After a virus movement protein interacts with the HIPP26 stress sensor, the complex relocalizes to the nucleus, activating the drought stress response and thereby facilitating virus long-distance movement.
Abstract
Virus movement proteins facilitate virus entry into the vascular system to initiate systemic infection. The potato mop-top virus (PMTV) movement protein, TGB1, is involved in long-distance movement of both viral ribonucleoprotein complexes and virions. Here, our analysis of TGB1 interactions with host Nicotiana benthamiana proteins revealed an interaction with a member of the heavy metal-associated isoprenylated plant protein family, HIPP26, which acts as a plasma membrane-to-nucleus signal during abiotic stress. We found that knockdown of NbHIPP26 expression inhibited virus long-distance movement but did not affect cell-to-cell movement. Drought and PMTV infection up-regulated NbHIPP26 gene expression, and PMTV infection protected plants from drought. In addition, NbHIPP26 promoter-reporter fusions revealed vascular tissue-specific expression. Mutational and biochemical analyses indicated that NbHIPP26 subcellular localization at the plasma membrane and plasmodesmata was mediated by lipidation (S-acylation and prenylation), as nonlipidated NbHIPP26 was predominantly in the nucleus. Notably, coexpression of NbHIPP26 with TGB1 resulted in a similar nuclear accumulation of NbHIPP26. TGB1 interacted with the carboxyl-terminal CVVM (prenyl) domain of NbHIPP26, and bimolecular fluorescence complementation revealed that the TGB1-HIPP26 complex localized to microtubules and accumulated in the nucleolus, with little signal at the plasma membrane or plasmodesmata. These data support a mechanism where interaction with TGB1 negates or reverses NbHIPP26 lipidation, thus releasing membrane-associated NbHIPP26 and redirecting it via microtubules to the nucleus, thereby activating the drought stress response and facilitating virus long-distance movement.
The systemic infection of plants by viruses results in major economic losses in yield and/or quality of harvested tissues (Pennazio et al., 1996; Waterworth and Hadidi, 1998). To achieve a systemic infection, viruses must move from the initial point of entry to neighboring cells and then long distance in the vasculature to other leaves and organs (for review, see Tilsner et al., 2014). Although there has been much research on the mechanisms of virus cell-to-cell movement, there is much less knowledge of the molecular details of how viruses access the vasculature and the phloem mass transport pathway for long-distance movement. A better understanding of this process is needed to develop new interventions for disease control.
The plant vasculature is a major conduit not only for nutrient and water transfer but also for the perception of and response to abiotic and biotic stresses (Kehr and Buhtz, 2008). Plant proteins destined for phloem transport are made in, or transferred to, companion cells prior to transfer into sieve elements through specialized plasmodesmata (PD) called pore plasmodesmal units (for review, see Lough and Lucas, 2006). The phloem contains many different macromolecules, including protein chaperones, calcium sensors, RNA-binding proteins (Giavalisco et al., 2006), and different species of RNAs (including mRNAs, microRNAs, and small interfering RNAs; Buhtz et al., 2008, 2010; for review, see Turgeon and Wolf, 2009). These phloem-mobile signaling molecules enable plants to coordinate their growth and development and respond to stress (Kehr and Buhtz, 2008). A recent report on the systemic movement of Tobacco mosaic virus (TMV) suggests that interaction with phloem-specific transcriptional regulators plays a major role in virus phloem loading in older tissues, suggesting that TMV reprogramming of gene expression enhances systemic spread (Collum et al., 2016).
In this article, we studied the long-distance movement of Potato mop-top virus (PMTV), the type member of the genus Pomovirus. PMTV is one of only a few plant viruses where, in addition to movement as encapsidated virions, the genome components also can move as ribonucleoprotein (vRNP) complexes, and the largest movement protein, TGB1, plays a key role in the long-distance movement of both (Torrance et al., 2009, 2011). Previous work has shown that the N-terminal domain of TGB1 contains two nucleolar localization signals, and TGB1 associates with importin-α, which is required for nuclear/nucleolar accumulation and virus long-distance movement (Lukhovitskaya et al., 2015). The nucleolus is a distinct region in the nucleus that was shown recently to play a role in the long-distance movement of some plant viruses (for review, see Taliansky et al., 2010). For example, Groundnut rosette virus (GRV) does not encode a coat protein but encodes the protein ORF3 that interacts with a nucleolar component, fibrillarin, to form movement-competent vRNP for long-distance transport (Kim et al., 2007).
Here, we describe the association of a virus movement protein, PMTV TGB1, with a vascular-expressed plant stress sensor, HIPP26. HIPP26 is one member of a family of proteins that contain heavy metal-binding and C-terminal isoprenylation motifs (heavy metal-associated isoprenylated plant protein [HIPP]). HIPPs are unique to vascular plants and form a large and diverse family in Arabidopsis (Arabidopsis thaliana; Dykema et al., 1999; Barth et al., 2009; Tehseen et al., 2010; de Abreu-Neto et al., 2013). They have been shown to act in heavy metal homeostasis and in regulating the transcriptional response to abiotic stress (Barth et al., 2009; Gao et al., 2009; de Abreu-Neto et al., 2013) and pathogens (de Abreu-Neto et al., 2013). Our results show that PMTV TGB1 interacts with HIPP26 and that the complex accumulates in the nucleus, which leads to the activation of the drought stress response pathway in vascular tissues, which, in turn, facilitates the access of PMTV to the transport phloem for long-distance movement.
RESULTS
PMTV TGB1 Interacts with Nicotiana benthamiana HIPP26
To investigate the interactions of TGB1 with host factors, an N. benthamiana cDNA library (Hybrigenics Services) was screened by the yeast two-hybrid (Y2H) system and 50 clones were identified as interacting with TGB1. The results were returned with each interacting clone provided with a predicted biological score (Formstecher et al., 2005) ranked in categories A to F, with A as the highest level of confidence in the specificity of the interaction. Twenty-nine clones were ranked A, and by using BLAST (Altschul et al., 1990), they were all identified as N. benthamiana homologs of Arabidopsis HIPP26. A representative clone, A44, which contained the nearly complete sequence (missing 33 5′ proximal bp coding for 11 N-terminal amino acid residues) of the N. benthamiana homolog of Arabidopsis HIPP26, was obtained from Hybrigenics. In addition, the complete open reading frame (ORF) was cloned from N. benthamiana plants and sequenced.
The cDNA sequence of the NbHIPP26 ORF was used to search the predicted proteins from the draft N. benthamiana genome (assembly version 1.01) using BLAST through the Sol Genomics Network Web server (http://solgenomics.net; Fernandez-Pozo et al., 2015). This revealed significant matches on two different scaffolds (Niben101Scf07109g05004.1 and Niben101Scf02621g04026.1) that have not yet been assigned to any chromosomes. The two proteins differed in only one amino acid residue at position 16 near the N terminus, which was either a Ser or Gly. Further genome assembly and annotation is required to confirm whether the proteins are coded by alleles or distinct genes. In this work, we have investigated the protein containing Ser-16, since Ser was the most common residue at that position in the interacting clones.
HIPPs have been variously clustered into five (de Abreu-Neto et al., 2013), six (Barth et al., 2009), and seven (Tehseen et al., 2010) major groups. The phylogeny created from the alignment of the interacting clone with full-length Arabidopsis HIPPs reveals five clusters similar to those identified by de Abreu-Neto et al. (2013; Fig. 1A; Supplemental Fig. S1). In this phylogeny, clone A44 clusters with AtHIPP26 in HIPP group II, which features proteins with a single heavy metal-associated (HMA) domain. Sequence alignments of NbHIPP26 with homologs from Arabidopsis, tomato (Solanum lycopersicum), and potato (Solanum tuberosum) revealed a high level of conservation, with 84.3% sequence identity between N. benthamiana and Arabidopsis, 95.4% identity between N. benthamiana and potato, and 94.8% identity between N. benthamiana and tomato sequences (Fig. 1, B and C).
Figure 1.
HIPP phylogeny. A, Diagrammatic representation of the phylogenetic relationship between A44 and five HIPP families (de Abreu-Neto et al., 2013). A44 is related to Arabidopsis HIPP subfamily II and most closely related to HIPP26 (for full phylogeny, see Supplemental Fig. S1). B, Multiple alignments of HIPP26 proteins from Arabidopsis (Arath), N. benthamiana (Benth), tomato, and potato with A44. The alignments were made using MUSCLE with default parameters (Edgar, 2004). The line below the alignment marks conserved positions: *, single, fully conserved residue; :, strong group, fully conserved, scoring > 0.5 in the Gonnet PAM 250 matrix; ., weaker group, fully conserved, scoring ≤ 0.5 in the Gonnet PAM 250 matrix. C, Percentage sequence identities for the four HIPP26 proteins and A44 shown in the alignment in B.
Analysis revealed that the C152VVM155 sequence at the C terminus of NbHIPP26 represents a CaaX amino acid motif (a = I, V, L, A, S, or T and X = I, L, M, Q, S, or A) characteristic of proteins that are posttranslationally lipid modified by the addition of a prenyl group to enable membrane association (for review, see Crowell, 2000). AtHIPP7 was shown previously to be prenylated in vitro through a similar CTVM motif (Dykema et al., 1999), while mammalian N-Ras is known to be prenylated at an identical CVVM motif in vivo and the CVVM motif alone fused to GFP is sufficient to target GFP to membranes (Choy et al., 1999). Plant proteins in the HIPP family all contain a CaaX motif and are all thought to be prenylated (de Abreu-Neto et al., 2013). Furthermore, a strong prenylation prediction was obtained when the NbHIPP26 sequence was analyzed using Prenylation Prediction Suite software (score = 2.514, P < 0.001; Maurer-Stroh and Eisenhaber, 2005). These data combined indicate that NbHIPP26 is likely prenylated; therefore, we term the HIPP26 CVVM motif a prenylation (or prenyl) motif.
The full-length NbHIPP26 ORF was cloned and tested again in the Y2H system, confirming the interaction with PMTV TGB1 (Fig. 2A). In addition, the movement proteins of four other viruses were tested to investigate whether the interaction with PMTV TGB1 was specific or whether other virus movement proteins also might interact with NbHIPP26. These proteins were chosen to represent common types of virus movement proteins (Verchot-Lubicz et al., 2010; Tilsner et al., 2014), including (1) BVQ TGB1, representing another pomovirus; (2) BSMV TGB1, a hordei-type TGB1 protein with a helicase domain and a large N-terminal (RNA-binding) extension, which does not share similarity with the N-terminal domain of PMTV TGB1; (3) PVX TGB1, representing a potex-type TGB, which contains a helicase domain but not an N-terminal extension; and (4) TMV 30K movement protein, representing the extensively studied 30K family. These were cloned into the Y2H system, but while PMTV TGB1 interacted with NbHIPP26, no interactions were obtained with the other virus movement proteins (Fig. 2A). The BSMV, BVQ, TMV, and PVX movement proteins were detected by immunoblot in the yeast clones, confirming that the proteins were expressed in yeast.
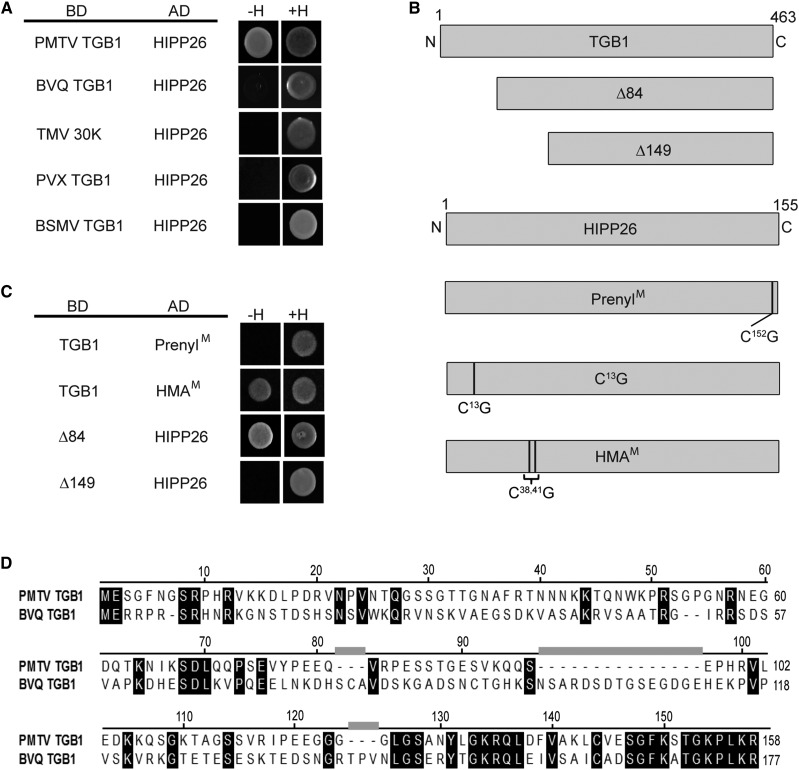
Figure 2.
