Alteration of photosynthetic electron transport, caused by a mitochondrial mutation, showed experimentally that algae better endure light damage targeting photosystem II instead of photosystem I.
Abstract
Photosynthetic organisms use sunlight as the primary source of energy to support their metabolism. In eukaryotes, reactions responsible of the conversion of light into chemical energy occur in specific organelles, the chloroplasts. In this study, we showed that mitochondria also have a seminal influence on cells’ energy metabolism and on photosynthetic reactions. This is illustrated by the observation that the strong photosensitivity of Chlamydomonas reinhardtii cells depleted of the chloroplast protein PGRL1 was rescued by the introduction of a mitochondrial mutation affecting respiratory complex I. Functional analysis showed that such a reduced respiratory activity influenced chloroplast electron transport with consequent overreduction of plastoquinone and donor-side limitation of photosystem I (PSI). As a consequence, damage due to excess light affected more photosystem II (PSII) rather than PSI. Double mutant cells are able to grow under excess illumination, while single pgrl1 are not, thanks to the presence of an efficient repair mechanism of PSII. These results also underline the seminal biological relevance of the regulation of electron transport reactions within the photosynthetic complexes. Photosynthetic organisms evolved a strategy to respond to excess light where damage is targeting preferentially to a specific complex, PSII. Cells are able to endure extensive damage targeting this complex thanks to an efficient repair mechanisms, while if PSI is affected, there are drastic consequences on growth.
Photosynthetic eukaryotic organisms rely on two organelles, chloroplasts and mitochondria, for the synthesis of the molecules fueling their metabolism, NAD(P)H and ATP. The two organelles share common features, such as membrane-bound enzymatic complexes implicated in electron transfer coupled to proton translocation and the generation of a proton motive force driving the synthesis of ATP.
In chloroplasts, light energy fuels the electron transport from water to NADP+ to generate NADPH via photosystem II (PSII), cytochrome b6f (Cyt b6f) complex, and photosystem I (PSI) in a pathway designated as linear electron transport. Linear electron transport is coupled to proton translocation from the chloroplast stroma into the thylakoid lumen and establishes a transmembrane electrochemical potential, exploited by the ATP synthase for the synthesis of ATP. However, the electrons can also follow alternative pathways, which play an essential role, especially at fluctuating light intensity, in conditions where demand/availability of ATP and NADPH are continuously changing (Kramer and Evans, 2011). Such alternative electron flow (AEF) pathways include as examples the plastid alternative oxidase, the water-water cycle, and the so-called cyclic electron flow (CEF) around PSI (Cardol et al., 2011). During CEF, electrons from PSI are recycled to the Cyt b6f complex, contributing to the proton translocation and ATP biosynthesis but not to synthesis of NADPH. In the model organism used in this work, the green alga Chlamydomonas reinhardtii, two pathways involved in CEF have been identified. One involving the NDA2 NADPH:plastoquinone oxidoreductase (Desplats et al., 2009; Jans et al., 2008) that uses NADPH to reduce plastoquinone (PQ) and the other relying on a PGR5-PGRL1 complex with putative ferredoxin:plastoquinone oxidoreductase activity (Dang et al., 2014; Hertle et al., 2013; Petroutsos et al., 2009; Tolleter et al., 2011; Yamori and Shikanai, 2016).
In mitochondria, the transfer of electrons from the substrates NADH and succinate is catalyzed by the enzymatic complexes I, II, III, and IV, with molecular oxygen being the final acceptor. Electron transfer is again coupled to the synthesis of ATP through an electrochemical transmembrane gradient generated by complexes I, III, and IV. As in chloroplasts, electrons can also follow alternative routes, bypassing complex III and complex IV activities via the alternative terminal oxidase (AOX; Dinant et al., 2001) or bypassing complex I activity via an alternative NADH dehydrogenase (such as Nda1; Lecler et al., 2012). These two routes are not coupled to the formation of electrochemical transmembrane gradient and they reduce the energy yield of respiration but also act as regulators in stress conditions (Zalutskaya et al., 2015), maintaining the ratio between reduced and oxidized forms of ubiquinone (Vanlerberghe, 2013) or NADH consumption (Lecler et al., 2012).
In photosynthetic organisms, both chloroplast and mitochondria respond to many different stimuli originating from the environment and are in continuous communication. This is well exemplified by C. reinhardtii mutants affected in respiratory complexes that experience a reorganization of the photosynthetic apparatus favoring ATP synthesis over NADP+ reduction in the chloroplast through activation of CEF (Cardol et al., 2003). On the other hand, alterations in chloroplast proteins were also shown to impact mitochondria. For example, the photosynthetic pgrl1 mutant altered in CEF showed an increased respiration (Dang et al., 2014; Petroutsos et al., 2009; Tolleter et al., 2011). The recent finding that in diatoms, optimized carbon fixation and growth require exchange of NADPH and ATP between plastids and mitochondria also points to an evolutionarily conserved seminal role of mitochondria-chloroplast cross-talk (Bailleul et al., 2015).
In this study, we investigated the mutual influence of respiratory and photosynthetic activity by analyzing a C. reinhardtii double mutant Δnd4pgrl1 that harbors mutations in both (1) the nuclear gene PGRL1 leading to an altered CEF and chloroplast ATP biosynthesis and (2) the mitochondrial complex I gene nd4 leading to a near complete loss of complex I activity. We show that, remarkably, the mitochondrial mutation is able to rescue the photosensitivity of pgrl1 mutant, further supporting the reciprocal influence of the two organelles. This phenotype is not due to increased photosynthetic capacity of the double mutant but rather to the fact that excess light damages PSII more than PSI with a much milder effect on growth due to the presence of efficient repair mechanisms.
RESULTS
A Mitochondrial Mutation Restores the Ability of the pgrl1 Mutant to Grow under Strong Illumination
The mutant strain affected in the expression of the chloroplast localized protein PGRL1, altered in photosynthetic electron transport rate (ETR), showed altered mitochondrial composition and activity (Dang et al., 2014; Massoz et al., 2017; Petroutsos et al., 2009). Considering that, in order to investigate a reciprocal influence of respiratory and photosynthetic activities, pgrl1 was crossed with several mitochondrial mutants (described in Salinas et al., 2014). In particular we selected (1) a complex III mutant, dum11, harboring a 0.7-kb mitochondrial DNA deletion and unable to grow under heterotrophic conditions; and (2) three complex I mutants with reduced respiration. These are dum5, presenting a thymine deletion in the 3′ untranslated region of the mitochondrial nd5 gene; dum25, in which two codons of nd1 are missing; and Δnd4 presenting an in frame deletion of codons 2 to 24 of the mitochondrial nd4 gene (Massoz et al., 2017).
As in C. reinhardtii, the mitochondrial DNA is inherited from the mating type minus parent (mt−; Matagne et al., 1989), we crossed an mt+ pgrl1 strain with the various mt− mitochondrial mutants. Progeny from all crosses was screened to ensure the selection of strains carrying both mutations. The Mendelian transmission of the pgrl1 mutation was assessed thanks to the resistance to paromomycin and was confirmed by PCR. The uniparental transmission of mitochondrial DNA mutations was instead confirmed by PCR and sequencing (Supplemental Fig. S1). A large number of double mutants were obtained from all crosses involving pgrl1 and complex I defective strains, while the cross between pgrl1 and complex III mutant (dum11) did not yield any viable double mutant progeny. This suggests the combination of these two mutations is lethal even under continuous illumination. This observation indicates that in the absence of PGRL1 cells can survive with a reduced respiration but cannot if this is completely inactivated.
Respiratory activity, measured as oxygen consumption in the dark, showed an insensitivity to complex I inhibitor rotenone in the complexI-pgrl1 double mutant (Δnd4pgrl1 strain) for cells grown mixotrophically in control light conditions, confirming that the contribution of complex I to the overall respiration is negligible in this strain, as it is for the single complex I mutant Δnd4 (Table I). These measurements also highlight that in Δnd4pgrl1, the overall respiration activity is increased by ∼50% compared to parental strain Δnd4, in agreement with previous findings showing an increased respiration in pgrl1 (Petroutsos et al., 2009). Respiratory activity was also evaluated after inactivating the mitochondrial electron transport at the level of cytochrome (involving complexes III and IV) or the AOX pathway, using the specific inhibitors KCN or salicylhydroxamic acid (SHAM), respectively. Both the cytochrome c pathway and the AOX-dependent respiration were increased in pgrl1 and Δnd4pgrl1 with respect to the wild type and Δnd4, respectively (Table I), suggesting that the absence of PGRL1 impacts the overall electron flux in the mitochondrial transport chain without specifically inducing one pathway with respect to the other.
