performed. Interestingly, the well-known abiotic stress-related marker genes such as PEROXIDASE RELATED GENES, STRESS-RESPONSE NAC, and NAM, ATAF1/2, AND CUC2 (Hou et al., 2009) were not changed in the STTM166 lines compared to those in the wild type (Supplemental Table S1).
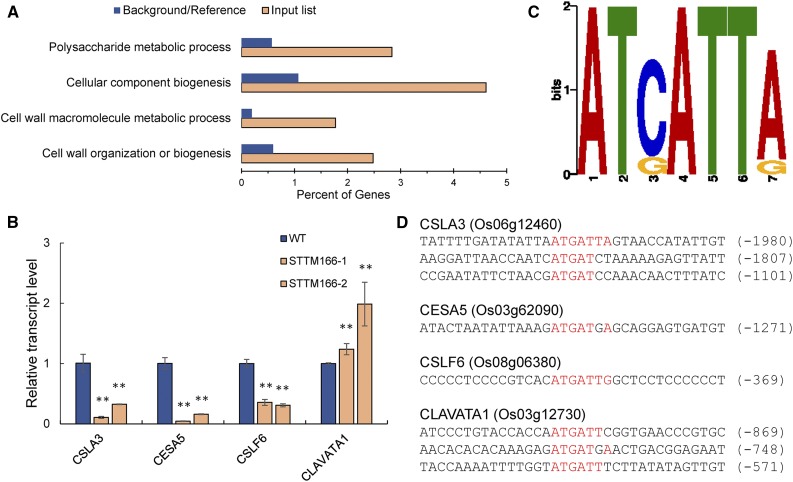
The up- and down-regulated genes in STTM166 lines were subjected to gene ontology (GO) analysis using AgriGO (Du et al., 2010). Among the differentially expressed genes, those involved in cell wall organization or biogenesis and polysaccharide metabolic process were strongly enriched (Fig. 7A). For example, these included several polysaccharide synthase genes and a CLAVATA1-related gene, which are known to be associated with cell wall biogenesis (Gardiner et al., 2003; Watanabe et al., 2015; Diévart et al., 2003; Supplemental Table S2). Significant changes in the expression levels of these three polysaccharide synthase genes and the CLAVATA1-related gene are shown in the heat map (Supplemental Fig. S10) and further confirmed by RT-qPCR (Fig. 7B).
Figure 7.
Differentially expressed genes in STTM166 lines. A, Enrichment of selected Gene Ontology (GO) categories of differentially expressed genes by RNA-seq analysis. Plotted is the percentage of genes (x axis) identified in the differentially expressed genes by RNA-seq analysis (diagonal bars) in comparison with the abundance in the whole transcriptome (black bars) for three significantly enriched GO categories. B, The expression patterns of the polysaccharide synthesis-related genes. CSLA3, Cellulose Synthase-like Protein A3, probably for mannan synthesis leading to the formation of noncellulosic polysaccharides of plant cell wall; CESA5, Cellulose Synthase A Catalytic Subunit5, a probable cellulose synthesis enzyme; CSLF6, Cellulose Synthase-like Protein F6, probably for synthesis of mixed-linked glucan; CLAVATA1, receptor protein kinase CLAVATA1. C, DNA sequence representing the predicted core of the in vitro HD-ZIP III binding site. D, The putative binding site (red-colored) of HD-ZIP III transcription factors in the genes related to cell wall organization and biogenesis. The numbers in brackets indicate the distance to the ATG start codon. Data are presented as means ± sd (n = 3 in B). **P < 0.01, two-tailed, two-sample Student’s t test.
To determine whether these polysaccharide synthase genes and the CLAVATA1-related gene are downstream targets of OsHB4, we first identified the putative cis-regulatory motif bound by the HD-Zip III proteins. The in vitro binding data for the HD-Zip III proteins from Sessa et al. (1998) was used, and the sequence motif AT[C/G]ATT[A/C] was identified using MEME (http://meme-suite.org; Fig. 7C), which was also identified as the REVOLUTA binding site in Arabidopsis (Brandt et al., 2012). This binding motif was found in the 5′ promoter region of these three polysaccharide synthase genes and the CLAVATA1-related gene (Fig. 7D). Together, our data suggest that miR166 mediates the cleavage of OsHB4 transcripts and the OsHB4 transcription factor directly regulates some cell wall and polysaccharide metabolism-related genes to influence cell wall formation and vascular development.
