Ribosome pausing in chloroplasts is caused by multiple features of mRNA and nascent peptide chain and influences transmembrane protein folding and cofactor integration.
Abstract
Many mRNAs contain pause sites that briefly interrupt the progress of translation. Specific features that induce ribosome pausing have been described; however, their individual contributions to pause-site formation, and the overall biological significance of ribosome pausing, remain largely unclear. We have taken advantage of the compact genome of chloroplasts to carry out a plastid genome-wide survey of pause sites, as a basis for studying the impact of pausing on posttranslational processes. Based on ribosomal profiling of Arabidopsis (Arabidopsis thaliana) chloroplast mRNAs, we demonstrate that a combination of factors—mRNA secondary structure, internal Shine-Dalgarno sequences, and positively charged amino acids in the nascent peptide chain—explains 95% of the major pause sites on plastid mRNAs, whereas codon usage has little impact. The distribution of the pause sites is nonrandom and conforms to distinct patterns in the vicinity of sequences coding for transmembrane domains, which depend on their orientation within the membrane as well as being next to sequences coding for cofactor binding sites. We found strong indications that the mechanisms causing ribosomal pausing and at least some of the ribosomes pause sites are conserved between distantly related plant species. In addition, the positions of features that cause pausing are well conserved in photoautotrophic plants, but less so in their nonphotosynthetic, parasitic relatives, implying that the synthesis and assembly of photosynthetic multiprotein complexes requires localized ribosome pausing.
A variety of methods has demonstrated that translational elongation occurs at a nonuniform rate in diverse organisms from different domains of life (Kim et al., 1991; Wen et al., 2008; Charneski and Hurst, 2013; Chadani et al., 2016). Recent research has revealed that structural features in both the mRNA and the nascent peptide chain are associated with ribosome pausing. In bacteria and eukaryotes, rare codons and codons for specific amino acids can cause pausing (Zhang et al., 2009; Artieri and Fraser, 2014; Mohammad et al., 2016; Weinberg et al., 2016), as do stable mRNA secondary structures (Wen et al., 2008; Tuller et al., 2011; Pop et al., 2014), which can even block elongation (Tholstrup et al., 2012). In bacterial-type systems, Shine-Dalgarno sequences, which facilitate start codon recognition and thus determine the efficiency of translation initiation, have also been reported to cause ribosome pausing (Li et al., 2012; Zoschke et al., 2013; Fluman et al., 2014; Nakahigashi et al., 2014), although these findings have recently been challenged (Mohammad et al., 2016). Besides these RNA-based features, clustered, and even single, positively charged amino acids in the nascent peptide chain have been described to induce pausing in both bacteria and eukaryotes (Lu and Deutsch, 2008; Tuller et al., 2011; Charneski and Hurst, 2013; Koutmou et al., 2015; Chen et al., 2015; Weinberg et al., 2016). This claim has also been disputed, at least for Saccharomyces cerevisiae (Artieri and Fraser, 2014). In addition, different short, often proline-containing, peptide sequences in the nascent peptide influence elongation by causing pausing or stalling (Woolstenhulme et al., 2013; Wilson et al., 2016; Buskirk and Green, 2017; Sabi and Tuller, 2017).
In most cases, the biological significance of ribosome pausing remains obscure, although there are reports that ribosome pause sites located at protein domain boundaries facilitate folding (Thanaraj and Argos, 1996; Zhang et al., 2009; Yona et al., 2013; Fluman et al., 2014; Hess et al., 2015; Weinberg et al., 2016) or are involved in membrane targeting in bacteria (Fluman et al., 2014).
The translation apparatus in plastids is of cyanobacterial origin, and now consists of a mosaic of conserved bacterial elements and regulatory elements that evolved in eukaryotes (Barkan, 2011; Tiller and Bock, 2014). Plastid ribosomes are of the bacterial 70S-type, and most of their components are strongly conserved. Some of the ribosomal proteins have extended termini, e.g. at the ribosome exit tunnel and the mRNA entry and exit site (Yamaguchi et al., 2000; Yamaguchi and Subramanian, 2000; Fleischmann et al., 2011; Bieri et al., 2017). There are four additional plastid-specific ribosomal proteins, which mainly compensate for structural changes in the rRNAs, but one is reported to alter the mRNA exit site (Sharma et al., 2007; Tiller et al., 2012; Ahmed et al., 2016, 2017; Bieri et al., 2017). Plastids use the standard genetic code, but have a strongly reduced tRNA set, which is not optimized for the A/U-rich codons in the plastid genome, but for decoding by wobbling and superwobbling (Alkatib et al., 2012). The cis-acting elements required for translation initiation are (1) Shine-Dalgarno sequences or (2) local minima of mRNA structure (Scharff et al., 2011; Zhang et al., 2012). In contrast, not much is known about the elements that influence rates of elongation, apart from some indications from a low-resolution analysis that Shine-Dalgarno sequences might be involved (Zoschke et al., 2013). Our aim here is to provide a comprehensive analysis of ribosome pausing in chloroplasts, focusing on what causes pausing, what is influenced by pausing, and whether pausing sites are conserved.
RESULTS
We analyzed translation elongation in chloroplasts of young leaves of the dicot plant Arabidopsis (Arabidopsis thaliana) using ribosome profiling (Ingolia et al., 2009). Using three biological replicates we mapped a total of 4.8 million reads to the 79 coding regions of the plastid genome (Supplemental Fig. S1A). Reads mapped to start codons were used to determine how to assign ribosome densities to specific codons as described previously (Woolstenhulme et al., 2015; Chotewutmontri and Barkan, 2016; Mohammad et al., 2016). Micrococcal nuclease (MNase), which was used to generate the footprints, produces a population of diverse ends (Woolstenhulme et al., 2015; Oh et al., 2011). In our data, the 5′ ends of the footprints around start codons were better defined (Supplemental Fig. S2), whereas in an MNase data set acquired from Escherichia coli RNAs, and a Zea mays data set produced with RNase I, the 3′ ends of the footprints were the more uniform (Woolstenhulme et al., 2015; Chotewutmontri and Barkan, 2016). Therefore, we used the 5′ ends to assign ribosome densities to specific stretches of the coding regions. The most frequently encountered distance between the 5′ ends of the reads and start codons was 23 nucleotides (Supplemental Fig. S2B). This distance is compatible with the asymmetric assignment of the reads observed in maize (Zea mays) chloroplasts (Chotewutmontri and Barkan, 2016) and was therefore used to assign the reads to codons. The asymmetric footprint observed in plastids contrasts with the symmetrical one seen in bacteria (Woolstenhulme et al., 2015; Hwang and Buskirk, 2017) and could be caused by differences in the mRNA entry and exit sites of the ribosome (Bieri et al., 2017). In our analysis of translation elongation, we focused on 28- to 40-nucleotide footprints (Supplemental Fig. S1A), because their ends were better defined (Supplemental Fig. S2A). For comparison, we also tested the assignment relative to 3′ ends of the footprints and included shorter footprint lengths (see below).
Features Associated with Ribosome Pausing in Chloroplasts
Ribosome profiling allows one to define pause sites on mRNAs by comparing the ribosome density on specific segments of an mRNA with the mean density across the coding region. The resulting pause score indicates the relative duration of pausing. This is based on the assumption that a reduction in the velocity of translation elongation will be reflected in an increase in local ribosome density (Li et al., 2012; Charneski and Hurst, 2013). For the analysis of pausing during translation elongation, reads that mapped to the first and last 51 nucleotides (17 codons) of each coding region were removed, to exclude influences arising from translation initiation and termination, as well as overlapping reading frames, from the analysis. The patterns of ribosome pausing detected in the three biological replicates were very similar (Supplemental Fig. S1B). We analyzed all pause sites on plastid mRNA with a pause score higher than 50, i.e. a ribosome density that is greater than 50 times higher than the mean density for the corresponding coding region.