Table I. Total respiration and respiration in presence of mitochondrial inhibitors for the wild type, pgrl1, Δnd4, and Δnd4pgrl1.
Total dark respiration (average ± sd, n > 3) expressed in nmol O2 × min−1 × 107 cells−1 ± sd and measured in the absence or in the presence of 100 μm rotenone, 1 mm KCN, or 1 mm SHAM for cells grown in CL 60 μE m−2 s−1.
| Wild Type | pgrl1 | Δnd4 | Δnd4pgrl1 | |
|---|---|---|---|---|
| Total Respiration CL | 37 ± 6 | 57 ± 6 | 21 ± 3 | 39 ± 10 |
| +Rotenone | 22 ± 6 | 33 ± 8 | 18 ± 4 | 36 ± 9 |
| +KCN | 13 ± 1 | 17 ± 8 | 10 ± 2 | 22 ± 5 |
| +SHAM | 27 ± 3 | 38 ± 15 | 16 ± 2 | 30 ± 10 |
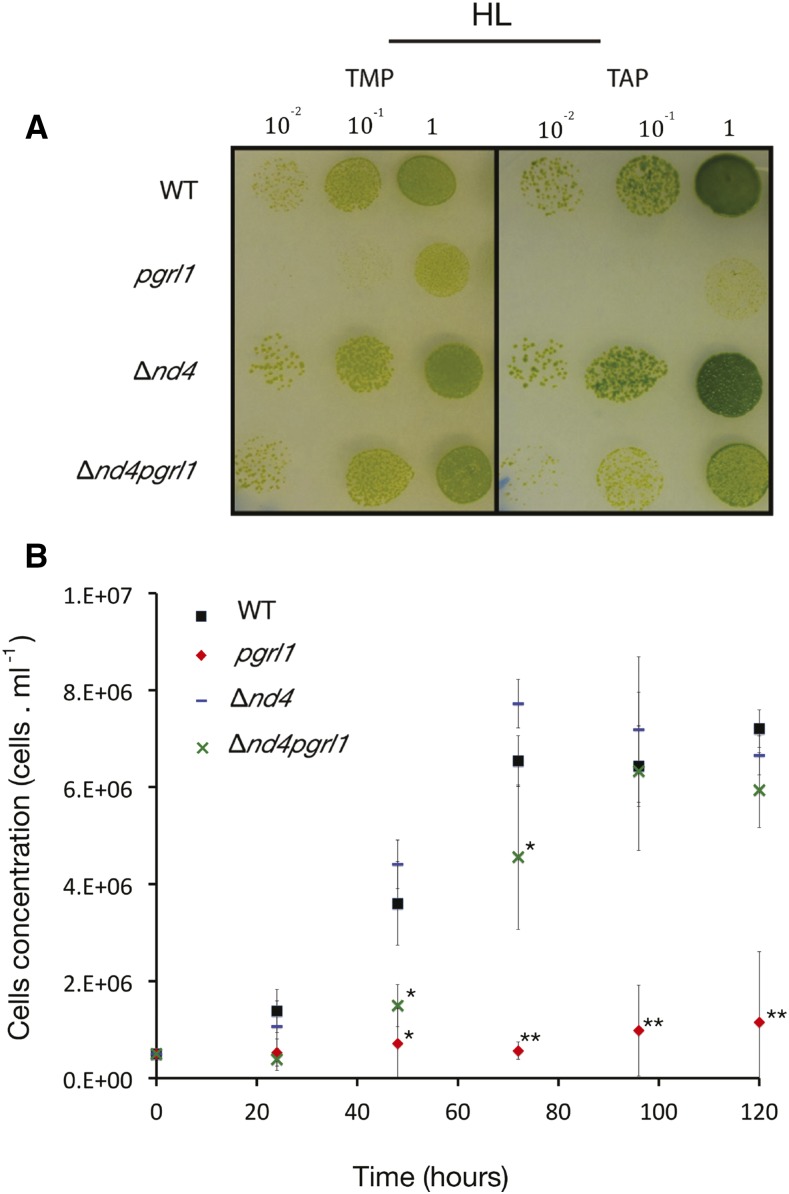
Interestingly, we observed that the combination of mutations affecting mitochondria and chloroplast resulted in a specific peculiar feature of double mutants. While single pgrl1 mutant shows a clear photosensitivity when exposed to a strong illumination, as previously reported (Dang et al., 2014; Kukuczka et al., 2014), all double mutants showed a rescued growth even if at a slower rate than the wild type and Δnd4 (Fig. 1A; Supplemental Fig. S2). The rescued growth was observed for all double mutants, dum5pgrl1, dum25pgrl1, and Δnd4pgrl1, independently of the specific complex I mutation (Supplemental Fig. S2A), confirming that the decreased photosensitivity was indeed due to the mitochondrial mutation (Fig. 1B; Supplemental Fig. S2B). This surprising growth phenotype was confirmed in liquid cultures (Fig. 2B; Supplemental Fig. S2) and in presence of an exogenous organic source (Supplemental Fig. S2B), demonstrating it is not correlated to specific growth conditions.
Figure 1.
Characterization of the wild type, Δnd4, pgrl1, and Δnd4pgrl1 growth under strong illumination. Impact of chloroplastic and mitochondrial mutation on growth of C. reinhardtii strains is assessed by spot test (A), i.e. cells plated in different dilutions on solid media in mixotrophic (TAP) or photoautotrophic (TMP) conditions for wild type, pgrl1, Δnd4, and Δnd4pgrl1 mutant strains. The cells were grown at HL intensity of 850 μE m−2 s−1. Photos were taken after 3 to 5 d. Impact of mutation was also assessed in liquid culture (B) in photoautotrophic conditions (TMP) for wild type, pgrl1, Δnd4, and Δnd4pgrl1 at a HL intensity of 1,500 μE m−2 s−1. Error bars indicate sd (n = 3–6 independent biological replicates). Statistically significant differences compared to the wild type are indicated with an asterisk. Two asterisks instead indicate significant differences with both the wild type and Δnd4pgrl1.
Figure 2.
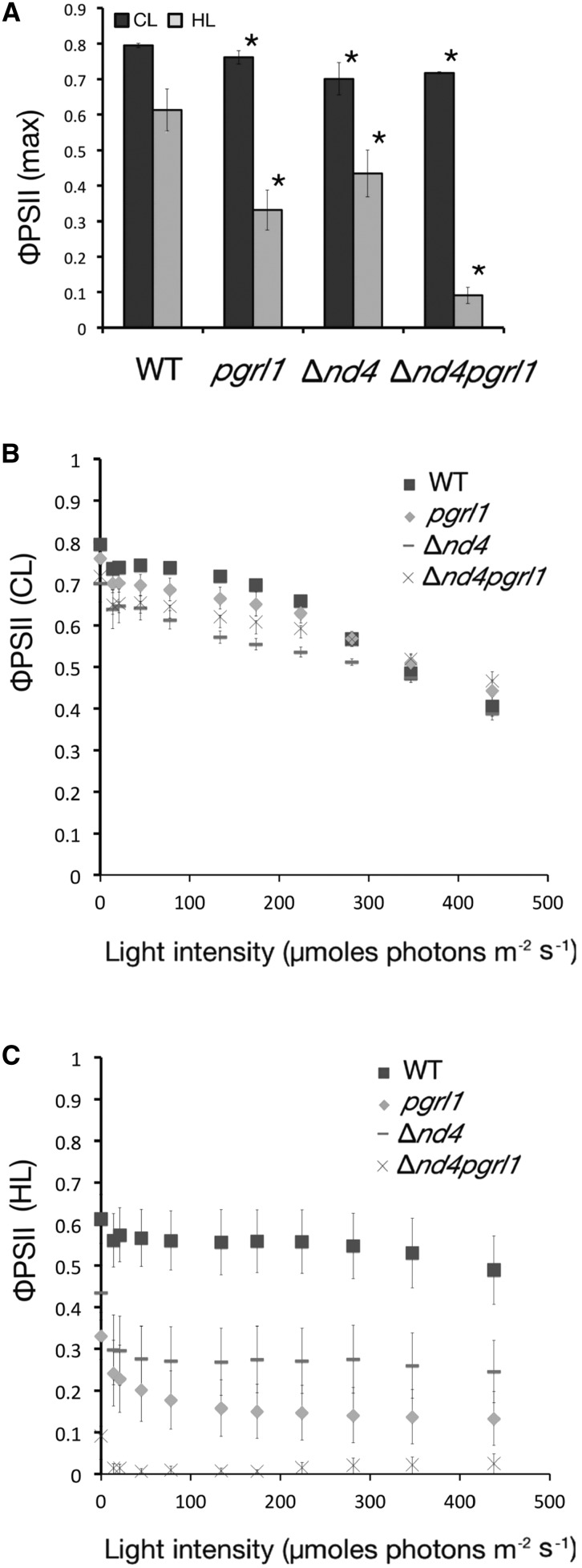
Analysis of PSII efficiency of the wild type, Δnd4, pgrl1, and Δnd4pgrl1. A, ΦPSII monitored with a PAM fluorimeter measured after 30 min of dark adaptation of wild-type, pgrl1, Δnd4, and Δnd4pgrl1 strains grown photoautotrophically in CL and HL conditions. Data are expressed as averages ± sd, n = 4. Values significantly different from the wild type are marked with an asterisk (ANOVA, P value < 0.05). B and C, PSII quantum yield dependence from illumination intensity for wild-type, pgrl1, Δnd4, and Δnd4pgrl1 strains grown photoautotrophically in CL (B) and HL (C) conditions. Measurements were performed using 1-min steps of increasing light intensity (n = 4 ± sd).