Y2H interaction analysis. A, Virus movement proteins, PMTV TGB1, beet virus Q (BVQ) TGB1, TMV 30K, potato virus X (PVX) TGB1, and barley stripe mosaic virus (BSMV) TGB1 proteins were fused to the LexA binding domain (BD) and tested for interaction with HIPP26 fused to the p6 activation domain (AD) on double dropout (+H) or triple dropout (−H) medium. B, Diagrammatic representation of the wild type and mutants in PMTV TGB1 (∆84 and ∆149 deleted by amino acids 1–84 and 1–149, respectively) and the wild type and mutants of HIPP26 showing the positions of the Cys residues changed to Gly in putative prenylation (PrenylM), S-acylation (C13G), and heavy metal-associated (HMAM) domains. C, TGB1 wild type and deletion mutants were tested for interaction with HIPP26, PrenylM, and HMAM. D, Amino acid sequence alignment of PMTV and BVQ N-terminal amino acids.
TGB1 Interacts with the C Terminus of NbHIPP26
To identify key residues involved in the interactions, mutations were introduced into PMTV TGB1 and the predicted functional domains of NbHIPP26. The TGB1 mutants were shown previously to produce N-terminally truncated TGB1 proteins lacking amino acid residues 1 to 84 and 1 to 149 (TGB1 Δ84 and Δ149, respectively; Wright et al., 2010; Lukhovitskaya et al., 2015). In NbHIPP26, the prenyl motif, C152VVM155, was altered by replacing Cys-152 with Gly, which would interrupt any possible prenylation, and was referred to as PrenylM. The HMA domain (M36DCEGC41) was altered by replacing the Cys-38 and Cys-41 residues with Gly residues to give HMAM (Fig. 2B).
In Y2H assays, TGB1 Δ84 interacted with NbHIPP26 but the truncated protein TGB1 Δ149 did not (Fig. 2C), suggesting that some or all of the interacting surface is within the TGB1 region spanning amino acid residues 85 to 149. This idea was further supported by the lack of interaction between NbHIPP26 and BVQ TGB1, the movement protein of a related virus that showed very little similarity in the TGB185-149 fragment (Fig. 2D). In general, the C-terminal part of pomoviral TGB1 proteins, such as those from PMTV and BVQ, was well conserved and comprised the helicase domain, whereas the N termini of the proteins showed very little similarity. The PrenylM altered protein did not interact with TGB1, whereas HMAM retained the interaction (Fig. 2C), indicating that the site containing the CVVM motif was important for binding to TGB1.
GFP-NbHIPP26 Localizes in Several Subcellular Domains
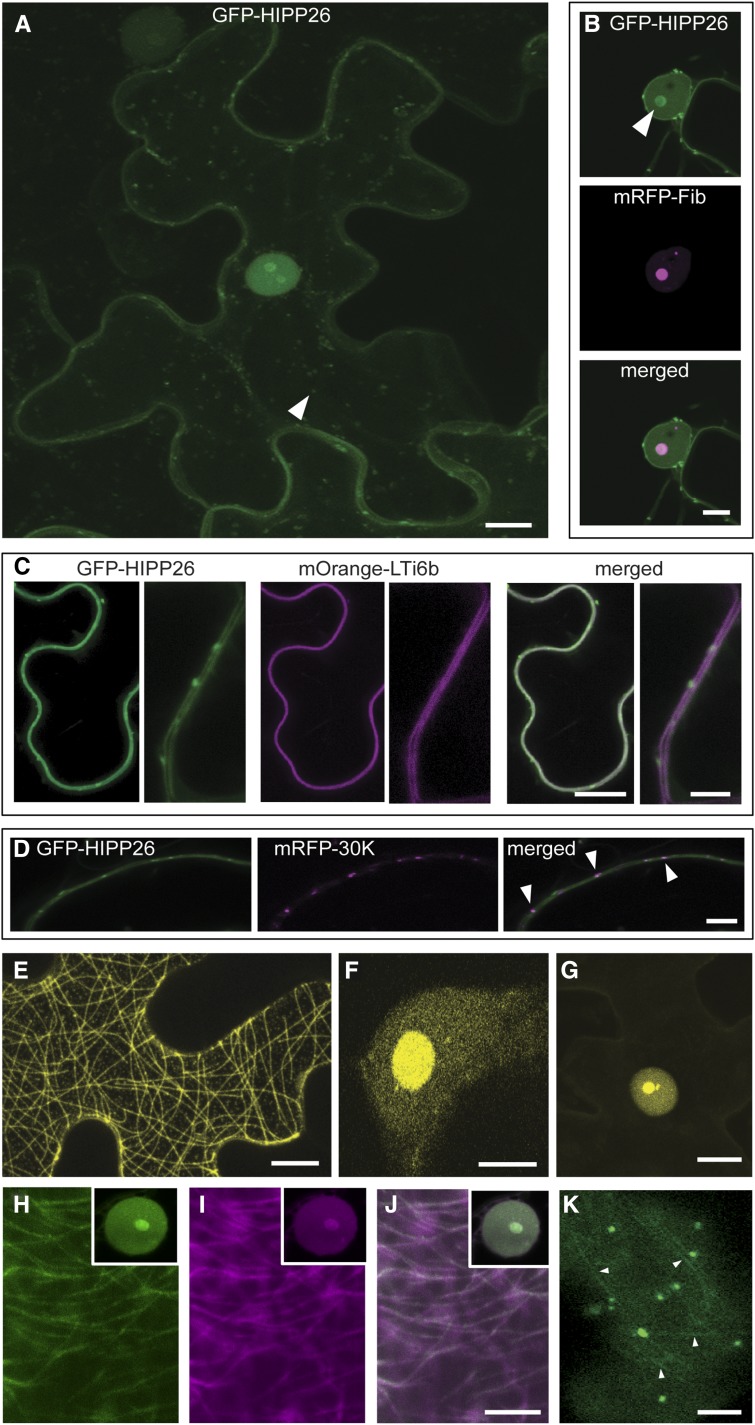
To investigate the localization of NbHIPP26, the protein was expressed as a translational fusion to GFP in N. benthamiana epidermal cells. Green fluorescence was observed in the plasma membrane, small motile vesicles (approximately 2 µm diameter; Supplemental Movie S1), the nucleoplasm, and the nucleoli (Fig. 3A). Coexpression with mRFP-fibrillarin (a marker for the nucleolus; Goodin et al., 2007) showed precise colocalization (Fig. 3B). Expression of GFP-NbHIPP26 in N. benthamiana plants transformed with the mOrange-LTi6b plasma membrane marker (Wright et al., 2010) showed strong colocalization of GFP and mOrange signal as well as punctate spots of GFP-NbHIPP26 around the cell periphery, which resembled PD (Fig. 3C). To determine whether these spots were PD, GFP-NbHIPP26 was coexpressed with the PD-localized TMV 30K movement protein fused to mRFP. Both fluorescent proteins colocalized in some punctate spots at the periphery (Fig. 3D), showing that NbHIPP26 localizes to a population of PDs. Thus, the results show that NbHIPP26 has several subcellular localizations, including PD, plasma membrane, motile vesicles, nucleoplasm, and nucleolus.
Figure 3.
Subcellular localizations of HIPP26 and the HIPP26-TGB1 complex. A, The GFP-HIPP26 fusion protein localized to the plasma membrane and a population of small (∼2 µm diameter), motile vesicles (arrowhead). A small amount of cytosolic labeling and faint transvacuolar strands were seen in some cells. GFP-HIPP26 also was present in the nucleus, both in the nucleoplasm and the nucleolus, and to levels greater than would be expected as a result of passive diffusion from the low levels in the cytosol. Therefore, HIPP26 was targeted to the nucleus. B and C, Coexpression with an mRFP-fibrillarin (a nucleolar marker) showed precise colocalization of GFP-HIPP26 in the nucleolus (B; indicated by the arrowhead), while coexpression and colocalization with mOrange-LTi6b (a plasma membrane marker) showed that HIPP26 also was targeted to the plasma membrane (C). D, In addition to the motile vesicles that were seen in the cytoplasm of the cell periphery (compare with A and Supplemental Movie S1), small nonmotile punctae also were observed at the cell periphery. Colocalization of these structures with an mRFP-tagged TMV 30K protein (a PD marker) showed that HIPP26 also was associated with a population of PD (indicated by arrowheads). E and F, BiFC analysis of PMTV TGB1 and NbHIPP26 in N. benthamiana epidermal cells showed coexpression of Y-HIPP26 and TGB1-FP and reconstitution of yellow fluorescence localized to microtubules (E), to the nucleoplasm, and strong accumulation in the nucleolus (F). G, Coexpression of Y-HIPP26 and ∆84TGB1-FP showed reconstitution of yellow fluorescence localized to the nucleoplasm and strong accumulation in the nucleolus with no microtubule labeling. No complementation was obtained in control experiments, where Y-HIPP26 or TGB1-FP was coexpressed with the complementary empty split YFP plasmid (no fluorescence was visible, so no images are shown). H to J, Colocalization of GFP-HIPP26 and mRFP-TGB1 (H, GFP; I, RFP; and J, merged) showed that both proteins localized to the nucleoplasm, nucleolus, and microtubules, as expected from previously published data and the BiFC results. K, Expression of GFP-HIPP26 in a PMTV-infected cell showed that GFP fluorescence accumulated on microtubules (indicated by arrowheads). Bars = = 10 µm in A, C (at left), D, E, G, and J (used for H–J) and 5 µm in B, C (at right), F, and K.
The TGB1-NbHIPP26 Complex Accumulates in the Nucleolus
To investigate the TGB1-NbHIPP26 interaction in plant cells, bimolecular fluorescence complementation (BiFC) experiments were conducted using split YFP constructs (Martin et al., 2009). NbHIPP26 and TGB1 genes were cloned to allow the expression of NbHIPP26 fused to the N-terminal domain of YFP (Y-NbHIPP26) and TGB1 fused to the C-terminal domain (TGB1-FP). When these constructs were coexpressed in N. benthamiana epidermal cells, yellow fluorescence was seen labeling microtubules and nucleoplasm, with particularly strong yellow fluorescence in the nucleolus (Fig. 3, E and F). GFP-HIPP26 never localized to microtubules when expressed alone. The TGB1 N-terminally truncated derivatives ∆84 and ∆149 also were tested in the BiFC system, and the results showed that yellow fluorescence was reconstituted in the nucleus and nucleolus but not on microtubules with the combination ∆84-FP and Y-NbHIPP26 (Fig. 3G), but no complementation was observed in tests with ∆149-FP and Y-NbHIPP26 (data not shown; no fluorescence was visible). No complementation (no fluorescence) was observed in control experiments with Y-NbHIPP26 or TGB1-FP and the corresponding empty vectors encoding Y or FP fragments. Both HIPP26 and TGB1 localize in the nucleus (Barth et al., 2009; Lukhovitskaya et al., 2015); however, the BiFC data show that the TGB1-NbHIPP26 complex also localizes to microtubules. This result, together with the absence of visible fluorescence at the plasma membrane and PD, indicated that the interaction with TGB1 caused a microtubule-guided relocalization of HIPP26 from the plasma membrane to the nucleus and nucleolus. This result recapitulates the TGB1 distribution pattern seen previously with YFP-TGB1 behind the leading edge of the virus infection (Wright et al., 2010).