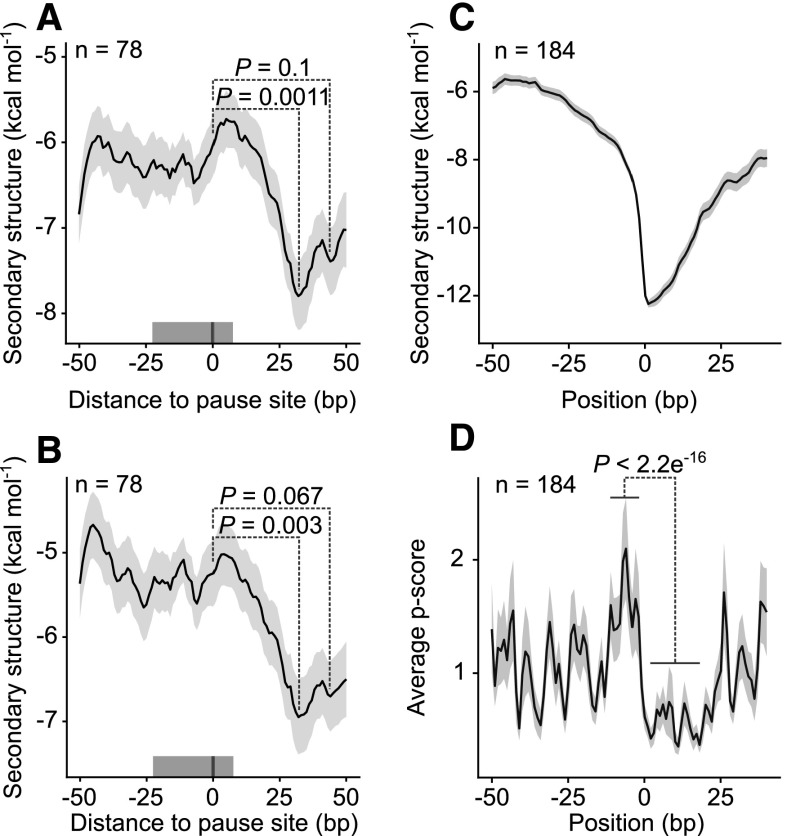
In cases where pausing is attributable to secondary structure in the mRNA, these structures would be expected to lie downstream of the pause site (Wen et al., 2008; Pop et al., 2014), and the duration of the pause should reflect the time taken for the ribosome’s helicase activity to remove the structural obstacle. On the basis of our profiling data, we found that the stability of mRNA secondary structure indeed peaks at a distance of 31 nucleotides downstream of pause sites (Fig. 1A), whereas no such correlation was found using random sequences (Supplemental Fig. S3A). These results were obtained using both predicted mRNA secondary structures and in vivo-determined mRNA secondary structures (Ding et al., 2014; Fig. 1B). This finding strongly indicates that mRNA structures indeed contribute to ribosome pausing. We also investigated whether the most stable 10% of these secondary structures (Supplemental Fig. S4) causes ribosome pausing. Here again, there is a statistically significant correlation with pause sites directly upstream of such structures (Fig. 1, C and D), which is not observed in random sequences (Supplemental Fig. S3, B and C). In this case, the distance is shorter than that indicated by the converse analysis, probably because the latter corresponds to the difference from the point of maximum (center) of structure stability (Fig. 1A), whereas the former is measured from the borders of the structure (Fig. 1D). We confirmed the correlation between ribosome pausing and predicted downstream mRNA structure by analyzing a published ribosome profiling data set from the monocot plant species maize (Chotewutmontri and Barkan, 2016; Supplemental Fig. S5). We also tested whether different methods for assigning reads to codons, and different footprint lengths, influence the results. In all cases an increase in the stability of mRNA structure was found downstream of the pause site (Supplemental Fig. S6). Only the inclusion of short footprints in the analysis weakened the correlation. This is likely attributable to the less sharply defined ends of short reads (Supplemental Fig. S2A). Generally, there is a clear influence of the pause score on the correlation. The longer the inferred pause, the more stable the mRNA structures found downstream. This argues that the two factors are functionally related and that internal mRNA secondary structure indeed causes pausing (Supplemental Fig. S6).
Figure 1.
Correlation of ribosome pause sites with the presence of stable mRNA secondary structures downstream. A, Predicted distribution of mRNA structures around 78 pause sites with a pause score calculated per nucleotide above 50 (position 0; see “Materials and Methods”). The line shows the running mean MFE; the shaded area around it indicates the mean ± se. The mean was calculated using a centered 51-nucleotide sliding window. The P values (Wilcoxon rank sum test) show that the stability of secondary structure is statistically significantly higher downstream of pause sites than at the sites themselves. The gray bar above the x axis represents the region occupied by the ribosome. B, The mRNA structure around the same pause sites, as determined in vivo (Ding et al., 2014). C, For the converse analysis, regions centered on the structure (10% strongest mRNA structures, MFE < −11.1, position 0; Supplemental Fig. S4) and with maximally two sites with MFE < −11.1 between positions −50 and −1 were selected (n = 189). The line shows the running mean of MFE; the shaded area around it is the mean ± se. D, Ribosome pausing measured by the pause score (p score) at the 10% most stable mRNA structures shown in C. There is statistically significantly more pausing at positions from −12 to −2 relative to region from 1 to 19 (Wilcoxon rank sum test). The solid line shows the mean of the pause score, with the shaded area representing the mean ± se.
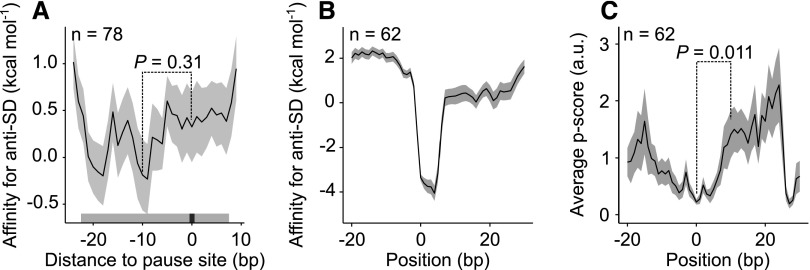
Internal Shine-Dalgarno sequences (SD) in coding regions were reported to cause pausing in E. coli (Li et al., 2012) and there are indications from an array-based ribosome profiling analysis that the same mechanism is also active in chloroplasts of maize (Zoschke et al., 2013). For E. coli, these results were recently questioned, and attributed to the preferential isolation of long footprints (Mohammad et al., 2016). If internal SDs indeed cause pausing, such sequences would be expected upstream of the pause site. In our data set, we observe a trend for binding of the anti-Shine-Dalgarno sequence (aSD) at the 3′ end of the 16S rRNA to sequences upstream of pause sites (Fig. 2A), which is statistically significant if weaker pause sites (pause score 40) or shorter reads (20–40 nucleotides instead of 28–40 nucleotides) are included and if only footprints of specific lengths are analyzed (Supplemental Fig. S7). We find the same trend in our analysis of the data from maize chloroplasts (Chotewutmontri and Barkan, 2016; Supplemental Fig. S8). In neither of the chloroplast data sets did we observe that the correlation between pausing and binding of the aSD to internal SDs was restricted to long footprints (Supplemental Figs. S7 and S8). No trend was found using assignment to 3′ ends because additional internal SDs were surprisingly detected at the pause site itself (Supplemental Fig. S7). However, when we analyzed pause sites downstream of the strongest 19.9% of internal SDs (Supplemental Fig. S9), we found a statistically significant correlation (Fig. 2, B and C). Using random sequences as control, neither of these correlations can be found (Supplemental Fig. S10). In addition, we observed a direct correlation between predicted stability of the aSD-mRNA interaction upstream of the pause site and the length of the inferred pause (i.e. the pause score; Supplemental Fig. S7), which strongly argues that internal SDs contribute to pausing.
Figure 2.
Correlation of ribosomal pause sites with internal SDs located upstream. A, Affinity of sequences in the vicinity of 78 ribosome pause sites with calculated pause scores per nucleotide > 50 (position 0; see “Materials and Methods”) to the aSD motif in the 16S rRNA. The P values (Wilcoxon rank sum test) demonstrate that this trend is not statistically significant. The gray bar above the x axis represents the region occupied by the ribosome. B, aSD/mRNA interaction near the strongest 19.9% of internal SDs (ΔG < −2 kcal mol−1; Supplemental Fig. S9). C, Ribosome pausing measured by the pause score (p score) at the strongest 19.9% of internal SDs. There is statistically significantly more pausing at position 10 relative to the 5′ end of the SD (Wilcoxon rank sum test).
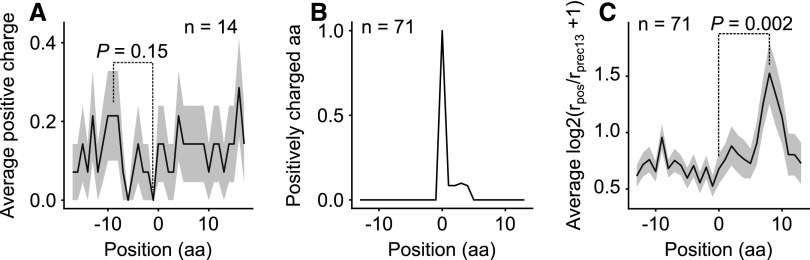
Besides features of the mRNA, the occurrence of positively charged amino acids in the newly synthesized protein, i.e. the nascent peptide chain, has been shown to cause pausing. Such residues interact with the negatively charged interior of the exit tunnel of the ribosome (Lu and Deutsch, 2008; Charneski and Hurst, 2013). However, we found only a weak tendency for positively charged amino acids to be enriched upstream of major pause sites (Fig. 3A). This is also true for weaker pause sites (Supplemental Fig. S11). Interestingly, a statistically significant correlation emerged when pausing was analyzed downstream of positively charged amino acids (Fig. 3, B and C). This indicates that, whereas positively charged amino acids alone can cause pausing, they are not sufficient to provoke long pauses. These results were confirmed by analyzing data from maize chloroplasts (Chotewutmontri and Barkan, 2016; Supplemental Fig. S12). Some of the codons for charged amino acids (i.e. AGA, AAG, and AGG) can form part of internal SDs. But these contribute less to the correlation than do the other codons for positively charged amino acids, which do not resemble SD sequences (Supplemental Fig. S13, A–D), and in addition they tend to be located further upstream than expected for an SD. We also excluded the possibility that AGA, AGG, AAG, and AAA sequences at the 5′ ends of the footprints (including A-ending codons for positively charged amino acids) might cause the correlation, thereby excluding the tendency of MNase to cut at As (Becker et al., 2013) as a contributory factor (Supplemental Fig. S13, E and F). In random sequences no correlation between pausing and positively charged amino acids was found (Supplemental Fig. 13, G–I).
Figure 3.
