Thioredoxin-dependent redox modification regulates plant regeneration via modulation of ROS homeostasis.
Abstract
Plant regeneration is fundamental to basic research and agricultural applications. The regeneration capacity of plants varies largely in different genotypes, but the reason for this variation remains elusive. Here, we identified a novel thioredoxin DCC1 in determining the capacity of shoot regeneration among Arabidopsis (Arabidopsis thaliana) natural variation. Loss of function of DCC1 resulted in inhibited shoot regeneration. DCC1 was expressed mainly in the inner tissues of the callus and encoded a functional thioredoxin that was localized in the mitochondria. DCC1 protein interacted directly with CARBONIC ANHYDRASE2 (CA2), which is an essential subunit of the respiratory chain NADH dehydrogenase complex (Complex I). DCC1 regulated Complex I activity via redox modification of CA2 protein. Mutation of DCC1 or CA2 led to reduced Complex I activity and triggered mitochondrial reactive oxygen species (ROS) production. The increased ROS level regulated shoot regeneration by repressing expression of the genes involved in multiple pathways. Furthermore, linkage disequilibrium analysis indicated that DCC1 was a major determinant of the natural variation in shoot regeneration among Arabidopsis ecotypes. Thus, our study uncovers a novel regulatory mechanism by which thioredoxin-dependent redox modification regulates de novo shoot initiation via the modulation of ROS homeostasis and provides new insights into improving the capacity of plant regeneration.
Plant cells have the capacity to regenerate new shoots from highly differentiated tissues or organs under suitable conditions, a process known as shoot regeneration (Birnbaum and Alvarado, 2008; Duclercq et al., 2011). Shoot regeneration normally includes two steps (Ikeuchi et al., 2016). The first step is callus formation, which is regulated by a number of transcription factors, such as WUSCHEL RELATED HOMEOBOX5 (WOX5), WOX11, and WOX12 (Sugimoto et al., 2010; Liu et al., 2014), PLETHORA (PLT; Kareem et al., 2015), LATERAL ORGAN BOUNDARIES DOMAIN (LBD; Fan et al., 2012), and WOUND INDUCED DEDIFFERENTIATION1 (WIND1; Iwase et al., 2011). The second step is shoot induction from the callus, which consists of several critical events, such as the appropriate distribution of phytohormones, shoot meristem initiation, and organ formation (Cheng et al., 2013; Ikeuchi et al., 2016). One of the most important events in the second step is the induction of the organizing center regulator WUSCHEL (WUS), which is controlled by the interaction between auxin and cytokinin (Gordon et al., 2007; Cheng et al., 2013). The correct distribution of auxin and cytokinin is essential for WUS induction during de novo shoot regeneration (Cheng et al., 2013). Importantly, two recent studies have shown that cytokinin directly activates WUS expression by the B-type ARABIDOPSIS RESPONSE REGULATORS (Meng et al., 2017; Zhang et al., 2017).
Shoot regeneration is very valuable in genetic engineering and agricultural applications, but most plant species cannot regenerate shoots from highly differentiated tissues or organs (Birnbaum and Alvarado, 2008). Even in the same species, the shoot regeneration capacity varies among different genotypes (Motte et al., 2014). However, the mechanisms underlying this natural variation in shoot regeneration capacity remain unclear. Several studies have identified a few quantitative trait loci related to natural variations in shoot regeneration (Schiantarelli et al., 2001; Lall et al., 2004; Velázquez et al., 2004). A recent study has shown that the RECEPTOR-LIKE PROTEIN KINASE1 gene is involved in natural variation in shoot regeneration capacity (Motte et al., 2014). Further identification of the critical genes that control shoot regeneration and how they vary among different genotypes are required for research in both the mechanisms and applications of shoot regeneration.
Reactive oxygen species (ROS) are crucial signaling molecules that can alter their target protein’s activity by oxidative posttranslational modifications (Waszczak et al., 2015). Many proteins that act in hormonal signal perception and response, MAPK signal transduction, and the influx channel pathway are redox sensitive to ROS (Waszczak et al., 2015; Kimura et al., 2017). Altered levels of ROS lead to changes in the activity of these target proteins and affect various developmental processes (Schippers et al., 2016). During root development, ROS homeostasis is critical for the transition from cell proliferation to cell differentiation (Tsukagoshi et al., 2010). High levels of ROS result in reduced root meristem activity (Tsukagoshi et al., 2010) and small leaf size (Lu et al., 2014). ROS also is involved in regulating the polar growth of pollen and root hairs (Mangano et al., 2016) and the senescence of leaf and flower (Rogers and Munné-Bosch, 2016). In addition, ROS is generated in various subcellular compartments. Chloroplasts (Dietz et al., 2016), mitochondria (Huang et al., 2016), and peroxisomes (Sandalio and Romero-Puertas, 2015) are the major producers of ROS in plant cells.
ROS homeostasis is governed by diverse antioxidant factors (Considine and Foyer, 2014; Lu and Holmgren, 2014). Thioredoxins (Trxs) are key actors in modulating ROS scavenging, and functional loss of Trx results in altered ROS levels (Dos Santos and Rey, 2006; Schippers et al., 2016). Trxs are widely distributed proteins with a main function to reduce a specific S-S group through the conserved motif CxxC (Gelhaye et al., 2005; Meyer et al., 2005). Trxs alter the activity of interacting target proteins via thiol-based redox modifications (Rouhier et al., 2015). A number of potential Trx targets have been identified using proteomic approaches (Montrichard et al., 2009). Trxs can affect diverse signaling pathways by modulating the activity of their target proteins, such as enzymes of peroxiredoxins and glutathione peroxidase for ROS scavenging (Dos Santos and Rey, 2006), transcription factors for gene expression (Murmu et al., 2010; Viola et al., 2013), and hormonal receptors for signal transduction (Tada et al., 2008). The mitochondrial respiratory chain NADH dehydrogenase complex (Complex I) consists of several essential subunits and is one of the major sites of electron entry into the electron transport chain in the process of ATP production (Braun et al., 2014). An altered redox state of the subunits results in reduced Complex I activity (Galkin et al., 2008). Proteomic analyses have revealed that many subunits of Complex I are potential targets of Trxs, suggesting a crucial role for Trxs in regulating mitochondrial Complex I function (Balmer et al., 2004; Yoshida et al., 2013; Braun et al., 2014; Dröse et al., 2014). However, the evidence that Trxs directly regulate these subunits remains missing. Redox of Trxs regulates diverse developmental processes, such as embryo formation, leaf development, and maintenance of meristem activity (Benitez-Alfonso et al., 2009; Bashandy et al., 2010; Meng et al., 2010). Here, we showed that Trx-dependent redox modification regulated de novo shoot initiation via the modulation of ROS homeostasis. This study provides novel information for understanding the mechanisms of natural variation in plant regeneration.
RESULTS
Loss of Function of DCC1 Results in the Inhibition of Shoot Regeneration
To determine the function of Trxs in shoot regeneration, seven T-DNA insertion mutants of Trx (Trx O1, Trx O2, Trx H1, Trx H2, Trx H5, AT5G50100, and AT1G52590) from the Arabidopsis Biological Resource Center were screened, but only the SALK_051222C (AT5G50100) mutant showed severe defects in shoot regeneration (Supplemental Table S1). This mutant contained a T-DNA insert in the fourth intron of AT5G50100 (Supplemental Fig. S1A). Reverse transcription-PCR analyses showed that the transcript level of this gene was decreased in this mutant (Supplemental Fig. S1B), suggesting that the T-DNA insertion leads to reduced levels of functional transcripts. AT5G50100 encodes a protein in the thiol-disulfide oxidoreductase family, whose members contain a conserved DxxCxxC motif in their N terminus (Ginalski et al., 2004). In Arabidopsis (Arabidopsis thaliana), there exist three members with a DxxCxxC motif, but their functions have not been determined. Thus, AT5G50100 was designated as DCC1.
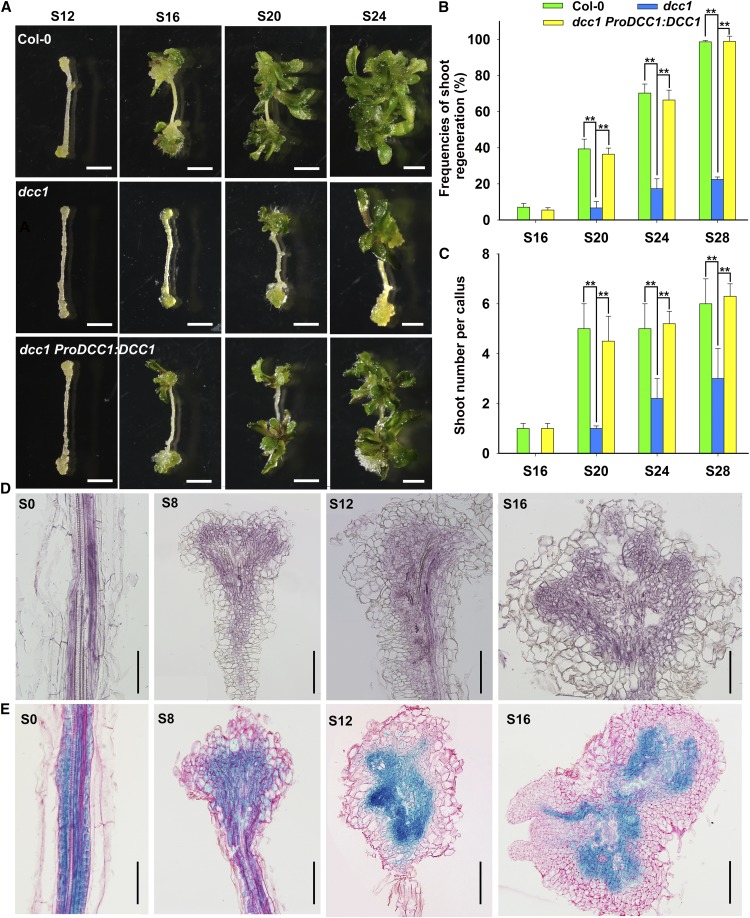
Loss of function of DCC1 resulted in a decreased capacity for shoot regeneration, including the low shoot regeneration frequency and the small number of shoots per callus (Fig. 1, A–C). Wild-type callus generated shoots on shoot induction medium (SIM) at 16 d (Fig. 1, A and B), but the dcc1 mutant callus took 20 d to produce shoots (Fig. 1, A and B). The shoot regeneration frequency in wild-type Columbia-0 (Col-0) was 100% at 28 d on SIM, while that of dcc1 was about 30% (Fig. 1, A and B). The number of shoots per callus was significantly lower in dcc1 than in the wild type (Fig. 1, A and C). Furthermore, the phenotypes of the frequencies of shoot regeneration and the shoot number per callus caused by mutation of DCC1 were both rescued by the transformation of the complementary construct ProDCC1:DCC1 into the dcc1 mutant (Fig. 1, A–C). Thus, these results indicate that mutation of DCC1 leads to an inhibition of shoot regeneration. Furthermore, we showed that DCC1 was expressed mainly in the inner region of the callus during shoot regeneration by both in situ hybridization (Fig. 1D) and GUS analyses (Fig. 1E).
Figure 1.
Shoot regeneration is inhibited in the dcc1 mutant. A to C, Callus morphology (A), frequencies of shoot regeneration from calli (B), and number of shoots per callus (C) in wild-type Col-0, dcc1, and dcc1 ProDCC1:DCC1 cultured on SIM at the indicated times. D, Expression pattern of DCC1 detected by in situ hybridization in calli of Col-0 at the indicated times on SIM. E, Expression pattern of DCC1 detected by GUS staining in calli of ProDCC1:DCC1-GUS transgenetic lines at the indicated times on SIM. S0 represents calli cultured on callus induction medium (CIM) at 6 d using roots as explants before transfer to SIM. S8, S12, S16, S20, S24, and S28 indicate calli cultured on SIM at 8, 12, 16, 20, 24, and 28 d, respectively. se values were calculated from three sets of biological replicates, and more than 100 calli were examined in each replicate. Asterisks indicate significant differences: **, P < 0.01 (Student’s one-tailed t test). Bars = 500 µm.