To support the BiFC data, GFP-HIPP26 also was coexpressed in epidermal cells with RFP-TGB1. In these experiments, green and red fluorescence colocalized precisely in the nucleoplasm and nucleolus and on microtubules (Fig. 3, H–J). We also expressed GFP-HIPP26 in PMTV-infected cells, and in these cells, GFP fluorescence was localized as described before when expressed in noninfected cells, but it also accumulated on microtubules (Fig. 3K). Note that GFP-HIPP26 was never seen on microtubules when expressed alone. Thus, we believe that these data support the BiFC results and provide confirmatory evidence of the interaction between HIPP26 and TGB1 in plants.
NbHIPP26 Is an S-Acylated Protein
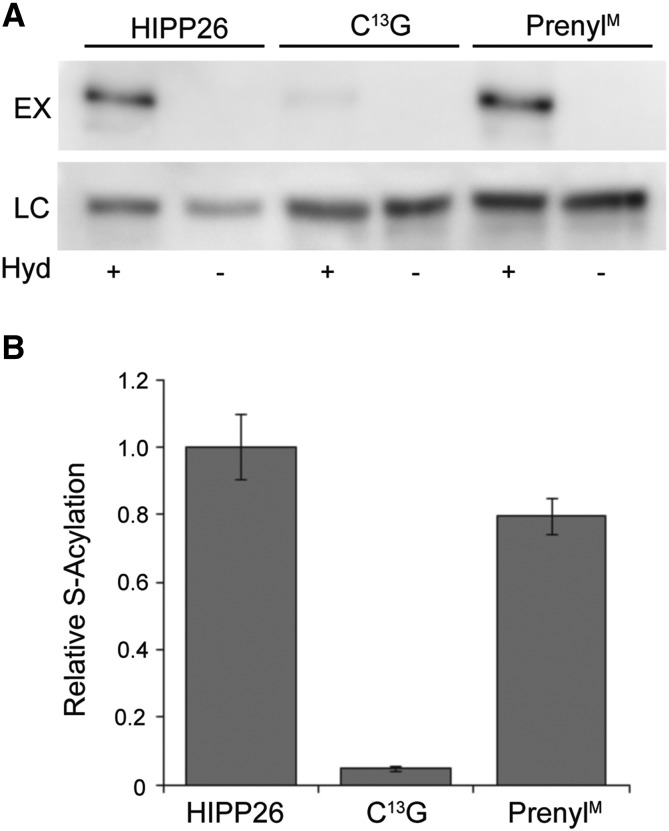
Proteomic studies have revealed that many Arabidopsis proteins involved in abiotic or biotic stresses can be reversibly lipid modified by S-acylation (Hemsley et al., 2013). S-Acylation involves the posttranslational addition of fatty acids, predominantly stearic or palmitic acid, to Cys residues within proteins through a reversible thioester linkage (Sorek et al., 2017). Various Arabidopsis HIPP family members were proposed as S-acylated based on proteomics data (Hemsley et al., 2013). Therefore, we tested whether GFP-NbHIPP26 expressed in N. benthamiana was S-acylated using a modified acyl-RAC assay (Hemsley et al., 2008; Forrester et al., 2011). The results revealed that NbHIPP26 was indeed S-acylated (Fig. 4). Substitution of candidate S-acylated Cys resides by Gly, Ala, or Ser is the most common way to identify S-acylation sites. NbHIPP26 contains four Cys residues: one as the prenyl acceptor in the CVVM motif, two in the HMA domain presumed to act as metal ion coordinators, and one of unknown function at the N terminus (C13). On the basis that C13 is the only Cys residue with no proposed function, we replaced it with Gly (C13G). Subsequent quantification of S-acylation assays comparing the C13G and PrenylM NbHIPP26 with the wild type (Figs. 2B and 4A) indicated that the C13G altered protein showed almost total loss of S-acylation signal and, therefore, that C13 was the likely site of S-acylation. The C13G altered protein is henceforth referred to as S-AcylM. Prior membrane association is thought to be required for maximal S-acylation efficiency by the integral membrane S-acylating enzymes. The small but apparent (n = 2, P = 0.103) decrease in the S-acylation state of NbHIPP26 altered protein PrenylM compared with the wild type (Fig. 4B) indicated that nonprenylated NbHIPP26 was still a substrate for S-acylation. This suggests that HIPP26 can interact with membranes through means other than prenylation and S-acylation.
Figure 4.
HIPP26 S-acylation. A, HIPP26 was S-acylated under normal plant growth conditions at Cys-13. S-Acylation was determined by the Acyl-RAC assay. Free Cys residues were blocked with N-ethylmaleimide. S-Acyl groups on Cys residues within proteins were cleaved by hydroxylamine (Hyd +) to reveal free sulfhydryls. Sulfhydryl-reactive thiopropyl Sepharose beads were used to capture free sulfhydryl-containing proteins. Negative controls lacked hydroxylamine (Hyd −). Signal strength in experimental lanes (EX Hyd+) showed the level of S-acylation. Loading controls (LC) demonstrated equal loading of hydroxylamine-treated and -untreated samples onto thiopropyl Sepharose beads. B, Relative quantification of blots shown in A. Mutation of the C-terminal CVVM motif Cys (PrenylM) had a small effect on the S-acylation state (n = 2; P = 0.103 by one-tailed Student’s t test), while changing Cys-13 to Gly effectively abolished S-acylation (n = 2; P = 0.05). Values were calculated as intensity in EX Hyd+ lanes divided by intensity in LC Hyd+ lanes. Values are means represented as the proportion of wild-type HIPP26 S-acylation. Error bars indicate sd.
NbHIPP26 Proteins Altered in the Lipidation Domains Show Depletion from Membranes and Nuclear Enrichment
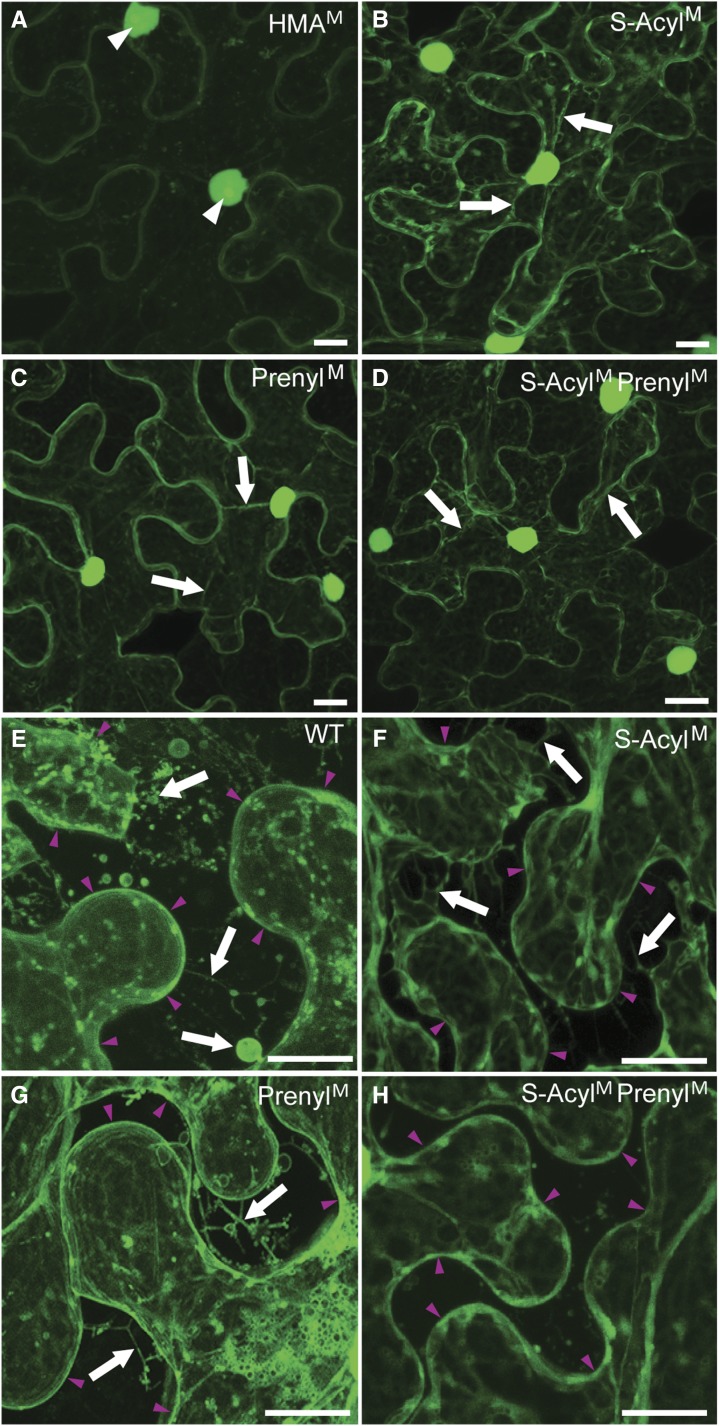
To determine whether lipidation is needed for NbHIPP26 binding to the plasma membrane and PD, the subcellular localizations of GFP-tagged NbHIPP26 wild-type and altered proteins S-AcylM, PrenylM, S-AcylM PrenylM, and HMAM were investigated (Fig. 5, A–D). Expression of HMAM gave a phenotype essentially the same as that of the wild type (compare Fig. 5A with Fig. 3A). In comparison with the wild-type GFP-HIPP26 and HMAM, GFP fluorescence in the cells expressing each of the other three altered proteins was observed to accumulate more strongly in the nucleus (Fig. 5, B–D). In addition, GFP labeling of the plasma membrane was reduced and substantial fluorescence was observed in the cytosol and in some motile vesicles. The phenotypes of the three altered proteins were similar, except that there were fewer motile vesicles and more cytosolic labeling in cells expressing the double modified protein, S-AcylM PrenylM. These experiments indicated that NbHIPP26 binding to the plasma membrane and PD was weaker in the altered (nonlipidated) proteins, suggesting that disruption of the lipid anchors results in release of the proteins to the cytoplasm and allows the subsequent marked increase in nuclear accumulation.
Figure 5.
Subcellular localization of GFP-tagged NbHIPP26-modified proteins. A to D, Four mutants expressing proteins with altered HMA (HMAM) or lipidation domains (S-AcylM, PrenylM, and S-AcylM PrenylM) were investigated. The HMA altered protein (A) was broadly similar to the wild-type HIPP26 (compare with Fig. 3A), showing plasma membrane labeling, low levels of cytosolic labeling, some small motile vesicle labeling, and accumulation in the nucleoplasm and nucleolus. In comparison, all three proteins altered in lipidation domains (B–D) showed significant increases in their nuclear accumulation (in both the nucleoli and nucleoplasm) and reductions in the amount of plasma membrane labeling accompanied by increases in the level of cytosolic fluorescence (seen most clearly in transvacuolar strands; arrows). Motile vesicles were seen in all three lipidation-deficient proteins, and the phenotypes were broadly similar, although cells expressing the doubly modified protein, S-AcylM PrenylM (D), showed fewer vesicles and more cytosolic localization. E to H, Plasmolysis of cells expressing wild-type HIPP26 (WT; E) and the three altered lipidation domain proteins (F–H) also were investigated. Here, the fluorescent cytosol was contained within the protoplast, which retracted from the cell walls, leaving a black space in the extracellular space/apoplast. Arrowheads situated in the extracellular space point to the plasma membrane of the retracted protoplasts. Hechtian strands (stretched extensions of plasma membrane that extend from the plasmolyzed protoplast to the cell wall), membrane blebs, and vesicles were clearly visible in the apoplast of the wild type (arrows). These structures were not easily detected in bright-field images, but GFP fluorescence associated with the remaining membrane made them clearly visible in the confocal micrographs shown here. There were fewer of these structures, and their morphology was somewhat altered in the cells expressing the proteins altered in a single lipid domain (F and G; arrows) and much less apparent in H, suggesting that removal of both lipidation domains reduced association with the plasma membrane in an additive way. Bars = 10 µm.
