New insights into the regulation of GUN1 uncover its function in early chloroplast biogenesis and suggest a role in developmental phase transitions.
Abstract
The exchange of signals between cellular compartments coordinates development and differentiation, modulates metabolic pathways, and triggers responses to environmental conditions. The proposed central regulator of plastid-to-nucleus retrograde signaling, GENOMES UNCOUPLED1 (GUN1), is present at very low levels, which has hampered the discovery of its precise molecular function. Here, we show that the Arabidopsis (Arabidopsis thaliana) GUN1 protein accumulates to detectable levels only at very early stages of leaf development, where it functions in the regulation of chloroplast biogenesis. GUN1 mRNA is present at high levels in all tissues, but GUN1 protein undergoes rapid degradation (with an estimated half-life of ∼4 h) in all tissues where chloroplast biogenesis has been completed. The rapid turnover of GUN1 is controlled mainly by the chaperone ClpC1, suggesting degradation of GUN1 by the Clp protease. Degradation of GUN1 slows under stress conditions that alter retrograde signaling, thus ensuring that the plant has sufficient GUN1 protein. We also find that the pentatricopeptide repeat motifs of GUN1 are important determinants of GUN1 stability. Moreover, overexpression of GUN1 causes an early flowering phenotype, suggesting a function of GUN1 in developmental phase transitions beyond chloroplast biogenesis. Taken together, our results provide new insight into the regulation of GUN1 by proteolytic degradation, uncover its function in early chloroplast biogenesis, and suggest a role in developmental phase transitions.
The exchange of signals between cellular compartments is essential to coordinate development and differentiation, optimize the output of metabolic pathways, and trigger appropriate responses to environmental stimuli and stresses (Parikh et al., 1987; Cottage et al., 2010; Estavillo et al., 2011; Xiao et al., 2012; Esteves et al., 2014). The plastids (chloroplasts) of plants and eukaryotic algae arose from a formerly free-living cyanobacterium through endosymbiosis. During the course of evolution, the majority of plastid genes were transferred to the nuclear genome; therefore, many chloroplast protein complexes (e.g. ribosomes and photosystems) are mosaics of subunits encoded by plastid genes and subunits encoded by nuclear genes. Consequently, the tightly coordinated expression of genes in both genomes by anterograde and retrograde signals is of fundamental importance, especially during plastid biogenesis and under stress conditions that damage chloroplast membranes and protein complexes and impair proper chloroplast function (Nott et al., 2006; Lee et al., 2007; Pogson et al., 2008; Woodson et al., 2013; Martín et al., 2016; Pornsiriwong et al., 2017). Known retrograde signaling pathways from plastids to the nucleus can be divided into two types: (1) pathways that optimize the cellular responses to environmental cues, also referred to as operational control, and (2) pathways that regulate chloroplast development (especially thylakoid biogenesis), referred to as biogenic control, by influencing the expression of photosynthesis-associated nuclear genes.
Stress responses regulated by retrograde signaling include the responses to high-light stress and drought stress, two conditions that can severely perturb photosynthesis (Wagner et al., 2004; Rossel et al., 2007; Estavillo et al., 2011; Xiao et al., 2012). A number of reactive oxygen species (ROS), including singlet oxygen, the superoxide anion, hydrogen peroxide, and the hydroxyl radical, are generated when plant cells suffer from environmental stresses. Therefore, it may be unsurprising that ROS generated in plastids are involved in retrograde signaling, especially under conditions of excess light (Apel and Hirt, 2004; Li et al., 2009). The identification and extensive characterization of the conditional fluorescent (flu) mutant (Meskauskiene et al., 2001) generated important insights into the role of singlet oxygen in retrograde signaling and the control of cell death (op den Camp et al., 2003). In addition to ROS, a few metabolites also were demonstrated to function as retrograde signaling molecules (Estavillo et al., 2011; Xiao et al., 2012). 3′-Phosphoadenosine 5′-phosphate can move between cellular compartments to regulate the retrograde responses of plants to drought, high light, programmed cell death, and abscisic acid signaling (Bruggeman et al., 2016; Pornsiriwong et al., 2017). Methylerythritol cyclodiphosphate, an intermediate of the isoprenoid biosynthetic pathway, regulates a specific set of nuclear genes upon high light and wounding stress and stimulates the unfolded protein response in the endoplasmic reticulum (Xiao et al., 2012; Walley et al., 2015).
Genetic screens for mutants defective in plastid-to-nucleus retrograde signaling identified several loci, known as GENOMES UNCOUPLED (GUN) genes, that regulate the expression of nuclear genes for chloroplast development and photosystem biogenesis in response to signals emanating from the plastid (Susek et al., 1993). Among the six gun mutants identified so far, five (GUN2–GUN6) are affected in genes for enzymes involved in tetrapyrrole biosynthesis (Mochizuki et al., 2001; Larkin et al., 2003; Strand et al., 2003; Woodson et al., 2011). By contrast, the sixth gene, GUN1, encodes a pentatricopeptide repeat (PPR) protein with a C-terminal small MutS-related (SMR) domain that, thus far, has not been implicated in tetrapyrrole metabolism. GUN1 was proposed to participate in multiple retrograde signaling pathways (Koussevitzky et al., 2007). Although initially proposed to interact with DNA (Koussevitzky et al., 2007), and although PPR proteins usually bind RNA in a sequence-specific manner, recent work has suggested that GUN1 is more likely to engage in protein-protein interactions than in interactions with nucleic acids (Colombo et al., 2016; Tadini et al., 2016) and possibly functions in the regulation of chloroplast protein homeostasis (Llamas et al., 2017), although the mechanism of its suspected role in fine-tuning protein homeostasis remains to be elucidated.
Interestingly, the GUN1 protein appears to be present in plant cells at very low levels and is hardly detectable by proteomic approaches (Plant Proteome DataBase; http://ppdb.tc.cornell.edu/). How its expression and its accumulation in relation to the activity of retrograde signaling pathways are regulated has remained largely unknown.
The abundance of proteins in the plastid compartment is regulated by the balance between their rate of synthesis and their degradation rate. The sequence motifs and/or structural features influencing the half-lives of plastid proteins are not well understood (Apel et al., 2010; Li et al., 2017). Using biochemical, genetic, bioinformatic, and proteomic approaches, more than 20 proteolytic activities have been identified in plastids, including the major bacteria-type ATP-dependent (Clp, Lon, and FtsH) and ATP-independent (e.g. members of the Deg family) proteases (Shanklin et al., 1995; Lindahl et al., 1996; Itzhaki et al., 1998; Adam et al., 2006; Rigas et al., 2014; van Wijk, 2015; Nishimura et al., 2016). The Clp protease system is the major stromal protease in chloroplasts (Nishimura and van Wijk, 2015). A small proportion of the Clp protease pool was found to associate with the inner envelope membrane of the chloroplast, where it acts as a quality control system to degrade damaged or otherwise unwanted proteins upon import (Feng et al., 2016; Flores-Pérez et al., 2016).
In view of the proposed central role of GUN1 in retrograde signaling and the conspicuous absence of proteomic evidence for GUN1 expression at the protein level, we decided to investigate the regulation of GUN1 protein abundance and its regulation in Arabidopsis (Arabidopsis thaliana). We find that GUN1 is present at high levels in differentiating chloroplasts of very young leaves and functions in the regulation of chloroplast biogenesis. In more mature leaves, GUN1 is present in very low abundance, but the GUN1 transcript is present at high levels. We report that GUN1 is a rapidly turned over protein with an estimated half-life of ∼4 h. The degradation of GUN1 is largely dependent on ClpC1 and, thus, is likely conducted by the Clp protease. Importantly, the degradation of GUN1 slows down under stress conditions known to alter retrograde signaling, thus leading to a larger protein pool that can function in triggering adaptation responses that are under retrograde control. Moreover, by establishing stable GUN1 overexpression lines, we reveal that the overaccumulation of GUN1 induces early flowering, suggesting a functional connection between retrograde communication and the regulation of plant development.
RESULTS
The GUN1 mRNA Is Expressed throughout Plant Development
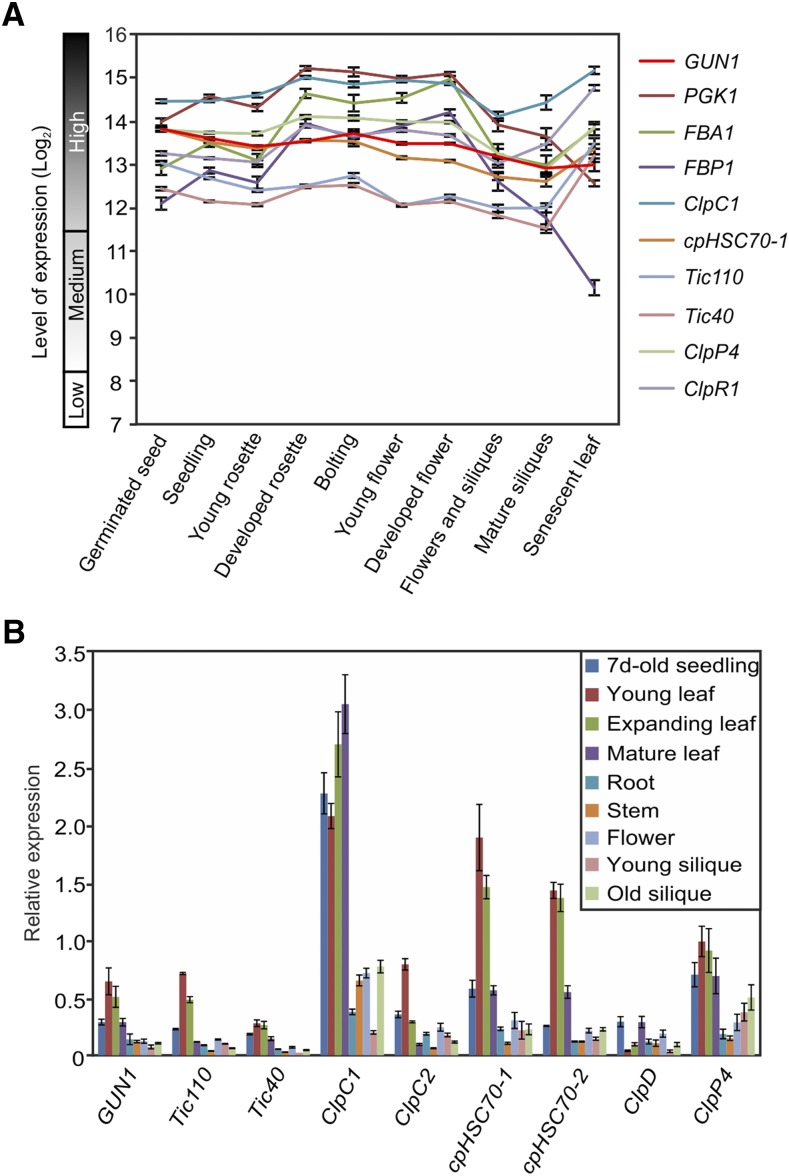
To understand the function of GUN1 in plant development, we first investigated the expression of GUN1 in different developmental stages using publicly available data sets (https://genevestigator.com/) and compared its expression with that of various highly expressed chloroplast genes from different pathways, including genes from the Calvin cycle (phosphoglycerate kinase1 [PGK1], Fru-bisphosphate aldolase1 [FBA1], and Fru-1,6-bisphosphate phosphatase1 [FBP1]), subunits of the Tic complex of the protein import apparatus (Tic110 and Tic40), chloroplast chaperones (ClpC1 and cpHSC70-1), and subunits of the Clp protease (ClpP4 and ClpR1). At the RNA level, GUN1 is highly expressed across all developmental stages and even higher than highly abundant subunits of the Tic complex (Tic110 and Tic40; Fig. 1A). To further verify this finding, we used RT-qPCR to analyze the expression level of GUN1 and selected other genes for comparison (Fig. 1B). While the GUN1 mRNA is highly expressed in young and expanding leaves, expression is substantially lower in seedlings and mature leaves and even lower in all other tissues investigated (Fig. 1B). When compared with the reference genes, the abundance of GUN1 transcripts is in the same order of magnitude as other highly expressed genes but somewhat lower than the expression of genes for highly abundant chloroplast chaperones (ClpC1 and cpHSC70-1; Fig. 1B).
Figure 1.
Relative expression levels of GUN1 and selected other nucleus-encoded genes for chloroplast proteins. A, Expression of the GUN1 mRNA in different developmental stages of wild-type Arabidopsis plants based on Genevestigator data (http://genevestigator.com/gv/). Chloroplast genes involved in various pathways were investigated for comparison, including enzymes in the Calvin cycle (PGK1, FBA1, and FBP1), chaperones (ClpC1 and cpHSC70-1), and subunits of the Tic complex (Tic110 and Tic40) and the Clp protease (ClpP4 and ClpR1). Gene expression data are represented as signal intensities (log2) on Affymetrix ATH1 genome arrays. B, RT-qPCR analysis of GUN1 expression in different tissues in comparison with other nuclear genes for chloroplast proteins. The relative expression of GUN1 in 7-d-old seedlings, young leaves (eighth and ninth leaves of a rosette 21 d after germination [DAG], with leaf length less than 1 cm), expanding leaves (seventh leaf of a rosette 21 DAG, with leaf length of 2–3 cm), mature leaves (fifth leaf of a rosette 21 DAG representing a fully expanded leaf), roots, stems, flowers, young siliques (less than 1 cm), and old siliques (eighth to 10th siliques from the top) was analyzed by RT-qPCR and normalized to actin gene expression. Nuclear genes encoding chloroplast proteins of the Tic complex (Tic110 and Tic40), chloroplast chaperones (cpHSC70-1 and cpHSC70-2), and chaperones or subunits of the Clp protease (ClpC1, ClpC2, ClpD, and ClpP4) were analyzed for comparison. Relative expression values were calculated for each target gene (including calculation of the amplification efficiencies of the different primers; Pfaffl, 2001) The data are from three biological replicates and presented as means ± sd.
GUN1 Localizes to the Chloroplast Stroma and Functions in Biogenic Control of Plastid Differentiation
To explore the function of GUN1, we constructed a GUN1-GFP fusion and generated transgenic Arabidopsis plants in both the wild-type and the gun1 mutant backgrounds (Fig. 2, A and B). Importantly, the fusion protein was fully functional in that it complemented the gun1 mutant and restored retrograde signaling (Fig. 2, C and D).