Correlation of ribosomal pause sites with the presence of positively charged amino acids (Arg, His, and Lys) upstream. A, The proportion of positively charged amino acids around all pause sites (n = 78) with a pause score > 50 calculated for codons (see Methods). B, The proportion of positively charged amino acids around a subset (n = 73) of such sites. All such amino acids were chosen if there were no other positively charged amino acids between positions −13 and −1 or 6 and 13. C, Ribosome occupancy around positively charged amino acids presented in (B) (centered on 0, n = 71). There is a statistically significant difference between positions 0 and 8 (Wilcoxon rank sum test).
We also tested whether codon usage in general or for specific amino acids can explain pausing, but detected no statistically significant influence (Supplemental Fig. S14). We also tested the impact of assignment to 3′ ends, the inclusion of short reads, and the specific analysis of especially well-defined footprint lengths (Supplemental Fig. S2A), but none of these approaches revealed any significant role of codon usage in ribosome pausing.
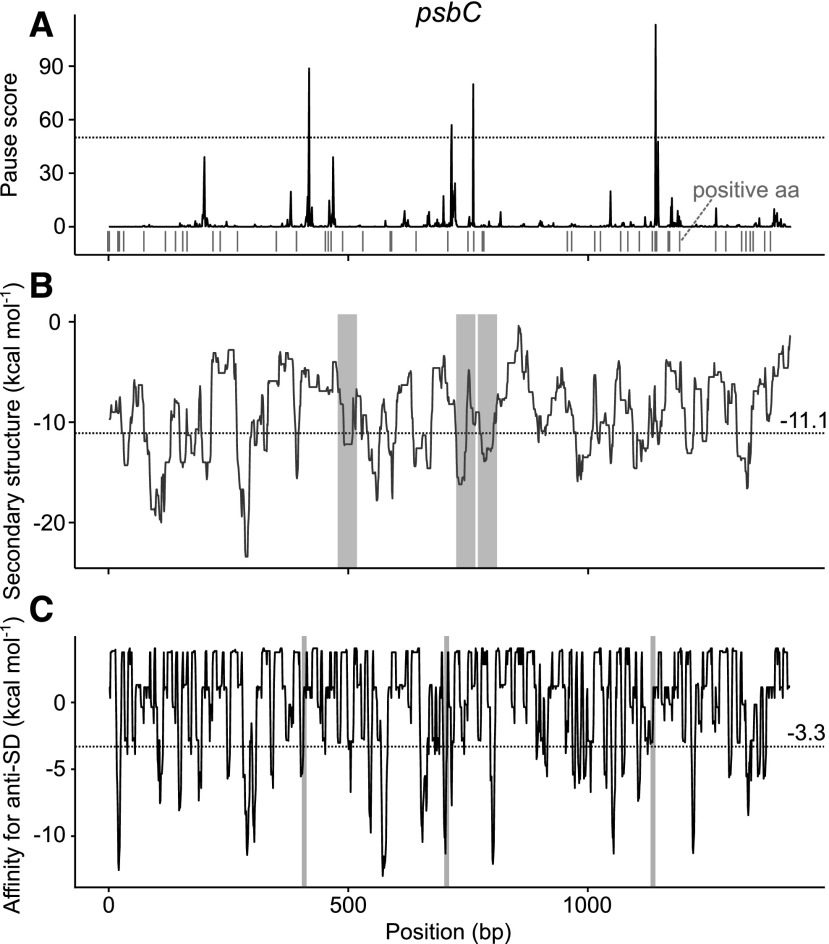
The results presented above show that mRNA structure, internal SDs, and the presence of positively charged amino acids in the nascent peptide chain can all cause plastid ribosome pausing. We checked how many pause sites with a pause score higher than 50 can be correlated with these features. It turned out that mRNA structure can explain 35.9% of these sites if only the most stable 10% of mRNA structures (Supplemental Fig. S4) are considered, and 68.0% when the strongest 30% are included. SDs explain 24.4% of the pause sites if only internal SDs with equal to or lower than −5 kcal mol−1 affinity for aSD are considered (at least 4-nucleotide-long SDs, such as GAGG, AGGA, and GGAG; see also Supplemental Fig. S9 for the distribution of kcal mol−1 in coding regions). It was found that 55.1% of pause sites are correlated with SDs with equal to or lower than −3.3 kcal mol−1 affinity for aSD (e.g. GGG), and 73.0% are explained if a 0 kcal mol−1 threshold (Scharff et al., 2011) for the aSD/SD hybridization is applied, which encompasses all stable base-pairing between the mRNA and the aSD. Positively charged amino acids explain 28.2% of pause sites if their average representation in all coding regions is used as the threshold. The average content of positively charged amino acids is 13.9%, which corresponds to 2.35 amino acids per 17-amino acid window used for an analysis. That means that here only clusters of at least three positively charged amino acids were included. But they explain 84.6% of pause sites if one assumes that one positively charged amino acid can be sufficient to induce pausing, as has been described for S. cerevisiae (Charneski and Hurst, 2013). In other words, depending on the choice of the thresholds given above, 32.1 to 91.0% of the pause sites correlate with more than one feature. In total, 55.1 to 94.9% of all pause sites can be explained. The psbC gene is an example for which all major pause sites can be explained on this basis (Fig. 4).
Figure 4.
All major ribosome pause sites in psbC are associated with pause-promoting elements. A, Pause sites in the psbC-coding region. The dotted line marks all pause sites with pause scores > 50. The positions of positively charged amino acids are indicated below the plot. B, Incidence of mRNA secondary structure in the psbC-coding region. The dotted line indicates the most stable 10% of mRNA structures (Supplemental Fig. S4). Shaded areas indicate mRNA structures that can explain the locations of major pause sites. C, The stability of aSD hybridization to internal SD sequences. The dotted line marks the strongest 10% of SD sequences (Supplemental Fig. S9). Shaded areas indicate SD sequences that explain major pause sites.
Plastid Ribosome Pausing and Transmembrane Domain Integration
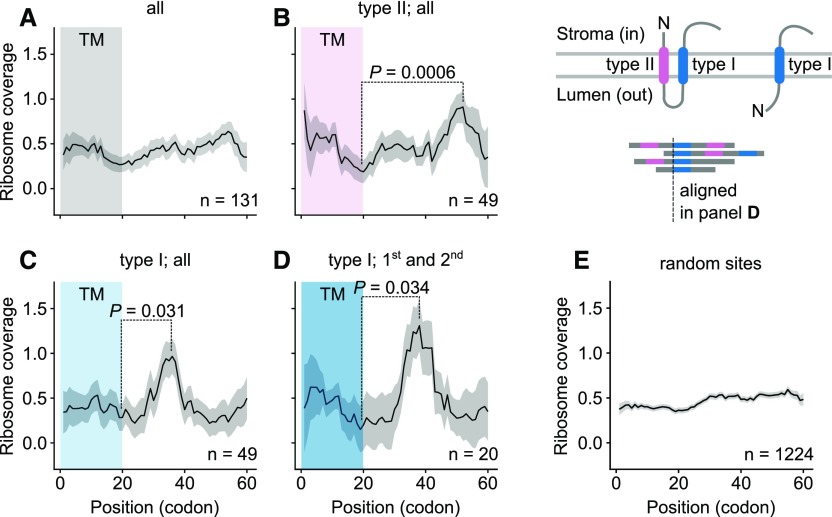
Ribosome pausing has been shown to be important for protein folding in several biological systems (Zhang et al., 2009; Yona et al., 2013; Fluman et al., 2014). The underlying idea is that a preceding structural domain must first be folded before translation of the protein can proceed. We therefore asked whether there is a correlation between ribosome pausing and the folding and integration of plastid-encoded membrane proteins. When all transmembrane domains (TMs) are considered, no clear correlation between their dispositions and pausing can be discerned (Fig. 5A). However, if the TMs are sorted according to their orientation in the membrane, a significant pausing signal at a position 52 amino acids downstream of the start of type II TMs (whose N termini project into the stroma) is found (Fig. 5B). This should allow for the emergence of the complete, ∼20-amino-acid-long TM from the exit tunnel of the ribosome. The exit tunnel is approximately 90 Å long (Bieri et al., 2017) and can therefore harbor a 25-residue peptide in the extended conformation (∼3.5 Å per amino acid) or 60 amino acids in an α-helical conformation (∼1.5 Å per amino acid; Voss et al., 2006; Yonath et al., 1987; Smith et al., 1978). In contrast, there is a statistically significant rise in the tendency to pause only 34 amino acids downstream of the start of type I TMs (whose N termini project to the outside/lumen; Fig. 5C). This difference becomes even more pronounced when only the first type I TM of each protein is considered (Fig. 5D). A pause at this distance from the N-terminal end of the TM probably would not allow the complete TM to leave the exit tunnel, but its N-terminal end will nevertheless be accessible for membrane targeting. This type of pausing has also been described for first TMs in E. coli (Fluman et al., 2014) and S. cerevisiae (Pechmann et al., 2014). We found a similar trend for type I TMs when we analyzed published data for maize (Chotewutmontri and Barkan, 2016; Supplemental Fig. S15). Thus, the positions of the pause sites relative to TMs depend on the particular TM’s orientation in the membrane (see also examples in Supplemental Figs. S16 and S17). This would facilitate insertion of TMs in the correct orientation, and thereby ensure correct assembly of the membrane complexes.
Figure 5.
Correlation of pause sites with TMs. A, Ribosome coverage downstream of all TMs. B, Statistically significant pausing occurs 52 amino acids after the start of type II TMs. See also the cartoon on the upper right for the distinction between type-I and type-II TMs. C, Statistically significant pausing occurs 35 amino acids after the start of type-I TMs. D, Locations of statistically significant pause sites relative to type-I TMs, if only the first type-I TM of each protein is included (see cartoon on upper right). In A to D, the line shows the running median of ribosome coverage; the shaded area around it represents the median ± se. Example genes can be found in Supplemental Figs. S16 and S17. E, Randomized sequences as control show no statistically significant differences in ribosome coverage. We selected random sites in proteins with at least one TM, and average ribosome profiling is calculated and plotted.
