A cotton pectin methylesterase inhibitor interacts with pectin methylesterases, rendering plants more resistant to fungal infection.
Abstract
Pectins are major components of the primary plant cell wall, which functions as the primary barrier against pathogens. Pectin methylesterases (PMEs) catalyze the demethylesterification of the homogalacturonan domains of pectin in the plant cell wall. Their activity is regulated by PME inhibitors (PMEIs). Here, we provide evidence that the pectin methylesterase-inhibiting protein GhPMEI3 from cotton (Gossypium hirsutum) functions in plant responses to infection by the fungus Verticillium dahliae. GhPMEI3 interacts with PMEs and regulates the expression of a specific fungal polygalacturonase (VdPG1). Ectopic expression of GhPMEI3 increased pectin methyl esterification and limited fungal disease in cotton, while also modulating root elongation. Enzymatic analyses revealed that GhPMEI3 efficiently inhibited the activity of cotton GhPME2/GhPME31. Experiments using transgenic Arabidopsis (Arabidopsis thaliana) plants expressing the GhPMEI3 gene under the control of the CaMV 35S promoter revealed that GhPMEI3 inhibits the endogenous PME activity in vitro. Moreover, the enhanced resistance to V. dahliae was associated with altered VdPG1 expression. Virus-induced silencing of GhPMEI3 resulted in increased susceptibility to V. dahliae. Further, we investigated the interaction between GhPMEI3 and GhPME2/GhPME31 using inhibition assays and molecular docking simulations. The peculiar structural features of GhPMEI3 were responsible for the formation of a 1:1 stoichiometric complex with GhPME2/GhPME31. Together, these results suggest that GhPMEI3 enhances resistance to Verticillium wilt. Moreover, GhPMEI3-GhPMEs interactions would be needed before drawing the correlation between structure-function and are crucial for plant development against the ever-evolving fungal pathogens.
Plant cell walls are highly heterogeneous extracellular structures that contain three major classes of polysaccharides—cellulose, hemicellulose, and pectin—as well as phenolic compounds and cell wall proteins (Dedeurwaerder et al., 2009; Nguyen et al., 2016). Pectin is the most complex of these polysaccharides; it is located in the primary cell wall and constitutes the principal component of the middle lamella (Johansson et al., 2002). It is composed of three fractions of galacturonans: homogalacturonan (HG), rhamnogalacturonan-I, and minor amounts of rhamnogalacturonan-II (Caffall and Mohnen, 2009). HG is a chain composed of α-1,4-linked-d-GalUA units; it is synthesized in the Golgi and then secreted to the cell wall in a highly methyl esterified (up to 80%) form (Sterling et al., 2001). There, pectin methylesterases (PMEs; EC 3.1.1.11), which are cell wall enzymes, deesterify HG, releasing methanol and protons (Dedeurwaerder et al., 2009).
In cotton (Gossypium hirsutum), PMEs are encoded by a large multigene family. Based on their structure, PMEs have been classified into two enzyme types, both of which possess a conserved PME domain (Pfam 01095; Sénéchal et al., 2015). PMEs that contain a PRO region at the N-terminal end of the catalytic domain have been designated as type I (Micheli, 2001). The PRO region shares similarity with the PME inhibitor domain (Pfam 04043; Pelloux et al., 2007) that is presumed to be cleaved from the mature catalytic portion of the protein during secretion (Dorokhov et al., 2006; Dedeurwaerder et al., 2009). The PRO domain mediates the retention of PMEs in the Golgi compartment, and regulates the enzymatic activity of PME through a posttranslational mechanism (Wolf et al., 2009; Sénéchal et al., 2015). PMEs lacking the PRO region are type II enzymes and are produced by bacteria, fungi, Physcomitrella patens, and higher plants (Dedeurwaerder et al., 2009).
Plant PMEs play an important role in preparing the substrate for processing by polygalacturonases and pectate lyases during cell wall metabolism (Johansson et al., 2002). Plant PMEs are typically multigene-encoded isoenzymes that are involved in plant defense against pathogens (Raiola et al., 2011; Bellincampi et al., 2014; Bethke et al., 2014; Lionetti et al., 2017). It is thought that plant PMEs remove methylesters in a block-wise fashion (single-chain mechanism) generating continuous, deesterified GalUA residue domains (Willats et al., 2001). However, PMEs also exist in bacteria and in fungal pathogens. There, the PME mode of action for removing methylesters is random (multiple-chain mechanism; Kohn et al., 1983; Limberg et al., 2000), except for the fungal PME from Trichoderma reesei (Markoviě and Kohn, 1984).
Moreover, the three-dimensional structures of the plant and microbial PMEs are very similar, as the proteins are all composed of right-handed β-helices. A cleft in the surface of the β-helix structure constitutes the active region of the protein. In plant PMEs, the active site cavity is relatively shallow, with no obvious steric barriers, while the active site cavity of the Erwinia chrysanthemi PME is relatively deep (Jenkins et al., 2001). Since PMEs contain neither an α/β hydrolase fold nor a catalytic Ser-His-Asp triad, they are considered a new type of hydrolases. They appear to be carboxylate hydrolases containing two Asp residues in the active site (Jenkins et al., 2001; Johansson et al., 2002).
PME inhibitors (PMEIs) belong to a large multigene-encoded protein family, PF04043, that includes invertase inhibitors (INHs). PMEIs harbor four conserved Cys residues engaged in disulfide bridge formation, and an up-and-down four-helical bundle fold, which is similar to INHs (Di Matteo et al., 2005; Lionetti et al., 2015). PME activity is efficiently regulated by endogenous PMEIs. PMEIs are targeted to the extracellular matrix and typically inhibit plant PMEs by forming a specific and stable complex with 1:1 stoichiometry (Lionetti et al., 2014). A high-resolution three-dimensional structure of a PME-PMEI complex revealed that PMEI covers the pectin-binding cleft of PME and conceals the putative catalytic sites. Thus, it prevents the substrate from approaching the cleft (Di Matteo et al., 2005; Hothorn et al., 2010).
It is thought that most of the important interacting residues are conserved in plant PMEs, but not in fungal and bacterial enzymes; hence, PMEIs are ineffective against microbial enzymes (Di Matteo et al., 2005). Nevertheless, the PMEI from pepper (Capsicum annuum) exhibits antifungal activity against Fusarium oxysporum f. sp. matthiole, Alternaria brassicicola, and Botrytis cinerea (An et al., 2008). Virus-induced gene silencing of CaPMEI1 results in enhanced susceptibility to Xanthomonas campestris pv vesicatoria (An et al., 2008). The effectiveness of PMEIs in controlling endogenous PME activity was first demonstrated in planta by overexpressing AtPMEI-1 and AtPMEI-2 in Arabidopsis. The disease symptoms caused by B. cinerea and Pectobacterium carotovorum were considerably reduced in transgenic plants (Lionetti et al., 2007). Later, kiwi (Actinidia deliciosa) PMEI was shown to limit fungal infections caused by Bipolaris sorokiniana, F. graminearum, and Claviceps purpurea in durum wheat (Triticum aestivum; Volpi et al., 2011, 2013). Moreover, PMEIs counteract the action of plant PMEs and affect the susceptibility of plants to viruses. For example, Tobacco mosaic virus symptoms are reduced, and its systemic movement is limited in tobacco (Nicotiana tabacum) that heterologously expresses kiwi PMEI. Moreover, the overexpression of AtPMEI-2 in Arabidopsis results in a substantial reduction in its susceptibility to the Turnip vein-clearing virus (Lionetti et al., 2014).
PMEI was first discovered in the kiwi fruit (Balestrieri et al., 1990), and later detected in other plants, including Arabidopsis, pepper, and tomato (Solanum lycopersicum; Raiola et al., 2004; An et al., 2008; Reca et al., 2012). Disulfide bond formation and ionic interactions may be important for PMEI stability. In particular, subtle pH changes in the microenvironment greatly impact the stability of partially buried ionic interactions (Bonavita et al., 2016). For instance, the interaction between the tomato PME and the kiwi PMEI grows rapidly weaker with an increase in pH, and no complex is observed at pH 8.0 (Jolie et al., 2010). Further, at a pH >7.0, the inhibition of banana (Musa nana), carrot (Daucus carota), and strawberry (Fragaria × ananassa) PMEs by the kiwi PMEI is also reduced (D’Avino et al., 2003). Since the enzymatic activity of PME is regulated by pH, the stability of the PMEI-PME interaction is also affected by pH (Denès et al., 2000).
Cotton is an important cash crop worldwide, and is widely cultivated for the economic value of its fibers (Xu et al., 2011). The cotton fiber is an ideal model for plant cell elongation and cell wall biogenesis studies, because of its highly elongated structure (Kim et al., 2001). Cotton Verticillium wilt is caused by Verticillium dahliae, a soil-borne plant pathogenic fungus. The disease is difficult to control in cotton as the hyphae reside in the vascular (xylem) tissues of the plant. Moreover, this pathogen can overwinter as a mycelium within perennial hosts or in tubers, bulbs, or seeds, as well as in other propagative organs of the plant. Moreover, resting structures known as microsclerotia support the durable vegetative resting structures of Verticillium, which survive in the soil for many years (Fradin and Thomma, 2006). Therefore, Verticillium wilt leads to severe cotton yield loss each year and represents a major concern for cotton producers (Gao et al., 2011). However, the physiology of plant resistance against Verticillium is largely unexplored. Moreover, even less is known about the crucial fungal components involved in its pathogenicity. Historically, the roles of cell wall degrading enzymes, such as endopolygalacturonase, have attracted much attention (Fradin and Thomma, 2006). Recently, the Ve locus has been bred in various tomato cultivars and was shown to be responsible for resistance to V. dahliae (Kawchuk et al., 2001). Nevertheless, not much is known about the genetic and molecular mechanisms that underlie cotton resistance to Verticillium infection.
In the current study, we investigated whether cotton GhPMEI3 is active against GhPME2 (a type I enzyme) and GhPME31 (a type II enzyme). To this end, we expressed and purified the three proteins and performed in vitro inhibition studies. Enzyme inhibition assays and a PMEI-PME interaction analysis indicated that GhPMEI3 very efficiently inhibits these cotton PMEs. Furthermore, heterologous expression of GhPMEI3 in Arabidopsis lowered pectin levels in the cell wall, markedly enhancing the plant’s resistance to the V. dahliae fungal pathogen. In silico protein modeling, docking, and electrostatic charge distribution analyses were also performed to clarify the structural basis of the PMEI-PME interaction. The current study highlights the regulation of the PMEI-PME interaction and pectin deesterification in plants. Since fungal pathogens are unable to utilize methyl esterified pectin, and this modification reduces the ability of other cell wall-degrading enzymes to hydrolyze pectin, the PMEI-PME interaction enhances plant resistance to pathogens.
RESULTS
Bioinformatics Analysis of GhPMEI3, GhPME2, and GhPME31
The inferred domain architecture of GhPMEI3 was characterized using multiple alignment and bioinformatics approaches. GhPMEI3 was predicted to be encoded by a 720-bp open reading frame encoding a 240-amino acid protein with no signal peptide. The theoretical Mr of GhPMEI3 is 25.96 kD (that of the purified protein was 43.96 kD, with the protein linked to an 18-kD Trx tag, S tag, and His tag), with a pI of 5.07. The Mrs of GhPME2 and GhPME31 are 82.96 kD and 61.53 kD (including a 26-kD GST-tag from the vector), with isoelectric points of 9.23 and 7.52, respectively.
