REI1-LIKE ribosome biogenesis factor homologs guide cold acclimation and 60S subunit accumulation for plant growth at alternating temperatures.
Abstract
Arabidopsis (Arabidopsis thaliana) REI1-LIKE (REIL) proteins, REIL1 and REIL2, are homologs of a yeast ribosome biogenesis factor that participates in late cytoplasmic 60S ribosomal subunit maturation. Here, we report that the inhibited growth of the reil1-1 reil2-1 mutant at 10°C can be rescued by the expression of amino-terminal FLUORESCENT PROTEIN (FP)-REIL fusions driven by the UBIQUITIN10 promoter, allowing the analysis of REIL function in planta. Arabidopsis REIL1 appears to be functionally conserved, based on the cytosolic localization of FP-REIL1 and the interaction of native REIL1 with the 60S subunit in wild-type plants. In contrast to its yeast homologs, REIL1 also was present in translating ribosome fractions. Systems analysis revealed that wild-type Arabidopsis remodels the cytosolic translation machinery when grown at 10°C by accumulating cytosolic ribosome subunits and inducing the expression of cytosolic ribosomal RNA, ribosomal genes, ribosome biogenesis factors, and translation initiation or elongation factors. In the reil1-1 reil2-1 mutant, all processes associated with inhibited growth were delayed, although the plants maintained cellular integrity or acquired freezing tolerance. REIL proteins also were implicated in plant-specific processes: nonacclimated reil1-1 reil2-1 exhibited cold-acclimation responses, including activation of the DREB/CBF regulon. In addition, acclimated reil1-1 reil2-1 plants failed to activate FLOWERING LOCUS T expression in mature leaves. Therefore, in the wild type, REIL function may contribute to temperature perception by suppressing premature cold responses during growth at nonstressful temperatures. In conclusion, we suggest that Arabidopsis REIL proteins influence cold-induced plant ribosome remodeling and enhance the accumulation of cytosolic ribosome subunits after cold shift either by de novo synthesis or by recycling them from the translating ribosome fraction.
In plants, cytosolic ribosomes are composed of the canonical eukaryotic small ribosomal subunit (40S SSU), which involves the 18S ribosomal RNA (rRNA) and approximately 32 ribosomal proteins (RPs). Cytosolic ribosomes also include the large ribosomal subunit (60S LSU) that involves the 28S, 5.8S, and 5S rRNAs and approximately 48 RPs (Barakat et al., 2001). Cytosolic RPs are highly conserved across the eukaryotic kingdoms, but plants have between two and seven paralogs of each RP (Barakat et al., 2001). Accordingly, the Arabidopsis (Arabidopsis thaliana) genome contains ∼221 genes that code for cytosolic RPs in addition to 71 mitochondrial and 77 plastid RP genes (Sormani et al., 2011). These multiple plant paralogs of each cytosolic RP family create the potential for highly diverse cytosolic ribosome species that may function redundantly (Creff et al., 2010). However, the paralogs of cytosolic RPs can differentially change phosphorylation status or abundance upon environmental or nutritional cues (Chang et al., 2005; Wang et al., 2013; Browning and Bailey-Serres, 2015) and apparently have specific developmental roles (Degenhardt and Bonham-Smith, 2008; Byrne, 2009; Horiguchi et al., 2012; Rosado et al., 2012).
The complex biogenesis of the 40S SSU and 60S LSU is initiated in the nucleolus, continues in the nucleoplasm, and is completed after export to the cytosol by late maturation and structural proofreading steps that render the subunits translationally competent. Similar to other eukaryotic organisms, a 43S preinitiation complex is formed prior to translation that contains the eukaryotic initiation factors (eIFs). Under the guidance of eIF3, the preinitiation complex recruits either the conserved eIF4F complex or a plant-specific eIFiso4F complex. These are circularized complexes composed of cap-bound mRNA, eIF4 or eIFiso4, and poly(A)-binding protein. The combination of 43S and eIF4F-/eIFiso4F complexes forms the start codon (AUG) scanning 48S complexes. AUG recognition triggers 60S LSU recruitment, the exit of eIF1, eIF2, eIF3, eIF5, and eIF6, and ultimately the formation of the mRNA decoding 80S monosomes or polysomes (Browning and Bailey-Serres, 2015).
While cytoplasmic mRNA translation is well investigated (Browning and Bailey-Serres, 2015), a functional analysis of plant ribosome biogenesis is lacking. Due to the many 60S and 40S maturation steps, a multitude of eukaryotic ribosome biogenesis factors (RBFs) function in the nucleolus, in the nucleoplasm, or in the cytosol (Pendle et al., 2005; Palm et al., 2016). Only 25% of the plant homologs of yeast (Saccharomyces cerevisiae) RBFs, the currently best understood model organism, have been characterized so far (Weis et al., 2015). Currently, the sparse knowledge of plant ribosome biogenesis and the investigated details of preribosomal RNA processing indicate the existence of plant-specific ribosome biogenesis mechanisms that may have evolved to adapt the nonhomeostatic plant system to environmental changes (Weis et al., 2015).
The Arabidopsis zinc finger proteins REI1-LIKE1 (REIL1) and REIL2 were named after their yeast homologs Rei1 (Required for isotropic bud growth1) and its paralog Reh1 (Rei1 homologue1) and recently were recognized as late-stage RBF homologs that likely participate in the cytoplasmic steps of cytosolic ribosomal maturation (Schmidt et al., 2013; Weis et al., 2015). REIL1 and REIL2 apparently share evolutionarily conserved functions with their yeast homologs, indicated by REIL1 and REIL2 transcript coexpression and protein-protein interaction studies and through partial complementation of the yeast ∆rei1 mutant by heterologous expression of REIL1 (Schmidt et al., 2013, 2014). Furthermore, the loss of one or both paralogs showed that they were nonessential at standard growth temperature but caused severe growth defects at suboptimal low temperature, namely 10°C for Arabidopsis. These defects were surprisingly similar comparing the Arabidopsis mutants (Schmidt et al., 2013) with the respective homologous yeast mutants (Iwase and Toh-e, 2004; Lebreton et al., 2006; Parnell and Bass, 2009).
