The soybean genome encodes over 6,000 long intergenic noncoding RNAs implicated in many biological processes, including transcription, development, and possibly influencing agronomic traits.
Abstract
Long intergenic noncoding RNAs (lincRNAs) are emerging as important regulators of diverse biological processes. However, our understanding of lincRNA abundance and function remains very limited especially for agriculturally important plants. Soybean (Glycine max) is a major legume crop plant providing over a half of global oilseed production. Moreover, soybean can form symbiotic relationships with Rhizobium bacteria to fix atmospheric nitrogen. Soybean has a complex paleopolyploid genome and exhibits many vegetative and floral development complexities. Soybean cultivars have photoperiod requirements restricting its use and productivity. Molecular regulators of these legume-specific developmental processes remain enigmatic. Long noncoding RNAs may play important regulatory roles in soybean growth and development. In this study, over one billion RNA-seq read pairs from 37 samples representing nine tissues were used to discover 6,018 lincRNA loci. The lincRNAs were shorter than protein-coding transcripts and had lower expression levels and more sample specific expression. Few of the loci were found to be conserved in two other legume species (chickpea [Cicer arietinum] and Medicago truncatula), but almost 200 homeologous lincRNAs in the soybean genome were detected. Protein-coding gene-lincRNA coexpression analysis suggested an involvement of lincRNAs in stress response, signal transduction, and developmental processes. Positional analysis of lincRNA loci implicated involvement in transcriptional regulation. lincRNA expression from centromeric regions was observed especially in actively dividing tissues, suggesting possible roles in cell division. Integration of publicly available genome-wide association data with the lincRNA map of the soybean genome uncovered 23 lincRNAs potentially associated with agronomic traits.
Recently, it has been elucidated that eukaryotic genomes, including plant genomes, encode a multitude of noncoding RNAs (ncRNAs; Chekanova et al., 2007; Kapranov et al., 2007). One class of ncRNAs are long noncoding RNAs (lncRNAs), which are defined as transcripts >200 bp in length and harboring no discernible coding potential (Jin et al., 2013; Wang et al., 2014a; Chekanova, 2015). The relative location of lncRNA loci to protein-coding genes identifies a further subgroup known as long intergenic noncoding RNAs (lincRNAs), which do not overlap protein-coding genes. lncRNAs were long considered little beyond transcriptional noise; however, current evidence points to important roles in diverse biological processes across eukaryotes (van Werven et al., 2012; Ulitsky and Bartel, 2013; Flynn and Chang, 2014). In Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), lncRNAs have been shown to be involved in flowering time regulation, reproduction, and root organogenesis (Swiezewski et al., 2009; Cifuentes-Rojas et al., 2011; Heo and Sung, 2011; Ariel et al., 2014; Bardou et al., 2014; Matzke et al., 2015; Wang et al., 2014b; Zhang et al., 2014; Berry and Dean, 2015; Khemka et al., 2016). lncRNAs are found both in the nucleus and cytoplasm, which suggests a diversity of modes of action, including chromatin modification (Heo et al., 2013); acting as decoys preventing access of regulatory proteins, including splicing machinery, and microRNAs to their true RNA and DNA targets (Franco-Zorrilla et al., 2007; Wu et al., 2013; Bardou et al., 2014); and acting as scaffolds for assembly of larger protein-RNA complexes (Lai et al., 2013; Pefanis et al., 2015). Recently, a large number of lncRNAs have been found to be associated with ribosomes and coexpressed with ribosomal proteins, although not translated, which suggests possible roles in translation regulation (Carlevaro-Fita et al., 2016). Despite increasing efforts (Jin et al., 2013; Szcześniak et al., 2016), plant-specific lncRNA databases are scarce, and lncRNA genome-wide discovery and especially functional annotation in agriculture important plant species remain unavailable.
Legumes are a large family of plant species characterized by butterfly-like flowers and pod-shaped shaped fruits. They provide an invaluable contribution to ecosystems due to their ability to form symbiotic relationships with Rhizobium bacteria. This symbiosis results in dinitrogen capture from the air and its subsequent fixation, making legumes one of the major sources of bioavailable nitrogen. Legume seeds are the second, after cereals, source of human and animal food and include soybeans (Glycine max), peanuts (Arachis hypogaea), garden peas (Pisum sativum), and broad beans (Vicia faba). Additionally, soybean is responsible for over a half of global oilseed production. Due to its economic importance as a source of food and oils, soybean has increasingly become a target of genomic and transcriptomic research efforts. Sequencing of the soybean genome revealed its complex paleopolyploid structure (Schmutz et al., 2010). Although comparison between soybean and the model plant species Arabidopsis can be drawn, the two species are suggested to have diverged from a common ancestor 92 million years ago (Zhu et al., 2003) and soybean has undergone at least two genome duplication events resulting in homeologous relationships between chromosomes and gene loci (Shoemaker et al., 2006). One of the most interesting questions from the genomics point of view is, “Which genomic features of soybean define its characteristics and are responsible for its vegetative and floral complexities?” Considering that many of the key developmental control genes in soybean exist in multiple copies, a complex interplay and additional control for fine-tuning are expected. Our recent awareness of prevalence and importance of lncRNAs highlight that these may play important regulatory roles in soybean growth and development. lncRNAs could provide the additional level of control and signal integration, which is missing when only protein-coding genes are considered.
This study presents genome-wide discovery, characterization, and functional annotation of lincRNAs in the soybean genome. Genome-wide lincRNA discovery was performed using a combination of de novo and reference-guided assembly approaches generating a most comprehensive lincRNA database. Comparative analysis between soybean lincRNAs and other legume species was performed to identify lincRNAs, which could play universal roles in all legumes and the lincRNAs, which are soybean-specific. Functional analysis was conducted to uncover biological processes that could be influenced by lincRNA action. Finally, publicly available genome-wide association data were used to further characterize the lincRNAs discovered and find potential links to agronomic traits.
RESULTS AND DISCUSSION
Genome-Wide Discovery of 6,018 Long Noncoding Intergenic Loci
LincRNAs are a class of RNA molecules that are >200 bp long and have no discernible coding potential. High-throughput technologies offer an opportunity for both coding and noncoding transcript detection and quantification. In total 1,025,323,161 read pairs from 37 soybean samples were used in the analysis. The soybean sampled tissues included 28 samples representing stem (germination and trefoil stage), flower (flower bud, unopened flower, florescence and 5 d after flowering), leaf bud (germination, trefoil, and differentiation stage), leaf (trefoil, flower bud differentiation stage, and senescent leaves), pod (3, 4, and 5 weeks), seed (3, 5, 6, 8, and 10 weeks), seed and pod (2, 3, and 4 weeks), shoot meristem (flower bud differentiation stage), cotyledon (germination and trefoil stage), and root (Shen et al., 2014). Additionally, nine samples (four from leaf tissue and five from shoot apical meristem tissue [SAM]) representing time points during the floral transition period following short-day treatment (Wong et al., 2013) were used. Both de novo and reference guided transcriptome assembly strategies were applied. StringTie reference guided assembly resulted in 68,190 loci and 160,337 transcripts. Trinity assemblies rendered 448,338 transcripts using de novo and 337,955 transcripts using reference guided approach. The PASA comprehensive transcript database built using StringTie and Trinity assemblies comprised 147,825 loci and 293,537 transcripts. Both StringTie and PASA annotations were subjected to lincRNA discovery pipeline and PASA-derived lincRNAs, which did not appear in StringTie annotation, were used to supplement StringTie-derived lincRNAs (Supplemental Fig. S1). Loci were considered to encode lincRNAs if they did not produce any protein-coding transcripts (open reading frame [ORF] size ≤100 amino acids and no similarity to protein-coding genes) and did not overlap any protein-coding loci. The lincRNAs were filtered to remove loci producing transcripts with similarity to tRNAs, rRNAs, and snoRNAs (58 loci) found in the Rfam database, transcripts that were nested (entirety contained) within other lincRNAs (63 loci), and transcripts that overlapped protein-coding genes in Gmax_275_v2.0 genome annotation (126 loci). LincRNAs are known to be expressed at low levels (Li et al., 2014; Zhang et al., 2014; Hao et al., 2015). Choosing an expression cutoff requires balancing a trade-off between retaining the largest possible set of lincRNAs and discarding the spurious transcription and mapping artifacts. Two lincRNA sets were generated. The larger set (9,766 loci) with a permissive cutoff > 0.1 fragments per kilobase per million mapped fragments (FPKM) in a least one of the samples (Supplemental Table S2) and a filtered set generated using more stringent FPKM cutoff (≥1.0 FPKM in at least one of samples or ≥0.5 FPKM in at least two samples or gene size of at least 1,000 bp). The filtered lincRNA set consisted of 6,018 lincRNA loci (6,134 transcripts), including 3,435 StringTie-derived and 2,583 PASA-derived loci (Supplemental Table S3). The full set is provided for the benefit of the readers but only the filtered lincRNA set was used in the analysis.