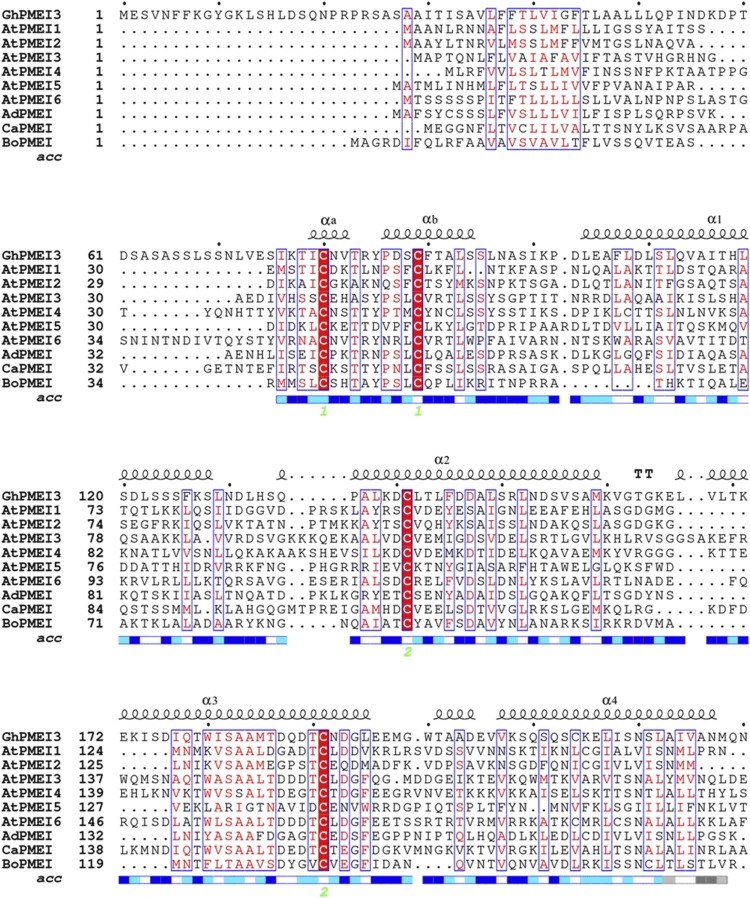
To functionally characterize GhPMEI3, multiple sequence alignment with various PMEIs was performed (Fig. 1). Similarly to other PMEIs, the structure of GhPMEI3 is mainly helical, with four long antiparallel helices (α1–4) arranged in a classic up-and-down four-helical bundle (Fig. 1; Di Matteo et al., 2005). GhPMEI3 has four Cys residues that form two disulfide bridges, one of which connects the α2 and α3 helices, stabilizing the interior of the bundle. The other disulfide bridge connects helices αa and αb in the N-terminal region (Di Matteo et al., 2005). PMEIs belong to the PF04043 family (Pfam database, http://pfam.xfam.org/) and share several structural properties with INH; however, their target enzymes are not related (Di Matteo et al., 2005; Lionetti et al., 2015).
Figure 1.
Sequence alignment of functionally characterized PMEIs from different plants. The predicted amino acid sequence of GhPMEI3 was compared with those of PMEIs from Arabidopsis (AtPMEI1, AAO39943.1; AtPMEI2, AEE75922.1; AtPMEI3, AAW704 08.1; AtPMEI4, AAO42834.1; AtPMEI5, NP_180701.1; and AtPMEI6, AEC10874.1), A. deliciosa (AdPMEI, P83326.2), C. annuum (CaPMEI, ABG47806.1), and Brassica oleracea (BoPMEI, Q45TJ7). The alignment was performed using ClustalW and drawn using ESPript 3.0; α1, α2, α3, and α4 indicate the four-helix bundle of GhPMEI3; the four conserved Cys residues marked with 1 and 2 at the bottom (in green) form two disulfide bonds. Similar residues are colored in red and boxed, the invariant residues are red shadowed. Numbers 1 and 2 with green under the blue rectangles denote disulfide bridges formed by the four conserved Cys residues, and the blue rectangles represent the region of four alpha helixes.
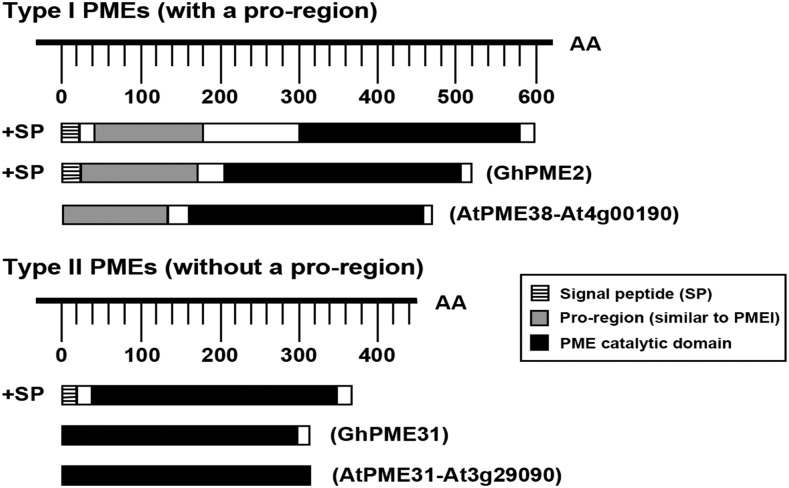
GhPME2 is predicted to encode a 25-amino acid signal peptide (SignalP 4.1 server), while GhPME31 does not encode a signal peptide (Fig. 2). GhPME2 and GhPME31 show similarity at the primary structure level with Arabidopsis PMEs (AtPME38 and AtPME31). GhPME2 contains an N-terminal extension PRO region and shares similarity with PMEIs, as well as a PME catalytic domain; however, GhPME31 contains only the PME catalytic domain (Fig. 2).
Figure 2.
Identification and schematic representation of the conserved domains of type-I and type-II pectin methylesterases. The PRO region of GhPME2 is designated in gray; the signal peptide is designated by a striped pattern; and the PME catalytic domain is designated in black. AA, Amino acid.
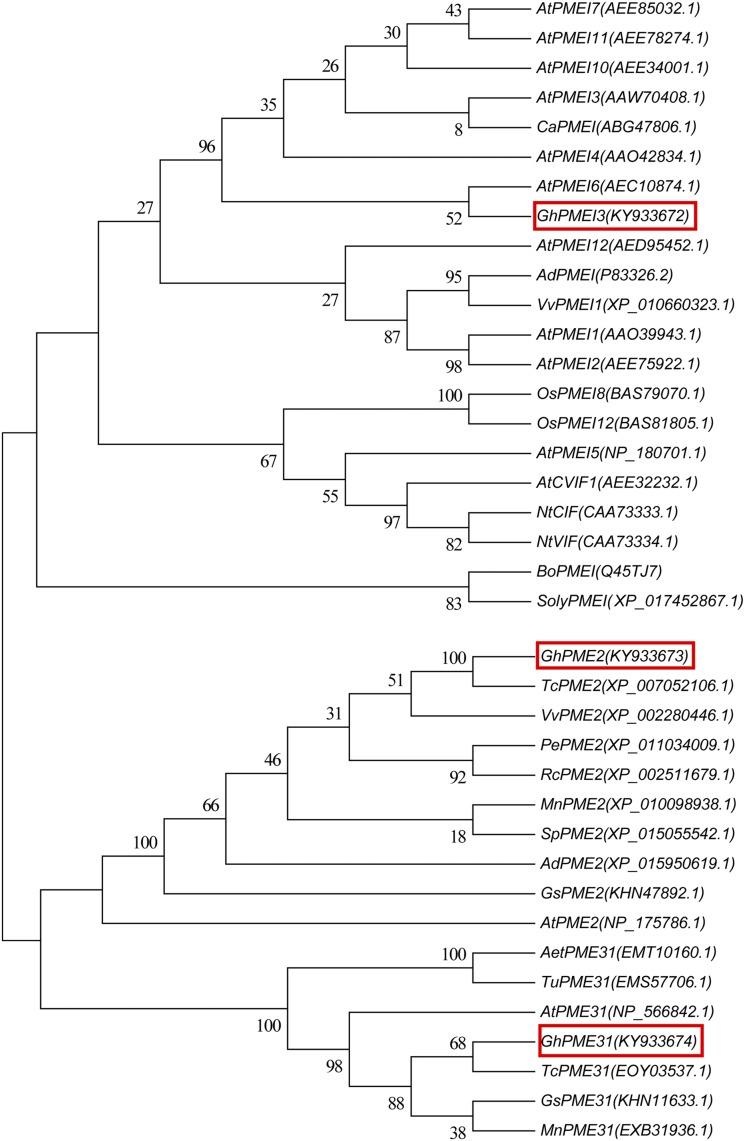
A phylogenetic tree for the amino acid sequences of GhPMEI3, GhPME2, and GhPME31 was generated, using sequences of proteins that had been functionally characterized in plants and deposited in the NCBI database (Fig. 3). To construct the phylogenetic tree, the neighbor-joining approach was employed. The phylogenetic analysis indicated that the inferred GhPMEI3 protein shared 63% identity with the PMEI from Vitis vinifera, while GhPME2 and GhPME31 shared 100% and 92% identity with PME2 and PME31, respectively, from the cacao tree (Theobroma cacao).
Figure 3.
Homology trees of PMEI, PME2, and PME31. Multiple-sequence alignments were performed using ClustalW, and neighbor-joining trees were built using MEGA5.1. The GenBank accession numbers are listed next to gene names; GhPMEI3, GhPME2, and GhPME31 from this study are framed by red boxes. The numbers at nodes represent the branch support values.
Expression of GhPMEI3, GhPME2, and GhPME31 in Response to Stress
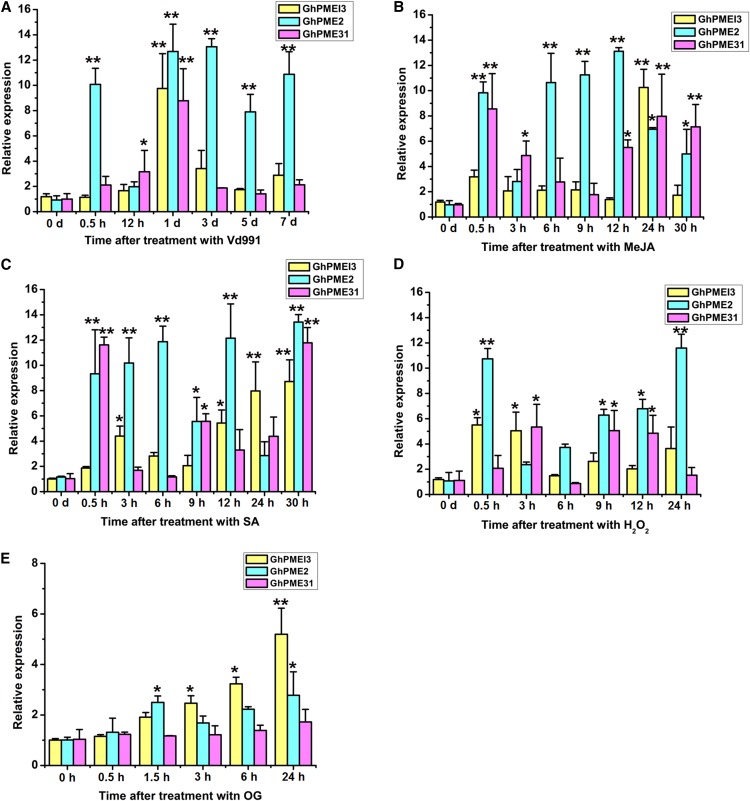
GhPMEI3 expression may possibly be induced as part of the immune response against V. dahliae. Therefore, the expression of GhPMEI3, GhPME2, and GhPME31 was evaluated following fungal infection in the presence of the methyl jasmonate (MeJA) and ethylene (ET) defense hormones, under hydrogen peroxide (H2O2) stress, and in response to oligogalacturonide (OG) treatment. Specific primers were used to assess the transcription of these genes via RT-qPCR. The expression of all three genes was up-regulated in response to the different stress factors (Fig. 4). The expression of GhPMEI3 and GhPME31 peaked 1 d postinoculation with Vd991 (a virulent strain of V. dahliae), while the expression of GhPME2 peaked 3 d postinoculation (Fig. 4A). GhPME31, GhPME2, and GhPMEI3 reached their maximum expression 0.5, 12, and 24 h following treatment with MeJA, respectively, but the expression of GhPMEI3 and GhPME31 dropped to near-basal levels after 9 h (Fig. 4B). Upon ET treatment, GhPME2 and GhPME31 were up-regulated after 0.5 h; the expression of GhPME2 continued to increase until it reached a maximum at 30 h, while the expression of GhPME31 decreased; the expression of GhPMEI3 increased slowly until peaking at 30 h (Fig. 4C). The expression of GhPMEI3, GhPME2, and GhPME31 in response to H2O2 showed different trends. The levels of GhPMEI3 and GhPME31 remained unchanged, while the expression of GhPME2 increased at 0.5 h, after which it plateaued, and finally reached a maximum at 24 h (Fig. 4D). GhPMEI3 expression is induced by exogenous OGs, an elicitor involved in various defense responses (Lionetti et al., 2017). Accordingly, the incubation of cotton seedlings with OGs caused a sustained increase in GhPMEI3 expression and a transient increase in GhPME2 expression, but did not affect GhPME31 levels. This increase in expression was detectable as early as 1.5 h after treatment, and basal transcript levels were observed after 24 h (Fig. 4E). These results were consistent with a previous report that showed that AtPMEI expression is strictly regulated by JA and ET (Lionetti et al., 2017). In that study, after induction with B. cinerea, JA and ET positively regulated AtPMEI10 and AtPMEI11 expression, while ET negatively regulated AtPMEI12 expression.