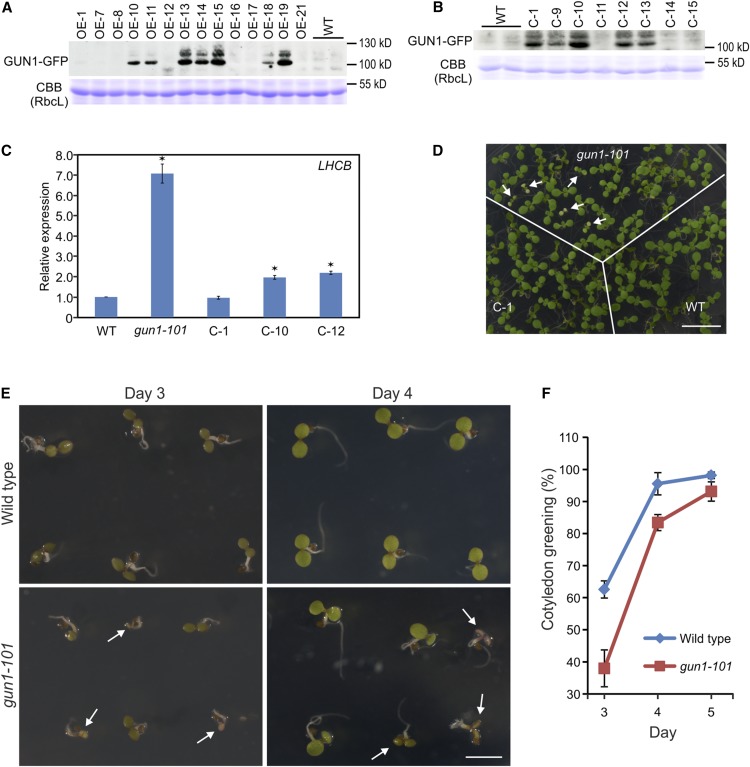
Figure 2.
Generation and characterization of GUN1-GFP-expressing transgenic lines and seedling phenotype of the gun1-101 mutant. A, Identification of transgenic lines overexpressing the GUN1-GFP fusion protein (OE lines). Total protein samples extracted from independent transgenic lines (generated in the wild-type [WT] background) were subjected to western-blot analysis with anti-GFP antibodies. Coomassie Brilliant Blue (CBB) staining of the gel region containing the large subunit of Rubisco (RbcL) served as a control for equal loading. B, Immunoblot identification of gun1-101 mutant lines complemented with the GUN1-GFP fusion gene (C lines). C, RT-qPCR analysis of LHCB1.2 expression under lincomycin treatment in different complemented lines (C-1, C-10, and C-12). Relative gene expression levels are compared with LHCB1.2 expression in the wild type. Note that line C-1 is fully complemented, while lines C-10 and C-12 are incompletely complemented. The data represent three biological replicates and are presented as means ± sd. Significant differences (*) were identified by a heteroscedastic Student’s t test (P < 0.001). D, Phenotypes of seedlings of the wild type, the gun1-101 mutant, and the complemented line C-1. Seedlings were grown under continuous light for 7 d. Note that some gun1-101 seedlings develop a pale green or variegated phenotype (white arrows), which is fully complemented in the C-1 line. Bar = 1 cm. E and F, Delayed cotyledon greening in the gun1 mutant compared with the wild type. E, Wild-type and gun1-101 seedlings 3 and 4 d after germination. Seedlings with delayed greening of the cotyledons in the gun1 mutant are indicated by white arrows. Bar = 2 mm. F, Quantitation was done on four independent experiments with ∼100 seeds per experiment and genotype. Data represent means ± sd.
Although genetically identical, a proportion of homozygous gun1 mutant seedlings showed a pale cotyledon phenotype upon germination, suggesting defective chloroplast biogenesis and the operation of a threshold mechanism that determines manifestation of the visible phenotype (Fig. 2D). To further explore the role of GUN1 in chloroplast development, we analyzed the speed of cotyledon greening in the wild type and the gun1 mutant during seed germination. While germination of the gun1 mutant is delayed only slightly compared with the wild type (Supplemental Fig. S1), cotyledon greening is much delayed in gun1 (Fig. 2, E and F). This phenotype is well in line with a function of GUN1 in the biogenic control of plastid differentiation.
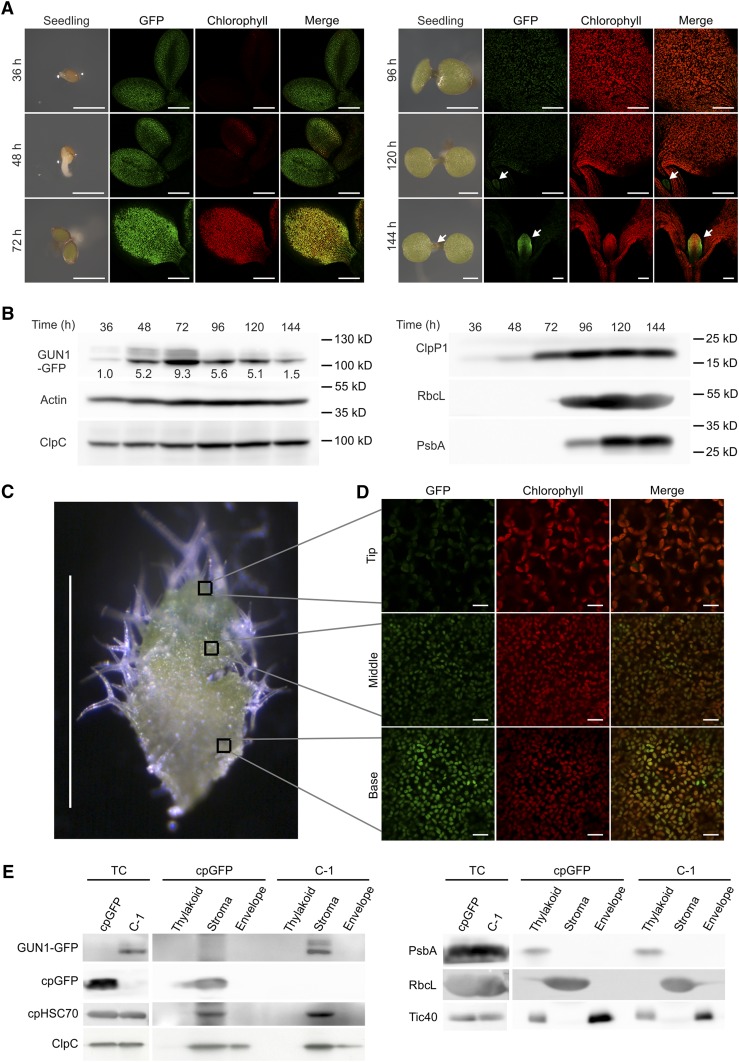
To test whether there is a correlation between GUN1 expression at the protein level and chloroplast biogenesis, we investigated the accumulation of the GUN1-GFP protein during the process of seedling development and cotyledon greening (Fig. 3, A and B). Photosynthesis-related proteins (RbcL and PsbA; Fig. 3B) started to accumulate strongly at 96 h. By contrast, GUN1-GFP is already expressed during the very early stages of seedling development, when the radicle just emerges from the testa and no chlorophyll can be detected yet (36 h). Compared with cpGFP (a chloroplast-localized unfused GFP targeted to plastids by the RBCS transit peptide), which gradually accumulates during seedling development (Supplemental Fig. S2, A and B), the expression of GUN1-GFP peaks around 72 h, when active chloroplast differentiation occurs, as evidenced by the light green cotyledons (72 h) turning green (96 h; Fig. 3, A and B). The accumulation of GUN1-GFP starts to decline at 96 h. Interestingly, this is when ClpP1, an essential catalytic subunit of the chloroplast Clp protease, becomes strongly expressed (Fig. 3B; Supplemental Fig. S2B). At later stages (120 and 144 h), GFP fluorescence is hardly detectable in the cotyledons, but remarkably, the newly emerging first true leaf at the shoot apical meristem now starts to express GUN1 (120 h). Expression becomes even stronger at 144 h, when active chloroplast differentiation occurs in the leaflet (Fig. 3A).
Figure 3.
The GUN1 protein is highly abundant in differentiating plastids and localizes to the chloroplast stroma. A, GUN1-GFP is highly expressed at the early stages of cotyledon greening. The complemented line C-1 was germinated, and the expression of GUN1-GFP was tracked at different time points. The plates were incubated at 22°C under long-day conditions. Arrows indicate the first true leaf emerging at the shoot apical meristem. To be directly comparable, all GFP images were taken with exactly the same settings, except for the 144-h time point, in which a lower magnification was used to include both true leaves and part of the cotyledons (for comparison of the GUN1-GFP signal in newly emerging true leaves and mature cotyledons). However, the microscopy settings were identical with those used for all other samples, and background fluorescence was similarly low as in the other time points (36–120 h). The experiments were repeated independently at least three times (analyzing more than 10 seedlings per time point in each replicate experiment), and representative images are shown here. Bars = 1 mm in the seedling images and 200 µm in the confocal microscopy images. B, Analysis of the expression of GUN1-GFP, subunits of the Clp protease complex (ClpC and ClpP1), and photosynthesis-related proteins (RbcL and PsbA) during germination and early seedling development. Material was harvested exactly as in A and analyzed by western blotting. Actin was used as a loading control. The relative intensity of the GUN1-GFP band was determined, normalized to actin, and is indicated below each band. C, Newly emerging rosette leaf. The basal part showing white to pale green color represents the zone where differentiation of proplastids to chloroplasts occurs, while the top part (tip region) of the leaflet is green and contains fully differentiated chloroplasts. The boxes indicate the areas from which the images in D were taken. The bottom box represents a region of early chloroplast differentiation, the top box represents a region with fully differentiated chloroplasts, and the middle box represents an intermediate stage. Bar = 1 mm. D, GUN1-GFP shows high expression in early differentiating plastids (i.e. in the basal part of the newly emerging leaflet; bottom box in C) of the complemented line C-1. Note that protein accumulation declines along the chloroplast developmental gradient, and no GFP fluorescence is detectable in the top region of the leaf that contains fully developed chloroplasts (top box in C). For direct comparison, all GFP images were taken with exactly the same settings. The experiments were independently repeated, and more than 10 plants were analyzed in each experiment. Representative images are shown here. Bars = 10 µm. E, Analysis of GUN1 localization by immunoblot analysis of protein fractions (thylakoids, stroma, and envelope membranes) purified from isolated chloroplasts of 7-d-old seedlings of the complemented line C-1 and a transgenic line expressing a chloroplast-localized unfused GFP in the wild-type background (cpGFP). cpHSC70 and ClpC were analyzed as controls. PsbA, RbcL, and Tic40 were used as marker proteins for thylakoid, stroma, and envelope membrane localization, respectively. Note that the Suc gradient centrifugation cannot completely separate envelope membranes from thylakoid membranes (as indicated by the presence of some Tic40 in the thylakoid fraction), whereas the stroma and envelope fractions are not contaminated by any of the other fractions. TC, Total chloroplast protein.
To further correlate the accumulation of the GUN1 protein with the process of chloroplast differentiation, we examined GUN1-GFP accumulation in the very young true leaves (approximately 1 mm in length) that show a steep gradient in chloroplast development, in that the leaf base is still largely unpigmented, whereas the leaf tip, the ontogenetically oldest part of the developing leaf, is visibly green and has completed chloroplast biogenesis (Fig. 3C). Confocal microscopy revealed that the GUN1-GFP signal is strongly present in the basal part of newly emerging leaflets that contain largely undeveloped plastids (Fig. 3, C and D). Interestingly, GUN1-GFP accumulation declines progressively toward the tip of the leaflet, correlating with the progression of chloroplast development (Fig. 3D). This pattern of protein accumulation is in stark contrast to GUN1 mRNA accumulation, which is similarly high across all developmental stages (Fig. 1). Considering that the strong and constitutively active cauliflower mosaic virus (CaMV) 35S promoter was used to drive the expression of the GUN1-GFP protein, this finding indicates that the regulation of GUN1 protein accumulation occurs mainly at the translational and/or posttranslational level. GUN1-GFP is already largely undetectable in the tip region of newly emerging leaves (Fig. 3D), where plastid differentiation into chloroplasts is largely complete, as evidenced by the green pigmentation of this region. GUN1-GFP also cannot be detected in expanding or mature leaves where differentiated chloroplasts are present. These observations suggest that, in plant development, GUN1 executes its function mainly during early chloroplast biogenesis, thus supporting a role of GUN1-mediated retrograde signaling in biogenic control. This conclusion is in line with the delayed greening and the pale or variegated cotyledon phenotypes displayed by some gun1 mutant seedlings (Fig. 2, D–F), which suggests partial failure to initiate chloroplast differentiation in a timely manner.
The GUN1-GFP protein shows a diffuse distribution in differentiating plastids (Fig. 3D) that would be consistent with stromal localization. Accumulation in the stroma was supported further by immunoblot analyses of purified chloroplast fractions (Fig. 3E). Although expression from the CaMV 35S promoter causes overexpression (and is needed to visualize the protein), the GUN1-GFP fusion protein fully complements the mutant phenotype and the retrograde signaling phenotype of the gun1 mutant (Fig. 2, C and D), indicating that the observed stromal localization likely represents the true localization of GUN1 in vivo. This localization is at odds with the reported punctate distribution of GUN1 in transient expression assays in tobacco (Nicotiana tabacum; which, for example, could come from aggregated proteins in tobacco cells) and the proposed colocalization with chloroplast nucleoids (Koussevitzky et al., 2007; Supplemental Fig. S2C).
The GUN1 Protein Is Present at Undetectably Low Levels in Differentiated Chloroplasts But Is Induced under Conditions That Affect Retrograde Signaling
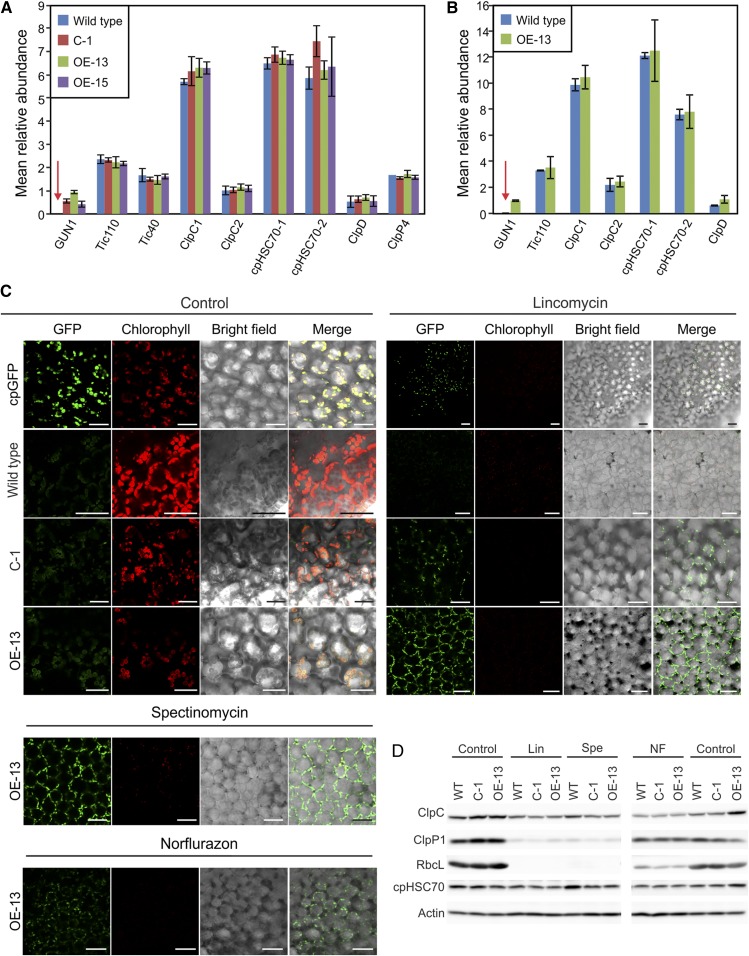
Since we were unable to detect GUN1-GFP in differentiated chloroplasts by confocal microscopy (Fig. 3, C and D), we next attempted to quantify the GUN1 protein using mass spectrometry-based proteomics. To this end, we investigated 7-d-old seedlings (Fig. 4A; Supplemental Data Set S1) and young rosette leaves (Fig. 4B; Supplemental Data Set S1) in three different genotypes: the wild type, the complemented gun1 mutant (line C-1), and two GUN1 overexpression lines (OE-13 and OE-15).