So far, the function of yeast Rei1 is the best characterized among the currently known REIL homologs that appear exclusively in eukaryotes (Schmidt et al., 2013). The cytosolic Rei1 protein was discovered through a mitotic proliferation defect of the ∆rei1 mutant (Iwase and Toh-e, 2004). This aspect was not further investigated when Rei1 was recognized as a late cytosolic RBF that associated with the nontranslating 60S LSU fraction (Hung and Johnson, 2006; Lebreton et al., 2006; Meyer et al., 2007). The current mechanistic model of late 60S biogenesis in yeast (Lo et al., 2010; Panse and Johnson, 2010; Greber et al., 2016; Ma et al., 2017) indicates a central role of Rei1 for the release of 60S nuclear export factors and triggering final maturation steps to full translation competence. In detail, prior to the Rei1 interaction, the 60S nuclear export factor Ribosomal-like protein24 (Rlp24) is replaced in the cytosol by Ribosomal protein of the large subunit24A (Rpl24A) or the almost identical Rpl24B paralog. Rpl24 proteins were suggested as binding partners of Rei1 that recruit Rei1 to the 60S LSU (Lebreton et al., 2006; Lo et al., 2010). Bound Rei1 cooperates with type III j-protein1 (Jjj1; Demoinet et al., 2007; Meyer et al., 2007) to activate Hsp70-type ATPases, Ssa1 or Ssa2, which in turn release an export factor complex that is composed of Associated with ribosomal export complex1 (Arx1) and ARX1 little brother1 (Alb1; Lo et al., 2010; Meyer et al., 2010). Arx1-Alb1 release is required to release the two final 60S RBFs. These RBFs are (1) the Nonsense-mediated mRNA decay3 protein, with a likely double function in 60S LSU structural proofreading and as a 60S LSU nuclear export factor (Johnson et al., 2002; Lo et al., 2010), and (2) the eukaryotic Translation initiation factor6 (or eIF6), an antiassociation factor that is located on the pre-60S LSU surface and interacts with the 40S SSU when forming the 80S ribosome (Basu et al., 2001; Klinge et al., 2011). Only the release of both final RBFs renders yeast 60S LSU translationally competent.
Recent cryo-electron microscopy structure analyses captured reconstituted and native yeast 60S-Arx1-Alb1-Rei1 complexes at near atomic resolution (Greber et al., 2012, 2016). In these complexes, the middle part of Rei1, amino acid residues 145 to 261 that contain two zinc fingers, is positioned close to the polypeptide tunnel exit. The C-terminal 95 amino acids, residues 299 to 393, of Rei1 are inserted fully into the tunnel and span the distance from tunnel exit to the peptidyl transferase active site (Greber et al., 2016). Prior to Rei1, the polypeptide tunnel is occupied by the ∼75 C-terminal amino acids of the nuclear pre-60S assembly and export factor Nucleolar G-protein1 Nog1 (Wu et al., 2016). In contrast to the well-structured Rei1 C terminus, the Rei1 N terminus (residues 1–144), which contains the first zinc finger, was not visualized and apparently is disordered in the captured complex. Jjj1 maps to the vicinity of the polypeptide tunnel exit close to Arx1 (Greber et al., 2012) and the Arx1-Alb1 complex is located in contact with Rei1 at the polypeptide tunnel exit (Greber et al., 2016). C-terminal fusions with GFP or the tandem affinity purification tag render Rei1 nonfunctional. C-terminal deletion studies demonstrated the requirement of up to 61 C-terminal amino acids, residues 333 to 393, and the presence of an Arx1-interacting helix of Rei1. These results suggest that Rei1 proofreading may test polypeptide tunnel integrity in the cytosol (Greber et al., 2016). Additional functions of yeast Rei1 remain to be discovered, such as the role of the disordered Rei1 N terminus (Greber et al., 2016) or a possible moonlighting function of non-ribosome-bound Rei1 that may be indicated by the initially discovered mitotic defect of ∆rei1 (Iwase and Toh-e, 2004). The role of the Rei1 paralog, Reh1, apparently is equivalent to that of Rei1. Reh1 stabilizes the 60S LSU in the absence of Rei1 (Parnell and Bass, 2009) and forms a complex with the 60S LSU with the Reh1 C terminus inserted into the polypeptide tunnel (Ma et al., 2017).
In contrast to yeast, the functional analyses of the likely Arabidopsis RBFs, REIL1 and REIL2, are in their initial stages. Our previous reports (Schmidt et al., 2013, 2014) supported the hypothesis of a partial conservation of Arabidopsis REIL function with yeast Rei1 and Reh1, but several observations indicate that the evolution of Arabidopsis REIL proteins diverged from that of yeast. (1) Heterologous expression only of Arabidopsis REIL1 but not of REIL2 complemented the temperature-dependent growth defect of the yeast ∆rei1 mutant (Schmidt et al., 2013). (2) Different gene duplication events apparently created the two paralogs in yeast and Arabidopsis independently (Schmidt et al., 2013). (3) In contrast to the four zinc finger domains present in Arabidopsis REIL1 and REIL2 that are arranged in two C2H2 zinc finger pairs, namely ZF1/ZF2 at the N terminus and ZF3/ZF4 in the middle of the protein (Schmidt et al., 2013), the yeast paralogs contain only three zinc fingers (Iwase and Toh-e, 2004; Greber et al., 2012).
To take the next step toward the functional analysis of Arabidopsis REIL proteins, we investigated both evidence of functional conservation and indications of plant-specific functions of the Arabidopsis REIL proteins, which may have been added on the evolutionary path toward the multicellular embryophyte Arabidopsis. For this purpose, we first established the previously created double mutant reil1-1 reil2-1 (Schmidt et al., 2013) as a valid plant system for the investigation of REIL function in Arabidopsis. We characterized the reil1-1 reil2-1 acclimation process to low temperature (10°C) in comparison with the Arabidopsis Columbia-0 (Col-0) wild type and found that this process likely involves ribosomal biogenesis and/or ribosome subunit recycling, processes that we cannot distinguish currently with our methods. We demonstrated functional complementation of the growth defect of reil1-1 reil2-1 in the cold and cytosolic protein localization by reintroducing N-terminal Fluorescent Protein (FP)-REIL fusion proteins. Furthermore, we demonstrated an association of REIL with cytosolic wild-type ribosomes and an enhancer role of REILs for 60S subunit accumulation during cold acclimation. Integrative metabolomic and transcriptomic systems analyses of reil1-1 reil2-1 linked REIL proteins to higher level plant functions, such as temperature perception. This association was indicated by premature triggering of cold-acclimation responses in reil1-1 reil2-1. Transcriptional compensation reactions in reil1-1 reil2-1 pointed toward a role of Arabidopsis REIL proteins for plant development at low temperature that may reach beyond ribosomal 60S LSU biogenesis.