LincRNAs Have Distinct Properties When Compared to Protein-Coding Genes
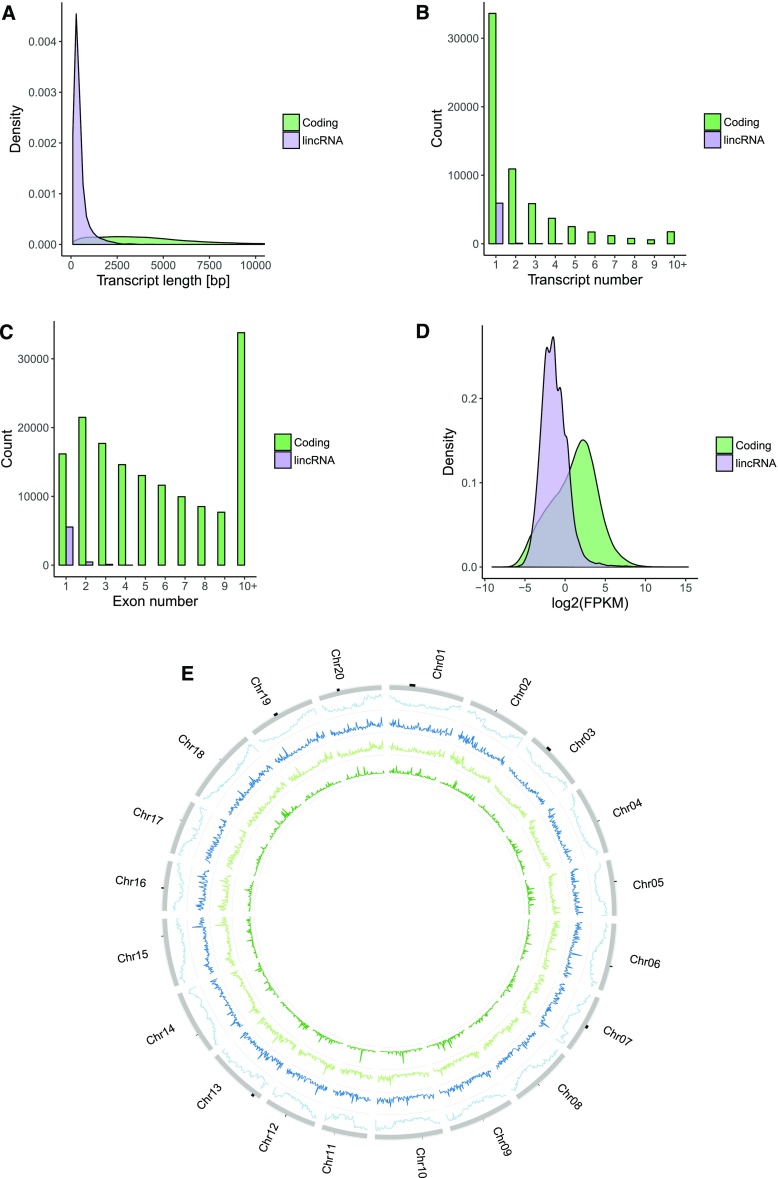
The lincRNA and protein-coding loci were examined for main gene characteristics. The lincRNA transcripts were on average shorter than protein-coding transcripts (Fig. 1A). The median length of lincRNA transcripts was 320 bp (mean: 467.3 bp), whereas the median length of protein-coding transcripts was 3,657 bp (mean: 4,450 bp). The lincRNA transcripts contained a lower number of exons than protein-coding transcripts (Fig. 1C). The majority of lincRNA transcripts (90.3%) contained a single exon. The maximum number of exons found in a lincRNA transcript was 4. The lincRNA genes had a lower number of isoforms compared to protein-coding genes (Fig. 1B). A vast majority of lincRNA genes (98.5%) had a single isoform. Finally, lincRNAs showed lower overall expression levels compared to coding genes (Fig. 1D). The observations are consistent with lincRNA studies in other plant species. LincRNAs in rice, cucumber (Cucumis sativus), and chickpea (Cicer arietinum) were reported to be shorter than protein-coding genes (Zhang et al., 2014; Hao et al., 2015; Khemka et al., 2016). LincRNAs in cucumber, maize (Zea mays), and chickpea were reported to have predominantly one exon only (Li et al., 2014; Hao et al., 2015; Khemka et al., 2016). Also, low expression levels of lincRNAs were observed in Arabidopsis, rice, and maize (Liu et al., 2012; Li et al., 2014; Zhang et al., 2014; Hao et al., 2015). Although usually lacking sequence homology (Hao et al., 2015; Mohammadin et al., 2015; Wang et al., 2015a), lincRNAs appear to share similar characteristics across different species that include short length and a low number of exons and splice variants.
Figure 1.
Comparison of properties of protein-coding and lincRNA genes. LincRNA genes differ from protein-coding genes with respect to transcript length, number of exons per transcript, number of transcripts per gene, and transcriptional profile. A, Comparison of transcript lengths of coding and noncoding genes. Noncoding genes have shorter transcripts. B, Comparison of the number of transcripts found in coding and noncoding genes. Noncoding genes have less isoforms. C, Comparison of the number of exons found in transcripts of coding and noncoding genes. Transcripts of noncoding genes have a lower number of exons. D, Comparison of log2 (FPKM) values of coding and noncoding genes. FPKM values calculated based on counts produced by featureCounts. Noncoding genes show lower expression levels compared to protein-coding genes. E, Plot presenting distribution of protein-coding and lincRNA loci across 20 soybean chromosomes. LincRNA loci are evenly distributed across chromosomes, whereas protein-coding genes show lower density in centromeric regions. Starting from the outer ring: (1) protein-coding genes, (2) all lincRNA genes, (3) non-TE lincRNA loci, and (4) lincRNA loci with transcripts harboring TEs.
Centromeric Regions of Soybean Chromosomes Show LincRNA Expression
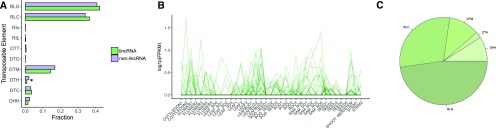
The distribution of lincRNAs across chromosomes can provide clues regarding possible functions and mechanisms of action. For example, lincRNAs located among protein-coding genes could modulate expression of their neighbors, while lincRNAs found close to centromeres or in gene deserts may act distally or have additional roles. Centromeric regions of soybean chromosomes are enriched in transposable elements (TEs) and depleted in protein-coding loci (Schmutz et al., 2010). In contrast, lincRNA loci display an even distribution across chromosomes (Fig. 1E), with active transcription from centromeric regions. LincRNAs transcribed from centromeric regions have been implicated to play roles in centromere maintenance and cellular division (Rošić and Erhardt, 2016). In total, 32 centromeric (as defined by transcription from regions delimited by GmCent-1 and GmCent-2 repeats) lincRNAs on chromosomes 1, 3, 5, 7, 13, 16, 17, and 19 were identified. The number of lincRNAs identified was weakly positively correlated with the identified centromere size (ρ = 0.25). No centromeric lincRNAs were expressed in all the samples, and the median number of samples showing centromeric lincRNA expression was seven. The median expression value in samples that expressed centromeric lincRNA (FPKM > 0.1) was 0.31 FPKM. Centromeric lincRNAs showed higher transcriptional activity in actively dividing tissues (flower bud, leaf bud, and SAM, Mann-Whitney U test, P value < 0.01; Fig. 2B). The most common transposable element type found within centromeric lincRNAs was LTR Gypsy retrotransposon (Fig. 2C), which is consistent with high prevalence of Gypsy transposable elements in the vicinity of centromeres (Schmutz et al., 2010).
Figure 2.
Transposable element composition of lincRNAs. A, Types of TEs found within lincRNA transcripts and in 50,000 randomly selected regions of soybean genome. TE composition of lincRNAs follows that of the soybean genome. RLG, LTR Gypsy; RLC, LTR Copia; RIu, LINE; RIL, LINE L1; DTT, Tc1-Mariner; DTO, PONG; DTM, Mutator; DTH, PIF-Harbinger; DTC, CACTA; DHH, Helitron; *P value < 0.01, Fisher’s test. B, Expression patterns of lincRNAs located in centromeric regions (n = 32). C, TE composition of centromeric lincRNAs.