DCC1 Encodes a Functional Trx Localized in Mitochondria
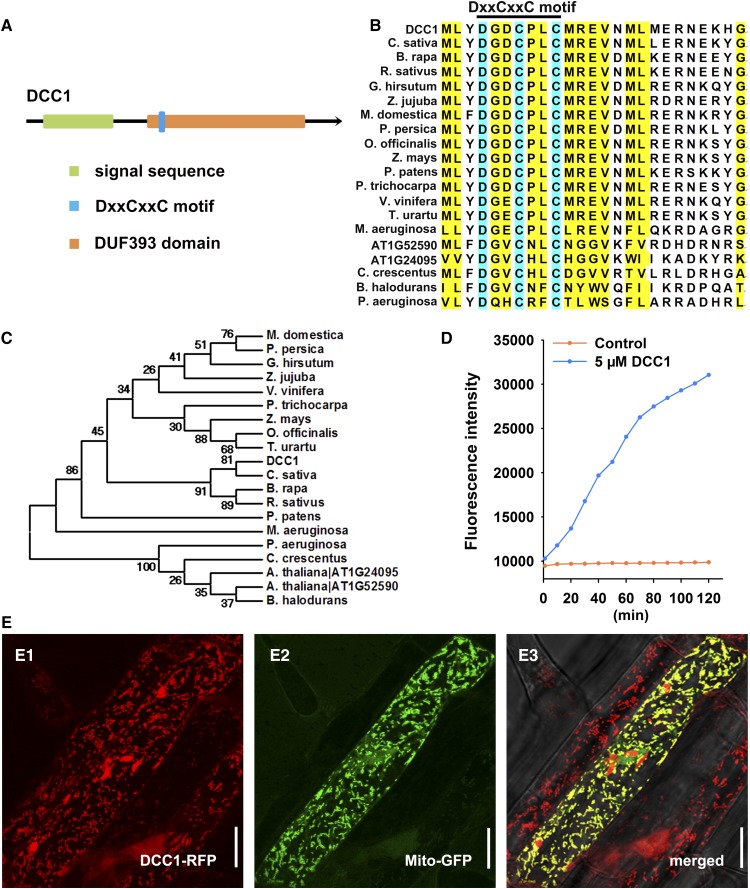
DCC1 contained an N-terminal DxxCxxC motif in a function-unknown DUF393 domain (in the Pfam database; Fig. 2A). The DxxCxxC motif is a conserved signature sequence for DCC family proteins in various species, but not in animals (Fig. 2B). Phylogenetic analysis indicated that DCC1 shared high homology with several proteins in species such as Camelina sativa, Brassica rapa, and Raphanus sativus (Fig. 2C). The presence of two Cys residues in the DxxCxxC motif implied a function for DCC1 in regulating thiol-disulfide exchange. Thus, we performed a Trx activity detection assay using fluorescein isothiocyanate-labeled insulin (FiTC-insulin). FiTC-insulin is a highly sensitive substrate for the measurements of Trx activity and displays higher fluorescence after disulfide reduction (Montano et al., 2014). The reduction assay was performed by incubation with purified DCC1 protein, and a high fluorescence intensity was achieved, whereas the control without DCC1 protein showed a low level of insulin reduction (Fig. 2D), confirming its activity as a functional Trx.
Figure 2.
DCC1 is a functional Trx localized in mitochondria. A, Model of the DCC1 protein with a mitochondria signal sequence predicted in the UniProt database and a conserved DxxCxxC motif in a function-unknown DUF393 domain analyzed in the Pfam database. B, Alignment of the amino acid sequences of different DCC family proteins. The DxxCxxC motif is a conserved signature sequence for DCC family proteins in various species. The National Center for Biotechnology Information accession numbers of proteins in different species are presented in “Materials and Methods.” C, Phylogenetic analyses of DCC1 with its homologs in various species. DCC1 shares high homology with several proteins in species such as C. sativa, B. rapa, and R. sativus. D, Insulin reduction by recombinant DCC1 proteins. Purified DCC1 proteins were subjected to a reduction assay by using FiTC-insulin as the substrate, which displayed higher fluorescence after disulfide reduction. The assay mixture lacking recombinant DCC1 proteins served as the control. Fluorescence intensity was recorded at 515- to 525-nm emission after 480- to 495-nm excitation for 120 min in a fluorescent plate reader at room temperature. E, Subcellular localization of DCC1. MT-GK is a well-established marker line specially expressing Mito-GFP. The construct 35S:DCC1-RFP was transformed into MT-GK. The T1 transgenetic line roots were excised for imaging by a confocal microscope. The DCC1-RFP signal (E1) was observed at 505- to 550-nm emission after 561-nm excitation, whereas the Mito-GFP signal (E2) was observed at 570- to 620-nm emission after 488-nm excitation. The merged signals of GFP and RFP showed yellow color (E3), indicating that DCC1 is localized in the mitochondria. Bars = 20 µm.
DCC1 contained a mitochondrial signal sequence, implying that it might be localized in the mitochondria (Fig. 2A). To test this hypothesis, we obtained a well-established marker line, MT-GK, specially expressing GFP in mitochondria (Mito-GFP; Nelson et al., 2007). Then, the construct 35S:DCC1-RFP was introduced into the MT-GK line. The roots of the T1 seedlings were used for imaging by laser confocal microscopy. The DCC1-RFP signal was colocalized with the Mito-GFP signal of the control mitochondrion marker (Fig. 2E), indicating that DCC1 is localized in mitochondria.
DCC1 Interacts Directly with CA2
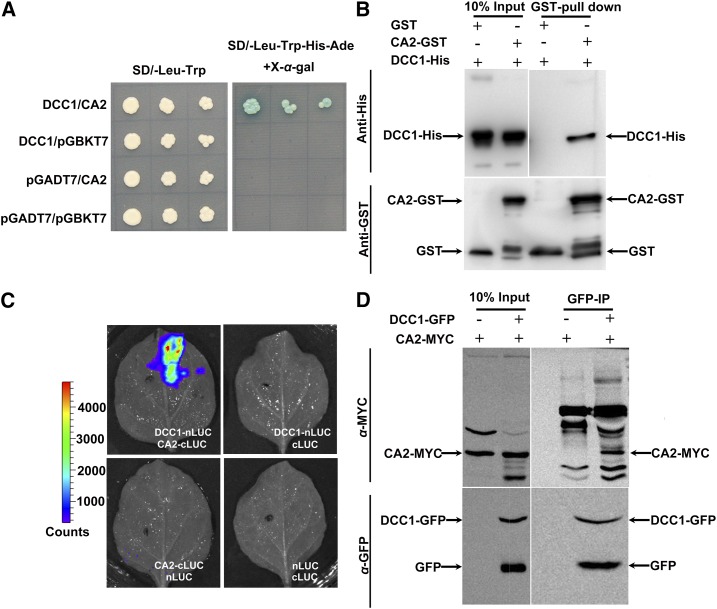
To uncover the functional pathway of DCC1 in shoot regeneration, we searched for its potential target(s). We analyzed the published proteome-wide binary protein-protein interaction map constructed by yeast two-hybrid assay in Arabidopsis and found a DCC1 potential interaction target, CARBONIC ANHYDRASE2 (CA2; Arabidopsis Interactome Mapping Consortium, 2011), which is a β-carbonic anhydrase (Soto et al., 2015). CA2, together with other four carbonic anhydrases (CA1, CA3, CAL1, and CAL2), constitutes a functional subunit of mitochondrial respiratory complex I (Soto et al., 2015). By a yeast two-hybrid assay, we found that only the yeast cotransformed with DCC1-AD and CA2-BD constructs could survive on selection medium (Fig. 3A). In pull-down assays using the purified proteins of DCC1 and CA2, an obvious lane of DCC1-His was observed (Fig. 3B). To further confirm the interaction between DCC1 and CA2 in vivo, we cotransformed the constructs DCC1-nLUC and CA2-cLUC into Nicotiana benthamiana leaves and observed high fluorescence intensity compared with that in the controls (Fig. 3C). In DCC1-GFP immunoprecipitation assays of the F1 generation of the 35S:DCC1-GFP line crossed with the 35S:CA2-MYC line, we detected an obvious lane of the CA2-MYC protein (Fig. 3D), indicating an interaction between DCC1 and CA2.
Figure 3.
DCC1 interacts with CA2. A, Interaction between DCC1 and CA2 in the yeast two-hybrid assay. Activation was observed at 3 d on selection plates (synthetic dextrose [SD]-Leu-Trp-His-Ade) with X-α-gal. B, Interaction between DCC1 and CA2 in the pull-down assay. The 10% input and GST pull-down proteins were detected by immunoblotting using anti-His antibody (top row). CA2-GST and GST proteins were detected by immunoblotting using anti-GST antibody (bottom row). C, Interaction between DCC1 and CA2 in firefly luciferase complementation assays in transiently transfected leaf of N. benthamiana. D, Interaction between DCC1 and CA2 in coimmunoprecipitation assays. The 10% input and immunoprecipitated proteins with anti-GFP (GFP-IP) were detected by immunoblotting using an anti-MYC antibody (top row). The 10% input and DCC1-GFP proteins were detected by immunoblotting using an anti-GFP antibody (bottom row).
Mutation of CA2 Causes the Decreased Shoot Regeneration Capacity
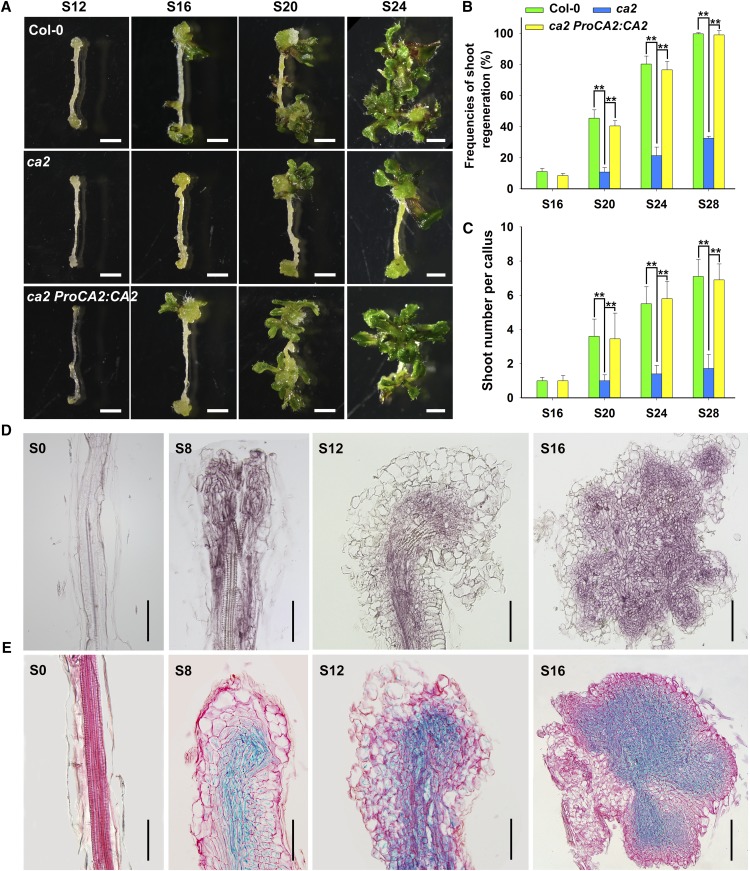
To determine the function of CA2 in shoot regeneration, we obtained the T-DNA insertion mutant ca2 and analyzed its shoot regeneration capacity. Mutation of CA2 led to a significantly decreased shoot regeneration capacity. During shoot regeneration, the shoots started to emerge at 16 d on SIM in the wild type, whereas the mutant callus generated shoots at 20 d on SIM (Fig. 4A). The shoot regeneration frequency of the wild type reached 100% at 28 d after transfer of calli onto SIM, while that of ca2 was only about 40% (Fig. 4B). The number of shoots per callus showed a similar inhibition (Fig. 4C). The frequencies of shoot regeneration and the shoot number per callus were completely rescued by the transformation of the complementary construct ProCA2:CA2 into the ca2 mutant (Fig. 4, A–C). Next, we crossed the two single mutants and obtained the homozygous double mutant dcc1ca2. The double mutant dcc1ca2 showed similar phenotypes to those in the dcc1 mutant in terms of inhibited shoot regeneration (Supplemental Fig. S2, A–C). Furthermore, we detected the spatiotemporal expression patterns of CA2 and showed that CA2 expression in the callus started at 8 d on SIM and increased gradually in the inner region of the callus (Fig. 4, D and E).
Figure 4.