In order to study the association of the wild type and altered NbHIPP26 proteins with the plasma membrane in more detail, cells expressing the GFP fusion proteins were plasmolyzed to pull the plasma membrane away from the cell wall (Fig. 5, E–H). It is known that, as the protoplast retracts away from the wall, the plasma membrane is stretched into thin Hechtian strands, where it appears as thin tubules with associated small blebs and vesicles along the strands. The wild-type NbHIPP26 protein was associated with a profusion of these extra-protoplast structures (Fig. 5E). In cells expressing either PrenylM or S-AcylM, GFP fluorescence was associated with some membrane-derived structures external to the protoplast (e.g. Hechtian strands, vesicles, and membrane blebs), which formed as the plasma membrane pulled off the cell wall as the protoplast retracted. The amount of apoplastic fluorescence was reduced compared with the wild type (Fig. 5, F and G), and the expression of S-AcylM PrenylM showed almost no association with remnants of the plasma membrane in the region between the protoplast and the cell wall (Fig. 5H). These results suggest that lipidation is required to retain NbHIPP26 in association with membranes supporting a mechanism whereby the interaction with TGB1 disturbs or disrupts NbHIPP26 lipidation, resulting in the dissociation of the NbHIPP26-TGB1 complex from the membranes at the cell periphery.
NbHIPP26 Is Expressed Specifically in the Vascular Parenchyma and Induced by Drought and PMTV Infection
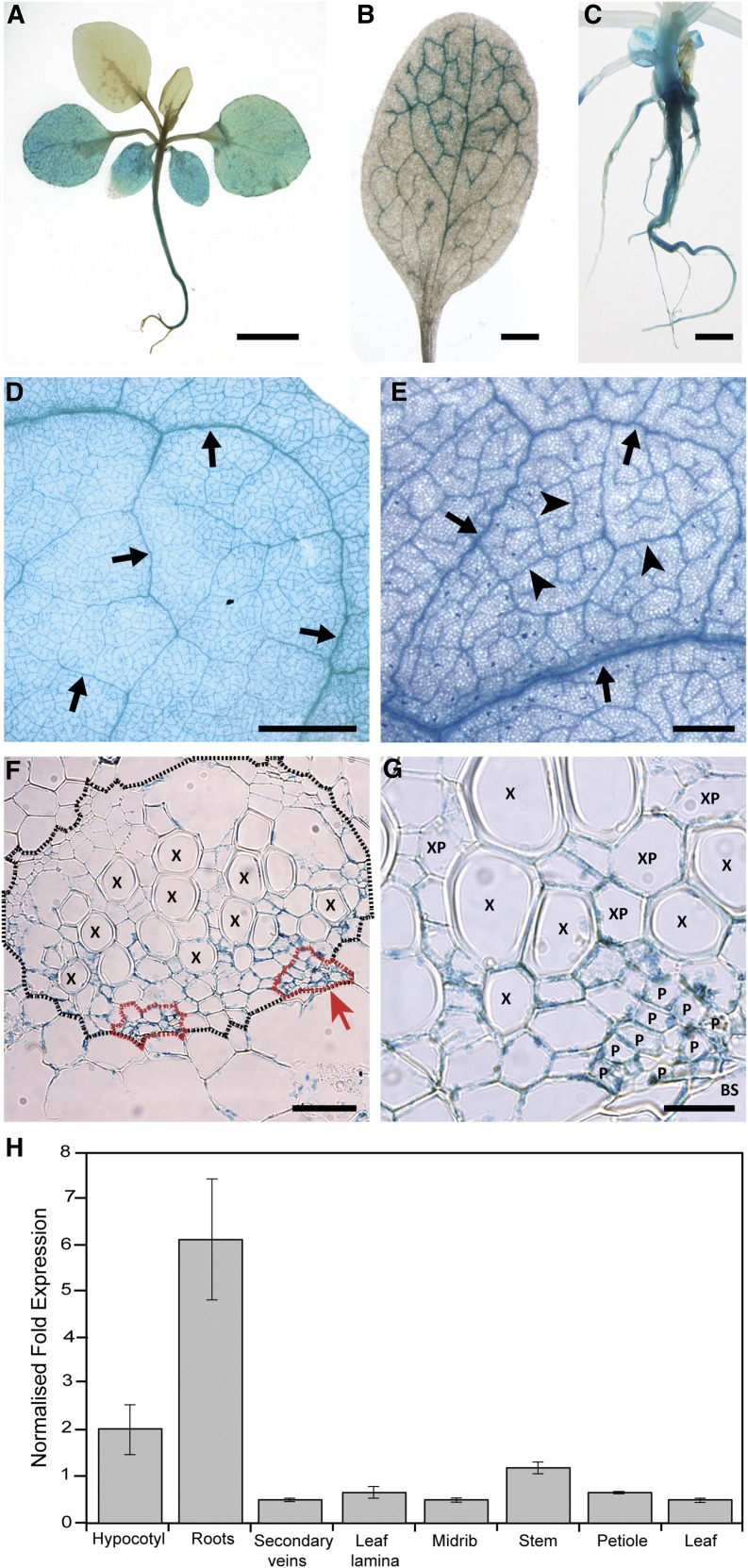
Analysis of the nucleotide sequence up to 2 kb upstream from the NbHIPP26 ORF showed the presence of multiple stress-responsive elements (Supplemental Fig. S2). A region of approximately 1 kb upstream of the predicted translational start site of NbHIPP26 thought to encompass the promoter sequence was cloned and fused to the uidA gene, and the resulting construct was used to transform N. benthamiana plants. Seven independent transgenic lines were tested for GUS expression, and staining was visibly concentrated in the vascular tissue of all plants. In whole plants, GUS staining was seen predominantly in the vasculature of lower leaves, stems, and roots (Fig. 6, A–C). At low magnification (Fig. 6, D and E), GUS activity was seen in all vein classes of mature N. benthamiana leaves. Transverse sections of major veins (Fig. 6, F and G) showed GUS activity to be concentrated in the periphery of cells within the vascular bundles. Vascular parenchyma cells (associated with both phloem and xylem tissue) showed blue staining, with the strongest promoter activity associated with clusters of phloem cells (Fig. 6, F [areas outlined in red] and G). Phloem tissue was identified as clusters of cells containing small cells (sieve elements) that lie immediately adaxial and abaxial to the xylem tissue. These results are supported by experiments using HIPP26pro:uidA constructs in Arabidopsis, which also show strong vascular expression of GUS activity (Barth et al., 2009), and are consistent with previous reports that stress-related genes are expressed preferentially in the vascular tissues (for review, see Turgeon and Wolf, 2009).
Figure 6.
Images of GUS expression in N. benthamiana transformed with NbHIPP26 predicted promoter sequences fused to the uidA gene. A to C, GUS staining in mature plants (A), mature leaves (B), and roots (C) was detected throughout the vasculature of the mature plant but was not present in young sink leaves (A). Expression was particularly strong in the roots and hypocotyl, and in leaves, GUS staining was more evident in older leaves. Expression in leaf veins appeared to follow the sink-source transition: GUS staining was evident in tip (source) veins, while the basal (sink) portion was clearer (B). D and E, When studied in more detail in leaf veins, GUS was localized specifically to cells within the vascular bundle of all vein classes in mature leaves (arrows point to major vein classes and arrowheads to minor veins). At low magnification, there appeared to be little or no GUS staining in ground tissues between the veins, and intact leaf petioles did not show staining. Once cut, however, GUS was visible in the vascular bundles of petioles. D and E show images of intact, mature N. benthamiana leaves photographed using a combination of bottom and top lighting. F and G, GUS staining in transverse sections through major veins. The vascular bundle (F) is delineated with a dotted black line, and phloem regions are outlined with red dotted lines. Some GUS staining was detected in cells surrounding the vascular bundles, but much less than inside the vascular system. G shows an enlargement of the phloem bundle marked in F with a red arrow. Blue GUS staining can be seen clearly in xylem parenchyma (XP) and phloem (P) cells but is generally absent from metaxylem vessels (X), and the bundle sheath (BS) also is shown. Bars = 2 mm in A and D, 500 µm in B and E, 50 µm in C and F, and 20 µm in G. H, Relative NbHIPP26 expression in different tissues measured by RT-qPCR. The histogram bars show mean values ± sd (n = 3, two independent experiments).
NbHIPP26 expression was measured in different tissues by reverse transcription-quantitative PCR (RT-qPCR) and was shown to be most strongly expressed in root tissues, followed by the hypocotyl, but was detected in all tissues sampled (Fig. 6H).
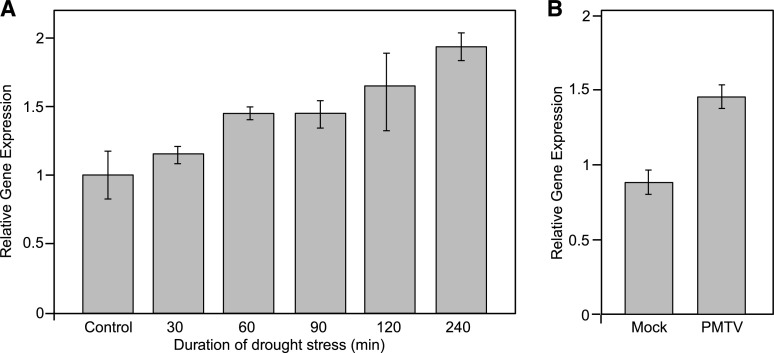
To establish whether NbHIPP26 is induced by drought, N. benthamiana plants were subjected to drought stress by removing whole plants from the growing medium and exposing them to air at room temperature for 30 to 240 min, during which NbHIPP26 expression was measured by RT-qPCR. Relative NbHIPP26 expression was compared against the endogenous genes PROTEIN PHOSPHATASE2A (PP2A) and F-Box protein (F-BOX) and was shown to increase over time in response to drought stress (Fig. 7A).
Figure 7.
Relative expression of NbHIPP26 in leaves of N. benthamiana. Relative expression is shown in N. benthamiana leaves after 30 to 240 min of drought stress (A; representative experiment showing means ± sd for each time point; n = 3) and in PMTV-infected leaves 12 d post inoculation (B; representative experiment showing means ± sd for each time point; n = 8), demonstrating that both drought stress and PMTV infection up-regulate NbHIPP26 expression.
To determine whether NbHIPP26 also is induced by virus infection, gene expression levels were investigated in PMTV-infected N. benthamiana. Total RNA was obtained from upper noninoculated leaves. Relative expression levels of NbHIPP26 were compared by RT-qPCR against three endogenous genes (PP2A, CYCLIN-DEPENDENT KINASE2 [CDC2], and F-BOX) in both virus-infected and mock-inoculated plants at different time points post inoculation. Relative NbHIPP26 expression increased in approximately 71% of PMTV-infected plants tested between 10 and 14 d post inoculation (dpi) compared with mock-inoculated plants (Fig. 7B shows a representative experiment at 12 dpi).
PMTV Infection Protects N. benthamiana from Drought Stress
Plants infected with some plant viruses are known to be drought tolerant (Xu et al., 2008). Therefore, we investigated whether infection with PMTV influenced drought tolerance. N. benthamiana plants were manually inoculated with PMTV or mock inoculated and then maintained in a glasshouse for 14 d to allow systemic spread. Water was then withheld, and plants were monitored for visible drought response. After a further 13 d, noninfected plants showed signs of wilting, but in marked contrast, the infected plants did not wilt at this time point (Fig. 8, A and B). We then tested whether PMTV could protect plants from drought by withholding water until both mock-infected (Fig. 8, C–E) and PMTV-infected (Fig. 8, F–H) plants were completely wilted and then rewatering the plants. The results were similar in two independent experiments; the numbers of plants surviving (as judged by the presence of new green shoots)/number tested were as follows: experiment 1, 2/10 mock-infected plants and 5/8 PMTV-infected plants; experiment 2, 2/5 mock-infected plants and 7/10 PMTV-infected plants. Thus, fewer mock-inoculated plants survived (27%) compared with plants infected with PMTV (67%).