Although centromeric lincRNA expression was observed, similar to rice and maize (Wang et al., 2015a), the majority of lincRNAs were found relatively close to neighboring protein-coding genes. The median distance from lincRNA to protein-coding gene was 1,064 bp (mean distance: 3,497 bp). LincRNAs found a short distance from protein-coding genes could modulate their expression by actively recruiting activators, repressors, and epigenetic modifiers or simply by transcription from the lincRNA locus (Wang and Chang, 2011; Kornienko et al., 2013).
Nearly a Fifth of lincRNA Transcripts Has Sequence Similarity to Transposable Elements
The relatively high abundance of lincRNAs proximal to centromeres sparked an investigation of the contribution of transposable elements to lincRNA transcript composition. In total, 18.3% of lincRNA transcripts were predicted to harbor TEs, and a higher proportion of lincRNAs than coding transcripts (10.8%) contained TEs. For transcripts, which harbored TEs, TEs contributed a larger amount of sequence to lincRNAs (median lincRNA coverage by TEs was 100%, mean: 82.8%) than to protein-coding transcripts (median coding transcript coverage by TEs: 19%, mean: 36.5%). A similar pattern was observed in the human genome, where two-thirds of mature noncoding transcripts showed similarity to TEs and TEs were found to contribute signals essential for biogenesis of many lncRNAs (Kapusta et al., 2013). The lincRNAs were found harbor more retrotransposons than DNA transposons (Fig. 2A), which reflects the overall TE landscape of the soybean genome (Du et al., 2010; Schmutz et al., 2010).
Soybean LincRNAs Have Low Levels of Sequence and Positional Conservation in Chickpea and Medicago truncatula
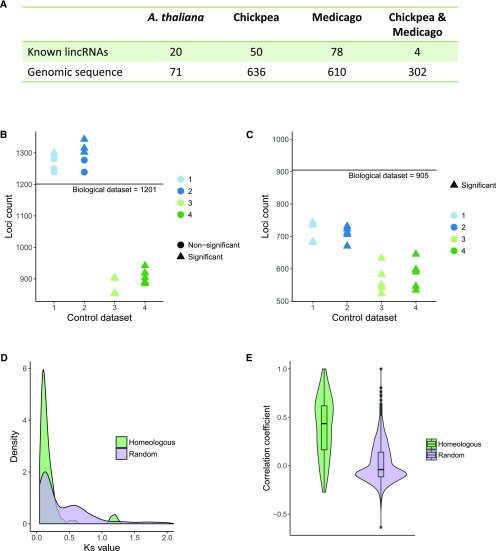
Information about conservation of lincRNAs across species can provide further inputs regarding their possible functions and the processes in which they are involved. If a lincRNA is well conserved in a number of species, it can be assumed to play a generally important role. Conversely, if a lincRNA is species specific, it may play a role unique to given organism or provide a modulatory function that alters the otherwise conserved system. It has been noted that the sequence conservation of lincRNAs is much lower than protein-coding genes (Hao et al., 2015; Mohammadin et al., 2015), but higher levels of positional based conservation have been postulated (Mohammadin et al., 2015; Wang et al., 2015a). In total, 6,018 soybean, 2,248 chickpea, 5,794 M. truncatula, and 6,480 Arabidopsis lincRNAs were available for analysis. Reciprocal best BLAST comparison uncovered 143 soybean lincRNAs that have sequence similarity to lincRNA in other species, with four lincRNAs showing similarity to lincRNAs in both chickpea and M. truncatula. Because different tissue samples and discovery pipelines were used, it is possible that some conserved lincRNA pairs were missed. To address this, soybean lincRNAs were compared against full genome assemblies, which resulted in the discovery of 787 additional loci with sequence similarity to genomes of other species (Fig. 3A). Those could correspond to unannotated noncoding transcripts. However, in the absence of evidence of transcription, their function remains unknown, and those loci were not considered in further analysis.
Figure 3.
Conservation of lincRNA loci in chickpea, M. truncatula, and Arabidopsis. A, Number of soybean lincRNA loci showing sequence similarity with lincRNAs or genomes of other species. B, Number of soybean lincRNA loci in biological and control datasets showing positional similarity with lincRNAs in other species. C, Number of soybean lincRNA loci in biological and control datasets showing positional similarity with other lincRNAs in soybean genome. D, Ks values calculated for protein-coding gene pairs flanking homeologous lincRNA loci and a random selection of homeologous protein-coding gene pairs (n = 444). The distribution of Ks values representing random selection has two peaks corresponding two duplication events. The protein pairs flanking homeologous loci mostly represent single, more recent duplication. E, Correlation of expression between homeologous lincRNA and a random selection of lincRNA pairs (n = 3,000). Homeologous loci have higher levels of coexpression.
Positional conservation between lncRNA loci has been suggested to extend across longer evolutionary distances than sequence conservation (Mohammadin et al., 2015; Wang et al., 2015a). A long noncoding RNA is often considered positionally conserved if found in the same orientation (upstream or downstream) relative to orthologous protein-coding gene in at least two species (Mohammadin et al., 2015; Wang et al., 2015a). If the direction of transcription of lincRNA is known, transcription from the same strand is also required. However, it is conceivable that if a large number of lincRNAs are considered, a number of those will show positional similarity across species (found in the same orientation relative to protein-coding genes) by chance only, rather than as a result of evolutionary conservation. To test this, the number of soybean lincRNAs that had positional similarity with chickpea, M. truncatula, and Arabidopsis lincRNAs was compared with control data sets constructed by random redistribution of lincRNAs across genomes of all four species. Two properties of lincRNA loci were considered while constructing the control datasets: (1) A proportion of lincRNA loci is found in clusters of two or more loci (mirroring this property the in control datasets will result in a more realistic distribution of lincRNA), and (2) lincRNA loci are enriched proximal to transcription factors (uneven distribution of lincRNAs relative to transcription factors could affect the results if transcription factors are preferentially retained or lost from syntenic regions). To accommodate those, four types of control data sets (5 simulations each, 20 data sets in total) were constructed: (1) random redistribution of lincRNA relative to protein-coding loci; (2) random redistribution of lincRNA relative to protein-coding loci, but maintaining the proportion of lincRNA found adjacent to transcription factors; (3) random redistribution of existing lincRNA clusters relative to protein-coding loci; and (4) random redistribution of existing lincRNA clusters relative to protein-coding loci, but maintaining the proportion of lincRNA found adjacent to transcription factors. The true biological lincRNA data set and the simulated control data sets were analyzed using the same positional conservation discovery pipeline. The number of positionally similar lincRNAs in the biological data set (1,201) and the simulation data sets 1 and 2 were not significantly different (Fisher’s test, P value > 0.01 for majority comparisons; Fig. 3B). However, more positionally similar lincRNAs were found in the biological data set when compared to simulation data sets 3 and 4 (Fisher’s test, P value < 0.01 for all comparisons). Although comparison with simulation data sets 3 and 4 suggests that positional similarity observed is somewhat higher than expected by chance alone, the difference is not large (Fig. 3B). Results of analyses of positional conservation ought to be interpreted with caution, especially across larger evolutionary distances, and considered in conjunction with sequence similarity and analysis of expression patterns.
Strong support for positional conservation of lincRNAs rather than chance positional similarity would be any sequence similarity between transcripts. Comparison of positionally similar transcript pairs uncovered 48 soybean lincRNAs that show positional similarity and sequence similarity with lncRNAs in other species. Sequence comparison of the positionally similar lincRNA pairs in simulated data sets (100 simulated data sets using random redistribution of lincRNA relative to protein-coding loci) showed them to have no sequence similarity (median number of pairs with sequence similarity per data set: 0), suggesting that the sequence similarity observed was not due to chance alone (permutation test, P value < 0.01). Subsequently, the 48 loci were analyzed in more detail. Protein-coding genes and short RNA primary transcripts are known to have higher conservation levels than long noncoding RNAs (Hezroni et al., 2015). Some lncRNAs are known to be sRNA precursors. The 48 putative conserved lincRNAs were inspected to check whether they (1) show similarity to TEs, (2) could encode small conserved peptides that would be missed by the lincRNA discovery pipeline (peptides <100 amino acids and with no similarity to proteins as evaluated by BLASTX), or (3) could be sRNA precursors. They were also compared against NCBI RefSeq-RNA database to check for similarity with any other known ncRNAs. Only one of the 48 lincRNAs had similarity to TEs (Supplemental Table S5). Three of the 48 lincRNAs had significant similarity to tens of sequences annotated as ncRNAs in the RefSeq database. However, a detailed analysis of the homologous region revealed it to contain a short 25 amino acid ORF encoding a peptide RPL41 (ribosomal protein L41), which was embedded within a much longer transcript. Because of the short length of the peptide, transcripts carrying RPL41 were annotated as lncRNAs by the pipeline used in this study as well as NCBI annotation pipeline. Following this discovery, the entire lincRNA data set was reanalyzed to check for presence of other RPL41 ORFs. However, only five lincRNA loci (including the three conserved ones) were carrying RPL41 ORF. This finding does suggest that some of the transcripts classified as lncRNAs based on the discovery algorithm parameters used could, in fact, encode small peptides (Niazi and Valadkhan, 2012; Ruiz-Orera et al., 2014; Nelson et al., 2016). Short of extremely well-conserved examples like RPL41, in the absence of proteomic data, these are impossible to discern. Six of the lincRNAs showed 100% percentage identity to microRNA, suggesting that they could be precursors of short RNAs. Reanalysis of the whole lincRNA data set suggested that 56 of lincRNAs could be microRNA precursors and the microRNA precursors were overrepresented in the positionally and sequence conserved lincRNAs (Fisher’s test, P value < 0.01). Finally, 19 lincRNAs showed similarity to other lncRNA transcripts in RefSeq and those represented other species.
