Serotonin biosynthesis is dramatically elevated, and closely linked with dark-brown color of the endosperm, in high-lysine rice.
Abstract
Cereal endosperms produce a vast array of metabolites, including the essential amino acid lysine (Lys). Enhanced accumulation of Lys has been achieved via metabolic engineering in cereals, but the potential connection between metabolic engineering and Lys fortification is unclear. In mature seeds of engineered High Free Lysine (HFL) rice (Oryza sativa), the endosperm takes on a characteristic dark-brown appearance. In this study, we use an integrated metabolomic and transcriptomic approach combined with functional validation to elucidate the key metabolites responsible for the dark-brown phenotype. Importantly, we found that serotonin biosynthesis was elevated dramatically and closely linked with dark-brown endosperm color in HFL rice. A functional connection between serotonin and endosperm color was confirmed via overexpression of TDC3, a key enzyme of serotonin biosynthesis. Furthermore, we show that both the jasmonate signaling pathway and TDC expression were strongly induced in the late stage of endosperm development of HFL rice, coinciding with serotonin accumulation and dark-brown pigmentation. We propose a model for the metabolic connection between Lys and serotonin metabolism in which elevated 2-aminoadipate from Lys catabolism may play a key role in the connection between the jasmonate signaling pathway, serotonin accumulation, and the brown phenotype in rice endosperm. Our data provide a deeper understanding of amino acid metabolism in rice. In addition, the finding that both Lys and serotonin accumulate in HFL rice grains should promote efforts to create a nutritionally favorable crop.
Plants are capable of producing more than 200,000 kinds of metabolites (Goodacre et al., 2004). All metabolites are produced by specific pathways, which form a complex metabolic network via direct or indirect connections (Fiehn et al., 2000; Fernie et al., 2004). Cereal seeds have numerous metabolites, some of which are related to yield and quality (Keurentjes et al., 2006) and play an important role in human nutrition and health (Saito and Matsuda, 2010). Unfortunately, the nutritional quality of cereals can suffer from deficiency in essential amino acids, especially Lys (FAO, 1991, 1993; Juliano and Gu, 1995; Zhu et al., 2007; Angelovici et al., 2010).
Among all living organisms, higher plants are considered to exhibit the highest complexity in metabolic networks, making it particularly challenging to enhance their value-added compositional traits via metabolic engineering (Dersch et al., 2016). Amino acid pathways are important targets for metabolic engineering in higher plants, to biofortify products with essential amino acids, such as Lys, Met, and Trp, or to enhance stress tolerance via elevated levels of Pro or Glu (Galili, 2002). Different amino acid metabolic pathways have been well elucidated, but the associations and connections between them are not clear (Less and Galili, 2009). Therefore, it is important to elucidate the metabolic flux and connections between the pathways of target amino acids for fortification and other pathways, especially in cereals as the main food for humans.
In higher plants, Lys is synthesized through a branch of the Asp family pathway and strongly regulated by a feedback inhibition loop (Galili, 1995). The activity of Asp kinase (AK), the first enzyme in the pathway, is inhibited by both Lys and Thr. Furthermore, Lys also inhibits dihydrodipicolinate synthase (DHPS), which is the first special enzyme committed to Lys biosynthesis (Galili, 2002). Additionally, Lys ketoglutaric acid reductase/saccharopine dehydropine dehydrogenase (LKR/SDH) is a bifunctional enzyme that plays a key role in the Lys degradation pathway (Galili et al., 2001). Efforts have been made to increase Lys content by altering Lys metabolism in plants, including in Arabidopsis (Arabidopsis thaliana; Zhu and Galili, 2003, 2004), maize (Zea mays; Frizzi et al., 2008), and rice (Oryza sativa; Long et al., 2013; Yang et al., 2016). However, the seed germination of high-Lys Arabidopsis was retarded significantly (Zhu and Galili, 2003, 2004). In QPM mutant maize, the endosperm was hardened and quality was compromised (Betrán et al., 2003; Gibbon et al., 2003). In soybean (Glycine max), seed-specific expression of feedback-insensitive DHPS and AK resulted in wrinkled seeds and low germination (Falco et al., 1995). In rice, morphological changes were observed in transgenic rice seeds upon expressing bacterial DHPS, although the free Lys content did increase 1.77-fold compared with the wild type (Lee et al., 2001). More recently, a combination of overexpressed feedback-insensitive bacterial AK and DHPS and inhibited LKR/SDH resulted in a 60-fold increase of free Lys in transgenic seeds compared with the wild type. Unlike in Arabidopsis, plant performance and seed germination were unchanged between the high-Lys transgenic rice and the wild type (Long et al., 2013). Therefore, it seems that differences in the metabolic flux and the connections associated with Lys metabolism may exist among plant species.
The metabolic connection of the Asp family pathway with the tricarboxylic acid cycle has been mapped in Arabidopsis (Galili, 2011; Less et al., 2011). The results from Galili’s group have shown that enhancing Lys synthesis and inhibiting its catabolism in Arabidopsis had a notable influence on the levels of several tricarboxylic acid cycle metabolites (Zhu and Galili, 2003, 2004; Angelovici et al., 2009, 2010; Galili and Amir, 2013). It was proposed that the amino acid catabolic flux within the Asp family pathway contributed significantly to cellular energy homoeostasis during both seed development and germination (Galili and Amir, 2013). Furthermore, in plants, the regulatory metabolic link of the Asp family pathway with the tricarboxylic acid cycle could further explore various abiotic stress responses (Galili, 2011). Previous studies have shown that the saccharopine pathway of Lys catabolism also may be involved in a stress-related metabolic pathway in both dicots and monocots. Yet, the metabolic linkage and stress response between Arabidopsis and maize differed (Moulin et al. 2000, 2006; Galili, 2002; Less et al. 2011; Kiyota et al., 2015).
Taken together, the previous studies indicate that the Asp family may have multiple roles in plant metabolism, and their biological functions in monocotyledons might be different from those in dicot plants. To date, most literature on the roles of the Asp family had focused on dicots but was limited in monocotyledons. Therefore, it is important to further understand the role of the Lys metabolic pathway and its metabolic network in diverse organisms, especially in cereals. This should help efforts to design an optimal strategy to improve nutritional quality (Galili, 2011; Neshich et al., 2013; Wang and Galili, 2016).
Recently, we generated a series of transgenic rice lines with overexpression of feedback-insensitive bacterial AK and DHPS (35S) or down-regulation of LKR/SDH (Ri or GR) with an aim to breed Lys-rich rice (Long et al., 2013). Furthermore, we bred High Free Lysine (HFL) rice lines by pyramiding the 35S and GR lines (Yang et al., 2016; Fig. 1A). The free Lys content in the mature endosperm of two HFL transgenic lines (HFL1 and HFL2) is increased by 25-fold compared with the wild type. Importantly, the Lys-biofortified rice contributed significantly to improved growth performance, food efficiency, and Lys availability in growing rats (Yang et al., 2017a, 2017b). However, the mature endosperm of these HFL rice lines had a dark-brown appearance (Yang et al., 2016; Fig. 1A), which has not been reported previously in Arabidopsis, tobacco (Nicotiana tabacum), and canola (Brassica napus; Shaul and Galili, 1992, 1993; Falco et al., 1995; Zhu and Galili, 2003).
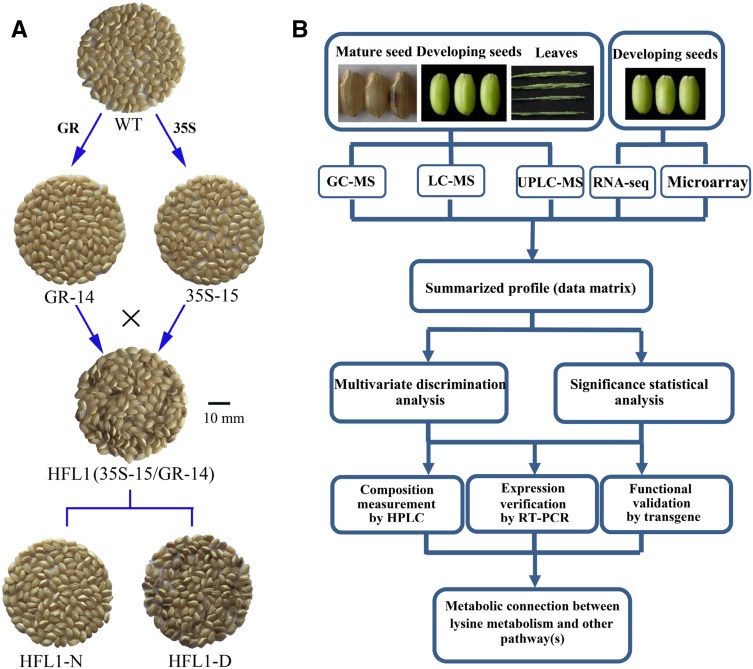
Figure 1.
Rice seed samples and schematic diagram of the experimental approaches used in this study. A, Appearance of dehulled mature seeds in various transgenic rice lines and their wild type (WT). GR-14 and 35S-15 transgenic lines were generated by transformation of the GR and 35S constructs into the wild type, respectively. The HFL1 pyramiding transgenic line was produced by crossing GR-14 and 35S-15 lines (35S-15/GR-14) and contained the above two transgenic constructs. HFL1-N and HFL1-D were mature seeds with normal or dark-brown appearance, respectively, from the same set of HFL1 transgenic lines. Bar = 10 mm. B, Schematic diagram of the experimental approaches used in this study.
This study aimed to elucidate the underlying connections of the dark-brown seed phenotype with Lys biofortification as well as the cause of the dark-brown color formation in mature endosperm of HFL rice, using an integrated approach based on metabolomic and transcriptomic analyses combined with functional validation (Fig. 1B). Our results indicated an underlying connection between Lys and serotonin metabolism in rice endosperm. We found that high accumulation of free Lys in HFL rice endosperm induced many plant stress responses, especially the jasmonate (JA) signing pathway, which resulted in the enhancement of serotonin biosynthesis and ultimately led to the observed dark-brown endosperm phenotype.
RESULTS
Dark-Brown Color Appearance in HFL Endosperm
The engineered rice, including 35S, GR, and their pyramiding HFL lines, grew normally in the field (Yang et al., 2016), and there was no significant difference in grain weight and size or husk appearance compared with the wild type (Supplemental Fig. S1). The dehulled mature seeds of 35S-15 and GR-14 lines showed comparable appearance to their wild type. However, the pyramid HFL1 rice (35S-15/GR-14) had some dehulled seeds with an apparent dark-brown endosperm phenotype (Fig. 1A). The same phenomenon appeared in another pyramiding HFL2 line derived from the cross between 35S-15 and GR-65 lines (Supplemental Fig. S2A). Under the normal summer growing conditions of Yangzhou, China, the strongly dark-brown grains of HFL1 and HFL2 accounted for more than 20% of the grains and consistently appeared in every generation of both HFL lines. From the cross-sectional observation of endosperm, the degree of dark-brown color was different among mature seeds derived from the same HFL line, and the dark-brown coloring existed not only in the aleurone layers and the outer endosperm but also in the inner endosperm (Fig. 2A). In addition, the dark-brown HFL1 seeds had a slight increase in chalkiness when compared with the wild type (Fig. 2A).