Loss of function of CA2 caused the decreased capacity of shoot regeneration. A to C, Callus morphology (A), frequencies of shoot regeneration from calli (B), and number of shoots per callus (C) in wild-type Col-0, ca2, and ca2 ProCA2:CA2 cultured on SIM at the indicated times. D, Expression pattern of CA2 detected by in situ hybridization in calli of Col-0 at the indicated times on SIM. E, Expression pattern of CA2 detected by GUS staining in calli of ProCA2:CA2-GUS transgenetic lines at the indicated times on SIM. S0 represents calli cultured on CIM at 6 d using roots as explants before transfer to SIM. S8, S12, S16, S20, S24, and S28 indicate calli cultured on SIM at 8, 12, 16, 20, 24, and 28 d, respectively. se values were calculated from three sets of biological replicates, and more than 100 calli were examined in each replicate. Asterisks indicate significant differences: **, P < 0.01 (Student’s one-tailed t test). Bars = 500 µm.
Mutation of DCC1 or CA2 Leads to Reduced Complex I Activity and Increased ROS Level
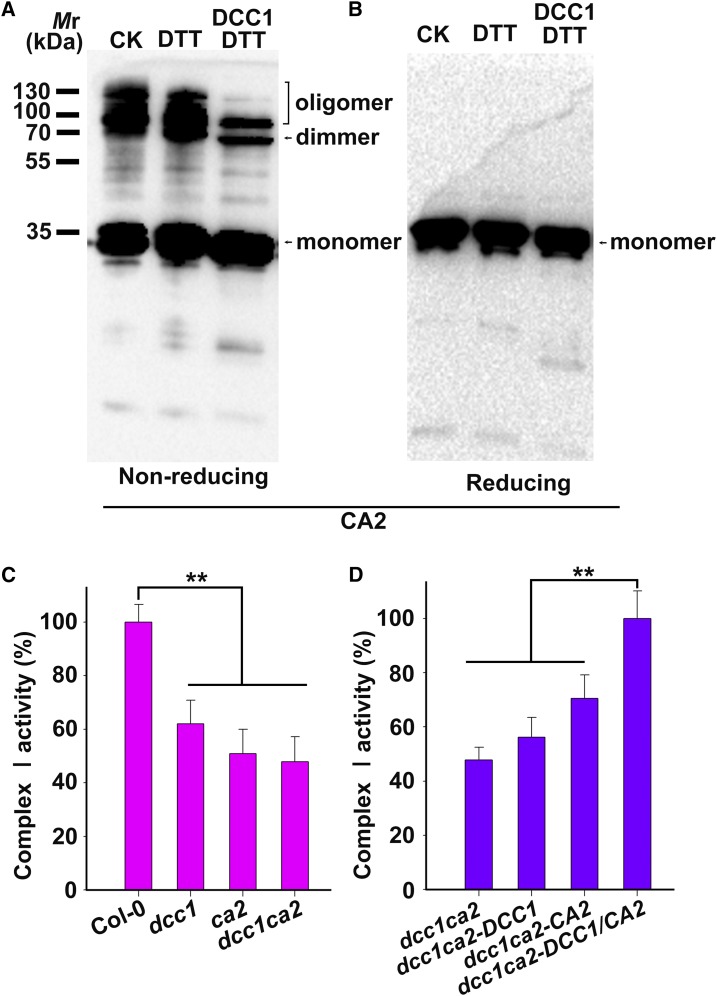
DCC1 was a functional Trx and interacted directly with CA2 protein, implying that CA2 might be reduced by DCC1. Therefore, we performed a reduction assay by using purified recombinant CA2 protein as the substrate. CA2 was incubated with or without GST-cleaved recombinant DCC1 protein. DTT (0.3 mm) was added to recycle DCC1 activity. The reduction of CA2 was monitored by the mobility shift in nonreducing SDS-PAGE. CA2 contained two Cys residues (Cys-83 and Cys-137) and formed dimer and oligomer complexes by intermolecular disulfide bonds (Fig. 5, A and B). The intensity of CA2 complex lanes was decreased significantly after incubation with DCC1 protein (Fig. 5, A and B), suggesting that DCC1 promotes monomer formation by reduction of the CA2 complex.
Figure 5.
DCC1 regulates Complex I activity via CA2 protein. A and B, CA2 dimer and oligomer were reduced by DCC1. Purified CA2-His protein was incubated in the assay mixture with or without DCC1 GST-cleaved protein (5 μm) for 30 min at room temperature and then subjected to nonreducing (A) or reducing (B) SDS-PAGE followed by immunoblot analysis. DTT (0.3 mm) was added to recycle DCC1 Trx activity. The positions of the monomer, dimer, and oligomer of CA2 are indicated. CA2 dimer and oligomer showed significantly reduced levels when incubated with DCC1 proteins (right lane). The reaction mixture without DCC1 proteins was used as the control (left and middle lanes). CK, Control check. C, Relative activity of mitochondrial respiratory Complex I. Mitochondria were isolated from calli (0.1 g) of wild-type Col-0, dcc1, ca2, and dcc1ca2 cultured on SIM at 16 d. Mitochondrial proteins (20 μg) were used to determine the Complex I activity. The absorbance of all samples was measured at 340 nm for 60 min using a plate reader. Complex I activity was calculated from the decrease in absorbance per minute. Functional loss of DCC1 or CA2 resulted in reduced activity of Complex I. D, Relative activity of Complex I in calli of dcc1ca2 after the addition of recombinant CA2 or DCC1 protein. Purified CA2-His proteins and DCC1-His proteins were added to the assay mixture. Only the addition of both DCC1 and CA2 proteins completely rescued the reduced Complex I activity. Asterisks indicate significant differences: **, P < 0.01 (Student’s one-tailed t test).
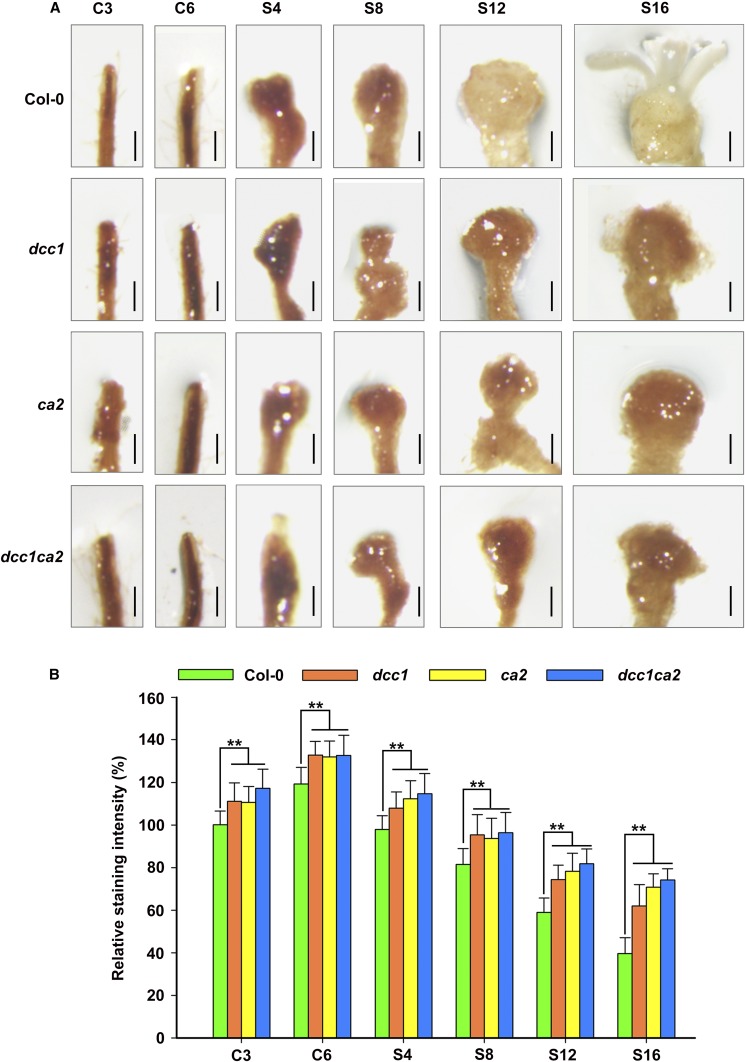
CA2 is a key subunit of the mitochondrial respiratory complex I, and defect in CA2 leads to decreased activity of complex I (Soto et al., 2015). Thus, we detected the activity of respiratory complex I in dcc1, ca2, and dcc1ca2 during shoot regeneration. Functional loss of DCC1 or CA2 caused a significantly decreased activity of the mitochondrial respiratory complex I (Fig. 5C). The complex I activity in the double mutant dcc1ca2 was not more affected than that in the single mutants, indicating that both proteins act in the same pathway. Furthermore, the reduced activity of complex I in the dcc1ca2 double mutant was completely rescued by the addition of both of DCC1 and CA2 proteins but not by the addition of only DCC1 or CA2 (Fig. 5D). These results suggest that DCC1 affects complex I activity via redox regulation of CA2 protein. A previous study has shown that the impaired activity of respiratory complex I results in increased ROS levels (Soto et al., 2015). Thus, we hypothesize that the reduced mitochondrial respiratory complex I activity triggers mitochondrial ROS production. To test this hypothesis, we performed 3,3′-diaminobenzidine (DAB) staining assays to detect the ROS levels in callus (Xie et al., 2014). During shoot regeneration, functional loss of DCC1 or CA2 resulted in increased ROS levels in calli cultured on CIM or SIM, respectively (Fig. 6). The ROS level in the double mutant dcc1ca2 was consistent with that in either single mutant (Fig. 6).
Figure 6.
Mutation of DCC1 or CA2 results in increased ROS levels. A, Levels of ROS in calli of Col-0, dcc1, ca2, and dcc1ca2 on CIM and SIM at the indicated times by DAB staining. B, Relative ROS staining intensity in calli of wild-type Col-0, dcc1, ca2, and dcc1ca2 on CIM and SIM at the indicated times. C3 and C6 indicate calli cultured on CIM at 3 and 6 d, respectively. S4, S8, S12, and S16 indicate calli cultured on SIM at 4, 8, 12, and 16 d, respectively. se values were calculated from three sets of biological replicates, and more than 100 calli were examined in each replicate. Asterisks indicate significant differences: **, P < 0.01 (Student’s one-tailed t test). Bars = 500 µm.
ROS Levels Mediate the Inhibition of Shoot Regeneration
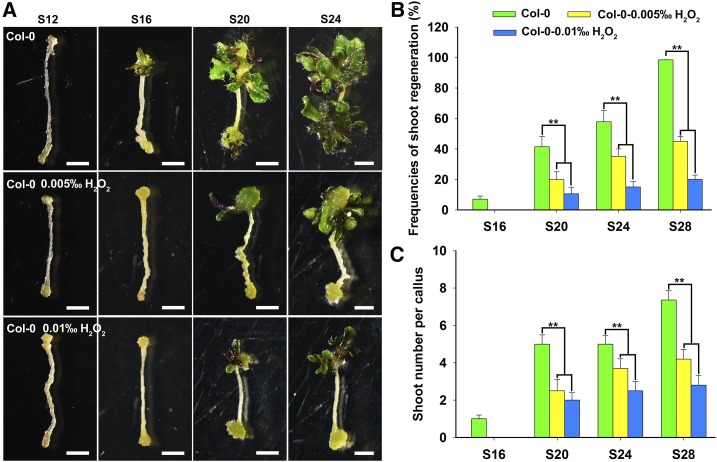
Since the loss of function of DCC1 and CA2 resulted in increased ROS levels, we thought that the inhibition of shoot regeneration in the dcc1 or ca2 mutant might be due to increased ROS levels. To test this hypothesis, we treated explants of the wild type with exogenous H2O2, a well-demonstrated regent responsible for increasing ROS levels (Yu et al., 2016), and found that shoot regeneration was inhibited significantly in the treated explants (Fig. 7). Moreover, higher levels of H2O2 led to lower shoot regeneration capacities (Fig. 7). The wild type without H2O2 treatment generated visible shoots at 16 d on SIM, whereas the wild type treated with 0.005‰ H2O2 and 0.01‰ H2O2 generated shoots at 20 d on SIM (Fig. 7A). At 28 d on SIM, the wild type without H2O2 treatment had a shoot regeneration frequency of 100% (Fig. 7B), whereas the wild type treated with 0.005‰ H2O2 and 0.01‰ H2O2 had shoot regeneration frequencies of 70% and 30%, respectively (Fig. 7B). The number of shoots regenerated from wild-type calli also was decreased when treated with exogenous H2O2 (Fig. 7C), indicating that ROS inhibits shoot regeneration.
Figure 7.