Figure 4.
GhPMEI3, GhPME2, and GhPME31 expression, as determined by RT-qPCR. A, GhPMEI3, GhPME2, and GhPME31 expression in cotton inoculated with Vd991 at 0 h, 0.5 h, 12 h, 1 d, 3 d, 5 d, and 7 d after infection. B, GhPMEI3, GhPME2, and GhPME31 expression in cotton after MeJA treatment (50 μm). Samples were analyzed 0, 0.5, 3, 6, 9, 12, 24, and 30 h after treatment. C, GhPMEI3, GhPME2, and GhPME31 expression after ET treatment (10 μL/L). Samples were analyzed 0, 0.5, 3, 6, 9, 12, 24, and 30 h after treatment. D, GhPMEI3, GhPME2, and GhPME31 expression after H2O2 treatment (1% [v/v]). E, Induction of GhPMEI3, GhPME2, and GhPME31 expression in the presence of 100 μg/mL OG, harvested at the indicated times. For the MeJA, ET, and H2O2 treatments, plants treated with double-distilled water were used as controls; for the treatment with Vd991, samples treated with Czapek medium were used as a control. Data were collected from three independent biological replicates. The data are presented as the means ± se (n = 3). Asterisks indicate a significant difference from the control (least significance difference, lsd, test; *P < 0.05, **P < 0.01).
Pectin-derived OGs are constituents of the cell wall. Modifications of pectins, such as pectin methyl esterification, may alter the plant defense response. The degree of pectin methyl esterification (DM) can influence the type of OGs present, in turn affecting plant resistance to pathogens, on account of their elicitor activity (Lionetti et al., 2007). Moreover, differences in methylester distribution are hypothesized to affect the type of OG elicitor released during plant-pathogen interactions (Wiethölter et al., 2003). Thus, it is possible that GhPMEI3 may affect the DM that is required for eliciting a defense response in the cotton plant.
Purification and Characterization of Recombinant GhPMEI3, GhPME2, and GhPME31 Proteins
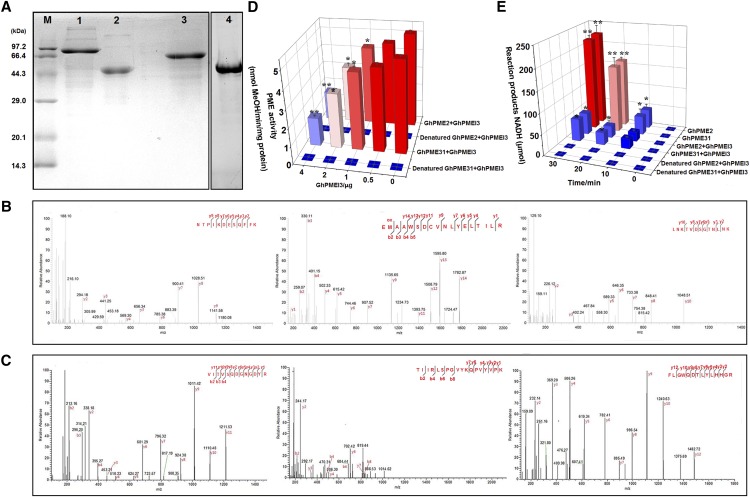
Recombinant GhPMEI3, GhPME2, and GhPME31 proteins were produced in the Escherichia coli BL21 DE3 strain (see “Materials and Methods”). The molecular masses of the three purified proteins, as determined by SDS-PAGE, were close to the values inferred from their gene sequences (Fig. 5A). The identity of the purified GhPMEI3 protein was further confirmed by western blotting, with its His6-tag recognized by a His-tag antibody. Moreover, the identities of GhPME2 and GhPME31 proteins were confirmed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (Fig. 5, B and C). Mascot search results revealed that the two proteins share a high amino acid sequence similarity with two pectinesterases: PE = 2 (SV = 1; Mascot score 33,384; 7 peptide matches; accession no. R9QQU5; Fig. 5B) and PE = 4 (SV = 1; Mascot score 18,432; 6 peptide matches; accession no. A0A0D2RVP9), respectively (Fig. 5C). Hence, the two purified proteins were confirmed as GhPME2 and GhPME31.
Figure 5.
Characterization of recombinant proteins and inhibition of GhPME2 and GhPME31 by GhPMEI3. A, Production of GhPMEI3, GhPME2, and GhPME31 in transgenic E. coli. Lane 1, purified GhPME2; lane 2, purified GhPMEI3; lane 3, purified GhPME31; lane 4, purified GhPMEI3 detected by western-blot analysis with anti-His antibodies. B, MALDI-TOF mass spectra of the GhPME2 PRO region. C, MALDI-TOF mass spectra of the GhPME31 PME catalytic domain. The obtained recombinant protein spots were digested by trypsin, and the resulting peptides were analyzed as described in “Materials and Methods.” D, Inhibition of GhPME2 and GhPME31 activity by GhPMEI3. The experiments were carried out using 1 mU of GhPME2 or GhPME31, with different amounts of purified GhPMEI3. The same reaction was performed with heat-denatured (95°C, 5 min) PME; water instead of PME was used as a negative control. The results are presented as the means ± se (n = 3). Asterisks indicate significant differences compared with reactions lacking GhPMEI3 (least significance difference, lsd, test; *P < 0.05, **P < 0.01). E, Inhibition of GhPME2 and GhPME31 by GhPMEI3; 2 μg of GhPMEI3 was added to the reaction mixture 10 min after the initiation of the reaction. The same reaction was performed with heat-denatured (95°C, 5 min) PME. Water instead of PME was used as a negative control. The results indicate the amounts of the reaction products (NADH). Means marked with the same letter were not significantly different, according to Tukey’s HSD test at P < 0.05.
Inhibition of the PME and Antifungal Activity of GhPMEI3
Purified GhPME2 and GhPME31 proteins were used to characterize the PME-inhibiting capacity of GhPMEI3. Thus, 1 mU of PMEs was incubated for 30 min in the presence of various amounts (0.5–4 μg) of purified GhPMEI3 (Fig. 5D). As shown, 0.5 μg of purified GhPMEI3 inhibited both GhPME2 and GhPME31 (∼10% inhibitory activity). The inhibitory activity was 40% when 2 μg of purified GhPMEI3 was used in the assay, and reached 80% when 4 μg of purified GhPMEI3 was used. Further, biochemical assays revealed that GhPMEI3 exhibits a similar inhibitory activity toward GhPME2 and GhPME31 (Fig. 5E).
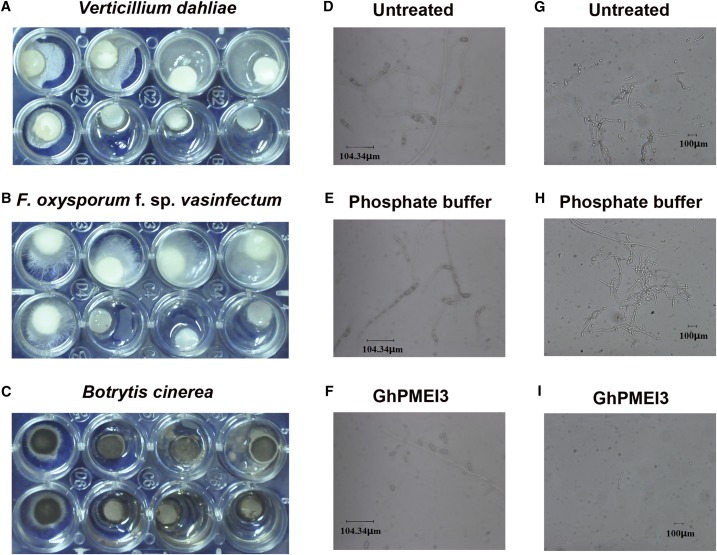
The purified GhPMEI3 inhibited the growth and development of fungal pathogens in vitro. At 50 μg/mL, it inhibited the mycelial growth of V. dahliae, F. oxysporum f. sp. vasinfectum, and B. cinerea (Fig. 6, A–C). Moreover, it inhibited spore germination of F. oxysporum f. sp. vasinfectum (Fig. 6, D–F) and V. dahliae (Fig. 6, G–I), at a concentration of 250 μg/mL.
Figure 6.
Antimicrobial activity of GhPMEI3. A, The effect of GhPMEI3 on the mycelial growth of V. dahliae. B, The effect of GhPMEI3 on the mycelial growth of F. oxysporum f. sp. vasinfectum. C, The effect of GhPMEI3 inhibitor on the mycelial growth of B. cinerea. In each plate, the upper wells were treated with PBS. Purified GhPMEI3 was added to the bottom wells at the following concentrations (μg/mL): 0, 50, 100, and 250. D and G, Spore germination and hyphal growth of F. oxysporum f. sp. vasinfectum and V. dahliae in PD broth. E and H, Spore germination and hyphal growth of F. oxysporum f. sp. vasinfectum and V. dahliae in PBS. F and I, Inhibition of spore germination and hyphal growth of F. oxysporum f. sp. vasinfectum and V. dahliae by purified GhPMEI3 (250 μg/mL). Results from three independent experiments are shown. Bar = 104.34 μm in D to F and 100 μm in G to I.
Altered Growth of Transgenic Arabidopsis Plants and Enhanced Resistance to V. dahliae
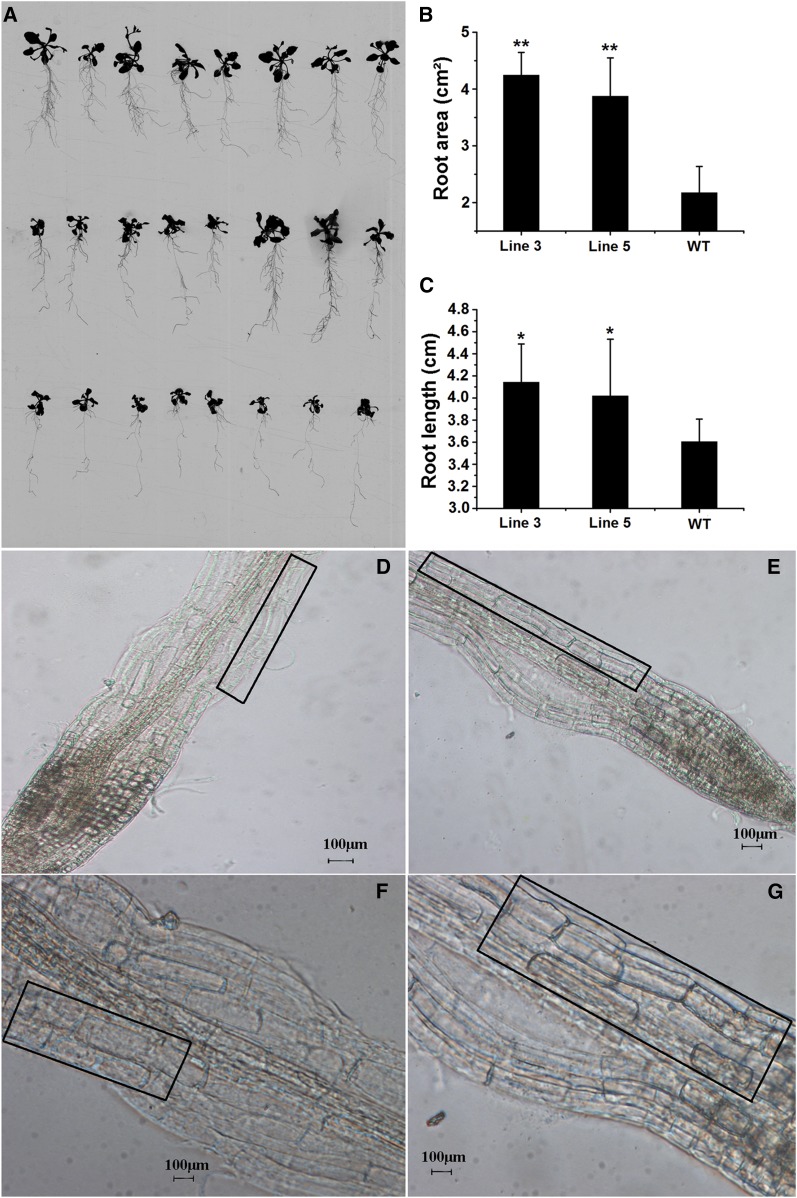
The growth and development of transformed plants grown on solid Murashige and Skoog medium were clearly different from the wild type, especially with respect to root length and the number of lateral roots. A significant 50% increase in the root area and 13% increase in root length were observed in transgenic plants when compared to wild-type plants (Fig. 7, A–C). The elongation zone of the root cells in transgenic Arabidopsis plants appeared obviously elongated when compared with the wild-type plants (Fig. 7, D–G). It has been established that methyl esterification of pectin is highest during the cell expansion stage and decreases in the cell elongation phase (Goldberg, 1984), playing a key role in plant growth. In transgenic Arabidopsis plants, the activities of PMEs are specifically inhibited by PMEIs, and the plants develop longer roots when grown in a vertical position, which is primarily attributed to lengthening in the cell elongation phase (Lionetti et al., 2007). Further, the fine-tuning of the DM of the homogalacturonan backbone seems crucial for plant growth and development (Hocq et al., 2017). Consistently, our observations suggested that an increased pectin DM elongated the cell expansion phase and positively affected plant growth (Supplemental Fig. S1).