Figure 4.
GUN1 is a very-low-abundance protein and is induced in conditions under which retrograde signaling can be demonstrated. A, Mass spectrometric quantification of GUN1 protein abundance in 7-d-old seedlings of the wild type, the complemented line C-1, and two overexpression lines (OE-13 and OE-15). B, Mass spectrometric quantification of GUN1 in rosette leaves of the wild type and the overexpression line OE-13. In A and B, selected other chloroplast proteins and subunits of multiprotein complexes were analyzed for comparison, including subunits of the Tic complex (Tic110 and Tic40), chaperones (cpHSC70-1 and cpHSC70-2), and proteins of the Clp protease system (ClpC1, ClpC2, ClpD, and ClpP4). The intensity-based relative protein abundance (label-free quantification) was normalized within each sample. The normalized relative abundance of each protein represents the proportion of each protein of all detected proteins in the given sample. Note that GUN1 is undetectable in the wild type both in 7-d-old seedlings and in rosette leaves (red arrows). C, Induction of GUN1-GFP accumulation when retrograde signaling is altered. When the plastid gene expression-dependent pathway of retrograde signaling is affected by lincomycin inhibition of plastid translation, GUN1-GFP (C-1 and OE-13) strongly accumulates compared with the cpGFP control, whose accumulation is even reduced. The OE-13 line also was investigated under spectinomycin and norflurazon treatment, where it also showed induction of GUN1-GFP accumulation as upon lincomycin treatment. For comparison, all GFP images were taken with exactly the same settings. The experiments were repeated independently at least three times (analyzing more than 20 seedlings for each treatment in each replicate experiment), and representative images are shown here. Note that chlorophyll fluorescence is still detectable at very low levels in the lincomycin and spectinomycin treatments but not in the norflurazon treatment. Bars = 25 µm. D, Western-blot analyses of the expression of the Clp protease components ClpP1 and ClpC under lincomycin (Lin), spectinomycin (Spe), and norflurazon (NF) treatments compared with untreated control plants. RbcL and cpHSC70 were included as control proteins, and actin was used as a loading control. WT, Wild type.
In the wild type, we were unable to detect GUN1 with our mass spectrometry workflow, consistent with its absence from most other proteomic studies conducted in chloroplasts (http://ppdb.tc.cornell.edu/). Considering the comparable mRNA accumulation levels of GUN1 to Tic110 and Tic40, two highly abundant chloroplast proteins (Fig. 4, A and B), this finding provides further evidence for GUN1 being largely regulated at the posttranscriptional level.
Although GUN1-GFP is not detectable by confocal microscopy in differentiated chloroplasts, the protein is detected by mass spectrometry in the complemented lines and the overexpression lines in both seedlings and rosette leaves (Fig. 4, A and B; Supplemental Data Set S1). However, the protein accumulation level is still substantially lower than that of Tic110 or Tic40 and much lower than that of the chaperone proteins ClpC1 and cpHSC70, indicating that GUN1 is either poorly translated or that the GUN1 protein is rather unstable and quickly degraded upon or after import into chloroplasts.
Since GUN1 was proposed to be the central regulator, and possibly the integrator, of the three classical retrograde signaling pathways (Koussevitzky et al., 2007), we next asked how GUN1 protein abundance responds to conditions under which retrograde signaling can be demonstrated. To this end, we treated a complemented line (C-1) and an overexpression line (OE-13) with lincomycin to repress plastid translation, thus inducing the plastid gene expression pathway of retrograde signaling, and then followed the changes in GUN1-GFP accumulation by confocal microscopy (using cpGFP as a control; Fig. 4C). As expected based on the results described above, GUN-GFP is undetectable in the (unstressed) control. However, when retrograde signaling is altered by treatment with lincomycin, GUN1-GFP accumulates strikingly in both lines, resulting in strong GFP fluorescence, while the unfused GFP in the control plants remains unchanged or is even reduced. A similar surge in GUN1 protein accumulation was seen upon treatment with another chloroplast translation inhibitor, spectinomycin. As translation of the essential plastid-encoded catalytic subunit of the Clp protease, ClpP1, is inhibited by lincomycin and spectinomycin (Fig. 4D), increased accumulation of GUN1-GFP under these conditions could be a consequence of reduced Clp protease activity, if GUN1 is a substrate of the Clp protease. To investigate whether the induction of GUN1 also occurs in other conditions that alter retrograde signaling, we analyzed GUN1-GFP accumulation under norflurazon treatment (which interferes with tetrapyrrole-based retrograde signaling). Increased GUN1-GFP accumulation also was seen in the presence of norflurazon (Fig. 4C) but was less pronounced than upon lincomycin or spectinomycin treatment. If one assumes that GUN1 is a substrate of the Clp protease, this difference could be readily explained by the strong repression of the essential plastid-encoded ClpP1 subunit by the chloroplast translation inhibitors lincomycin and spectinomycin (Fig. 4D).
From the above results, it seems reasonable to conclude that the induction of GUN1 accumulation at the protein level enables its central function in the regulation of nuclear gene expression.
GUN1 Is Actively Translated and Its Translation Is Not Stimulated under Conditions That Alter Retrograde Signaling
Although GUN1 is highly expressed at the transcriptional level (Fig. 1), the GUN1 protein accumulates only to extremely low levels (Fig. 4, A and B; Supplemental Data Set S1). Indeed, GUN1 is within the bottom 2% of low-abundance proteins in the protein abundance database PaxDb (ranked 5,649 out of 5,731 proteins measured in juvenile leaves; Wang et al., 2012). By contrast, Tic110 and ClpC1 are within the top 3% of highly abundant proteins, and Tic40 is within the top 8%.
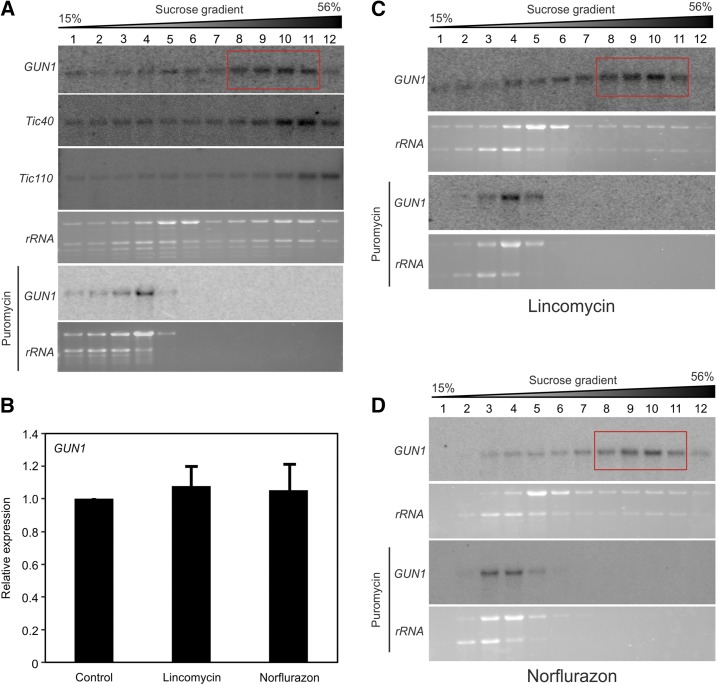
In order to better understand this apparently very strong posttranscriptional regulation of GUN1 expression, we analyzed GUN1 translation by determining polysome-loading profiles (Fig. 5A). The bulk of GUN1 transcripts were found in fractions 8 to 11 of the Suc gradient, which, as evidenced by control gradients centrifuged in the presence of the ribosome-releasing drug puromycin, contain polysomal complexes. Strong ribosome association indicates active translation of the GUN1 mRNA. For comparison, we also determined the polysome profiles of the Tic110 and Tic40 mRNAs. Both transcripts also showed strong association with ribosomes (Fig. 5A), consistent with the encoded proteins being highly abundant.
Figure 5.
Active translation of the GUN1 mRNA and unaltered transcription and translation of GUN1 in the presence of lincomycin or norflurazon. A, Polysome profiles reveal that GUN1 transcripts accumulate mainly in polysome fractions 8 to 11 (red box), indicating that GUN1 is actively translated under unstressed conditions. The polysome profiles of the Tic40 and Tic110 mRNAs are shown for comparison. B, GUN1 mRNA accumulation is not induced upon induction of retrograde signaling by lincomycin or norflurazon treatment. GUN1 expression levels were analyzed by RT-qPCR, and the expression in the presence of lincomycin or norflurazon was compared with the expression in untreated control plants. Data are presented as means ± sd from three biological replicates. C, Polysome profiles of GUN1 upon lincomycin treatment. Compared with untreated plants (A), GUN1 translation is not stimulated by lincomycin and GUN1 transcripts accumulate in the same gradient fractions (with the bulk of the mRNA being in fractions 8–11; red box). D, Polysome profiles of GUN1 upon norflurazon treatment show that GUN1 translation also is not stimulated by norflurazon treatment. In A, C, and D, puromycin-treated samples were analyzed as a control to identify polysome-containing gradient fractions. Images of the rRNAs visualized by ethidium bromide staining of the agarose gels prior to blotting also are shown. The wedges above the blots indicate the increasing Suc concentration across the gradient.
Since the GUN1-GFP protein accumulated under conditions that alter retrograde signaling (Fig. 4C), we wanted to examine whether this induction results from the up-regulation of GUN1 expression at the transcriptional or translational level. RT-qPCR assays revealed that GUN1 transcription is not induced by lincomycin or norflurazon treatment (Fig. 5B). Likewise, when the translation of GUN1 upon lincomycin (Fig. 5C) or norflurazon (Fig. 5D) treatment was analyzed, the distribution of GUN1 transcripts across the fractions of the polysome gradient was found to be very similar to that seen in the untreated control (Fig. 5A). These results suggest that the induction of GUN1 does not occur through up-regulated transcription or increased translation.
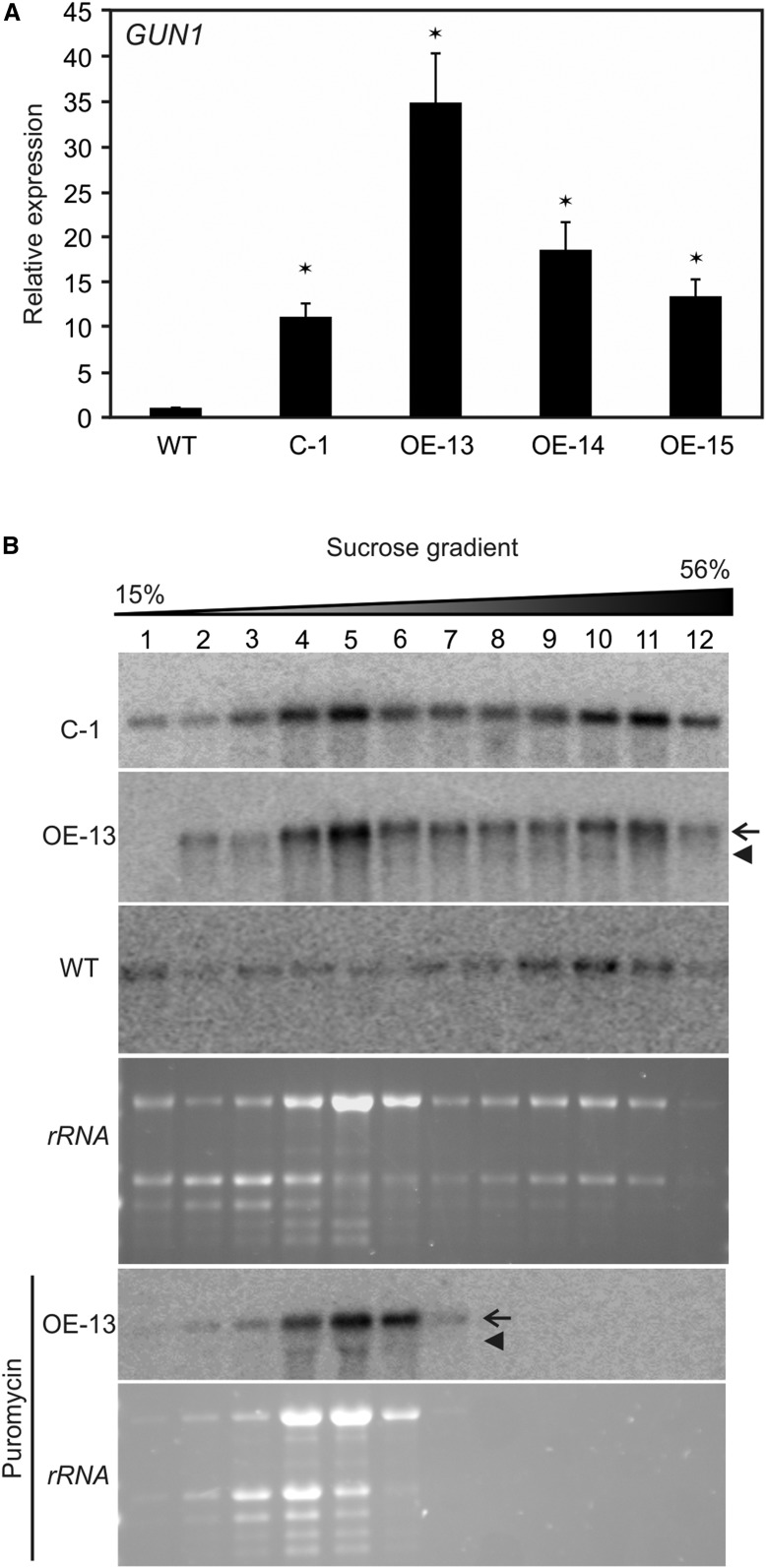
The CaMV 35S promoter was used to drive GUN1-GFP expression in the complementation and overexpression lines, resulting in high transcription of the GUN1-GFP gene (Fig. 6A). The polysome profiles determined for the C-1 and OE-13 lines revealed that a large proportion of the GUN1-GFP transcripts accumulated in gradient fractions 4 and 5, which contain mainly free mRNA that is not loaded with ribosomes (as evidenced by the puromycin control) and, hence, is translationally inactive (Fig. 6B). The presence of a large pool of untranslated mRNAs is even more pronounced in OE-13 than in C-1 (Fig. 6B), which is consistent with the stronger overexpression of GUN1-GFP at the transcript level in line OE-13 (Fig. 6A).
Figure 6.