RESULTS
The reil1-1 reil2-1 Mutant Stops Growth and Development When Exposed to Low Temperature
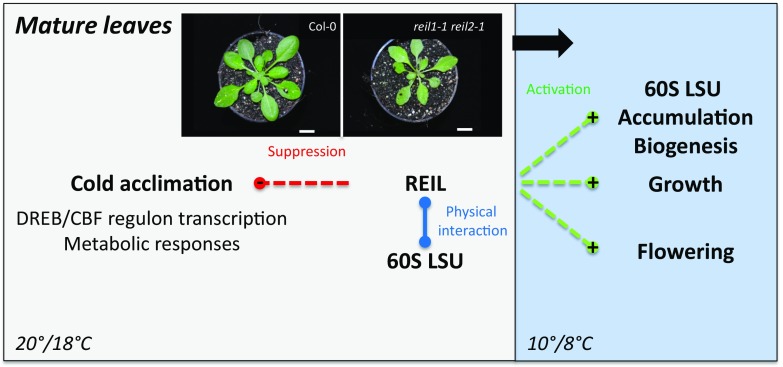
At temperature conditions that were approximated to standard Arabidopsis laboratory cultivations, namely 20°C to 21°C with ∼150 µmol m–2 s–1 photons during the day and 17°C to 18°C during the night (referred to in the following as standard conditions), reil1-1 reil2-1 germinated, grew, and developed almost indistinguishably compared with Col-0 (Fig. 1). Mutant plants at the 10-leaf stage (i.e. stage ∼1.10; Boyes et al., 2001) were slightly but not significantly smaller than Col-0 (Fig. 2, A and B). We confirmed the previous observation (Fig. 1A) that the young developing rosette leaves had a mild pointed-leaves phenotype (Schmidt et al., 2013). This leaf phenotype is a characteristic of many cytosolic ribosomal mutants (Van Lijsebettens et al., 1994; Horiguchi et al., 2012).
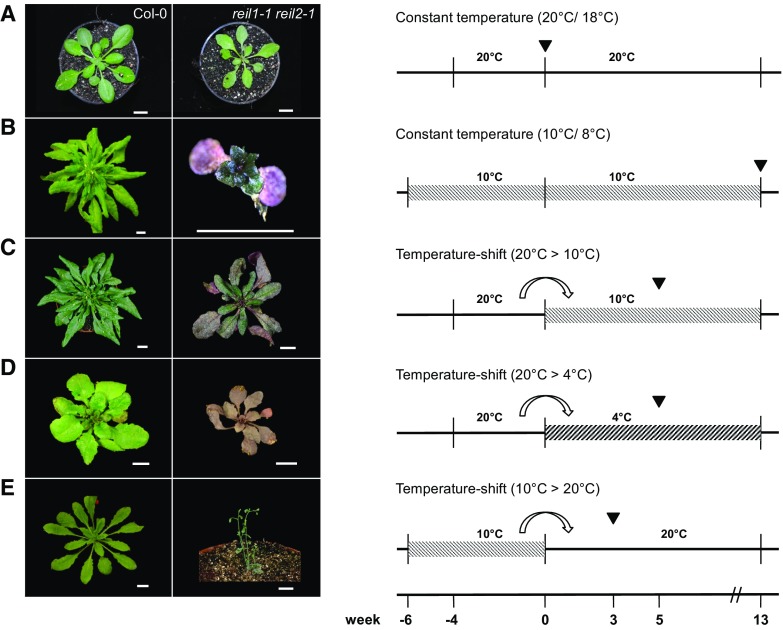
Figure 1.
Developmental phenotypes of the reil1-1 reil2-1 mutant under standard and suboptimal temperature regimes. reil1-1 reil2-1 and Arabidopsis Col-0 wild-type plants were compared under constant temperature and temperature-shift regimes. A, Constant standard temperature conditions at 20°C/18°C (day/night). Mutant and wild-type plants reached vegetative developmental stage ∼1.10 (Boyes et al., 2001) approximately 4 weeks after transfer to soil. Note that the mutant had a mild pointed-leaves phenotype (Van Lijsebettens et al., 1994; Horiguchi et al., 2012). B, Constant low-temperature conditions at 10°C/8°C (day/night). Mutant plants survived at least 13 weeks after germination and transfer to soil but remained extremely dwarfed, with final rosette diameters less than 1 cm and only five to seven visible leaves. C, Temperature shift from the 20°C to the 10°C regime. In contrast to the acclimating wild type, growth and development of the mutant were arrested. Mutant plants survived at least 13 weeks at low temperatures. D, Temperature shift from the 20°C to a 4°C/4°C (day/night) regime. Mutant plants and the wild type were growth arrested. Mutant survival was not tested. E, Inverse temperature shift from the 10°C to the 20°C regime. In contrast to the deacclimated wild type, mutant plants entered a rapid flowering program reminiscent of stress-induced early flowering (Xu et al., 2014). Temperature shifts in C and D were performed at developmental stage ∼1.10 of wild-type and mutant plants at the stages shown in A at week 0. Note that plant age is given by week prior to or post developmental stage ∼1.10. Plants were germinated under sterile conditions (Schmidt et al., 2013) and transferred to soil at stage 1.02-1.03 (i.e. at −6 weeks [10°C] or −4 weeks [20°C]). The inverse temperature shift from 10°C to 20°C (E) was performed when the wild type reached stage ∼1.10 at 10°C. Cocultivated mutant plants had stage 1.02-1.03. Arrowheads within the experimental schemes indicate the time at which representative photographs of n ≥ 10 plants per experiment were taken. Bars = 1 cm.
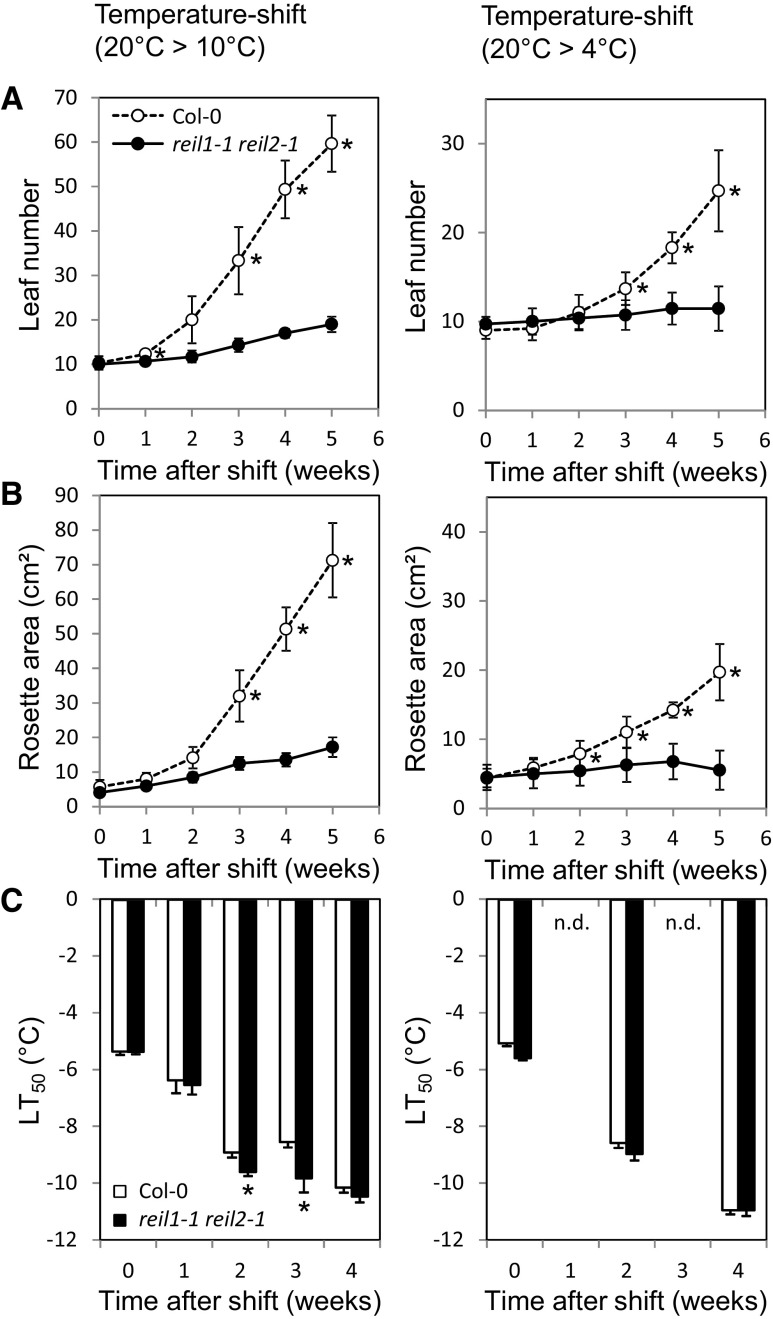
Figure 2.