Almost 200 Homeologous LincRNA Loci Can Be Traced to a Soybean Lineage-Specific Whole-Genome Duplication That Occurred ∼13 Million Years Ago
The soybean genome has a paleopolyploid structure resulting in extensive homeology across chromosomes (Shoemaker et al., 2006). It has undergone two rounds of whole-genome duplications, a more ancient event that occurred ∼59 million years ago (MYA) and soybean lineage-specific paleotetraploidization, which took place ∼13 MYA. As a result, the soybean genome is composed of large blocks of homeologous regions (Schmutz et al., 2010). It is possible that akin to protein-coding loci, homeologous lincRNA loci in soybean genome exist. Following a similar procedure for lincRNA positional similarity analysis performed between species, analysis of positional similarity of lincRNA loci within soybean genome was performed. Again, control data sets 1, 2, 3, and 4 were used to compare the number of positionally similar lincRNA loci found to the number that would be expected by chance alone. The number of positionally similar lincRNA loci in the true biological data set was significantly larger than the number found in any of the control datasets (Fisher’s test, P value < 0.01 for all comparisons; Fig. 3C). The difference between biological and control data sets suggested that at least 200 to 300 lincRNA loci with homeologs in the soybean genome were to be expected. Sequences of the lincRNA pairs with positional similarity within soybean genome were compared, which allowed identification of 103 pairs of homeologous loci (Supplemental Table S6). Sequence comparison of the positionally similar lincRNA pairs in simulated data sets (100 simulated data sets using random redistribution of lincRNA relative to protein-coding loci) showed them to have no sequence similarity (median number of pairs with sequence similarity per data set: 0), again suggesting that that the sequence similarity observed was not due to chance alone (permutation test, P value < 0.01). The number also roughly corresponds to the predictions based on comparison of positional similarity in biological and control data sets.
The age of homeologous blocks can be established using pairwise synonymous distance (Ks values) of paralogs (Schlueter et al., 2004; Pfeil et al., 2005; Schmutz et al., 2010). In case of soybean the Ks values of 0.06 to 0.39 correspond to 13-million year genome duplication and the Ks values of 0.40 to 0.80 to the 59-million year genome duplication (Schmutz et al., 2010). The vast majority of Ks values of protein-coding gene pairs flanking homeologous lincRNA loci fall within the 0.06 to 0.39 range (Fig. 3D), suggesting a more recent origin resulting from the soybean-lineage-specific paleotetraploidization. It is also possible that some homeologous loci representing the ∼59 MYA duplication do exist, but sequence divergence prevents their identification. Taken together, results of inter- and intraspecies comparisons suggest that while a lifespan of soybean lincRNA can exceed 15 million years it is unlikely to extend over 60 million years.
Functional enrichment of proteins flanking homeologous loci revealed overrepresentation of genes involved in response to abiotic stimuli including cellular response to phosphate starvation and response to absence of light (Supplemental Table S7). Finally, the coexpression of homeologous lincRNA loci was significantly higher (Fig. 3E, Mann-Whitney U test, P value < 0.01) when compared to a randomly selected lincRNA loci pairs, suggesting at least partial conservation of expression patterns.
The LincRNAs Show Highly Tissue-Specific Expression
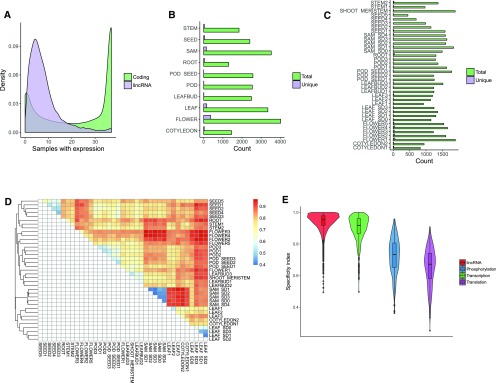
Expression of lincRNAs across all tissues was investigated using a combination of straightforward counting method and Tau specificity index, which were recently shown to be most successful methods of expression characterization (Kryuchkova-Mostacci and Robinson-Rechavi, 2017). LincRNAs displayed more tissue-specific expression than protein-coding genes (Fig. 4A). Any given lincRNA was on average expressed in eight samples (median: 6.0), whereas any given protein-coding gene was on average expressed in 23 samples (median: 30). Only 27 lincRNAs were expressed in all the samples. The tissue with the highest number of lincRNAs expressed (FPKM > 0.1) was floral tissue, followed by shoot apical meristem and leaf, suggesting an active role of lincRNAs in flowering and developmental processes. The sample with the highest number of lincRNAs expressed in total and uniquely was flower bud (flower1; 1,891 lincRNAs expressed in total, 51 expressed uniquely; Fig. 4, B and C; Supplemental Table S3). A large number of lincRNAs expressed in the SAM are consistent with previous observations in chickpea and other plants (Khemka et al., 2016). Overall, samples from the same tissue show similar expression patterns (Fig. 4D). Samples representing SAM, leaf, flower, and seed are grouped together. Nine of the samples from two tissues (leaf and SAM) represent floral transition period following short-day treatment. In total, 366 lincRNAs were uniquely expressed in the floral transition samples, and of these, 363 (99% of all lincRNAs) were expressed following short-day treatment, with 89, 128, and 149 lincRNAs expressed in leaf only, SAM only, and leaf and SAM, respectively. These lincRNAs represent an interesting target for the study of the mechanism of soybean floral transition.
Figure 4.
Sample-specific lincRNA expression. A, Comparison of the number of samples showing expression of coding and noncoding genes. Expression of noncoding genes is more sample specific. B, Number of total and unique lincRNA genes expressed in each tissue (samples from different time points for the same tissue were combined). Tissues with the highest total number of lincRNAs expressed and the highest number of uniquely expressed lincRNAs are floral tissue and shoot apical meristem. C, Number of lincRNAs expressed in each sample. The sample with most lincRNAs expressed is flower 1. D, Heat map showing relationships between samples. Samples from the same tissue have similar lincRNA expression profiles and cluster together. The color corresponds to distance calculated as 1-cor(log1p(FPKM)). Clustering was performed using hclust, method = complete. E, Tau expression specificity index calculated for lincRNA loci and three groups of protein coding genes representing different biological processes. Higher values of Tau correspond to more sample specific expression. Cotyledon 1, germination stage; cotyledon 2, trefoil stage; flower 1, flower bud differentiation stage; flower 2, flowering stage, bud before flowering; flower 3, flowering stage, florescence; flower 4, flowering stage, 5 d after flowering; flower 5, flowering stage, florescence, different stage; leaf 1, trefoil stage; leaf 2, flower bud differentiation stage; leaf 3, senescent leaves; leaf bud 1, germination stage; leaf bud 2, trefoil stage; leaf bud 3, flower bud differentiation stage; pod seed 1, 2 weeks; pod seed 2, 3 weeks; pod seed 3, 4 weeks; pod 1, 3 weeks; pod 2, 4 weeks; pod 3, 5 weeks; seed 1, 3 weeks; seed 2, 5 weeks; seed 3, 6 weeks; seed 4, 8 weeks; seed 5, 10 weeks; shoot meristem, flower bud differentiation stage; stem 1, germination; stem 2, trefoil stage; root, germination stage; sam sd0, SAM before short-day treatment; sam sd1-4, SAM short days 1 to 4; leaf sd0, leaf before short-day treatment; leaf sd1-3, leaf short days 1 to 3.