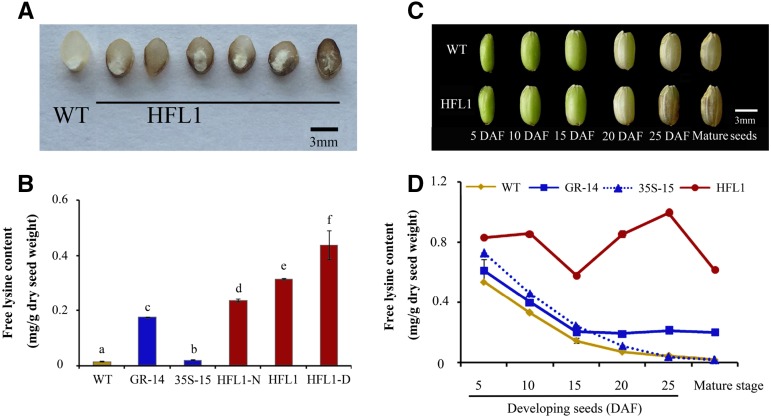
Figure 2.
Appearance and free Lys content of mature and developing seeds from different transgenic lines and their wild type (WT). A, Cross section of mature grains showing the aleurone layer and the mature endosperm. B, Free Lys content in mature seeds from the wild type, the two parental lines (GR-14 and 35S-15), and the pyramid HFL1 lines. HFL1-D and HFL1-N were mature seeds from the same HFL1 line with normal and dark-brown color appearance, respectively. C, Appearance of caryopsis at different developmental stages. D, Dynamic change of free Lys content during seed development. Error bars represent sd of three biological replicates, and different letters indicate statistical significance among transgenic plants and the wild type at P < 0.05. Bars = 3 mm.
Based on the dark-brown color, all the dehulled rice kernels from the same HFL1 (also called HFL1-M, which included dark-brown color and normal appearance of seeds and not to distinguish differences according to the seed phenotype) line could be divided into two types, HFL1-D, with strong dark-brown color, and HFL1-N, with normal appearance consistent with the 35S-15 and GR-14 parents or the wild type (Fig. 1A). The free Lys content in the different mature seed types was measured separately, and HFL1 as well as its two types of endosperm contained more free Lys than either the wild type or the parents, 35S-15 and GR-14 (Fig. 2B). The level of free Lys was closely related to the endosperm color among the seeds from the same HFL1 line; that is, the HFL1-D endosperm accumulated the highest level of free Lys while HFL1-N contained the lowest, and HFL1 contained moderate levels (Fig. 2B). The same phenomenon also existed in another pyramid line, HFL2, derived from the cross between 35S-15 and GR-65 (Supplemental Fig. S2B). Therefore, it is obvious that the dark-brown color was closely related to the accumulation of free Lys in mature endosperm of HFL rice.
During endosperm development (Fig. 2C), the dark-brown substance in HFL1 caryopsis was observed mainly in the later stage (more than 25 d after flowering [DAF]). Dynamic analysis revealed that the free Lys level was high in the wild-type rice during the early stages of seed development (5 DAF) yet declined rapidly along with grain filling and was reduced to a very low level after seed maturation (Fig. 2D). A similar decrease in the free Lys content also was detected in the 35S-15 and GR-14 lines (Fig. 2D). By contrast, during endosperm development in HFL1 rice, the free Lys accumulation always remained highly active, leading to a dramatic difference in free Lys levels of the mature seeds between HFL1 and its parents or the wild type (Fig. 2D). There was a decline of free Lys from 10 to 15 DAF relative to other periods, which might be related to the expansion via deposition of storage proteins in developing seeds about 12 DAF (Qu and Takaiwa, 2004). Thus, there was a tight association between the dark-brown color emerging and Lys accumulation at the late stage of endosperm development in HFL rice.
Global Comparison of the Metabolome between HFL Dark-Brown Endosperm and Related Samples
To elucidate the global metabolome change in HFL rice seeds, a metabolomic platform integrated with ultra-high-performance liquid chromatography (UHPLC)-tandem mass spectrometry (MS) and gas chromatography (GC)-MS was applied as described above (Fig. 1B). A total of 16 samples were subjected to metabolomic analyses, including four mature leaf samples collected at 10 DAF, four dehulled developing seed samples at 10 DAF, and eight dehulled mature seed samples (Supplemental Table S1). Among the 285 identified metabolites with known structures in rice, 232, 244, and 223 molecules or metabolites were detected in the leaf, developing seeds, and mature seeds, respectively (Supplemental Table S1). Details of all detected metabolites are listed in Supplemental Data Sets S1 to S4.
To obtain a global perspective on these detected metabolites, we first performed a principal component analysis (PCA) based on correlations of metabolite levels in mature seeds (Fig. 3A), leaf, or developing seeds (Supplemental Fig. S3). There was no significant difference in the levels of metabolites between wild-type and transgenic rice from either leaf or developing seeds at 10 DAF (Supplemental Fig. S3). However, the first and second principal components (PCs) accounted for 76.6% and 8.3% of the total variance among the eight mature seed samples, respectively (Fig. 3A). There were three intensive positions of metabolomes from mature seeds among eight samples, which could be distinguished clearly from each other based on combinations of PC1 and PC2 (Fig. 3A). The first position (position I) contained the wild type, 35S-15, and two GR samples, indicating a high similarity in metabolites in the mature seeds of 35S and GR transgenic rice compared with the wild type. The second position (position II) included samples from both pyramid HFL lines, except the HFL1-D sample with strong dark-brown color. This sample belonged to the third position (position III; Fig. 3A). There was a major difference in the metabolome of HFL mature seeds, especially in HFL1-D mature seeds, compared with their parents or the wild type.
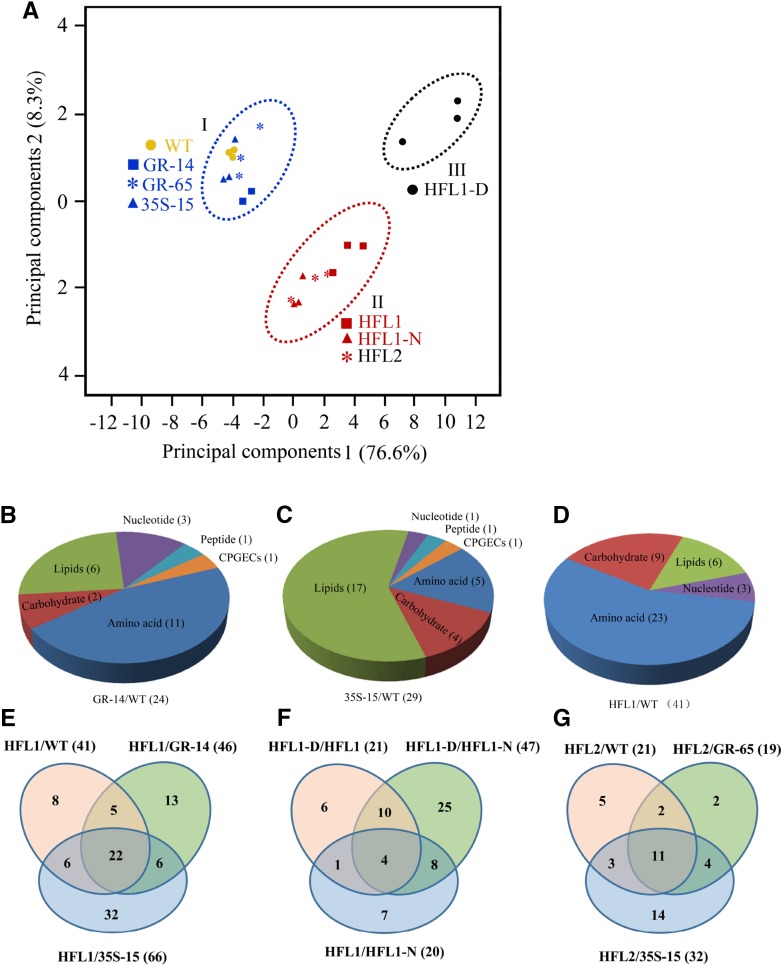
Figure 3.
Overview of the metabolomic changes in mature seeds among different comparisons. A, PCA of detected metabolomic data in mature seeds. B to D, Pie charts representing the distribution of DMs in mature seeds from GR-14, 35S-15, and HFL1 compared with the wild type (WT), respectively. E to G, Venn diagrams of DMs detected in mature seeds from different comparisons.
The relative content of all detected metabolites was compared between transgenic rice and the wild type, and the number of statistically differential metabolites (DMs) for each comparison is shown in Supplemental Table S1. In the developing seeds at 10 DAF, there were several metabolites detected at different levels in either 35S-15 (nine) or GR-14 (one) or their pyramid HFL1 rice (seven) compared with the wild type. In the leaf samples, GR-14 presented only two DMs, but 35S-15 and the HFL1 lines contained many DMs (26 and 21, respectively). This difference could result from the constitutive expression of foreign AK and DHPS in 35S-15 and HFL1 rice, while the ectopic expression of transgenes in GR-14 is driven by an endosperm-specific promoter (Yang et al., 2016). In contrast to the limited number of DMs detected in the mature leaf and developing seed at 10 DAF, more DMs were detected in the mature seeds among the transgenic rice when compared with the wild type. In HFL1-D mature seeds, the number of DMs (46) detected accounted for more than 20% of the total detected compounds (223), and most were up-regulated (41 out of 46). Furthermore, the simple clustering analyses of DMs showed that most of the DMs in the mature seeds belonged to amino acids, followed by lipids, carbohydrates, peptides, nucleotides, and cofactors (prosthetic group, electronic carrier), in Lys accumulation of GR and HFL lines, not in 35S-15 (Fig. 3, B–D). Taken together, the global metabolomic analyses revealed a dramatic change in the metabolome in HFL mature seeds, especially in the HFL1-D mature seeds with a strong dark-brown color phenotype.