Morphometric and electrolyte leakage assays of reil1-1 reil2-1 at low temperatures. reil1-1 reil2-1 and Arabidopsis Col-0 wild-type plants were compared after the shift from 20°C to either 10°C (left) or 4°C (right). A, Leaf number. Mutant and wild-type plants differed significantly after 2 weeks at either 10°C or 4°C (*, P < 0.05; means ± sd of n = 3–10 plants). B, Rosette area. Mutant and wild-type plants differed significantly after 2 weeks at either 10°C or 4°C (*, P < 0.05; means ± sd of n = 3–10 plants). C, Electrolyte leakage assay for the quantitative assessment of freezing tolerance (LT50, temperature of 50% normalized electrolyte leakage). The mutant did not show increased electrolyte leakage compared with Col-0 (*, P < 0.05; means ± se of n = 4–8 pools of leaves; n.d., not determined). Experimental designs were according to schemes C and D in Figure 1. Mutant and wild-type plants were shifted at developmental stage ∼1.10 (Boyes et al., 2001). Week-0 plants were cultivated at 20°C and assayed immediately before the temperature shift. Significance was tested using the heteroscedastic Student’s t test.
At constant 10°C temperature (called low temperature in the following), the mutant germinated and reached the cotyledon stage but stopped to produce regular rosette leaves (Schmidt et al., 2013). Populations of reil1-1 reil2-1, nevertheless, survived at least 13 weeks of cultivation on soil with temperatures set to 10°C during the day and 8°C at night (Fig. 1B). The mutation was apparently not lethal but caused a cold-conditional and extremely dwarfed phenotype with few stunted leaves. The cold-arrested mutant was rescued by a shift from low to standard temperature (Fig. 1E), but in contrast to the deacclimating wild type, reil1-1 reil2-1 entered a rapid flowering program.
The temperature-dependent growth arrest that we first observed directly after germination in the cold also occurred at the vegetative developmental stage ∼1.10 (Boyes et al., 2001) after a shift from standard to suboptimal cultivation temperatures, namely to both 10°C/8°C and 4°C/4°C during day/night, respectively (Fig. 1, C and D). The mutant and Col-0 wild type stopped rosette expansion growth and production of new leaves immediately after the shift to 10°C or 4°C (Fig. 2, A and B), but only Col-0 resumed both processes after a lag phase of approximately 1 or 2 weeks, respectively. In contrast, reil1-1 reil2-1 maintained only minimal extension growth and produced few leaves even 13 weeks after the shift to 10°C. Residual extension growth and leaf formation of the mutant stopped completely at 4°C. The mutant but not Col-0 accumulated intense purple-violet pigmentation that is caused by the activation of Arabidopsis flavonol and anthocyanin metabolism (Schulz et al., 2015). Because changes of flavonol and anthocyanin pool sizes in some flavonol biosynthesis pathway mutants cause significant differences in freezing tolerance (Schulz et al., 2016), we applied an electrolyte leakage-based freezing tolerance assay (Rohde et al., 2004; Thalhammer et al., 2014) to assess the maintenance of reil1-1 reil2-1 cellular integrity in nonacclimated and cold-acclimating mature rosette leaves (Fig. 2C). reil1-1 reil2-1 did not differ from Col-0 in the nonacclimated state and, furthermore, did not lose cellular integrity even after weeks of growth arrest at 10°C or 4°C (Fig. 2C). Small differences in the mutant compared with the wild type indicated a marginally faster acquisition of freezing tolerance (Fig. 2C).
N-terminal FP Fusions of REIL1 and REIL2 Localize to the Cytosol and Restore reil1-1 reil2-1 Growth at Low Temperature
To demonstrate complementation of the temperature-dependent reil1-1 reil2-1 phenotypes, we introduced N-terminal fusion proteins of REIL1 and REIL2 with the GFP or the red fluorescent protein (RFP) into the mutant under the control of the constitutive UBIQUITIN10 (UBQ10; AT4G05320) promoter (Supplemental Fig. S1). Attempts to introduce analogous C-terminal fusion proteins failed, in agreement with the known requirement of an unaltered and intact C terminus for yeast Rei1 function (Greber et al., 2016).
Introduction of FP-REIL2 reporter proteins into the reil1-1 reil2-1 mutant restored the expected phenotype of the reil1-1 mutant that is highly similar to the Col-0 wild type (Schmidt et al., 2013). Two independently generated reporter lines, UBQ10::GFP-REIL2 (line 41) and UBQ10::RFP-REIL2 (line 109), that carried either of these expression cassettes in the reil1-1 reil2-1 mutant background and line UBQ10::REIL2 (line 95) that expressed REIL2 without reporter gene fusion in reil1-1 reil2-1 exemplify our observations (Fig. 3). In a previously established in vitro germination assay (Schmidt et al., 2013), UBQ10::GFP-REIL2 (41) and UBQ10::RFP-REIL2 (109) germinated at constant 10°C and generated first rosette leaves. Germination kinetics were similar to those of the reil1-1 mutant that was slightly delayed compared with Col-0 (Fig. 3, A and C). Line UBQ10::REIL2 (95) without an FP fusion also was indistinguishable from reil1-1 (Fig. 3C). After transfer to soil and continued cultivation at 10°C/8°C (day/night), both UBQ10::GFP-REIL2 (41) and UBQ10::RFP-REIL2 (109) developed rosettes that were similar to those of reil1-1 and the wild type (Fig. 3B). Line UBQ10::GFP-REIL2 (41) had a mild pointed-leaves phenotype in the cold (Fig. 3Bb). The timing of reil1-1 reil2-1 and Col-0 germination in vitro at 20°C was indistinguishable and approximately twice as fast as at 10°C (Supplemental Fig. S2). The presence of the expression cassettes UBQ10::GFP-REIL2, UBQ10::RFP-REIL2, and UBQ10::REIL2 in the reil1-1 reil2-1 mutant background did not affect the timing of germination at 20°C.