Modulation of Photosynthetic Apparatus and Electron Transport in Δnd4pgrl1
Photosynthetic activity in Δnd4pgrl1 was investigated in more detail to clarify the reason for the rescued ability to grow under strong illumination. The maximal photochemical efficiency of PSII in dark-adapted cells (ΦPSII max) was first estimated exploiting chlorophyll fluorescence showing a slight but significant decrease with respect to the wild type in all three mutants grown in phototrophy at control light (CL) intensity (Fig. 2A), as already observed for the pgrl1 mutant (Dang et al., 2014; Kukuczka et al., 2014; Massoz et al., 2017; Petroutsos et al., 2009). In cells grown at high light (HL) intensities, all genotypes, including the wild type, showed a substantial decrease in ΦPSII due to photoinhibition. This effect was more evident in the mutants with the largest impact on Δnd4pgrl1, revealing an additive effect of the two mutations on PSII photosensitivity (Fig. 2A). Again, this observation did not depend on the growth medium, even if the impact was larger in mixotrophy (Supplemental Fig. S3). PSII efficiency did not recover even if cells were treated with far-red light, confirming that its strong decrease in Δnd4pgrl1 is attributable to photoinhibition (Supplemental Fig. S3). PSII efficiency was also measured in light-adapted cells (ΦPSII’). CL-grown cells exposed to increasing light intensities in all genotypes showed a progressive PSII saturation (Fig. 2B). In HL-grown cells, instead, Δnd4pgrl1 PSII efficiency showed no light-dependent change, confirming that this photosystem is inhibited and is unable to sustain a strong light-dependent activity (Fig. 2C). Such a strong PSII photoinhibition is not attributable to defects in NPQ that is activated in all strains even more rapidly than in the wild type (Supplemental Fig. S5).
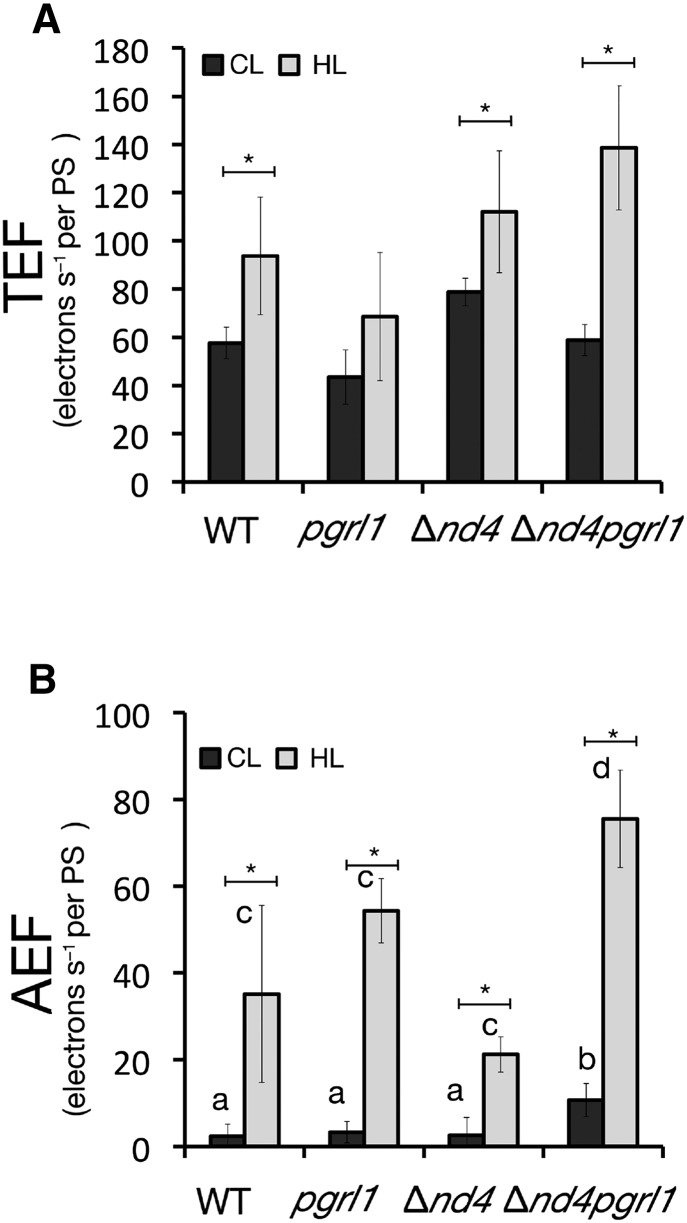
Photosynthetic ETR was evaluated exploiting the carotenoid electrochromic shift signal (ECS; Bailleul et al., 2010). This signal originates from the transmembrane potential; thus, it responds to the activity of both PSII and PSI, at difference from fluorescence analysis that is mostly emitted by the former. Total and alternative electron flows (TEF and AEF, respectively) were increased in all genotypes in response to HL (Fig. 3). In Δnd4pgrl1, alternative electron transports independent from PSII were found more active than in the other genotypes, both at CL and HL (Fig. 3B). Interestingly, HL contribution of AEF, determined by inhibiting PSII activity with DCMU, reached even ∼50% of the TEF for Δnd4pgrl1.
Figure 3.
Photosynthetic electron transport rate of the wild type, Δnd4, pgrl1, and Δnd4pgrl1. TEF (A) and AEF (B) electron transport rates evaluated from ECS relaxation kinetics. Rates are normalized to the total PSI and PSII amount quantified using a saturating flash. Data refer to the rates measured in wild-type, pgrl1, Δnd4, and Δnd4pgrl1 strains grown photoautotrophically in CL (n = 6 ± sd) and in HL condition (n = 4 ± sd). Asterisks indicate statistically significant differences between the same genotype in CL and HL conditions. Different letters instead indicate statistically significant differences between genotypes in the same light conditions (ANOVA, P value < 0.05).