Treatment with exogenous ROS inhibits shoot regeneration. Callus morphology (A), frequencies of shoot regeneration from calli (B), and number of shoots per callus (C) in wild-type Col-0 after treatment with H2O2 at different concentrations during shoot regeneration are shown. S12, S16, S20, S24, and S28 indicate calli cultured on SIM at 12, 16, 20, 24, and 28 d, respectively. se values were calculated from three sets of biological replicates, and more than 100 calli were examined in each replicate. Asterisks indicate significant differences: **, P < 0.01 (Student’s one-tailed t test). Bars = 500 µm.
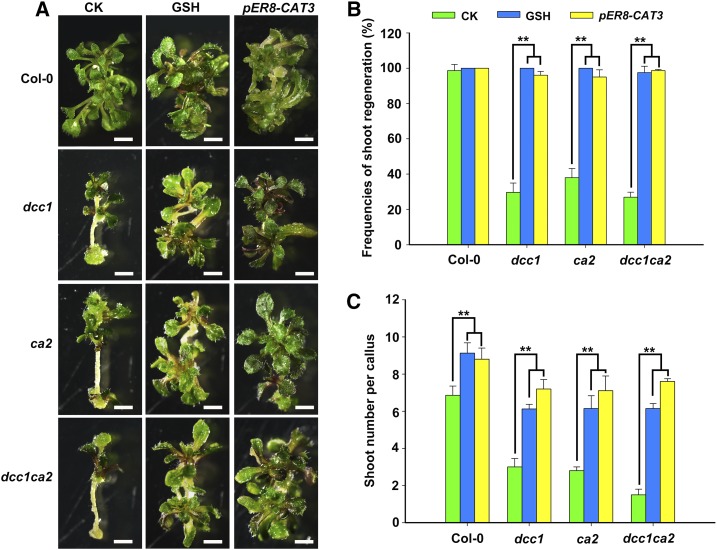
Next, we determined whether GSH, the main endogenous antioxidant responsible for ROS removal (Lu and Holmgren, 2014), could rescue the phenotypes of dcc1 and ca2 by decreasing the ROS levels. The frequencies of shoot regeneration in the dcc1, ca2, and dcc1ca2 mutants were increased with gradually increasing concentrations of GSH (Supplemental Fig. S3A). The number of shoots per callus showed similar increases under GSH treatment (Supplemental Fig. S3B). The dcc1, ca2, and dcc1ca2 mutants were completely rescued by GSH at 600 µm (Fig. 8). Although shoot regeneration was promoted in the wild type by GSH, the shoot regeneration in the dcc1, ca2, and dcc1ca2 mutants by GSH was increased significantly compared with that in the wild type (Fig. 8). These results indicate that the reduction of ROS levels by GSH promotes shoot regeneration. To further confirm the function of ROS in shoot regeneration, we transformed the construct pER8-CAT3, which overexpressed CATALASE3 (CAT3) when induced with estradiol, into Col-0 and the dcc1, ca2, and dcc1ca2 mutants. CAT3 encodes a catalase that catalyzes the breakdown of H2O2 into water and oxygen, thereby reducing the H2O2 level (Zou et al., 2015). The CAT3 overexpression transgenic lines were obtained in the backgrounds of Col-0, dcc1, ca2, and dcc1ca2 (Supplemental Fig. S4). The shoot regeneration frequencies and number of shoots per callus were increased in the CAT3-overexpressing transgenic lines (Fig. 8), indicating that the reduction in ROS levels by overexpression of CAT3 rescued the phenotypes of the mutants. CAT3 overexpression in Col-0 led to about two more shoots per callus than Col-0, whereas CAT3 overexpression in dcc1, ca2, and dcc1ca2 mutants showed about five more shoots compared with their mutants. Thus, these results confirm that the ROS level is critical for regulating the shoot regeneration capacity.
Figure 8.
Decreased ROS level rescues phenotypes of the dcc1ca2 double mutant. Callus morphology (A), frequencies of shoot regeneration from calli (B), and number of shoots per callus (C) in wild-type Col-0, dcc1, ca2, and dcc1ca2 after treatment with GSH (600 μm) or overexpression of CAT3 on SIM at 28 d are shown. GSH is the main endogenous antioxidant responsible for ROS removal. CAT3 encodes a catalase that catalyzes the breakdown of H2O2 into water and oxygen, thereby reducing the H2O2 level. CK, Control check. pER8-CAT3 represents the overexpression of CAT3 in the dcc1, ca2, and dcc1ca2 backgrounds when induced with estradiol. se values were calculated from three sets of biological replicates, and more than 100 calli were examined in each replicate. Asterisks indicate significant differences: **, P < 0.01 (Student’s one-tailed t test). Bars = 500 µm.
ROS Regulates Shoot Regeneration by Modulating Multiple Pathways
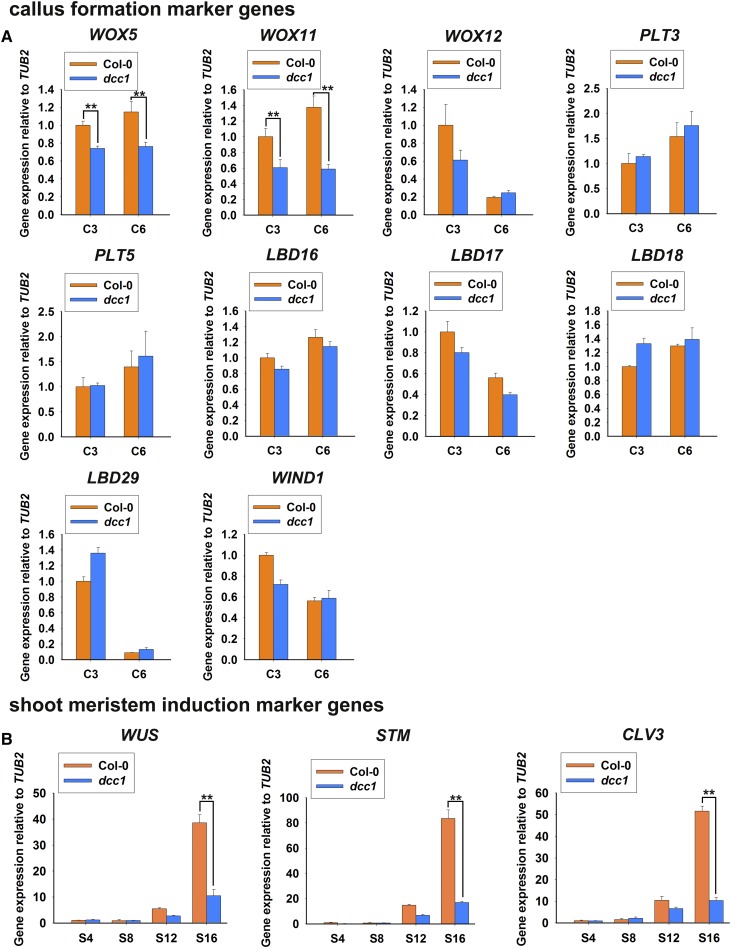
Shoot regeneration includes callus formation and shoot meristem induction. To determine whether ROS was involved in the regulation of callus formation by master regulators, we analyzed the expression of WOXs, PLTs, LBDs, and WIND1 during this process. Only WOX5 and WOX11 showed reduced levels in the dcc1 mutant (Fig. 9A), suggesting that ROS affects callus formation by the modulation of WOX5 and WOX11 expression. Next, we detected WUS, SHOOT MERISTEMLESS (STM), and CLAVATA3 (CLV3), which are involved in shoot meristem induction (Ikeuchi et al., 2016). The transcript levels of all three of these genes were decreased in the dcc1 mutant cultured on SIM at 12 and 16 d (Fig. 9B), suggesting that ROS regulates shoot induction by modulating the expression of WOXs, WUS, CLV3, and STM.
Figure 9.
Transcript levels of master genes involved in shoot regeneration. Transcript levels of master genes for callus formation (A) and shoot meristem induction (B) during shoot regeneration are shown. C3 and C6 represent calli cultured on CIM at 3 and 6 d, respectively, using roots as explants before transfer to SIM. S4, S8, S12, and S16 indicate calli cultured on SIM at 4, 8, 12, and 16 d, respectively. se values were calculated from three sets of biological replicates. Asterisks indicate significant differences: **, P < 0.01 (Student’s one-tailed t test).
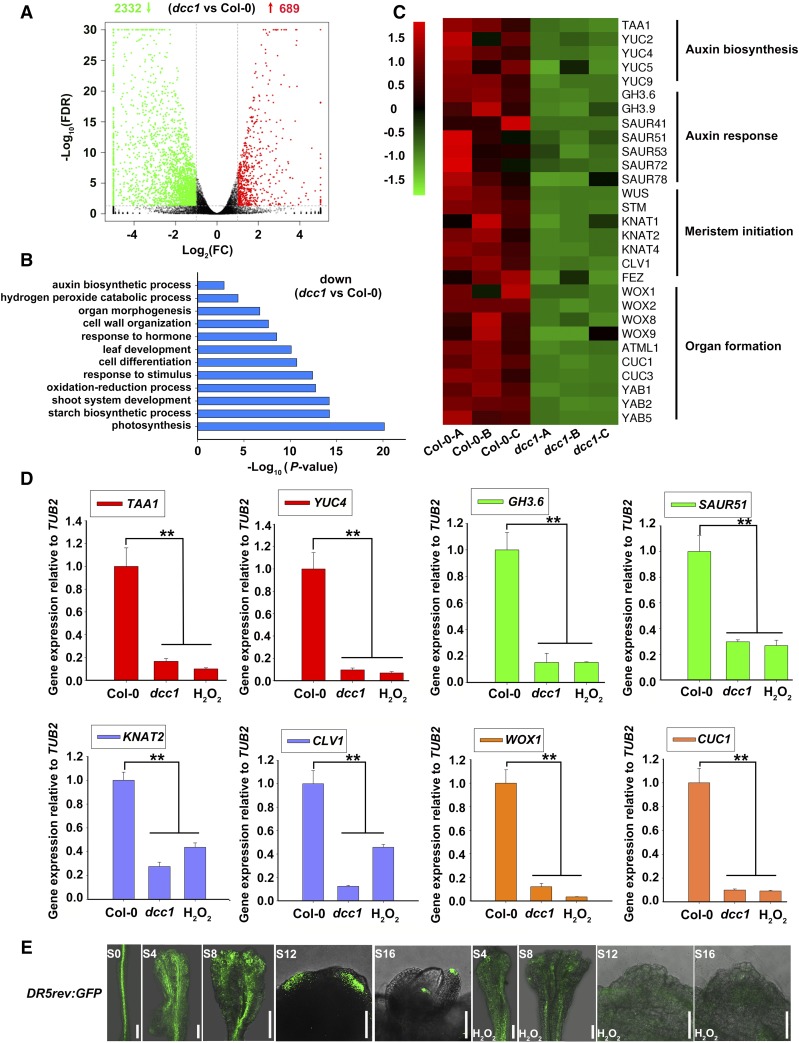
To uncover the functional pathways of ROS in detail, we performed an RNA-seq analysis using the calli of wild-type Col-0 and dcc1 cultured on SIM at 16 d. Three high-quality biological repeats of the wild type (Col-0-A, Col-0-B, and Col-0-C) and dcc1 (dcc1-A, dcc1-B, and dcc1-C) were used for RNA-seq analyses (Supplemental Fig. S5). Using a 2-fold change in the gene transcript levels between wild-type Col-0 and the mutant dcc1 with an adjusted P value (false discovery rate [FDR]) cutoff of 0.05 as the criterion for differential expression, we identified many differentially expressed genes (Fig. 10A). Among them, 689 genes were up-regulated and 2,332 genes were down-regulated in dcc1 (Fig. 10A; Supplemental Tables S2 and S3). Gene Ontology (GO) analyses showed that DCC1 regulated multiple processes, such as auxin biosynthetic process (P = 1.38 × 10−3), shoot system development (P = 6.33 × 10−15), oxidation-reduction process (P = 1.83 × 10−13), cell differentiation (P = 1.98 × 10−11), response to hormones (P = 2.88 × 10−9), cell wall organization (P = 2.24 × 10−8), organ morphogenesis (P = 1.94 × 10−7), and H2O2 catabolic process (P = 4.35 × 10−5; Fig. 10B). In addition to WUS and STM, the meristem master genes KNOTTED-LIKE FROM ARABIDOPSIS THALIANA1 (KNAT1), KNAT2, and KNAT4 (Scofield et al., 2007), WOXs (van der Graaff et al., 2009), and CUP-SHAPED COTYLEDON (CUCs; Ikeuchi et al., 2016) were down-regulated in dcc1 (Fig. 10, B and C). Next, we performed quantitative reverse transcription (qRT)-PCR to confirm the transcript levels of genes identified in the RNA-seq analyses. As expected, the transcript levels of these genes were low in the dcc1 mutant and in the presence of exogenous H2O2 (Fig. 10D; Supplemental Fig. S6). These results suggest that the DCC1-mediated ROS level regulates shoot regeneration by the genes involved in shoot meristem initiation.