RT-qPCR analysis of GUN1 expression and polysome profile analysis of complemented (C) and overexpression (OE) lines reveals a negative correlation of GUN1 translation efficiency with mRNA abundance. A, RT-qPCR analysis of GUN1 expression in complemented line C-1 and three overexpression lines (OE-13, OE-14, and OE-15). The expression of GUN1 was compared with that in the wild type (WT) and is presented as means ± sd from three biological replicates. Significant differences from the wild type are indicated (*, P < 0.01). B, Polysome profiles of GUN1 in complemented line C-1 and overexpression line OE-13 reveal down-regulated translation efficiency in these transgenic lines. A representative puromycin-treated sample (from OE-13) is shown as a control to identify polysome-containing gradient fractions. Arrows and arrowheads mark the GUN1-GFP mRNA and the native GUN1 mRNA, respectively. Representative images of rRNAs visualized by ethidium bromide staining of agarose gels prior to blotting also are shown. The wedge above the blots indicates the increasing Suc concentration across the gradient. Note that the bulk of the GUN1-GFP transcripts in the strongest overaccumulator (OE-13) remains untranslated and accumulates in the light gradient fractions (4–6) that contain ribosome-free mRNAs.
GUN1 Is an Unstable Protein with a Very Short Half-Life, and Its Degradation Depends on ClpC1
Our data described above prompted us to investigate the turnover of the GUN1 protein to examine whether GUN1 abundance is regulated at the level of protein stability.
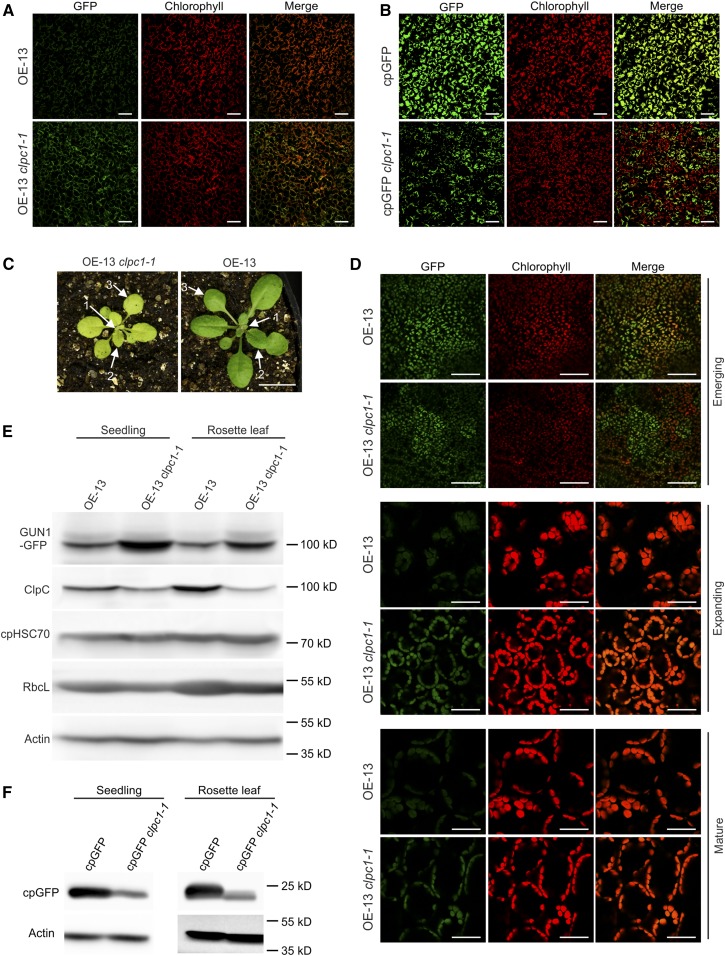
Considering the pivotal role of the Clp protease in protein homeostasis in the chloroplast stroma (van Wijk, 2015) and the fact that GUN1-GFP strongly accumulates upon lincomycin or spectinomycin treatment (conditions that inhibit the translation of clpP1; Fig. 4, C and D), GUN1 could be a substrate protein of the Clp protease. To explore this possibility, we crossed the GUN1-GFP-expressing line OE-13 with the clpc1-1 mutant and, as a control, also crossed the cpGFP transgenic line with clpc1-1. ClpC1 is the major chaperone that unfolds and delivers protein substrates to the Clp protease core complex for degradation (Sjögren et al., 2014). Interestingly, overaccumulation of GUN1-GFP in the clpc1 background was observed by confocal microscopy (in cotyledons of 7-d-old seedlings; Fig. 7A). By contrast, cpGFP is down-regulated in the clpc1 background (Fig. 7B). This finding was further confirmed by western-blot analyses (Fig. 7, E and F). Due to the up-regulation of ClpC2 in the clpc1 mutant (Sjögren et al., 2014), the mutant still accumulates ClpC (to ∼25% of the wild-type level; Fig. 7E). GUN1-GFP overaccumulates in the clpc1-1 mutant, while RbcL is down-regulated and cpHSC70 is not changed significantly, either in seedlings or in rosette leaves (Fig. 7E). Interestingly, both GUN1-GFP and cpGFP are not expressed visibly in some mesophyll cells of the clpc1-1 mutant (Fig. 7, A and B). Considering the dual function of ClpC1 in substrate delivery to the Clp protease and in protein import, these nonexpressing cells might suffer from an import defect (leading to rapid degradation of the unimported precursor proteins by the cytosolic protein quality control system). Furthermore, we analyzed GUN1-GFP accumulation at different stages of rosette leaf development in both the wild-type background (OE-13) and the clpc1-1 background (OE-13 clpc1-1; Fig. 7, C and D). While the accumulation of GUN1-GFP is similarly high in the basal part of newly emerging leaves (Fig. 7D, top section), it is hardly detectable in expanding and mature OE-13 leaves. By contrast, GUN1-GFP still accumulates to substantial levels in expanding and mature OE-13 clpc1-1 leaves (Fig. 7D, middle and bottom sections), indicating that it cannot be degraded efficiently in the absence of ClpC1.
Figure 7.
GUN1 overaccumulates in the clpc1 mutant. A, Confocal microscopy analysis of GUN1-GFP accumulation in cotyledons from 7-d-old seedlings in the wild-type background (OE-13) or the clpc1-1 background (OE-13 clpc1-1). GFP images were taken with the same setting in both genotypes to allow for direct comparison. Bars = 100 µm. B, Analysis of the accumulation of chloroplast-localized unfused GFP (cpGFP; targeted to plastids by the RBCS transit peptide) in cotyledons from 7-d-old seedlings in the wild-type background (cpGFP) or the clpc1-1 background (cpGFP clpc1-1). The cpGFP expressed in the wild-type background was crossed into the clpc1-1 mutant to generate exactly the same transgenic insertion. GFP images were taken with the same setting in both genotypes to allow for direct comparison. Note that cpGFP is not visibly expressed in all cells of the clpc1-1 mutant. Bars = 100 µm. C, OE-13 and OE-13 clpc1-1 plants at the age of 3 weeks. Numbers and arrows indicate the rosette leaves from which the images in D were taken. Bar = 1 cm. D, Confocal microscopy analysis of GUN1-GFP expression in rosette leaves at different developmental stages in OE-13 and OE-13 clpc1-1. Three different developmental stages of rosette leaves were analyzed: a newly emerging leaf (leaf 1 in C), an expanding leaf (leaf 2 in C), and a mature leaf (leaf 3 in C). The images were taken from the basal part of each leaf. More than 10 individual plants were analyzed with comparable results, and representative images are shown. Bars = 25 µm. E, Immunoblot analysis of GUN1-GFP accumulation in 7-d-old seedlings and rosette leaves of OE-13 and OE-13 clpc1-1. Rosette leaves similar to leaf 3 in C were harvested and subjected to western-blot analysis. ClpC is still detectable in the clpc1-1 mutant due to expression of the ClpC2 gene. The expression of RbcL and cpHSC70 also was analyzed. Actin was used as a loading control. F, Immunoblot analysis of cpGFP accumulation in 7-d-old seedlings or rosette leaves in the wild-type background (cpGFP) and in the clpc1-1 background (cpGFP clpc1-1). The accumulation of cpGFP in clpc1-1 is reduced, because not all cells faithfully express cpGFP (compare with B).
Taken together, the induction of GUN1 accumulation in response to ClpC1 deficiency may suggest that GUN1 is a genuine substrate of the Clp protease in that GUN1 degradation requires recognition and/or unfolding by ClpC1.
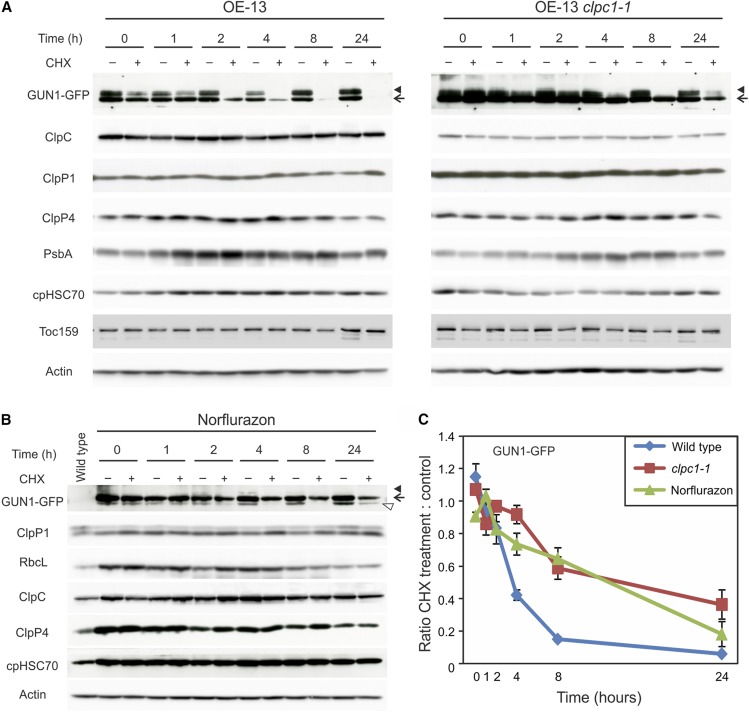
To further investigate the turnover of GUN1 and its possible control by ClpC1 and the Clp protease, we performed cycloheximide chase assays to determine the half-life of the GUN1 protein (Supplemental Fig. S3). The degradation of GUN1 was followed by western blotting upon the inhibition of cytosolic translation with cycloheximide (resulting in blocked de novo synthesis of GUN1) and comparison of the wild-type background with the clpc1-1 mutant background. The level of the GUN1 protein started to decrease substantially from the 2-h time point on in the cycloheximide-treated plants, fell to approximately 40% of the untreated control after 4 h, and was undetectably low 24 h after the inhibition of de novo protein synthesis (Fig. 8, A, left, and C). The clpc1 mutant still accumulates ∼25% of ClpC protein, due to the known up-regulation of ClpC2 in response to the loss of ClpC1 (Fig. 8A; Sjögren et al., 2014). Interestingly, the degradation of GUN1 in the clpc1-1 background is much slower than in the wild type. Protein levels were still largely unchanged after 4 h of cycloheximide treatment and decreased relatively mildly to approximately 60% after 8 h and to 40% after 24 h (Fig. 8, A, right, and C). Interestingly, in addition to the main signal, a larger double band was detected for GUN1-GFP in all blots. The identities of the less abundant larger bands are currently unknown and will need to be investigated further. Other nucleus-encoded chloroplast proteins (cpHSC70, ClpP4, and Toc159) analyzed as controls were relatively stable and did not show pronounced changes within the 24-h time course (Fig. 8A). Most importantly, free cpGFP also is stable and did not show differences between the wild type and the clpc1 mutant over the 24-h time course, arguing against a possible destabilizing effect of the GFP fusion on GUN1 protein stability (Supplemental Fig. S4A). Also, the fact that the GUN1-GFP fusion protein fully complements the gun1 mutant phenotype and the retrograde signaling deficiency of the mutant (Fig. 2, C and D) indicates that the fusion protein is functional and faithfully regulated in planta.
Figure 8.
GUN1 is a short-lived protein, and its degradation is controlled by ClpC1. A, Cycloheximide (CHX) chase assays show that GUN1 is a protein with a very short half-life. Seven-day-old seedlings grown on plates were transferred to liquid medium with or without CHX, and samples were collected at different time points (compare with Supplemental Fig. S3). GUN1-GFP and various other proteins were then analyzed by western blotting in the wild-type and the clpc1-1 mutant backgrounds with (+) or without (−) CHX treatment. As CHX inhibits only cytosolic translation but not protein synthesis in the chloroplast, plastid genome-encoded proteins (PsbA and ClpP1) can serve as loading controls. Nucleus-encoded chloroplast proteins (cpHSC70, ClpP4, and Toc159) and a cytosolic housekeeping protein (Actin) also were analyzed in comparison with GUN1-GFP. Note that some ClpC protein is still detected in the clpc1-1 background because of the presence of a second gene copy (ClpC2). Note that, in addition to the main band of GUN1-GFP (marked with arrows), a larger double band (arrowheads) is detected (see Fig. 10), the identity of which is currently unknown. The extra bands are seen in all western blots for the full-length GUN1-GFP and the different truncations (with the exception of the GT-5 line; compare with Fig. 9B) and, therefore, represent specific signals. They could, for example, represent posttranslationally modified forms of GUN1. Only the main band was used for quantification. B, The degradation of GUN1 is slowed down under norflurazon treatment, a condition known to alter retrograde signaling. Seeds were germinated and grown on plates containing 5 µm norflurazon for 5 d in the dark followed by 2 d in continuous light and then transferred to liquid medium supplemented with 5 µm norflurazon or 5 µm norflurazon and 10 µm CHX for different times. The larger double band of GUN1-GFP (arrowhead) is less abundant under norflurazon treatment; instead, an additional smaller band (marked with an open triangle) is seen below the main band (arrow), which could be a degradation product. Only the main band was used for quantification. C, Comparison of the degradation rates of GUN-GFP in the wild type, the clpc1-1 mutant, and the wild type under norflurazon treatment. The band intensities in western-blot analyses of three biological replicates (as in A and B) were determined and normalized to the analyzed chloroplast-encoded proteins (PsbA or, in the norflurazon experiment, RbcL, since PsbA is destabilized by the norflurazon-mediated block in carotenoid biosynthesis). The ratio in protein abundance upon CHX treatment to the untreated control at each time point is shown as means ± se from three biological replicates.
A recent study employing 15N labeling determined the degradation rate of 1,228 proteins in Arabidopsis rosette leaves (Li et al., 2017). We analyzed the half-lives of different chloroplast proteins in the data set and compared them with the results of our cycloheximide chase assays (Supplemental Fig. S4, B and C). Tic110, Tic40, ClpC1, and cpHSC70-1 have similar half-lives of 2 to 3 d, while actin (∼5 d) and RbcL (∼18 d) were more stable (Li et al., 2017). Consistent with these data, Tic40 and cpHSC70 did not show a visible decrease in our cycloheximide chase assays over 24 h, and Tic110 showed a moderate decrease only after 24 h (Supplemental Fig. S4C). The proteins with the highest turnover identified by Li et al. (2017) were the chloroplast thiamine biosynthesis enzyme THI1, β-amylase3 (BAM3), and the D1 protein of PSII, with half-lives of 8 to 12 h (0.3–0.5 d; Supplemental Fig. S4B). The half-life of GUN1 of ∼4 h determined by our cycloheximide chase assay (Fig. 8C) is even shorter, indicating an exceptionally high turnover of GUN1 in chloroplasts and identifying GUN1 as one of the most unstable plant proteins known to date.