Identification of Specific Metabolites Responsible for the Dark-Brown Phenotype in Mature Seeds
To identify the major substance(s) responsible for the dark-brown endosperm phenotype, we compared the metabolites in HFL mature seeds with their corresponding parents (35S and GR) and the wild type (Fig. 3; Supplemental Data Set S1). In the HFL1 mature seeds, 22 DMs were found to accumulate differently from three related samples, 35S-15, GR-14, and the wild type (Fig. 3E). In the case of HFL2, with relatively weaker dark-brown appearance than HFL1 (Supplemental Fig. S2), 11 compounds were altered significantly compared with 35S-15, GR-65, and the wild type (Fig. 3G). These DMs are involved in the metabolism of amino acids, carbohydrates, and nucleic acids (Table I; Supplemental Table S2). Moreover, a combined multiple comparison was performed between HFL1-D with HFL1-M (HFL1) or HFL1-N (Fig. 3F), and the results showed that only four compounds were altered specifically in HFL1-D mature seeds (Fig. 3F). These were 2-aminoadipate, Lys, serotonin, and Trp (Table I), and they differed greatly in HFL1 and HFL2 mature seeds compared with their parents (Table I; Supplemental Table S2).
Table I. Differential metabolites and their fold change in mature seeds among multiple comparisons within HFL1 and its derived samples.
Dark gray-shaded cells indicate that the mean values were significantly higher or lower for that comparison, respectively, at the level of P ≤ 0.05; light gray-shaded cells indicate the same at the level of 0.05 < P < 0.1.
| Biochemical Name | Fold Change (Ratios of Group Means from Scaled Imputed Data) |
|||||||
|---|---|---|---|---|---|---|---|---|
| GR-14/Wild Type | 35S-15/Wild Type | HFL1/Wild Type | HFL1/GR-14 | HFL1/35S-15 | HFL1-D/HFL1-M | HFL1-D/HFL1-N | HFL1-M/HFL1-N | |
| 2-Aminoadipate | 1.55 | 1.88 | 5.28 | 3.41 | 2.81 | 1.41 | 2.04 | 1.45 |
| Lys | 12.52 | 1.62 | 194.77 | 15.56 | 120.2 | 1.93 | 3.07 | 1.59 |
| Serotonin | 2.16 | 1.51 | 8 | 3.7 | 5.3 | 1.75 | 3.03 | 1.72 |
| Trp | 0.81 | 1.14 | 1.91 | 2.35 | 1.68 | 1.61 | 2.38 | 1.47 |
| Nδ-Acetyl-Lys | 2.99 | 1 | 11.16 | 3.73 | 11.16 | 1.15 | 1.47 | 1.28 |
| Gln | 1.49 | 1.06 | 2.49 | 1.67 | 2.36 | 1.49 | 3.03 | 2.04 |
| Arg | 1.31 | 1.05 | 2.97 | 2.27 | 2.82 | 1.25 | 1.49 | 1.20 |
| Pseudouridine | 1.36 | 0.98 | 1.92 | 1.41 | 1.95 | 1.30 | 1.32 | 1.01 |
| Phe | 1.05 | 0.88 | 1.35 | 1.29 | 1.54 | 1.15 | 1.45 | 1.25 |
| Nδ-Trimethyl-Lys | 1.65 | 1.19 | 1.99 | 1.21 | 1.66 | 1.04 | 1.03 | 0.99 |
| Pipecolate | 5.94 | 1.84 | 12.43 | 2.09 | 6.74 | 0.96 | 0.96 | 1.00 |
| Saccharopine | 22.19 | 1.41 | 67.15 | 3.03 | 47.49 | 1.08 | 1.18 | 1.09 |
| Nδ-Acetyl-Orn | 9.21 | 1.75 | 23.11 | 2.51 | 13.21 | 1.20 | 1.47 | 1.20 |
| γ-Aminobutyrate | 1.15 | 0.55 | 2.06 | 1.78 | 3.76 | 1.49 | 2.38 | 1.61 |
| Putrescine | 1.14 | 1.93 | 6.97 | 6.14 | 3.62 | 1.05 | 1.96 | 1.85 |
| Ribose | 0.98 | 0.84 | 1.21 | 1.24 | 1.44 | 1.06 | 1.32 | 1.23 |
| Xylonate | 0.86 | 0.73 | 1.72 | 1.99 | 2.34 | 1.16 | 1.59 | 1.37 |
| Uracil | 1.21 | 0.78 | 2.35 | 1.95 | 3.01 | 1.23 | 1.72 | 1.39 |
| Squalene | 1.06 | 1.08 | 1.5 | 1.42 | 1.4 | 1.06 | 1.16 | 1.09 |
| γ-Glutamyl-Gln | 1.01 | 1 | 2.04 | 2.03 | 2.04 | 1.59 | 2.94 | 1.85 |
| Citramalate | 1.21 | 3.19 | 2.56 | 2.12 | 0.8 | 2.38 | 3.70 | 1.52 |
| Adenine | 0.81 | 0.71 | 1.23 | 1.52 | 1.73 | 1.49 | 2.22 | 1.49 |
| GlcNAc | 0.52 | 0.63 | 0.21 | 0.39 | 0.32 | 0.63 | 0.75 | 1.20 |
| Glucosaminate | 0.92 | 0.76 | 0.54 | 0.59 | 0.71 | 0.89 | 0.96 | 1.08 |
| Pantothenate | 0.87 | 0.93 | 0.65 | 0.74 | 0.7 | 0.93 | 0.84 | 0.90 |
Among them, 2-aminoadipate is a degradation product of saccharopine from the Lys catabolism pathway. Among previous reports of mutated or transgenic plants with altered Lys metabolism, no evidence had been shown that elevated Lys and/or 2-aminoadipate could result in brown-colored seeds. In higher plants, serotonin is derived from Trp via the intermediate tryptamine, and all three compounds are involved in Trp metabolism as well as aromatic amino acid catabolism (Fig. 4A; Ishihara et al., 2008). A previous study showed that increasing serotonin biosynthesis could result in the dark-brown phenotype in transgenic rice (Kanjanaphachoat et al., 2012). Therefore, Trp and serotonin, especially the latter, might be responsible for the dark-brown endosperm in HFL rice.
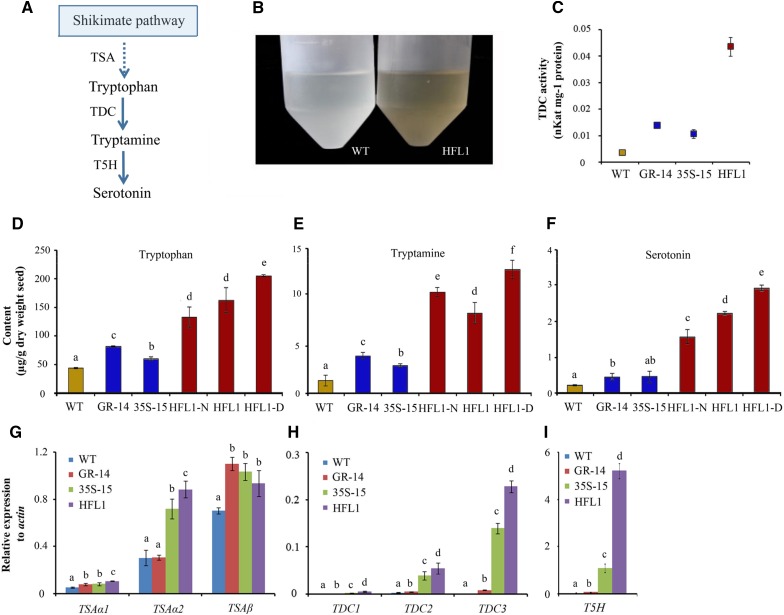
Figure 4.
Differential expression of Trp and serotonin biosynthesis pathway among different transgenic lines and the wild type (WT). A, Trp and serotonin biosynthesis pathway in higher plants (modified from Dubouzet et al., 2013). B, Serotonin extraction solution from mature seeds of HFL1 and the wild type. C, TDC enzyme activity in developing seeds at 15 DAF. D to F, Contents of free Trp (D), tryptamine (E), and serotonin (F) in mature seeds detected using LC-MS. G to I, Relative expression of TSA (G), TDC (H), and T5H (I) genes in developing seeds at 10 DAF. Error bars represent sd of three biological replicates, and different letters indicate significant differences among transgenic plants and the wild type at P < 0.05.
According to the metabolomic data (Table I; Supplemental Table S2; Supplemental Fig. S4, A–C), the contents of Trp, serotonin, and tryptamine in mature seeds are similar among the 35S and GR transgenic lines and the wild type, but they showed a dramatic increase in the HFL samples, especially in HFL1-D seeds. Interestingly, their levels also showed close correlation with the free Lys level among the seeds from the same HFL1 line; that is, the HFL1-D endosperms exhibited the highest level of serotonin-related metabolites as well as of free Lys (Supplemental Fig. S4). In contrast to mature seeds, the above three compounds showed no significant difference in accumulation in mature leaves or developing seeds among all the transgenic rice lines (GR, 35S, and HFL1) when compared with the wild-type plants (Supplemental Data Sets S3 and S4).
The concentrations of serotonin, Trp, and tryptamine were quantified further using HPLC-MS. It is interesting that extracts from mature seeds showed a dark-brown color, in contrast to the clear-colored extracts from wild-type seeds (Fig. 4B). Using liquid chromatography (LC)-MS, while a similar or small difference in Trp, tryptamine, and serotonin accumulation was detected among 35S-15, GR-14, and their wild-type lines, a dramatic increase of these metabolites was detected in the HFL1 mature seed extracts (Fig. 4, D–F). Specifically, the HFL1-D seeds with the dark-brown phenotype showed the highest metabolite accumulation among all samples tested, with a 14-fold increase in serotonin accumulation (Fig. 4F). Thus, these data further support the notion that serotonin enrichment in mature seeds is closely linked to the dark-brown endosperm in HFL mature seeds.
Enhancement of the Serotonin Biosynthesis Pathway in Developing Seeds of HFL Rice
The serotonin biosynthesis pathway was further compared between HFL and other samples. Trp, synthesized via Trp synthase (TSA) from chorismate, is decarboxylated by TDC to produce tryptamine, which is then utilized by tryptamine 5-hydroxylase (T5H) to generate serotonin (Fig. 4A). Along this pathway, TDC serves as a bottleneck in regulating serotonin biosynthesis (Kang et al., 2007a, 2007b). In the rice genome, there are three copies of TSA genes (TSAα1, TSAα2, and TSAβ), three TDC genes (TDC1, TDC2, and TDC3), and only one T5H gene (Byeon et al., 2014).
Using reverse transcription-quantitative PCR (RT-qPCR), we revealed that the transcription levels of the above seven genes were very low in developing seeds of wild-type rice (Fig. 4, G–I; Supplemental Fig. S5). However, most of these genes were highly expressed in the developing seeds of transgenic rice, especially TDC and T5H in HFL1 rice (Fig. 4, H and I; Supplemental Fig. S5). Interestingly, the transcript elevation of the above genes existed only at the medium and late stages in the developing seeds of transgenic rice but not in the early stage of seed development (Supplemental Fig. S5). Accordingly, the TDC activity in the developing seeds of pyramid HFL1 rice at 15 DAF was much higher than that of its two parents, 35S-15 and GR-14, and 6-fold more than that of the wild type (Fig. 4C). Therefore, the pathway for serotonin biosynthesis was greatly enhanced in developing seeds of HFL transgenic rice, especially in pyramid HFL rice with simultaneous engineering of Lys biosynthesis and catabolism.