DISCUSSION
Plants possess 20- to 24-nucleotide microRNAs (miRNAs), which mediate sequence-specific posttranscriptional gene silencing through transcript cleavage or translational repression. A single miRNA could target multiple genes and regulate diverse processes, and thus miRNAs are considered as genetic tools to manipulate agronomic traits of crop plants (Shriram et al., 2016; Li et al., 2017; Li and Zhang, 2016). With the ultimate goal of improving important agronomic traits by manipulating key miRNAs, we attempted a large-scale knockdown of miRNA families in rice using the STTM technology (Zhang et al., 2017). Independent STTM lines from the knockdown of 37 miRNAs families were generated and phenotyped under natural paddy field conditions. The STTM166 plants, showing a down-regulation of miR166, exhibited multiple morphological changes including reduced plant height (Fig. 1A) and adaxially rolled leaf (Fig. 1B). Since moderate leaf rolling may be associated with drought resistance, STTM166 lines were selected for further characterization in this study.
Using the combination of anatomical and physiological analyses, we showed that the morphological changes of the bulliform cells at the adaxial side of the leaf blade may be responsible for the rolled-leaf phenotype of STTM166 plants (Fig. 1E). The rolled-leaf phenotype of STTM166 plants mimics the response of leaves to drought conditions in rice. Under drought conditions, the bulliform cells deflate due to water deficit thus losing turgor pressure, resulting in leaf rolling. Leaf rolling is thought to be a protective mechanism by reducing water loss through minimizing exposure of the leaves to sunlight and adjusting transpiration in monocots (De Micco and Aronne, 2012). Currently, more than 30 mutants showing rolled-leaf phenotype have been reported in rice and the candidate genes responsible for the phenotype are either enzymes (Fang et al., 2012; Xiang et al., 2012) or transcription factors (Li et al., 2016b; Zhang et al., 2009) that are broadly associated with the establishment of adaxial-abaxial leaf polarity. In STTM166 plants, abnormality of sclerenchymatous cells at abaxial side were also observed (Fig. 1E), which could be another factor contributing to the rolled-leaf phenotype. A previously characterized rice mutant named shallot-like1 with an extreme leaf rolling phenotype was associated with defective sclerenchymatous cells (Zhang et al., 2009).
As a protective mechanism in rice, the rolled-leaf phenotype of STTM166 plants was expected to confer drought resistance. This was indeed the case, as indicated by the increased survival rates of STTM166 plants under drought stress (Fig. 2B). The drought resistant phenotype of STTM166 plants is consistent with reduced transpiration rate (Fig. 3, A and B), decreased stomatal conductance (Fig. 3C), increased leaf water potential (Fig. 3D), and decreased water loss in STTM166 plants (Fig. 2C). A recent report has shown that miR165/166 in Arabidopsis targets the transcription factor PHABULOSA, which binds to the BETA-GLUCOSIDASE1 promoter, and knockdown of miR165/166 resulted in increased expression of BETA-GLUCOSIDASE1, thus higher levels of ABA and improved drought resistance (Yan et al., 2016). However, our study showed that knockdown of miRNA166 in rice did not result in changes in ABA contents under either control or drought-stress conditions (Supplemental Fig. S6, A and B), which suggests that the functions of miR165/166 have diverged between Arabidopsis and rice.