Since GUN1 is induced under conditions when retrograde signaling can be demonstrated (Fig. 4C) and its increased protein accumulation is neither caused by elevated mRNA accumulation nor by enhanced translation (Fig. 5), we next wanted to examine whether altered retrograde signaling would slow down GUN1 degradation. Therefore, we analyzed the stability of GUN1-GFP under norflurazon treatment (Fig. 8, B and C; Supplemental Fig. S3B). Indeed, compared with normal growth conditions (Fig. 8A, left), GUN1 degradation is strongly reduced upon treatment of seedlings with norflurazon (Fig. 8B) and the degradation rate of GUN1 is now similar to that in the clpc1 mutant background (Fig. 8, A and C).
The PPR Motifs Are Key Determinants of GUN1 Instability
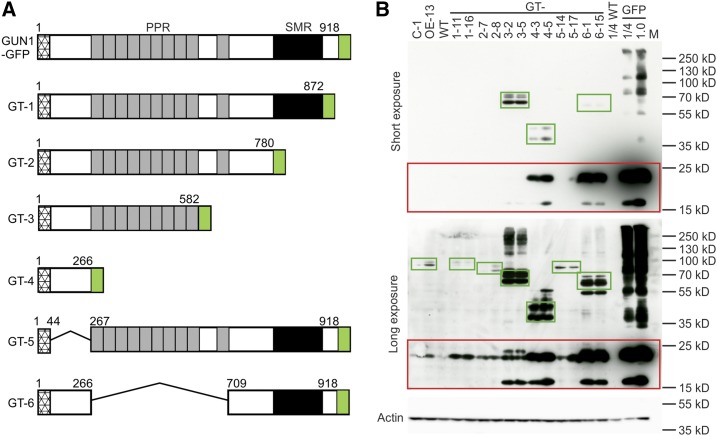
In order to identify which part of the GUN1 protein harbors the degradation signals, a series of truncations were constructed and tested in vivo by stable transformation into the gun1 mutant background (Fig. 9A). Transgenic lines expressing the different truncations were screened by western blotting, and three independent lines per construct were selected for further analysis (Supplemental Fig. S5). Evaluation of protein accumulation in the different transgenic lines (referred to as GT-1 [for GUN1 Truncation1] to GT-6; Fig. 9A) revealed not only substantial differences in protein accumulation levels but also the appearance of free GFP as a stable degradation product of the fusion proteins. Accumulation of free GFP was particularly high in the GT-4 and GT-6 transgenic lines (Fig. 9B).
Figure 9.
Serial truncations of GUN1 identify domains determining protein instability. A, Schematic representation of the GUN1-GFP fusion protein and the six different truncations constructed. The hatched boxes indicate the transit peptide as predicted by ChloroP. The PPR motifs are represented as gray boxes, and the SMR domain is shown as black boxes. GFP was fused to the C terminus of each protein variant and is shown in green. The numbers above each diagram indicate the number of amino acids included in the truncated protein (e.g. for GT-1, amino acids 1–872 were fused to GFP and amino acids 873–918 were deleted). B, Immunoblot analysis of the expression of the different GUN1 truncations. Two independent transgenic lines for each truncation construct were analyzed. C-1, OE-13, and the wild type (WT) were included as controls. The GT-GFP fusion protein and the bands representing free GFP (that likely arises from degradation of the fusion protein, especially in the GT-4 and GT-6 lines) are boxed in green and red, respectively. A chloroplast-localized GFP (targeted to plastids by the RBCS transit peptide; GFP) was loaded as a control for the free GFP released upon GUN1 degradation. An overexposure of the blot (at bottom) is shown to visualize low-abundance fusion proteins. As a control for equal loading, the membrane was stripped and reprobed with an anti-actin antibody.
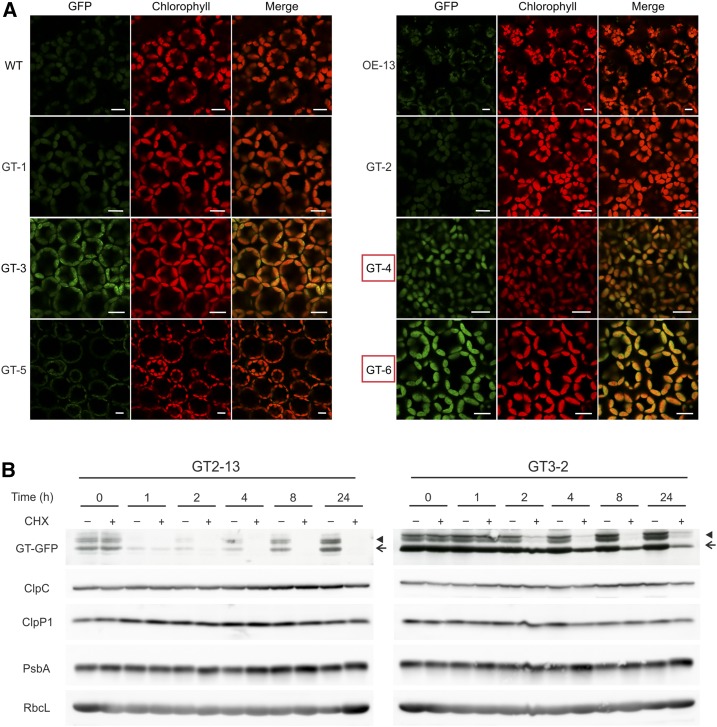
The truncations GT-1 (lacking the C-terminal region downstream of the SMR domain), GT-2 (additionally lacking the SMR domain), and GT-5 (lacking the N-terminal domain downstream of the transit peptide for protein import into chloroplasts and upstream of the PPR motifs; Fig. 9A) do not have pronounced effects on protein stability, in that protein accumulation was similarly low to that of the full-length GUN1-GFP fusion protein (which is undetectable by confocal microscopy in differentiated chloroplasts but detectable as a weakly hybridizing band by western blotting; Figs. 9B and 10A). Although a signal for free GFP as a likely degradation intermediate of the GUN1-GFP fusion protein can be detected in the C-1 and OE-13 lines (as well as in the GT-1, GT-2, and GT-5 lines) by immunoblotting, no GFP fluorescence can be seen in these lines (Fig. 10A), ruling out the possibility that the GUN1-GFP signal during early chloroplast biogenesis (Fig. 3, A and B) and in conditions that alter retrograde signaling (Fig. 4C) comes from the degradation intermediates.
Figure 10.
Expression and chloroplast localization of different truncations of the GUN1-GFP fusion protein and cycloheximide chase assays for GT-2 and GT-3. A, Confocal microscopy images show the expression and chloroplast localization of different truncations of the GUN1-GFP fusion protein. For GT-1, GT-2, and GT-5 as well as the full-length GUN1-GFP fusion (OE-13), no above-background GFP fluorescence was observed, although the fusion proteins can be weakly detected on western blots (compare with Fig. 9). The protein in GT-3 (truncated after the tandem PPR motifs) shows a punctate distribution. GFP fluorescence in GT-4 and GT-6 (boxed in red) comes mainly from free (soluble) GFP that accumulates as a degradation intermediate of the fusion protein (compare with Fig. 9B). WT, Wild type. Bars = 10 µm. B, Cycloheximide (CHX) chase assay of GT-2 and GT-3 transgenic lines. Compared with the GT-2 line (expressing a GUN1 protein variant that lacks the SMR domain and the downstream C-terminal sequence), the instability of the truncated GUN1-GFP protein in line GT-3 (additionally lacking the last PPR motif) is reduced. ClpC and ClpP1 also were analyzed to demonstrate that there is no obvious difference in the abundance of the Clp protease between the different samples. Chloroplast-encoded proteins (PsbA and RbcL) were included as loading controls, as cycloheximide only inhibits cytosolic translation. The arrows and arrowheads mark the main GUN1-GFP band and the additional larger double band, respectively.
By contrast, further truncation removing the last PPR motif and the spacer sequence to the upstream PPR tracts (GT-3) strongly reduces the instability of the protein. It accumulates to much higher levels than the full-length protein (Fig. 9B) and is readily detectable by confocal microscopy (Fig. 10A). Complete removal of the PPR motifs in GT-4 and GT-6 lines also led to stabilization of the protein (Figs. 9 and 10), thus suggesting an important role of the PPR motifs in determining the turnover rate and, in this way, regulating GUN1 accumulation in vivo. Large amounts of free GFP accumulate as degradation products in the GT-4 and GT-6 lines (Fig. 9B) and are likely responsible for the strong GFP signal in these plants (Fig. 10A). In agreement with this assumption, the GFP signal intensity in GT-4 and GT-6 (Fig. 10A) is positively correlated with the intensity of the free GFP bands but negatively correlated with the abundance of the fusion protein (Fig. 9B).
To further prove the key role of the PPR motifs in determining GUN1 protein stability, we performed cycloheximide chase assays with GT-2 (lacking the SMR domain and the downstream C-terminal sequence) and GT-3 (harboring C-terminally truncated PPR motifs) to track their degradation (Fig. 10B). While GT-2 shows a very similar degradation pattern to the full-length GUN1 (Fig. 8A, left), protein instability is reduced in GT-3 (Fig. 10B). This finding is consistent with an important role of the PPR motifs in conferring instability to the GUN1 protein.
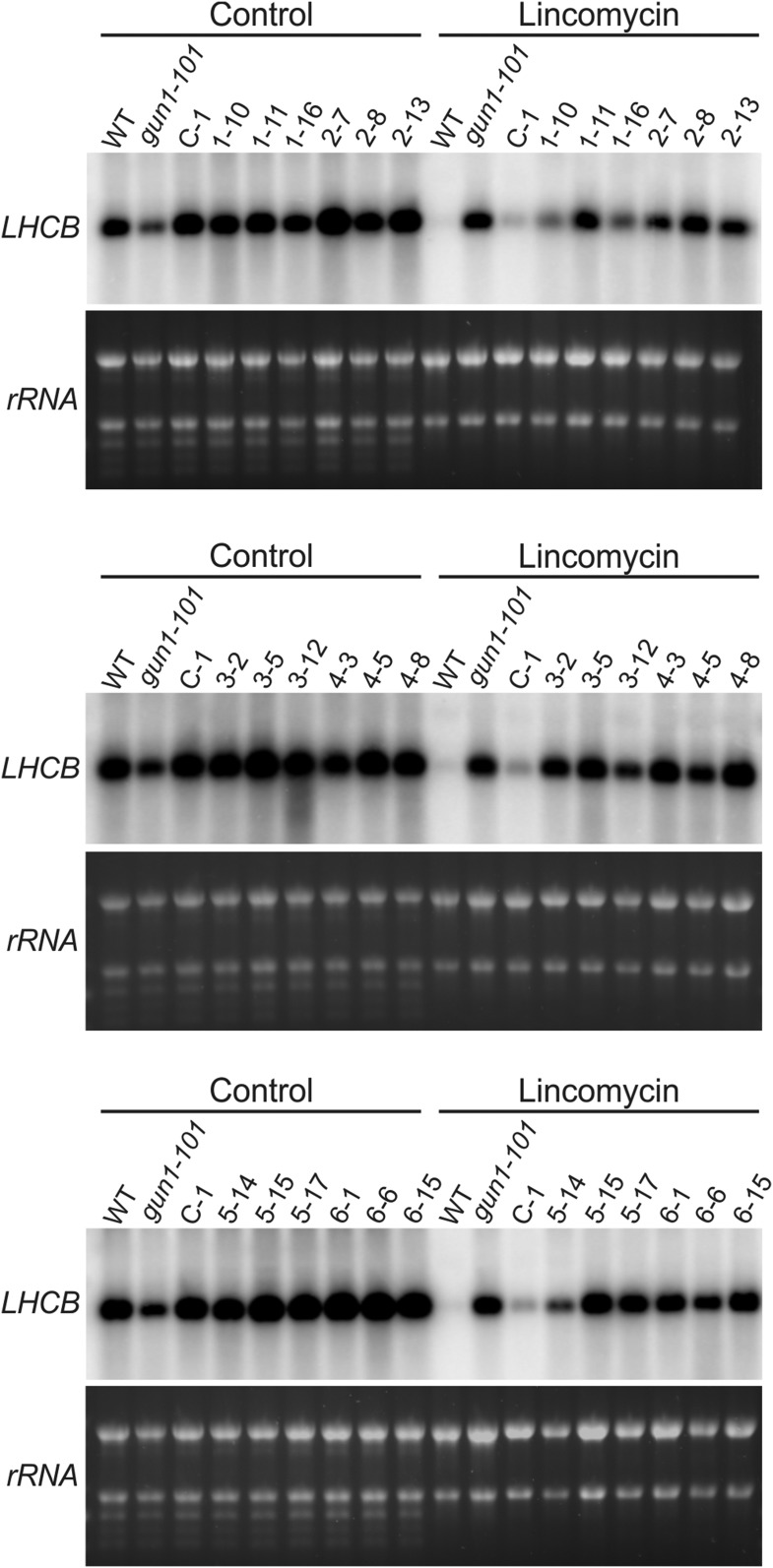
To assess the contribution of the different protein domains to the activity of GUN1 in the regulation of retrograde signaling, three independent transgenic lines from each truncation construct were subjected to northern-blot analysis (Fig. 11) to test for restored repression of LHCB expression upon lincomycin treatment. In contrast to the full-length GUN1-GFP that fully complemented the defective retrograde signaling phenotype of the gun1 mutant, none of the truncations was able to complement the mutant, with the exception of GT-1, which displayed partial complementation in two independent lines (1-10 and 1-16; Fig. 11). This result indicates that both the PPR motifs and the SMR domain are required for GUN1 function in retrograde signaling.
Figure 11.
Complementation analyses of the LHCB expression phenotype of the gun1 mutant with the different truncation constructs. Three transgenic lines for each GUN1 truncation construct (indicated by asterisks in Supplemental Fig. S5) were grown in the presence or absence of lincomycin, and the expression of the LHCB gene was analyzed by northern blotting. The gun1-101 mutant and the complemented line C-1 were used as negative and positive controls, respectively. The rRNA bands on the ethidium bromide-stained agarose gel prior to blotting are shown as controls for equal loading. WT, Wild type.