Plastid Ribosome Pausing and Cofactor Integration
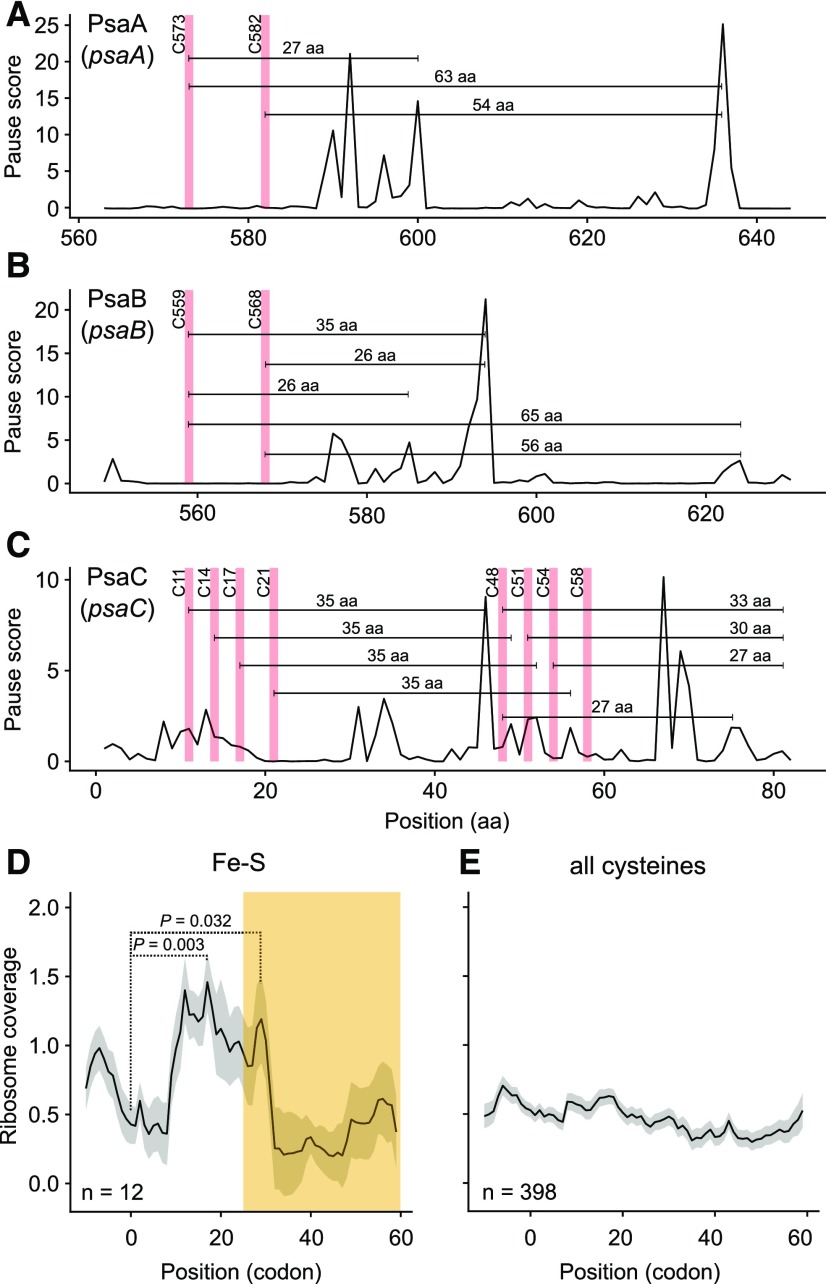
A second process related to protein folding is cofactor integration. The FeS clusters in photosystem I are bound by PsaA, PsaB, and PsaC (Qin et al., 2015). These clusters are located on the stroma side of the thylakoid membrane, i.e. facing the same compartment as the plastid ribosomes. There are ribosome pause sites suitably placed to allow the correctly positioned cysteines in all three subunits to bind—after they appear from the ribosome exit tunnel—the FeS clusters in the stroma (Fig. 6, A–C; Supplemental Fig. S16). We confirmed these results by comparing ribosome pausing behind cysteines binding FeS clusters and all other cysteines. Behind FeS cluster binding sites there is statistically significant ribosome pausing in a suitable distance (Fig. 6D), which cannot be seen downstream of all other cysteines (Fig. 6E). Pause sites are also located at appropriate positions in psbA (coding for the D1 subunit of photosystem II) to enable the transfer of the first binding sites of the Mn4CaO5 cluster in D1 (Wei et al., 2016) to the lumen, where they can bind the Mn4CaO5 cluster (Supplemental Fig. S17). These pause sites are not only found in the dicot species Arabidopsis, but were also detected in the monocot species barley (Hordeum vulgare) using a primer-extension inhibition (toeprinting) assay (Kim et al., 1991; Supplemental Fig. S18). Moreover, the positions of the mRNA secondary structures and internal SD sequences responsible for these pause sites in psbA are predominantly conserved between Arabidopsis and barley (Supplemental Fig. S18). Taken together, these findings indicate a role for ribosome pausing in cofactor integration.
Figure 6.
Ribosome pausing at the FeS cluster-binding sites in photosystem I subunits. A, Ribosome pause sites in PsaA. The cysteines binding the FeS cluster are depicted with reddish lines. The region around the FeS cluster binding sites is magnified to the same scale as for PsaB (B) and PsaC (C). For the full sequence of PsaA, see Supplemental Fig. S16A. The FeS clusters are located on the stroma side of the thylakoid membrane, i.e. in the same compartment as the plastid ribosomes. Therefore, the distance between the binding site and the pause site is expected to be larger than the length of the peptide within the exit tunnel of the ribosome: 25 to 60 amino acids (Bieri et al., 2017; see main text). One FeS cluster is bound by two cysteines in PsaA and two in PsaB. There are two pause sites located at the expected distances after C573 and one pause site after C582 in PsaA. B, Ribosome pausing in PsaB; magnified view of the region around the FeS cluster binding sites (the full PsaB is shown in Supplemental Fig. S16B). Downstream of C559 are three pause sites; downstream of C568 are two. C, Ribosome pausing in PsaC. PsaC has eight cysteines, which together bind two FeS clusters. After the first group of four cysteines (C11, C14, C17, and C21) there is a group of four pause sites, each at a distance of 35 amino acids from one of the cysteines. Similarly, after the second Cys group (C48, C51, C54, and C58), there are four pause sites near the C terminus. The distance is this case is only 19 to 22 amino acids, i.e. each Cys would still be in the exit tunnel when the immediately adjacent pause site is encountered, but each would emerge from the ribosome at a more downstream pause site (with the exception of the very last Cys, which is still in the exit tunnel when translation terminates). D, Ribosome coverage around cysteines of photosystem I subunits binding the FeS clusters. The yellowish area indicates the region with a suitable distance between Cys and pause site. There is statistically significant ribosome pausing at a suitable distance (Wilcoxon rank sum test), but also pausing occurring more closely, likely related to more upstream FeS binding sites (see the Cys pairs in A and B and the Cys quartets in C). E, Ribosome coverage around all other plastid-encoded cysteines. There is no ribosome pausing detected.
Conservation of Features Linked to Ribosome Pausing
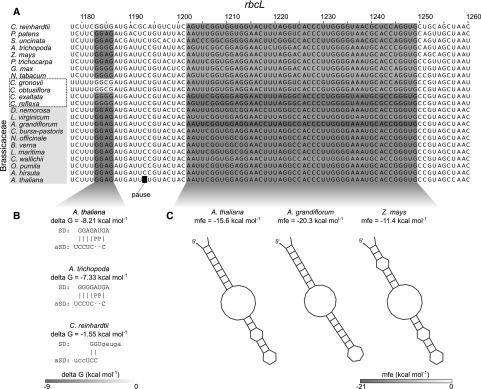
Pause sites that are of real biological significance are likely to be conserved. In the mRNA for the large subunit of Rubisco, rbcL, a major pause site, is found at the codon for Ser-398 (Fig. 7; Supplemental Fig. S19C). This may allow for recruitment of chaperones and/or the folding of most of the protein before synthesis of its C-terminal part, whose folding back to the active site is crucial for the enzymatic activity (Andersson, 2008; Bracher et al., 2017). Both the internal SD ahead of the pause site and the mRNA structure beyond it are strongly conserved across land plants. The latter element is even conserved in the green alga Chlamydomonas reinhardtii (Fig. 7). Interestingly, there is a correlation between lack of conservation of the internal SD and the gradual loss of photosynthetic capacity in obligate parasitic plant species of the genus Cuscuta. Here, we compared two parasitic species with reduced photosynthetic capacity (C. reflexa and C. exalata) to two with very low photosynthetic capacity (C. gronovii and C. obtusiflora; Funk et al., 2007; McNeal et al., 2007). In the first pair of species, the internal SD is still present, whereas both members of the second pair have lost it (Fig. 7). This correlation indicates a relaxation of selection pressure in species that are no longer dependent on photosynthesis. The positions of features associated with pausing in psbC are also well conserved over a broad range of plant species, but less so in parasitic Cuscuta species (Supplemental Fig. S20). The conservation of features leading to ribosome pausing in photosynthetic plants and their loss in parasitic plants represents a further argument for the importance of pause sites for the efficient folding, membrane insertion, and assembly of the photosynthetic complexes.