In addition to the rolled-leaf phenotype, we also observed morphological changes in the vasculature of the stem of STTM166 plants (Fig. 4A), but the changes were not observed in the vasculature of roots and leaves of STTM166 plants (Fig. 4, C and D). This suggests that miR166 is somewhat specific to vascular development in the stem in rice, distinct from the Arabidopsis miR165/166, which were shown to affect vascular patterning in roots (Miyashima et al., 2011). The reduced diameter of xylem conduits in the stem of STTM166 rice plants reduced water conductance in STTM166 plants as compared to that of the wild type (Fig. 4B). The decreased water conductivity of the shoot may contribute to the reduced stomatal conductance and lower transpiration in the leaf, since changes in water conductivity are known to be a systemic signal in plants (Zhu, 2016). Therefore, we demonstrate that miR166 regulates stem vascular development and affects water conductivity in rice. The preferential expression of miR166 in the stem vascular tissue (Fig. 5, A and B) further supports its role in the vascular development of the stem.
How miR166 regulates stem vasculature and leaf morphology was studied by identifying the key targets of miR166 and downstream genes in rice. Based on the expression and mRNA cleavage studies, OsHB4 was identified as the major target of miR166 (Fig. 5, C and D). This was supported by the genetic evidence showing that overexpression of miR166-resistant version of OsHB4 (rOsHB4) under the control of the OsActin promoter phenocopies the STTM166 phenotypes such as rolled leaves and drought resistance (Fig. 6). A recent report also showed that overexpression of the native OsHB4 (also known as OsHox32) under the control of 35S promoter resulted in rolled-leaf phenotype in rice (Li et al., 2016a). Interestingly, overexpression of miR166-resistant versions of OsHB1, OsHB3, and OSHB5 also resulted in a rolled-leaf phenotype (Itoh et al., 2008). This suggests that these OsHB transcription factors may have overlapping functions in leaf development. This notion is supported by our observations that a loss-of-function mutant of OsHB4 and plants with overexpressing the miR166 precursor did not show morphological phenotypes (Supplemental Fig. S9). Higher-order mutants should be generated in the future to understand the potentially overlapping and distinct functions of OsHB genes in rice.
Identification of putative downstream genes regulated by miR166-OsHB4 was attempted by using transcriptomic analysis. Differentially expressed genes and GO term analyses revealed strong enrichment of genes involved in cell wall organization or biogenesis and polysaccharide metabolic process, which further supports the role of miR166-OsHB4 in the more specialized cell types such as the xylem vessels and bulliform cells. Among the enriched genes, three polysaccharide synthase genes and a CLAVATA1-related gene containing the putative HD-Zip III binding motif AT[C/G]ATT[A/C] in their promoter regions were identified. Therefore, OsHB4 is likely to directly bind to the promoters of some cell wall and polysaccharide metabolism related genes to regulate cell wall formation and vascular development in rice.
In summary, we provided genetic and molecular evidence that miR166 targets the OsHB4 transcription factor transcripts and regulates leaf morphology and stem vascular development. We also showed that drought resistance in rice could be improved by creating rolled leaves and reducing the diameter of xylem conduit in the stem through down-regulating the level of miR166.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Transgenic lines were generated in rice (Oryza sativa) variety Nipponbare (ssp. japonica) in this study. Wild-type and transgenic plants were grown under natural-field conditions in Shanghai, China (30°N, 121°E) during summer season from mid-May to mid-October and Lingshui, China (18°N, 110°E) during winter season from mid-December to mid-April. The phytotron, with a 30°C/24°C ± 1°C day/night temperature, 50% to 70% relative humidity, and a light/dark period of 14 h/10 h, were used to culture rice seedlings before they were transplanted to paddy fields.
Drought-Stress Treatment and Measurement of Water Loss and Leaf Water Potential
Drought resistance assay was performed using pot-grown plants in a phytotron growth chamber. Seedlings were established under normal growth conditions for 15 d and drought was then initiated by withholding water. On the 10th day after the drought-stress treatment, recovery of plants was attempted by rewatering, and the survival rate was recorded on the eighth day after rewatering. To measure water loss, each pot with same amount of soil was periodically weighed during the drought-stress treatment by using an electronic balance. Water loss was calculated as a percentage of the decreased weight to the initial weight of the pot. The experiment was performed three times (n = 3), each time with two STTM lines (36 seedlings of each lines). In paddy field, drought stress was initiated at the panicle development stage by discontinuing watering of plants, and the plants were recovered with irrigation at the flowering and seed maturation stages. Leaf water potential was measured in plants under normal growth condition and at the fifth day after drought-stress treatment by using WP4C Dewpoint PotentiaMeter (Decagon Devices). Five plants were measured for each line (n = 5).