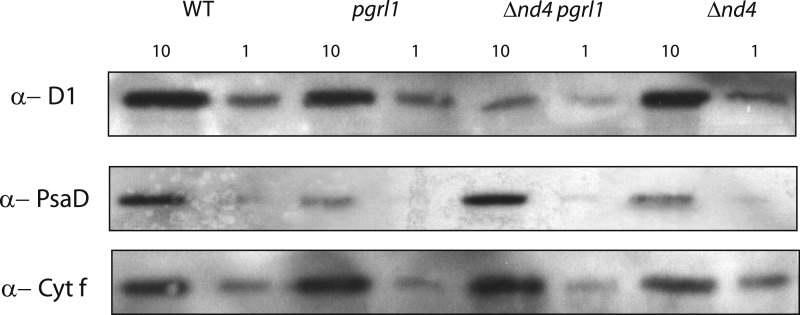
The composition of the photosynthetic apparatus was investigated in these strains by western-blot analysis (Fig. 4; Supplemental Fig. S4A). The relative amount of PSII in HL, quantified using anti-D1 and D2 antibodies, was drastically decreased in Δnd4pgrl1 with respect to the other mutants and the wild type (Fig. 4; Supplemental Fig. S4), in accordance with the observed reduction in ΦPSII. PSI content assessed by the anti-PsaD antibody was instead strongly reduced in pgrl1 grown in HL, consistent with previous observations (Dang et al., 2014), while this was less affected in the other strains. Introduction of the mitochondrial mutation in Δnd4pgrl1 thus caused light damage to affect more PSII content while PSI recovered. At the same time, there was no compensating increase in NDA2 (Supplemental Fig. S4A), involved in another mechanisms for AEF. The relative accumulation of PSI and II was further verified using 77K fluorescence where they can be detected from their specific emissions at 685 and 715 nm, respectively. Δnd4pgrl1 cells showed a strong relative increase in PSI emission compared to the other genotypes both in CL and HL conditions (Fig. 5; Supplemental Fig. S4B), consistent with western-blot results.
Figure 4.
Immunodetection of total extract membrane for the wild type, Δnd4, pgrl1, and Δnd4pgrl1. Immunodetection of total extract of wild type, pgrl1, Δnd4, and Δnd4pgrl1 culture grown photoautotrophically in HL using an antibody against PSII (D1), PSI (PsaD), and cytochrome f (Cyt f); 10 or 1 μg of chlorophyll was loaded (n > 6).
Figure 5.
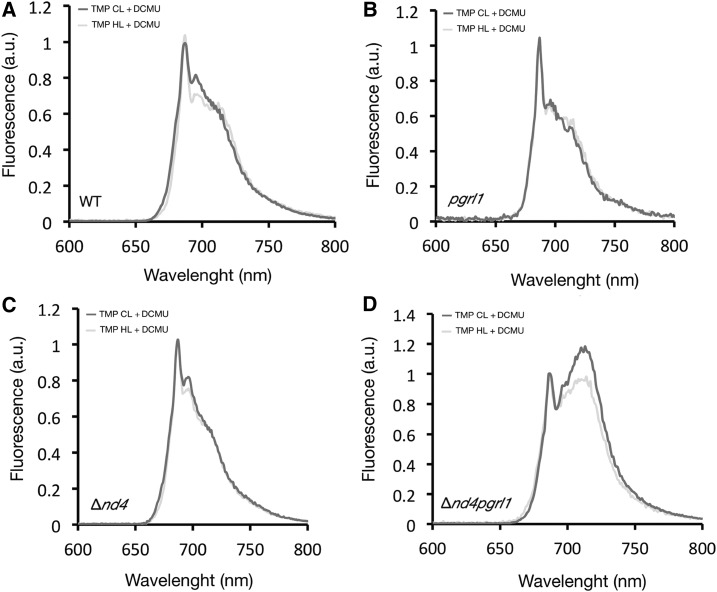
The 77K fluorescence analyses of the wild type, Δnd4, pgrl1, and Δnd4pgrl1. Relative content of PSI and PSII monitored with low-temperature emission spectra (77K fluorescence) normalized to their amplitude at 685 nm for the wild type (A), pgrl1 (B), Δnd4 (C), and Δnd4pgrl1 (D) grown photoautotrophically in CL or HL and DCMU-poisoned before freezing. Two mains peaks are resolved at 685 nm and in the 710-nm regions, which correspond to fluorescence by PSII and PSI, respectively.
The above measurements were performed in cells incubated with DCMU in order to minimize state transitions, since PSI fluorescence could increase because of LHCII association. The comparison of the same spectra in the absence of DCMU (Supplemental Fig. S4B) can provide also additional information on the possible activation of state transition. Indeed, Δnd4pgrl1 showed a stronger activation of state transitions with evidence of LHCII associated with PSI even in HL conditions (Supplemental Fig. S4B).
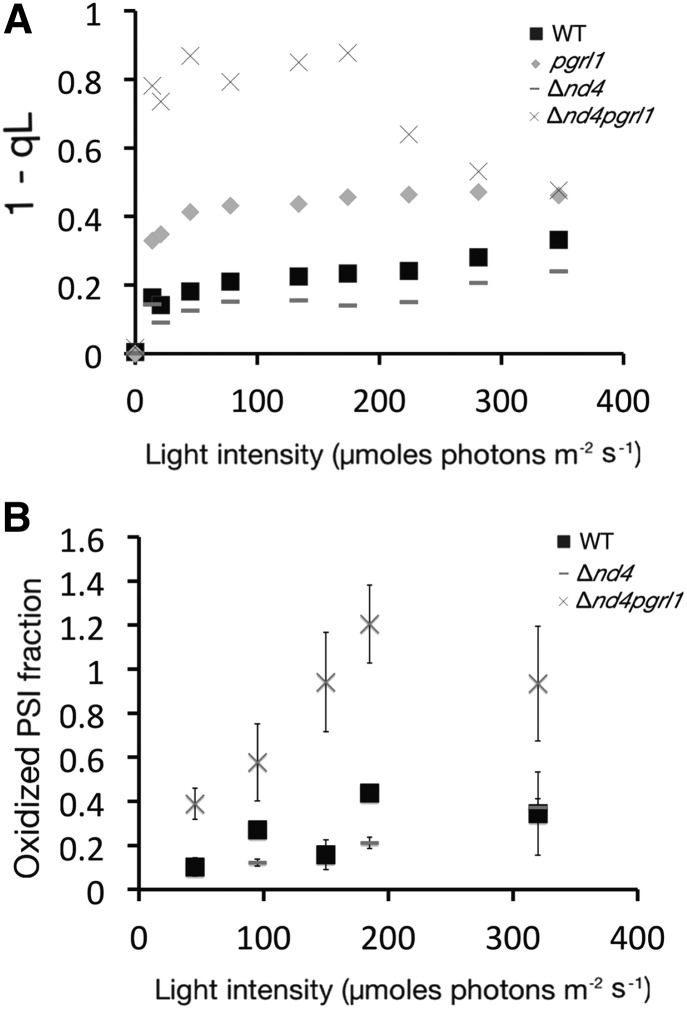
The above results suggest an altered balance between PSI and PSII in Δnd4pgrl1 with respect to the parental strains. This hypothesis was further strengthened by estimating the plastoquinone (Qa) redox state (Hertle et al., 2013). In Δnd4pgrl1, different from all other strains, Qa is very easily reduced even using dim light (Fig. 6A), suggesting that PSII is easily saturated in this strain. This observation also explains the stronger state transition activation that is promoted by accumulation of reduced plastoquinone (Vener et al., 1995; Zito et al., 1999). An independent evidence of altered balance of electron transport can be obtained by estimating the oxidization of PSI (P700+/total P700 ratio) in illuminated cells. The fraction of oxidized PSI is clearly higher for Δnd4pgrl1 with respect to the wild type and Δnd4, reaching a complete oxidation already under relatively dim illumination (150 µmol photons m−2 s−1), suggesting that PSI electron transport is strongly limited from its donor side (Fig. 6B).
Figure 6.
Redox state of Qa pool and PSI analysis. A, PQ redox state assessed by the fluorescence parameter 1- qL in wild-type, pgrl1, Δnd4, and Δnd4pgrl1 cells grown in HL conditions. B, Oxidized PSI fraction both in function of increasing light intensities in cells grown in HL conditions. The fraction is calculated as the ratio between the amount of oxidized P700 at a specific light and the total P700 quantified in the same sample using a saturating light (2,050 μmol of photons m−2 s−1) in the presence of DCMU and DBMIB. pgrl1 is not shown because its photosensitivity impairs obtaining a good signal from HL-grown cells. All measurements were performed using 25 × 106 cells/mL resuspended in TMP medium supplemented with 20% Ficoll.