Overexpressing TDC Promotes Serotonin and Lys Accumulation and Dark-Brown Phenotype Development in Mature Endosperms
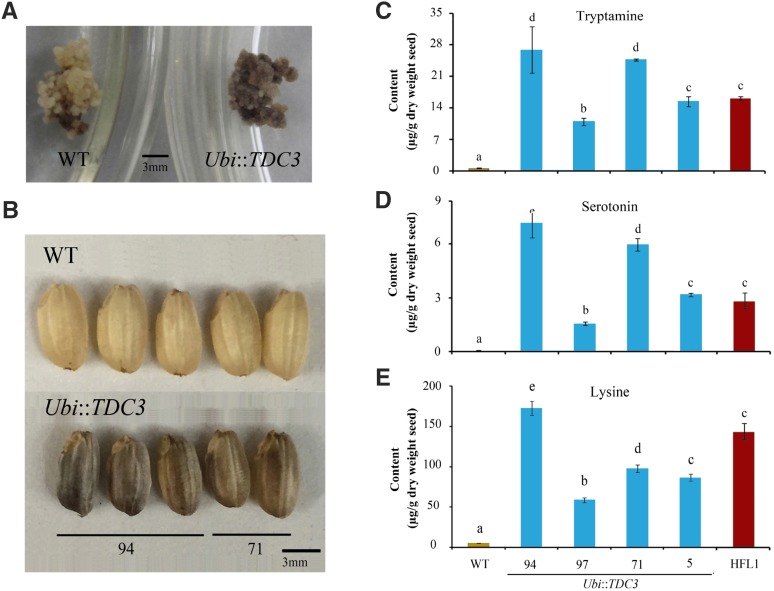
To further verify the relationship between serotonin accumulation and the dark-brown phenotype, a maize Ubiquitin (Ubi) promoter-driven expression of the rice TDC3 overexpression construct (Ubi::TDC3) was introduced into the same wild-type cv Wuxiangjing 9 (Supplemental Fig. S6A). More than 20 independent transformants were confirmed (Supplemental Fig. S6). Compared with the wild type, some transgenic calli showed the dark and/or brown color phenotype (Fig. 5A), and the levels of tryptamine and serotonin were found to accumulate to much higher levels in the Ubi::TDC3 transgenic calli than those of the wild type (Supplemental Fig. S6, B and C). In the developing seeds, the expression of TDC3, as well as of TDC1, TDC2, and T5H, obviously increased in homozygous Ubi::TDC3 transgenic rice (Supplemental Fig. S6, E–G). In the mature seeds of these transformants, the content of serotonin and tryptamine was enhanced significantly, and their levels were comparable to or higher than those in the HFL1 mature seeds (Fig. 5, C and D).
Figure 5.
Appearance of calli and mature seeds and the profile of seed metabolites from TDC3-overexpressing transgenic rice. A, Appearance of transformed callus with Ubi::TDC3 or untransformed wild type (WT). B, Appearance of mature seeds from transgenic Ubi::TDC3 or untransformed wild type. C to E, Concentrations of tryptamine (C), serotonin (D), and free Lys (E) in mature seeds of Ubi::TDC3 and HFL1 transgenic rice. Error bars represent sd of three biological replicates, and different letters indicate statistical significance among transgenic plants and the wild type at P < 0.05.
It is worth noting that the Ubi::TDC3 transgenic mature seeds also exhibited the dark-brown appearance in the endosperm (Fig. 5B), which was similar to previous reports (Kanjanaphachoat et al., 2012; Dubouzet et al., 2013). Although TDC overexpression varied among different transformants, the levels of serotonin, tryptamine, and the dark-brown phenotype of mature seeds were closely related (Fig. 5; Supplemental Fig. S6). Furthermore, the free Lys content also increased dramatically in mature seeds of TDC3-overexpressed rice (Fig. 5E). Therefore, it is clear that there is a close connection between serotonin accumulation and Lys metabolism, and serotonin accumulation is responsible for the dark-brown color in mature endosperms of either serotonin- or Lys-engineered rice.
Differential Transcriptome in Developing Seeds of HFL Rice
We compared the global transcriptome in developing seeds at 10 or 15 DAF among the transgenic lines and the wild type (Supplemental Fig. S7). PCA of the transcriptomes showed that there was little difference in transcriptome among the tested samples (Supplemental Fig. S7). Similar numbers of differentially expressed genes (DEGs) were detected in the developing seeds (10 DAF) of the transgenic rice lines GR-14 (1,203 DEGs; P < 0.05), 35S-15 (954 DEGs; P < 0.05), and the pyramid HFL1 (1,289 DEGs; P < 0.05) when compared with the wild type (Supplemental Table S3). Among these DEGs, the number of up- or down-regulated genes was similar for each comparison (Supplemental Table S3). Interestingly, the number of DEGs (P < 0.05) in the developing seeds at 15 DAF between HFL1 and the wild type was much less (only 41) than in the developing seeds at the early stage (10 DAF; Supplemental Table S3; Supplemental Fig. S7).
In the developing seeds at 10 DAF, the identified DEGs in HFL1 were classified further according to their physiological functions. The results showed that these DEGs are associated mainly with plant stress response, amino acid metabolism, carbohydrate metabolism, and lipid metabolism (Supplemental Data Set S5). In the late stage of endosperm development (15 DAF), Gene Ontology (GO) enrichment analysis revealed that 41 DEGs were attributed mainly to plant stress response, such as endoplasmic reticulum metabolism, regulation and control of enzyme activity, cell wall composition, stress response, protein metabolism, and cell differentiation (Supplemental Data Set S6; Supplemental Fig. S7). Therefore, these data suggested that engineering Lys metabolism could elicit stress responses during grain filling, and many genes relating to stress responses were differentially expressed in the developing seeds, especially at the late stage, of the HFL high-Lys transgenic rice.
Induced JA Signaling Pathway in Developing Seeds of HFL Rice
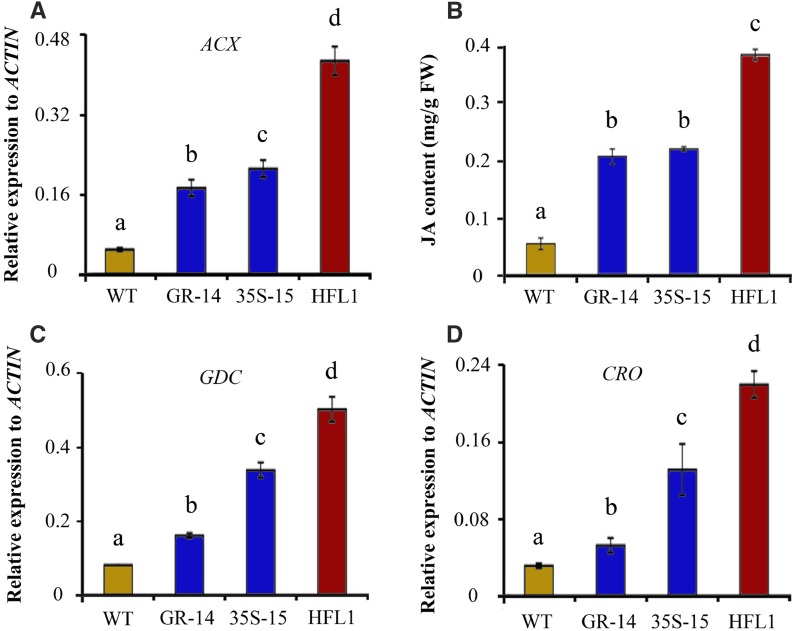
Based on our results, several metabolic pathways associated with stress responses were induced or enhanced in HFL transgenic rice (Supplemental Fig. S8; Supplemental Table S4). RNA sequencing (RNA-seq) analysis further revealed an increase in the expression of genes related to plant stress response (Supplemental Data Set S6). Among the DMs and DEGs involved in stress response, most were involved in the JA pathway. For example, acyl-CoA oxidase or β-oxidase (ACX) and lipoxygenase are two key enzymes involved in JA biosynthesis (Li et al., 2005). The transcripts of ACX (LOC_Os11g39220) and LIPOXYGENASE (LOC_Os03g08220) genes were increased significantly (1.35- and 2.09-fold higher, respectively) in developing seeds at 10 DAF of HFL1 rice (Supplemental Fig. S8; Supplemental Data Set S5). The elevated levels of the ACX transcript were further confirmed in HFL1 developing endosperm by RT-qPCR (Fig. 6A). When compared with the wild type, two specific genes within the JA signal pathway, a γ-thionin family domain-containing protein gene (LOC_Os02g07628) and a defense in DEFL family gene (LOC_Os07g41290), showed 5.09- and 3.2-fold increases, respectively, in expression in developing seeds of HFL1 (Supplemental Fig. S8).
Figure 6.
Expression of the JA pathway in different transgenic rice lines and their wild type (WT). A, Relative expression of GLUTAMATE DECARBOXYLASE (GDC) genes in developing seeds at 15 DAF. B, JA content in developing seeds at 15 DAF of transgenic rice and the wild type. FW, Fresh weight. C and D, Relative expression of GDC (C) and the common enzyme CROTONASE (CRO; D) genes in developing seeds at 15 DAF. Error bars represent sd of three biological replicates, and different letters indicate significant differences among transgenic plants and the wild type at P < 0.05.
The JA content in developing seeds at 15 DAF was significantly higher in HFL1 than in the wild type (Fig. 6B), and the increased concentration (4 µg g−1 fresh weight) is considered high enough to promote the JA signaling pathway (Truman et al., 2007). Besides, Nδ-acetyl-Orn also is a marker within the JA signaling pathway (Adio et al., 2011), and its relative content also was increased by 20-fold in HFL mature seeds (Supplemental Fig. S8D). Yet, the levels of other stress-related metabolites associated with Lys metabolism, such as salicylic acid (SA), indoleacetate (IAA), Pro, and homoserine, were detected to be not significantly different in both leaves and seeds among different transgenic rice lines and the wild type (Supplemental Fig. S9).