Trait Measurements
Plant height, tiller number, grain length, grain width, 1,000-grain weight, panicle length, grain number per panicle, internode length, and diameter of the third internode were measured at full maturity of the plants. Plant height was measured in the paddy fields. Grain length and width were measured using an SC-A grain analysis system (Wseen Company). The 1,000-grain weight was measured using an SC-A grain analysis system (Wseen Company) after fully filled grains were dried at 42°C for 2 weeks. Full grains and total grains of each panicle were counted for spikelet fertility in paddy field. Grain yield (gram/plant) was measured by harvesting all full grains of each plant. Twenty-four plants were measured for each line (n = 24).
Phenotyping and Histological Experiments
Most plant materials were photographed with a Canon EOS7D digital camera and an OLYMPUS BX53 microscope. Grain length and width photographs were generated using an SC-A grain analysis system (Wseen Company).
Rice stems and leaves were collected and fixed overnight at 4°C in FAA solution and dehydrated in a graded ethanol series. The samples were then embedded in Technovit 7100 resin (Hereaus Kulzer), and 2-μm sections were made using a Leica RM 2265 programmable rotary microtome (Leica Microsystems). After being stained with 0.05% toluidine blue, transverse sections were photographed using an Olympus BX53 microscope.
Transpiration Rate and Hydraulic Conductance Measurements
Wild-type and transgenic plants were grown under natural field conditions in Shanghai or Lingshui regions. Li-Cor 6400 Portable Photosynthesis System was used to measure the transpiration and photosynthesis rate of 2-month-old seedlings grown in paddy fields, with adequate irrigation and other growing conditions. The measurements were conducted at three time points (10:00 am, 12:00 pm, and 16:00 pm) during sunny days.
At approximately 2 months of growth, when rice plants have matured and have elongated stems, an HPFM (Dynamax) was used to measure the conductivity of water through the vascular bundle of shoot in wild-type and STTM166 plants. In brief, the aerial part of the plant grown in paddy fields was covered with aluminum foil to prevent transpiration before the plant was measurement. The aerial part was then removed for measurement by cutting at the site of joint between stem and root under water to prevent air from being sucked into xylem, and the cutting end of the aerial part was then inserted quickly and tightly into the HPFM instrument adaptor and the conductivity and flow of water were measured using quasi-steady-state flow meter. All leaves of each plant were collected and weighed, and the total leaf area of each plant was calculated based on the leaf weight per unit area. “Cond (kg/s/MPa/m)” represents the conductivity of water, kilograms of water per second per mega-Pascal normalized by leaf area.
Vector Construction and Transformation of Rice
STTM vectors were constructed as described by Tang et al. (2012). All constructs were introduced into the Agrobacterium tumefaciens strain EHA105 and subsequently transferred into the japonica variety Nipponbare by A. tumefaciens-mediated rice transformation as previously described (Hiei et al., 1994). Sequences of the primers used are listed in Supplemental Table S3. For CRISPR/Cas9 vector, guide RNAs were inserted into the Cas9 expression vector downstream of the OsU6 promoter, and the constructs were transformed into the EHA105 A. tumefaciens cells for rice transformation.
Small RNA Northern Analysis
Total RNA was isolated using the Trizol Reagent (Invitrogen) according to the manufacturer’s instructions. About 40 μg of total RNA were analyzed on a denaturing 19% polyacrylamide gel, transferred to Nytran Super Charge Nylon Membranes (Schleicher & Schuell BioScience) and cross-linked using a Stratagene UV Crosslinker. DNA oligonucleotides complementary to different sequences of miRNAs were synthesized and labeled with [32P]γ-ATP (Perkin-Elmer) using T4 polynucleotide kinase (TaKaRa). The membranes were prehybridized with PerfectHyb (Sigma-Aldrich) hybridization solution and then hybridized with the labeled probes. After several times of washing, the membranes were autoradiographed using an x-ray film (Carestream, X-OMAT BT Film). U6 was used as a loading control. The probe sequences are listed in Supplemental Table S3.