Figure 7.
Conservation of features linked to pausing in rbcL. A, Alignment of diverse rbcL sequences around a major pause site at position 1193 found in different Brassicaceae species. The other sequences listed are from dicot species of different families (Nicotiana tabacum, Glycine max, and Populus trichocarpa), a monocot (maize), a basal flowering plant (Amborella trichopoda), a member of the Lycopodiopsida (Selaginella uncinata), a moss (Physcomitrella patens), and a green alga (C. reinhardtii). The four Cuscuta species in the dashed box are obligate parasites. Their photosynthesis capacity ranges from reduced (C. reflexa and C. exalata) to very low levels (C. gronovii and C. obtusiflora; Funk et al., 2007; McNeal et al., 2007). Upstream of the pause site is an internal SD sequence, whose strength is indicated by the shade of gray. For examples and the scale bar, see B. Downstream of the pause site is a secondary structure, whose stability is indicated. For examples and the scale bar, see C. In addition to the mRNA-encoded features, there are two His codons outside of the sequence shown. These histidines are located 12 and 15 amino acids upstream of the pause site, respectively, and can also contribute to pausing when positively charged.
DISCUSSION
The aim of this study was to analyze translation elongation in chloroplasts and to determine what causes ribosome pausing and what biological functions it may have. For this we produced a ribosome profiling data set. Our results support that ribosome pausing is mainly caused by mRNA secondary structure, internal SDs, and positively charged amino acids in the nascent peptide chain. In all, 94.9% of major pause sites can be correlated with at least one of these features. At most major pause sites (91.0%), multiple features apparently work together. For instance, the strongest pause site we found, in rbcL, is marked by the presence of an internal SD, a structured region in the mRNA and a positively charged segment in the nascent protein, indicating that this combination explains why this particular site is especially effective (Fig. 7).
It might be assumed that ribosomes could remove all secondary structure from mRNA being translated, but the analysis of in vivo mRNA secondary structure has demonstrated that coding regions contain structure and can therefore influence translation elongation (Ding et al., 2014; Fig. 1B). Not only that, in E. coli, the structure found in the coding regions is actually a major determinant of translation efficiency (Burkhardt et al., 2017). The finding that internal SDs cause pausing in E. coli (Li et al., 2012; Fluman et al., 2014; Nakahigashi et al., 2014) was attributed in recent studies (Mohammad et al., 2016; Hwang and Buskirk, 2017) to a bias introduced by the exclusive analysis of long footprints. However, we were able to detect a correlation between SDs and pausing not only by analyzing the more sharply defined longer reads (Fig. 2), but also when short reads were included (Supplemental Fig. S7). In maize chloroplasts, ribosome pausing is also found to be correlated with the presence of internal SDs when shorter reads are included in the analysis (Supplemental Fig. S8). In addition, the stability of the anti-Shine-Dalgarno interactions with the mRNA is positively correlated with inferred pause duration (Supplemental Fig. S7), strongly indicating that internal SDs can cause ribosome pausing.
Ribosome pause sites indicate only local differences in the speed of translation. We could not observe differences in ribosome coverage before and after pause sites; therefore, these sites are not related to ribosome stalling causing the abortion of translation elongation. Whereas local differences in ribosome coverage are caused by pausing, the overall loading of the mRNA with ribosomes is likely to mainly depend on the efficiency of translation initiation as described for bacteria (Duval et al., 2015). In general, ribosome coverage of coding regions is in good agreement with other measurements of translation (Liu et al., 2017), and we have no indications that ribosome pausing is interfering with measurements of translation efficiencies.
Our protocol did not include any treatment with translation elongation inhibitors before extraction of the ribosomes, which can result in biases (Nakahigashi et al., 2014; Artieri and Fraser, 2014; Weinberg et al., 2016). An elongation inhibitor (chloramphenicol) is only added during extraction, and therefore the data obtained should reflect the in vivo situation more closely.
We found no evidence for any direct influence of codon usage on ribosome pausing (Supplemental Fig. S14). However, it is not surprising that specific codons do not cause pausing in plastids. The reduced tRNA set in plastids is not optimized for the many A/U-rich codons used in plastid mRNAs, but for decoding by wobbling and superwobbling (Alkatib et al., 2012). This makes it unlikely that rare codons could have an influence on elongation rate, given that even abundant codons can only be read by superwobbling, which is thought to be less efficient than standard base-pairing or wobbling (Rogalski et al., 2008). Interestingly, in contrast to the wild-type genes analyzed here, for proteins heterologously expressed in plastids, pausing or stalling at Ala and a pair of CUC codons has been described (Kwon et al., 2016).
The location of pause sites relative to TMs differs depending on the orientation of the TM concerned. Pause sites near TMs of type I (with the N terminus in the thylakoid lumen or outside the inner envelope membrane) are positioned downstream of their coding sequences, but too close to them for the TM to have fully emerged from the ribosome (Fig. 5C). The disposition could allow for the targeting of the TM to the membrane. However, in the case of type II TMs (with the N terminus exposed to the stroma), translation pauses only after the TM has completely left the exit tunnel of the ribosome (Fig. 5B). This suggests two alternative models: (1) the TM is first synthesized and inserted posttranslationally into the membrane; or (2) the TM is inserted cotranslationally like a type I TM, but the pause gives its N terminus time to be reinserted into the membrane from the “outside.” Afterward, the N terminus is transferred back to the inside and establishes the correct orientation, with the TM projecting into the stroma (Devaraneni et al., 2011).
For maize chloroplasts, it has been determined which gene products are inserted cotranslationally into the thylakoid membranes and which posttranslationally (Zoschke and Barkan, 2015). The posttranslationally inserted transmembrane proteins are very short and the majority of these have only one, or at most two, TMs. Their TMs have not yet emerged fully from the ribosome by the time translation is completed. Except for Cyt f (encoded by petA), we observed no clear pattern of pausing in this group. This protein is cotranslationally inserted, even though its TM is very close to the C terminus. Moreover, Cyt f is the only plastid-encoded protein with a cleavable signal peptide, and is targeted to the membrane by SecA (Röhl and van Wijk, 2001). Interestingly, this protein has a minor pause site in the signal peptide sequence (Supplemental Fig. S19B). D1 (encoded by psbA) is the only plastid-encoded protein reported to be signal-recognition-particle (SRP)-dependently targeted to the membrane (Nilsson et al., 1999). It has a major pause site, which lies upstream of the first TM and could facilitate targeting (Supplemental Fig. S19A). Recently, SRP-dependent targeting was found not to be related to ribosome pausing in E. coli (Schibich et al., 2016) or yeast (Chartron et al., 2016), although in yeast, pausing can at least facilitate SRP binding (Pechmann et al., 2014; Chartron et al., 2016).
Our results indicate a link between ribosome pausing and cofactor integration into the photosystems (Supplemental Figs. S16 and S17). We cannot distinguish whether this is caused by features of the mRNA and/or nascent chain or is itself dependent on the incorporation of cofactors. A recent analysis found no changes in translation elongation in mutants of chlorophyll synthesis, indicating that at least chlorophyll supply does not influence ribosome pausing (Zoschke et al., 2017). It is known that Mn4CaO5 cluster binds to D1 and CP43. We observed pausing near Mn4CaO5 cluster binding sites of D1 but not of the one of CP43 (Supplemental Fig. S17). There are currently two models of the Mn4CaO5 cluster incorporation into photosystem II. One describes it as a late step of photosystem II (PSII) assembly and the PsbP subunit to deliver the Mn2+ ions (Bondarava et al., 2007; Schmidt et al., 2016). The other is based on findings in cyanobacteria where PratA delivers Mn2+ ions to D1 possibly cotranslationally. Thus, the loading with Mn2+ is an early step of PSII assembly (Stengel et al., 2012; Nickelsen and Rengstl, 2013). In line with the later model, there is in contrast to D1 no ribosome pausing in a fitting distance to the Mn4CaO5 cluster binding site of CP43 (Supplemental Fig. S17). CP43 is added in a later step during PSII assembly (Nickelsen and Rengstl, 2013). Alternatively, it is also possible that ribosome pausing near Mn4CaO5 cluster binding sites could only be relevant during PSII repair, for which D1 has to be de novo synthesized (Theis and Schroda, 2016).
A reliable protocol for transformation of Arabidopsis plastids would enable us to mutate some of these features and test their influence on pausing and protein assembly but, unfortunately, no such method is currently available (Yu et al., 2017). Therefore, we decided to validate our results by comparing them with those for pause sites detected in very distantly related monocot species (Kim et al., 1991; Chotewutmontri and Barkan, 2016; Supplemental Figs. S5, S8, S12, S15, and S18). In addition, we analyzed whether the features linked to pausing in genes for photosynthetic subunits are conserved in land plants, and less so in parasitic species that do not depend on their functionality (Fig. 7; Supplemental Fig. S20). We found both conserved pause sites as well as conservation of related features in photosynthetic species, strongly indicating that positive selection pressure maintains the pattern of pausing. Accordingly, in obligate parasitic plant species of the genus Cuscuta, the extent of conservation correlates with the remaining photosynthetic capacity. Hence, in species that display very little photosynthetic activity, the features typically associated with pause sites in genes for photosystem components in Arabidopsis are less well conserved. This strongly suggests a degree of coevolution between ribosome pausing in plastids and the assembly of photosynthetic protein complexes.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) wild-type plants (ecotype Col-0) were grown in short-day conditions (8 h light, 140 μE m−2 s−1 to 160 μE m−2 s−1, 20°C) for 7 weeks. All young leaves with a maximal length of 20 mm (diameter of rosettes at this growth stage was 68 ± 3 mm) were harvested at noon into liquid nitrogen. See Supplemental Fig. S1 for an image and the photosynthetic performance of the harvested leaves. Chlorophyll a fluorescence parameters were measured in triplicates using the MAXI IMAGING-PAM M-series (Walz). Before F0 and Fm determination, plants were dark-acclimated for 30 min. Next, plants were exposed to 5 min of blue (450 nm) actinic light illumination (81 μE m−2 s−1) while saturating light pulses were applied at 20-s intervals. Results presented in Supplemental Fig. S1 were calculated for the last saturating pulse during the actinic light period. Fv/Fm, Y(II), Y(NPQ), and Y(NO) parameters were calculated as described by Klughammer and Schreiber (2008). Three biological replicates were analyzed. For each replicate, leaves from at least three plants were pooled.