Figure 8.
Representative images of PMTV- or mock-inoculated N. benthamiana plants after drought stress. A and B, Mock-inoculated (A) and PMTV-inoculated (B) plants after water was withheld for 13 d, showing that PMTV-infected plants are more resilient to wilting compared with the mock-infected plants. C to H, Representative mock-inoculated (C–E) and PMTV-inoculated (F–H) plants before treatment (C and F), after withholding water (D and G), and after rewatering (E and H), showing that PMTV-infected plants produced green shoots and show recovery from drought treatment in contrast to the mock-inoculated plants (E).
Knockdown of NbHIPP26 Expression Inhibits PMTV Long-Distance Movement
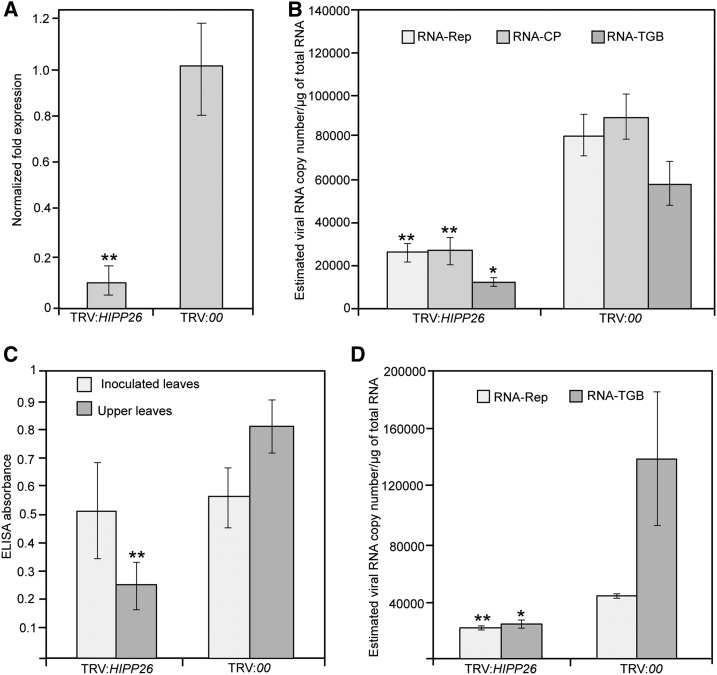
To examine the role of HIPP26 in PMTV movement, a tobacco rattle virus (TRV)-based system was used to knock down the expression of HIPP26 in N. benthamiana plants. A 120-bp fragment of NbHIPP26 was cloned into TRV in an antisense orientation (TRV:HIPP26), and the results showed that expression of the targeted HIPP26 gene was reduced specifically in TRV-RNA1 + TRV:HIPP26 inoculated plants compared with the empty vector controls (TRV-RNA1 + TRV:00; Fig. 9A). The NbHIPP26-silenced and control plants were challenged with PMTV and analyzed 2 weeks post inoculation. RT-qPCR of RNA samples isolated from the upper noninoculated leaves of PMTV-inoculated plants revealed the accumulation of all three genomic RNA components (Fig. 9B). However, the accumulation of all three virus genomic components in upper leaves of the plants silenced for NbHIPP26 was decreased markedly as compared with TRV:00 control plants (Fig. 9B). Quantitative analysis of PMTV accumulation by ELISA (to detect virus particles) 2 weeks post inoculation revealed similar levels of PMTV accumulation in inoculated leaves of plants silenced for NbHIPP26 and in nonsilenced plants inoculated with TRV:00 (Fig. 9C), indicating that the cell-to-cell movement of virus particles and accumulation in inoculated leaves were not affected. However, in the upper leaves of plants silenced for NbHIPP26, PMTV particles accumulated in much lower amounts as compared with the control plants (absorbance values were ∼30% of control; P < 0.05 by two-tailed Student’s t test; Fig. 9C). Seven of 36 (19%) NbHIPP26-silenced plants challenged with PMTV over six experiments were completely negative for the presence of virus particles in ELISA, while all control plants were infected with PMTV.
Figure 9.
Effects of the knockdown of NbHIPP26 expression on PMTV accumulation in upper noninoculated leaves. A, Gene silencing of the NbHIPP26 gene in TRV:HIPP26-infected plants. Total RNAs were extracted from the upper leaves 14 dpi and were used for RT-qPCR. Data are means ± sd from three experiments (n = 14 for each treatment). NbF-BOX and NbPP2A were used as normalization controls. B, Effect of HIPP26 gene silencing on viral RNA levels in upper noninoculated leaves. Total RNAs were extracted from the upper leaves 14 dpi with PMTV and used for absolute RT-qPCR. Ten-fold serial dilutions of full-length infectious cDNA clones of RNA-Rep, RNA-CP, and RNA-TGB were used to obtain standard curves and determine viral RNA copy number. NbF-BOX and NbPP2A were used as normalization controls. Data are means ± sd from four independent experiments with three or four biological replicates each. C, ELISA was used to determine PMTV infection in inoculated and upper noninoculated leaves of treated plants. Absorbance values (indicating the presence of PMTV capsid protein) in samples of inoculated leaves were similar in both treatments but were more than 50% greater in the upper noninoculated leaves of plants inoculated with empty vector, showing suppression of the long-distance movement of PMTV particles in NbHIPP26 knockdown plants. D, RT-qPCR quantification of RNA-Rep and RNA-TGB in upper noninoculated leaves of plants challenged with these two PMTV genomic components. Data are means ± sd; n = 6, two independent experiments. Asterisks indicate statistically significant differences compared with TRV:00: *, P < 0.01 and **, P ≤ 0.05, by Student’s two-tailed t test.
As PMTV moves long distance both as virus particles and vRNPs, we next asked whether the systemic movement of vRNPs was affected in the plants silenced for NbHIPP26. To address this question, NbHIPP26-silenced and control plants were inoculated with RNA-Rep and RNA-TGB in vitro-generated transcripts and analyzed 2 weeks post inoculation. RT-qPCR of RNA samples isolated from the upper noninoculated leaves of plants challenged with these two genomic components revealed the accumulation of RNA-Rep and RNA-TGB (Fig. 9D). However, the accumulation of these two genomic components in upper leaves of the plants silenced for NbHIPP26 was decreased markedly as compared with TRV:00 control plants (P < 0.05; Fig. 9D). Hence, collectively, the results show that the systemic movement of both virus particles and vRNPs but not cell-to-cell movement in N. benthamiana plants silenced for NbHIPP26 was inhibited. Overall, these results show that the interaction of PMTV TGB1 with NbHIPP26 is essential for efficient systemic movement of the virus.
DISCUSSION
TGB1 plays a central role in the long-distance movement of PMTV. TGB1 accumulates in the nucleolus, and we previously showed that nucleolar accumulation by an importin-α-dependent mechanism contributes to long-distance movement (Lukhovitskaya et al., 2015). Here, we further investigated the interactions of TGB1 with plant proteins and the role of TGB1 nucleolar accumulation in systemic virus movement. Y2H experiments showed that TGB1 interacted with the N. benthamiana homolog of the Arabidopsis protein HIPP26, and sequence analysis revealed a high level of sequence identity (84%) between the two proteins. HIPP26 is a member of a family of proteins that contain heavy metal-binding and C-terminal isoprenylation motifs.
Previously, NbHIPP26 was found to be present in the PD proteome (Fernandez-Calvino et al., 2011) and the plasma membrane proteome (Marmagne et al., 2007), and YFP-TGB1 accumulated in PD when expressed from a virus clone (Wright et al., 2010). HIPP proteins, like many membrane-associated proteins, are modified posttranslationally by the attachment of lipid moieties. HIPP26 contains a C-terminal motif, CVVM, identical to the known prenylation motif of N-Ras, and we showed that NbHIPP26 is lipid modified by S-acylation through residue C13. Both modifications were shown to be important for the observed localization of NbHIPP26 at PD and the plasma membrane. Mutation of both S-acylation and CVVM lipidation sites led to a marked accumulation of HIPP26 in the nucleus and nucleolus and loss from the cell periphery. S-Acylation provides a membrane association strength many times greater than a prenyl group (more akin to a transmembrane domain) but, critically, is reversible. This provides a mechanism to change the membrane association of proteins, as has been shown for active and inactive type I ROP GTPases (Sorek et al., 2017). While lipid modifications usually promote protein-membrane interaction, they also can mediate protein-protein interactions, as illustrated by the prenyl group-mediated interaction of K-Ras with Galectin-1 and -3 (Ashery et al., 2006). Given that TGB1 binding to NbHIPP26 requires an intact prenylation site, it is possible that TGB1 binds NbHIPP26 at least partly through the prenyl moiety. This may mask the ability of the prenyl group to interact with the membrane and promote the dissociation of NbHIPP26 from the plasma membrane environment. Consistent with this idea, BiFC showed that, on interaction with TGB1, the NbHIPP26-TGB1 complex localized predominantly to microtubules and the nucleolus with a small quantity in the nucleoplasm. The following model is consistent with our data: TGB1 binds NbHIPP26, most likely at PD, through the NbHIPP26 prenyl group, leading to NbHIPP26 conformational change and exposure of the S-acyl thioester. The thioester is now accessible for cleavage and releases NbHIPP26 from the plasma membrane, allowing trafficking of the NbHIPP26-TGB1 complex to the nucleus via microtubules.
N. benthamiana plants expressing HIPP26 promoter-reporter fusions revealed vascular tissue-specific expression similar to that found for AtHIPP26, suggesting a similar role for NbHIPP26 in drought tolerance. In addition, PMTV infection helps to protect N. benthamiana plants against drought, presumably due to the increased expression of NbHIPP26 during the systemic infection process.
Virus-induced gene silencing (VIGS) of NbHIPP26 expression resulted in decreased accumulation of PMTV in upper (noninoculated) leaves. In many of the NbHIPP26 knockdown plants, there was approximately 50% less virus than in noninoculated plants; in 19% of plants, no particles were detected, whereas particles were detected in all of the nonsilenced control plants.
The eukaryotic cell nucleolus contains the factors needed for its main function in ribosome subunit production, and this is where rRNA transcription, processing, and ribosomal RNP assembly occur. In mammalian cells, major changes occur in the organization and protein composition of the nucleolus in response to stress, where it plays a key role in responding to abiotic and biotic stress signaling (Boulon et al., 2010). Several positive-strand RNA viruses encode proteins that localize to the nucleolus (Taliansky et al., 2010), including the GRV ORF3 movement protein. The GRV ORF3 movement protein enters the nucleolus and associates with the nucleolar protein fibrillarin before the fibrillarin-ORF3 complex returns to the cytoplasm, enabling the formation of vRNP needed for long-distance movement (Kim et al., 2007).
The TMV replication protein interacts with three vascular-expressed auxin/indole acetic acid (Aux/IAA) transcriptional regulators, including IAA26, which is expressed in phloem companion cells (Padmanabhan et al., 2006). These Aux/IAA proteins are thought to function in the regulation of genes involved in phloem loading. Recently, it was shown that TMV infection inhibits the nuclear localization of Aux/IAA proteins, leading to transcriptional reprogramming of mature vascular tissue, which correlates with enhanced TMV movement and spread in the phloem of older leaves (Collum et al., 2016).
Our results indicate that the nuclear and nucleolar association of PMTV TGB1 has a different function in virus long-distance movement from that described for other viruses. They suggest that the nuclear association of plant RNA virus components can manipulate transcriptional regulation and reprogram gene expression in vascular tissue and that this may be a more general strategy used by plant viruses to facilitate long-distance movement than previously supposed (Solovyev and Savenkov, 2014; Collum et al., 2016).