Figure 7.
Morphology of transgenic and wild-type plants. A, Wild-type plants (bottom) and GhPMEI3 transgenic lines (line 3, top; line 5, middle) are shown. B, Root areas in the wild-type and transgenic plants. C, Root lengths in the wild-type and transgenic plants. The results are shown as the means ± se, and each experiment (n = 20 for wild-type and transgenic plants) was carried out in triplicate. The asterisks indicate a significant difference from the wild type (Student’s t test; *P < 0.05, **P < 0.01). D and E, Microscopy observations of wild-type and transgenic Arabidopsis epidermal root cells. The elongation zones are marked by black boxes. F and G, Magnification of elongation areas in D and E, respectively. The black boxes in F and G were the enlargement of the elongation zones in D and E. Scale bars, 100 μm.
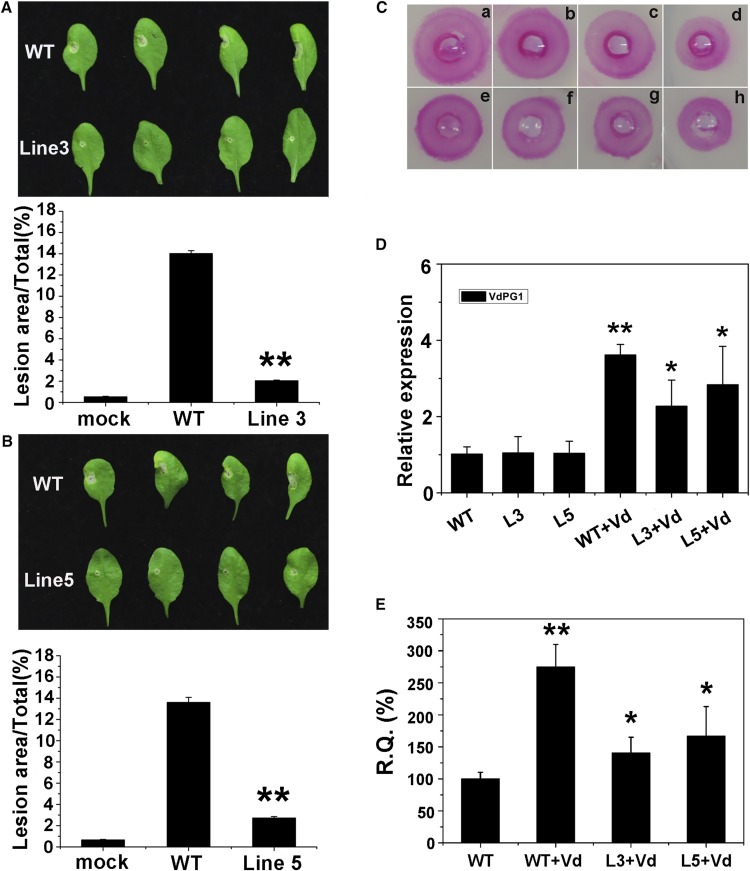
The resistance of transgenic Arabidopsis plants to V. dahliae was next investigated. Leaves from the wild-type plant and two transgenic lines were inoculated with V. dahliae conidia, and the lesion areas were monitored 5 d postinoculation. Substantial differences were observed in the areas of expanding lesions in transformed lines and wild-type plants. Compared with the wild type, the lesion areas were reduced in lines 3 and 5 (Fig. 8, A and B). Proteins from wild-type and transgenic plants were extracted to examine the inhibition of GhPME2 and GhPME31 in these different contexts. In vitro assays revealed that proteins from transgenic plants clearly inhibited the activity of GhPME2 and GhPME31 (Fig. 8C). To test whether the reduced disease symptoms were associated with the overexpression of GhPMEI3, the relative VdPG1 levels in wild-type and transgenic plants were analyzed following inoculation with V. dahliae. Gene transcription was significantly higher in the wild-type plants than in transgenic plants. No VdPG1 expression was detected in noninoculated plants (Fig. 8D). In addition, plant phenotypes correlated with the degree of V. dahliae colonization. The fungal biomass was determined by RT-qPCR in each transgenic line and normalized to the biomass, with respect to the wild-type plants, to determine the GhPMEI3-mediated biomass reduction. The accumulation of fungal biomass was substantially reduced in the GhPMEI3 transgenic lines in comparison with nontransgenic plants (Fig. 8E).
Figure 8.
Reduction of disease symptoms following V. dahliae infiltration, and detection of GhPMEI3 activity in transgenic Arabidopsis plants. A and B, Disease symptoms (top) and disease lesion size (bottom) of wild-type and transgenic Arabidopsis plants after inoculated with V. dahliae at 5 d postinoculation. The inoculum (10 μL) was placed on each leaf: the right half of the leaf was inoculated with 10 μL of Czapek liquid medium; the left half of the leaf was inoculated with 10 μL of V. dahliae. The lesion size was quantified using ImageJ software. The experiment was repeated three times (n = 20) with similar results. Asterisks indicate a significant difference from the wild type (Student’s t test, **P < 0.01). C, Inhibitory activity of GhPMEI3 in transgenic plants. a, 1 mU of purified GhPME2; b to d, GhPME2 with 15 μg of crude GhPMEI3 preparation from wild-type, or transgenic line 5 and 3 plants; e, 1 mU of purified GhPME31; f to h, GhPME31 with 15 μg of crude GhPMEI3 preparation from wild-type, or transgenic line 5 and 3 plants. D, Relative VdPG1 levels in wild-type and transgenic plants. The data are presented as the means ± se (n = 20). The experiment was repeated three times with similar results. Asterisks indicate a significant difference from the wild type (Student’s t test; *P < 0.05, **P < 0.01). E, RT-qPCR quantification of fungal biomass in 100 mg of transgenic Arabidopsis tissues compared to untransformed wild type. All experiments were performed three times, each time in triplicate (n = 20). Data from a representative experiment were analyzed using Student’s t test *P < 0.05, **P < 0.01.
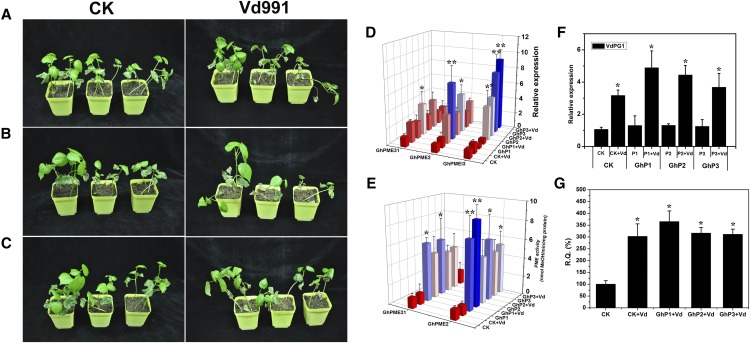
Silencing of GhPMEI3 Increases the Susceptibility of Cotton to V. dahliae
Wild-type cotton plants and cotton plants in which GhPMEI3 was silenced with virus-induced gene silencing (VIGS) were treated with Czapek medium as a control, or V. dahliae. Two weeks later, the disease progression following V. dahliae infection was monitored based on the degree of stunting. In the control check (CK) treated plants, little stunting was observed, while the silenced plants inoculated with V. dahliae exhibited evident and consistent stunting (Fig. 9, A–C). The transcript levels of GhPMEI3, GhPME2, and GhPME31 in silenced plants were determined by RT-qPCR. In the completely and partially silenced cotton plants, the transcript levels were considerably higher than those in the CK plants (Fig. 9D). Moreover, the possible contribution of GhPMEI3 in controlling GhPME2 and GhPME31 activity in silenced cotton plants during V. dahliae infection was evaluated. The GhPME2 and GhPME31 activity levels were determined in CK and in GhP1, GhP2, and GhP3 silenced plants after V. dahliae inoculation, using biochemical assays (Lionetti et al., 2015). The increase in PME activity observed in silenced plants was higher than in CK plants (Fig. 9E). This demonstrated that GhPMEI3 functioned as an inhibitor to control the activity of PMEs during fungal infection. To further investigate the correlation between PMEI, PMEs, and PG, the expression of VdPG1 in CK and silenced plants infected with V. dahliae was monitored. VdPG1 transcript levels were higher in silenced plants than in CK plants, and no significant differences were observed between the three silenced plants (Fig. 9F). To confirm the observed phenotypes, the fungal biomass was determined by RT-qPCR in each silenced plant and normalized to the biomass of CK plants. This analysis revealed that the basal defense against V. dahliae was compromised in the GhP1 line, with the GhP2 and GhP3 lines showing levels of susceptibility that were similar to CK plants (Fig. 9G).
Figure 9.
Silencing of GhPMEI3 enhances cotton susceptibility to V. dahliae infection. A, Wild-type cotton plants infiltrated with Czapek liquid medium or a V. dahliae spore suspension. B, Plants in which GhPMEI3 was silenced by VIGS, named GhP1, were infiltrated with Czapek liquid medium or a V. dahliae spore suspension. C, Another VIGS-silenced cotton line, named GhP2, was infiltrated with Czapek liquid medium or a V. dahliae spore suspension. Whole plants are shown 21 d postinfection. D, RT-qPCR analysis of GhPMEI3, GhPME2, and GhPME31 expression in the control and silenced cotton plants postinoculation with V. dahliae, with GhUBQ7 as an endogenous control. The experiments were performed in triplicate and yielded similar results. E, The activity of GhPME2 and GhPME31 in the control and silenced plants following inoculation with V. dahliae. F, Relative VdPG1 levels in the control and silenced plants after inoculation with V. dahliae. G, Fungal biomass, as determined by RT-qPCR (R.Q.) in the control and silenced plants following inoculation with V. dahliae. The experiments were performed in triplicate and yielded similar results. Asterisks indicate a significant difference from the CK (Student’s t test; *P < 0.05, **P < 0.01).
Docking Analysis of GhPMEI3 with GhPME2 and GhPME31
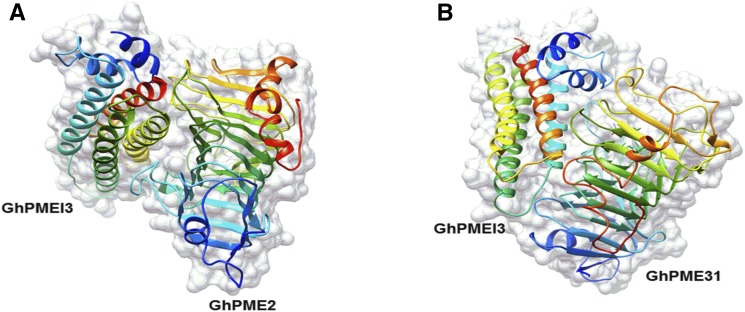
PMEIs contain four long helices (α1–4), packed in an antiparallel orientation, in a classic up-and-down helical bundle (Di Matteo et al., 2005). As previously reported, GhPME2 and GhPME31 form a 1:1 stoichiometric complex with GhPMEI3, with GhPMEI3 covering most of the shallow cleft of the enzyme where the ligand-binding site is located. Docking analyses revealed that GhPMEI3 interacts with GhPME2 and GhPME31 in the putative active site region via its helices. Furthermore, the helices of GhPMEI3 were positioned perpendicularly to the β-helix in either GhPME (Fig. 10, A and B).