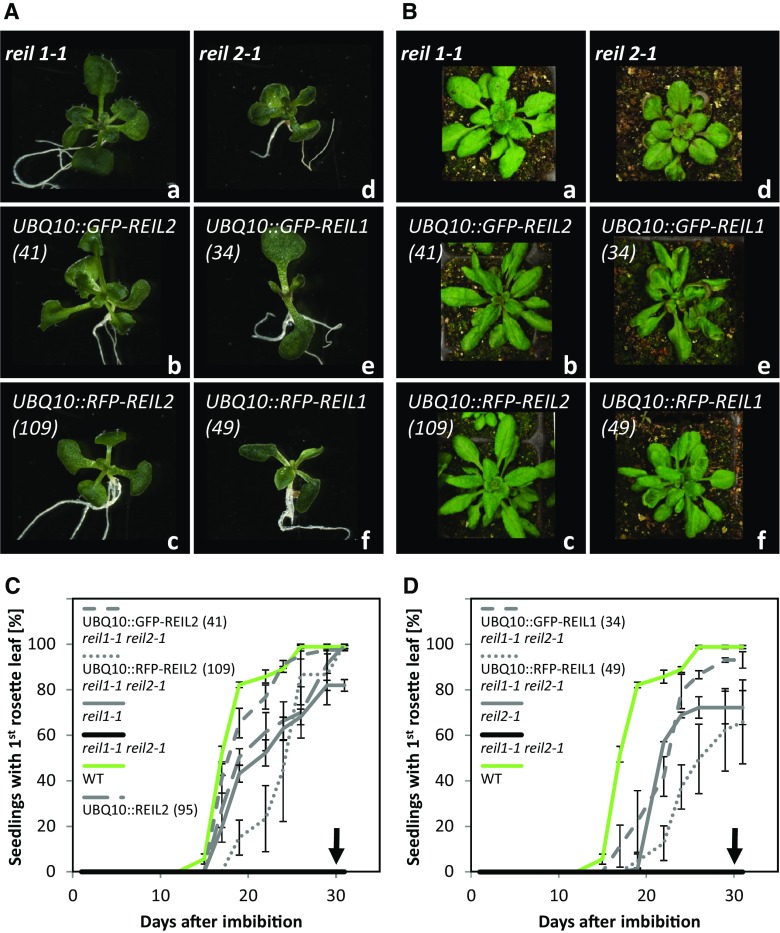
Figure 3.
Constitutive expression of GFP-REIL and RFP-REIL fusion proteins under the control of the UBQ10 promoter in reil1-1 reil2-1 restores single mutant morphology and development. Seedling development and vegetative rosette morphology of representative reil1-1 reil2-1 transformation events using the constructs UBQ10::GFP-REIL2 (b), UBQ10::RFP-REIL2 (c), UBQ10::GFP-REIL1 (e), and UBQ10::RFP-REIL1 (f) were compared with reil1-1 (a) and reil2-1 (d) plants. A, Seedlings after in vitro germination and cultivation at 10°C. Representative photographs of n ≥ 10 in vitro-grown plants per genotype were taken 30 d after imbibition (see C and D). B, Rosette plants after in vitro germination at 10°C and constant-temperature cultivation on soil at 10°C/8°C (day/night) according to cultivation scheme B in Figure 1. Representative photographs of n ≥ 10 plants per genotype were taken 6 to 7 weeks after transfer to soil. Racks of 6-cm-square pots were used. C, Appearance of the first juvenile rosette leaves after the introduction of REIL2 fusion proteins into reil1-1 reil2-1 (means ± sd of n = 4–5 plates per genotype; each plate had ∼80 seeds). D, Appearance of the first juvenile rosette leaves after the introduction of REIL1 fusion proteins into reil1-1 reil2-1 (means ± sd of n = 4–5 plates per genotype; each plate had ∼80 seeds). Leaf appearance was scored using the 10°C in vitro germination and cultivation assay described by Schmidt et al. (2013). The analysis included the reil1-1 reil2-1 mutant that is strongly delayed for the appearance and development of rosette leaves. Arrows indicate the absence of the first rosette leaf (compare with Fig. 1B), Arabidopsis Col-0 wild type (WT), and a nonfusion protein control, namely UBQ10::REIL2, transformed into reil1-1 reil2-1 (see C). The introduction of REIL1 restored reil2-1 morphology, and the introduction of REIL2 restored reil1-1 morphology. Note that reil1-1 morphology is virtually indistinguishable from that of the wild type (Schmidt et al., 2013).
Introduction of FP-REIL1 reporter proteins into the reil1-1 reil2-1 mutant restored the reil2-1 mutant phenotype. This phenotype is highly similar to the allelic reil2-2 mutant phenotype and is characterized by a delayed appearance of the first rosette leaves at 10°C and spoon-shaped leaves (Schmidt et al., 2013). The reporter lines UBQ10::GFP-REIL1 (line 34) and UBQ10::RFP-REIL1 (line 49) that carried the UBQ10::GFP-REIL1 and UBQ10::RFP-REIL1 expression cassettes in the reil1-1 reil2-1 mutant background germinated with the expected delay compared with Col-0 (Fig. 3, A and D) and, as expected, developed spoon-shaped leaves after continued cultivation on soil at 10°C/8°C (day/night; Fig. 3B). The timing of in vitro germination at 20°C was delayed slightly by the presence of the expression cassettes UBQ10::GFP-REIL1 and UBQ10::RFP-REIL1 in the reil1-1 reil2-1 mutant background (Supplemental Fig. S2).