The specificity of lincRNA expression can be better contextualized when compared with different groups of protein-coding genes. The lincRNA tissue expression patterns were compared with expression patterns of protein-coding genes representing different specificity groups (transcription factors, high specificity; protein phosphorylation, medium specificity; translation, low specificity). LincRNAs have higher tissue specificity than any of the protein-coding gene groups, but the expression pattern is closest to the transcription factors (Fig. 4E). Transcription factors are known master regulators of gene expression and the parallels observed can suggest similar roles of lincRNAs. The high tissue-specific lincRNA expression supports the idea of their highly specialized, possible regulatory functions. It also allows for the possibility of using lincRNAs as tissue type and state markers.
The LincRNA-Protein-Coding Gene Coexpression Network and Position of lincRNAs Relative to Protein-Coding Neighbors Allows Functional Annotation of Noncoding RNAs
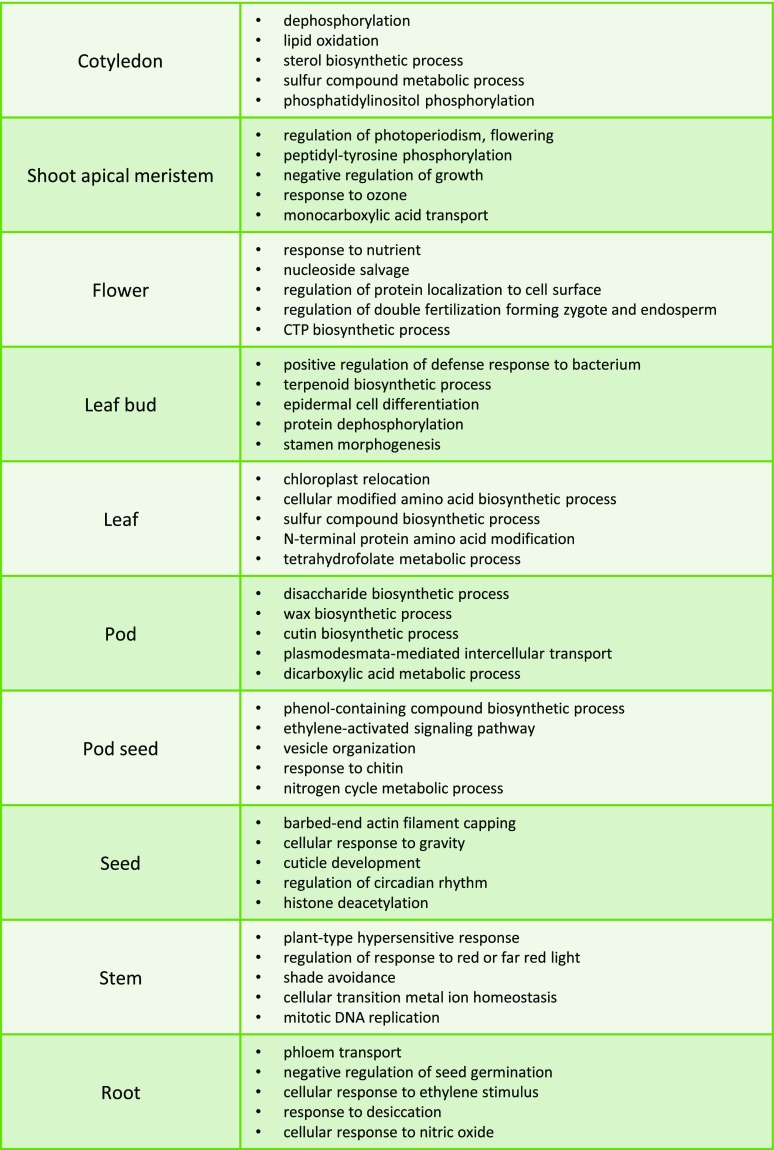
Functional annotation of long noncoding RNAs poses a considerable challenge. In the case of protein-coding genes, often extensive information about the function of a gene in a model organism is available, and sequence homology can be used to transfer existing annotation to newly discovered loci. In the case of lincRNAs, very few functional assignments exist, and lack of sequence homology hampers interspecies comparisons (Rinn and Chang, 2012; Smith and Mattick, 2017). The primary form of annotation involves a construction of coexpression network and using a method of so-called “guilt-by-association.” Correlation of expression between lincRNAs and protein-coding genes can imply involvement in common biological processes. Spearman correlation between expression of lincRNA and protein-coding loci was calculated. Only significant correlations were used in the analysis (P value < 0.05, P value adjusted for multiple comparisons using method “holm”). The resulting distribution of correlation coefficients is presented in Supplemental Figure S2A. The minimum absolute value of correlation coefficient used in the analysis was 0.84. A higher number of positive than negative correlations was observed and a large number of perfect correlations (ρ = 1) were observed. A similar observation was made in a human lncRNA annotation project, noting a higher number of positive correlations (Derrien et al., 2012). The high number of perfect correlations was due to high tissue specificity of lincRNA expression. LincRNAs were annotated using a hub-based approach (Liao et al., 2011). Gene Ontology (GO) enrichment analysis of protein-coding first-degree neighbors resulted in functional annotation of 1,574 lincRNAs (Supplemental Table S8). The summary of the GO annotation mapped to GOslim terms is presented in Supplemental Figure S2B. Overall, lincRNAs are annotated with a range of functions including stress response, signal transduction, and DNA methylation. Genes that are specifically or highly expressed in a given tissue are considered likely to contribute to relevant biological processes (Boyle et al., 2017). Clustering of lincRNAs based on their expression across tissues showed that genes that have peak expression in a given tissue are likely to have overall similar expression profiles (Supplemental Fig. S3), implying involvement in common biological process. The lincRNAs have been divided based on the tissue with peak expression (each set contained lincRNAs with peak expression in a given tissue) and GO enrichment for each of the lincRNA sets (peak expression in cotyledon, SAM, flower, leaf, leaf bud, pod, pod seed, seed, stem, and root; Fig. 5) was calculated. The enrichment of highly or specifically expressed lincRNA functions correlated well with the tissue-associated biological processes. For example, functionally annotated lincRNAs expressed in SAM, floral tissue, and root were highly enriched with processes associated with regulation of photoperiodism, sexual reproduction, and phloem transport, respectively. The results suggest possible involvement of lincRNAs in tissue-specific biological processes.
Figure 5.
Top significantly enriched biological processes among lincRNAs that show peak expression in a given tissue. Enrichment calculated using topGO, adjustment for multiple comparisons using method ‘weight’. P value < 0.01.
Finally, lincRNAs often exert their function on neighboring protein-coding genes; therefore, analysis of overrepresentation of classes of protein-coding genes flanking lincRNA loci provides additional source of functional annotation. The genes flanking lincRNAs were enriched in functions associated with transcription and development, suggesting possible lincRNA involvement in these processes (Supplemental Table S9).
Several LincRNAs Are Potentially Related to Agronomic Traits
Genome-wide association studies (GWAS) have been successful in uncovering the genetic basis of trait variation and linking casual loci to phenotypic traits. However, only a portion of variants identified by GWA studies can be assigned to protein-coding genes (Sonah et al., 2015; Zhang et al., 2015; Zhou et al., 2015a; Zhou et al., 2015b). Some of the remaining intergenic trait-associated variants can potentially be assigned to lincRNAs and serve as an additional source of functional annotation. In total, 316 single-nucleotide polymorphisms (SNPs) identified as associated with agronomic traits were used in the analysis. A lincRNA was identified as potentially related to a trait if the SNP was found either within the lincRNA locus or the locus was closer to the SNP than any other protein-coding gene. In total, 23 lincRNA candidates have been identified (Supplemental Fig. S4). Six of the lincRNAs overlapped trait-associated SNPs, and the remainder were found in close proximity (median distance: 981 bp). The putative trait-related lincRNAs are enriched in multiexon loci (Fisher’s test, P value < 0.01). The SNPs proximal to candidate lincRNA loci were related to traits such as number of days to flowering, number of days from flowering to maturity, and number of seeds per pod.