Low-Temperature Stress Enhances the Dark-Brown Phenotype and Serotonin Accumulation in HFL Endosperms
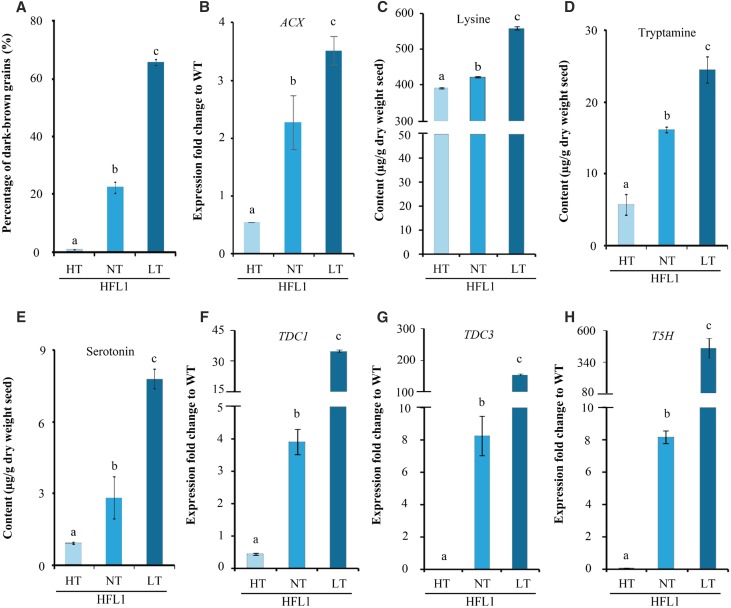
JA biosynthesis could be up-regulated by low temperature (Hu et al., 2013). We investigated the effect of temperature treatment stress on dark-brown endosperm in HFL rice. The growing rice plants were moved into the greenhouse for grain filling and maturation under different air temperatures: high (33°C ± 2°C [HT]), low (20°C ± 2°C [LT]), and middle (26°C ± 2°C, similar to normal conditions [NT]). For the wild type, the harvested mature endosperms had identical grain color phenotypes among three temperature treatments. The proportion of dark-brown mature endosperms was significantly higher under LT treatment (up to 66.7%) than HT or NT treatment in the two HFL rice lines, HFL1 and HFL2 (Fig. 7A; Supplemental Fig. S10A). Furthermore, ACX transcript expression also increased dramatically in HFL1 developing seeds under LT treatment, which is consistent with the observed change in endosperm color (Fig. 7B).
Figure 7.
Grain color, metabolite content, and gene expression of HFL1 in comparison with wild-type values (WT) under different temperature treatments during seed development. A, Percentage of dark-brown grains in total mature seeds of HFL1. B, Relative expression of ACX genes in developing seeds at 15 DAF. C to E, Levels of free Lys (C), tryptamine (D), and serotonin (E) in HFL1 mature seeds. F to H, Relative expression of TDC1 (F), TDC3 (G), and T5H (H) genes in developing seeds at 15 DAF. Error bars represent sd of three biological replicates, and different letters indicate statistical significance among transgenic plants at P < 0.05.
Moreover, the contents of Lys, serotonin, and tryptamine in mature endosperms also were changed similarly under different temperature treatments (Fig. 7, C–E; Supplemental Fig. S10B). The accumulation of both Lys and serotonin in HFL mature endosperm was enhanced under low temperature. As expected, an increase in the expression of key genes (TDC and T5H) also was detected in developing seeds of HFL rice under low temperature (Fig. 7, F–H; Supplemental Fig. S10, C–E).
Connection between Lys and Serotonin Metabolism in Rice Endosperm
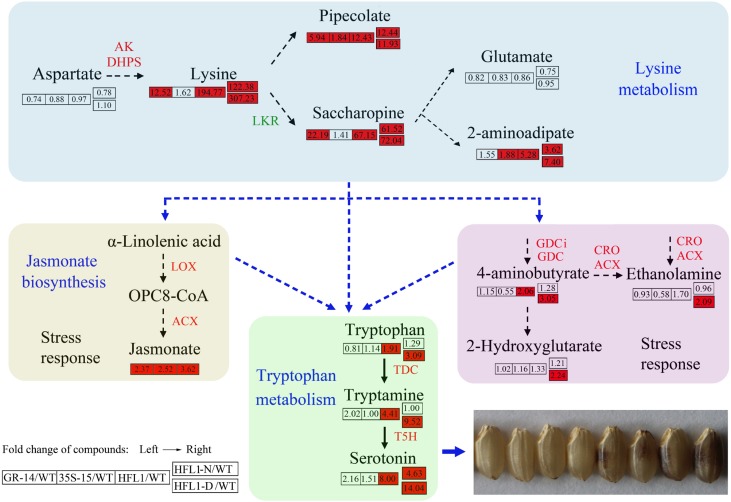
Taken together, the above data suggest a strong positive correlation among the levels of Lys, serotonin, and dark-brown color in mature endosperm. They also imply that JA stress responses may play a major role in the connection between different metabolic pathways and endosperm appearance in HFL rice. Figure 8 is a proposed model for the connection between Lys and serotonin. In HFL1-D rice endosperm, 2-aminoadipate, a product of Lys catabolism, increased significantly compared with the wild type, 35S-15, GR-14, and HFL1-N (Table I; Supplemental Table S2). Somehow, JA biosynthesis as well as associated stress responses were greatly induced by the elevated Lys and/or its metabolites. Subsequently, these elevations might lead to the activation of TDC and T5H expression for enhanced Trp metabolism and serotonin production, eventually resulting in the dark-brown phenotype in the HFL rice endosperm (Fig. 8).
Figure 8.
Proposed model for the metabolic connection between Lys metabolism and serotonin biosynthesis in endosperm of HFL rice. The data within shaded cells represent relative values (ratios) comparing the level of each metabolite detected in transgenic rice with that in the wild type (WT), and the red background indicates significantly increased ratios compared with the wild type at P < 0.05. Mean values of three biological replicates were used for comparative analysis.
In HFL rice endosperm, the expression of ACX and related enzymes was induced and enhanced, which may result in higher JA and serotonin synthesis. The data from RT-qPCR analysis showed that the transcription of related genes, including those encoding ACX, CRO (LOC_Os06g39344), GLU DECARBOXYLASE ISOZYME1 (GDCi; LOC_Os03g51080), and GDC (LOC_Os08g36320), obviously increased in developing seeds of HFL1 rice at 15 DAF compared with the wild type (Fig. 6, A, C, and D; Supplemental Fig. S8C). To confirm the relationship between JA levels and serotonin synthesis in rice, we further treated the seedlings of wild-type rice with exogenous JA and subsequently tested the effect on serotonin biosynthesis (Supplemental Fig. S11). The results showed that the expression of TDC3 and T5H, two key genes involved in serotonin biosynthesis, were obviously induced by JA treatment (Supplemental Fig. S11, A and B). Meanwhile, the serotonin content in the seedlings upon JA treatment was significantly higher compared with the untreated wild type (Supplemental Fig. S11C). These results implied that JA treatment could promote the expression of TDC3 and T5H genes and the accumulation of serotonin in rice.
DISCUSSION
How Were JA and Serotonin Induced in HFL Rice?
In this study, the integrated analyses of both metabolome and transcriptome revealed that, in seeds of HFL rice, there were major changes in the pathways of Asp and aromatic amino acids and minor changes in the pathways associated with the metabolism of other amino acids, tricarboxylic acid cycle, glycolysis, starch and Suc, lipid, purine, pyrimidine, and several secondary metabolites (Supplemental Fig. S12). Interestingly, JA-related stress responses and serotonin accumulation were detected in HFL rice endosperm (Figs. 4 and 6).
In higher plants, Lys is converted into 2-aminoadipate via saccharopine by the LKR/SDH enzyme, accompanied by the synthesis of Glu (Arruda et al., 2000; Kiyota et al., 2015). In bacteria, the saccharopine pathway is associated with stress responses (Less et al., 2011; Neshich et al., 2013). In Arabidopsis, 2-aminoadipate and pipecolate can induce JA accumulation and prime the plant for effective local resistance induction (Návarová et al., 2012). Recent studies have shown that Lys catabolites, especially pipecolate, have multiple functions in dicots, including positive regulation of SA biosynthesis, induction of effective local resistance against pathogens, and establishment of salicylic acid responses (Vogel-Adghough et al., 2013; Zeier, 2013; Yang and Ludewig, 2014). In this study, although pipecolate accumulated in the mature seeds of all HFL transgenic rice (Supplemental Data Sets S1 and S2), there was no significant correlation with the dark-brown color phenotype. Moreover, the SA content and the transcript of the ICS1 gene involved in SA biosynthesis did not change significantly in leaves, developing seeds, and mature seeds of all tested transgenic lines compared with the wild type (Supplemental Fig. S9, C–F), suggesting a different functional role for pipecolate in monocots and dicots. Therefore, in our HFL rice endosperm, our data implied that 2-aminoadipate, rather than pipecolate, may induce JA accumulation in HFL rice, with the JA signaling pathway also induced (Fig. 8). However, more experiments are needed to test this relationship.
In higher plants, serotonin, usually derived from Trp metabolism, plays an important role in innate immunity and can induce the expression of defense genes and cell death and enhance blast resistance (Fujiwara et al., 2010; Ramakrishna et al., 2012). Trp belongs to the aromatic amino acids and is synthesized through the shikimic acid pathway (Supplemental Fig. S12). After a series of enzymatic reactions, three aromatic amino acids, Tyr, Trp, and Phe, are synthesized (Maeda and Dudareva, 2012). Among them, several key enzymes play a major role in Trp synthesis, including 3-dehydroquinate synthase (3DS), chorismic acid synthase (CS), and anthranilic acid synthase (AS). In our HFL1 transgenic rice, the Trp level increased slightly compared with the wild type, but no increase was observed in the expression of the genes encoding the above three key enzymes in the HFL developing seeds at 10 or 15 DAF compared with the wild type (Supplemental Fig. S13). In addition, Trp is the precursor of IAA in higher plants (Hull et al., 2000). Although Trp increased in HFL rice seeds, the IAA level in the developing seeds and mature seeds was not significantly different between the HFL rice and the wild type (Supplemental Fig. S9, A and B). The pathway for IAA biosynthesis may not be differentially regulated in HFL transgenic rice. Therefore, the increase of Trp and serotonin in the HFL mature seeds may not be due to the shikimic acid pathway but, rather, to other reasons, such as stress responses (Rai, 2002; Less and Galili, 2008).
What Is the Bridge for the Metabolic Connection?
In Arabidopsis, the metabolic connection of the Asp family pathway with the tricarboxylic acid cycle and seed germination has been mapped (Galili, 2011; Less et al., 2011). The saccharopine pathway of Lys catabolism also is involved in a stress-related metabolic pathway (Less et al. 2011; Kiyota et al., 2015). In our study, there was a clear connection between Lys metabolism and serotonin metabolism (Fig. 8).
Previous studies have shown that JA can induce the expression of TDC and T5H genes and then the accumulation of serotonin in rice and other plants (Ishihara et al., 2008; Park et al., 2009; Byeon et al., 2015). More recently, it was reported that the Arabidopsis JA-related mutants, including coronatine insensitive1 and jasmonic acid resistant1, show tolerance to serotonin (Pelagio-Flores et al., 2016). The data from our JA treatment experiment in seedlings revealed that serotonin biosynthesis could be induced by JA in rice (Supplemental Fig. S12), and it gave some evidence supporting our proposed model for the relationship between the serotonin and JA pathways in HFL rice (Fig. 8).