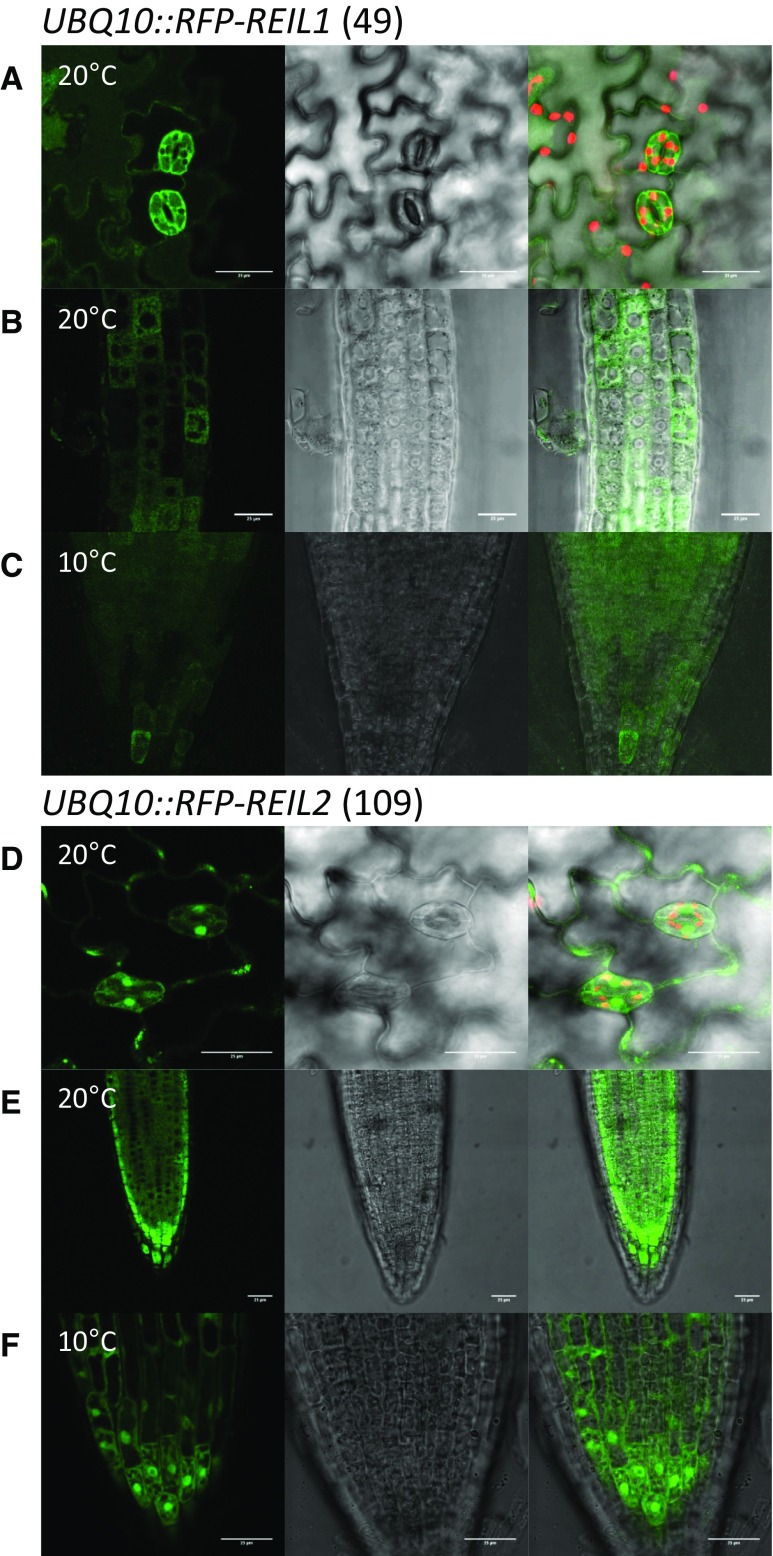
For localization studies, we selected lines UBQ10::RFP-REIL1 (49) and UBQ10::RFP-REIL2 (109) that expressed stable RFP-REIL reporter proteins in the reil1-1 reil2-1 mutant background (Supplemental Fig. S1C). In contrast, preparations of GFP-REIL reporter proteins contained varying amounts of cleaved GFP that could not be completely suppressed by protease inhibitor mixtures (data not shown). RFP-REIL1 was located in the cytosol at 20°C and 10°C (Fig. 4, A–C). The fluorescence signal was homogenous in the cytosol and excluded from vacuoles, nuclei, and the chloroplasts. RFP-REIL2 also located to the cytosol both at 20°C and 10°C (Fig. 4, D–F). In addition, the fluorescence signal also was present in the nuclei of guard cells (Fig. 4D) and at 10°C also in the nuclei of cells of the root tip (Fig. 4F).
Figure 4.
Subcellular fluorescence localization of RFP-REIL fusion proteins. The UBQ10::RFP-REIL1 (49) and UBQ10::RFP-REIL2 (109) transformation events of reil1-1 reil2-1 that restored respective single mutant phenotypes (see Fig. 3) were analyzed using seedlings 7 to 10 d after germination at standard temperature (20°C) and after 1 d or more in the cold (10°C). A, Cotyledon epidermis of UBQ10::RFP-REIL1 (49) after in vitro germination and cultivation at 20°C. B, Root tip meristem to transition zone of UBQ10::RFP-REIL1 (49) after in vitro germination and cultivation at 20°C. C, Root tip of UBQ10::RFP-REIL1 (49) after in vitro germination at 20°C and cold shift. D, Cotyledon epidermis of UBQ10::RFP-REIL2 (109) after in vitro germination and cultivation at 20°C. E, Root tip of UBQ10::RFP-REIL2 (109) after in vitro germination and cultivation at 20°C. F, Root tip of UBQ10::RFP-REIL2 (109) after in vitro germination at 20°C and cold shift. Representative analyses include (left to right) false-color image of RFP fluorescence (green), Nomarski differential interference contrast image, and overlay including false-color image of chlorophyll autofluorescence (red). All images are projection stacks of multiple confocal sections. Note the absence of RFP-REIL1 fluorescent signal from vacuolar, chloroplast, and nuclear lumen (A–C). Bars = 25 µm.
The reil1-1 reil2-1 Mutation Delays Accumulation of the 60S Ribosomal Subunit and of Cytosolic rRNA after a 10°C Cold Shift
The preceding morphometric and complementation studies established the reil1-1 reil2-1 mutant as a viable system for the study of Arabidopsis REIL protein function. Furthermore, we demonstrated that a shift to 10°C was sufficient to turn off extension growth of preexisting mature leaves and the development of new leaves in the reil1-1 reil2-1 mutant without apparent effects on the cell integrity of mature leaf tissue (Fig. 2). Based on these observations, we performed an integrated system analysis of the reil1-1 reil2-1 mutant and investigated in detail the differential response of the mutant and the Col-0 wild type before and after the cold shift. Throughout the following studies, we applied the 20°C-to-10°C cold acclimation experimental design C (Fig. 1C) and focused our experiments on mature leaf tissue. This choice facilitated combined differential analyses of relative transcript and metabolite levels and, importantly, provided sufficient leaf material for ribosome analyses.