However, another consideration is whether the interaction of TGB1 with NbHIPP26 may have a role in pathogenicity. As stated before, HIPPs are a large family of plant proteins that play roles in response to both abiotic and biotic stresses (de Abreu-Neto et al., 2013). For example, the cytoplasmically located rice (Oryza sativa) protein OsHIPP05 is a recessive susceptibility factor in rice blast disease; the resistant allele carries deletions in Pro-rich motifs thought to be targeted by pathogen protein(s) to facilitate infection (Fukuoka et al., 2009). While we cannot completely rule out that the interaction of NbHIPP26 with PMTV-TGB1 may suppress a putative role of HIPP26 in host defense (e.g. by transporting HIPP26 on microtubules for degradation), we think that this is unlikely, since, logically, knock down of NbHIPP26 would result in increased spread of infection and not inhibition, as we observed.
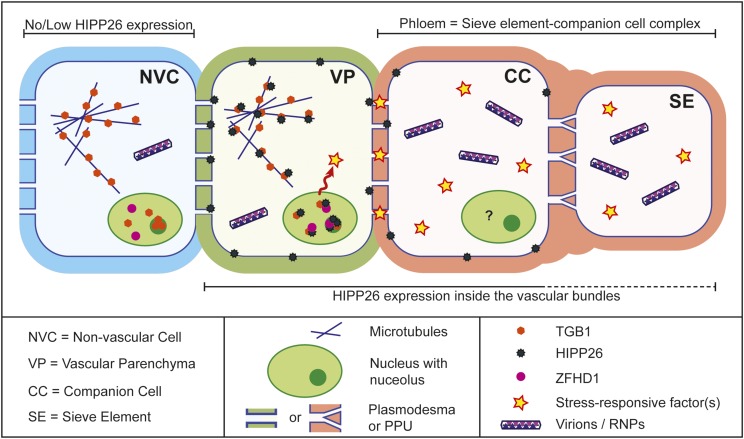
Our results showed that NbHIPP26 moves as a complex with TGB1 and accumulates in the nucleus. By analogy to AtHIPP26, which interacts in the nucleus with the transcriptional activator ZFHD1 (Tran et al., 2004, 2007; Barth et al., 2009), we suggest that the N. benthamiana TGB1 interaction leads to the promotion of drought tolerance by activating the expression of dehydration-inducible genes. Loss of function of HIPP26 in an Arabidopsis mutant inhibits the expression of stress-responsive genes regulated by ZFHD1, and Barth et al. (2009) proposed a mechanism where the heavy metal carried by HIPP26 inactivates or displaces the ZF-HD repressor and allows the activation of the stress-inducible genes. Our data suggests a model where, once the PMTV infection reaches the vascular tissue, TGB1 binds to NbHIPP26. The NbHIPP26/TGB1 complex is then transferred from the plasma membrane or PDs via microtubule-directed transfer to the nucleus and nucleolus, where HIPP26 accumulation leads to the up-regulation of dehydration-responsive gene expression in the vasculature and the establishment of a drought-tolerant state in the plant. The changes in vascular gene expression also allow virions or RNPs to enter the sieve element/companion cell complex for long-distance movement (Fig. 10). The PMTV genome is tripartite, and intact virus particles are essential for vector transmission (Cowan et al., 1997; Reavy et al., 1998). A mechanism allowing the systemic movement of virions or RNPs (including all genome components) ensures genome integrity for successful future vector-based transmission of fully infective virus from tissues far removed from the initial infection site.
Figure 10.
Model of the mechanism of long-distance movement. In a PMTV infection, TGB1 localizes to microtubules and traffics to the nucleus, where it is localized in the nucleoplasm and nucleolus (as shown in the nonvascular cell [NVC]; blue). HIPP26 is expressed mainly inside the vascular bundle, predominantly in regions of the bundle containing phloem cells (phloem parenchyma, companion cells, and sieve elements), and also in other vascular parenchyma cells, which can sometimes be identified as xylem parenchyma due to their proximity to metaxylem elements. Once the virus reaches a vascular bundle where HIPP26 is expressed, the TGB1 binds near the CVVM (prenyl domain) of NbHIPP26 at the plasma membrane, most likely in the vicinity of PD. This de-S-acylates HIPP26, which releases the NbHIPP26/TGB1 complex from the plasma membrane into the cytosol, from where it localizes to microtubules and subsequently moves via importin-α-directed transport to the nucleus and nucleolus, where HIPP26 interacts with the transcriptional activator ZFHD1. Interaction with ZFHD1 stimulates host proteins and/or stress response factors such as NAC transcription factors, along with molecules that increase the size exclusion limit of the PD at the vascular parenchyma (VP)-companion cell (CC) interface. These PDs are seen as a key control point for entry into the sieve element (SE)/companion cell complex and an important barrier to systemic spread for viruses. The pore-plasmodesma units (PPU) that connect companion cells and sieve elements are known to have a large size exclusion limit, so the sieve element/companion cell complex has been proposed to be a single domain where even large molecules can move relatively freely into the sieve element. It is difficult to differentiate between what may occur in the phloem parenchyma and companion cells; protein expression and interactions may be different or the same in these cell types, indicated by the question mark in the companion cell nucleus. The key point is that we believe the virus exploits the systemic signaling response to drought in order to gain entry into the phloem. Once inside the phloem tissue, in the presence of host proteins that gate PD, virions or RNPs can enter the sieve element for systemic spread.
MATERIALS AND METHODS
Y2H Analysis
Y2H screening was performed by Hybrigenics Services. The Hybrigenics system employs plasmids pB27 (containing the LexA binding domain) and pP6 (containing the P6 activation domain) derived from the original pBTM116 (Vojtek and Hollenberg, 1995) and pGADGH (Bartel et al., 1993) plasmids, respectively. The coding sequence for the full-length protein TGB1 (GenBank accession no. AJ277556) was PCR amplified and cloned into pB27 as a C-terminal fusion to LexA. The construct was confirmed by sequencing the entire insert and used as a bait to screen a randomly primed Nicotiana benthamiana cDNA library constructed into pP6. A total of 125 million clones (12-fold the complexity of the library) were screened using a mating approach with YHGX13 (Y187 ade2-101::loxP-kanMX-loxP, mata) and L40∆Gal4 (mata) yeast strains as described previously (Fromont-Racine et al., 1997). A total of 186 colonies were selected on a medium lacking Trp, Leu, and His. The prey fragments of the positive clones were amplified by PCR and sequenced at their 5′ and 3′ junctions. The resulting sequences were used to identify the corresponding interacting proteins in the GenBank database (National Center for Biotechnology Information) using a fully automated procedure.
A confidence score (PBS, for Predicted Biological Score) was attributed to each interaction as described previously (Formstecher et al., 2005). The PBS relies on two different levels of analysis. First, a local score takes into account the redundancy and independence of prey fragments as well as the distribution of reading frames and stop codons in overlapping fragments. Second, a global score takes into account the interactions found in all the screens performed at Hybrigenics using the same library. This global score represents the probability of an interaction being nonspecific. For practical use, the scores were divided into four categories, from A (highest confidence) to D (lowest confidence). A fifth category (E) specifically flags interactions involving highly connected prey domains previously found several times in screens performed on libraries derived from the same organism. Finally, several of these highly connected domains were confirmed as false positives of the technique and were tagged as F. The PBS scores have been shown to positively correlate with the biological significance of interactions (Rain et al., 2001; Wojcik et al., 2002).
Plasmids of representative interacting clone pP6-A44 were obtained from Hybrigenics and tested again at the James Hutton Institute. The interaction between Gal4 and LexA protein fusions was examined in the yeast L40 strain [MATa his3D200 trp1-901 leu 2-3,112 ade2 lys2-801am URA3::(lexAop)8-lacZ, LYS2::(lexAop)4-HIS3]. Yeast cells were cotransformed by the small-scale lithium acetate yeast transformation method as described in Clontech’s Yeast Protocols Handbook. Transformants were selected on synthetic dropout minimal medium base containing 2% (w/v) Glc and dropout supplements lacking Leu and Trp (+H) or lacking Leu, Trp, and His (−H). In experiments with LexA-IMP and LexA-PVXTGB1, a small amount of autoactivation was observed, which was suppressed by the addition of 10 and 50 mm 3-aminotriazole, respectively.
Primer sequences used for cloning are given in Supplemental Table S1. The coding sequence for NbImportin-α1 (ABMO5487; GenBank accession no. EF137253.1) was PCR amplified using primers IMPFOR and IMPREV (Lukhovitskaya et al., 2015) with template plasmid eGFP-NbIMP-α1 and cloned into pP6 (Hybrigenics). The virus movement proteins were cloned into the bait vector pB27. PVX TGB1 was PCR amplified from pPVX201 (Baulcombe et al., 1995) using primers PVXTGB1FOR and PVXTGB1REV, TMV 30K was PCR amplified from TMV-30B (Shivprasad et al., 1999) using primers TMV30KFOR and TMV30KREV, and BSMV TGB1 was PCR amplified from a BSMV RNA β cDNA clone (Torrance et al., 2006) using primers BSMVTGB1FOR and BSMVTGB1REV.
Sequence Alignments and Phylogenies
To place the clone A44 (NbHIPP26) within the full family repertoire of HIPP proteins identified previously in Arabidopsis (Arabidopsis thaliana), the full-length protein sequences of 45 HIPP proteins identified by de Abreu-Neto et al. (2013) were downloaded from GenBank (release 205; Benson et al., 2013). A multiple alignment of these 45 proteins along with the clone sequence A44 was made using MUSCLE with default parameters (Edgar, 2004), and a phylogeny was created using the neighbor-joining method within MEGA (version 6.06). The phylogenetic tree was rendered as a midpoint-rooted tree with proportional branch lengths using FigTree version 1.4.2 (Rambault, 2012).
To assess the percentage sequence similarity between the NbHIPP26 protein and its homologs in potato (Solanum tuberosum), tomato (Solanum lycopersicum), and Arabidopsis, a multiple sequence alignment was made using MUSCLE with default parameters (Edgar, 2004). A sequence identity matrix of protein-protein pairs was calculated from the alignment.
Plant Material
N. benthamiana plants were grown in a glasshouse (21°C day and 16°C night, 18-h daylength with supplementary lighting of 250 W m−2). N. benthamiana plants expressing mRFP:AtFib1 fusions (RFP-labeled fibrillarin) were provided by Michael Goodin. N. benthamiana plants expressing mOrange-LTi6b (mOrange-labeled plasma membrane) were created at the James Hutton Institute (Wright et al., 2017).
To Check Gene Expression
N. benthamiana plants (35 d post germination) were removed from growth medium. The roots were rinsed free of compost and plants were left in air on filter paper for the designated time. Tissues were then processed for RT-qPCR as described below.
To Test the Effect of PMTV Infection
In two independent experiments, N. benthamiana plants were mechanically inoculated with PMTV or water (mock) and maintained in the glasshouse (as described above). The upper noninoculated leaves were tested by ELISA 12 to 14 dpi to confirm systemic infection. Water was then withheld until the plants reached the wilting point, and then watering was restored.
Cloning of Plasmids and Mutants
All of the plasmids were constructed so that fluorescent protein expression was driven by the 35S promoter. Primer sequences are provided in Supplemental Table S1.
Constructs Used for Confocal Microscopy
The NbHIPP26-encoding sequence was amplified from N. benthamiana total RNA using attB adapter-flanked primers HIPP26FOR and HIPP26REV and recombined into pDONR207 using Gateway BP Clonase II. This entry clone was recombined with pB7WGF2 (Karimi et al., 2002) using Gateway LR Clonase II to create plasmid eGFP-HIPP26.
Constructs Used for BiFC
pDONR207 entry clone constructs containing the sequences coding for NbHIPP26 and TGB1 were recombined into pSITE-cEYFP-N1 (Martin et al., 2009) using Gateway LR Clonase II. pDONR207 constructs containing the sequences coding for TGB1, NbHIPP26C174G, and TGB1∆84 were recombined into pSITE-nEYFP-C1 (Martin et al., 2009) using Gateway LR Clonase II (Invitrogen).