RNA Extraction and RT-qPCR
Total RNA was isolated from roots, stem, leaf, leaf sheath, flowers, and seeds from different developmental stages of rice plants using the Trizol Reagent (Invitrogen) according to the manufacturer’s instructions. Shoot samples were also collected from plants with or without drought-stress treatment for 5 d for RNA isolation. After treatment with RNase-free DNase I (Promega), total RNA (1 μg) was reverse transcribed using the TransScript II One-Step gDNA Removal and cDNA Synthesis SuperMix kits (TransGen Biotech). The reverse transcription products were used as templates for quantitative real-time PCR performed on a CFX96 real-time PCR system (Bio-Rad) using SYBR Premix EX Taq (TaKaRa) according to the manufacturer’s protocol. ACTIN1 was used to normalize samples; relative expression levels were measured using the 2−ΔΔCt analysis method. The sequences used in RT-qPCR are listed in Supplemental Table S3.
In Situ Hybridization Analysis
The stems of rice seedling were fixed in 4% paraformaldehyde in 0.1 m sodium phosphate buffer by vacuuming for 10 min and then dehydrated in a graded ethanol series. The dehydrated samples in 100% ethanol were replaced with Histo-Clear and embedded in Paraplast Plus (Sigma-Aldrich). Paraffin sections (5 µm thick) were placed onto microscope slides coated with poly-l-Lys (Sigma-Aldrich). LNA-miR166a antisense probes labeled with Digoxigenin were used for in situ hybridization and immunological detection of miR166 as previously described (Javelle and Timmermans, 2012). The purple signals in localized tissue cells were photographed using an Olympus BX53 microscope.
RNA-Seq Analysis
Total RNA was extracted using RNeasy Plant mini Kit (Qiagen) according to the manufacturer’s instructions. Two independent replicates were used for each sample. The cDNA synthesis, purification, and labeling were then performed using standard protocols. The RNA-seq data were analyzed with TopHat and Cufflinks software. Genes with at least 2-fold up- and down-regulation in the STTM166 plants compared with those in wild-type plants were considered as significantly changed genes. The expression patterns of some differential expressed genes were further confirmed by real-time PCR.
RLM-RACE
RLM-RACE was conducted following the protocol of FirstChoice RLM-RACE kit (Ambion). In brief, total RNA was extracted, and the first and second PCRs were performed by using the primers of HB-inner and HB-outer (listed in Supplemental Table S3), respectively. The products from the second PCR were purified by using agarose gel electrophoresis and then cloned for sequencing.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Plant height and seed size of wild-type and STTM166 plants.
Supplemental Figure S2. Grain yield under normal growth conditions and spikelet fertility under normal and drought conditions.
Supplemental Figure S3. STTM166 plants exhibited drought-resistance phenotype.
Supplemental Figure S4. The stomatal density in STTM166 and wild-type leaves.
Supplemental Figure S5. Stomatal size in leaves of wild-type and STTM166 plants.
Supplemental Figure S6. ABA content in STTM166 and wild-type plants.
Supplemental Figure S7. The expression profiles of aquaporin genes in STTM166 and wild-type plants.
Supplemental Figure S8. The expression of OsHB genes in different tissues and in response to drought stress in wild-type plants.
Supplemental Figure S9. Overexpression of miR166 and the mutation of OsHB4 in rice.
Supplemental Figure S10. The heat map of expression pattern of the polysaccharide synthesis-related genes from the RNA-seq data.
Supplemental Table S1. The differentially expressed genes in STTM166 plants.
Supplemental Table S2. Functional annotation of differentially expressed genes related to cell wall organization and biogenesis.