Overexpression of GUN1 Confers Early Flowering
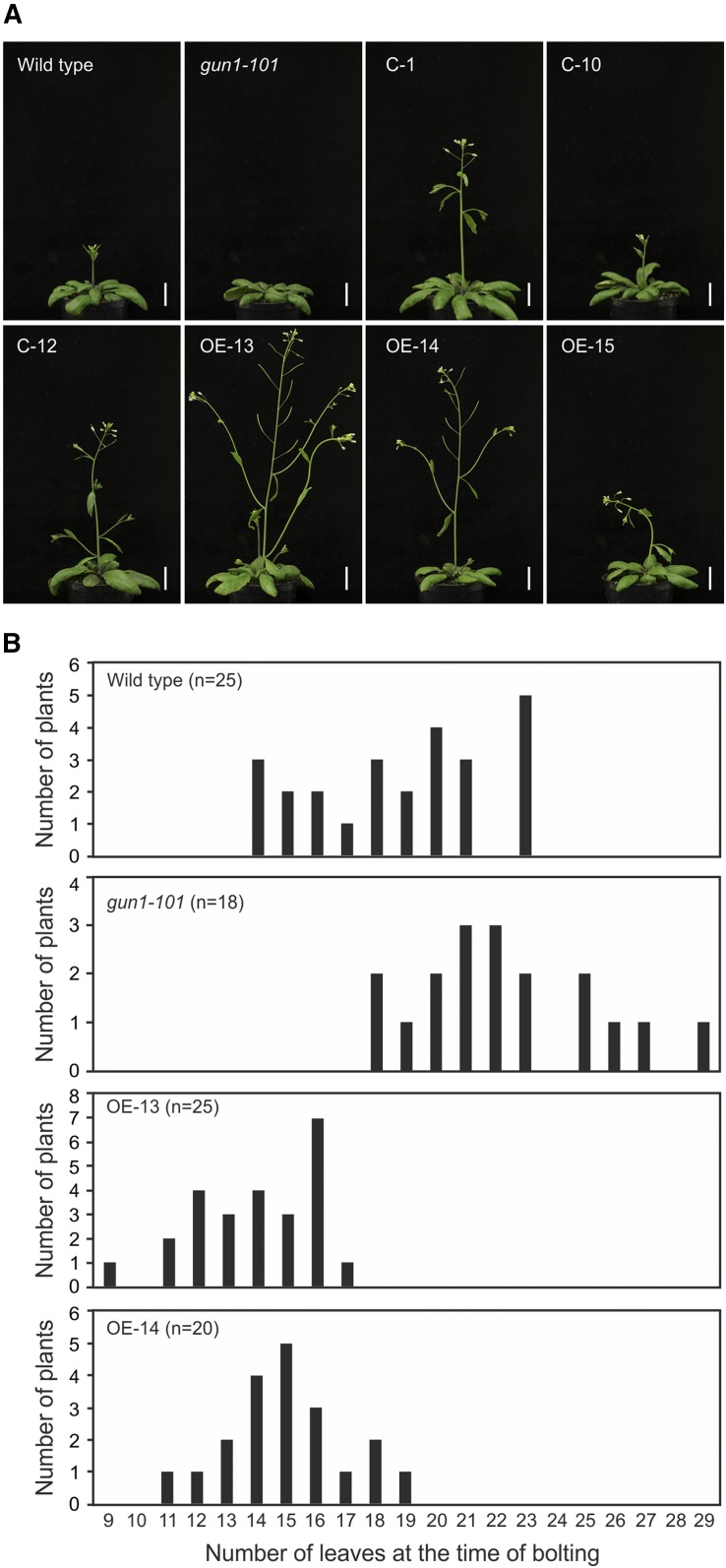
Having established overexpression lines for GUN1, we became interested in determining whether GUN1 participates in other developmental processes besides the regulation of early chloroplast biogenesis. This question was motivated by our observation that gun1 mutant plants showed a mild delay in flowering (Fig. 12).
Figure 12.
Overexpression of GUN1 causes an early flowering phenotype. A, Flowering phenotypes of the wild type, the gun1-101 mutant, three complemented lines (C), and the overexpression lines (OE) of GUN1. Plants were grown for 32 d in long-day conditions and then photographed. The CaMV 35S promoter was used to drive GUN1 transgene expression in both the C and OE lines. Note that the early flowering phenotype is correlated with the strength of the GUN1-GFP overexpression (compare with Fig. 6A). Bars = 2 cm. B, Histograms showing the flowering times of the wild type, the gun1-101 mutant, and two strong overexpression lines (OE-13 and OE-14; Fig. 6A). The y axis gives the number of plants that flowered with a given number of leaves. The x axis indicates the number of leaves at the time of bolting.
Interestingly, overexpression of GUN1 caused an early flowering phenotype in Arabidopsis (Fig. 12). Both overexpression lines analyzed (OE-13 and OE-14) flowered significantly earlier than the wild type (Fig. 12A), a phenotype that was further confirmed by their lower number of leaves upon bolting (Fig. 12B). The strength of the phenotype was correlated with the expression level of GUN1 (Fig. 6A), in that the strongest overexpressor (OE-13) showed the strongest acceleration of floral induction (Fig. 12B). The complemented lines also flowered earlier than the wild type (Fig. 12A), because the strong CaMV 35S promoter drives the GUN1 expression also used in these lines.
Abiotic stresses, such as nutrient deficiency, high light, high temperature, and continuous light, can trigger early flowering, presumably to ensure reproductive success also under adverse environmental conditions (Levy and Dean, 1998). Although chloroplast retrograde signals have been implicated in the promotion of flowering by high light (Feng et al., 2016), these results were recently refuted by the demonstration that the suggested functional connection (the transcription factor PTM) plays no role in retrograde signaling (Page et al., 2017). In an attempt to resolve this controversy, we sought to examine whether GUN1 participates in high-light-induced early flowering. To this end, we treated the wild type, the gun1 knockout mutant, and two strong overexpression lines (OE-13 and OE-14) with high light (Supplemental Fig. S6). Although high light accelerated flowering in all genotypes relative to standard light conditions (Supplemental Fig. S6), the differences between the genotypes under standard conditions (Fig. 12) remained unchanged, indicating that GUN1-mediated retrograde signaling does not specifically function in high-light-induced early flowering.
DISCUSSION
While the nuclear control of organelle function, also referred to as anterograde communication, is a long-established concept, the retrograde control of nuclear gene expression by the organelles was recognized only much later as a general phenomenon in eukaryotes. All nonphotosynthetic eukaryotes, from unicellular fungi to humans, utilize retrograde signaling pathways to adjust nuclear gene expression to mitochondrial physiology (Jia et al., 1997; Liu et al., 2003; Butow and Avadhani, 2004; Battersby and Richter, 2013). Likewise, all photosynthetic eukaryotes, from unicellular algae to seed plants, have implemented additional retrograde pathways to tune nuclear gene expression not only to mitochondrial but also to plastid demands (von Gromoff et al., 2008; Woodson and Chory, 2008; Kleine et al., 2009; Van Aken and Whelan, 2012; De Clercq et al., 2013; Ng et al., 2013).
Chloroplast retrograde signals regulate photomorphogenesis via the CRY1 and PIF-GLK1 transcriptional networks (Ruckle et al., 2007; Waters et al., 2009; Martín et al., 2016) and also fine-tune responses to environmental perturbations such as drought and high light (Lee et al., 2007; Estavillo et al., 2011; Xiao et al., 2012; Jung et al., 2013; Pornsiriwong et al., 2017). Out of the six classical gun mutants, five (gun2–gun6) are affected in genes for enzymes or coenzymes in the tetrapyrrole biosynthetic pathways (Mochizuki et al., 2001; Larkin et al., 2003; Strand et al., 2003; Woodson et al., 2011). These findings suggested very strongly that defects in photosynthetic pigment accumulation affect chloroplast retrograde signaling to adjust nuclear gene expression and development. Genetic analyses and identification of the sixth GUN gene, GUN1, led to the hypothesis that GUN1 represents (or is part of) a central integrator at which the three classical retrograde signaling pathways converge (Koussevitzky et al., 2007). More recently, it was suggested that GUN1 might play a role in regulating chloroplast protein homeostasis (Colombo et al., 2016; Llamas et al., 2017).
GUN1 is a very low-abundance protein (http://ppdb.tc.cornell.edu/). To better understand the role of GUN1 in the control of retrograde signaling and protein homeostasis, we here studied the regulation of the expression of the GUN1 gene and the possible role of GUN1 in the regulation of chloroplast biogenesis and phase transitions during plant development. GUN1 is transcribed at high levels in different tissues and across developmental stages (Fig. 1), with expression being highest in young rosette leaves, a finding that would be compatible with a function of GUN1 in early chloroplast development (Fig. 1B). Interestingly, when labeled with GFP, the GUN1 protein can only be detected in cotyledons during the early stages of seedling development, in leaf primordia of the shoot apical meristem, and in the base of newly emerging, very small leaflets (of approximately 1 mm length), where active plastid differentiation into chloroplasts takes place (Fig. 3, A–D). The GFP signal decreases along the gradient of chloroplast differentiation and becomes undetectable in green cotyledons, in the (more mature) tip region of the very young leaflets, and in all other tissues. This expression pattern is compatible with GUN1 functioning in the regulation of chloroplast development, a conclusion that gains further support from (1) the observed defects in chloroplast biogenesis in cotyledons of seedlings challenged by mild environmental stress (Fig. 2D), (2) the delayed greening of cotyledons during germination (Fig. 2, E and F), and (3) the retarded chloroplast differentiation during the early stages of greening upon deetiolation of dark-grown seedlings (Mochizuki et al., 1996). Taken together, these observations support an important role of GUN1 in the biogenic control exerted by retrograde signaling.
GUN1 was demonstrated to be a very-low-abundance protein under unstressed conditions. It is hardly detectable by proteomics (Fig. 4, A and B) and within the bottom 2% of the Arabidopsis proteome sorted by protein abundance (PaxDb; Wang et al., 2012). However, the GUN1 gene is highly expressed at the transcriptional level (Fig. 1) and also is actively translated (Fig. 5A). This suggested that GUN1 protein abundance is regulated mainly posttranslationally, at the level of protein turnover. Therefore, we determined the half-life of GUN1 by cycloheximide chase assays and found it to be very short, ∼4 h. Li et al. (2017) previously characterized the degradation rate of 1,228 proteins in Arabidopsis rosette leaves using progressive 15N labeling and linked protein abundance positively with protein half-life. Among the 1,228 proteins measured, the most unstable protein was THI1, an enzyme in vitamin metabolism, with a half-life of 0.36 d (8.6 h). Although a different approach was used in our study to determine protein half-lives, the protein degradation rates measured with our cycloheximide chase assays are comparable with those measured in 15N-labeling experiments (Supplemental Fig. S4, B and C). Compared with the most labile protein identified previously (THI1; Li et al., 2017), GUN1 is even less stable, thus providing a likely explanation for its low abundance (Fig. 4, A and B; Supplemental Data Set S1). GUN1 remains low in abundance under unstressed conditions when its function may be dispensable, as also suggested by the lack of a visible phenotype of mature gun1 mutant plants under normal growth conditions. The low abundance of GUN1 is likely achieved by its rapid degradation, which is likely initiated by its delivery to the Clp protease complex through ClpC1, as evidenced by the slowed-down degradation (Fig. 8, A and C) and overaccumulation of GUN1-GFP in the clpc1 mutant (Fig. 7). This is well in line with the stronger accumulation of GUN1-GFP in the presence of lincomycin or spectinomycin compared with norflurazon (as revealed by confocal microscopy; Fig. 4C), because both lincomycin and spectinomycin inhibit the synthesis of the chloroplast-encoded essential catalytic subunit of the Clp protease, ClpP1.
Interestingly, GUN1 protein accumulation is strongly induced under conditions in which retrograde signaling can be demonstrated (Fig. 4C). This does not occur through transcriptional or translational up-regulation (Fig. 5, B–D) but, instead, through slowed-down protein degradation (Fig. 8, B and C). Chloroplasts have been proposed to act as a central sensor of environmental stress conditions (Lee et al., 2007; Padmanabhan and Dinesh-Kumar, 2010; Šimková et al., 2012; de Torres Zabala et al., 2015). Employing the posttranslational regulation of protein stability as a mechanism of stress adaptation offers the potential advantage of facilitating very fast responses to environmental stimuli. Ceased degradation of short-lived proteins may be the most rapid way of readjusting the cellular concentration of important regulator proteins without the need for de novo induction of gene expression. This would enable GUN1 to quickly generate the retrograde signals required to tune nuclear gene expression and adjust chloroplast protein homeostasis (Colombo et al., 2016; Llamas et al., 2017).
The expression of a series of truncated GUN1 variants in planta has provided first insights into the determinants of GUN1 protein (in)stability (Figs. 9 and 10). Removal of the last PPR motif (in GT-3) or complete removal of the PPR domains (GT-4 and GT-6; Fig. 9) results in increased accumulation of the protein (Fig. 9B), while all deletions that do not affect the PPR motifs (GT-1, GT-2, and GT-5) do not result in protein stabilization. These results indicate that the PPR motifs may mediate the rapid degradation of GUN1, perhaps by being the domains recognized by ClpC1, a chaperone crucially involved in unfolding and delivering substrate proteins to the Clp protease. This conclusion was further supported by comparison of the degradation of GT-2 and GT-3 in cycloheximide chase assays. While GT-2 (which lacks the SMR domain and the downstream C-terminal region) is degraded at a very similar rate to the full-length GUN1 protein, additional removal of the last PPR motif stabilizes the protein (GT-3; Fig. 10B).
Most PPR proteins characterized thus far act as regulators of organellar gene expression (Lurin et al., 2004; Schmitz-Linneweber and Small, 2008; Barkan and Small, 2014). Although PPRs usually confer RNA-binding activity (Small and Peeters, 2000), accumulating evidence suggests the ability of at least some PPR-like domains to mediate protein-protein interactions (Bentolila et al., 2012; Spåhr et al., 2016; Andrés-Colás et al., 2017; Guillaumot et al., 2017), similar to the related tetratricopeptide repeat motifs (Goebl and Yanagida, 1991; Das et al., 1998; Allan and Ratajczak, 2011). Since GUN1 is most likely not a nucleic acid-binding protein (Tadini et al., 2016), its PPR tracts may represent another example of PPR motifs engaging in protein-protein interactions (Bentolila et al., 2012; Spåhr et al., 2016; Andrés-Colás et al., 2017).
The gun1 mutant shows no visible phenotype under normal growth conditions but displays a delay in cotyledon greening during germination (Fig. 2, E and F), develops a pale cotyledon phenotype under mild stress (Fig. 2D), and is hypersensitive to Suc and the plant stress hormone abscisic acid (Cottage et al., 2010). These phenotypes are in line with a role of GUN1 in the regulation of plant development and responses to abiotic stress. Interestingly, the gun1 mutant displays delayed floral induction, while the overexpression of GUN1 causes early flowering (Fig. 12). These reciprocal phenotypes of the gun1 mutant and the overexpression lines suggest an involvement of GUN1 in the timing of the vegetative-to-reproductive phase transition. Whether the involvement of GUN1 also is indicative of an involvement of retrograde signaling in the regulation of flowering time remains to be determined. Although a role of chloroplast retrograde signals in the regulation of high light-induced early flowering was proposed and the activation of FLOWERING LOCUS C was suggested as a possible mechanism (Feng et al., 2016), the connection between retrograde signaling and high light-induced flowering was refuted by a recent study (Page et al., 2017) and also is not supported by our data presented here (Supplemental Fig. S6).