In summary, our data suggest that the double mutant does not show an increased ability to use strong illumination for photochemistry. With respect to both parental strains, it in fact shows a decrease in PSII content and efficiency. However, this is paralleled by a higher PSI content, especially if compared to pgrl1. As a consequence, ETR is easily saturated at the level of PQ that becomes the limiting step causing a donor side limitation to PSI. A way to mimic the restriction of electron transport at the level of PQ and to mimic PSI donor side limitation induced by the mitochondrial mutation is to artificially inhibit electron transport using DCMU. Under these conditions, when pgrl1 cells were exposed to strong illumination, PSI showed reduced light sensitivity, as observed by immunodetection (Fig. 7).
Figure 7.
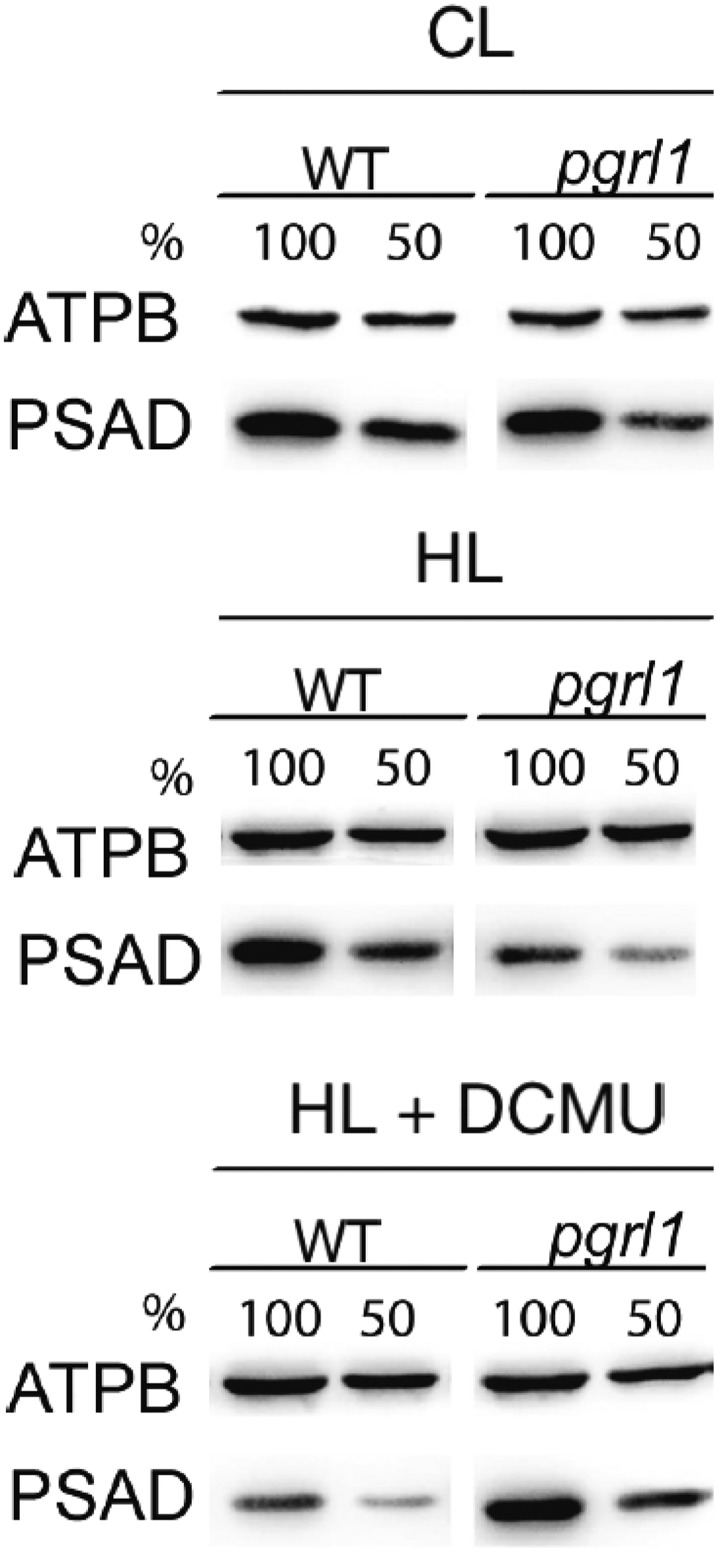
PSI photoinhibition in pgrl1 cells exposed to phototrophic high light conditions can be rescued by the addition of DCMU. Cells acclimated to low-light (CL) conditions (TAP medium at 20 µmol photons m−2 s−1 illumination) were shifted to HL (200 µmol photons m−2 s−1)/photoautotrophic conditions for 24 h. A control culture was kept in low light/photoautotrophic conditions. For DCMU treatment (indicated as + DCMU), 10 µm DCMU was added upon shift to phototrophic conditions. Changes in protein abundance of the PSI subunit PSAD in pgrl1 and the wild type at CL and HL in the absence and presence of DCMU. Detection of the chloroplastic ATPase ATPB subunit was utilized as a loading control. Cells were adjusted to a total chlorophyll amount of 2.5 mg (100%) or 1.25 mg (50%) prior to protein separation by SDS-PAGE.
DISCUSSION
Chloroplasts and mitochondria are organelles with a central role in energetic metabolism of photosynthetic organisms. Their activity in photosynthetic eukaryotes is tightly linked (Raghavendra and Padmasree, 2003) and undergoes both short- and long-term modulation in response to environmental stimuli. The strong interconnection between these organelles is evidenced also in this work where the introduction of complex I mitochondrial mutation (Δnd4, dum5, and dum25) in the pgrl1 background causes significant, although somewhat unexpected physiological effects. Surprisingly, reduction of complex I function altered photosensitivity in pgrl1 and allowed the survival of double mutants under strong illumination. Investigation of the alterations in photosynthetic apparatus in the double mutant Δnd4pgrl1 evidenced an altered PSI and PSII content with respect to parental strains, particularly in HL conditions. In Δnd4pgrl1, PSII is in fact more prone to photoinhibition while PSI content is higher, especially under strong illumination, with an increased capacity for electron transport that is independent from PSII (Figs. 4–6).
Δnd4pgrl1 also showed altered balance of electron transport with a more reduced PQ pool (Fig. 7) that stimulates state transitions with the association of LHCII to PSI even under strong illumination (Supplemental Fig. S4). In order to understand how a mitochondrial mutation can cause this phenotype, it should be considered that mitochondria use reducing power exported from the chloroplast to synthesize ATP (Taniguchi and Miyake, 2012). This activity is particularly important in pgrl1 as shown by their increased respiratory activity (Table I; Dang et al., 2014; Petroutsos et al., 2009) and by our finding on the inability to isolate pgrl1 mutants depleted of mitochondrial complex III. In Δnd4pgrl1, the reduction of mitochondrial respiration likely affects efficiency of reducing power export from the chloroplast. Those reducing equivalents accumulated in the stroma thus would be available to NADPH-PQ reductase enzymes like NDA2 that, in the absence of PGRL1, are the expected responsible players of the high AEF activity in the double mutant (Fig. 4). NDA2 protein content is not increased in pgrl1 (Dang et al., 2014) nor in Δnd4pgrl1 (Fig. 5), but this does not exclude an increased activity, nor the possibility that other NAD(P)H:PQ oxidoreductase present in the C. reinhardtii genome (Terashima et al., 2011) can be responsible for the increased AEF activity in Δnd4pgrl1. Independently from the molecular mechanisms responsible, this increased AEF in the double mutant in turn leads to reduction of the PQ pool even though PSII efficiency is much reduced.
One striking observation was that the introduction of a mitochondrial mutation in Δnd4pgrl1 showed a positive effect on the fitness under strong illumination even if PSII was strongly damaged. This finding can be explained by the above-described limitation of electron transport at the level of PQ. It is interesting to observe that this damage to PSII is paralleled by a higher stability of PSI, which is more abundant and active in Δnd4pgrl1 with respect to pgrl1 in HL conditions (Figs. 5 and 6). This can be explained by the observation that PSI is limited from the donor side with an accumulation of P700+ under illumination. P700+ is well known to be an efficient quencher of excitation energy (Shubin et al., 2008); thus, donor-limited PSI can safely dissipate the excess absorbed energy as heat. On the contrary, in pgrl1 electrons are accumulated at the PSI acceptor side, a condition where PSI is likely to be photoinhibited (reviewed in Chaux et al., 2015). This picture thus suggests that the double mutant Δnd4pgrl1 does not have an increased fitness because of a more efficient photosynthetic ETR, but because the damage from excess illumination in the double mutant targets prevalently PSII instead of PSI as it occurs in the parental strain pgrl1.