Figure 10.
Docking analysis of GhPMEI3 onto cotton PMEs. A, Interaction between GhPMEI3 and GhPME2. B, Interaction between GhPMEI3 and GhPME31. The α2, α3, and α4 GhPMEI3 α-helices fit into the pectin-binding cleft of GhPMEs.
Electrostatic Surface Potentials of the Individual GhPMEI3, GhPME2, and GhPME31 Proteins, and Protein Complexes
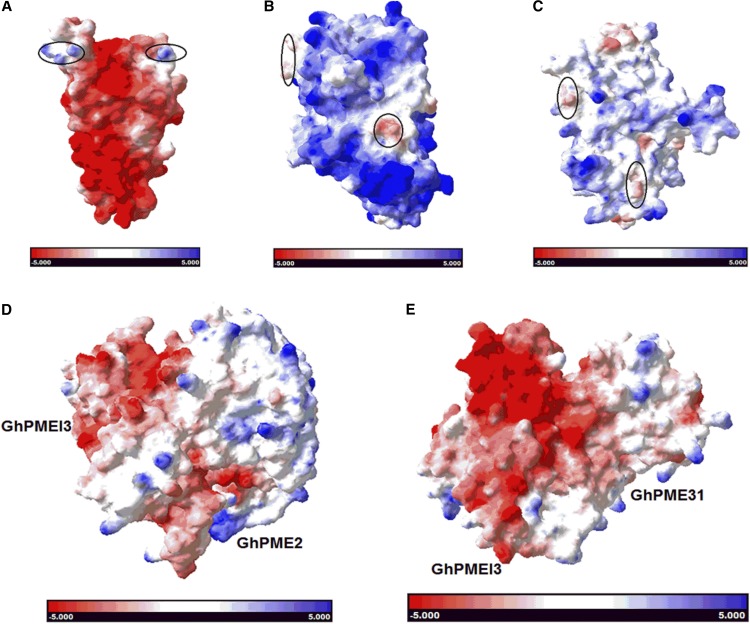
The electrostatic surface potential of the GhPMEI3, GhPME2, and GhPME31 proteins was then predicted to analyze their surface charge distributions (Fig. 11, A–C). The mode of assembly of the two protein complexes, that is GhPMEI3-GhPME2 and GhPMEI3-GhPME31, is presented in Figure 11, D and E, respectively. The surface charge distributions over the individual proteins clearly differed, particularly for GhPME2 and GhPME31, over the concave interaction site. However, the surface potential of the two protein complexes was similar, as was the mode of binding.
Figure 11.
Electrostatic potential of individual proteins and protein complexes. Electrostatic surface potential maps of solvent-accessible surfaces of GhPMEI3 (A), GhPME2 (B), GhPME31 (C), GhPMEI3-GhPME2 complex (D), and GhPMEI3-GhPME31 complex (E). The surface potential is designated in red (−5) or blue (+5); the regions of individual proteins marked in A to C possess distinctly different surface electrostatic potentials.
DISCUSSION
In this study, we provided insights into how PMEI modulates the activity of PME through the formation of the PME-PMEI complex. We chose GhPMEI3 as a possible pathogen resistance-associated PMEI, and GhPME2 and GhPME31 as representatives of two different types of PMEs. The alignment of the GhPMEI3 sequence with sequences of functionally characterized PMEIs showed that GhPMEI3 has two INH sequences (Fig. 1). The structure of an INH from tobacco (Nt-CIF) has been previously elucidated (Hothorn et al., 2004a). KwPMEI and Nt-CIF are strikingly similar from a structural point of view, but recognize different target enzymes, as key amino acids that are involved in the formation of intermolecular H-bonds with PMEs are only conserved in PMEIs. Therefore, the structural view of the PMEI-PME complex provided insights into the specific binding between GhPMEI3 and GhPME2/ GhPME31. Since the highly conserved INHs lack the PKF motif in the α3 helix, GhPMEI3 may be unable to participate in interactions with INHs, but might participate in the formation of enzyme-inhibitor complexes (Lionetti et al., 2015). Many PME proteins are predicted to contain an N-terminal PRO region, which shares moderate homology with PMEIs (Wolf et al., 2009). The PRO region is considered to be crucial for protein targeting to the endoplasmic reticulum (Micheli, 2001), and is thought to play an auto-inhibitory role in PMEs during secretion to the apoplast (Giovane et al., 2004; Lionetti et al., 2007). Consistent with the PME classification, GhPME2 was characterized as a type-I PME and GhPME31 as a type-II PME (Fig. 2).
Previous studies suggested that PMEI can markedly inhibit the activity of plant PMEs and that pectin methyl esterification can play a role in plant resistance to bacterial and fungal pathogens (An et al., 2008; Volpi et al., 2011). The ability of GhPMEI3 proteins to inhibit the activities of cotton GhPME2 and GhPME31 was tested in enzymatic assays (Fig. 5, B and C). In addition, GhPMEI3 exhibited antifungal activity against V. dahliae, F. oxysporum f. sp. vasinfectum, and B. cinerea in vitro (Fig. 6, A–C), suggesting that GhPMEI3 might directly interfere with the plant pathogen during infection. It was initially thought that PMEIs are unable to inhibit the PMEs of fungal pathogens (Giovane et al., 2004; Di Matteo et al., 2005). However, more recently, the overexpression of AtPMEI-1 or AtPMEI-2 inhibitors in Arabidopsis was shown to increase the DM of the pectin cell wall and reduce plant susceptibility to fungal and bacterial necrotrophs (Lionetti et al., 2007; Raiola et al., 2011). In addition, plant resistance to fungal pathogens is compromised in PMEI mutants where PMEI expression is impaired (Lionetti et al., 2017). In agreement with these previous findings, GhPMEI3 was able to restrict fungal mycelial growth. Hence, it may function to reinforce the cell wall barrier and promote the antifungal resistance in cotton.
To investigate the role of GhPMEI3 in plant growth and plant pathogen interactions, we heterologously expressed GhPMEI3 in Arabidopsis. GhPMEI3-expressing transgenic plants displayed reduced activities of GhPME2 and GhPME31, and alterations in the pectin DM and in the plant growth response (Fig. 7). In addition, the transformed plants showed a reduced susceptibility to V. dahliae (Fig. 8, A and B), since the methyl esterification of pectin may be associated with pectin-degrading enzymes, such as endo-polygalacturonase (Lionetti et al., 2007). Thus, it is likely that the higher DM of pectins in transgenic plants hampered the activity of VdPG1, produced by V. dahliae, thus further delaying the colonization of the fungus and increasing resistance to pathogens (Fig. 8, D and E). Recently, a significant reduction in disease symptoms caused by B. sorokiniana and F. graminearum was reported in transgenic wheat (Volpi et al., 2011). Further, the ability of PMEI to restrict the progression of Tobacco mosaic virus infection in tobacco and Turnip vein-clearing virus infection in Arabidopsis was recently demonstrated (Tundo et al., 2016). PMEIs in tobacco and transgenic Arabidopsis plants not only affect the PMEs already present in the plant, but also inhibit the viral or fungal induction of PME activity. Therefore, they further prevent the pathogen from exploiting the host’s susceptibility factors that are required for infection (Lionetti et al., 2014). The enhanced resistance of these transgenic plants to viral and fungal pathogens is attributed to the methyl esterification of pectin, which obstructs the growth of these pathogens and reduces the capacity of their PGs to hydrolyze methyl-esterified pectin (Tundo et al., 2016).
In the current study, virus-induced gene silencing was used to investigate the effects of GhPMEI3 loss of function in cotton. The experiments revealed that GhPMEI3 silencing compromised cotton resistance to V. dahliae (Fig. 9, A–C). These observations were similar to those in a previous study with PMEI mutants (Lionetti et al., 2017). In addition, following inoculation with V. dahliae, the relative expression of GhPME2 and GhPME31, and also of VdPG1, was higher in the GhPMEI3-silenced plants than in the wild type. This suggested that GhPMEI3 controls pectin methyl esterification through PME during V. dahliae infection. Further, the comparison of CK and GhPMEI3-silenced plants revealed a significant reduction of DM in the latter (Supplemental Fig. S2).
Plant pathogens produce cell wall-degrading enzymes, such as polygalacturonase and pectin lyase, during infection (An et al., 2008). Among these, PGs play a primary role, particularly because they act as virulence factors during plant–pathogen interactions (Volpi et al., 2011), for example B. cinerea and tomato (ten Have et al., 1998), and Alternaria citri and citrus (Isshiki et al., 2001). Previous studies have demonstrated that highly methyl esterified pectins are less susceptible to the action of PGs from B. sorokiniana and F. graminearum and to other fungal PGs than pectins with lower DM (Limberg et al., 2000; Volpi et al., 2011). This suggested that the increased level of pectin methyl esterification in transgenic plants impairs the ability of the fungus to colonize host tissues and hampers the activity of endopolygalacturonases produced by fungal pathogens (Lionetti et al., 2007; Volpi et al., 2011). The observation that VdPG1 was expressed at a relatively low level in lines 3 and 5 further indicates that pectin methyl esterification, associated with PMEI overexpression in plants, results in a structure that is inaccessible to PGs secreted by fungal pathogens, which hydrolyze pectin during infection and colonization of host tissues.
PMEI and PME form a 1:1 stoichiometric complex and the inhibitor masks the putative active site of the enzyme. The four-helix bundle of GhPMEI3 is perpendicular to the parallel PME ligand-binding cleft structure, and three helices (α2, α3, and α4, but not α1) interact with the enzyme in the proximity of the shallow cleft (Fig. 10, A and B). Masking of the pectin-binding cleft of PMEs masks the substrate-binding and catalytic sites, further preventing the access of the substrate (Di Matteo et al., 2005; Sénéchal et al., 2015). By exposing its N-terminal α-hairpin, PMEI interacts with the C-terminal helix at the PME surface (Hothorn et al., 2004b). The N-terminal region of AtPMEI1 was proposed to be important for the interaction with PMEs. This region may play a crucial role in the structural stability of PMEIs, but is not extensively involved in the formation of the PMEI-PME complex (Hothorn et al., 2004a, 2004b). According to previous docking studies, GhPMEI3 binds in an open conformation at the N terminus and interacts with the C terminus of GhPMEs, which is consistent with a report proposing that PMEI may easily bind to homologous type-I PMEs (Micheli, 2001). It is likely that the putative binding site of the inhibitor on a bacterial enzyme is much deeper than in plant PMEs, thus leading to steric hindrance that prevents their interaction with PMEIs (Di Matteo et al., 2005). The residues that are crucial for the PMEI-PME interaction are deemed to be conserved in plant PMEs, but not in the fungal PMEs, which might provide another possible explanation for the lack of inhibition (Di Matteo et al., 2005).