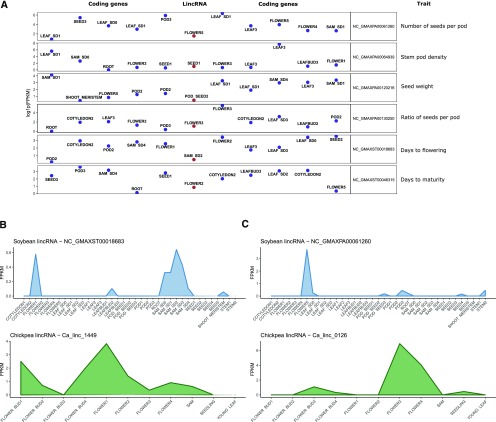
Several loci are typically found in the vicinity of a trait associated SNP and it is usually not immediately obvious which one may contribute to the trait. Accordingly, although the 23 lincRNAs were found closer to the SNP than any other protein-coding gene, it possible that a more distal coding gene contributes to the trait instead of the lincRNA (an interaction between the lincRNA and neighboring protein-coding gene is also possible). To add more confidence to the functional predictions, the genomic-position-only-based analysis was supplemented with investigation of expression patterns of neighboring genes. For each of the 23 putative, trait-related lincRNAs, the samples with peak expression for the lincRNA as well as five upstream and downstream protein-coding genes were investigated. The lincRNAs were considered more likely to influence the trait if they showed peak expression in a relevant tissue (for example, lincRNA associated with days to flowering being highly expressed in shoot apical meristem upon short day treatment; Supplemental Table S10; Supplemental Fig. S4). As a result, the top six lincRNAs that were found in the vicinity of trait-associated SNPs and showed consistent expression patterns were analyzed in more detail (Fig. 6A). Interestingly, four out of six had positionally similar lincRNAs in other species. Two of them showed expression in similar tissue types across species (NC_GMAXST00018683 and NC_GMAXPA00061260); for the remaining two, expression data in relevant tissue in chickpea and M. truncatula were unavailable. One of the lincRNAs (NC_GMAXST00018683) that overlapped SNP associated with the number of days to flowering and had peak expression in shoot apical meristem upon short-day treatment had positional similarity with lincRNA in chickpea (Fig. 6B). Comparison of expression patterns across samples in soybean and chickpea showed the lincRNAs to be expressed in flower buds and SAM in both species (Fig. 6B). The other lincRNA (NC_GMAXPA00061260) was found 223 bp from a SNP associated with number of seeds per pod and again had a positionally similar lincRNA in chickpea. Both lincRNAs showed peak expression in mature flowers (Fig. 6C). The proximity to trait-associated SNPs, expression in relevant tissues, and conservation of expression patterns across species makes them likely candidate for trait related lincRNAs. Combination of proximity to trait-associated SNPs and expression profile, as well as interspecies conservation, has been successfully used for functional annotation of lncRNAs in other species, including human, zebra fish, rice, and maize (Ulitsky et al., 2011; Gong et al., 2015; Wang et al., 2015a; Hon et al., 2017). In human, the study incorporating expression and genetic data found that lncRNAs that harbored trait-associated SNPs were also specifically expressed in tissues relevant to the trait, leading the authors to conclude the lncRNAs are likely functional and play important roles in disease (Hon et al., 2017). Furthermore, the putative functional lncRNAs also exhibited higher levels of conservation (Hon et al., 2017). Similarly, in maize, SNPs associated with leaf morphological traits were significantly enriched in genomic loci encoding maize lincRNAs, leading the authors to suggest roles of lincRNAs in control of agronomic traits (Wang et al., 2015a). Even without the support of GWAS data, lncRNA conservation itself was also found to be indicative of functionality. In zebra fish, lincRNAs selected based on their tissue-specific expression and synteny with mammalian lincRNAs were shown to be important for developmental processes (Ulitsky et al., 2011). Taken together, the availability of evidence from several sources and earlier studies suggesting that the GWAS, expression profile, and conservation evidence are highly indicative of lncRNA functionality further supports the functional predictions.
Figure 6.
Analysis of potential trait-related lincRNAs. A, For each of the putative trait-related lincRNAs, the plot presents 11 genes found in close vicinity of trait-associated SNP (1 putative trait related lincRNA + 5 downstream protein-coding genes + 5 upstream protein-coding genes). Each dot point represents a gene (red, lincRNA; blue, protein-coding gene) and is labeled with sample with peak expression (y axis represents actual expression value). B and C, Expression of lincRNA in soybean samples and its positional ortholog in chickpea. The x axis corresponds to samples. The y axis corresponds to the FPKM values.
CONCLUSION
The soybean genome encodes several thousand of lincRNAs, and several lincRNAs may be related to agronomic traits. Further investigations on detailed function and regulation, including identification of interacting partners and regulators of the lincRNAs, will elucidate their mechanism of action. This study also provides evidence that the network controlling and implementing biological processes in soybean involves complex interactions between proteins and long and short noncoding RNAs. Furthermore, this study presents a comprehensive atlas of lincRNAs in the soybean genome and paves the way for future research.
MATERIALS AND METHODS
Data
RNA-seq sequence data corresponding to Sequence Read Archive projects SRP020868 and PRJNA238493 were downloaded (full list of accessions can be found in Supplemental Table S1). The soybean (Glycine max) genome assembly (Gmax_275_v2.0) and corresponding annotation (Gmax_275_Wm82.a2.v1) were downloaded from Phytozome v11.
LincRNA Annotation
Reads were mapped to the reference genome using HISAT2 v2.0.5 (Kim et al., 2015; –min-intronlen 20–max-intronlen 2000). For each accession, transcripts were assembled and subsequently merged using StringTie v1.3.0 (Pertea et al., 2015; –merge -F 0.5 -T 0.5 -G Gmax_275_Wm82.a2.v1.gene_exons.gff3). Reads were also assembled using Trinity v2.3.2 (Grabherr et al., 2011). Both de novo (–seqType fq –max_memory 50G –verbose –normalize_reads –trimmomatic –CPU 16) and reference-guided (–genome_guided_bam –genome_guided_max_intron 10000 –max_memory 50G –verbose –CPU 16, reads trimmed and normalized during de novo Trinity run were used) assemblies were performed. The resulting StringTie and Trinity assemblies were supplied to PASA (Haas et al., 2003) in order to build comprehensive transcriptome database using procedure as described in PASA user guide (http://pasapipeline.github.io/). The aligner used was BLAT and MAX_INTRON_LENGTH was set to 2000. StringTie only and PASA transcripts were processed in parallel to identify potential lncRNAs (Supplemental Fig. S1). Transcripts >200 bp in length were subjected to ORF discovery using OrfPredictor v3.0 (Min et al., 2005). Transcripts with ORFs >300 bp (100 amino acids) were considered coding. Remaining transcripts were extracted and subjected to DIAMOND v0.8.25 (Buchfink et al., 2015) BLASTX search (–more-sensitive –evalue 0.01) against the NCBI nr database (obtained on the 23.10.2016). Transcripts that had a significant BLASTX match were considered coding. The remaining transcripts fulfilling the three criteria (1) length >200 bp, (2) ORF size ≤300 bp, and (3) no significant BLASTX hit were considered putative lncRNAs. A gene was considered coding if at least one transcript was coding. A gene was considered noncoding if none of the transcripts were coding. The positions of noncoding genes from StringTie and PASA annotations were compared against positions of coding genes in both annotations. If the putative lncRNA gene did not overlap any coding loci, it was considered a lincRNA gene. LincRNA loci from both annotations were merged. If lincRNA loci from both annotations had positional overlap, StringTie annotation was kept. Finally, reads mapping to gene were counted using Subread v1.5.1 featureCounts (Liao et al., 2014; -p -B -P -d 0 -d 1000) and FPKM values were calculated for each gene (109*fragments mapped to exons/assigned fragments*total length of exons). LincRNAs that did not have FPKM values larger than 0.1 in one of the samples were discarded.
LincRNA Functional Annotation
LincRNA functional annotation was performed by building lincRNA-protein-coding gene coexpression network. Coexpression was measured between identified lincRNA loci and protein-coding loci from Gmax_275_Wm82.a2.v1 annotation updated by StringTie. FPKM values were used for Spearman correlation calculation. Correlation coefficients and corresponding P values were calculated using corr.test function of R package Psych. Adjustment for multiple comparisons was performed using method ‘holm’. Only lincRNA-protein-coding gene pairs with P values < 0.05 were retained. All the protein partners were functionally annotated using Blast2GO (Conesa et al., 2005; nr subset corresponding to “Arabidopsis” [porgn] OR “Oryza” [porgn] OR “Sorghum” [porgn] OR “Glycine” [porgn] OR “Medicago” [porgn] OR “Brachypodium” [porgn]). For each of the lincRNAs, all the proteins that were significantly correlated were gathered and GO enrichment of biological processes category was calculated using topGO v2.22.0 (Alexa et al., 2006). All proteins in correlation with lincRNAs were used as background. Adjustment for multiple comparisons was performed using method ‘weight’. GO terms that were significantly enriched were assigned to the corresponding lincRNA as functional annotation (P value cutoff 0.05). The GO terms were mapped to the plant GOslim terms using Map2Slim option of owltools.