Supplemental Table S3. List of primers used in this study.
Acknowledgments
We thank Dr. Minjie Cao and Dr. Chunbo Miao for their constructive discussion and assistance. We also thank Dr. Lijun Luo and Dr. Shunwu Yu from Shanghai Academy of Agricultural Science (SAAS) for help with drought-stress treatment in the paddy field.
Footnotes
This work was supported by the Chinese Academy of Sciences.
References
- Agalou A, Purwantomo S, Overnäs E, Johannesson H, Zhu X, Estiati A, de Kam RJ, Engström P, Slamet-Loedin IH, Zhu Z, et al. (2008) A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol Biol 66: 87–103 [DOI] [PubMed] [Google Scholar]
- Ariel FD, Manavella PA, Dezar CA, Chan RL (2007) The true story of the HD-Zip family. Trends Plant Sci 12: 419–426 [DOI] [PubMed] [Google Scholar]
- Brandt R, Salla-Martret M, Bou-Torrent J, Musielak T, Stahl M, Lanz C, Ott F, Schmid M, Greb T, Schwarz M, et al. (2012) Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J 72: 31–42 [DOI] [PubMed] [Google Scholar]
- Brodribb TJ, Feild TS (2000) Stem hydraulic supply is linked to leaf photosynthetic capacity: Evidence from New Caledonian and Tasmanian rainforests. Plant Cell Environ 23: 1381–1388 [Google Scholar]
- Candela H, Johnston R, Gerhold A, Foster T, Hake S (2008) The milkweed pod1 gene encodes a KANADI protein that is required for abaxial/adaxial patterning in maize leaves. Plant Cell 20: 2073–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Xie Q, Gao J, Wang W, Sun B, Liu B, Zhu H, Peng H, Zhao H, Liu C, et al. (2015) Characterization of Rolled and Erect Leaf 1 in regulating leave morphology in rice. J Exp Bot 66: 6047–6058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DH, Nogueira FT, Howell MD, Montgomery TA, Carrington JC, Timmermans MC (2009) Pattern formation via small RNA mobility. Genes Dev 23: 549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Micco V, Aronne G. 2012. Morpho-anatomical traits for plant adaptation to drought. In Aroca R, ed, Plant Responses to Drought Stress. Springer, Berlin, pp 37–61 [Google Scholar]
- De Rybel B, Mähönen AP, Helariutta Y, Weijers D (2016) Plant vascular development: From early specification to differentiation. Nat Rev Mol Cell Biol 17: 30–40 [DOI] [PubMed] [Google Scholar]
- Diévart A, Dalal M, Tax FE, Lacey AD, Huttly A, Li J, Clark SE (2003) CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development. Plant Cell 15: 1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13: 1768–1774 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL (2001) Establishment of polarity in lateral organs of plants. Curr Biol 11: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Izhaki A, Baum SF, Floyd SK, Bowman JL (2004) Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131: 2997–3006 [DOI] [PubMed] [Google Scholar]
- Fang L, Zhao F, Cong Y, Sang X, Du Q, Wang D, Li Y, Ling Y, Yang Z, He G (2012) Rolling-leaf14 is a 2OG-Fe (II) oxygenase family protein that modulates rice leaf rolling by affecting secondary cell wall formation in leaves. Plant Biotechnol J 10: 524–532 [DOI] [PubMed] [Google Scholar]
- Floyd SK, Bowman JL (2004) Gene regulation: Ancient microRNA target sequences in plants. Nature 428: 485–486 [DOI] [PubMed] [Google Scholar]
- Fouracre JP, Poethig RS (2016) The role of small RNAs in vegetative shoot development. Curr Opin Plant Biol 29: 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner JC, Taylor NG, Turner SR (2003) Control of cellulose synthase complex localization in developing xylem. Plant Cell 15: 1740–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacke UG, Spicer R, Schreiber SG, Plavcová L (2017) An ecophysiological and developmental perspective on variation in vessel diameter. Plant Cell Environ 40: 831–845 [DOI] [PubMed] [Google Scholar]