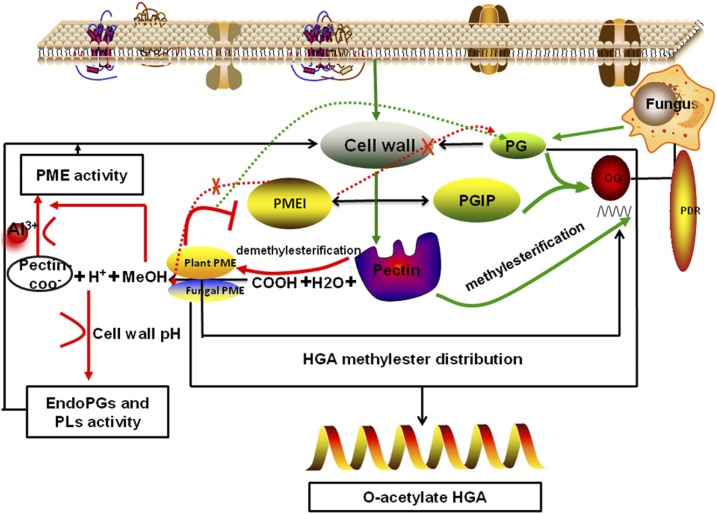
Taken together, the inhibition of cotton PMEs by GhPMEI3 might explain how the enzymatic activity of PME is coordinated within the cell wall. In Figure 12, we propose a model of PMEI involvement in plant immunity against fungal pathogens. Above all, PME is required for the release of MeOH, H+, and pectin-COO− during infection. MeOH, as a damage-associated molecular pattern (DAMP)-like alarm signal, down-regulates the expression of the pathogen-related GhPMEI3. Moreover, H+ ions affect the pH of the plant cell wall and promote the hydrolysis of VdPG1 to homogalacturonan. Pectin-COO−, which contains exposed negative charge groups, can bind cations; Al3+ affects the methylation status of pectin in the root cell walls and regulates PME activity. OGs are oligomers of α-1,4-linked galacturonosyl residues that can be generated by a partial hydrolysis of polygalacturonic acid or released from plant cell walls through partial degradation of homogalacturonan; they are recognized as DAMPs, leading to the activation of the plant immune response (Nothnagel et al., 1983; Ferrari et al., 2013). The degree and pattern of pectin methyl esterification influences the release and type of OGs (Bethke et al., 2014). Moreover, polygalacturonase-inhibiting proteins (PGIPs) interact with PG and lead to the accumulation of OGs. OGs can be detected by members of the wall-associated kinase family and are responsible for the constitutive activation of pathogen-related defense responses and other signal transduction pathways in plant cells, including reactive oxygen species production, ET production, and callose deposition (Brutus et al., 2010; Ferrari et al., 2013; Bethke et al., 2014). Finally, PMEI may directly inhibit the activity of fungal PMEs and fungal PGs. However, it is possible that GhPMEI3 interacts with plant GhPME2 and GhPME31, leading to pectin methyl esterification and influencing the physicochemical properties of the cell wall. Ultimately, this impairs the ability of V. dahliae to grow on methyl esterified pectin and reduces the capacity of VdPG1 to hydrolyze methyl esterified pectin, thus protecting the plant cell wall from degradation. The current study provides mechanistic insights into the regulation of enzyme activity by their inhibitors and the interaction between PMEI and PME using molecular modeling. Broadening our knowledge of how plants regulate cell wall metabolism is of key importance for improving plant resistance to fungal pathogens.
Figure 12.
Model of PMEI involvement in plant immunity against fungal pathogens. The plant cell wall is the first barrier against pathogen attack. Above all, PME is required for the release of MeOH, H+, and pectin-COO− during pathogen infection. MeOH, a DAMP-like alarm signal, down-regulates the expression of the pathogen-related GhPMEI3. H+ ions affect the pH of the plant cell wall and promote the hydrolysis of VdPG1 to homogalacturonan. Pectin-COO−, with exposed negatively charged groups, binds cations. Al3+ ions affect the methylation status of pectin in root cell walls and regulate PME activity. Pectin methyl esterification affects the release and the type of OGs. Moreover, PGIP interacts with PG, which leads to the accumulation of OGs. OGs elicit an antipathogen defense and promote signal transduction in plant cells. Lastly, PMEI is unable to directly inhibit the activity of fungal PMEs and fungal PGs. However, it is possible that GhPMEI3 interacts with plant GhPME2 and GhPME31, reducing the capacity of VdPG1 to hydrolyze methyl esterified pectin and protecting the plant cell wall from degradation.
MATERIALS AND METHODS
Plant Growth and Preparation of Fungal Pathogens
Seeds of the state cotton (Gossypium hirsutum), isolate 2006001 (original strain no. GK44), were provided by the Cotton Research Institute, Chinese Academy of Agricultural Sciences. Plump seeds were selected and sterilized using a 4% (v/v) NaClO solution, 75% (v/v) ethanol, and then rinsed with sterile water. The seeds were planted in a mixture of vermiculite and nutrient soil (1:2, w/w), and cultured in a growth chamber with a light intensity of 150 μmol/m2/s and a photoperiod of 16 h/8 h.
Wild-type Arabidopsis (Arabidopsis thaliana, Col-0) and transgenic Arabidopsis plants were grown in a controlled environmental chamber maintained at 22°C, 70% relative humidity, with a 16-h photoperiod. For vertical growth, the seeds were sterilized and germinated for 12 d on agar plates containing 0.8% (w/v) agar and 1% (w/v) Suc, with a 16-h photoperiod, in a vertical position (Lionetti et al., 2007). Root lengths and areas were measured using a root scanner (Expression 11000XL; Epson). Root elongation zones were observed under a microscope and photographed under a 4×/0.25 numerical aperture objective using a Nikon eclipse Ti microscope.
Virulent strains of Verticillium dahliae Vd991, Fusarium oxysporum f. sp. matthiole, Alternaria brassicicola, and Botrytis cinerea were prepared from cultures grown for 5 d at 25°C in Czapek liquid medium; the experiments were performed using conidial suspensions of 106 conidia/mL.
Sequence and Phylogenetic Analyses
A homology search using sequences deposited in the NCBI database was conducted to verify that the obtained sequences encode GhPMEI3, GhPME2, and GhPME31. The SMART software (http://smart.embl-heidelberg.de/) was used for domain structure prediction and the sequence alignment was prepared using the ESPript program (Robert and Gouet, 2014). The phylogenetic analysis was carried out in MEGA 5.1 (Bednarek et al., 2009).
Determining the Effects of Exposure to Fungal Pathogens and Mediators of Stress Responses on Gene Expression in Plants Using RT-qPCR
Cotton seedlings were cultured in a growth chamber at a day/night temperature of 25°C/22°C. The seedlings were infected with V. dahliae and the leaves were sprayed with 10 μL/L ET, 50 μm MeJA (Sigma-Aldrich), or 1% (v/v) H2O2 (Tapia et al., 2005). For OG treatment, the seeds were placed in a culture flask containing MS medium with 0.5% (w/v) Suc and incubated for 10 d; then, the medium was replaced with fresh medium containing 100 μg/mL OGs with a degree of polymerization of 10 to 15, and the incubation was continued under the same conditions before harvesting (Ferrari et al., 2003).
Total RNA was extracted from the cotton complete stool using an RNA extraction kit, according to the manufacturer’s instructions (RA103-01; Biomed, Enzo Life Sciences). The isolated RNA was treated with RNase-free DNase I (Sigma-Aldrich) and reverse-transcribed into cDNA using the Fast Quant cDNA reverse kit (Tiangen Biotech), as per manufacturers’ instructions.
Specific primers for the detection of the GhPMEI3, GhPME2, and GhPME31 transcripts and primers for the detection of the endogenous control (EF1ɑ from cotton) are listed in Supplemental Table S1. The expression levels of target genes (GhPMEI3, GhPME2, and GhPME31) relative to the reference gene (EF1ɑ) were analyzed using the comparative CT (2−ΔΔCT) method. The RT-qPCR conditions were as follows: an initial denaturation step of 30 s at 95°C, followed by 40 cycles of denaturation for 5 s at 95°C and annealing for 34 s at 60°C.
Purification of the Recombinant GhPMEI3, GhPME2, and GhPME31 Proteins, and Testing the Inhibition of PME Activity
The GhPMEI3, GhPME2, and GhPME31 open reading frames were amplified using the primers listed in Supplemental Table S1, and cloned into the PET-32a or pGEX-4T-1 vectors for the production of fusion proteins harboring a His6-tag or a GST-tag, respectively (Supplemental Fig. S3). The transformants were grown in LB medium at 37°C, with shaking at 200 rpm. At OD600 = 0.6–1.2, isopropyl-β-d-1-thiogalactopyranoside was added to a final concentration of 1.0 mm. The cultures were grown at 28°C for an additional 4 h with shaking at 200 rpm, and the cells were harvested by centrifugation at 10,000g for 15 min. The pellets were resuspended in PBS and the cells were disrupted using an ultrasonic cell breaker. The proteins were purified by precipitation under denaturing and refolding conditions, as previously described (Wang et al., 2013). Recombinant GhPMEI3 was purified using a His6-tagged protein purification kit (CW BIO), and GhPME2 and GhPME31 were purified using a GST purification kit (Takara). The thrombin cleavage capture kit (Novagen) was used to remove the tags. The purified GhPMEI3, GhPME2, and GhPME31 proteins were resolved by SDS-PAGE and their concentrations determined using the Bradford method (Bradford, 1976), with bovine serum albumin as a standard.
The resolved GhPMEI3 proteins were blotted onto a polyvinylidene difluoride immobilon-p membrane (Millipore) following the manufacturer’s protocol (100 V for 30–60 min). The membranes were blocked in 1× TBS-Tween containing 5% (w/v) fat-free skimmed milk for 2 h at room temperature. The membranes were then incubated in a solution of anti-His6-tag monoclonal antibody (1:10,000, diluted in a blocking solution; Proteintech Group) at 4°C, overnight. They were then washed five times (10 min each) with 1× TBS-Tween with shaking. Finally, the membranes were washed with peroxidase-conjugated affinipure goat antimouse IgG (1:8,000, diluted in the blocking solution; Origene). The signal was detected using the ECL system (LI-COR Biosciences). The GhPME2 and GhPME31 protein spots were excised from the 2D gel and digested with trypsin, as reported previously (Granvogl et al., 2007). The MS analysis was carried out according to the method described by Prabhu (Prabhu et al., 2015).
The PME activity (mU or nmol/min) was determined using an alcohol oxidase-coupled microassay, as described previously (Klavons and Bennett, 1986). The reaction solution contained 90% methyl esterified Citrus pectin (Sigma-Aldrich P9561; 0.8 mg/mL) and 0.005 U of Pichia alcohol oxidase (Sigma-Aldrich A2404) in 98 μL of 50 mm phosphate (Na) buffer at pH 8.0 (Dedeurwaerder et al., 2009). The reaction was initiated by the addition of 2 μL of diluted extract and stopped by transferring the reaction to a plate containing 100 μL of the developing solution. The plate was incubated at 67°C for 15 min, and sample absorbance was measured at 420 nm using a UV spectrophotometer (TU-1900). The amount of MeOH released by PME was calculated by comparing the OD420 values to the standards (containing 0.3 mm methanol; Dedeurwaerder et al., 2009). The PME enzyme assays using NADH were performed as previously described (Grsic-Rausch and Rausch, 2004). As a negative control, the same reaction was performed with heat-denatured (95°C, 5 min) PME or water instead of PME.
Determination of the in Vitro Antifungal Activity of GhPMEI3
The antifungal activity of GhPMEI3 was determined as previously reported (An et al., 2008). The plant pathogens V. dahliae, F. oxysporum f. sp. vasinfectum, and B. cinerea were used in the current study. Briefly, the fungi were incubated on potato dextrose agar at 25°C for 1 week; they were then transferred to 24-well tissue culture plates (TCP20-24; Bio Basic) containing 4× concentrated potato dextrose broth (100 μL) and 0, 50, 100, or 250 μg/mL of GhPMEI3. The spore suspensions were cultured for an additional 4 to 7 d. Spore germination and hyphal growth were examined as previously described (An et al., 2008). Briefly, the spore suspensions were placed on glass slides and incubated for 12 h at 25°C; spore germination and the hyphae were then photographed under a Nikon eclipse Ti microscope using a 4×/0.25 numerical aperture objective.
Extraction of Alcohol-Insoluble Residues and Determination of DM
The alcohol-insoluble residues were extracted as described elsewhere (Lionetti et al., 2017). Briefly, wild-type, transgenic, or silenced plants (uninfected, mock, and V. dahliae-treated leaves of 6-week-old plants) were collected and homogenized. They were washed twice with 70% (v/v) ethanol preheated to 70°C, vortex-mixed, and centrifuged at 14,000g for 10 min to collect the precipitate. The precipitate was resuspended in a chloroform:methanol mixture (1:1), shaken at room temperature for 30 min, and collected at 14,000g; it was then resuspended in 1 mL of 80% (v/v) acetone, twice, and the supernatant was discarded after centrifugation. The pellet that contained the alcohol-insoluble residues was dried overnight in a fume hood at room temperature, and starch was removed by α-amylase treatment (Lionetti et al., 2017).