Sequence-Based Similarity of LincRNAs
Sequence-based similarity of lincRNAs was measured using reciprocal best BLAST (BLAST+ v2.5.0; -task blastn –evalue 1e-3). Best hits were identified by lowest e-value. Coordinates of chickpea (Cicer arietinum) lincRNAs were obtained from Khemka et al. (2016), Medicago truncatula lincRNAs from Wang et al. (2015b), and Arabidopsis (Arabidopsis thaliana) lincRNA from http://chualab.rockefeller.edu/gbrowse2/homepage.html. The lincRNA sequences were extracted from genome assemblies (chickpea Cicer_arietinum_GA_v1.0, Medicago Mt4.0v1, and Arabidopsis TAIR9). Comparisons against genome sequence were performed using BLAST+ v2.5.0 (-task dc-megablast –evalue 1e-3). In order to remove spurious hits due to presence of transposable elements or repetitive sequences, lincRNAs that had more than three matches in the genome were excluded. Additionally, the most significant high-scoring pair between lincRNAs and the genome was required to cover at least 10% of the lincRNA.
TE Composition of LincRNAs
The soybean TE database was obtained from SoyBase (SoyBase_TE_Fasta.txt). The lincRNA transcripts were compared against the TE database using BLAST+ v2.2.30 (blastn -task megablast –evalue 1e-5). The 50,000 random nonoverlapping intervals that did not overlap lincRNAs were identified in the soybean genome using regioneR (Gel et al., 2016). The corresponding sequences were extracted and compared against the TE database with the same BLAST parameters as for lincRNAs.
Centromeric LincRNA Identification
Centromeres were identified by presence of two soybean centromere-specific repeats: CentGm-1 and CentGm-2. CentGm-1 and CentGm-2 were compared against soybean genome (Gmax_275_v2.0) using BLAST+ v2.2.30 (blastn -task megablast). The coordinates of a centromere for a given chromosome corresponded to first and third quartile of CentGm-1 and CentGm-2 match coordinates. LincRNAs that fell within centromeres were identified as centromeric lincRNAs.
Position-Based Similarity of LincRNAs
Syntenic blocks between genomes of soybean (Gmax_275_v2.0), chickpea (Cicer_arietinum_GA_v1.0), Medicago (Mt4.0v1), and Arabidopsis (TAIR10) were identified using MCScanX (Wang et al., 2012). The syntenic blocks were used to identify positional similarity between soybean lincRNAs and lincRNAs from other species. For each lincRNA, five protein-coding neighbors upstream and downstream were extracted. The neighbors were then compared with collinear blocks identified by MCScanX. The lincRNA was said to belong to a collinear block if at least 3 out of 10 protein-coding neighbors were found in the block. LincRNAs from two species were said to be positionally similar if they belonged to the same collinear block, at least one of the two pairs of flanking protein-coding genes was identified as orthologous, and the lincRNAs shared the same relative position (upstream or downstream) with respect to the orthologous gene/genes. The lincRNA loci that shared positional similarity were compared using BLAST+ v2.5.0 (-task blastn –evalue 1e-3). Comparison against the RefSeq RNA database (downloaded on: 27.06.2017) was also performed with BLAST+ v2.5.0 (-task blastn –evalue 1e-3).
Generation of Control Data Sets
The control data sets were generated by assigning existing lincRNA to new protein-coding neighbors, taken from the pool of all protein-coding genes found in the genome. For data sets 1 and 2, a coordinate sorted full list of protein-coding genes was shuffled using Linux shuf function, which generates random permutations, and first n genes corresponding to the number of lincRNAs in a given data set were assigned to existing lincRNAs. The assigned protein-coding gene became new downstream protein-coding neighbor and the new lincRNA position was immediately upstream of the protein-coding gene assigned. For data sets 3 and 4, the procedure was similar, but the existing lincRNA clusters were kept together.
Calculation of Synonymous Substitution Rate
The synonymous substitution rates were computed between pairs of genes identified as homeologous by MCScanX. Proteins were aligned by Clustal Omega v1.2.0 (Sievers et al., 2011). The protein alignments were converted to nucleotide alignments using PAL2NAL v14 (Suyama et al., 2006). The Ks values were calculated using PAML v4.7 (yn00) (Yang, 2007).
Selections of Protein Groups for Comparison of Tissue-Specific Expression with LincRNAs
The protein-coding genes were divided into three categories. Genes expressed in no more than 15 samples (high specificity expression pattern), genes expressed in 16 to 35 samples (medium specificity expression pattern), and genes expressed in more than 35 samples (low specificity expression pattern). For each group GO biological process term enrichment was performed using topGO (Alexa et al., 2006), using all protein-coding genes as background. Adjustment for multiple comparisons was performed using method ‘weight’. For each category, a representative process was chosen (process with the highest number of significant genes among top 10 enriched GO terms). All the genes from a given category annotated with representative process were gathered and Tau specificity indices were calculated (Yanai et al., 2005).
Identification of LincRNAs Potentially Related to Agronomic Traits
The positions of SNPs associated with agronomic traits identified by Zhou et al. (2015a), Zhang et al. (2015), Zhou et al. (2015b), Sonah et al. (2015), and Fang et al. (2017) were obtained. Some of the SNPs were originally discovered against an older version of soybean genome (NCBI accession GCA_000004515.1); therefore, their coordinates were transferred to the Gmax_275_v2.0 genome assembly using NCBI remap tool (https://www.ncbi.nlm.nih.gov/genome/tools/remap). The lincRNA was consider potentially related to agronomic trait if it either harbored a SNP identified in association studies or it was closer to a SNP than any protein-coding gene and no further than 10 kb.
Code and Data Availability
The code used for generation of all the figures can be found at https://github.com/agolicz/lncRNAs-Plots. The data set described in the manuscript can be downloaded from https://osf.io/d7qz2/.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers SRP020868 and PRJNA238493.
Supplemental Data
Supplemental Figure S1. Workflow combining StringTie and PASA annotation to create the final nonredundant set of putative lincRNAs.
Supplemental Figure S2. Functional annotation of lincRNAs.
Supplemental Figure S3. Clustering of lincRNAs based on expression across all tissues.
Supplemental Figure S4. Candidate lincRNAs associated with agronomic traits.
Supplemental Table S1. Sequencing and mapping statistics for the libraries used.
Supplemental Table S2. Full list of putative lincRNA loci.
Supplemental Table S3. List of confident lincRNA loci used in the analysis and their expression.
Supplemental Table S4. Number of conserved lincRNAs identified using different e-value cutoffs.
Supplemental Table S5. List of conserved lincRNA loci.
Supplemental Table S6. List of homeologous lincRNA loci.
Supplemental Table S7. GO enrichment of coding genes flanking homeologous lincRNAs.
Supplemental Table S8. GO annotation of lincRNAs.
Supplemental Table S9. GO enrichment of all coding genes flanking lincRNAs.
Supplemental Table S10. Tissues considered relevant to a given trait.
Footnotes
This work was supported by Australian Research Council Discovery Grant ARC DP0988972 and by Melbourne Bioinformatics at the University of Melbourne (project UOM0033).
Articles can be viewed without a subscription.