Two other phenotypic hallmarks of Rei1p deficiency are known in yeast besides cold sensitivity of growth. The deletion of yeast Rei1p causes a reduction of the relative amount of the cytosolic 60S LSU under cold conditions (Hung and Johnson, 2006). In addition, so-called polysome half-mers appear in the cold (i.e. mRNAs with one or more 80S ribosomes and an additional 40S subunit attached; Parnell and Bass, 2009; Greber et al., 2016). To investigate these phenotypes, we started our system analysis with a comparison of reil1-1 reil2-1 and Col-0 ribosome composition.
Microfluidic electrophoresis of total RNA preparations from mature leaf tissue enabled the relative quantitation of cytosolic and chloroplast rRNAs, specifically cytosolic 18S and 25S rRNAs compared with plastid 16S and 23S rRNAs (Supplemental Fig. S3). 5S and 5.8S rRNAs were detectable but not clearly separated. We analyzed 23S rRNA by summation of the two previously described postmaturation fragments, 23S′ and 23S′′, that result from hidden breaks of the mature 23S rRNA (Nishimura et al., 2010; Tiller et al., 2012). Fragments 23S′ and 23S′′ were abundant in all analyzed samples. A third small 23S′′′ hidden break product and the potential full-size rRNA precursor (Tiller et al., 2012) were detectable only at very low intensity and disregarded in this study.
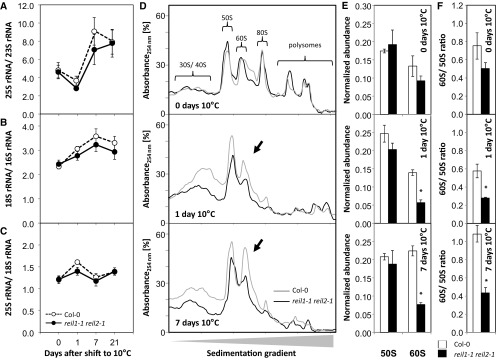
Ratios of cytosolic compared with chloroplast rRNAs changed during Col-0 cold acclimation (Fig. 5, A–C) after the shift to the 10°C cultivation regime (Fig. 1C). The 25S/23S rRNA and the 18S/16S rRNA ratios both increased 1 week after the cold shift (Fig. 5, A and B). This observation indicated enhanced de novo synthesis of cytosolic rRNA relative to chloroplast rRNA. The ratio of the cytosolic large subunit 25S to the cytosolic small subunit 18S rRNA remained unchanged (Fig. 5C). The accumulation of cytosolic rRNA relative to chloroplast rRNA preceded the resumption of growth and development of Col-0 (Fig. 2, A and B). We concluded that cold acclimation causes an altered balance between cytosolic and chloroplast ribosome biogenesis and, therefore, represents a condition that is suitable for the analysis of potential cytosolic ribosome biogenesis factors, such as the Arabidopsis REIL proteins (Schmidt et al., 2013; Weis et al., 2015). Cytosolic rRNA accumulation in the reil1-1 reil2-1 mutant after exposure to cold was not fully abolished but delayed compared with cold-acclimating Col-0 (Fig. 5, A and B). Therefore, we hypothesized that REIL proteins have a role in enhancing the kinetics of cytosolic ribosome biogenesis in the cold.
Figure 5.
Relative abundance of rRNAs within total RNA preparations and of 60S and 50S large ribosomal subunits from total ribosome preparations before and after the shift to 10°C. Complete reil1-1 reil2-1 rosettes were compared with wild-type (Col-0) rosettes in the nonacclimated state (i.e. at 0 d) and at 1, 7, and 21 d after transfer to the cold according to cultivation scheme C in Figure 1. A, Ratio of the cytosolic 25S large subunit rRNA relative to chloroplast 23S rRNA (P = 0.001). B, Ratio of the cytosolic small subunit 18S rRNA relative to chloroplast 16S rRNA (P = 0.003). C, Ratio of the cytosolic 25S large subunit rRNA and cytosolic 18S small subunit rRNA. For A to C, data are means ± se (n = 3–4 preparations from independent pools of mature rosette leaves). D, Representative sedimentation profiles of total ribosome preparations from ∼100 mg fresh weight of rosettes sampled at days 0, 1, and 7 of acclimation to 10°C. The sedimentation analysis was optimized for the separation of the 50S to 60S fractions. Indicated fractions were monitored by blank gradient subtracted absorbance (A254). Fraction identity was verified by rRNA analysis (Supplemental Fig. S3). E, Analysis of ribosome sedimentation profiles shown in D by calculating the normalized A254 abundance of the 60S and 50S fractions relative to the sum of all observed fractions (means ± se of n = 3 preparations from independent pools of mature rosette leaves). F, Ratio of the 60S fraction relative to the 50S fraction calculated from the data sets of E and F (means ± se of n = 3 preparations from independent pools of mature rosette leaves). The experimental design was according to scheme C in Figure 1. Mutant and wild-type plants were shifted at developmental stage ∼1.10. Nonacclimated rosettes were cultivated at 20°C and assayed immediately before the temperature shift. Peak areas of rRNA were determined from total RNA extracts by microfluidic electrophoresis. 23S rRNA was determined by the sum of two naturally occurring postmaturation cleavage products. For A to C, significance (P) of the time effect was tested by two-way ANOVA. For E and F, asterisks indicate significant changes of the mutant compared with the wild type (P < 0.05, heteroscedastic Student’s t test).
To further validate this hypothesis, we analyzed the ribosome composition of mutant and Col-0 leaves by differential ultracentrifugation (Fig. 5, D–F). Conventional ultracentrifugation methods of plant ribosome/polysome preparations (Mustroph et al., 2009) were initially adapted from technologies developed for organisms, such as yeast, that contain only small and negligible amounts of prokaryotic-type mitochondrial ribosomes (Parnell and Bass, 2009; Greber et al., 2016). Plant leaves, however, contain large amounts of chloroplast ribosomes that cannot be neglected (Tiller et al., 2012). The chloroplast ribosomes were not separated from cytosolic ribosomes by current methods (Mustroph et al., 2009). For our study, we developed a differential ultracentrifugation protocol with a sedimentation gradient that was optimized for the separation of the cytosolic 60S and the chloroplast 50S large subunits (Supplemental Fig. S3). The modified sedimentation gradient included the 30S and 40S small subunits, the 50S and 60S large subunits, the 70S and 80S monosomes, and, in addition, a mixed chloroplast and cytosolic polysome fraction (Supplemental Fig. S3, A and B). Detailed microfluidic electrophoretic analysis of RNA preparations from respective fractions of the sedimentation gradient revealed that the cytosolic 60S fraction separated from the chloroplast 50S fraction and from the 80S monosomes (Supplemental Fig. S3, C and D). The 70S monosomes were present at low abundance between the 60S and 80S fractions. The 30S and 40S subunit fractions were clearly separated from the 50S subunits but overlapped and formed a 30S-enriched fraction followed by a 40S-enriched fraction. Cytoplasmic and chloroplast polysome fractions also overlapped and did not allow unambiguous diagnosis of the presence of cold-induced cytosolic polysome half-mers (Parnell and Bass, 2009; Greber et al., 2016).