- Heo JO, Blob B, Helariutta Y (2017) Differentiation of conductive cells: A matter of life and death. Curr Opin Plant Biol 35: 23–29 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hou X, Xie K, Yao J, Qi Z, Xiong L (2009) A homolog of human ski-interacting protein in rice positively regulates cell viability and stress tolerance. Proc Natl Acad Sci USA 106: 6410–6415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J, Hibara K, Sato Y, Nagato Y (2008) Developmental role and auxin responsiveness of Class III homeodomain leucine zipper gene family members in rice. Plant Physiol 147: 1960–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javelle M, Timmermans MC (2012) In situ localization of small RNAs in plants by using LNA probes. Nat Protoc 7: 533–541 [DOI] [PubMed] [Google Scholar]
- Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MC (2004) microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428: 84–88 [DOI] [PubMed] [Google Scholar]
- Lang Y, Zhang Z, Gu X, Yang J, Zhu Q (2003) Physiological and ecological effects of crimpy leaf character in rice (Oryza sativa L.) II. Photosynthetic character, dry mass production and yield forming. Zuo Wu Xue Bao 30: 883–887 [Google Scholar]
- Li S, Castillo-González C, Yu B, Zhang X (2017) The functions of plant small RNAs in development and in stress responses. Plant J 90: 654–670 [DOI] [PubMed] [Google Scholar]
- Li YY, Shen A, Xiong W, Sun QL, Luo Q, Song T, Li ZL, Luan WJ (2016a) Overexpression of OsHox32 results in pleiotropic effects on plant type architecture and leaf development in rice. Rice (N Y) 9: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Zhang B (2016) MicroRNAs in control of plant development. J Cell Physiol 231: 303–313 [DOI] [PubMed] [Google Scholar]
- Li C, Zou X, Zhang C, Shao Q, Liu J, Liu B, Li H, Zhao T (2016b) OsLBD3-7 overexpression induced adaxially rolled leaves in rice. PLoS One 11: e0156413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li M, Liu K, Tang D, Sun M, Li Y, Shen Y, Du G, Cheng Z (2016) Semi-Rolled Leaf2 modulates rice leaf rolling by regulating abaxial side cell differentiation. J Exp Bot 67: 2139–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Guo Z, Li L (2013) Evolutionary conservation of microRNA regulatory programs in plant flower development. Dev Biol 380: 133–144 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP (2004) MicroRNA control of PHABULOSA in leaf development: Importance of pairing to the microRNA 5′ region. EMBO J 23: 3356–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK (2001) Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713 [DOI] [PubMed] [Google Scholar]
- McCulloh K, Sperry JS, Lachenbruch B, Meinzer FC, Reich PB, Voelker S (2010) Moving water well: comparing hydraulic efficiency in twigs and trunks of coniferous, ring-porous, and diffuse-porous saplings from temperate and tropical forests. New Phytol 186: 439–450 [DOI] [PubMed] [Google Scholar]
- Merelo P, Ram H, Pia Caggiano M, Ohno C, Ott F, Straub D, Graeff M, Cho SK, Yang SW, Wenkel S, et al. (2016) Regulation of MIR165/166 by class II and class III homeodomain leucine zipper proteins establishes leaf polarity. Proc Natl Acad Sci USA 113: 11973–11978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashima S, Koi S, Hashimoto T, Nakajima K (2011) Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138: 2303–2313 [DOI] [PubMed] [Google Scholar]
- Moon J, Hake S (2011) How a leaf gets its shape. Curr Opin Plant Biol 14: 24–30 [DOI] [PubMed] [Google Scholar]
- Nagasaki H, Itoh J, Hayashi K, Hibara K, Satoh-Nagasawa N, Nosaka M, Mukouhata M, Ashikari M, Kitano H, Matsuoka M, et al. (2007) The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc Natl Acad Sci USA 104: 14867–14871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JM, Lane B, Freeling M (2002) Expression of a mutant maize gene in the ventral leaf epidermis is sufficient to signal a switch of the leaf’s dorsoventral axis. Development 129: 4581–4589 [DOI] [PubMed] [Google Scholar]