Furthermore, our data have shown that the increased serotonin level and the dark-brown seed phenotype in HFL rice were correlated under low-temperature treatment (Fig. 7; Supplemental Fig. S10). Since JA positively regulates resistance against cold stress in Arabidopsis, it is expected that the low-temperature treatment can induce the expression of enzymes related to JA biosynthesis and then increase endogenous JA and regulate plant secondary metabolism and ICE-CBF/DREB1 signaling pathways (Hu et al., 2013). In this study, in the developing endosperm of HFL rice, low-temperature treatment may further induce JA biosynthesis and signaling and eventually lead to the enhanced serotonin accumulation and dark-brown pigmentation. Even so, further studies are required to elucidate the molecular mechanism and the connection between the JA signaling pathway and serotonin biosynthesis in plants, especially to identify which are responsible for the connection in rice endosperm.
Why Did the Endosperm Color Segregate within HFL Mature Seeds?
As shown in Figures 1 and 2, within the harvested seeds from the same HFL lines, the endosperm appearance was quite different, and their color was segregated. In the normal condition, only 20% of the seeds were strongly dark brown (HFL-D). As all tested transgenic rice lines were homozygous for their transgenes, the segregation was not due to a genetic factor but, rather, must have environmental causes. The data from our temperature treatment experiments showed obvious evidence for this conclusion, as the proportion of HFL dark-brown mature endosperms was significantly higher under low-temperature conditions (Fig. 7). The reasons might be as follows. First, the time and processing of grain filling can be quite different among the caryopses from the same panicle (Chen et al., 2014; Gratani, 2014); thus, the environment, such as temperature, differs during grain filling. Unfortunately, no orderliness was observed between the location of caryopsis and their endosperm appearance within the same panicle (data not shown). Second, as our data show, the dark-brown phenotype formed in the late period of seed development. Thus, different grains of the same plant were experiencing diverse environmental conditions during the process of seed development, likely resulting in the different phenotypes. Third, but not finally, the metabolism during seed development is quite complex, and most of the metabolites, especially JA and serotonin, are sensitive to stresses, including temperature. More importantly, our data demonstrate that the dark-brown phenotype is tightly associated with the serotonin level (Fig. 4). Therefore, in some seeds from HFL rice, the serotonin accumulation is not sufficiently high to cause the dark-brown appearance. In brief, the segregation of endosperm color among HFL mature seeds might be considered a response to environmental factors (Gratani, 2014). However, its detailed mechanism needs more study.
Functional and Health Aspects of Serotonin and Trp
Serotonin, as a neurotransmitter, hormone, and mitogenic factor, is a ubiquitous monoamine that plays multiple roles and mediates a series of activities in animal and human cells (Kang et al., 2009). Furthermore, serotonin is present in many plant species and is involved in an array of physiological functions in plants, such as growth regulation, flowering, and morphogenesis (Ramakrishna and Ravishankar, 2011). Trp is an essential amino acid for protein formation in many protein-based foods and dietary proteins (Friedman and Levin, 2012; Jenkins et al., 2016). The lack of Trp and serotonin can cause health problems (Vlaev et al., 2017). In this study, one beneficial effect in HFL rice was the high accumulation of serotonin, which has not been reported previously in other high-Lys plants. Interestingly, the phenotype was observed only in late and mature stages of endosperm development (after 20 DAF; Fig. 2), suggesting that there was no effect on seed development or seed size and yield. Therefore, the accumulation of Lys, serotonin, and Trp in HFL rice grains may provide a nutritionally favorable crop.
MATERIALS AND METHODS
Plant Materials and Growth
Six rice (Oryza sativa) samples were used in this study, including five transgenic lines (GR-14, GR-65, 35S-15, HFL1, and HFL2) with altered expression of Lys metabolism and their wild-type cv Wuxiangjing 9, an elite japonica rice cultivar from China. The relationship between these rice samples is shown in Figure 1A and Supplemental Figure S2. All transgenic rice lines are free of the selectable marker gene, and their generation, molecular characterization, nutritional evaluation, and field performance have been well reported (Yang et al., 2016, 2017a, 2017b). Rice plants were grown under the same climate and management conditions during the summer of years 2013 to 2015 in a paddy field at the Wenhui Road Campus of Yangzhou University in Jiangsu Province, China. Each line was grown in triplicate and planted randomly in each plot. The seedlings were grown in round containers with 60-cm diameter under normal climate and management conditions. Each container housed three plants. After most of the caryopses had pollinated, the plants were transferred to artificial climate chambers with different air temperatures: HT (33°C ± 2°C), NT (26°C ± 2°C), and LT (20°C ± 2°C). The growth and JA treatment of rice seedlings were performed as described by Byeon et al. (2015). In brief, 4-week-old seedlings of wild-type cv Wuxiangjing 9 were treated with 100 µm JA solution. After 48 h, the leaves were collected from JA-treated (named JA) or nontreated (wild-type) plants and stored at −80°C until use for analysis. Each line in each treatment was in triplicate.
Experimental Design and Sample Preparation
The experimental design is shown in Figure 1B, where three types of rice samples were collected for analyses. The developing seeds were collected at different DAF; those at 10 and 15 DAF were used for transcriptomic analysis and those at 10 DAF were used for metabolomics analysis. At 10 DAF, the upper three leaves (flag, second, and third upper leaves) of each plant were collected for both transcriptomic and metabolomic analyses. After maturity, the mature seeds were harvested for metabolomic and compound analyses. For the analyses of transcriptome, transcripts, and enzyme activity, the collected samples were immediately frozen with liquid nitrogen and stored at −80°C. For the metabolome and compound measurements, the seed glume was removed on ice and the dehulled seed samples and leaf samples were immediately frozen with liquid nitrogen and then dried under vacuum. The glume was removed from the HFL mature seeds and then divided into HFL1-D with dark-brown color and HFL-N with normal color, as the wild type. All samples were stored at −80°C until experimental analyses. The above-mentioned leaf samples were harvested from corresponding rice lines at the same time in a certain plot as one biological replicate, and three random plots were used as three biological replicates. Each biological replicate of leaf sample was extracted from mixed leaves of one rice line that contained 20 plants. The same harvesting method was carried out for developing seeds and mature seeds.
Metabolomic Analysis
A total of 16 samples, including eight mature seeds, four developing seeds, and four leaf samples (Supplemental Table S1), were prepared using the automated MicroLab STAR system from Hamilton and analyzed by GC-MS and UHPLC-MS (positive and negative mode; Hu et al., 2016). The metabolomic analyses were carried out on the metabolome platform at Shanghai Jiaotong University. Detailed information concerning GC-MS and UHPLC-MS analysis can be found in previous studies, with data acquisition and processing being performed with TagFinder software (Hu et al., 2014, 2016). Briefly, the samples destined for GC-MS analysis were redried under vacuum desiccation for a minimum of 24 h prior to derivatization under dried nitrogen using bistrimethyl-silyl-triflouroacetamide. Samples were analyzed on a Thermo-Finnigan Trace DSQ fast-scanning single-quadrupole mass spectrometer using electron-impact ionization. LC was performed using a Waters ACQUITY ultra-performance liquid chromatograph and a Thermo Scientific Q-Exactive high-resolution/accurate mass spectrometer interfaced with a heated electrospray ionization source and an Orbitrap mass analyzer operated at 35,000 mass resolution. Raw data analysis for rice metabolites was conducted as described previously, and metabolic differences within tested samples were determined using Welch’s t test in R packages (Hu et al., 2016).
Transcriptomic and Transcriptional Analyses
Total RNA extraction and preparation of each sample (in three biological replicates) were performed as described previously (Yang et al., 2016). The developing seeds at 10 DAF from three independent transgenic lines, HFL1, GR-14, and 35S-15, and the wild type were collected and used for microarray analysis using the Affymetrix Rice Genome array. The developing seeds at 15 DAF from HFL1 transgenic rice and wild-type rice were used for RNA-seq by generating 100-nucleotide-long paired-end sequence reads on an Illumina platform (HiSeq 2000). Microarray and RNA-seq preliminary analyses were performed by Shanghai Biotechnology. The analysis of DEGs was performed using the limma package (Smyth, 2004). Total RNA was purified and reverse transcribed into cDNA for reverse transcription (RT)-PCR and RT-qPCR analyses. The rice housekeeping gene ACTIN served as an internal control. The gene-specific primers for RT-PCR were designed using Primer Premier 5 according to the coding sequences or previous reports and are listed in Supplemental Table S5.
Enzyme Activity Assay
TDC activity was measured according to the method of Ishihara et al. (2008). Developing seeds at 15 DAF were ground into fine powder and then extracted with 50 mm K-Pi buffer (pH 6.9) containing 5 mm β-mercaptoethanol. After purification and incubation, the resulting supernatant was subjected to LC-MS analysis as described below.
Compound Measurement
The content of free Lys was measured according to a method from our previous study (Yang et al., 2016). Free Trp, tryptamine, and serotonin were extracted and purified using the methods described by Comai et al. (2007), Kang et al. (2007a), and Dubouzet et al. (2013), respectively, with minor adjustments. The extracting solution was injected into an Agilent 1200-6460 QQQ with Cadenza CD-C18 column (150 × 4.6 mm; particle size, 5 μm; Agilent). Elution was performed with 0.1% formic acid:acetonitrile (100:0 [v/v] at 0 min, 99:1 at 10 min, 96:4 at 30 min, 70:30 at 60 min, 0:100 at 75 min, and 100:0 at 80 min) at a flow rate of 1 mL min−1 and was monitored at a wavelength of 280 nm. The extraction and purification of JA were performed according to the methods described by Li et al. (2002) and Pan et al. (2010) with minor modifications. The extracting solution was subjected to HPLC using a Waters e2695 with reverse C18 column; solvent A was acetonitrile and solvent B was water containing 0.1% formic acid. Elution was performed as follows: 15% A at 0 min, 50% A at 2 min, 70% A at 5 min, 100% A at 10 min, and 15% A at 16 min, with a flow rate of 0.3 mL min−1. JA content was analyzed in the negative ion and selected ion monitoring mode. The retention time of JA was 7.35 min. Each value represents the mean ± sd from three experiments. All the standards were purchased from Sigma.
Transgenic Construct and Rice Transformation
A full-length cDNA fragment encoding rice TDC3 (1,572 bp) was isolated by RT-PCR from the total RNA of wild-type rice cv Wuxiangjing 9. The primer pairs are shown in Supplemental Table S5. After sequencing confirmation, the TDC3 cDNA with BamHI and KpnI restriction sites was inserted between the maize Ubi promoter and Nopaline Synthase terminator in binary vector pCAMBIA1300. The resultant vector, p1300-Ubi-TDC3 (Supplemental Fig. S6A), was transferred into Agrobacterium tumefaciens strain EHA105, and rice transformation was performed as reported previously (Liu et al., 1998).