In order to understand why this difference in the damage localization has such a strong effect on growth, it should be considered that in all photosynthetic organisms PSII is damaged even under dim illumination (Roach and Krieger-Liszkay, 2014) and a very efficient system for PSII repair evolved to replace damaged D1 subunits with high turnover rates (Andersson and Aro, 2001). This mechanism allows cells to maintain a sufficient photosynthetic electron transport and growth even if they sustain a high level of damage. To our knowledge, a mechanism with equivalent efficiency to repair PSI has not been described, and here recovery from light damage is much less efficient since it requires the whole supercomplex to be degraded and then reassembled (Kudoh and Sonoike, 2002; Martin et al., 1997; Sonoike, 1996; Tikkanen and Grebe, 2018). In the absence of an efficient repair, PSI, if damaged, accumulates within the cell. Damaged PSI clearly has a negative impact on photosynthetic efficiency but also directly produce ROS species with a toxic effect on cells (Hippler et al., 2000). The impact of the damage localization is further supported by the observation that inhibition of PSII using a specific inhibitor (DCMU) protects PSI from light damage in pgrl1 (Fig. 7). DCMU addition clearly does not enhance photosynthesis but, again, has a beneficial effect to PSI in pgrl1 by limiting electron excess (Fig. 7). Thus, limitation of PSII function, either by DCMU or by complex I mutation, equally leads to increased overall fitness by reducing PSI damage.
The results presented here suggest that evolution selected a strategy to stand excess light by concentrating damage to a specific complex, PSII, and in particular its subunit D1 that is efficiently resynthesized without the need of dismantling the whole complex (Pokorska et al., 2009). With this strategy, PSII inhibition acts as a safety valve for all other components of the photosynthetic apparatus that are thus protected and can have a much higher stability (Tikkanen et al., 2014; Tiwari et al., 2016), avoiding a costly high turnover for all components. This work clearly demonstrates the impact of such as strategy, since the double mutant Δnd4pgrl1 did not show an increased photosynthesis with respect to parental strains but still was able to survive at high light intensity in contrast to pgrl1, because the damage was localized to PSII and not to PSI. In this strain, damage to PSII shows a protective effect on PSI by reducing electron flux to PSI avoiding overexcitation (Tiwari et al., 2016).
Regulation of photosynthetic electron flux can provide a significant contribution to PSI protection even when PSII damage is not as extensive, as observed here in Δnd4pgrl1. The so-called photosynthetic control, in fact, involves a decrease of the Cyt b6f complex turnover rate (Finazzi and Rappaport, 1998) activated under excess illumination by low lumenal pH (Colombo et al., 2016; Tikhonov, 2014). Such a decrease in lumenal pH slows down the rate limiting step in linear electron flow, namely, the oxidation of PQH2 at the Q0 site of the b6f complex (Stiehl and Witt, 1969), thus increasing the saturation of PQ pool. Observations presented here clearly suggest that under strong illumination, such a state is much safer for the cells since any eventual light damage is concentrated in PSII while PSI is instead safeguarded with a major positive impact on the cells ability to survive under strong illumination.
MATERIALS AND METHODS
Strains and Growth Conditions
The wild-type strain used in this study is derived from the wild-type 137c strain of the green algae Chlamydomonas reinhardtii. The pgrl1 mutant used in this study is the paromomycin-resistant insertional mutant described by Tolleter et al. (2011), which was crossed with our wild-type strain to obtain both mating types and to be able to reduce the genetic background difference with our mitochondrial mutants. The dum11, the complex III-deficient mutant described by Dorthu et al. (1992), and the complex I-deficient mutants dum25, dum5, and Δnd4 are described (Cardol et al., 2002; Massoz et al., 2017; Remacle et al., 2001, 2006).
The double mutants were obtained by crossing an mt+ pgrl1 with mt− mitochondrial mutants.
Cells were routinely grown at 25°C in Tris-acetate phosphate medium (TAP; Harris, 1989) or with no acetate (TMP) with 50 μmol photons m−2 s−1 illumination for CL illumination. Spot tests were performed plating 20 µL of a 106 cell culture on solid media in mixotrophic (TAP) or photoautotrophic (TMP) conditions. Cells were grown under HL intensities at 850 (solid medium) and 1,500 (liquid medium) μmol photons m−2 s−1.
Algal growth was measured through daily changes in the cell number monitored using a Cellometer Auto X4 cell counter (Nexcelom Bioscience). For cultivation in the presence of inhibitors, the wild type and pgrl1 were treated with 10 μm DCMU.
Whole-Cell Respiration and Oxygen Evolution Measurement
Whole cells from mixotrophic culture (TAP) grown at CL and HL were harvested by centrifugation and suspended in TAP medium at a concentration of 1 × 107 cells. Their respiratory rate was measured in the dark at 25°C using a Clark-type O2 electrode (Hansatech). Measurements were made in 2-mL total volume of cells culture. For the inhibition studies, 1 mm KCN, 1 mm SHAM, and 100 μm rotenone were used.
Chlorophyll Fluorescence Measurements
Chlorophyll fluorescence was measured in vivo on C. reinhardtii cells, using a Dual PAM 100 fluorometer (Walz). Cells were collected during the exponential growth phase and dark-adapted for 30 min keeping them gently mixed. After the dark adaptation, photosynthetic parameters were assessed using saturating pulses (200 ms, 9,000 μE m−2 s−1) during an actinic light treatment (12 min, starting from 0 to 438 μE m−2 s−1).
After 30 s of illumination, saturating pulse is given to fully reduce PSII acceptors and reach the maximal fluorescence yield (Fm). These three values allow the calculation of the maximal quantum yield of PSII photochemistry ΦPSII max [Fv/Fm = (Fm − F0)/Fm] and the PSII quantum yield at a given light intensity ΦPSII’ [(Fm’ − Fs)/Fm’] (Demmig-Adams et al., 1996; Maxwell and Johnson, 2000). The QA redox state was estimated by the parameter 1-qL (Kramer et al., 2004). NPQ measurements were obtained after maintaining cells for 10 generations under HL.
The 77K fluorescence spectra between 650 and 800 nm were recorded in a buffer containing 60% (w/v) glycerol, 10 mm HEPES (pH 7.5) with an excitation at 440 nm (LS 50 luminescence spectrometer; Perkin-Elmer).
Spectroscopic Measurements
The spectroscopic analysis was performed in vivo using a Joliot-type spectrophotometer (JTS-10; Biologic). Photosystems stoichiometry and photosynthetic electron flows were performed measuring the electrochromic shift (ECS) spectral change in intact cells, representing a shift in the pigment absorption bands associated with changes in the membrane potential and consequently photosystems activity (Bailleul et al., 2010).
ECS was measured as the difference between the signals at 520 and 546 nm (the positive and negative peaks of the ECS signal, respectively) to eliminate minor additional spectral changes that were associated with the membrane potential.
For these measurements, the final cellular concentration was 7 × 106 cell/mL for CL-adapted cells and 25 × 106 cell/mL for HL-adapted cells. DCMU (20 µm) and hydroxylamine (1.5 mm) inhibitors were employed to irreversibly block PSII charge separation, acting as acceptor and donor side inhibitors of PSII, respectively.
TEF and AEF were measured assessing the relaxation kinetics of the ECS signal in the dark after a steady-state illumination (Joliot and Joliot, 2002; Sacksteder and Kramer, 2000). AEF was estimated as the residual photochemical activity insensitive to a 20 µm DCMU treatment. In detail, cells were illuminated with saturating actinic light until the achievement of an ECS steady-state level (15 s at 940 μE m−2 s−1). The signal at steady state is proportional to transmembrane potential generated by PSII, Cyt b6f complex, PSI, and from transmembrane potential dissipation by the plastid ATP synthase.
When light is switched off, photosystem activities stop immediately, while ATP synthase and Cyt b6f complex activities remain unchanged. The difference between the slopes of the ECS signal measured in the light and after the light is switched off is proportional to the rate of PSI and PSII photochemistry (Dang et al., 2014; Joliot and Joliot, 2002).