Figure 10.
DCC1 regulates shoot regeneration through modulating multiple signaling by RNA-seq analyses. A, Differential gene expression detected by edgeR. A 2-fold change in gene transcript levels between wild-type Col-0 and the mutant dcc1 with an adjusted P value (FDR) cutoff of 0.05 was considered as differential expression: 2,332 genes were down-regulated and 689 genes were up-regulated in the dcc1 mutant. B, GO analyses of down-regulated genes in the dcc1 mutant. C, Cluster analysis of transcript levels of genes involved in auxin signaling, meristem initiation, and organ formation. Col-0-A, Col-0-B, and Col-0-C are three repeats of Col-0, and dcc1-A, dcc1-B, and dcc1-C are three repeats of dcc1. D, Transcript levels of TAA1, YUC4, GH3.6, SAUR51, KNAT2, CLV1, WOX1, and CUC1 in calli of Col-0, dcc1, and H2O2 (wild-type Col-0 treated with 0.01‰ H2O2 during shoot regeneration) on SIM at 16 d, as determined by qRT-PCR. TUBULIN2 (TUB2) was the reference gene. Asterisks indicate significant differences: **, P < 0.01 (Student’s one-tailed t test). E, Response patterns of DR5rev:GFP in calli with or without H2O2 treatment on SIM at the indicated times. S0 represents calli cultured on CIM at 6 d using roots as explants before transfer to SIM. S4, S8, S12, and S16 indicate calli cultured on SIM at 4, 8, 12, and 16 d, respectively. Bars = 500 μm.
Interestingly, several auxin biosynthetic genes, including TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1), YUCCA2 (YUC2), YUC4, YUC5, and YUC9 (Zhao, 2010), were significantly down-regulated in dcc1 (Fig. 10, C and D; Supplemental Fig. S6). The transcript levels of auxin response genes, such as GRETCHEN HAGEN3.6 (GH3.6) and SMALL AUXIN UPREGULATED51 (SAUR51; Hagen and Guilfoyle, 2002; Staswick et al., 2005), also were decreased in dcc1 (Fig. 10, C and D; Supplemental Fig. S6). Furthermore, we used the well-demonstrated auxin marker DR5rev:GFP to detect the auxin response signal distribution in regenerating shoots (Cheng et al., 2013). The GFP signal was weakened by H2O2 treatment during shoot regeneration (Fig. 10E). Auxin is a critical determinant in shoot regeneration, and the impaired auxin biosynthesis and signaling repress shoot regeneration (Gordon et al., 2007; Cheng et al., 2013). Moreover, the functional loss of DCC1 led to increased ROS levels (Fig. 6, A and B), and ROS repressed auxin signaling (Fig. 10E). Thus, we propose that the DCC1-mediated ROS level regulates shoot regeneration, at least in part, through a modulation of the auxin biosynthesis and signaling pathway.
DCC1 Is a Major Determinant of Natural Variation in Shoot Regeneration
To determine whether DCC1 was a factor in the natural variation in shoot regeneration, 48 ecotypes of Arabidopsis were used in shoot regeneration analyses using the roots as explants. The shoot regeneration capacity varied widely among the different ecotypes of Arabidopsis. The shoot regeneration frequencies ranged from 5% to 100% (Supplemental Fig. S7A). The ecotypes Gu-0, Nc-1, Wa-1, and Kelsterbach-4 showed very low shoot regeneration frequencies on SIM at 28 d, while the ecotypes Col-0, Ws-0, Pu2-7, Lan-0, and Bu-0 showed high shoot regeneration frequencies (Supplemental Fig. S7A). These diverse frequencies indicated the wide natural variation in regeneration capacity among Arabidopsis genotypes. There was also a wide range among genotypes in the number of shoots per callus (Supplemental Fig. S7B). Each callus of Col-0, Ws-0, Pu2-7, Bu-0, Tsu-1, and Ct-1 generated more than five shoots, while calli of other ecotypes generated four or fewer. These results confirm that there are diverse shoot regeneration capacities among different ecotypes of Arabidopsis.
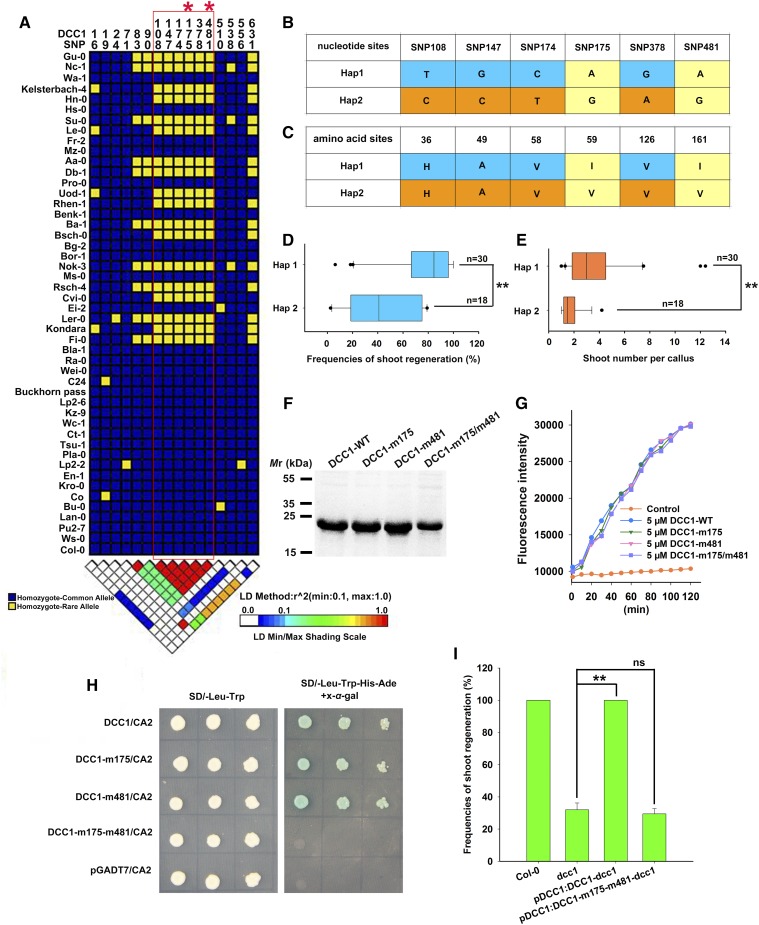
We further performed linkage disequilibrium analyses using the nucleotide sequences of DCC1 in 48 different ecotypes and found that six single-nucleotide polymorphisms (SNP 108, 147, 174, 175, 378, and 481) were highly associated with the shoot regeneration frequency (Fig. 11A). Two haplotypes (Hap1 and Hap2) of DCC1 were identified among 48 natural variants of Arabidopsis. Hap1 included 30 ecotypes such as Col-0, Ws-0, Pu2-7, Lan-0, and Bu-0, and Hap2 included 18 ecotypes such as Gu-0, Nc-1, Keleterbach-4, Hn-0, and Su-0 (Fig. 11, A–C). The shoot regeneration frequencies and the number of shoots per callus were much higher in the Hap1 ecotypes than in the Hap2 ecotypes (Fig. 11, D and E), suggesting that this natural variation in DCC1 is related to the regulation of shoot regeneration. Among the six SNPs, only two (SNP 175 and 481) resulted in the sense mutation (I59V and I161V) for the corresponding amino acids (Fig. 11, B and C). To identify the critical SNPs in DCC1 among the natural variants, we detected the Trx activity by reduction of FiTC-insulin. Mutations at both positions 175 and 481 did not affect Trx activity of DCC1 (Fig. 11, F and G). Next, we performed yeast two-hybrid assays using DCC1 with mutations at positions 175 and 481. We found that DCC1 did not interact with CA2 when there were mutations at both positions 175 and 481 (Fig. 11H). We also transformed a pDCC1:DCC1-m175-m481 construct with DCC1 mutated at positions 175 and 481 to rescue the dcc1 mutant. The shoot regeneration frequencies in the pDCC1:DCC1-m175-m481-dcc1 transgenic lines were not significantly different from those in dcc1 (Fig. 11I), implying that SNP 175 and 481 of DCC1 are critical for shoot regeneration.
Figure 11.
DCC1 regulates the capacity of shoot regeneration among different Arabidopsis ecotypes. A, Linkage disequilibrium analyses of DCC1 in 48 Arabidopsis ecotypes indicated six critical SNPs (SNP 108, 147, 174, 175, 378, and 481). B, Two haplotypes (Hap1 and Hap2) of DCC1 were characterized based on six SNPs of nucleotide sequences in Arabidopsis natural variants. C, Amino acid sequences of Hap1 and Hap2. Two SNPs (SNP 175 and 481) resulted in amino acid mutations (I59V and I161V). D and E, Shoot regeneration frequencies from calli (D) and number of shoots per callus (E) in each haplotype group (n, number of genotypes belonging to each haplotype group). F, Purified proteins were subjected to SDS-PAGE and stained by Coomassie Brilliant Blue R-250. DCC1-WT from Col-0 was used as the control. DCC1-m175, DCC1-m481, and DCC1-m175/m481 were mutated according to Hap2 SNPs (SNP 175 and 481). G, Insulin reduction assay by the various proteins. Purified proteins were subjected to a reduction assay by using FiTC-insulin as the substrate, which displayed higher fluorescence after disulfide reduction. The assay mixture lacking recombinant proteins served as the control. Fluorescence intensity was recorded at 515- to 525-nm emission after 480- to 495-nm excitation for 120 min in a fluorescent plate reader at room temperature. H, Both SNP 175 and SNP 481 were critical for the interaction of DCC1 and CA2 by yeast two-hybrid assay. I, Frequencies of shoot regeneration from calli of wild-type Col-0, dcc1, pDCC1:DCC1-dcc1, and pDCC1:DCC1-m175-m481-dcc1 on SIM at 28 d. Significant differences are indicated: **, P < 0.01 and ns, P > 0.05 (Student’s one-tailed t test).
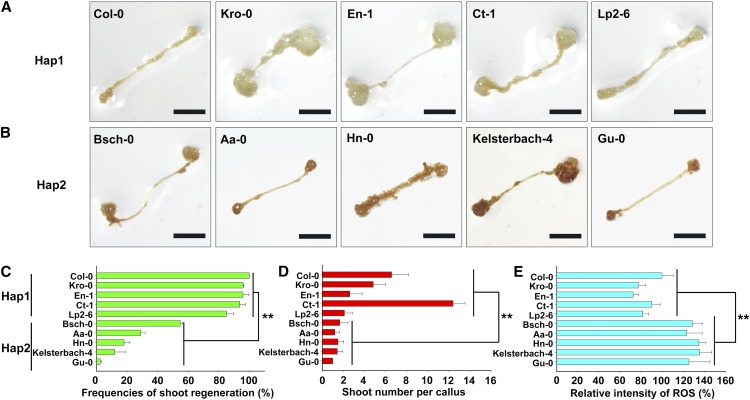
Because DCC1 regulated shoot regeneration through ROS level, we assumed that DCC1-mediated ROS homeostasis might be involved in the natural variation in shoot regeneration among Arabidopsis ecotypes. To test this hypothesis, we detected the ROS levels in the Hap1 and Hap2 ecotypes by DAB staining (Fig. 12, A and B). The shoot regeneration frequencies and the number of shoots per callus were higher in the Hap1 ecotypes (Col-0, Kro-0, En-1, Ct-1, and Lp2-6) than in the Hap2 ecotypes (Bsch-0, Aa-0, Hn-0, Kelsterbach-4, and Gu-0; Fig. 12, C and D). Interestingly, the ROS levels were significantly lower in the Hap1 ecotypes than in the Hap2 ecotypes (Fig. 12E). These results suggest that DCC1-mediated ROS homeostasis is critical to the natural variation in shoot regeneration among Arabidopsis ecotypes.
Figure 12.