In view of the important role of GUN1 in retrograde signaling from the plastid to the nucleus and, probably, in regulating chloroplast protein homeostasis (Koussevitzky et al., 2007; Colombo et al., 2016; Llamas et al., 2017), an increased understanding of the regulation of the GUN1 protein will provide important clues about how GUN1 executes its proposed role as a central integrator of retrograde signaling pathways. Our work reported here reveals that GUN1 is regulated mainly posttranslationally, at the level of protein stability. Its extraordinarily high turnover is likely mediated by the Clp protease. How this turnover is slowed down when the GUN1 protein is needed will be interesting to investigate. From our work reported here, it also appears clear that GUN1 participates in biogenic control during early chloroplast development. Finally, our study links GUN1 with flowering control in Arabidopsis, and the molecular mechanism underlying this connection also will be interesting, and challenging, to explore.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The Columbia-0 strain of Arabidopsis (Arabidopsis thaliana) was used as the wild-type genetic background in all experiments. The gun1-101 (SAIL_33_D01) and clpc1-1 (SALK_014058) mutants were obtained from the European Arabidopsis Stock Centre. Primers used for the verification of T-DNA insertions are listed in Supplemental Table S1.
For plant growth under aseptic conditions, seeds were surface sterilized with 1.2% NaOCl for 10 min and then washed five times with sterile water. Sterilized seeds were sown on 0.5× Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) with 1% (w/v) Suc and stratified for 48 h at 4°C in the dark prior to germination. For the selection of transgenic lines, 0.5× MS medium was supplemented with 20 µg mL−1 hygromycin. For lincomycin, spectinomycin, and norflurazon treatments for the induction of retrograde signaling, 0.5× MS medium containing 1% (w/v) Suc was supplemented with 220 μg mL−1 lincomycin, 10 μg mL−1 spectinomycin, or 5 µm norflurazon. Seedlings were initially grown for 5 d in the dark, followed by growth for 2 d under continuous light. For cycloheximide chase assays, seeds were germinated and grown for 7 d on 0.5× MS medium containing 1% (w/v) Suc with netting (pore size, 500 µm) to avoid wounding upon transfer. Seedlings were then transferred to 500-mL flasks with 0.5× liquid MS medium containing 1% (w/v) Suc and 0.03% DMSO or 10 µm cycloheximide and incubated under slow rotation.
To determine the flowering time of wild-type plants, mutants, complemented lines, and overexpression lines, seeds were sown directly into soil and stratified at 4°C for 48 h prior to growth in long-day conditions (16 h of 130 µmol m−2 s−1 light at 20°C, 8 h of dark at 16°C, and 70% relative humidity). For measurements of high light-stimulated early flowering, plants were grown in long-day conditions under normal light (as described above) for 2 weeks, then transferred to high light (16 h of high light of 1,000 µmol m−2 s−1 and 8 h of darkness) and grown for another 2 weeks. Control plants stayed in normal light.
Construction of Transformation Vectors and Generation of Transgenic Plants
To produce a GUN1-GFP fusion, the complete coding region of GUN1 was amplified from cDNA adding EcoRI and BamHI restriction sites with the primer sequences (Supplemental Table S1). The amplified PCR products were cloned into the pEZR(H)-LN binary vector, generating an in-frame fusion to GFP (with GFP forming the C-terminal part of the resulting fusion protein). Expression of the fusion is driven by the CaMV 35S promoter. For expression of an unfused cpGFP, the RBCS1B transit peptide was amplified (introducing EcoRI and BamHI restriction sites) and cloned into vector pEZR(H)-LN. To generate cpGFP and GUN1-GFP lines in the clpc1-1 background, the cpGFP and GUN1-GFP (OE-13) transgenic lines produced by transformation of the wild type were crossed with the clpc1-1 mutant followed by selection of homozygous lines.
To produce a series of truncated versions of GUN1 fused to GFP, different parts of the GUN1 coding region were amplified from cDNA with specific primers (Supplemental Table S1). The amplified PCR products were cloned into the pEZR(H)-LN binary vector using the In-Fusion HD Cloning kit (Clontech) to generate in-frame fusions to GFP (Fig. 9A). Expression of the truncated variants is driven by the CaMV 35S promoter. The constructs were transformed into Agrobacterium tumefaciens strain GV3101 and introduced into both the wild type and the gun1-101 mutant using the flower-dipping method (Clough and Bent, 1998).
Seed Germination and Cotyledon Greening Assays
Seed batches harvested from plants grown under identical conditions were used for seed germination assays and cotyledon greening assays. Seeds of the different genotypes were sown on 0.5× MS medium with 1% (w/v) Suc and stratified for 48 h at 4°C in the dark to synchronize germination. The plates were then incubated in a growth chamber with long-day conditions. Seed germination was defined as visible radicle protrusion out of the testa (analyzed with a stereomicroscope), and the germination rate was scored at different time points. Seedlings developing green cotyledons were counted each day 1 h after the start of the light period. The assays were done in four independent experiments with different batches of seeds. GUN1-GFP expression in complemented line C-1 was analyzed at different time points by confocal microscopy and western blotting.
RNA Extraction, RT-qPCR, and RNA Gel-Blot Analyses
Total RNA was extracted using the NucleoSpin RNA Plant kit (Macherey-Nagel). To eliminate contaminating genomic DNA, RNA was treated with DNase (TURBO DNA-free Kit; Ambion). For RT-qPCR analysis, first-strand cDNA was synthesized using SuperScript III Reverse Transcriptase (Invitrogen) and qPCR was performed with the ABI 7900HT system (Applied Biosystems) using SYBR Green detection (Applied Biosystems) and actin gene expression (AT5G09810) as an internal standard. The amplification efficiency of the primers for each gene was tested using a dilution series of cDNA as PCR template. Primer pairs with differences in the slope of the standard curve for amplification of less than 0.1 were selected. Relative expression values were calculated for each target gene (including calculation of the amplification efficiencies of the different primers; Pfaffl, 2001).
For northern-blot analysis, RNA samples were separated on 1% (w/v) denaturing agarose gels containing formaldehyde, then blotted onto nylon membranes (Hybond-XL; GE Healthcare) and subsequently cross-linked by UV light. cDNA sequences were amplified with gene-specific primers to generate hybridization probes, which were labeled with [α-32P]dCTP using the Megaprime DNA labeling system (GE Healthcare). Hybridizations were performed at 65°C according to standard protocols. Primers used for RT-qPCR and the generation of hybridization probes are listed in Supplemental Table S1.
Polysome Analyses
Plant material for polysome analyses was generated as described above by growing seedlings in the presence of lincomycin or norflurazon or in untreated control conditions. Polysome analyses and puromycin controls were performed essentially as described previously (Barkan, 1998; Rogalski et al., 2008). RNA was extracted from gradient fractions, the RNA pellet was resuspended in 30 µL of 10 mm Tris-HCl, pH 8.1, and aliquots of 5 µL per fraction were analyzed by northern blotting.
Protein Extraction and Western-Blot Analysis
Total cellular protein was isolated according to a published procedure (Cahoon et al., 1992) and quantified with the BCA method according to the manufacturer’s instructions (Thermo Fisher Scientific). Protein samples were separated by 10% (w/v) SDS-PAGE and blotted onto PVDF membranes using standard protocols. Immunoblots were performed with specific antibodies, and chemiluminescence-based signal detection was done with the G:BOX Chemi Imaging System (Syngene) or x-ray films (GE Healthcare). Antibodies were obtained from commercial suppliers (GFP from Clonetech; Actin from Sigma; and Toc159, cpHSC70, Tic110, Tic40, RbcL, and PsbA from Agrisera), except for the ClpC, ClpP1, and ClpP4 antibodies, which were kindly provided by Dr. Adrian Clarke. Band intensities were determined using the GeneTools software (Syngene).
Chloroplast Isolation and Fractionation
For isolation of chloroplasts, leaf material harvested from the different genotypes was ground in chloroplast isolation buffer (0.3 m sorbitol, 5 mm MgCl2, 5 mm EGTA, 5 mm EDTA, 10 mm NaHCO3, and 20 mm HEPES-KOH, pH 8), and the released chloroplasts were loaded onto a linear Percoll gradient (Aronsson and Jarvis, 2011) and centrifuged in a swing-out rotor for 10 min at 7,800g (at 4°C). Intact chloroplasts were collected from the lower green band.
For the separation of chloroplasts into soluble, thylakoid, and envelope fractions, freshly isolated chloroplasts were resuspended in hypotonic buffer (25 mm HEPES-KOH, pH 8, supplemented with protease inhibitor cocktail) and fractionated according to published methods (Flores-Pérez et al., 2016). The different fractions were analyzed by western blotting in equivalent amounts (30% of the total isolate).
Label-Free Quantification by Mass Spectrometric Analyses
For the quantification and comparison of GUN1 with other chloroplast-localized proteins at the seedling stage, total protein was extracted and on-column digestion with trypsin was performed according to published procedures (Wiśniewski et al., 2009) with the following modifications: 100 mg of plant material was ground in liquid nitrogen and extracted with 200 µL of SDS buffer (4% SDS, 5% glycerol in 40 mm Tris-HCl, pH 6.8, and 2× protease inhibitor from Roche). After protein quantification, 100 µg of total protein was digested (Microcon 30-kD filter units; Merck Millipore) by trypsin overnight at 37°C in a 1:25 ratio to the total protein (i.e. 4 µg of trypsin for 100 µg of protein). The peptides were eluted from the column, cleaned up with a Sep-Pak column (Finisterre C18 SPE Column; Teknokroma), and then separated by reverse-phase chromatography with an 89-min linear gradient of 4% to 48% (v/v) acetonitrile.
For the quantification of GUN1 in rosette leaves, the seventh, eighth, and ninth rosette leaves from 3-week-old plants were collected (with the ninth rosette leaf typically being the leaf that just emerged). Total protein was extracted as above using SDS buffer and quantified. Samples of 100 µg of protein were resolved on 10% (w/v) SDS-PAGE gels and stained with colloidal Coomassie Blue. The gel region of 70 to 130 kD from each lane was excised and subjected to in-gel tryptic digestion (Walz et al., 2002).
For mass spectrometric analysis, peptides were separated by reverse-phase chromatography with a nanoflow HPLC device (Proxeon Biosystems) using a Chromolith CapRod RP-18e 150-0.2 column (Merck) with an 84-min (seedlings) or a 160-min (rosette leaves) linear gradient of 4% to 48% (v/v) acetonitrile, followed by a final peptide elution step for 5 min (seedlings) or 10 min (rosette leaves) with 64% (v/v) acetonitrile. The HPLC device was coupled via a nano-electrospray ionization ion source to a high-resolution Orbitrap hybrid mass spectrometer (LTQ-Orbitrap; Thermo Fisher Scientific). Spectral acquisition for full-scan mass spectrometric spectra was performed at a full-width half-maximum resolution of 70,000 in the Orbitrap section of the mass spectrometer, while the data-dependent tandem mass spectrometry spectra, with up to 15 spectra per preceding full scan, were obtained in the linear ion trap of the LTQ.
Protein identification and ion intensity quantitation were performed using MaxQuant software (Cox and Mann, 2008). The parameters were as follows: fixed modification, carbamidomethylation of Cys; variable modification, oxidation of Met, protein N-terminal acetylation; multiplicity set to 1 for label-free quantitation; trypsin specified as protease allowing one missed cleavage; razor and unique peptides (also including modified peptides) were used for quantification; all other parameters were set as default. Spectra were matched against the Arabidopsis proteome (TAIR10; 35,386 entries). Common contaminations (e.g. trypsin and keratin) were included in the database searches. The protein intensity was normalized and scaled with the cRacker software (Zauber and Schulze, 2012). For each peptide ion species (i.e. each m/z value), the ion intensity within each sample was normalized to the total ion intensity of all peptide ions in that sample. Protein abundance ratios were calculated by averaging the respective peptide ion intensity ratios. Subsequently, normalized protein intensities were median scaled and mean averaged. The normalized relative abundance of each protein represents the proportion of each protein of all detected proteins in the given sample.
Cycloheximide Chase Assays
Plant material for cycloheximide chase assays (to determine the half-lives of nucleus-encoded proteins) was produced by germinating seeds and growing the resulting seedlings for 7 d in long-day conditions in petri dishes on 0.5× MS medium with netting. Seedlings were then transferred to flasks with liquid 0.5× MS medium and incubated under slow agitation. The medium was supplemented with 10 µm cycloheximide or, in control experiments, 0.03% DMSO. Samples were collected at different time points and snap frozen in liquid nitrogen for further analyses.
For cycloheximide chase assay under conditions of altered retrograde signaling, seeds were germinated and grown in the presence of 5 μm norflurazon for 5 d in the dark, followed by 2 d in continuous light in petri dishes with 0.5× MS medium with netting. Seedlings were then transferred into flasks with liquid 0.5× MS medium and cultured under slow agitation. The medium was supplemented with either 5 µm norflurazon or 5 µm norflurazon + 10 µm cycloheximide. Samples were collected at different time points and snap frozen in liquid nitrogen for further analyses.
Confocal Microscopic Analyses
Confocal laser-scanning microscopy (TCS SP5; Leica) was used to determine the subcellular localization of GFP and chlorophyll fluorescence. For GFP fluorescence, the excitation wavelength was 488 nm and emission was detected with a 495- to 530-nm filter. For detection of chlorophyll fluorescence, a 650- to 702-nm filter was used. 4′,6-Diamidino-2-phenylindole (DAPI) fluorescence was excited at 358 nm with UV light, and the fluorescence emission was observed at 430 to 470 nm.
Nucleoid Visualization by DAPI Staining
For DAPI staining to visualize chloroplast nucleoids, the DAPI stock solution was diluted to 10 µg mL−1 in HS buffer (50 mm HEPES-KOH, pH 8, and 0.33 m sorbitol) and mixed 1:1 with isolated intact chloroplasts to reach a final DAPI concentration of 5 µg mL−1. Staining was allowed to proceed for 10 min in the dark prior to examination by confocal laser-scanning microscopy.
Accession Numbers
The sequence data from this article can be found in The Arabidopsis Information Resource or GenBank/EMBL database under the following accession numbers: GUN1 (At2g31400), ClpC1 (At5g50920), ClpC2 (At3g48870), cpHSC70-1 (At4g24280), cpHSC70-2 (At5g49910), Tic110 (At1g06950), Tic40 (At5g16620), Toc159 (At4g02510), PGK1 (At3g12780), FBA1 (At2g21330), FBP1 (At3g54050), LhcB1.2 (At1g29910), ClpD (At5g51070), clpP1 (AtCg00670), ClpP4 (At5g45390), ClpR1 (At1g49970), psbA (AtCg00020), rbcL (AtCg00490), THI1 (At5g54770), and BAM3 (At4g17090).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Germination assay of wild-type and gun1-101 mutant seeds.
Supplemental Figure S2. cpGFP protein accumulation during germination and DAPI staining of chloroplast nucleoids.
Supplemental Figure S3. Schematic representation of the experimental design of the cycloheximide chase assays.
Supplemental Figure S4. Cycloheximide chase assays to compare cpGFP degradation in the wild-type and the clpc1-1 backgrounds, half-lives of different chloroplast proteins, and cycloheximide chase assays for Tic110 and Tic40.
Supplemental Figure S5. Western-blot analyses to identify transgenic lines expressing the different GUN1 truncations (GT-1 to GT-6).
Supplemental Figure S6. GUN1 does not function in high-light-stimulated early flowering.