- Nogueira FT, Madi S, Chitwood DH, Juarez MT, Timmermans MC (2007) Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev 21: 750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole JC, Cruz RT (1980) Response of leaf water potential, stomatal resistance, and leaf rolling to water stress. Plant Physiol 65: 428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE (2001) REVOLUTA regulates meristem initiation at lateral positions. Plant J 25: 223–236 [DOI] [PubMed] [Google Scholar]
- Price A, Young E, Tomos A (1997) Quantitative trait loci associated with stomatal conductance, leaf rolling and heading date mapped in upland rice (Oryza sativa). New Phytol 137: 83–91 [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M, Mizutani M, Sakata K, Takatsuto S, Yoshida S, et al. (2006) Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol 24: 105–109 [DOI] [PubMed] [Google Scholar]
- Sessa G, Steindler C, Morelli G, Ruberti I (1998) The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol Biol 38: 609–622 [DOI] [PubMed] [Google Scholar]
- Shriram V, Kumar V, Devarumath RM, Khare TS, Wani SH (2016) MicroRNAs as potential targets for abiotic stress tolerance in plants. Front Plant Sci 7: 817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Reinhart BJ, Bartel DP, Zamore PD (2003) A biochemical framework for RNA silencing in plants. Genes Dev 17: 49–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Yan J, Gu Y, Qiao M, Fan R, Mao Y, Tang X (2012) Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods 58: 118–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretter EM, Alvarez JP, Eshed Y, Bowman JL (2008) Activity range of Arabidopsis small RNAs derived from different biogenesis pathways. Plant Physiol 147: 58–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Meents MJ, McDonnell LM, Barkwill S, Sampathkumar A, Cartwright HN, Demura T, Ehrhardt DW, Samuels AL, Mansfield SD (2015) Visualization of cellulose synthases in Arabidopsis secondary cell walls. Science 350: 198–203 [DOI] [PubMed] [Google Scholar]
- Wu X. (2009) Prospects of developing hybrid rice with super high yield. Agron J 101: 688–695 [Google Scholar]
- Xiang JJ, Zhang GH, Qian Q, Xue HW (2012) Semi-rolled leaf1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiol 159: 1488–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Chen H, Fan T, Huang Y, Yu S, Chen S, Hong X (2012a) Rice flag leaf physiology, organ and canopy temperature in response to water stress. Plant Prod Sci 15: 92–99 [Google Scholar]
- Yan J, Gu Y, Jia X, Kang W, Pan S, Tang X, Chen X, Tang G (2012b) Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24: 415–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Zhao C, Zhou J, Yang Y, Wang P, Zhu X, Tang G, Bressan RA, Zhu JK (2016) The miR165/166 mediated regulatory module plays critical roles in ABA homeostasis and response in Arabidopsis thaliana. PLoS Genet 12: e1006416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SQ, Li WQ, Miao H, Gan PF, Qiao L, Chang YL, Shi CH, Chen KM (2016) REL2, a gene encoding an unknown function protein which contains DUF630 and DUF632 domains controls leaf rolling in rice. Rice (N Y) 9: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JJ, Wu SY, Jiang L, Wang JL, Zhang X, Guo XP, Wu CY, Wan JM (2015) A detailed analysis of the leaf rolling mutant sll2 reveals complex nature in regulation of bulliform cell development in rice (Oryza sativa L.). Plant Biol (Stuttg) 17: 437–448 [DOI] [PubMed] [Google Scholar]
- Zhang G-H, Xu Q, Zhu X-D, Qian Q, Xue H-W (2009) SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 21: 719–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang J, Yan J, Gou F, Mao Y, Tang G, Botella JR, Zhu JK (2017) Short tandem target mimic rice lines uncover functions of miRNAs in regulating important agronomic traits. Proc Natl Acad Sci USA 114: 5277–5282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. (2016) Abiotic stress signaling and responses in plants. Cell 167: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]