Ribosome Profiling
Ribosomal footprint profiling was carried out in accordance with published protocols (Oh et al., 2011; Zoschke et al., 2013), with the following modifications: ground and frozen leaf material (400 mg) was thawed on ice in 5 mL of extraction buffer (200 mm Tris/HCl, pH 8.0, 200 mm KCl, 35 mm MgCl2, 200 mm sucrose, 1% Triton X-100, 2% polyoxyethylen-10-tridecyl-ether, 5 mm DTT, 100 µg/mL chloramphenicol, and 50 μg/mL cycloheximide). The extract was centrifuged for 5 min at 13,200g and 4°C to remove insoluble materials and nuclei, and the resulting supernatant was centrifuged for 10 min at 15,000g and 4°C. After the addition of CaCl2 (to a final concentration of 5 mm) and 750 units of micrococcal nuclease (Thermo Fisher Scientific), the extract was incubated for 1 h at room temperature and then loaded on a 2-mL sucrose cushion (40 mm Tris-acetate, pH 8.0, 100 mm KCl, 15 mm MgCl2, 1 m sucrose, 5 mm DTT, 100 μg/mL chloramphenicol, and 50 μg/mL cycloheximide). After a 3-h centrifugation at 55,000g at 4°C in a Type 70.1 Ti rotor (Beckman), the pellet containing the ribosomes was solubilized in 1% SDS, 10 mm Tris/HCl (pH 8.0), and 1 mm EDTA. Small RNAs including mRNA fragments corresponding to ribosomal footprints were purified from the ribosome fraction using the PureLink miRNA Isolation Kit (Invitrogen), and the 16- to 42-nucleotide fraction was isolated by electrophoresis. This fraction was treated with T4 polynucleotide kinase to dephosphorylate the 3′ ends before library preparation using the TruSeq Small RNA Library Preparation Kit and sequencing was done on the HiSeq 4000 platform (Illumina; performed by BGI).
Data
The genome sequence of Arabidopsis Col-0 and annotation files (TAIR 10 version 28) was downloaded from Ensembl Plants (http://plants.ensembl.org). Sequences of genes of nonparasitic species used for conservation analysis were retrieved from the Chloroplast Genome Database (Cui et al., 2006). Sequences of genes from four Cuscuta species were downloaded from the NCBI (NC_009963, NC_009765, NC_009949, and NC_009766). Sequence and annotation of the maize (Zea mays) plastid genome was downloaded from NCBI (X86563.2). Ribosome profiling data for maize (Chotewutmontri and Barkan, 2016) was downloaded from the NCBI Sequence Read Archive (accession no. SRP070787). We analyzed a subset of these ribosome footprints: those from maize leaf segment 9. We removed replicate 1 because it was significantly different from replicates 2 and 3.
Reads Preprocessing and Mapping
Quality and adapter trimming was performed using the software Trim Galore! (version 0.4.0; http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) with settings of quality = 20 and length = 18. Sequencing quality was assessed based on reports generated by Trim Galore! Next, reads were mapped to the Arabidopsis plastid coding sequences (86 genes) with an additional 50 nucleotides at both 5′ and 3′ ends with the software STAR Aligner (version 2.4.2; Dobin et al., 2013). We used the following STAR settings: outFilterMismatchNmax = 2; outMultimapperOrder = Random; outSAMmultNmax = 1; alignIntronMax = 1; alignIntronMin = 2). Using these settings, reads mapping to more than one target sequence were randomly assigned to only one target. Unless noted otherwise, we analyzed reads mapped to the “forward” strand of length between 28 and 40 bp. Maize footprints (Chotewutmontri and Barkan, 2016; see “Data”) were analyzed in a similar way, but reads of all lengths were included.
Identification of Pause Sites
All analyses were performed in the R (R Development Core Team, 2009) and Bioconductor packages (www.bioconductor.org). To select a method for ribosome occupancy assignment, we first performed an analysis using all ribosome footprints without subsetting by length (Supplemental Fig. S2A). However, this kind of analysis of MNase-produced footprints of plastid genes may be biased because of (1) significant differences in translation levels observed in plastid genes, (2) overlapping genes, and (3) MNase cutting bias. Thus, to determine the method for assignment, we selected 13 genes with evident peaks at start codons that did not overlap with other features and used only the better-defined footprint lengths (28–40 bp). To reduce the influence of differences in gene translation levels, we normalized the ribosome occupancies by dividing them by the mean occupancy for the analyzed region (Supplemental Fig. S2, B and C). Because 5′-assigned reads yield better alignments with the ribosomal P-site in our data sets than the 3′ assignment strategy (Supplemental Fig, S2), the density was shifted downstream of the 5′ end by 23 bp. Pause scores were calculated by taking the ribosome density at each nucleotide of a gene and dividing it by the mean ribosome density for that gene. Unless otherwise stated, genes with fewer than three reads per codon were excluded. Ribosome occupancy in the first and last 51 bp (17 codons) was excluded from the analysis of ribosome pausing so as to eliminate any influence of translation initiation and termination, and of overlapping genes. To calculate ribosome pause scores for individual codons in a gene (Figs. 3 and 5, E and F; Supplemental Figs. S11–S14, S16, S17, and S19), we summed the ribosome density for each nucleotide of a codon and divided the sum by the mean ribosome occupancy of all codons in that gene. The ribosome occupancies presented in Figure 5 were calculated by dividing the coverage of whole reads (without P-site assignment) mapped around a transmembrane domain by the mean coverage of the region analyzed. In all cases, we present the average of three biological replicates. Maize ribosome footprints that were produced using RNase I (Chotewutmontri and Barkan, 2016) were assigned using the 3′ assignment strategy and the density was shifted upstream by 7 bp.
Analysis of mRNA Secondary Structure
mRNA secondary structures in plastid protein-coding transcripts were predicted by calculating the minimum free energy (MFE) of a 51-bp-centered window using the RNAfold program in the Vienna RNAfold package (Gruber et al., 2008) as done previously (Scharff et al., 2011), but with a 51-nucleotide window instead of a 50-nucleotide window to facilitate centering. To analyze mRNA structure in vivo, we used published data (Ding et al., 2014): quality and adapter trimming was performed using Trim Galore! with the following settings: quality = 20, length = 18. Reads were mapped to the Arabidopsis plastid-coding sequences with an additional 50 nucletodies at both 5′ and 3′ ends using STAR Aligner (version 2.4.2; Dobin et al., 2013). We used the following STAR settings: clip5pNbases = 3; alignIntronMax = 1; alignIntronMin = 2; outFilterMismatchNmax = 2; outMultimapperOrder = Random; outSAMmultNmax = 1. Reverse transcriptase stop counts and DMS reactivity were calculated using the Galaxy RNA Structure tool (https://usegalaxy.org/). Next, we calculated the MFE of a 51-bp-centered window using the RNAfold program with constraints generated by considering any base with DMS reactivity under 0.3 as paired, and any value above 0.7 as unpaired.
Prediction of SD/aSD Hybridization
The aSD sequence (5′-CCUCCU-3′) was computationally hybridized in an 8-nucleotide window to coding regions at 20°C (31°C in maize) using the free2bind RNA-RNA hybridization algorithm (Starmer et al., 2006). The ∆G value was assigned to the first nucleotide of the analyzed window.
Analysis of Influence of Positively Charged Amino Acid on Ribosome Pausing
The role of positively charged amino acids in ribosome pausing was analyzed using the methodology already described in Charneski and Hurst (2013), with the following modifications: (1) instead of analyzing a 61-amino acid window, we analyzed a 27-residue window with a centered positively charged amino acid at position 0. This modification was prompted by the limited number of genes available for analysis in the plastid genome. (2) Positively charged amino acids were allowed at positions between 0 and 4 (e.g. see Fig. 3B). (3) We observed that use of the original methodology (Charneski and Hurst, 2013) results in a bias that suggests ribosome pausing even in random data. Similar observations have already been reported by others (Artieri and Fraser, 2014). Therefore, we used a logarithmic transformation of the rpos/rprec13 value (log2(rpos/rprec13 +1)), which eliminates this bias (Supplemental Fig. S5D).
Codon Bias Analysis
Codon bias analysis was performed as previously described by Artieri and Fraser (2014) with the following modifications: (1) we did not normalize ribosome sequencing results with respect to parallel mRNA sequencing; (2) we did not log-transform the values obtained; (3) in addition to codon bias, we also calculated the ribosome occupancy for amino acids (Supplemental Fig. S14); and (4) we used different methods and read lengths to assign the P-site of the ribosome (Supplemental Fig. S14).