References
- Alexa A, Rahnenführer J, Lengauer T (2006) Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607 [DOI] [PubMed] [Google Scholar]
- Ariel F, Jegu T, Latrasse D, Romero-Barrios N, Christ A, Benhamed M, Crespi M (2014) Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol Cell 55: 383–396 [DOI] [PubMed] [Google Scholar]
- Bardou F, Ariel F, Simpson CG, Romero-Barrios N, Laporte P, Balzergue S, Brown JWS, Crespi M (2014) Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev Cell 30: 166–176 [DOI] [PubMed] [Google Scholar]
- Berry S, Dean C (2015) Environmental perception and epigenetic memory: mechanistic insight through FLC. Plant J 83: 133–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EA, Li YI, Pritchard JK (2017) An expanded view of complex traits: from polygenic to omnigenic. Cell 169: 1177–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH (2015) Fast and sensitive protein alignment using DIAMOND. Nat Methods 12: 59–60 [DOI] [PubMed] [Google Scholar]
- Carlevaro-Fita J, Rahim A, Guigó R, Vardy LA, Johnson R (2016) Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA 22: 867–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova JA. (2015) Long non-coding RNAs and their functions in plants. Curr Opin Plant Biol 27: 207–216 [DOI] [PubMed] [Google Scholar]
- Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, et al. (2007) Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 131: 1340–1353 [DOI] [PubMed] [Google Scholar]
- Cifuentes-Rojas C, Kannan K, Tseng L, Shippen DE (2011) Two RNA subunits and POT1a are components of Arabidopsis telomerase. Proc Natl Acad Sci USA 108: 73–78 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676 [DOI] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22: 1775–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Grant D, Tian Z, Nelson RT, Zhu L, Shoemaker RC, Ma J (2010) SoyTEdb: a comprehensive database of transposable elements in the soybean genome. BMC Genomics 11: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang C, Ma Y, Wu S, Liu Z, Wang Z, Yang R, Hu G, Zhou Z, Yu H, Zhang M, et al. (2017) Genome-wide association studies dissect the genetic networks underlying agronomical traits in soybean. Genome Biol 18: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RA, Chang HY (2014) Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell 14: 752–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Gel B, Díez-Villanueva A, Serra E, Buschbeck M, Peinado MA, Malinverni R (2016) regioneR: an R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics 32: 289–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Liu W, Zhang J, Miao X, Guo A-Y (2015) lncRNASNP: a database of SNPs in lncRNAs and their potential functions in human and mouse. Nucleic Acids Res 43: D181–D186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. (2011) Trinity: reconstructing a full-length transcriptome without a genome from RNA-seq data. Nat Biotechnol 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK Jr, Hannick LI, Maiti R, Ronning CM, Rusch DB, Town CD, Salzberg SL, White O (2003) Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res 31: 5654–5666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Z, Fan C, Cheng T, Su Y, Wei Q, Li G (2015) Genome-wide identification, characterization and evolutionary analysis of long intergenic noncoding RNAs in cucumber. PLoS One 10: e0121800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JB, Lee YS, Sung S (2013) Epigenetic regulation by long noncoding RNAs in plants. Chromosome Res 21: 685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JB, Sung S (2011) Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79 [DOI] [PubMed] [Google Scholar]
- Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP, Ulitsky I (2015) Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Reports 11: 1110–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon C-C, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJL, Gough J, Denisenko E, Schmeier S, Poulsen TM, Severin J, et al. (2017) An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 543: 199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Liu J, Wang H, Wong L, Chua NH (2013) PLncDB: plant long non-coding RNA database. Bioinformatics 29: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, et al. (2007) RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488 [DOI] [PubMed] [Google Scholar]
- Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C (2013) Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet 9: e1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khemka N, Singh VK, Garg R, Jain M (2016) Genome-wide analysis of long intergenic non-coding RNAs in chickpea and their potential role in flower development. Sci Rep 6: 33297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornienko AE, Guenzl PM, Barlow DP, Pauler FM (2013) Gene regulation by the act of long non-coding RNA transcription. BMC Biol 11: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryuchkova-Mostacci N, Robinson-Rechavi M (2017) A benchmark of gene expression tissue-specificity metrics. Brief Bioinform 18: 205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R (2013) Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494: 497–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Eichten SR, Shimizu R, Petsch K, Yeh CT, Wu W, Chettoor AM, Givan SA, Cole RA, Fowler JE, et al. (2014) Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biol 15: R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Q, Liu C, Yuan X, Kang S, Miao R, Xiao H, Zhao G, Luo H, Bu D, Zhao H, et al. (2011) Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res 39: 3864–3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930 [DOI] [PubMed] [Google Scholar]
- Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, Arenas-Huertero C, Chua N-H (2012) Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24: 4333–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Kanno T, Matzke AJ (2015) RNA-directed DNA methylation: the evolution of a complex epigenetic pathway in flowering plants. Annu Rev Plant Biol 66: 243–267 [DOI] [PubMed] [Google Scholar]
- Min XJ, Butler G, Storms R, Tsang A (2005) OrfPredictor: predicting protein-coding regions in EST-derived sequences. Nucleic Acids Res 33: W677–W680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadin S, Edger PP, Pires JC, Schranz ME (2015) Positionally-conserved but sequence-diverged: identification of long non-coding RNAs in the Brassicaceae and Cleomaceae. BMC Plant Biol 15: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu F, Reese AL, McAnally JR, Chen X, Kavalali ET, et al. (2016) A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351: 271–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niazi F, Valadkhan S (2012) Computational analysis of functional long noncoding RNAs reveals lack of peptide-coding capacity and parallels with 3′ UTRs. RNA 18: 825–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu ZP, et al. (2015) RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 161: 774–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33: 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeil BE, Schlueter JA, Shoemaker RC, Doyle JJ (2005) Placing paleopolyploidy in relation to taxon divergence: a phylogenetic analysis in legumes using 39 gene families. Syst Biol 54: 441–454 [DOI] [PubMed] [Google Scholar]
- Rinn JL, Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81: 145–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rošić S, Erhardt S (2016) No longer a nuisance: long non-coding RNAs join CENP-A in epigenetic centromere regulation. Cell Mol Life Sci 73: 1387–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Orera J, Messeguer X, Subirana JA, Alba MM (2014) Long non-coding RNAs as a source of new peptides. eLife 3: e03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlueter JA, Dixon P, Granger C, Grant D, Clark L, Doyle JJ, Shoemaker RC (2004) Mining EST databases to resolve evolutionary events in major crop species. Genome 47: 868–876 [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Shen Y, Zhou Z, Wang Z, Li W, Fang C, Wu M, Ma Y, Liu T, Kong L-A, Peng D-L, Tian Z (2014) Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell; 26: 996–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker RC, Schlueter J, Doyle JJ (2006) Paleopolyploidy and gene duplication in soybean and other legumes. Curr Opin Plant Biol 9: 104–109 [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Mattick JS (2017) Structural and functional annotation of long noncoding RNAs. In JM Keith, ed, Bioinformatics: Volume II: Structure, Function, and Applications. Springer, New York, pp 65–85 [DOI] [PubMed] [Google Scholar]
- Sonah H, O’Donoughue L, Cober E, Rajcan I, Belzile F (2015) Identification of loci governing eight agronomic traits using a GBS-GWAS approach and validation by QTL mapping in soya bean. Plant Biotechnol J 13: 211–221 [DOI] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P (2006) PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34: W609–W612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462: 799–802 [DOI] [PubMed] [Google Scholar]
- Szcześniak MW, Rosikiewicz W, Makałowska I (2016) CANTATAdb: A collection of plant long non-coding RNAs. Plant Cell Physiol 57: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP (2013) lincRNAs: genomics, evolution, and mechanisms. Cell 154: 26–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP (2011) Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147: 1537–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven FJ, Neuert G, Hendrick N, Lardenois A, Buratowski S, van Oudenaarden A, Primig M, Amon A (2012) Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell 150: 1170–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Chung PJ, Liu J, Jang IC, Kean MJ, Xu J, Chua NH (2014a) Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res 24: 444–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Niu QW, Wu HW, Liu J, Ye J, Yu N, Chua NH (2015a) Analysis of non-coding transcriptome in rice and maize uncovers roles of conserved lncRNAs associated with agriculture traits. Plant J 84: 404–416 [DOI] [PubMed] [Google Scholar]
- Wang KC, Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43: 904–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T-Z, Liu M, Zhao M-G, Chen R, Zhang W-H (2015b) Identification and characterization of long non-coding RNAs involved in osmotic and salt stress in Medicago truncatula using genome-wide high-throughput sequencing. BMC Plant Biol 15: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Fan X, Lin F, He G, Terzaghi W, Zhu D, Deng XW (2014b) Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc Natl Acad Sci USA 111: 10359–10364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, Kissinger JC, Paterson AH (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40: e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CE, Singh MB, Bhalla PL (2013) The dynamics of soybean leaf and shoot apical meristem transcriptome undergoing floral initiation process. PLoS One 8: e65319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Wang ZM, Wang M, Wang XJ (2013) Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol 161: 1875–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, Benjamin H, Shmoish M, Chalifa-Caspi V, Shklar M, Ophir R, Bar-Even A, Horn-Saban S, Safran M, Domany E, Lancet D, Shmueli O (2005) Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 21: 650–659 [DOI] [PubMed] [Google Scholar]
- Yang Z. (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Zhang J, Song Q, Cregan PB, Nelson RL, Wang X, Wu J, Jiang G-L (2015) Genome-wide association study for flowering time, maturity dates and plant height in early maturing soybean (Glycine max) germplasm. BMC Genomics 16: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YC, Liao JY, Li ZY, Yu Y, Zhang JP, Li QF, Qu LH, Shu WS, Chen YQ (2014) Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol 15: 512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Wang S-B, Jian J, Geng Q-C, Wen J, Song Q, Wu Z, Li G-J, Liu Y-Q, Dunwell JM, et al. (2015a) Identification of domestication-related loci associated with flowering time and seed size in soybean with the RAD-seq genotyping method. Sci Rep 5: 9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Jiang Y, Wang Z, Gou Z, Lyu J, Li W, Yu Y, Shu L, Zhao Y, Ma Y, et al. (2015b) Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat Biotechnol 33: 408–414 [DOI] [PubMed] [Google Scholar]
- Zhu YL, Song QJ, Hyten DL, Van Tassell CP, Matukumalli LK, Grimm DR, Hyatt SM, Fickus EW, Young ND, Cregan PB (2003) Single-nucleotide polymorphisms in soybean. Genetics 163: 1123–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]