Construction of HIPP26 Mutants
Mutations in the NbHIPP26 sequence were made using the QuikChange Site-Directed Mutagenesis kit (Stratagene). The Cys-152 codon was changed to GGA (Gly codon) using primers CVVMFOR and CVVMREV to create NbHIPP26C152G. The Cys-38 and Cys-41 codons within the HMA domain were changed to GGA using primers HMAmutFOR and HMAmutREV to create NbHIPP26G38C,G41C. The Cys-13 codon was mutated to GGA using primers HIPP26cysmutFOR and HIPP26cysmutREV to create NbHIPP26C13G. A double mutant was created where the Cys-13 and Cys-152 codons were both mutated to GGA using primers HIPP26cysmutFOR and HIPP26cysmutREV with template GFP-NbHIPP26C152G .
Live-Cell Imaging of Fluorescent Proteins
Unless otherwise stated, all constructs were delivered into N. benthamiana leaves by agroinfiltration through the abaxial stomata. Agrobacterium tumefaciens (strain LB4404) cultures carrying the plasmid constructs used for live-cell imaging were grown overnight (16 h) at 28°C in Luria-Bertani medium supplemented with rifampicin (50 mg L−1) and spectinomycin (100 mg L−1). Cells were collected by centrifugation at 3,500 rpm for 15 min and then resuspended in infiltration buffer (10 mm MgCl2, 10 mm MES, and 150 μm acetosyringone) to a final OD600 of 0.1 before infiltration into the abaxial side of N. benthamiana leaves. All confocal imaging was conducted using a Leica TCS-SP2 AOBS spectral confocal scanner (Leica Microsystems). Unless otherwise stated, images were obtained using a Leica HCX APO x63/0.9W water-dipping lens and whole lesions using an HCX PL Fluotar ×1.6/0.05 lens. GFP and YFP were imaged sequentially: GFP excitation at 488 nm, emission at 490 to 510 nm; YFP excitation at 514 nm, emission at 535 to 545 nm. GFP and mRFP were imaged sequentially: GFP excitation at 488 nm, emission at 500 to 530 nm; mRFP excitation at 561 nm, emission at 590 to 630 nm. Images of GUS localization were taken on a Zeiss AxioImager microscope (Carl Zeiss) equipped with a Zeiss Axiocam HRC camera. Images were prepared using Adobe Photoshop CS5.
RT-qPCR
Five-week-old seedlings of N. benthamiana were manually inoculated with leaf extracts from PMTV-infected N. benthamiana or mock-inoculated plants and grown in a Snijders growth cabinet providing a 12-h day (with a light intensity setting of 375 µmol m−2 s−1) and 12-h night at constant 14°C. Total RNA was isolated from leaf samples using the RNeasy Plant Minikit with the on-column DNaseI treatment for the removal of genomic DNA contamination according to the manufacturer’s protocol (Qiagen). Reverse transcription of 5 µg of total RNA was performed using Ready-To-Go-You-Prime-First Strand Beads (GE Healthcare) according to the manufacturer’s protocol. cDNA was used as a template for real-time PCR using the Universal Probe Library (UPL) system (https://www.roche-applied-science.com/sis/rtpcr/upl/index.jsp). Reactions were performed in 25 μL containing 1× FastStart TaqMan Probe Master (supplemented with ROX reference dye). Gene-specific primers and probes were used at concentrations of 0.2 and 0.1 μm, respectively. Thermal cycling conditions were 95°C for 10 min followed by 40 cycles of 15 s at 94°C and 60 s at 60°C. Relative expression levels were calculated and the primers validated using the ΔΔCt method (Livak, 1997) using data obtained from the reference controls: PP2A (accession no. TC21939), F-BOX (accession no. Niben.v0.3.Ctg24993647), and CDC2 (accession no. Q40482). The UPL primer pairs and probe sequences were as follows: cdc2FOR/cdc2REV with UPL probe number 19, F-BOXFOR/F-BOXREV with UPL probe number 1, PP2AFOR/PP2AREV with UPL probe number 22, and HIPP26QFOR/HIPP26QREV with UPL probe number 112. Primer sequences are provided in Supplemental Table S1.
TRV-Mediated VIGS
To facilitate the use of TRV-mediated VIGS, the 0.12-kb fragment of NbHIPP26 cDNA was cloned into TRV:00 (TRV RNA2 infectious cDNA clone) in the antisense orientation. Two weeks post TRV-RNA1 + TRV:HIPP26 infection, RT-qPCR tests were carried out on upper (noninoculated infected) leaves using primers designed to amplify endogenous NbHIPP26 RNA transcripts.
A. tumefaciens (strain C58C1) cultures carrying the TRV-RNA1 plasmid and TRV-RNA2 cassettes were grown overnight, collected by centrifugation at 3,500 rpm for 10 min, and then resuspended in infiltration buffer to a final OD600 of 0.5. Equal volumes of TRV-RNA2-expressing cells and the TRV-RNA1 cultures were mixed and infiltrated into the abaxial side of two N. benthamiana leaves on seedlings at the four-leaf stage. Plants were grown for 2 weeks prior to challenging with PMTV. The uppermost leaves were collected and assayed for NbHIPP26 silencing by RT-qPCR.
RT-qPCR for PMTV
Results were obtained from two RT-qPCR experiments (three plants per treatment, either TRV:HIPP26 or TRV:00). Reverse transcription of 1 µg of total RNA was performed using the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer’s instructions. Four microliters of 10-fold diluted cDNA was used as a template for real-time PCR. Reactions were performed in a 20-µL reaction containing 2× DyNAmo Flash SYBR Green master mix supplemented with ROX passive reference dye, along with 0.2 µm primers specific to the respective gene of interest. Data analysis was performed using the Bio-Rad CFX manager 3.1.
In order to obtain a standard curve, 10-fold serial dilutions for respective plasmids of RNA-Rep, RNA-CP, and RNA-TGB were made from 100 to 0.01 pg µL−1. Four microliters of the diluted plasmid was used as a template for the real-time PCR. The total number of viral copies was calculated using the following formula: number of molecules = (ng of dsDNA) × (number of molecules/mol) × (1/number of bases) × (1/660 g) × (1/109 ng/g). The Avogadro constant of 6,023 × 1023 molecules/mol was used to calculate the number of copies, and the formula weight of bp in dsDNA was considered as 660 g. See also the copy number calculator for real-time PCR (http://scienceprimer.com/copy-number-calculator-for-realtime-pcr).
S-Acylation Assays
Protein S-acylation state was assessed as described by Hemsley et al. (2008) and Forrester et al. (2011) with some modifications. GFP-tagged NbHIPP26 constructs were transiently expressed in N. benthamiana using agroinfiltration along with the p19 silencing suppressor at OD600 of 0.1 for both. Flash-frozen tissue was ground in liquid nitrogen to a fine powder and solubilized in 4 volumes of TENS buffer (100 mm Tris-HCl, pH 7.2, 5 mm EDTA, 150 mm NaCl, and 2.5% SDS) with protease inhibitors (Sigma P9599; 5 μL mL−1) and 25 mm N-ethylmaleimide at 20°C for 10 min with constant mixing. Lysates were clarified for 5 min at 3,000g, passed through two layers of Miracloth, and recentrifuged at 16,000g for 10 min, all at 20°C. The clarified supernatant was retained, and protein concentration was calculated using a bicinchoninic acid assay. One milligram of protein was diluted to 1-mL final volume in TENS buffer with protease inhibitors and 25 mm N-ethylmaleimide and incubated with constant mixing for 1 h at 20°C. Samples were precipitated with chloroform/methanol (Wessel and Flügge, 1984), briefly air dried, and resuspended in 1 mL of buffer (100 mm Tris-HCl, pH 7.2, 5 mm EDTA, 150 mm NaCl, 6 m urea, and 2% SDS) with heating to 37°C and gentle mixing. Samples were then split into two 0.5-mL aliquots in 1.5-mL microfuge tubes. One aliquot was treated with 0.5 mL of 1 m hydroxylamine-HCl (pH 7.2 with 10 m NaOH, made fresh), while the other (negative control) was treated with 0.5 mL of 1 m NaCl. After brief mixing, 100 µL was removed from each as a loading control and incubated at 20°C for 1 h before precipitation with chloroform/methanol. To the remainder, 40 µL of 50% thiopropyl Sepharose 6B bead suspension in TENS buffer was added to capture S-acylated proteins and gently mixed at 20°C for 1 h. Beads were washed three times with 1 mL of TENS buffer for 5 min each and then dried by aspiration. Loading controls and bead samples were resuspended in a 2× SDS-PAGE sample buffer containing 6 m urea and 50 µL mL−1 β-mercaptoethanol and heated at 37°C for 30 min with frequent mixing. Samples were subsequently run on 12.5% Laemmli SDS-PAGE gels and blotted to a PVDF membrane. EGFP-HIPP26 variants were detected using an anti-GFP monoclonal antibody (Abgent AM1009a). Blots were imaged using GeneSys on a G:BOX XT4 GelDoc system and quantified using GeneTools (Syngene).
Preparation of the NbHIPP26 Promoter Construct and Production of Transgenic Plants
The 1,128-bp region upstream of the translational start site of the NbHIPP26 gene was amplified using primers HIPProFOR and HIPProREV on genomic DNA of N. benthamiana (Nb-1). The resulting PCR fragment was cloned between SacII-XbaI sites of a binary vector, pORE-R2 (Coutu et al., 2007), in front of the uidA gene. The resulting vector was named pORE-R2NbHIPP26. This vector was transferred to A. tumefaciens (strain AGL1). Wild-type N. benthamiana (Sainsbury) plants were transformed, and transgenic plants were regenerated as described by Clemente (2006).
GUS-Staining of Transgenic Plants
The HIPP26pro:uidA transgenic N. benthamiana plants were grown in a glasshouse (21°C day and 16°C night, 18-h daylength with supplementary lighting of 250 W m−2). Samples of leaf and petiole were taken from plants 10 weeks post germination and embedded in 3% agar (Oxoid No. 1; Oxoid) to allow suitable orientation in a further block of agar mounted on a vibroslice (model 752M; Campden Instruments) to allow transverse sections to be cut through major veins in the leaf lamina. Sections of fresh tissue were cut approximately 200 µm thick and dropped immediately into GUS assay buffer [28 mm NaH2PO4, 72 mm Na2HPO4 (pH 7.2) containing 0.2% (w/v) Triton X-100, and 2 mm each of potassium hexacyanoferrate(III) and potassium hexacyanoferrate(III) trihydrate] before being vacuum infiltrated and incubated overnight in the dark at 37°C. Sections were washed subsequently in several changes of 70% ethanol and stored in 70% ethanol before being mounted in water and imaged using a Leica DMFLS light microscope. Photographs were collected using a Zeiss Axiocam to show the distribution of GUS stain.
Accession Numbers
Accession numbers are as follows: Arabidopsis HIPP26, At4g38580; and N. benthamiana HIPP26, Niben101Scf02621g04026.1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Arabidopsis HIPP family phylogeny.
Supplemental Figure S2. Cis-regulatory elements upstream of the HIPP26 ORF.
Supplemental Table S1. List of primers and sequences.
Supplemental Movie S1. Motile vesicles tagged with GFP-HIPP26.
Acknowledgments
We thank Michael Goodin for the RFP-labeled fibrillarin plants and Indu Sama and Nina Lukovitskaya for technical assistance with VIGS.
Footnotes
The work of L.T., G.H.C., S.J., and A.G.R. is funded by the Scottish Government’s Rural and Environmental Science and Analytical Services (RESAS) Division, P.A.H. by the BBSRC (grant BB/M024911/1) and The Royal Society, and E.I.S. by the Swedish Research Council Formas and the Carl Tryggers Foundation.