ROS levels are associated with the shoot regeneration capacities of different ecotypes in Arabidopsis. A, DAB staining showing ROS levels in calli of different ecotypes of Hap1 cultured on SIM at 16 d. B, DAB staining showing ROS levels in calli of different ecotypes of Hap2 cultured on SIM at 16 d. C, Shoot regeneration frequencies from calli of different ecotypes cultured on SIM at 28 d. D, Number of shoots per callus of different ecotypes cultured on SIM at 28 d. E, Relative staining intensity of ROS in calli of different ecotypes cultured on SIM at 16 d. se values were calculated from three sets of biological replicates, and more than 100 calli were examined in each replicate. Asterisks indicate significant differences: **, P < 0.01 (Student’s one-tailed t test). Bars = 500 µm.
DISCUSSION
Trxs belong to small proteins with a conserved CxxC motif throughout all organisms (Gelhaye et al., 2005; Meyer et al., 2005). Classic Trxs, such as Trx m, Trx o, Trx h, and Trx f, contain a motif of WCGPC or WCPPC, and atypical Trxs, such as APS REDUCTASE1 and HIGH CHLOROPHYLL FLUORESCENCE164, have a motif of WCPFC, HCGPC, or WCEVC (Gelhaye et al., 2005; Meyer et al., 2005). Trxs catalyze the reduction of the disulfide bond through these motifs and are critical to change the redox state of their target proteins (Rouhier et al., 2015). The redox state of ND3 (an important subunit of Complex I) determines the activity of Complex I (Galkin et al., 2008). Many subunits of mitochondrial respiratory Complex I (such as NAD7, NAD9, NDS8A, and NDUV2) are identified as potential targets of Trxs, but the direct evidence of redox regulation of these subunits by Trxs is missing (Balmer et al., 2004; Yoshida et al., 2013). In this study, we characterized a novel atypical Trx DCC1 that belongs to the DCC family of proteins. These proteins contain a conserved DxxCxxC motif (such as DGDCPLC, DGECPLC, or DGVCHLC) that differs from motifs of classic and atypical Trxs (Fig. 2B). DCC1 had the Trx activity determined by the reduction of insulin (Fig. 2D). CA2 is an essential subunit of Complex I (Soto et al., 2015) and was identified as a direct target of DCC1 (Fig. 5, A and B). DCC1 regulated Complex I activity by reduction of the CA2 complex (Fig. 5). Inhibition of Complex I activity caused by the mutation of DCC1 or CA2 triggered increased ROS production (Fig. 6), suggesting that DCC1 regulates Complex I activity and ROS homeostasis by redox regulation of CA2.
Many studies show that ROS can act as signaling factors regulating diverse processes (Rouhier et al., 2015; Schmidt and Schippers, 2015; Schippers et al., 2016). NADPH-DEPENDENT THIOREDOXIN REDUCTASE A (NTRA), NADPH-DEPENDENT THIOREDOXIN REDUCTASE B (NTRB), and the glutathione biosynthesis gene CADMIUM SENSITIVE2 (CAD2) function to maintain ROS homeostasis, and the triple mutant ntra ntrb cad2 generated an abnormal shoot meristem (Bashandy et al., 2010). Higher levels of ROS caused by the overexpression of UP BEAT1 led to a short root meristem (Tsukagoshi et al., 2010). Elevated levels of ROS by the mutation of KUODA1 led to a decreased leaf cell size (Lu et al., 2014). Here, we showed that increasing levels of ROS resulted in the inhibition of shoot regeneration (Figs. 6 and 7), whereas decreasing levels of ROS increased the capacity of shoot regeneration in the dcc1, ca2, and dcc1ca2 mutants (Fig. 8). Thus, ROS homeostasis is critical for the regulation of shoot regeneration.
Different mechanisms have been proposed to mediate the perception of ROS (Wrzaczek et al., 2013). An important mechanism is the one-component redox signaling system, which is based on redox-sensitive transcription factors (Schmidt and Schippers, 2015). Recent studies have identified a number of crucial redox-sensitive transcription factors, such as PHAVOLUTA for meristem development and organ polarity, BASIC REGION/LEUCINE ZIPPER TRANSCRIPTION FACTOR16 for light signaling, and TEOSINTE BRANCHED1-CYCLOIDEAPCF15 (TCP15) for auxin signaling (Schmidt and Schippers, 2015). Our results indicated that the increased ROS levels resulted in repressed expression of master genes for callus formation (WOX5 and WOX11) and shoot meristem initiation (WUS, CLV3, and STM; Fig. 9). Moreover, auxin biosynthesis genes (YUC4 and TAA1) were repressed by ROS (Fig. 10, C and D). Previously, it was shown that auxin was critical for shoot induction, and mutation of the auxin biosynthetic YUC genes repressed shoot regeneration (Gordon et al., 2007; Cheng et al., 2013). The redox-sensitive transcription factors TCPs regulate YUC expression and auxin biosynthesis (Viola et al., 2013; Lucero et al., 2015; Challa et al., 2016). Thus, it is likely that ROS regulates shoot regeneration by the redox modification of transcription factors involved in auxin biosynthesis and signaling.
The evolutionary adaptation of plants to environmental changes leads to diverse natural variation, which is critical for plant diversity (Mitchell-Olds and Schmitt, 2006). Natural variation causes a wide range of biological phenotypes and provides rich resources with which to analyze many important complex traits (Mitchell-Olds and Schmitt, 2006). A number of factors involved in natural variations have been identified in diverse biological processes, including regulation of flowering, seed development, and drought resistance (Johanson et al., 2000; Li et al., 2011b; Wang et al., 2016). FRIGIDA is a major determinant of the natural variation in Arabidopsis flowering time (Johanson et al., 2000). Natural variations in GS5 can explain variations in rice (Oryza sativa) grain size and yield (Li et al., 2011b), and those in ZmVPP1 contribute to the drought tolerance trait in maize (Zea mays; Wang et al., 2016). Here, we identified a major regulator, DCC1, that explains the variation in shoot regeneration capacity among natural variants of Arabidopsis (Fig. 11, A–E). The polymorphisms of DCC1 were used to classify the different ecotypes of Arabidopsis into two groups: Hap1 and Hap2 (Fig. 11B). Interestingly, mutations at both critical SNPs (SNP 175 and 481) did not affect the Trx activity of DCC1 (Fig. 11G) but blocked the interaction between DCC1 and CA2 (Fig. 11H), indicating that the two SNPs are critical for the function of DCC1, which is a major player in determining shoot regeneration capacity among natural variations.
MATERIALS AND METHODS
Plant Materials
The mutants dcc1 (SALK_051222C) and ca2 (SALK_010194C), line MT-GK (CS16263), and different ecotypes of Arabidopsis (Arabidopsis thaliana) were obtained from the Arabidopsis Biological Resource Center. DR5rev:GFP lines were generously provided by Dr. Jian Xu (Department of Biological Science, National University of Singapore; Xu et al., 2006).
Shoot Regeneration
The shoot regeneration experiment was performed as described previously (Li et al., 2011a). Seeds were placed on germination medium after sterilization in 70% (v/v) ethanol for 5 min and 2.6% (v/v) sodium hypochlorite for 10 min. All the seeds were stratified at 4°C for 3 d and then grown for 12 d in a growth chamber at 22°C under a 16-h-light/8-h-dark photoperiod. Root segments (5–10 mm) were cut and placed on CIM (Gamborg’s B5 medium supplemented with 0.5 g L−1 MES, 2% [w/v] Glc, 0.2 µm kinetin, 2.2 µm 2,4-dichlorophenoxyacetic acid, and 0.8% [w/v] agar). After the explants were incubated in continuous light for 6 d, the calli were transferred onto SIM (Gamborg’s B5 medium supplemented with 0.5 g L−1 MES, 2% [w/v] Glc, 0.9 µm 3-indoleacetic acid, 0.5 µm 2-isopentenyladenine, and 0.8% [w/v] agar) and incubated in continuous light. The calli were transferred onto fresh medium every 6 d. Images of calli were acquired using an Olympus DP72 microscope. The shoots on each callus were defined as being at least 2 mm long. Shoot regeneration frequency (%) was calculated as follows: number of calli with at least one shoot/total number of calli cultured on SIM × 100. Three sets of biological replicates were used in the calculations, and more than 100 calli were examined in each replicate.
Plasmid Construction
The oligonucleotide primers for all constructs are given in Supplemental Table S4. For phenotypic complementation and GUS signal analyses, DCC1 or CA2 genomic fragments including the promoter and gene were introduced into pMDC99 to construct ProDCC1:DCC1 and ProCA2:CA2, respectively, and into pMDC163 to obtain ProDCC1:DCC1-GUS and ProCA2:CA2-GUS, respectively. For subcellular localization, DCC1 cDNA was cloned into pH7RWG2 to generate 35S:DCC1-RFP by Gateway technology (Karimi et al., 2002). For the yeast two-hybrid constructs, cDNAs of DCC1 and CA2 without the signal peptide sequence were cloned into pGADT7 and pGBKT7 to generate DCC1-AD and CA2-BD, respectively. For the pull-down assay, cDNAs of DCC1 and CA2 without the signal peptide sequence were cloned into pET28a and pGEX-4T-1 to generate DCC1-His and CA2-GST, respectively. For the firefly luciferase complementation assay, gene sequences of DCC1 and CA2 were cloned into p1300-nLUC and p1300-cLUC to generate 35S:DCC1-nLUC and 35S:CA2-cLUC, respectively. For the coimmunoprecipitation, gene sequences of DCC1 and CA2 were introduced into pEarlyGate103 and pEarlyGate203 to generate 35S:DCC1-GFP and 35S:CA2-MYC, respectively. For the reduction of CA2, cDNA of DCC1 without the signal peptide sequence was cloned into pGEX-4T-1 to generate DCC1-GST. For CAT3 overexpression, CAT3 cDNA was amplified and cloned into pER8 to construct pER8-CAT3.
For the construct pDCC1:DCC1-m175-m481, the DCC1 promoter sequence was amplified using the primer DCC1-F7/R7 and cloned into pBI121. DCC1-m175-m481 (m175 indicates mutation at position 175 of the DCC1 gene) fragments were amplified using primers (DCC1-F8/R8, DCC1-F9/R9, and DCC1-F10/R10) by overlap-extension two-step PCR mutagenesis and introduced into pBI121 containing the DCC1 promoter. For DCC1 Trx activity determination, DCC1-m175-His (with primer DCC1-F11/R11), DCC1-m481-His (with primers DCC1-F12/R12 and DCC1-F13/R13), and DCC1-m175-m481-His (with primers DCC1-F11/R12 and DCC1-F13/R13) were generated by overlap-extension two-step PCR mutagenesis and inserted into pET28a for protein purification. For yeast hybrid interaction, DCC1-m175-AD (with primer DCC1-F14/R14), DCC1-m481-AD (with primers DCC1-F15/R15 and DCC1-F16/R16), and DCC1-m175-m481-AD (with primers DCC1-F14/R15 and DCC1-F16/R16) were amplified by overlap-extension two-step PCR mutagenesis and inserted into pGADT7 for yeast hybrid interaction.
GUS Staining Assay
We conducted GUS staining assays as described elsewhere (Sieburth and Meyerowitz, 1997; Su et al., 2009). Briefly, calli were incubated in GUS assay buffer at 37°C for 10 h and then fixed in 70% (v/v) ethanol at room temperature. The calli were embedded in paraffin and cut into 10-μm sections. Ruthenium Red (0.2 g L−1) was used to stain cell walls.
Confocal Microscopy
MT-GK is a reported marker line specially expressing Mito-GFP (Nelson et al., 2007). To determine the subcellular localization of DCC1, the construct 35S:DCC1-RFP was introduced into MT-GK. The roots of the T1 transgenetic lines were excised for imaging by a Leica TCS SP5 confocal microscope. The DCC1-RFP signal was observed at 505- to 550-nm emission after 561-nm excitation, whereas the Mito-GFP signal was observed at 570- to 620-nm emission after 488-nm excitation. The merged signals of GFP and RFP showed yellow color. To detect the auxin response pattern, approximately 60 calli cultured on SIM at different times were sampled to observe the expression of DR5rev:GFP. The calli were cut into 1- to 2-mm sections along the longitudinal axis, and the sections were viewed with a Leica TCS SP5 confocal microscope. The GFP signal was observed at 505- to 550-nm emission after 488-nm excitation.