The DM of pectin in transgenic and silenced plants was determined using the alcohol oxidase-coupled assay (Klavons and Bennett, 1986). The reaction solution contained 90% methyl esterified Citrus pectin (0.8 mg/mL; Sigma-Aldrich P9561) and 0.005 U of Pichia alcohol oxidase (Sigma-Aldrich A2404) in 98 μL of 50 mm sodium phosphate buffer (pH 8). The mixture was preincubated for 3 min at 30°C; then, 2 μL of the diluted extract was added and the incubation was continued at 30°C for 15 min; 100 μL of the developing solution was added to stop the reaction (Klavons and Bennett, 1986; Dedeurwaerder et al., 2009). The experiment was performed after preincubating the proteins at 30°C for 3 h, and the reaction was stopped by boiling at 100°C for 10 min; 0.03 U of alcohol oxidase was added, and the mixture was incubated for 15 min at room temperature with shaking. This was followed by the addition of 100 μL of the developing solution (Lionetti et al., 2015). The mixture was then incubated at 67°C for 15 min, and OD420 was determined using a TU-1900 spectrophotometer. Standards containing 0.3 mm methanol were included in each batch of assays. The amount of methanol released by PME was quantified based on the measured OD420 values (Dedeurwaerder et al., 2009).
Extraction of Total Proteins and Determination of PME Activity in Transgenic Plants Inoculated with Vd991
Total protein extraction from transgenic Arabidopsis specimens was performed as previously described (Lionetti et al., 2014). Briefly, 2 mL of extraction buffer (1 m NaCl, 20 mm sodium acetate, 0.02% [w/v] sodium azide, and protease inhibitor, 1:100, pH 5.5) was added per 1 g of plant tissue. The samples were shaken at 4°C for 1.5 h, and centrifuged for 15 min at 15,000g; the supernatant was then collected. Protein concentrations were determined using the Bradford method, with bovine serum albumin as a standard. The proteins were resolved using SDS-PAGE, loading 5 μg of protein extract per lane. The proteins were identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (Prabhu et al., 2015).
Arabidopsis plants were inoculated with V. dahliae by leaf dipping (5 × 106 conidia/mL). Plants that were mock-inoculated with Czapek-Dox medium were used as a control (Lionetti et al., 2007). The disease symptoms were quantified with ImageJ software. The fragments were inserted into pCAMBIA-1304 vector with NcoI as restriction site (Supplemental Fig. S4), and the GhPMEI3 transgenic lines were selected (Supplemental Fig. S5). The PME activity was detected using the radical gel diffusion assay in wild-type and transgenic GhPMEI3 Arabidopsis plants after inoculation with Vd991 (Downie et al., 1998; Lionetti et al., 2007). Briefly, gels were prepared using 0.1% of 81% methyl esterified lime pectin, 1% (w/v) agarose, 12.5 mm citric acid, and 50 mm Na2HPO4 (pH 7.0). Wells (9-mm diameter) were punched, and protein samples were loaded into each well. The plates were then incubated at 30°C for 16 h; the gels were stained with 0.05% (w/v) ruthenium red for 45 min, and washed thoroughly with distilled water. The diameter of the red-stained areas was indicative of the hydrolysis of esterified pectin in the gels.
VIGS and Determination of PME Activity in Plants Inoculated with Vd991
VIGS vectors were constructed and Agrobacterium tumefaciens-mediated gene silencing in cotton was performed as previously described (Gao et al., 2011). GhCLA1, GhPMEI3, GhPME2, and GhPME31 cDNAs were PCR-amplified using the primers listed in Supplemental Table S1. The amplified fragments were cloned into pTRV1 and pTRV2 vectors (a kind gift from Professor Yule Liu, Tsinghua University, China) to construct the pTRV1-pTRV2:GhCLA1, pTRV1-pTRV2:GhPMEI3, pTRV1-pTRV2:GhPME2, and pTRV1-pTRV2:GhPME31 vectors, accordingly. These vector derivatives were used to transform A. tumefaciens GV3101 strain. The cultures and cells were handled as described earlier (Gao et al., 2011). Cultures of bacteria harboring pTRV1 and pTRV2 vectors were mixed in a 1:1 ratio, and the cotyledons of 2-week-old cotton seedlings were infiltrated with a needleless syringe. Small holes were punched on the underside of the cotyledon to facilitate the infiltration. The assays were performed with at least six plants for each constructed vector, and the experiments were repeated at least three times.
V. dahliae Vd991 was grown on potato dextrose agar at 25°C for 7 d. Spore suspensions were prepared at 1 × 106 conidia/mL in a solution containing 0.001% (v/v) Tween 20 (Gao et al., 2011). Both the control and VIGS-silenced plants were stem-inoculated using a syringe needle ∼1 cm below the cotyledons (Bolek et al., 2005).
In Planta Fungal Biomass Assessment and PG Expression Analysis
V. dahliae biomass was assessed as previously described (Zambounis et al., 2007; Ellendorff et al., 2009). For Verticillium inoculations, 2- to 3-week-old wild-type and single insert transgenic Arabidopsis lines were uprooted. The roots were cleaned with water and then dipped in a suspension of 106 conidia/mL for 3 min (Fradin et al., 2011). Wild-type and GhPMEI3-silenced cotton were inoculated with Verticillium as previously described (Gao et al., 2011). The control plants were treated with potato dextrose broth without the conidia. Arabidopsis samples were collected on day 21 after treatment, and cotton samples were collected 18 d after the inoculation. To estimate the amount of Verticillium biomass, DNA was extracted from 100 mg of liquid nitrogen-ground sample powder and further analyzed by RT-qPCR. The primer sequences (ITS1-F/ST-Ve1-R and FOV-3F/FOV-4R) are listed in Supplemental Table S1. To evaluate the expression of PG-encoding genes in Arabidopsis and cotton tissues, total RNA was extracted from whole plants and then reverse-transcribed into cDNA (Tiangen Biotech). The RT-qPCR reactions were performed as described above, with primers listed in Supplemental Table S1.
Homology Modeling and Protein Docking
Three-dimensional structure homology modeling of GhPMEI3, GhPME2, and GhPME31 was performed using Phyre2 software in the normal modeling mode (Kelley et al., 2015). The structure of kiwi PMEI (PDB ID 1XG2, chain B) was used as a template to construct a homology model of GhPMEI3, based on score and rank; the models of GhPME2 and GhPME31 were built using the complex (PDB ID 1XG2.1.A) and carrot PME (PDB ID 1GQ8) as structural templates with high scores (Supplemental Fig. S6). The postrefinement and visualization of the structural models were carried out as previously described (Sénéchal et al., 2015; Liu et al., 2017; Supplemental Table S2). The interactions of GhPMEI3 with GhPME2, and GhPMEI3 with GhPME31 were assessed by docking analysis using the PatchDock (Schneidman-Duhovny) molecular docking algorithm, based on the shape complementarity principles (Liu et al., 2017). Model data of the top 20 best models were then refined and minimized in FiberDock; the highest-scoring ranked models were selected as the most probable prediction candidates.
Electrostatic Potential Distribution Analysis
The DeepView-Swiss-PdbViewer (Guex and Peitsch, 1997) was employed to compute the distributions of the electrostatic potentials for GhPMEI3, GhPME2, and GhPME31, and for the GhPMEI3-GhPME2 and GhPMEI3-GhPME31 complexes. The surface charge was visualized using UCSF Chimera v. 1.10.1 software (Pettersen et al., 2004).
Accession Numbers
Sequence data for the proteins analyzed in the current article may be accessed in the GenBank library under the following accession numbers: KY933672, for GhPMEI3; KY933673, for GhPME2; and KY933674, for GhPME31.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. GhPMEI3 regulates the level of pectin methyl esterification during fungal infection.
Supplemental Figure S2. Determination of the DM in CK and GhPMEI3-silenced cotton lines.
Supplemental Figure S3. Schematic representation of PET-32a-GhPMEI3 and pGEX-4T-1-GhPME2/GhPME31 bacterial expression plasmids.
Supplemental Figure S4. Construction of the overexpression vector pCAMBIA1304-GhPMEI3.
Supplemental Figure S5. Transcript level determination in T1-transgenic Arabidopsis.
Supplemental Figure S6. Homology modeling of GhPMEI3, GhPME2, and GhPME31.
Supplemental Table S1. Primers used in RT-PCR and RT-qPCR.
Supplemental Table S2. Refinement of GhPMEI3-GhPME2 and GhPMEI3- GhPME31 complexes.
Acknowledgments
We thank Professor Yule Liu (Tsinghua University, China) for the kind gift of pTRV1 and pTRV2 vectors for the gene silencing experiments.
Footnotes
This work was sponsored by the “Seven Crop Breeding” National Major Project (grant no. 2016YFD0101006), the Genetically Modified Organism Breeding Major Project (grant no. 2018ZX08005001-002), and the State Key Laboratory of Cotton Biology Open Fund (grant no. CB2017B03).
Articles can be viewed without a subscription.
References
- An SH, Sohn KH, Choi HW, Hwang IS, Lee SC, Hwang BK (2008) Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta 228: 61–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrieri C, Castaldo D, Giovane A, Quagliuolo L, Servillo L (1990) A glycoprotein inhibitor of pectin methylesterase in kiwi fruit (Actinidia chinensis). Eur J Biochem 193: 183–187 [DOI] [PubMed] [Google Scholar]
- Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al. (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323: 101–106 [DOI] [PubMed] [Google Scholar]
- Bellincampi D, Cervone F, Lionetti V (2014) Plant cell wall dynamics and wall-related susceptibility in plant-pathogen interactions. Front Plant Sci 5: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke G, Grundman RE, Sreekanta S, Truman W, Katagiri F, Glazebrook J (2014) Arabidopsis PECTIN METHYLESTERASEs contribute to immunity against Pseudomonas syringae. Plant Physiol 164: 1093–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolek Y, Elzik KM, Pepper AE, Bell AA, Magill CW, Thaxton PM, Ouk R (2005) Mapping of verticillium wilt resistance genes in cotton. Plant Sci 168: 1581–1590 [Google Scholar]
- Bonavita A, Carratore V, Ciardiello MA, Giovane A, Servillo L, D’Avino R (2016) Influence of pH on the structure and function of kiwi pectin methylesterase inhibitor. J Agric Food Chem 64: 5866–5876 [DOI] [PubMed] [Google Scholar]
- Bradford MMA. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G (2010) A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci USA 107: 9452–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffall KH, Mohnen D (2009) The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr Res 344: 1879–1900 [DOI] [PubMed] [Google Scholar]
- D’Avino R, Camardella L, Christensen TM, Giovane A, Servillo L (2003) Tomato pectin methylesterase: modeling, fluorescence, and inhibitor interaction studies-comparison with the bacterial (Erwinia chrysanthemi) enzyme. Proteins 53: 830–839 [DOI] [PubMed] [Google Scholar]
- Dedeurwaerder S, Menu-Bouaouiche L, Mareck A, Lerouge P, Guerineau F (2009) Activity of an atypical Arabidopsis thaliana pectin methylesterase. Planta 229: 311–321 [DOI] [PubMed] [Google Scholar]
- Denès JM, Baron A, Renard CMGC, Péan C, Drilleau JF (2000) Different action patterns for apple pectin methylesterase at pH 7.0 and 4.5. Carbohydr Res 327: 385–393 [DOI] [PubMed] [Google Scholar]
- Di Matteo A, Giovane A, Raiola A, Camardella L, Bonivento D, De Lorenzo G, Cervone F, Bellincampi D, Tsernoglou D (2005) Structural basis for the interaction between pectin methylesterase and a specific inhibitor protein. Plant Cell 17: 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorokhov YL, Skurat EV, Frolova OY, Gasanova TV, Ivanov PA, Ravin NV, Skryabin KG, Mäkinen KM, Klimyuk VI, Gleba YY, et al. (2006) Role of the leader sequence in tobacco pectin methylesterase secretion. FEBS Lett 580: 3329–3334 [DOI] [PubMed] [Google Scholar]
- Downie B, Dirk LM, Hadfield KA, Wilkins TA, Bennett AB, Bradford KJ (1998) A gel diffusion assay for quantification of pectin methylesterase activity. Anal Biochem 264: 149–157 [DOI] [PubMed] [Google Scholar]