Articles can be viewed without a subscription.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Ashery U, Yizhar O, Rotblat B, Elad-Sfadia G, Barkan B, Haklai R, Kloog Y (2006) Spatiotemporal organization of Ras signaling: rasosomes and the galectin switch. Cell Mol Neurobiol 26: 471–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel PL, Chien CT, Sternglanz R, Fields S (1993) Using the two-hybrid system to detect protein-protein interactions. In Hartley DA, ed, Cellular Interactions in Development: A Practical Approach. Oxford University Press, Oxford, pp 153–179 [Google Scholar]
- Barth O, Vogt S, Uhlemann R, Zschiesche W, Humbeck K (2009) Stress induced and nuclear localized HIPP26 from Arabidopsis thaliana interacts via its heavy metal associated domain with the drought stress related zinc finger transcription factor ATHB29. Plant Mol Biol 69: 213–226 [DOI] [PubMed] [Google Scholar]
- Baulcombe DC, Chapman S, Santa Cruz S (1995) Jellyfish green fluorescent protein as a reporter for virus infections. Plant J 7: 1045–1053 [DOI] [PubMed] [Google Scholar]
- Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2013) GenBank. Nucleic Acids Res 41: D36–D42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI (2010) The nucleolus under stress. Mol Cell 40: 216–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhtz A, Pieritz J, Springer F, Kehr J (2010) Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant Biol 10: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhtz A, Springer F, Chappell L, Baulcombe DC, Kehr J (2008) Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J 53: 739–749 [DOI] [PubMed] [Google Scholar]
- Choy E, Chiu VK, Silletti J, Feoktistov M, Morimoto T, Michaelson D, Ivanov IE, Philips MR (1999) Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell 98: 69–80 [DOI] [PubMed] [Google Scholar]
- Clemente T. (2006) Nicotiana (Nicotiana tabacum, Nicotiana benthamiana). Methods Mol Biol 343: 143–154 [DOI] [PubMed] [Google Scholar]
- Collum TD, Padmanabhan MS, Hsieh YC, Culver JN (2016) Tobacco mosaic virus-directed reprogramming of auxin/indole acetic acid protein transcriptional responses enhances virus phloem loading. Proc Natl Acad Sci USA 113: E2740–E2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutu C, Brandle J, Brown D, Brown K, Miki B, Simmonds J, Hegedus DD (2007) pORE: a modular binary vector series suited for both monocot and dicot plant transformation. Transgenic Res 16: 771–781 [DOI] [PubMed] [Google Scholar]
- Cowan GH, Torrance L, Reavy B (1997) Detection of potato mop-top virus capsid readthrough protein in virus particles. J Gen Virol 78: 1779–1783 [DOI] [PubMed] [Google Scholar]
- Crowell DN. (2000) Functional implications of protein isoprenylation in plants. Prog Lipid Res 39: 393–408 [DOI] [PubMed] [Google Scholar]
- de Abreu-Neto JB, Turchetto-Zolet AC, de Oliveira LF, Zanettini MH, Margis-Pinheiro M (2013) Heavy metal-associated isoprenylated plant protein (HIPP): characterization of a family of proteins exclusive to plants. FEBS J 280: 1604–1616 [DOI] [PubMed] [Google Scholar]
- Dykema PE, Sipes PR, Marie A, Biermann BJ, Crowell DN, Randall SK (1999) A new class of proteins capable of binding transition metals. Plant Mol Biol 41: 139–150 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Calvino L, Faulkner C, Walshaw J, Saalbach G, Bayer E, Benitez-Alfonso Y, Maule A (2011) Arabidopsis plasmodesmal proteome. PLoS ONE 6: e18880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Pozo N, Menda N, Edwards JD, Saha S, Tecle IY, Strickler SR, Bombarely A, Fisher-York T, Pujar A, Foerster H, et al. (2015) The Sol Genomics Network (SGN): from genotype to phenotype to breeding. Nucleic Acids Res 43: D1036–D1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formstecher E, Aresta S, Collura V, Hamburger A, Meil A, Trehin A, Reverdy C, Betin V, Maire S, Brun C, et al. (2005) Protein interaction mapping: a Drosophila case study. Genome Res 15: 376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester MT, Hess DT, Thompson JW, Hultman R, Moseley MA, Stamler JS, Casey PJ (2011) Site-specific analysis of protein S-acylation by resin-assisted capture. J Lipid Res 52: 393–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M, Rain JC, Legrain P (1997) Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet 16: 277–282 [DOI] [PubMed] [Google Scholar]
- Fukuoka S, Saka N, Koga H, Ono K, Shimizu T, Ebana K, Hayashi N, Takahashi A, Hirochika H, Okuno K, et al. (2009) Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 325: 998–1001 [DOI] [PubMed] [Google Scholar]
- Gao W, Xiao S, Li HY, Tsao SW, Chye ML (2009) Arabidopsis thaliana acyl-CoA-binding protein ACBP2 interacts with heavy-metal-binding farnesylated protein AtFP6. New Phytol 181: 89–102 [DOI] [PubMed] [Google Scholar]
- Giavalisco P, Kapitza K, Kolasa A, Buhtz A, Kehr J (2006) Towards the proteome of Brassica napus phloem sap. Proteomics 6: 896–909 [DOI] [PubMed] [Google Scholar]
- Goodin MM, Chakrabarty R, Banerjee R, Yelton S, Debolt S (2007) New gateways to discovery. Plant Physiol 145: 1100–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley PA, Weimar T, Taylor L, Lilley K, Dupree P, Grierson CS (2013) A proteomic approach identifies many novel palmitoylated proteins in Arabidopsis. New Phytol 197: 805–814 [DOI] [PubMed] [Google Scholar]
- Hemsley PA, Taylor L, Grierson CS (2008) Assaying protein palmitoylation in plants. Plant Methods 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kehr J, Buhtz A (2008) Long distance transport and movement of RNA through the phloem. J Exp Bot 59: 85–92 [DOI] [PubMed] [Google Scholar]
- Kim SH, Ryabov EV, Kalinina NO, Rakitina DV, Gillespie T, MacFarlane S, Haupt S, Brown JW, Taliansky M (2007) Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J 26: 2169–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ. (1997) Relative Quantitation of Gene Expression. PE Applied Biosystems, Foster City, CA, pp 11–15 [Google Scholar]
- Lough TJ, Lucas WJ (2006) Integrative plant biology: role of phloem long-distance macromolecular trafficking. Annu Rev Plant Biol 57: 203–232 [DOI] [PubMed] [Google Scholar]
- Lukhovitskaya NI, Cowan GH, Vetukuri RR, Tilsner J, Torrance L, Savenkov EI (2015) Importin-α-mediated nucleolar localization of potato mop-top virus TRIPLE GENE BLOCK1 (TGB1) protein facilitates virus systemic movement, whereas TGB1 self-interaction is required for cell-to-cell movement in Nicotiana benthamiana. Plant Physiol 167: 738–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmagne A, Ferro M, Meinnel T, Bruley C, Kuhn L, Garin J, Barbier-Brygoo H, Ephritikhine G (2007) A high content in lipid-modified peripheral proteins and integral receptor kinases features in the Arabidopsis plasma membrane proteome. Mol Cell Proteomics 6: 1980–1996 [DOI] [PubMed] [Google Scholar]
- Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM (2009) Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59: 150–162 [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh S, Eisenhaber F (2005) Refinement and prediction of protein prenylation motifs. Genome Biol 6: R55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan MS, Shiferaw H, Culver JN (2006) The Tobacco mosaic virus replicase protein disrupts the localization and function of interacting Aux/IAA proteins. Mol Plant Microbe Interact 19: 864–873 [DOI] [PubMed] [Google Scholar]
- Pennazio S, Roggero P, Conti M (1996) Yield losses in virus-infected crops. Arch Phytopathol Pflanzenschutz 30: 283–296 [Google Scholar]
- Rain JC, Selig L, De Reuse H, Battaglia V, Reverdy C, Simon S, Lenzen G, Petel F, Wojcik J, Schächter V, et al. (2001) The protein-protein interaction map of Helicobacter pylori. Nature 409: 211–215 [DOI] [PubMed] [Google Scholar]
- Rambault A. (2012) FigTree 1.4.2. http://tree.bio.ed.ac.uk/software/figtree/
- Reavy B, Arif M, Cowan GH, Torrance L (1998) Association of sequences in the coat protein/readthrough domain of potato mop-top virus with transmission by Spongospora subterranea. J Gen Virol 79: 2343–2347 [DOI] [PubMed] [Google Scholar]
- Shivprasad S, Pogue GP, Lewandowski DJ, Hidalgo J, Donson J, Grill LK, Dawson WO (1999) Heterologous sequences greatly affect foreign gene expression in tobacco mosaic virus-based vectors. Virology 255: 312–323 [DOI] [PubMed] [Google Scholar]
- Solovyev AG, Savenkov EI (2014) Factors involved in the systemic transport of plant RNA viruses: the emerging role of the nucleus. J Exp Bot 65: 1689–1697 [DOI] [PubMed] [Google Scholar]
- Sorek N, Poraty L, Sternberg H, Buriakovsky E, Bar E, Lewinsohn E, Yalovsky S (2017) Activation status coupled transient S-acylation determines membrane partitioning of a plant Rho-related GTPase. Mol Cell Biol 27: 2144–2154 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Taliansky ME, Brown JW, Rajamäki ML, Valkonen JP, Kalinina NO (2010) Involvement of the plant nucleolus in virus and viroid infections: parallels with animal pathosystems. Adv Virus Res 77: 119–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehseen M, Cairns N, Sherson S, Cobbett CS (2010) Metallochaperone-like genes in Arabidopsis thaliana. Metallomics 2: 556–564 [DOI] [PubMed] [Google Scholar]
- Tilsner J, Taliansky ME, Torrance L (2014) Plant virus movement. eLS. [Google Scholar]
- Torrance L, Cowan GH, Gillespie T, Ziegler A, Lacomme C (2006) Barley stripe mosaic virus-encoded proteins triple-gene block 2 and gammab localize to chloroplasts in virus-infected monocot and dicot plants, revealing hitherto-unknown roles in virus replication. J Gen Virol 87: 2403–2411 [DOI] [PubMed] [Google Scholar]
- Torrance L, Lukhovitskaya NI, Schepetilnikov MV, Cowan GH, Ziegler A, Savenkov EI (2009) Unusual long-distance movement strategies of Potato mop-top virus RNAs in Nicotiana benthamiana. Mol Plant Microbe Interact 22: 381–390 [DOI] [PubMed] [Google Scholar]
- Torrance L, Wright KM, Crutzen F, Cowan GH, Lukhovitskaya NI, Bragard C, Savenkov EI (2011) Unusual features of pomoviral RNA movement. Front Microbiol 2: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Osakabe Y, Qin F, Simpson SD, Maruyama K, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K (2007) Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J 49: 46–63 [DOI] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon R, Wolf S (2009) Phloem transport: cellular pathways and molecular trafficking. Annu Rev Plant Biol 60: 207–221 [DOI] [PubMed] [Google Scholar]
- Verchot-Lubicz J, Torrance L, Solovyev AG, Morozov SY, Jackson AO, Gilmer D (2010) Varied movement strategies employed by triple gene block-encoding viruses. Mol Plant Microbe Interact 23: 1231–1247 [DOI] [PubMed] [Google Scholar]
- Vojtek AB, Hollenberg SM (1995) Ras-Raf interaction: two-hybrid analysis. Methods Enzymol 255: 331–342 [DOI] [PubMed] [Google Scholar]
- Waterworth HE, Hadidi A (1998) Economic losses due to plant viruses. In Hadidi A, Khetarpal RK, Koganezawa N, eds, Plant Virus Disease Control. APS Press, St Paul, MN, pp 1–13 [Google Scholar]
- Wessel D, Flügge UI (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 138: 141–143 [DOI] [PubMed] [Google Scholar]
- Wojcik J, Boneca IG, Legrain P (2002) Prediction, assessment and validation of protein interaction maps in bacteria. J Mol Biol 323: 763–770 [DOI] [PubMed] [Google Scholar]
- Wright KM, Cowan GH, Lukhovitskaya NI, Tilsner J, Roberts AG, Savenkov EI, Torrance L (2010) The N-terminal domain of PMTV TGB1 movement protein is required for nucleolar localization, microtubule association, and long-distance movement. Mol Plant Microbe Interact 23: 1486–1497 [DOI] [PubMed] [Google Scholar]
- Xu P, Chen F, Mannas JP, Feldman T, Sumner LW, Roossinck MJ (2008) Virus infection improves drought tolerance. New Phytol 180: 911–921 [DOI] [PubMed] [Google Scholar]