Phylogenetic Analysis
Phylogenetic analysis used DCC1 (NP_568719.1) with its homologs in Camelina sativa (XP_010441588.1), Brassica rapa (XP_010441588.1), Raphanus sativus (XP_018455194.1), Gossypium hirsutum (XP_016755751.1), Ziziphus jujuba (XP_015882648.1), Malus domestica (XP_008393058.1), Prunus persica (XP_007209569.1), Oryza officinalis (XP_015633461.1), Zea mays (XP_008663715.1), Physcomitrella patens (XP_001777984.1), Populus trichocarpa (XP_002322151.2), Vitis vinifera (XP_002283569.1), Triticum urartu (EMS60538.1), Microcystis aeruginosa (WP_002792509.1), Arabidopsis (AT1G52590 [NP_564611.1] and AT1G24095 [NP_001185076.1]), Caulobacter crescentus (NP_422377.1), Bacillus halodurans (WP_010897025.1), and Pseudomonas aeruginosa (NP_249825.1). The analysis was performed by MEGA 5.1 software. The protein sequences were downloaded from the National Center for Biotechnology Information database.
Trx Activity Assay
The construct pET28a-DCC1 was transferred into Rosetta competent cells. DCC1-His proteins were induced by incubation in 1 mm IPTG at 22°C with shaking at 120 rpm for 16 h. The cell pellet was resuspended in buffer (25 mm Tris, pH 7.5, and 0.5 mm DTT; Motohashi et al., 2003). Protein purification was performed using Ni Sepharose 6 Fast Flow (GE Healthcare; catalog no. 28-9355-97). Then, the proteins were dialyzed against buffer (25 mm Tris, pH 7.5) to remove imidazole using Slide-A-Lyzer Dialysis Cassettes (Thermo Fisher Scientific; catalog no. 66810) for 24 h. Trx activity was measured using the Thioredoxin Activity Fluorescent Assay Kit (Cayman Chemicals; catalog no. 11527). The FiTC-insulin in the kit was used as the substrate of DCC1. FiTC displays higher fluorescence after disulfide reduction (Montano et al., 2014). The reduction assay was performed by incubation with purified DCC1 protein (10 μg). NADPH and TrxR were used to recycle the DCC1 Trx activity. The reaction mixture without DCC1 proteins was used as the control. The fluorescence intensity of each sample was recorded at 515- to 525-nm emission after excitation at 480 to 495 nm for 60 min using the BioTek Synergy 2 Multi-Mode Reader (BioTek) at ambient room temperature. Three sets of biological replicates were performed for each reaction.
Yeast Two-Hybrid Assay
The yeast two-hybrid assay was performed using the Matchmaker Gold Yeast Two-Hybrid System (Clontech; catalog no. 630489) according to the manufacturer’s instructions. The constructs DCC1-AD and CA2-BD were cotransferred into Y2H Gold competent cells. Activation was observed at 3 d on selection plates (SD/-Leu-Trp-His-Ade) with X-α-gal.
Firefly Luciferase Complementation Assay
Firefly luciferase complementation assays were conducted as described previously (Li et al., 2014). The constructs DCC1-nLUC and CA2-cLUC were transformed into leaves of Nicotiana benthamiana for transient expression. Transfected leaves were sprayed with buffer (1 mm d-luciferin sodium salt and 0.1% [v/v] Triton X-100) 5 min before being imaged using the IVIS Lumina II system (Caliper Life Sciences).
Pull-Down Assay
For the pull-down assay, 10 μg of CA2-GST protein was incubated with 10 μL of glutathione-Sepharose-4B beads (GE Healthcare; catalog no. 17-0756-01) in blocking buffer (140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, 0.1% [w/v] Nonidet P-40, and 1 g of BSA) under agitation at 4°C for 2 h. Then, 1 μg of DCC1-His protein was added, and the mixture was incubated for 2 h in binding buffer (140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, and 0.1% [w/v] Nonidet P-40). The beads were washed 10 times with washing buffer (400 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, and 0.1% [v/v] Nonidet P-40). Finally, 10% input and pull-down proteins were separated by 12% SDS-PAGE and detected by immunoblotting using the antibodies anti-His, diluted 1:3,000 (Sigma; catalog no. H1029), or anti-GST, diluted 1:3,000 (Sigma; catalog no. G1160).
Coimmunoprecipitation Assay
We used transgenic seedlings of 35S:DCC1-GFP and the F1 generation of 35S:DCC1-GFP × 35S:CA2-MYC for immunoprecipitation analyses. Proteins were extracted from 2 g of seedlings with extraction buffer (140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, 0.1 mm EDTA, 0.1% [v/v] Nonidet P-40, 5 mm DTT, and 0.5 mm PMSF). Then, 60 μL of beads was incubated with 10 μL of anti-GFP antibody with agitation for 2 h at 4°C. Then, supernatants were added and incubated with agitation for 8 h at 4°C. After incubation, beads were washed 10 times with washing buffer (400 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, 0.1 mm EDTA, 0.1% [v/v] Nonidet P-40, 5 mm DTT, and 0.5 mm phenylmethylsulfonyl fluoride). Finally, 10% input and immunoprecipitation proteins were separated by 12% SDS-PAGE and detected by immunoblotting using the antibodies anti-MYC, diluted 1:1,000 (Sigma; catalog no. M4439), or anti-GFP, diluted 1:1,000 (Roche; catalog no. 11814460001).
Reduction of CA2 by DCC1
The reduction assay was performed as described previously with slight modifications (Yoshida et al., 2015). The GST tag of the recombinant DCC1-GST proteins was cleaved by a thrombin (Sigma; catalog no. T9326). Purified CA2-His protein (1 μm) was incubated with GST-cleaved DCC1 protein in the assay mixture containing 50 mm Tris-HCl (pH 7.5) and 50 mm NaCl. DTT (0.3 mm) was added to recycle DCC1 Trx activity. The mixture without DCC1 proteins was used as the control. After incubation for 30 min at 25°C, proteins were subjected to nonreducing (without 2-mercaptoethanol) or reducing (with 2-mercaptoethanol) SDS-PAGE followed by immunoblot analysis using anti-His antibody diluted 1:5,000 (Sigma; catalog no. H1029).
Mitochondrial Respiratory Complex I Activity Assay
Calli (0.1 g) of wild-type Col-0, dcc1, ca2, and dcc1ca2 cultured on SIM at 16 d were collected, and then the mitochondria were isolated using the Plant Mitochondria Isolation Kit (Biohao; catalog no. P0045) according to the manufacturer’s instructions. Mitochondrial respiratory Complex I activity was determined using 20 μg of mitochondrial protein with the MitoCheck Complex I Activity Assay Kit (Cayman Chemicals; catalog no. 700930). The absorbance of all samples was measured at 340 nm for 60 min using a plate reader. Complex I activity was calculated from the decrease in absorbance per minute. se values were calculated from three sets of biological replicates.
Chemical Treatments
Chemical reagents were added to the medium before transfer of calli, and calli were transferred to new medium with fresh reagents every 6 d. Different concentrations of H2O2 (0.005‰ and 0.01‰) or GSH (100, 300, and 600 μm) were added to the shoot regeneration system, and 10 μm estradiol was added to induce the transcription of CAT3 to complement the phenotypes of dcc1, ca2, and dcc1ca2.
DAB Staining Assay
Calli were incubated in DAB staining buffer (50 mm Tris, pH 7.6, and 0.6 mg mL−1 DAB) at room temperature in darkness and then fixed in 70% (v/v) ethanol at room temperature. The calli were observed with an Olympus DP72 microscope until the chlorophyll disappeared. The relative DAB staining intensity was analyzed by ImageJ software. Three sets of biological replicates were calculated, and more than 30 calli were examined in each replicate.
Quantitative Real-Time PCR
Total RNA was isolated using TRI reagent (Sigma; catalog no. T9424) and treated with DNase Ι (Thermo Fisher Scientific; catalog no. EN0521). Then, cDNAs were synthesized using M-MLV reverse transcriptase (Promega; catalog no. M1701). The qRT-PCR assays were conducted using the CFX96 Real-Time PCR Detection System (Bio-Rad) in a 20-µL reaction volume with 45 cycles. The cDNA levels were normalized against those of two housekeeping genes, TUB2 and ACTIN2. The relative expression level of each gene was standardized to TUB2 and calculated using the comparative CT method (Schmittgen and Livak, 2008). Mean values were calculated from three biological replicates. The sequences of primers used for qRT-PCR are listed in Supplemental Table S4.
RNA Sequencing and Data Analyses
Three biological repeats of the wild type (Col-0-A, Col-0-B, and Col-0-C) and the dcc1 mutant (dcc1-A, dcc1-B, and dcc1-C) were used for RNA sequencing. Total RNAs were isolated from the calli of wild-type Col-0 and dcc1 cultured on SIM at 16 d with TRI reagent (Sigma; catalog no. T9424). RNA was sequenced using the Illumina 2500 instrument in Gene Denovo Biotechnology. The raw reads were aligned to the genome sequences of TAIR10 (www.arabidopsis.org) using Tophat2 software (version 2.0.3.12; Kim et al., 2013). The gene expression levels were measured in FPKM using Cufflinks software (Trapnell et al., 2012). Statistical analyses were performed using the edgeR package (https://www.r-project.org/). Differentially expressed genes were those with fold change ≥ 2 and FDR < 0.05 between the wild-type Col-0 and the mutant dcc1. A GO enrichment analysis was performed in the GO database (http://www.geneontology.org/; Ashburner et al., 2000). The RNA sequencing data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under the accession number E-MTAB-5236.
Linkage Disequilibrium Analyses
Linkage disequilibrium analyses were performed using Genome Variation Server (Carlson et al., 2004) with DCC1 nucleotide sequences from 48 ecotypes of Arabidopsis.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AT5G50100 (DCC1), AT1G47260 (CA2), AT3G11260 (WOX5), AT3G03660 (WOX11), AT5G17810 (WOX12), AT5G10510 (PLT3), AT5G57390 (PLT5), AT2G42430 (LBD16), AT2G42440 (LBD17), AT2G45420 (LBD18), AT3G58190 (LBD29), AT1G78080 (WIND1), AT2G17950 (WUS), AT2G27250 (CLV3), and AT1G62360 (STM).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Identification of the dcc1 mutant.
Supplemental Figure S2. Phenotypes of the double mutant dcc1ca2 during shoot regeneration.
Supplemental Figure S3. Exogenous GSH promotes shoot regeneration.
Supplemental Figure S4. Transcript level of CAT3 in CAT3-overexpressing transgenic lines.
Supplemental Figure S5. Correlations among three biological repeats of RNA-seq analyses.
Supplemental Figure S6. Analyses of differentially expressed genes identified by RNA-seq were confirmed by qRT-PCR.
Supplemental Figure S7. Shoot regeneration capacity of different Arabidopsis ecotypes.
Supplemental Table S1. Frequencies of shoot regeneration in the Trx mutants.
Supplemental Table S2. Up-regulated genes in the dcc1 mutant as determined by RNA-seq analyses.
Supplemental Table S3. Down-regulated genes in the dcc1 mutant as determined by RNA-seq analyses.
Supplemental Table S4. Sequences of primers used in this study.
Acknowledgments
We thank all who generously provided plant materials in this research. We thank Dr. Min Ni (Department of Plant Biology, University of Minnesota) for critical reading of the article.
Footnotes
The work was supported by the National Natural Science Foundation of China (31270231 and 91217308).
Articles can be viewed without a subscription.