Data Analyses
Statistical analyses (ANOVA) were performed using SPSS version 17.0. The values presented are means or means ± sd of three biological replicates. The DEGs were classified according to their physiological functions using the enrichment and functional annotation clustering tools embedded in the DAVID bioinformatics software (Cohen et al., 2014). All transcriptomic data from three biological replicates of both lines grown under both conditions were analyzed by GO enrichment analysis using the SBC Analysis System. PCA was performed by SIMCA-P+11 (UMETRICS). The simplified metabolic pathways were constructed according to KEGG (the Kyoto Encyclopedia of Genes and Genomes) and MetaCyc and were generated using KEGG software. Heat maps of relative metabolite levels in rice were generated using Multi-Experiment Viewer version 4.8 with a normalized median value of each metabolite. Venn diagrams of DMs were analyzed by Venn Diagram Generator.
Accession Numbers
Gene sequence information from this article can be found in the RAP-DB (http://rapdb.dna.affrc.go.jp/) as follows: TSAα1 (LOC_Os03g58260), TSAα2 (LOC_Os07g08430), TSAβ (LOC_Os08g04180), TDC1 (LOC_Os08g04540), TDC2 (LOC_Os07g25590), TDC3 (LOC_Os08g04560), T5H (LOC_Os12g16720), ACX (LOC_Os11g39220), GDC (LOC_Os08g36320), CRO (LOC_Os06g39344), THIONIN (LOC_Os02g07628), DEFL (LOC_Os07g41290), LOX (LOC_Os03g08220), GDCi (LOC_Os03g51080), ICS (LOC_Os09g19734), 3DS (LOC_Os09g36800), CS1 (LOC_Os03g14990), ASI (LOC_Os03g15780), ASα1 (LOC_Os03g61120), ASβ1 (LOC_Os03g50880), ASβ2 (LOC_Os04g38950), and ACTIN (LOC_Os03g50885). The raw data set from RNA-seq has been deposited in the SRA (http://www.ncbi.nlm.nih.gov/sra/) database with accession number SRP130266.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phenotypic comparison of mature seeds from different transgenic rice lines and their wild type.
Supplemental Figure S2. Appearance and free Lys content of dehulled mature seeds from the pyramiding HFL2 line and its parents and wild type.
Supplemental Figure S3. PCA of detected metabolomic data in developing seeds at 10 DAF and in leaves.
Supplemental Figure S4. Relative content of metabolites in mature seeds detected by metabolomic analysis, and the chromatograms for serotonin measurement.
Supplemental Figure S5. Dynamic changes of transcriptional expression of TDC and T5H genes during seed development.
Supplemental Figure S6. Generation and characterization of Ubi::TDC3 transgenic rice.
Supplemental Figure S7. PCA and GO enrichment analyses of transcriptomes in developing seeds.
Supplemental Figure S8. Relative contents of metabolites and transcriptional expression levels of genes associated with JA metabolism.
Supplemental Figure S9. Relative contents of IAA, SA, Pro, and homoserine in leaves, developing seeds, and mature seeds.
Supplemental Figure S10. Percentage of dark-brown grain, Lys content in mature seeds, and gene transcriptional levels in developing seeds at 10 DAF of HFL2 under different temperature treatments during seed development.
Supplemental Figure S11. Relative expression of TDC3 and T5H genes and serotonin content in the seedlings of the wild type or under JA treatment.
Supplemental Figure S12. Changes of metabolite content within some main metabolic pathways in mature seeds of HFL1 compared with the wild type.
Supplemental Figure S13. Transcriptional levels of key genes involved in aromatic amino acid biosynthesis through the shikimic acid pathway.
Supplemental Table S1. List of detected and differential metabolic compounds between transgenic rice and the wild type.
Supplemental Table S2. Differential metabolites and their fold change in mature seeds among multiple comparisons within HFL2 and its derived samples.
Supplemental Table S3. Statistical analysis of the number of DEGs in developing seeds.
Supplemental Table S4. Metabolites associated with stress response in mature seeds of HFL1 transgenic rice compared with the wild type.
Supplemental Table S5. List of primers used for vector construction and RT-qPCR analyses.
Supplemental Data Set S1. Heat map of statistically significant biochemicals profiled in mature seeds of HFL1, their parents, and the wild type.
Supplemental Data Set S2. Heat map of statistically significant biochemicals profiled in mature seeds of HFL2, their parents, and the wild type.
Supplemental Data Set S3. Heat map of statistically significant biochemicals profiled in leaves of HFL1, their parents, and the wild type.
Supplemental Data Set S4. Heat map of statistically significant biochemicals profiled in developing seeds at 10 DAF of HFL1, their parents, and the wild type.
Supplemental Data Set S5. Differentially regulated transcripts in developing seeds at 10 DAF of HFL1 compared with the wild type.
Supplemental Data Set S6. Differentially regulated transcripts in developing seeds at 15 DAF of HFL1 compared with the wild type.
Acknowledgments
We thank Drs. Jian-Xin Shi, Chao-Yang Hu, and Danny Ng for comments on the article.
Footnotes
This work was supported by the National Natural Science Foundation (31471459), Ministry of Agriculture of the People’s Republic of China (grants 2016ZX08001006 and 2016ZX08009003-004), and Jiangsu PAPD and 333 Project to Q.Q.L., The Bill and Melinda Gates Foundation, The State Key Laboratory of Agrobiotechnology, The Lo KweeSeong Foundation, and The Lee Hysan Foundation to S.S.-M.S.
Articles can be viewed without a subscription.
References
- Adio AM, Casteel CL, De Vos M, Kim JH, Joshi V, Li B, Juéry C, Daron J, Kliebenstein DJ, Jander G (2011) Biosynthesis and defensive function of Nδ-acetylornithine, a jasmonate-induced Arabidopsis metabolite. Plant Cell 23: 3303–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelovici R, Fait A, Zhu X, Szymanski J, Feldmesser E, Fernie AR, Galili G (2009) Deciphering transcriptional and metabolic networks associated with lysine metabolism during Arabidopsis seed development. Plant Physiol 151: 2058–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelovici R, Galili G, Fernie AR, Fait A (2010) Seed desiccation: a bridge between maturation and germination. Trends Plant Sci 15: 211–218 [DOI] [PubMed] [Google Scholar]
- Arruda P, Kemper EL, Papes F, Leite A (2000) Regulation of lysine catabolism in higher plants. Trends Plant Sci 5: 324–330 [DOI] [PubMed] [Google Scholar]
- Betrán FJ, Bockholt A, Fojt F, Rooney L, Waniska R (2003) Registration of Tx802. Crop Sci 43: 1891–1892 [Google Scholar]
- Byeon Y, Lee HY, Hwang OJ, Lee HJ, Lee K, Back K (2015) Coordinated regulation of melatonin synthesis and degradation genes in rice leaves in response to cadmium treatment. J Pineal Res 58: 470–478 [DOI] [PubMed] [Google Scholar]
- Byeon Y, Park S, Lee HY, Kim YS, Back K (2014) Elevated production of melatonin in transgenic rice seeds expressing rice tryptophan decarboxylase. J Pineal Res 56: 275–282 [DOI] [PubMed] [Google Scholar]
- Chen D, Neumann K, Friedel S, Kilian B, Chen M, Altmann T, Klukas C (2014) Dissecting the phenotypic components of crop plant growth and drought responses based on high-throughput image analysis. Plant Cell 26: 4636–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen H, Israeli H, Matityahu I, Amir R (2014) Seed-specific expression of a feedback-insensitive form of CYSTATHIONINE-γ-SYNTHASE in Arabidopsis stimulates metabolic and transcriptomic responses associated with desiccation stress. Plant Physiol 166: 1575–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai S, Bertazzo A, Bailoni L, Zancato M, Costa C, Allegri G (2007) The content of proteic and nonproteic (free and protein-bound) tryptophan in quinoa and cereal flours. Food Chem 100: 1350–1355 [Google Scholar]
- Dersch LM, Beckers V, Wittmann C (2016) Green pathways: metabolic network analysis of plant systems. Metab Eng 34: 1–24 [DOI] [PubMed] [Google Scholar]
- Dubouzet JG, Matsuda F, Ishihara A, Miyagawa H, Wakasa K (2013) Production of indole alkaloids by metabolic engineering of the tryptophan pathway in rice. Plant Biotechnol J 11: 1103–1111 [DOI] [PubMed] [Google Scholar]
- Falco SC, Guida T, Locke M, Mauvais J, Sanders C, Ward RT, Webber P (1995) Transgenic canola and soybean seeds with increased lysine. Biotechnology (N Y) 13: 577–582 [DOI] [PubMed] [Google Scholar]
- FAO (1991) Protein Quality Evaluation: Report of Joint FAO/WHO Expert Consultation. Food and Agriculture Organization, Rome [Google Scholar]
- FAO (1993) Rice in Human Nutrition. Food and Agriculture Organization, Rome [Google Scholar]
- Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L (2004) Metabolite profiling: from diagnostics to systems biology. Nat Rev Mol Cell Biol 5: 763–769 [DOI] [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey RN, Willmitzer L (2000) Metabolite profiling for plant functional genomics. Nat Biotechnol 18: 1157–1161 [DOI] [PubMed] [Google Scholar]
- Friedman M, Levin CE (2012) Nutritional and medicinal aspects of D-amino acids. Amino Acids 42: 1553–1582 [DOI] [PubMed] [Google Scholar]
- Frizzi A, Huang S, Gilbertson LA, Armstrong TA, Luethy MH, Malvar TM (2008) Modifying lysine biosynthesis and catabolism in corn with a single bifunctional expression/silencing transgene cassette. Plant Biotechnol J 6: 13–21 [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Maisonneuve S, Isshiki M, Mizutani M, Chen L, Wong HL, Kawasaki T, Shimamoto K (2010) Sekiguchi lesion gene encodes a cytochrome P450 monooxygenase that catalyzes conversion of tryptamine to serotonin in rice. J Biol Chem 285: 11308–11313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili G. (1995) Regulation of lysine and threonine synthesis. Plant Cell 7: 899–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili G. (2002) New insights into the regulation and functional significance of lysine metabolism in plants. Annu Rev Plant Biol 53: 27–43 [DOI] [PubMed] [Google Scholar]
- Galili G. (2011) The aspartate-family pathway of plants: linking production of essential amino acids with energy and stress regulation. Plant Signal Behav 6: 192–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili G, Amir R (2013) Fortifying plants with the essential amino acids lysine and methionine to improve nutritional quality. Plant Biotechnol J 11: 211–222 [DOI] [PubMed] [Google Scholar]
- Galili G, Tang G, Zhu X, Gakiere B (2001) Lysine catabolism: a stress and development super-regulated metabolic pathway. Curr Opin Plant Biol 4: 261–266 [DOI] [PubMed] [Google Scholar]