Analysis of Ribosome Pausing in the Vicinity of TM Domains and Cofactor-Binding Sites
The annotation of the transmembrane domains was downloaded from UniProt (http://www.uniprot.org). The orientation of the transmembrane domains was predicted using the TMHMM Server v. 2.0 (Krogh et al., 2001). The orientation of the transmembrane domains was verified with structures obtained from the Molecular Modeling Database for ATP synthase (Zhou et al., 2015), the cytochrome b6f complex (Stroebel et al., 2003), photosystem I (Qin et al., 2015), and photosystem II (Wei et al., 2016), using the Cn3D viewer (Wang et al., 2000) for the subunits of these complexes. The positions of transmembrane domains in D1 and CP43 were corrected in accordance with the structures obtained from MMDB. NdhB and NdhD were omitted from the analysis, because there are different annotations in the Ensembl Plants and UniProt databases. The putative transmembrane protein Ycf2 was not included either, because no TM domains could be detected by TMHMM. Ribosome occupancies were calculated by dividing the coverage of whole reads (without P-site assignment) mapped around TM domains by the mean coverage of the region analyzed.
Conservation Analysis
mRNA sequences from analyzed species were aligned with MUSCLE (Edgar, 2004). Internal SD-like sequences and mRNA secondary structures were computationally predicted in all sequences using the free2bind (Starmer et al., 2006) and RNAfold (Gruber et al., 2008) programs, respectively.
Accession Numbers
The raw data were uploaded to the Sequence Read Archive under BioProject number PRJNA328073.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Quality control of reads.
Supplemental Figure S2. Assignment of reads to codons.
Supplemental Figure S3. Distribution of mRNA structure and ribosome pause sites in randomly selected regions of chloroplast-coding sequences.
Supplemental Figure S4. Distribution of MFE (kcal mol−1) values in all plastid transcripts.
Supplemental Figure S5. Correlation of ribosome pause sites with the occurrence of stable mRNA secondary structure downstream in Zea mays.
Supplemental Figure S6. Correlation of ribosome pause sites with the presence of stable mRNA secondary structures downstream: analysis of the influence of the strength of pausing (pause score), footprint length, and codon assignment strategy (compare with Fig. 1A).
Supplemental Figure S7. Correlation of ribosomal pause sites with internal SD sequences located upstream: analysis of the influence of the strength of pausing (pause score), footprint length, and codon assignment strategy (compare with Fig. 2A).
Supplemental Figure S8. Correlation of ribosomal pause sites with internal SD sequences located upstream in chloroplasts of maize depends on pause score.
Supplemental Figure S9. Distribution of ΔG (kcal mol−1) values, representing relative affinities of all plastid transcripts in Arabidopsis for the aSD.
Supplemental Figure S10. Distribution of SD sequences and ribosome pause sites in randomly selected regions of chloroplast coding sequences.
Supplemental Figure S11. Correlation of ribosome pause sites with the presence of codons for positively charged amino acids upstream: analysis of the influence of the strength of pausing (pause score).
Supplemental Figure S12. Correlation of ribosomal pause sites in chloroplast mRNAs of maize with the presence of positively charged amino acids (Arg, His, and Lys) upstream.
Supplemental Figure S13. Controls for the correlation of ribosomal pause sites with the presence of positively charged amino acids (Arg, His, and Lys) upstream (Fig. 3).
Supplemental Figure S14. Lack of correlation between ribosome pausing and codon usage.
Supplemental Figure S15. Correlation of pause sites with TMs in chloroplasts of maize.
Supplemental Figure S16. Ribosome pausing at the FeS cluster-binding sites in photosystem I subunits.
Supplemental Figure S17. Ribosome pausing at the manganese cluster-binding sites of PSII subunits.
Supplemental Figure S18. Conservation of ribosomal pause sites in psbA of Arabidopsis and H. vulgare.
Supplemental Figure S19. Pause sites are correlated with targeting to the thylakoid membranes and the folding of the large subunit of Rubisco.
Supplemental Figure S20. Conservation of features causing pausing in psbC.
Acknowledgments
We thank Paul Hardy and Stefanie Zintl for commenting on the manuscript.
Footnotes
[OPEN]Articles can be viewed without a subscription.
References
- Ahmed T, Shi J, Bhushan S (2017) Unique localization of the plastid-specific ribosomal proteins in the chloroplast ribosome small subunit provides mechanistic insights into the chloroplastic translation. Nucleic Acids Res 45: 8581–8595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed T, Yin Z, Bhushan S (2016) Cryo-EM structure of the large subunit of the spinach chloroplast ribosome. Sci Rep 6: 35793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkatib S, Scharff LB, Rogalski M, Fleischmann TT, Matthes A, Seeger S, Schöttler MA, Ruf S, Bock R (2012) The contributions of wobbling and superwobbling to the reading of the genetic code. PLoS Genet 8: e1003076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson I. (2008) Catalysis and regulation in Rubisco. J Exp Bot 59: 1555–1568 [DOI] [PubMed] [Google Scholar]
- Artieri CG, Fraser HB (2014) Accounting for biases in riboprofiling data indicates a major role for proline in stalling translation. Genome Res 24: 2011–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (2011) Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol 155: 1520–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker AH, Oh E, Weissman JS, Kramer G, Bukau B (2013) Selective ribosome profiling as a tool for studying the interaction of chaperones and targeting factors with nascent polypeptide chains and ribosomes. Nat Protoc 8: 2212–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieri P, Leibundgut M, Saurer M, Boehringer D, Ban N (2017) The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EMBO J 36: 475–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondarava N, Un S, Krieger-Liszkay A (2007) Manganese binding to the 23 kDa extrinsic protein of Photosystem II. Biochim Biophys Acta 1767: 583–588 [DOI] [PubMed] [Google Scholar]
- Bracher A, Whitney SM, Hartl FU, Hayer-Hartl M (2017) Biogenesis and metabolic maintenance of Rubisco. Annu Rev Plant Biol 68: 29–60 [DOI] [PubMed] [Google Scholar]
- Burkhardt DH, Rouskin S, Zhang Y, Li G-W, Weissman JS, Gross CA (2017) Operon mRNAs are organized into ORF-centric structures that predict translation efficiency. eLife 6: e22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskirk AR, Green R (2017) Ribosome pausing, arrest and rescue in bacteria and eukaryotes. Philos Trans R Soc Lond B Biol Sci 372: 20160183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadani Y, Niwa T, Chiba S, Taguchi H, Ito K (2016) Integrated in vivo and in vitro nascent chain profiling reveals widespread translational pausing. Proc Natl Acad Sci USA 113: E829–E838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charneski CA, Hurst LD (2013) Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol 11: e1001508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartron JW, Hunt KCL, Frydman J (2016) Cotranslational signal-independent SRP preloading during membrane targeting. Nature 536: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Coakley A, O’Connor M, Petrov A, O’Leary SE, Atkins JF, Puglisi JD (2015) Coupling of mRNA structure rearrangement to ribosome movement during bypassing of non-coding regions. Cell 163: 1267–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotewutmontri P, Barkan A (2016) Dynamics of chloroplast translation during chloroplast differentiation in maize. PLoS Genet 12: e1006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Veeraraghavan N, Richter A, Wall K, Jansen RK, Leebens-Mack J, Makalowska I, dePamphilis CW (2006) ChloroplastDB: the Chloroplast Genome Database. Nucleic Acids Res 34: D692–D696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraneni PK, Conti B, Matsumura Y, Yang Z, Johnson AE, Skach WR (2011) Stepwise insertion and inversion of a type II signal anchor sequence in the ribosome-Sec61 translocon complex. Cell 146: 134–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM (2014) In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505: 696–700 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval M, Simonetti A, Caldelari I, Marzi S (2015) Multiple ways to regulate translation initiation in bacteria: mechanisms, regulatory circuits, dynamics. Biochimie 114: 18–29 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann TT, Scharff LB, Alkatib S, Hasdorf S, Schöttler MA, Bock R (2011) Nonessential plastid-encoded ribosomal proteins in tobacco: a developmental role for plastid translation and implications for reductive genome evolution. Plant Cell 23: 3137–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluman N, Navon S, Bibi E, Pilpel Y (2014) mRNA-programmed translation pauses in the targeting of E. coli membrane proteins. eLife 3: e03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk HT, Berg S, Krupinska K, Maier UG, Krause K (2007) Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol 7: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL (2008) The Vienna RNA website. Nucleic Acids Res 36: W70–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess A-K, Saffert P, Liebeton K, Ignatova Z (2015) Optimization of translation profiles enhances protein expression and solubility. PLoS One 10: e0127039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang JY, Buskirk AR (2017) A ribosome profiling study of mRNA cleavage by the endonuclease RelE. Nucleic Acids Res 45: 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Klein PG, Mullet JE (1991) Ribosomes pause at specific sites during synthesis of membrane-bound chloroplast reaction center protein D1. J Biol Chem 266: 14931–14938 [PubMed] [Google Scholar]
- Klughammer C, Schreiber U (2008) Complementary PS II quantum yields calculated from simple fluorescence parameters measured by PAM fluorometry and the saturation pulse method. PAM Appl Notes 1: 27–35 [Google Scholar]