The amplitude of the ECS was normalized on the total amount of PSI obtained with hydroxylamine and DCMU treatment corresponding to one charge transferred across the membrane per PSI center (Joliot and Joliot, 2002). Data are directly expressed in electrons transferred per second per photosystem I (e−1 s−1 PSI−1).
The PSI content was evaluated based on the maximum change in the absorption of P700+ in cells treated with DCMU and DBMIB (dibromothymoquinone; 300 μm) at a saturating actinic light (2,050 μmol of photons m−2 s−1, 630 nm) in an equal number of cells. Under these conditions, rereduction of P700+ through photosynthetic electron flow is largely slowed down, thereby allowing to evaluate the full extent of photooxidizable P700.
The fraction of oxidizable PSI at a specific light intensity (45, 150, and 320 µmol of photons m−2 s−1) is calculated as the ratio between the change in the absorption of P700+ at a specific light and the maximum change in the absorption of P700+ achievable in the presence of DCMU, DBMIB, and a saturating light of 2050 μmol of photons m−2 s−1. The measurements were performed using 25 × 106 cells/mL resuspended in TMP medium supplemented with 20% Ficoll.
Western-Blot Detection
SDS-PAGE was performed on total protein extract as described (Emonds-Alt et al., 2017; Remacle et al., 2010). Polyclonal antibodies directed against C. reinhardtii Psb1 (D1, Agrisera; 1:10,000), PsaD (PSI, Agrisera; 1:2,000), and PetA (CytF, Agrisera; 1:50,000) and antibodies against NDA2 (1:1,000; Jans et al., 2008), D2 (1:1,000), RbcL (1:5,000), and ATPA subunits were used.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Determination of the double mutation in Δnd4pgrl1.
Supplemental Figure S2. Impact of chloroplastic and mitochondrial mutation on growth of Chlamydomonas reinhardtii strains.
Supplemental Figure S3. Analysis of photosystem II efficiency of the wild type, Δnd4, pgrl1, and Δnd4pgrl1.
Supplemental Figure S4. 77K fluorescence analysis of the wild type, Δnd4, pgrl1, and Δnd4pgrl1.
Supplemental Figure S5. Nonphotochemical quenching monitored with PAM fluorometry.
Acknowledgments
We thank Anna Segalla (Department of Biology, University of Padova), Claire Remacle, Nadine Coosemans, and Michèle Radoux (Genetics and Physiology of Microalgae Laboratory, Univerisity of Liège) for support and Daniel Dannhausen for editing advice on the manuscript.
Footnotes
Articles can be viewed without a subscription.
V.L. is a Chargé de Recherche of the Belgian FNRS in mobility at Padova University thanks to a Marie-Curie fellowship (PISCOPIA mobility grant). M.H. acknowledges support from the German Science Foundation (Deutsch Forschungsgemeinschaft). I.S. and T.M. are grateful to Human Frontier Science Program (RGP0052) for support.
References
- Andersson B, Aro EM (2001) Photodamage and D1 protein turnover in photosystem II. In EM Aro, B Andersson, eds, Regulation of Photosynthesis: Advances in Photosynthesis and Respiration, Vol 11. Springer, Dordrecht, The Netherlands, pp 377–393 [Google Scholar]
- Bailleul B, Berne N, Murik O, Petroutsos D, Prihoda J, Tanaka A, Villanova V, Bligny R, Flori S, Falconet D, et al. (2015) Energetic coupling between plastids and mitochondria drives CO2 assimilation in diatoms. Nature 524: 366–369 [DOI] [PubMed] [Google Scholar]
- Bailleul B, Cardol P, Breyton C, Finazzi G (2010) Electrochromism: a useful probe to study algal photosynthesis. Photosynth Res 106: 179–189 [DOI] [PubMed] [Google Scholar]
- Cardol P, Forti G, Finazzi G (2011) Regulation of electron transport in microalgae. Biochim Biophys Acta 1807: 912–918 [DOI] [PubMed] [Google Scholar]
- Cardol P, Gloire G, Havaux M, Remacle C, Matagne R, Franck F (2003) Photosynthesis and state transitions in mitochondrial mutants of Chlamydomonas reinhardtii affected in respiration. Plant Physiol 133: 2010–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardol P, Matagne RF, Remacle C (2002) Impact of mutations affecting ND mitochondria-encoded subunits on the activity and assembly of complex I in Chlamydomonas. Implication for the structural organization of the enzyme. J Mol Biol 319: 1211–1221 [DOI] [PubMed] [Google Scholar]
- Chaux F, Peltier G, Johnson X (2015) A security network in PSI photoprotection: regulation of photosynthetic control, NPQ and O2 photoreduction by cyclic electron flow. Front Plant Sci 6: 875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M, Suorsa M, Rossi F, Ferrari R, Tadini L, Barbato R, Pesaresi P (2016) Photosynthesis control: An underrated short-term regulatory mechanism essential for plant viability. Plant Signal Behav 11: e1165382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang KV, Plet J, Tolleter D, Jokel M, Cuiné S, Carrier P, Auroy P, Richaud P, Johnson X, Alric J, Allahverdiyeva Y, Peltier G (2014) Combined increases in mitochondrial cooperation and oxygen photoreduction compensate for deficiency in cyclic electron flow in Chlamydomonas reinhardtii. Plant Cell 26: 3036–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig-Adams B, Adams WW III, Barker DH, Logan BA, Bowling DR, Verhoeven AS (1996) Using chlorophyll fluorescence to assess the fraction of absorbed light allocated to thermal dissipation of excess excitation. Physiol Plant 98: 253–264 [Google Scholar]
- Desplats C, Mus F, Cuiné S, Billon E, Cournac L, Peltier G (2009) Characterization of Nda2, a plastoquinone-reducing type II NAD(P)H dehydrogenase in Chlamydomonas chloroplasts. J Biol Chem 284: 4148–4157 [DOI] [PubMed] [Google Scholar]
- Dinant M, Baurain D, Coosemans N, Joris B, Matagne RF (2001) Characterization of two genes encoding the mitochondrial alternative oxidase in Chlamydomonas reinhardtii. Curr Genet 39: 101–108 [DOI] [PubMed] [Google Scholar]
- Dorthu MP, Remy S, Michel-Wolwertz MR, Colleaux L, Breyer D, Beckers MC, Englebert S, Duyckaerts C, Sluse FE, Matagne RF (1992) Biochemical, genetic and molecular characterization of new respiratory-deficient mutants in Chlamydomonas reinhardtii. Plant Mol Biol 18: 759–772 [DOI] [PubMed] [Google Scholar]
- Emonds-Alt B, Coosemans N, Gerards T, Remacle C, Cardol P (2017) Isolation and characterization of mutants corresponding to the MENA, MENB, MENC and MENE enzymatic steps of 5′-monohydroxyphylloquinone biosynthesis in Chlamydomonas reinhardtii Plant J 89: 141–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finazzi G, Rappaport F (1998) In vivo characterization of the electrochemical proton gradient generated in darkness in green algae and its kinetic effects on cytochrome b6f turnover. Biochemistry 37: 9999–10005 [DOI] [PubMed] [Google Scholar]
- Harris EH. editor (1989) The Chlamydomonas Sourcebook. Academic Press, San Diego, CA [Google Scholar]
- Hertle AP, Blunder T, Wunder T, Pesaresi P, Pribil M, Armbruster U, Leister D (2013) PGRL1 is the elusive ferredoxin-plastoquinone reductase in photosynthetic cyclic electron flow. Mol Cell 49: 511–523 [DOI] [PubMed] [Google Scholar]
- Hippler M, Biehler K, Krieger-Liszkay A, van Dillewjin J, Rochaix JD (2000) Limitation in electron transfer in photosystem I donor side mutants of Chlamydomonas reinhardtii. Lethal photo-oxidative damage in high light is overcome in a suppressor strain deficient in the assembly of the light harvesting complex. J Biol Chem 275: 5852–5859 [DOI] [PubMed] [Google Scholar]