Supplemental Table S1. List of oligonucleotides used in this study.
Supplemental Data Set S1. Mass spectrometry data related to Figure 4, A and B.
Acknowledgments
We thank the Max-Planck-Institut für Molekulare Pflanzenphysiologie Green Team for help with plant transformation and Dr. Adrian Clarke (Gothenburg University) for providing antibodies against Clp proteins.
Footnotes
This research was financed by the Max Planck Society and grants from the Deutsche Forschungsgemeinschaft to R.B. (FOR 804, BO 1482/15-2, SFB-TR 175).
Articles can be viewed without a subscription.
References
- Adam Z, Rudella A, van Wijk KJ (2006) Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Curr Opin Plant Biol 9: 234–240 [DOI] [PubMed] [Google Scholar]
- Allan RK, Ratajczak T (2011) Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones 16: 353–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés-Colás N, Zhu Q, Takenaka M, De Rybel B, Weijers D, Van Der Straeten D (2017) Multiple PPR protein interactions are involved in the RNA editing system in Arabidopsis mitochondria and plastids. Proc Natl Acad Sci USA 114: 8883–8888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Apel W, Schulze WX, Bock R (2010) Identification of protein stability determinants in chloroplasts. Plant J 63: 636–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronsson H, Jarvis RP (2011) Rapid isolation of Arabidopsis chloroplasts and their use for in vitro protein import assays. Methods Mol Biol 774: 281–305 [DOI] [PubMed] [Google Scholar]
- Barkan A. (1998) Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol 297: 38–57 [Google Scholar]
- Barkan A, Small I (2014) Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65: 415–442 [DOI] [PubMed] [Google Scholar]
- Battersby BJ, Richter U (2013) Why translation counts for mitochondria: retrograde signalling links mitochondrial protein synthesis to mitochondrial biogenesis and cell proliferation. J Cell Sci 126: 4331–4338 [DOI] [PubMed] [Google Scholar]
- Bentolila S, Heller WP, Sun T, Babina AM, Friso G, van Wijk KJ, Hanson MR (2012) RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing. Proc Natl Acad Sci USA 109: E1453–E1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman Q, Mazubert C, Prunier F, Lugan R, Chan KX, Phua SY, Pogson BJ, Krieger-Liszkay A, Delarue M, Benhamed M, et al. (2016) Chloroplast activity and 3′phosphadenosine 5′phosphate signaling regulate programmed cell death in Arabidopsis. Plant Physiol 170: 1745–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butow RA, Avadhani NG (2004) Mitochondrial signaling: the retrograde response. Mol Cell 14: 1–15 [DOI] [PubMed] [Google Scholar]
- Cahoon EB, Shanklin J, Ohlrogge JB (1992) Expression of a coriander desaturase results in petroselinic acid production in transgenic tobacco. Proc Natl Acad Sci USA 89: 11184–11188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colombo M, Tadini L, Peracchio C, Ferrari R, Pesaresi P (2016) GUN1, a jack-of-all-trades in chloroplast protein homeostasis and signaling. Front Plant Sci 7: 1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottage A, Mott EK, Kempster JA, Gray JC (2010) The Arabidopsis plastid-signalling mutant gun1 (genomes uncoupled1) shows altered sensitivity to sucrose and abscisic acid and alterations in early seedling development. J Exp Bot 61: 3773–3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Das AK, Cohen PW, Barford D (1998) The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J 17: 1192–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq I, Vermeirssen V, Van Aken O, Vandepoele K, Murcha MW, Law SR, Inzé A, Ng S, Ivanova A, Rombaut D, et al. (2013) The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 25: 3472–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres Zabala M, Littlejohn G, Jayaraman S, Studholme D, Bailey T, Lawson T, Tillich M, Licht D, Bölter B, Delfino L, et al. (2015) Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nat Plants 1: 15074. [DOI] [PubMed] [Google Scholar]
- Estavillo GM, Crisp PA, Pornsiriwong W, Wirtz M, Collinge D, Carrie C, Giraud E, Whelan J, David P, Javot H, et al. (2011) Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23: 3992–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves P, Pecqueur C, Ransy C, Esnous C, Lenoir V, Bouillaud F, Bulteau AL, Lombès A, Prip-Buus C, Ricquier D, et al. (2014) Mitochondrial retrograde signaling mediated by UCP2 inhibits cancer cell proliferation and tumorigenesis. Cancer Res 74: 3971–3982 [DOI] [PubMed] [Google Scholar]
- Feng P, Guo H, Chi W, Chai X, Sun X, Xu X, Ma J, Rochaix JD, Leister D, Wang H, et al. (2016) Chloroplast retrograde signal regulates flowering. Proc Natl Acad Sci USA 113: 10708–10713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Pérez Ú, Bédard J, Tanabe N, Lymperopoulos P, Clarke AK, Jarvis P (2016) Functional analysis of the Hsp93/ClpC chaperone at the chloroplast envelope. Plant Physiol 170: 147–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebl M, Yanagida M (1991) The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem Sci 16: 173–177 [DOI] [PubMed] [Google Scholar]
- Guillaumot D, Lopez-Obando M, Baudry K, Avon A, Rigaill G, Falcon de Longevialle A, Broche B, Takenaka M, Berthomé R, De Jaeger G, et al. (2017) Two interacting PPR proteins are major Arabidopsis editing factors in plastid and mitochondria. Proc Natl Acad Sci USA 114: 8877–8882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhaki H, Naveh L, Lindahl M, Cook M, Adam Z (1998) Identification and characterization of DegP, a serine protease associated with the luminal side of the thylakoid membrane. J Biol Chem 273: 7094–7098 [DOI] [PubMed] [Google Scholar]
- Jia Y, Rothermel B, Thornton J, Butow RA (1997) A basic helix-loop-helix-leucine zipper transcription complex in yeast functions in a signaling pathway from mitochondria to the nucleus. Mol Cell Biol 17: 1110–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HS, Crisp PA, Estavillo GM, Cole B, Hong F, Mockler TC, Pogson BJ, Chory J (2013) Subset of heat-shock transcription factors required for the early response of Arabidopsis to excess light. Proc Natl Acad Sci USA 110: 14474–14479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T, Voigt C, Leister D (2009) Plastid signalling to the nucleus: messengers still lost in the mists? Trends Genet 25: 185–192 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Larkin RM, Alonso JM, Ecker JR, Chory J (2003) GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299: 902–906 [DOI] [PubMed] [Google Scholar]
- Lee KP, Kim C, Landgraf F, Apel K (2007) EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 10270–10275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy YY, Dean C (1998) The transition to flowering. Plant Cell 10: 1973–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Nelson CJ, Trösch J, Castleden I, Huang S, Millar AH (2017) Protein degradation rate in Arabidopsis thaliana leaf growth and development. Plant Cell 29: 207–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wakao S, Fischer BB, Niyogi KK (2009) Sensing and responding to excess light. Annu Rev Plant Biol 60: 239–260 [DOI] [PubMed] [Google Scholar]
- Lindahl M, Tabak S, Cseke L, Pichersky E, Andersson B, Adam Z (1996) Identification, characterization, and molecular cloning of a homologue of the bacterial FtsH protease in chloroplasts of higher plants. J Biol Chem 271: 29329–29334 [DOI] [PubMed] [Google Scholar]
- Liu Z, Sekito T, Spírek M, Thornton J, Butow RA (2003) Retrograde signaling is regulated by the dynamic interaction between Rtg2p and Mks1p. Mol Cell 12: 401–411 [DOI] [PubMed] [Google Scholar]
- Llamas E, Pulido P, Rodriguez-Concepcion M (2017) Interference with plastome gene expression and Clp protease activity in Arabidopsis triggers a chloroplast unfolded protein response to restore protein homeostasis. PLoS Genet 13: e1007022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín G, Leivar P, Ludevid D, Tepperman JM, Quail PH, Monte E (2016) Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network. Nat Commun 7: 11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K (2001) FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 98: 12826–12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J (2001) Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA 98: 2053–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Susek R, Chory J (1996) An intracellular signal transduction pathway between the chloroplast and nucleus is involved in de-etiolation. Plant Physiol 112: 1465–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Ng S, Ivanova A, Duncan O, Law SR, Van Aken O, De Clercq I, Wang Y, Carrie C, Xu L, Kmiec B, et al. (2013) A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25: 3450–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Kato Y, Sakamoto W (2016) Chloroplast proteases: updates on proteolysis within and across suborganellar compartments. Plant Physiol 171: 2280–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, van Wijk KJ (2015) Organization, function and substrates of the essential Clp protease system in plastids. Biochim Biophys Acta 1847: 915–930 [DOI] [PubMed] [Google Scholar]
- Nott A, Jung HS, Koussevitzky S, Chory J (2006) Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol 57: 739–759 [DOI] [PubMed] [Google Scholar]
- op den Camp RGL, Przybyla D, Ochsenbein C, Laloi C, Kim CH, Danon A, Wagner D, Hideg E, Gobel C, Feussner I, et al. (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15: 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan MS, Dinesh-Kumar SP (2010) All hands on deck: the role of chloroplasts, endoplasmic reticulum, and the nucleus in driving plant innate immunity. Mol Plant Microbe Interact 23: 1368–1380 [DOI] [PubMed] [Google Scholar]
- Page MT, Kacprzak SM, Mochizuki N, Okamoto H, Smith AG, Terry MJ (2017) Seedlings lacking the PTM protein do not show a genomes uncoupled (gun) mutant phenotype. Plant Physiol 174: 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh VS, Morgan MM, Scott R, Clements LS, Butow RA (1987) The mitochondrial genotype can influence nuclear gene expression in yeast. Science 235: 576–580 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Woo NS, Förster B, Small ID (2008) Plastid signalling to the nucleus and beyond. Trends Plant Sci 13: 602–609 [DOI] [PubMed] [Google Scholar]
- Pornsiriwong W, Estavillo GM, Chan KX, Tee EE, Ganguly D, Crisp PA, Phua SY, Zhao C, Qiu J, Park J, et al. (2017) A chloroplast retrograde signal, 3′-phosphoadenosine 5′-phosphate, acts as a secondary messenger in abscisic acid signaling in stomatal closure and germination. eLife 6: e23361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigas S, Daras G, Tsitsekian D, Alatzas A, Hatzopoulos P (2014) Evolution and significance of the Lon gene family in Arabidopsis organelle biogenesis and energy metabolism. Front Plant Sci 5: 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski M, Schöttler MA, Thiele W, Schulze WX, Bock R (2008) Rpl33, a nonessential plastid-encoded ribosomal protein in tobacco, is required under cold stress conditions. Plant Cell 20: 2221–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossel JB, Wilson PB, Hussain D, Woo NS, Gordon MJ, Mewett OP, Howell KA, Whelan J, Kazan K, Pogson BJ (2007) Systemic and intracellular responses to photooxidative stress in Arabidopsis. Plant Cell 19: 4091–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckle ME, DeMarco SM, Larkin RM (2007) Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19: 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I (2008) Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci 13: 663–670 [DOI] [PubMed] [Google Scholar]
- Shanklin J, DeWitt ND, Flanagan JM (1995) The stroma of higher plant plastids contain ClpP and ClpC, functional homologs of Escherichia coli ClpP and ClpA: an archetypal two-component ATP-dependent protease. Plant Cell 7: 1713–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šimková K, Kim C, Gacek K, Baruah A, Laloi C, Apel K (2012) The chloroplast division mutant caa33 of Arabidopsis thaliana reveals the crucial impact of chloroplast homeostasis on stress acclimation and retrograde plastid-to-nucleus signaling. Plant J 69: 701–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren LL, Tanabe N, Lymperopoulos P, Khan NZ, Rodermel SR, Aronsson H, Clarke AK (2014) Quantitative analysis of the chloroplast molecular chaperone ClpC/Hsp93 in Arabidopsis reveals new insights into its localization, interaction with the Clp proteolytic core, and functional importance. J Biol Chem 289: 11318–11330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small ID, Peeters N (2000) The PPR motif: a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci 25: 46–47 [DOI] [PubMed] [Google Scholar]
- Spåhr H, Rozanska A, Li X, Atanassov I, Lightowlers RN, Chrzanowska-Lightowlers ZMA, Rackham O, Larsson NG (2016) SLIRP stabilizes LRPPRC via an RRM-PPR protein interface. Nucleic Acids Res 44: 6868–6882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand A, Asami T, Alonso J, Ecker JR, Chory J (2003) Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421: 79–83 [DOI] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J (1993) Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799 [DOI] [PubMed] [Google Scholar]
- Tadini L, Pesaresi P, Kleine T, Rossi F, Guljamow A, Sommer F, Mühlhaus T, Schroda M, Masiero S, Pribil M, et al. (2016) GUN1 controls accumulation of the plastid ribosomal protein S1 at the protein level and interacts with proteins involved in plastid protein homeostasis. Plant Physiol 170: 1817–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O, Whelan J (2012) Comparison of transcriptional changes to chloroplast and mitochondrial perturbations reveals common and specific responses in Arabidopsis. Front Plant Sci 3: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk KJ. (2015) Protein maturation and proteolysis in plant plastids, mitochondria, and peroxisomes. Annu Rev Plant Biol 66: 75–111 [DOI] [PubMed] [Google Scholar]
- von Gromoff ED, Alawady A, Meinecke L, Grimm B, Beck CF (2008) Heme, a plastid-derived regulator of nuclear gene expression in Chlamydomonas. Plant Cell 20: 552–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Przybyla D, op den Camp R, Kim C, Landgraf F, Lee KP, Würsch M, Laloi C, Nater M, Hideg E, Apel K (2004) The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306: 1183–1185 [DOI] [PubMed] [Google Scholar]
- Walley J, Xiao Y, Wang JZ, Baidoo EE, Keasling JD, Shen Z, Briggs SP, Dehesh K (2015) Plastid-produced interorgannellar stress signal MEcPP potentiates induction of the unfolded protein response in endoplasmic reticulum. Proc Natl Acad Sci USA 112: 6212–6217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz C, Juenger M, Schad M, Kehr J (2002) Evidence for the presence and activity of a complete antioxidant defence system in mature sieve tubes. Plant J 31: 189–197 [DOI] [PubMed] [Google Scholar]
- Wang M, Weiss M, Simonovic M, Haertinger G, Schrimpf SP, Hengartner MO, von Mering C (2012) PaxDb, a database of protein abundance averages across all three domains of life. Mol Cell Proteomics 11: 492–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6: 359–362 [DOI] [PubMed] [Google Scholar]
- Woodson JD, Chory J (2008) Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet 9: 383–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Chory J (2011) Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr Biol 21: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Schmitz RJ, Ecker JR, Chory J (2013) Sigma factor-mediated plastid retrograde signals control nuclear gene expression. Plant J 73: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Savchenko T, Baidoo EEK, Chehab WE, Hayden DM, Tolstikov V, Corwin JA, Kliebenstein DJ, Keasling JD, Dehesh K (2012) Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149: 1525–1535 [DOI] [PubMed] [Google Scholar]
- Zauber H, Schulze WX (2012) Proteomics wants cRacker: automated standardized data analysis of LC-MS derived proteomic data. J Proteome Res 11: 5548–5555 [DOI] [PubMed] [Google Scholar]