- Koutmou KS, Schuller AP, Brunelle JL, Radhakrishnan A, Djuranovic S, Green R (2015) Ribosomes slide on lysine-encoding homopolymeric A stretches. eLife 4: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Kwon K-C, Chan H-T, León IR, Williams-Carrier R, Barkan A, Daniell H (2016) Codon optimization to enhance expression yields insights into chloroplast translation. Plant Physiol 172: 62–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G-W, Oh E, Weissman JS (2012) The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 484: 538–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T-Y, Huang HH, Wheeler D, Xu Y, Wells JA, Song YS, Wiita AP (2017) Time-resolved proteomics extends ribosome profiling-based measurements of protein synthesis dynamics. Cell Syst 4: 636–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Deutsch C (2008) Electrostatics in the ribosomal tunnel modulate chain elongation rates. J Mol Biol 384: 73–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeal JR, Kuehl JV, Boore JL, de Pamphilis CW (2007) Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol 7: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F, Woolstenhulme CJ, Green R, Buskirk AR (2016) Clarifying the translational pausing landscape in bacteria by ribosome profiling. Cell Reports 14: 686–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahigashi K, Takai Y, Shiwa Y, Wada M, Honma M, Yoshikawa H, Tomita M, Kanai A, Mori H (2014) Effect of codon adaptation on codon-level and gene-level translation efficiency in vivo. BMC Genomics 15: 1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickelsen J, Rengstl B (2013) Photosystem II assembly: from cyanobacteria to plants. Annu Rev Plant Biol 64: 609–635 [DOI] [PubMed] [Google Scholar]
- Nilsson R, Brunner J, Hoffman NE, van Wijk KJ (1999) Interactions of ribosome nascent chain complexes of the chloroplast-encoded D1 thylakoid membrane protein with cpSRP54. EMBO J 18: 733–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Becker AH, Sandikci A, Huber D, Chaba R, Gloge F, Nichols RJ, Typas A, Gross CA, Kramer G, Weissman JS, Bukau B (2011) Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell 147: 1295–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann S, Chartron JW, Frydman J (2014) Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo. Nat Struct Mol Biol 21: 1100–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop C, Rouskin S, Ingolia NT, Han L, Phizicky EM, Weissman JS, Koller D (2014) Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Mol Syst Biol 10: 770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Suga M, Kuang T, Shen J-R (2015) Photosynthesis. Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex. Science 348: 989–995 [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2009). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- Rogalski M, Karcher D, Bock R (2008) Superwobbling facilitates translation with reduced tRNA sets. Nat Struct Mol Biol 15: 192–198 [DOI] [PubMed] [Google Scholar]
- Röhl T, van Wijk KJ (2001) In vitro reconstitution of insertion and processing of cytochrome f in a homologous chloroplast translation system. J Biol Chem 276: 35465–35472 [DOI] [PubMed] [Google Scholar]
- Sabi R, Tuller T (2017) Computational analysis of nascent peptides that induce ribosome stalling and their proteomic distribution in Saccharomyces cerevisiae. RNA 23: 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharff LB, Childs L, Walther D, Bock R (2011) Local absence of secondary structure permits translation of mRNAs that lack ribosome-binding sites. PLoS Genet 7: e1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibich D, Gloge F, Pöhner I, Björkholm P, Wade RC, von Heijne G, Bukau B, Kramer G (2016) Global profiling of SRP interaction with nascent polypeptides. Nature 536: 219–223 [DOI] [PubMed] [Google Scholar]
- Schmidt SB, Jensen PE, Husted S (2016) Manganese deficiency in plants: the impact on photosystem II. Trends Plant Sci 21: 622–632 [DOI] [PubMed] [Google Scholar]
- Sharma MR, Wilson DN, Datta PP, Barat C, Schluenzen F, Fucini P, Agrawal RK (2007) Cryo-EM study of the spinach chloroplast ribosome reveals the structural and functional roles of plastid-specific ribosomal proteins. Proc Natl Acad Sci USA 104: 19315–19320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WP, Tai PC, Davis BD (1978) Interaction of secreted nascent chains with surrounding membrane in Bacillus subtilis. Proc Natl Acad Sci USA 75: 5922–5925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starmer J, Stomp A, Vouk M, Bitzer D (2006) Predicting Shine-Dalgarno sequence locations exposes genome annotation errors. PLOS Comput Biol 2: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel A, Gügel IL, Hilger D, Rengstl B, Jung H, Nickelsen J (2012) Initial steps of photosystem II de novo assembly and preloading with manganese take place in biogenesis centers in Synechocystis. Plant Cell 24: 660–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroebel D, Choquet Y, Popot J-L, Picot D (2003) An atypical haem in the cytochrome b6f complex. Nature 426: 413–418 [DOI] [PubMed] [Google Scholar]
- Thanaraj TA, Argos P (1996) Ribosome-mediated translational pause and protein domain organization. Protein Sci 5: 1594–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis J, Schroda M (2016) Revisiting the photosystem II repair cycle. Plant Signal Behav 11: e1218587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholstrup J, Oddershede LB, Sørensen MA (2012) mRNA pseudoknot structures can act as ribosomal roadblocks. Nucleic Acids Res 40: 303–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller N, Bock R (2014) The translational apparatus of plastids and its role in plant development. Mol Plant 7: 1105–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller N, Weingartner M, Thiele W, Maximova E, Schöttler MA, Bock R (2012) The plastid-specific ribosomal proteins of Arabidopsis thaliana can be divided into non-essential proteins and genuine ribosomal proteins. Plant J 69: 302–316 [DOI] [PubMed] [Google Scholar]
- Tuller T, Veksler-Lublinsky I, Gazit N, Kupiec M, Ruppin E, Ziv-Ukelson M (2011) Composite effects of gene determinants on the translation speed and density of ribosomes. Genome Biol 12: R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss NR, Gerstein M, Steitz TA, Moore PB (2006) The geometry of the ribosomal polypeptide exit tunnel. J Mol Biol 360: 893–906 [DOI] [PubMed] [Google Scholar]
- Wang Y, Geer LY, Chappey C, Kans JA, Bryant SH (2000) Cn3D: sequence and structure views for Entrez. Trends Biochem Sci 25: 300–302 [DOI] [PubMed] [Google Scholar]
- Wei X, Su X, Cao P, Liu X, Chang W, Li M, Zhang X, Liu Z (2016) Structure of spinach photosystem II-LHCII supercomplex at 3.2 Å resolution. Nature 534: 69–74 [DOI] [PubMed] [Google Scholar]
- Weinberg DE, Shah P, Eichhorn SW, Hussmann JA, Plotkin JB, Bartel DP (2016) Improved ribosome-footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Reports 14: 1787–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J-D, Lancaster L, Hodges C, Zeri A-C, Yoshimura SH, Noller HF, Bustamante C, Tinoco I (2008) Following translation by single ribosomes one codon at a time. Nature 452: 598–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN, Arenz S, Beckmann R (2016) Translation regulation via nascent polypeptide-mediated ribosome stalling. Curr Opin Struct Biol 37: 123–133 [DOI] [PubMed] [Google Scholar]
- Woolstenhulme CJ, Guydosh NR, Green R, Buskirk AR (2015) High-precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell Reports 11: 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolstenhulme CJ, Parajuli S, Healey DW, Valverde DP, Petersen EN, Starosta AL, Guydosh NR, Johnson WE, Wilson DN, Buskirk AR (2013) Nascent peptides that block protein synthesis in bacteria. Proc Natl Acad Sci USA 110: E878–E887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Subramanian AR (2000) The plastid ribosomal proteins. Identification of all the proteins in the 50 S subunit of an organelle ribosome (chloroplast). J Biol Chem 275: 28466–28482 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, von Knoblauch K, Subramanian AR (2000) The plastid ribosomal proteins. Identification of all the proteins in the 30 S subunit of an organelle ribosome (chloroplast). J Biol Chem 275: 28455–28465 [DOI] [PubMed] [Google Scholar]
- Yona AH, Bloom-Ackermann Z, Frumkin I, Hanson-Smith V, Charpak-Amikam Y, Feng Q, Boeke JD, Dahan O, Pilpel Y (2013) tRNA genes rapidly change in evolution to meet novel translational demands. eLife 2: e01339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonath A, Leonard KR Wittmann HG (1987) A tunnel in the large ribosomal subunit revealed by three-dimensional image reconstruction. Science 236: 813–816 [DOI] [PubMed] [Google Scholar]
- Yu Q, Lutz KA Maliga P (2017) Efficient plastid transformation in Arabidopsis. Plant Physiol 29: 00857.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Hubalewska M, Ignatova Z (2009) Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol 16: 274–280 [DOI] [PubMed] [Google Scholar]
- Zhang J, Ruf S, Hasse C, Childs L, Scharff LB, Bock R (2012) Identification of cis-elements conferring high levels of gene expression in non-green plastids. Plant J 72: 115–128 [DOI] [PubMed] [Google Scholar]
- Zhou A, Rohou A, Schep DG, Bason JV, Montgomery MG, Walker JE, Grigorieff N, Rubinstein JL (2015) Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM. eLife 4: e10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R, Barkan A (2015) Genome-wide analysis of thylakoid-bound ribosomes in maize reveals principles of cotranslational targeting to the thylakoid membrane. Proc Natl Acad Sci USA 112: E1678–E1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R, Chotewutmontri P, Barkan A (2017) Translation and co-translational membrane engagement of plastid-encoded chlorophyll-binding proteins are not influenced by chlorophyll availability in maize. Front Plant Sci 8: 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R, Watkins KP, Barkan A (2013) A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo. Plant Cell 25: 2265–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]