References
- Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000) Gene Ontology: tool for the unification of biology. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, Tanaka CK, Hurkman WJ, Gelhaye E, Rouhier N, Jacquot JP, Manieri W, Schürmann P, Droux M, et al. (2004) Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc Natl Acad Sci USA 101: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashandy T, Guilleminot J, Vernoux T, Caparros-Ruiz D, Ljung K, Meyer Y, Reichheld JP (2010) Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling. Plant Cell 22: 376–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Alfonso Y, Cilia M, San Roman A, Thomas C, Maule A, Hearn S, Jackson D (2009) Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc Natl Acad Sci USA 106: 3615–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum KD, Alvarado AS (2008) Slicing across kingdoms: regeneration in plants and animals. Cell 132: 697–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun HP, Binder S, Brennicke A, Eubel H, Fernie AR, Finkemeier I, Klodmann J, König AC, Kühn K, Meyer E, et al. (2014) The life of plant mitochondrial complex I. Mitochondrion 19: 295–313 [DOI] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA (2004) Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet 74: 106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challa KR, Aggarwal P, Nath U (2016) Activation of YUCCA5 by the transcription factor TCP4 integrates developmental and environmental signals to promote hypocotyl elongation in Arabidopsis. Plant Cell 2016: 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng ZJ, Wang L, Sun W, Zhang Y, Zhou C, Su YH, Li W, Sun TT, Zhao XY, Li XG, et al. (2013) Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytokinin biosynthesis by AUXIN RESPONSE FACTOR3. Plant Physiol 161: 240–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Considine MJ, Foyer CH (2014) Redox regulation of plant development. Antioxid Redox Signal 21: 1305–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz KJ, Turkan I, Krieger-Liszkay A (2016) Redox- and reactive oxygen species-dependent signaling into and out of the photosynthesizing chloroplast. Plant Physiol 171: 1541–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos CV, Rey P (2006) Plant thioredoxins are key actors in the oxidative stress response. Trends Plant Sci 11: 329–334 [DOI] [PubMed] [Google Scholar]
- Dröse S, Brandt U, Wittig I (2014) Mitochondrial respiratory chain complexes as sources and targets of thiol-based redox-regulation. Biochim Biophys Acta 1844: 1344–1354 [DOI] [PubMed] [Google Scholar]
- Duclercq J, Sangwan-Norreel B, Catterou M, Sangwan RS (2011) De novo shoot organogenesis: from art to science. Trends Plant Sci 16: 597–606 [DOI] [PubMed] [Google Scholar]
- Fan M, Xu C, Xu K, Hu Y (2012) LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res 22: 1169–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin A, Meyer B, Wittig I, Karas M, Schägger H, Vinogradov A, Brandt U (2008) Identification of the mitochondrial ND3 subunit as a structural component involved in the active/deactive enzyme transition of respiratory complex I. J Biol Chem 283: 20907–20913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelhaye E, Rouhier N, Navrot N, Jacquot JP (2005) The plant thioredoxin system. Cell Mol Life Sci 62: 24–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginalski K, Kinch L, Rychlewski L, Grishin NV (2004) DCC proteins: a novel family of thiol-disulfide oxidoreductases. Trends Biochem Sci 29: 339–342 [DOI] [PubMed] [Google Scholar]
- Gordon SP, Heisler MG, Reddy GV, Ohno C, Das P, Meyerowitz EM (2007) Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 134: 3539–3548 [DOI] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49: 373–385 [PubMed] [Google Scholar]
- Huang S, Van Aken O, Schwarzländer M, Belt K, Millar AH (2016) The roles of mitochondrial reactive oxygen species in cellular signaling and stress response in plants. Plant Physiol 171: 1551–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Ogawa Y, Iwase A, Sugimoto K (2016) Plant regeneration: cellular origins and molecular mechanisms. Development 143: 1442–1451 [DOI] [PubMed] [Google Scholar]
- Iwase A, Mitsuda N, Koyama T, Hiratsu K, Kojima M, Arai T, Inoue Y, Seki M, Sakakibara H, Sugimoto K, et al. (2011) The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr Biol 21: 508–514 [DOI] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347 [DOI] [PubMed] [Google Scholar]
- Kareem A, Durgaprasad K, Sugimoto K, Du Y, Pulianmackal AJ, Trivedi ZB, Abhayadev PV, Pinon V, Meyerowitz EM, Scheres B, et al. (2015) PLETHORA genes control regeneration by a two-step mechanism. Curr Biol 25: 1017–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Waszczak C, Hunter K, Wrzaczek M (2017) Bound by fate: the role of reactive oxygen species in receptor-like kinase signaling. Plant Cell 29: 638–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lall S, Nettleton D, DeCook R, Che P, Howell SH (2004) Quantitative trait loci associated with adventitious shoot formation in tissue culture and the program of shoot development in Arabidopsis. Genetics 167: 1883–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Liu H, Cheng ZJ, Su YH, Han HN, Zhang Y, Zhang XS (2011a) DNA methylation and histone modifications regulate de novo shoot regeneration in Arabidopsis by modulating WUSCHEL expression and auxin signaling. PLoS Genet 7: e1002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XR, Li HJ, Yuan L, Liu M, Shi DQ, Liu J, Yang WC (2014) Arabidopsis DAYU/ABERRANT PEROXISOME MORPHOLOGY9 is a key regulator of peroxisome biogenesis and plays critical roles during pollen maturation and germination in planta. Plant Cell 26: 619–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Fan C, Xing Y, Jiang Y, Luo L, Sun L, Shao D, Xu C, Li X, Xiao J, et al. (2011b) Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet 43: 1266–1269 [DOI] [PubMed] [Google Scholar]
- Liu J, Sheng L, Xu Y, Li J, Yang Z, Huang H, Xu L (2014) WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 26: 1081–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Wang T, Persson S, Mueller-Roeber B, Schippers JH (2014) Transcriptional control of ROS homeostasis by KUODA1 regulates cell expansion during leaf development. Nat Commun 5: 3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Holmgren A (2014) The thioredoxin antioxidant system. Free Radic Biol Med 66: 75–87 [DOI] [PubMed] [Google Scholar]
- Lucero LE, Uberti-Manassero NG, Arce AL, Colombatti F, Alemano SG, Gonzalez DH (2015) TCP15 modulates cytokinin and auxin responses during gynoecium development in Arabidopsis. Plant J 84: 267–282 [DOI] [PubMed] [Google Scholar]
- Mangano S, Juárez SPD, Estevez JM (2016) ROS regulation of polar growth in plant cells. Plant Physiol 171: 1593–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Wong JH, Feldman LJ, Lemaux PG, Buchanan BB (2010) A membrane-associated thioredoxin required for plant growth moves from cell to cell, suggestive of a role in intercellular communication. Proc Natl Acad Sci USA 107: 3900–3905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng WJ, Cheng ZJ, Sang YL, Zhang MM, Rong XF, Wang ZW, Tang YY, Zhang XS (2017) Type-B ARABIDOPSIS RESPONSE REGULATORs specify the shoot stem cell niche by dual regulation of WUSCHEL. Plant Cell 29: 1357–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer Y, Reichheld JP, Vignols F (2005) Thioredoxins in Arabidopsis and other plants. Photosynth Res 86: 419–433 [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T, Schmitt J (2006) Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature 441: 947–952 [DOI] [PubMed] [Google Scholar]
- Montano SJ, Lu J, Gustafsson TN, Holmgren A (2014) Activity assays of mammalian thioredoxin and thioredoxin reductase: fluorescent disulfide substrates, mechanisms, and use with tissue samples. Anal Biochem 449: 139–146 [DOI] [PubMed] [Google Scholar]
- Montrichard F, Alkhalfioui F, Yano H, Vensel WH, Hurkman WJ, Buchanan BB (2009) Thioredoxin targets in plants: the first 30 years. J Proteomics 72: 452–474 [DOI] [PubMed] [Google Scholar]
- Motohashi K, Koyama F, Nakanishi Y, Ueoka-Nakanishi H, Hisabori T (2003) Chloroplast cyclophilin is a target protein of thioredoxin: thiol modulation of the peptidyl-prolyl cis-trans isomerase activity. J Biol Chem 278: 31848–31852 [DOI] [PubMed] [Google Scholar]
- Motte H, Vercauteren A, Depuydt S, Landschoot S, Geelen D, Werbrouck S, Goormachtig S, Vuylsteke M, Vereecke D (2014) Combining linkage and association mapping identifies RECEPTOR-LIKE PROTEIN KINASE1 as an essential Arabidopsis shoot regeneration gene. Proc Natl Acad Sci USA 111: 8305–8310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murmu J, Bush MJ, DeLong C, Li S, Xu M, Khan M, Malcolmson C, Fobert PR, Zachgo S, Hepworth SR (2010) Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiol 154: 1492–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Rogers H, Munné-Bosch S (2016) Production and scavenging of reactive oxygen species and redox signaling during leaf and flower senescence: similar but different. Plant Physiol 171: 1560–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Cerveau D, Couturier J, Reichheld JP, Rey P (2015) Involvement of thiol-based mechanisms in plant development. Biochim Biophys Acta 1850: 1479–1496 [DOI] [PubMed] [Google Scholar]
- Sandalio LM, Romero-Puertas MC (2015) Peroxisomes sense and respond to environmental cues by regulating ROS and RNS signalling networks. Ann Bot 116: 475–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiantarelli E, De la Peña A, Candela M (2001) Use of recombinant inbred lines (RILs) to identify, locate and map major genes and quantitative trait loci involved with in vitro regeneration ability in Arabidopsis thaliana. Theor Appl Genet 102: 335–341 [Google Scholar]
- Schippers JH, Foyer CH, van Dongen JT (2016) Redox regulation in shoot growth, SAM maintenance and flowering. Curr Opin Plant Biol 29: 121–128 [DOI] [PubMed] [Google Scholar]
- Schmidt R, Schippers JH (2015) ROS-mediated redox signaling during cell differentiation in plants. Biochim Biophys Acta 1850: 1497–1508 [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Scofield S, Dewitte W, Murray JAH (2007) The KNOX gene SHOOT MERISTEMLESS is required for the development of reproductive meristematic tissues in Arabidopsis. Plant J 50: 767–781 [DOI] [PubMed] [Google Scholar]
- Sieburth LE, Meyerowitz EM (1997) Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9: 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto D, Córdoba JP, Villarreal F, Bartoli C, Schmitz J, Maurino VG, Braun HP, Pagnussat GC, Zabaleta E (2015) Functional characterization of mutants affected in the carbonic anhydrase domain of the respiratory complex I in Arabidopsis thaliana. Plant J 83: 831–844 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YH, Zhao XY, Liu YB, Zhang CL, O’Neill SD, Zhang XS (2009) Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis. Plant J 59: 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Jiao Y, Meyerowitz EM (2010) Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev Cell 18: 463–471 [DOI] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X (2008) Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 321: 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN (2010) Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143: 606–616 [DOI] [PubMed] [Google Scholar]
- van der Graaff E, Laux T, Rensing SA (2009) The WUS homeobox-containing (WOX) protein family. Genome Biol 10: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velázquez I, Valencia S, López-Lera A, de la Peña A, Candela M (2004) Analysis of natural allelic variation in in vitro organogenesis of Arabidopsis thaliana. Euphytica 137: 73–79 [Google Scholar]
- Viola IL, Güttlein LN, Gonzalez DH (2013) Redox modulation of plant developmental regulators from the class I TCP transcription factor family. Plant Physiol 162: 1434–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang H, Liu S, Ferjani A, Li J, Yan J, Yang X, Qin F (2016) Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat Genet 48: 1233–1241 [DOI] [PubMed] [Google Scholar]
- Waszczak C, Akter S, Jacques S, Huang J, Messens J, Van Breusegem F (2015) Oxidative post-translational modifications of cysteine residues in plant signal transduction. J Exp Bot 66: 2923–2934 [DOI] [PubMed] [Google Scholar]
- Wrzaczek M, Brosché M, Kangasjärvi J (2013) ROS signaling loops: production, perception, regulation. Curr Opin Plant Biol 16: 575–582 [DOI] [PubMed] [Google Scholar]
- Xie HT, Wan ZY, Li S, Zhang Y (2014) Spatiotemporal production of reactive oxygen species by NADPH oxidase is critical for tapetal programmed cell death and pollen development in Arabidopsis. Plant Cell 26: 2007–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Hofhuis H, Heidstra R, Sauer M, Friml J, Scheres B (2006) A molecular framework for plant regeneration. Science 311: 385–388 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Hara S, Hisabori T (2015) Thioredoxin selectivity for thiol-based redox regulation of target proteins in chloroplasts. J Biol Chem 290: 14278–14288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Noguchi K, Motohashi K, Hisabori T (2013) Systematic exploration of thioredoxin target proteins in plant mitochondria. Plant Cell Physiol 54: 875–892 [DOI] [PubMed] [Google Scholar]
- Yu Q, Tian H, Yue K, Liu J, Zhang B, Li X, Ding Z (2016) A P-loop NTPase regulates quiescent center cell division and distal stem cell identity through the regulation of ROS homeostasis in Arabidopsis root. PLoS Genet 12: e1006175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TQ, Lian H, Zhou CM, Xu L, Jiao Y, Wang JW (2017) A two-step model for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell 29: 1073–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2010) Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol 61: 49–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou JJ, Li XD, Ratnasekera D, Wang C, Liu WX, Song LF, Zhang WZ, Wu WH (2015) Arabidopsis CALCIUM-DEPENDENT PROTEIN KINASE8 and CATALASE3 function in abscisic acid-mediated signaling and H2O2 homeostasis in stomatal guard cells under drought stress. Plant Cell 27: 1445–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]