- Jans F, Mignolet E, Houyoux PA, Cardol P, Ghysels B, Cuiné S, Cournac L, Peltier G, Remacle C, Franck F (2008) A type II NAD(P)H dehydrogenase mediates light-independent plastoquinone reduction in the chloroplast of Chlamydomonas. Proc Natl Acad Sci USA 105: 20546–20551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot P, Joliot A (2002) Cyclic electron transfer in plant leaf. Proc Natl Acad Sci USA 99: 10209–10214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer DM, Evans JR (2011) The importance of energy balance in improving photosynthetic productivity. Plant Physiol 155: 70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer DM, Johnson G, Kiirats O, Edwards GE (2004) New fluorescence parameters for the determination of QA redox state and excitation energy fluxes. Photosynth Res 79: 209. [DOI] [PubMed] [Google Scholar]
- Kudoh H, Sonoike K (2002) Irreversible damage to photosystem I by chilling in the light: cause of the degradation of chlorophyll after returning to normal growth temperature. Planta 215: 541–548 [DOI] [PubMed] [Google Scholar]
- Kukuczka B, Magneschi L, Petroutsos D, Steinbeck J, Bald T, Powikrowska M, Fufezan C, Finazzi G, Hippler M (2014) Proton Gradient Regulation5-Like1-mediated cyclic electron flow is crucial for acclimation to anoxia and complementary to nonphotochemical quenching in stress adaptation. Plant Physiol 165: 1604–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecler R, Vigeolas H, Emonds-Alt B, Cardol P, Remacle C (2012) Characterization of an internal type-II NADH dehydrogenase from Chlamydomonas reinhardtii mitochondria. Curr Genet 58: 205–216 [DOI] [PubMed] [Google Scholar]
- Martin RE, Thomas DJ, Tucker DE, Herbert SK (1997) The effects of photooxidative stress on photosystem I measured in vivo in Chlamydomonas. Plant Cell Environ 20: 1451–1461 [Google Scholar]
- Massoz S, Hanikenne M, Bailleul B, Coosemans N, Radoux M, Miranda-Astudillo H, Cardol P, Larosa V, Remacle C (2017) In vivo chlorophyll fluorescence screening allows the isolation of a Chlamydomonas mutant defective for NDUFAF3, an assembly factor involved in in mitochondrial complex I assembly. Plant J 92: 584–595 [DOI] [PubMed] [Google Scholar]
- Matagne RF, Michel-Wolwertz MR, Munaut C, Duyckaerts C, Sluse F (1989) Induction and characterization of mitochondrial DNA mutants in Chlamydomonas reinhardtii. J Cell Biol 108: 1221–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN (2000) Chlorophyll fluorescence--a practical guide. J Exp Bot 51: 659–668 [DOI] [PubMed] [Google Scholar]
- Petroutsos D, Terauchi AM, Busch A, Hirschmann I, Merchant SS, Finazzi G, Hippler M (2009) PGRL1 participates in iron-induced remodeling of the photosynthetic apparatus and in energy metabolism in Chlamydomonas reinhardtii. J Biol Chem 284: 32770–32781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokorska B, Zienkiewicz M, Powikrowska M, Drozak A, Romanowska E (2009) Differential turnover of the photosystem II reaction centre D1 protein in mesophyll and bundle sheath chloroplasts of maize. Biochim Biophys Acta 1787: 1161–1169 [DOI] [PubMed] [Google Scholar]
- Raghavendra AS, Padmasree K (2003) Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci 8: 546–553 [DOI] [PubMed] [Google Scholar]
- Remacle C, Baurain D, Cardol P, Matagne RF (2001) Mutants of Chlamydomonas reinhardtii deficient in mitochondrial complex I: characterization of two mutations affecting the nd1 coding sequence. Genetics 158: 1051–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remacle C, Cardol P, Coosemans N, Gaisne M, Bonnefoy N (2006) High-efficiency biolistic transformation of Chlamydomonas mitochondria can be used to insert mutations in complex I genes. Proc Natl Acad Sci USA 103: 4771–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remacle C, Coosemans N, Jans F, Hanikenne M, Motte P, Cardol P (2010) Knock-down of the COX3 and COX17 gene expression of cytochrome c oxidase in the unicellular green alga Chlamydomonas reinhardtii. Plant Mol Biol 74: 223–233 [DOI] [PubMed] [Google Scholar]
- Roach T, Krieger-Liszkay A (2014) Regulation of photosynthetic electron transport and photoinhibition. Curr Protein Pept Sci 15: 351–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacksteder CA, Kramer DM (2000) Dark-interval relaxation kinetics (DIRK) of absorbance changes as a quantitative probe of steady-state electron transfer. Photosynth Res 66: 145–158 [DOI] [PubMed] [Google Scholar]
- Salinas T, Larosa V, Cardol P, Maréchal-Drouard L, Remacle C (2014) Respiratory-deficient mutants of the unicellular green alga Chlamydomonas: a review. Biochimie 100: 207–218 [DOI] [PubMed] [Google Scholar]
- Shubin VV, Terekhova IN, Kirillov BA, Karapetyan NV (2008) Quantum yield of P700+ photodestruction in isolated photosystem I complexes of the cyanobacterium Arthrospira platensis Photochem Photobiol Sci 7: 956–962 [DOI] [PubMed] [Google Scholar]
- Sonoike K. (1996) Photoinhibition of photosystem I: Its physiological significance in chilling sensitivity of plants. Plant Cell Physiol 37: 239–247 [Google Scholar]
- Stiehl HH, Witt HT (1969) Quantitative treatment of the function of plastoquinone in phostosynthesis. Z Naturforsch B 24: 1588–1598 [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Miyake H (2012) Redox-shuttling between chloroplast and cytosol: integration of intra-chloroplast and extra-chloroplast metabolism. Curr Opin Plant Biol 15: 252–260 [DOI] [PubMed] [Google Scholar]
- Terashima M, Specht M, Hippler M (2011) The chloroplast proteome: a survey from the Chlamydomonas reinhardtii perspective with a focus on distinctive features. Curr Genet 57: 151–168 [DOI] [PubMed] [Google Scholar]
- Tikhonov AN. (2014) The cytochrome b6f complex at the crossroad of photosynthetic electron transport pathways. Plant Physiol Biochem 81: 163–183 [DOI] [PubMed] [Google Scholar]
- Tikkanen M, Grebe S (2018) Switching off photoprotection of photosystem I - a novel tool for gradual PSI photoinhibition. Physiol Plant http://dx.doi.org/10.1111/ppl.12618 [DOI] [PubMed]
- Tikkanen M, Mekala NR, Aro EM (2014) Photosystem II photoinhibition-repair cycle protects photosystem I from irreversible damage. Biochim Biophys Acta 1837: 210–215 [DOI] [PubMed] [Google Scholar]
- Tiwari A, Mamedov F, Grieco M, Suorsa M, Jajoo A, Styring S, Tikkanen M, Aro EM (2016) Photodamage of iron-sulphur clusters in photosystem I induces non-photochemical energy dissipation. Nat Plants 2: 16035. [DOI] [PubMed] [Google Scholar]
- Tolleter D, Ghysels B, Alric J, Petroutsos D, Tolstygina I, Krawietz D, Happe T, Auroy P, Adriano JM, Beyly A, et al. (2011) Control of hydrogen photoproduction by the proton gradient generated by cyclic electron flow in Chlamydomonas reinhardtii. Plant Cell 23: 2619–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC. (2013) Alternative oxidase: a mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int J Mol Sci 14: 6805–6847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vener AV, Van Kan PJ, Gal A, Andersson B, Ohad I (1995) Activation/deactivation cycle of redox-controlled thylakoid protein phosphorylation. Role of plastoquinol bound to the reduced cytochrome bf complex. J Biol Chem 270: 25225–25232 [DOI] [PubMed] [Google Scholar]
- Yamori W, Shikanai T (2016) Physiological functions of cyclic electron transport around photosystem I in sustaining photosynthesis and plant growth. Annu Rev Plant Biol 67: 81–106 [DOI] [PubMed] [Google Scholar]
- Zalutskaya Z, Lapina T, Ermilova E (2015) The Chlamydomonas reinhardtii alternative oxidase 1 is regulated by heat stress. Plant Physiol Biochem 97: 229–234 [DOI] [PubMed] [Google Scholar]
- Zito F, Finazzi G, Delosme R, Nitschke W, Picot D, Wollman FA (1999) The Qo site of cytochrome b6f complexes controls the activation of the LHCII kinase. EMBO J 18: 2961–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]