- Ellendorff U, Fradin EF, de Jonge R, Thomma BPHJ (2009) RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J Exp Bot 60: 591–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Savatin DV, Sicilia F, Gramegna G, Cervone F, Lorenzo GD (2013) Oligogalacturonides: plant damage-associated molecular patterns and regulators of growth and development. Front Plant Sci 4: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Vairo D, Ausubel FM, Cervone F, De Lorenzo G (2003) Tandemly duplicated Arabidopsis genes that encode polygalacturonase-inhibiting proteins are regulated coordinately by different signal transduction pathways in response to fungal infection. Plant Cell 15: 93–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin EF, Abd-El-Haliem A, Masini L, van den Berg GC, Joosten MH, Thomma BP (2011) Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol 156: 2255–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin EF, Thomma BP (2006) Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol Plant Pathol 7: 71–86 [DOI] [PubMed] [Google Scholar]
- Gao X, Wheeler T, Li Z, Kenerley CM, He P, Shan L (2011) Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J 66: 293–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovane A, Servillo L, Balestrieri C, Raiola A, D’Avino R, Tamburrini M, Ciardiello MA, Camardella L (2004) Pectin methylesterase inhibitor. Biochim Biophys Acta 1696: 245–252 [DOI] [PubMed] [Google Scholar]
- Goldberg R. (1984) Changes in the properties of cell wall pectin methylesterase along the Vigna radiata hypocotyl. Physiol Plant 61: 58–63 [Google Scholar]
- Granvogl B, Plöscher M, Eichacker LA (2007) Sample preparation by in-gel digestion for mass spectrometry-based proteomics. Anal Bioanal Chem 389: 991–1002 [DOI] [PubMed] [Google Scholar]
- Grsic-Rausch S, Rausch T (2004) A coupled spectrophotometric enzyme assay for the determination of pectin methylesterase activity and its inhibition by proteinaceous inhibitors. Anal Biochem 333: 14–18 [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18: 2714–2723 [DOI] [PubMed] [Google Scholar]
- Hocq L, Sénéchal F, Lefebvre V, Lehner A, Domon JM, Mollet JC, Dehors J, Pageau K, Marcelo P, Guerineau F (2017) Combined experimental and computational approaches reveal distinct pH-dependence of pectin methyl esterase inhibitors. Plant Physiol 173: 1075–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M, D’Angelo I, Márquez JA, Greiner S, Scheffzek K (2004a) The invertase inhibitor Nt-CIF from tobacco: a highly thermostable four-helix bundle with an unusual N-terminal extension. J Mol Biol 335: 987–995 [DOI] [PubMed] [Google Scholar]
- Hothorn M, Van den Ende W, Lammens W, Rybin V, Scheffzek K (2010) Structural insights into the pH-controlled targeting of plant cell-wall invertase by a specific inhibitor protein. Proc Natl Acad Sci USA 107: 17427–17432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M, Wolf S, Aloy P, Greiner S, Scheffzek K (2004b) Structural insights into the target specificity of plant invertase and pectin methylesterase inhibitory proteins. Plant Cell 16: 3437–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki A, Akimitsu K, Yamamoto M, Yamamoto H (2001) Endopolygalacturonase is essential for citrus black rot caused by Alternaria citri but not brown spot caused by Alternaria alternata. Mol Plant Microbe Interact 14: 749–757 [DOI] [PubMed] [Google Scholar]
- Jenkins J, Mayans O, Smith D, Worboys K, Pickersgill RW (2001) Three-dimensional structure of Erwinia chrysanthemi pectin methylesterase reveals a novel esterase active site. J Mol Biol 305: 951–960 [DOI] [PubMed] [Google Scholar]
- Johansson K, El-Ahmad M, Friemann R, Jörnvall H, Markovič O, Eklund H (2002) Crystal structure of plant pectin methylesterase. FEBS Lett 514: 243–249 [DOI] [PubMed] [Google Scholar]
- Jolie RP, Duvetter T, Houben K, Vandevenne E, Loey AMV, Declerck PJ, Hendrickx ME, Gils A (2010) Plant pectin methylesterase and its inhibitor from kiwi fruit: interaction analysis by surface plasmon resonance. Food Chem 121: 207–214 [Google Scholar]
- Kawchuk LM, Hachey J, Lynch DR, Kulcsar F, van Rooijen G, Waterer DR, Robertson A, Kokko E, Byers R, Howard RJ, et al. (2001) Tomato Ve disease resistance genes encode cell surface-like receptors. Proc Natl Acad Sci USA 98: 6511–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Triplett BA (2001) Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiol 127: 1361–1366 [PMC free article] [PubMed] [Google Scholar]
- Klavons JA, Bennett RD (1986) Determination of methanol using alcohol oxidase and its application to methyl ester content of pectins. J Agric Food Chem 34: 597–599 [Google Scholar]
- Kohn R, Markovič O, Machová E (1983) Deesterification mode of pectin by pectin esterases of Aspergillus foetidus. Collect Czech Chem Commun 48: 790–797 [Google Scholar]
- Limberg G, Körner R, Buchholt HC, Christensen TM, Roepstorff P, Mikkelsen JD (2000) Analysis of different de-esterification mechanisms for pectin by enzymatic fingerprinting using endopectin lyase and endopolygalacturonase II from A. niger. Carbohydr Res 327: 293–307 [DOI] [PubMed] [Google Scholar]
- Lionetti V, Fabri E, De Caroli M, Hansen AR, Willats WG, Piro G, Bellincampi D (2017) Three pectin methylesterase inhibitors protect cell wall integrity for Arabidopsis immunity to botrytis. Plant Physiol 173: 1844–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V, Raiola A, Camardella L, Giovane A, Obel N, Pauly M, Favaron F, Cervone F, Bellincampi D (2007) Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol 143: 1871–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V, Raiola A, Cervone F, Bellincampi D (2014) Transgenic expression of pectin methylesterase inhibitors limits tobamovirus spread in tobacco and Arabidopsis. Mol Plant Pathol 15: 265–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti V, Raiola A, Mattei B, Bellincampi D (2015) The grapevine VvPMEI1 gene encodes a novel functional pectin methylesterase inhibitor associated to grape berry development. PLoS One 10: e0133810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Zhang X, Sun Y, Wang P, Li X, Pei Y, Li F, Hou Y (2017) Molecular evidence for the involvement of a polygalacturonase-inhibiting protein, GhPGIP1, in enhanced resistance to Verticillium and Fusarium wilts in cotton. Sci Rep 7: 39840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markoviě O, Kohn R (1984) Mode of pectin deesterification by Trichoderma reesei pectinesterase. Experientia 40: 842–843 [Google Scholar]
- Micheli F. (2001) Pectin methylesterases: cell wall enzymes with important roles in plant physiology. Trends Plant Sci 6: 414–419 [DOI] [PubMed] [Google Scholar]
- Nguyen HP, Jeong HY, Kim H, Kim YC, Lee C (2016) Molecular and biochemical characterization of rice pectin methylesterase inhibitors (OsPMEIs). Plant Physiol Biochem 101: 105–112 [DOI] [PubMed] [Google Scholar]
- Nothnagel EA, McNeil M, Albersheim P, Dell A (1983) Host-pathogen interactions: XXII. A galacturonic acid oligosaccharide from plant cell walls elicits phytoalexins. Plant Physiol 71: 916–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelloux J, Rustérucci C, Mellerowicz EJ (2007) New insights into pectin methylesterase structure and function. Trends Plant Sci 12: 267–277 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Prabhu SA, Wagenknecht M, Melvin P, Gnanesh Kumar BS, Veena M, Shailasree S, Moerschbacher BM, Kini KR (2015) Immuno-affinity purification of PglPGIP1, a polygalacturonase-inhibitor protein from pearl millet: studies on its inhibition of fungal polygalacturonases and role in resistance against the downy mildew pathogen. Mol Biol Rep 42: 1123–1138 [DOI] [PubMed] [Google Scholar]
- Raiola A, Camardella L, Giovane A, Mattei B, De Lorenzo G, Cervone F, Bellincampi D (2004) Two Arabidopsis thaliana genes encode functional pectin methylesterase inhibitors. FEBS Lett 557: 199–203 [DOI] [PubMed] [Google Scholar]
- Raiola A, Lionetti V, Elmaghraby I, Immerzeel P, Mellerowicz EJ, Salvi G, Cervone F, Bellincampi D (2011) Pectin methylesterase is induced in Arabidopsis upon infection and is necessary for a successful colonization by necrotrophic pathogens. Mol Plant Microbe Interact 24: 432–440 [DOI] [PubMed] [Google Scholar]
- Reca IB, Lionetti V, Camardella L, D’Avino R, Giardina T, Cervone F, Bellincampi D (2012) A functional pectin methylesterase inhibitor protein (SolyPMEI) is expressed during tomato fruit ripening and interacts with PME-1. Plant Mol Biol 79: 429–442 [DOI] [PubMed] [Google Scholar]
- Robert X, Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42: W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sénéchal F, L’Enfant M, Domon JM, Rosiau E, Crépeau MJ, Surcouf O, Esquivel-Rodriguez J, Marcelo P, Mareck A, Guérineau F, et al. (2015) Tuning of pectin methylesterification: PECTIN METHYLESTERASE INHIBITOR 7 modulates the processive activity of co-expressed PECTIN METHYLESTERASE 3 in a pH-dependent manner. J Biol Chem 290: 23320–23335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling JD, Quigley HF, Orellana A, Mohnen D (2001) The catalytic site of the pectin biosynthetic enzyme alpha-1,4-galacturonosyltransferase is located in the lumen of the Golgi. Plant Physiol 127: 360–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia G, Verdugo I, Yañez M, Ahumada I, Theoduloz C, Cordero C, Poblete F, González E, Ruiz-Lara S (2005) Involvement of ethylene in stress-induced expression of the TLC1.1 retrotransposon from Lycopersicon chilense Dun. Plant Physiol 138: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Have A, Mulder W, Visser J, van Kan JA (1998) The endopolygalacturonase gene Bcpg1 is required for full virulence of Botrytis cinerea. Mol Plant Microbe Interact 11: 1009–1016 [DOI] [PubMed] [Google Scholar]
- Tundo S, Kalunke R, Janni M, Volpi C, Lionetti V, Bellincampi D, Favaron F, D’Ovidio R (2016) Pyramiding PvPGIP2 and TAXI-III but not PvPGIP2 and PMEI enhances resistance against Fusarium graminearum. Mol Plant Microbe Interact 29: 629–639 [DOI] [PubMed] [Google Scholar]
- Volpi C, Janni M, Lionetti V, Bellincampi D, Favaron F, D’Ovidio R (2011) The ectopic expression of a pectin methyl esterase inhibitor increases pectin methyl esterification and limits fungal diseases in wheat. Mol Plant Microbe Interact 24: 1012–1019 [DOI] [PubMed] [Google Scholar]
- Volpi C, Raiola A, Janni M, Gordon A, O’Sullivan DM, Favaron F, D’Ovidio R (2013) Claviceps purpurea expressing polygalacturonases escaping PGIP inhibition fully infects PvPGIP2 wheat transgenic plants but its infection is delayed in wheat transgenic plants with increased level of pectin methyl esterification. Plant Physiol Biochem 73: 294–301 [DOI] [PubMed] [Google Scholar]
- Wang X, Zhu X, Tooley P, Zhang X (2013) Cloning and functional analysis of three genes encoding polygalacturonase-inhibiting proteins from Capsicum annuum and transgenic CaPGIP1 in tobacco in relation to increased resistance to two fungal pathogens. Plant Mol Biol 81: 379–400 [DOI] [PubMed] [Google Scholar]
- Wiethölter N, Graessner B, Mierau M, Mort AJ, Moerschbacher BM (2003) Differences in the methyl ester distribution of homogalacturonans from near-isogenic wheat lines resistant and susceptible to the wheat stem rust fungus. Mol Plant Microbe Interact 16: 945–952 [DOI] [PubMed] [Google Scholar]
- Willats WG, Orfila C, Limberg G, Buchholt HC, van Alebeek GJ, Voragen AG, Marcus SE, Christensen TM, Mikkelsen JD, Murray BS, et al. (2001) Modulation of the degree and pattern of methyl-esterification of pectic homogalacturonan in plant cell walls. Implications for pectin methyl esterase action, matrix properties, and cell adhesion. J Biol Chem 276: 19404–19413 [DOI] [PubMed] [Google Scholar]
- Wolf S, Rausch T, Greiner S (2009) The N-terminal pro region mediates retention of unprocessed type-I PME in the Golgi apparatus. Plant J 58: 361–375 [DOI] [PubMed] [Google Scholar]
- Xu L, Zhu L, Tu L, Liu L, Yuan D, Jin L, Long L, Zhang X (2011) Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J Exp Bot 62: 5607–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambounis AG, Paplomatas E, Tsaftaris AS (2007) Intergenic spacer-RFLP analysis and direct quantification of Australian Fusarium oxysporum f. sp. vasinfectum isolates from soil and infected cotton tissues. Plant Dis 91: 1564–1573 [DOI] [PubMed] [Google Scholar]