- Gibbon BC, Wang X, Larkins BA (2003) Altered starch structure is associated with endosperm modification in Quality Protein Maize. Proc Natl Acad Sci USA 100: 15329–15334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodacre R, Vaidyanathan S, Dunn WB, Harrigan GG, Kell DB (2004) Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol 22: 245–252 [DOI] [PubMed] [Google Scholar]
- Gratani L. (2014) Plant phenotypic plasticity in response to environmental factors. Adv Bot 2014: 208747 [Google Scholar]
- Hu C, Shi J, Quan S, Cui B, Kleessen S, Nikoloski Z, Tohge T, Alexander D, Guo L, Lin H, et al. (2014) Metabolic variation between japonica and indica rice cultivars as revealed by non-targeted metabolomics. Sci Rep 4: 5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C, Tohge T, Chan SA, Song Y, Rao J, Cui B, Lin H, Wang L, Fernie AR, Zhang D, et al. (2016) Identification of conserved and diverse metabolic shifts during rice grain development. Sci Rep 6: 20942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Jiang L, Wang F, Yu D (2013) Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25: 2907–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull AK, Vij R, Celenza JL(2000) Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc Natl Acad Sci USA 97: 2379–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara A, Hashimoto Y, Tanaka C, Dubouzet JG, Nakao T, Matsuda F, Nishioka T, Miyagawa H, Wakasa K (2008) The tryptophan pathway is involved in the defense responses of rice against pathogenic infection via serotonin production. Plant J 54: 481–495 [DOI] [PubMed] [Google Scholar]
- Jenkins TA, Nguyen JCD, Polglaze KE, Bertrand PP (2016) Influence of tryptophan and serotonin on mood and cognition with a possible role of the gut-brain axis. Nutrients 8: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano BO, Gu C (1995) Rice and Human Nutrition. China Agricultural University Press, Beijing, pp 31–34 [Google Scholar]
- Kang K, Kim YS, Park S, Back K (2009) Senescence-induced serotonin biosynthesis and its role in delaying senescence in rice leaves. Plant Physiol 150: 1380–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Kang K, Lee K, Back K (2007a) Characterization of rice tryptophan decarboxylases and their direct involvement in serotonin biosynthesis in transgenic rice. Planta 227: 263–272 [DOI] [PubMed] [Google Scholar]
- Kang S, Kang K, Lee K, Back K (2007b) Characterization of tryptamine 5-hydroxylase and serotonin synthesis in rice plants. Plant Cell Rep 26: 2009–2015 [DOI] [PubMed] [Google Scholar]
- Kanjanaphachoat P, Wei BY, Lo SF, Wang IW, Wang CS, Yu SM, Yen ML, Chiu SH, Lai CC, Chen LJ (2012) Serotonin accumulation in transgenic rice by over-expressing tryptophan decarboxylase results in a dark brown phenotype and stunted growth. Plant Mol Biol 78: 525–543 [DOI] [PubMed] [Google Scholar]
- Keurentjes JJB, Fu J, de Vos CH, Lommen A, Hall RD, Bino RJ, van der Plas LH, Jansen RC, Vreugdenhil D, Koornneef M (2006) The genetics of plant metabolism. Nat Genet 38: 842–849 [DOI] [PubMed] [Google Scholar]
- Kiyota E, Pena IA, Arruda P (2015) The saccharopine pathway in seed development and stress response of maize. Plant Cell Environ 38: 2450–2461 [DOI] [PubMed] [Google Scholar]
- Lee SI, Kim HU, Lee YH, Suh SC, Lim YP, Lee HY, Kim HI (2001) Constitutive and seed-specific expression of a maize lysine-feedback-insensitive dihydrodipicolinate synthase gene leads to increased free lysine levels in rice seeds. Mol Breed 8: 75–84 [Google Scholar]
- Less H, Angelovici R, Tzin V, Galili G (2011) Coordinated gene networks regulating Arabidopsis plant metabolism in response to various stresses and nutritional cues. Plant Cell 23: 1264–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Less H, Galili G (2008) Principal transcriptional programs regulating plant amino acid metabolism in response to abiotic stresses. Plant Physiol 147: 316–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Less H, Galili G (2009) Coordinations between gene modules control the operation of plant amino acid metabolic networks. BMC Syst Biol 3: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Schilmiller AL, Liu G, Lee GI, Jayanty S, Sageman C, Vrebalov J, Giovannoni JJ, Yagi K, Kobayashi Y, et al. (2005) Role of β-oxidation in jasmonate biosynthesis and systemic wound signaling in tomato. Plant Cell 17: 971–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li C, Lee GI, Howe GA (2002) Distinct roles for jasmonate synthesis and action in the systemic wound response of tomato. Proc Natl Acad Sci USA 99: 6416–6421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QQ, Zhang JL, Wang ZY, Hong MM, Gu MH (1998) A highly efficient transformation system mediated by Agrobacterium tumefaciens in rice (Oryza sativa L.). Acta Phytophysiol Sin 24: 259–271 [Google Scholar]
- Long X, Liu Q, Chan M, Wang Q, Sun SSM (2013) Metabolic engineering and profiling of rice with increased lysine. Plant Biotechnol J 11: 490–501 [DOI] [PubMed] [Google Scholar]
- Maeda H, Dudareva N(2012) The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu Rev Plant Biol 63: 73–105 [DOI] [PubMed] [Google Scholar]
- Moulin M, Deleu C, Larher F (2000) L-Lysine catabolism is osmo-regulated at the level of lysine-ketoglutarate reductase and saccharopine dehydrogenase in rapeseed leaf discs. Plant Physiol Biochem 38: 577–585 [DOI] [PubMed] [Google Scholar]
- Moulin M, Deleu C, Larher F, Bouchereau A (2006) The lysine-ketoglutarate reductase-saccharopine dehydrogenase is involved in the osmo-induced synthesis of pipecolic acid in rapeseed leaf tissues. Plant Physiol Biochem 44: 474–482 [DOI] [PubMed] [Google Scholar]
- Návarová H, Bernsdorff F, Döring AC, Zeier J (2012) Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24: 5123–5141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neshich IAP, Kiyota E, Arruda P (2013) Genome-wide analysis of lysine catabolism in bacteria reveals new connections with osmotic stress resistance. ISME J 7: 2400–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Q, Takaiwa F (2004) Evaluation of tissue specificity and expression strength of rice seed component gene promoters in transgenic rice. Plant Biotechnol J 2: 113–125 [DOI] [PubMed] [Google Scholar]
- Pan X, Welti R, Wang X (2010) Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat Protoc 5: 986–992 [DOI] [PubMed] [Google Scholar]
- Park S, Kang K, Lee K, Choi D, Kim YS, Back K (2009) Induction of serotonin biosynthesis is uncoupled from the coordinated induction of tryptophan biosynthesis in pepper fruits (Capsicum annuum) upon pathogen infection. Planta 230: 1197–1206 [DOI] [PubMed] [Google Scholar]
- Pelagio-Flores R, Ruiz-Herrera LF, López-Bucio J (2016) Serotonin modulates Arabidopsis root growth via changes in reactive oxygen species and jasmonic acid-ethylene signaling. Physiol Plant 158: 92–105 [DOI] [PubMed] [Google Scholar]
- Rai VK. (2002) Role of amino acids in plant responses to stresses. Biol Plant 45: 481–487 [Google Scholar]
- Ramakrishna A, Giridhar P, Sankar KU, Ravishankar GA (2012) Melatonin and serotonin profiles in beans of Coffea species. J Pineal Res 52: 470–476 [DOI] [PubMed] [Google Scholar]
- Ramakrishna A, Ravishankar GA (2011) Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal Behav 6: 1720–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Matsuda F (2010) Metabolomics for functional genomics, systems biology, and biotechnology. Annu Rev Plant Biol 61: 463–489 [DOI] [PubMed] [Google Scholar]
- Shaul O, Galili G (1992) Increased lysine synthesis in transgenic tobacco plants expressing a bacterial dihydrodipicolinate synthase in their chloroplasts. Plant J 2: 203–209 [DOI] [PubMed] [Google Scholar]
- Shaul O, Galili G (1993) Concerted regulation of lysine and threonine synthesis in tobacco plants expressing bacterial feedback-insensitive aspartate kinase and dihydrodipicolinate synthase. Plant Mol Biol 23: 759–768 [DOI] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: e3. [DOI] [PubMed] [Google Scholar]
- Truman W, Bennett MH, Kubigsteltig I, Turnbull C, Grant M (2007) Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc Natl Acad Sci USA 104: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlaev I, Crockett MJ, Clark L, Müller U, Robbins TW (2017) Serotonin enhances the impact of health information on food choice. Cogn Affect Behav Neurosci 17: 542–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel-Adghough D, Stahl E, Návarová H, Zeier J (2013) Pipecolic acid enhances resistance to bacterial infection and primes salicylic acid and nicotine accumulation in tobacco. Plant Signal Behav 8: e26366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Galili G (2016) Transgenic high-lysine rice: a realistic solution to malnutrition? J Exp Bot 67: 4009–4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Ludewig U (2014) Lysine catabolism, amino acid transport, and systemic acquired resistance. Plant Signal Behav 9: e28933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang QQ, He XY, Wu HY, Zhang CQ, Zou SY, Lang TQ, Sun SSM, Liu QQ (2017a) Subchronic feeding study of high-free-lysine transgenic rice in Sprague-Dawley rats. Food Chem Toxicol 105: 214–222 [DOI] [PubMed] [Google Scholar]
- Yang QQ, Suen PK, Zhang CQ, Mak WS, Gu MH, Liu QQ, Sun SSM (2017b) Improved growth performance, food efficiency, and lysine availability in growing rats fed with lysine-biofortified rice. Sci Rep 7: 1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang QQ, Zhang CQ, Chan ML, Zhao DS, Chen JZ, Wang Q, Li QF, Yu HX, Gu MH, Sun SSM, et al. (2016) Biofortification of rice with the essential amino acid lysine: molecular characterization, nutritional evaluation, and field performance. J Exp Bot 67: 4285–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeier J. (2013) New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant Cell Environ 36: 2085–2103 [DOI] [PubMed] [Google Scholar]
- Zhu C, Naqvi S, Gomez-Galera S, Pelacho AM, Capell T, Christou P (2007) Transgenic strategies for the nutritional enhancement of plants. Trends Plant Sci 12: 548–555 [DOI] [PubMed] [Google Scholar]
- Zhu X, Galili G (2003) Increased lysine synthesis coupled with a knockout of its catabolism synergistically boosts lysine content and also transregulates the metabolism of other amino acids in Arabidopsis seeds. Plant Cell 15: 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Galili G (2004) Lysine metabolism is concurrently regulated by synthesis and catabolism in both reproductive and vegetative tissues. Plant Physiol 135: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]