In the nonacclimated state, reil1-1 reil2-1 ribosome preparations were indistinguishable from those of Col-0 (Fig. 5D). One day after the cold shift, cytosolic and chloroplast ribosome subunits increased relative to the strongly reduced polysome fractions. Both the mutant and Col-0 had increased amounts of ribosome subunits after the cold shift, but reil1-1 reil2-1 clearly accumulated fewer ribosome subunits than Col-0. One week after the cold shift, Col-0 increased the 60S fraction relative to the 50S fraction, whereas reil1-1 reil2-1 lagged behind and still had significantly reduced amounts of 60S LSUs. This observation was confirmed by analyses of relative peak abundances of the ribosome fractions after blank gradient subtraction of absorbance (A254) and normalization to total absorbance of all observed ribosome fractions (Fig. 5E). A ratiometric analysis of normalized 60S to 50S abundance (Fig. 5F) corresponded to our 25S/23S rRNA ratio data (Fig. 5A). In Col-0, the amount of the 60S subunit fraction increased relative to the 50S subunits during cold acclimation. In contrast, the accumulation of 60S subunits was delayed significantly in the mutant.
The REIL1 Protein Is Associated with 60S, 80S, and Polysome Fractions
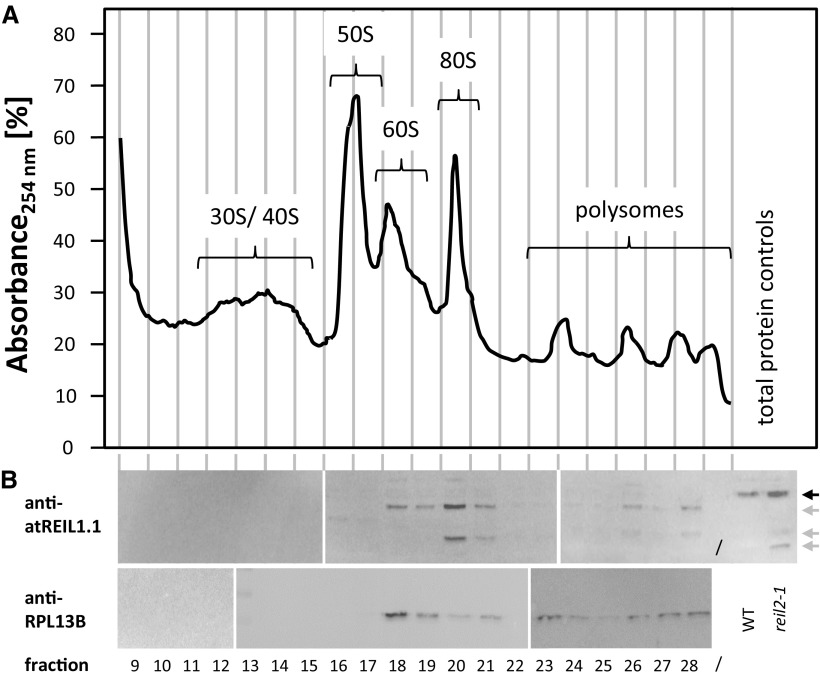
Peptide antibodies, anti-atREIL1.1 and anti-atREIL2.1, were directed against variable regions at the C terminus of REIL1 and REIL2. These peptides detected the proteins REIL1, 45,998 D with 404 amino acids, and REIL2, 44,987 D with 395 amino acids (https://apps.araport.org/thalemine/begin.do), in total protein preparations of 20°C/18°C (day/night) cultivated Col-0 at an electrophoretic mobility of approximately 35 kD. The proteins were not detectable in 20°C/18°C (day/night) cultivated reil1-1 reil2-1 (Supplemental Fig. S1). Western blots of Col-0 ribosome fractions from the modified Suc density ultracentrifugation gradients using anti-atREIL1.1 and anti-atREIL2.1 revealed the presence of REIL1 in the nontranslating 60S fraction but also in the 80S and polysome fractions (Fig. 6). The anti-atREIL1.1 antibody detected only very weak signals of the 35-kD protein. Instead, up to three cleavage products of REIL1 were present after preparation of the ribosome proteins from the ultracentrifugation fractions. These fragments were present at low abundance in total protein preparations of the reil2-1 mutant (Fig. 6; Supplemental Fig. S1). The anti-atREIL2.1 western analysis showed very weak signals of a 35-kD protein and did not reveal REIL cleavage products (data not shown).
Figure 6.
Western-blot analysis of Col-0 ribosome fractions. Ribosome protein fractions from the Col-0 wild type were compared with total protein preparations from Col-0 (WT) and reil2.1. All preparations were from rosette plants of stage ∼1.10 that were cultivated at 20°/18°C (day/night). A, Absorbance profile of the Suc density sedimentation gradient analysis at wavelength λ = 254 (A254) after blank gradient subtraction. Vertical lines indicate the approximate positions of the collected protein fractions. B, Western-blot analyses of the indicated fractions using anti-atREIL1.1 (top) and anti-RPL13B (bottom) antibodies. Note that anti-atREIL1.1 was directed against a variable region at the REIL1 C terminus. This antibody detects REIL1 in total protein extracts (black arrow) and cleavage products (gray arrows) in protein preparations after Suc density gradient centrifugation. Cleavage products also are detectable at low abundance in total protein from the reil2.1 mutant. The western-blot analysis was performed by three parallel-processed blots of fractions from the sedimentation gradient shown in A. The anti-RPL13B analyses were of an independent sedimentation gradient. Splice sites of blots are indicated by white bars.
Metabolic Systems Analysis of reil1-1 reil2-1 Reveals a Metabolic Phenotype in the Nonacclimated State and Deviations from Col-0 after Long-Term Acclimation to 10°C
To gain further insights into the consequences of the reil1-1 reil2-1 mutation and the absence of REIL proteins from the Arabidopsis system, we profiled the metabolome changes of the nonacclimated and the cold-acclimating mutant with high temporal resolution on day 1 after the cold shift and at longer intervals during the following 4 weeks in the cold (Supplemental Table S1). Phenotyping of the primary metabolome revealed 50 relevant either genotype-differential or cold-responsive metabolites (Fig. 7A) that were retrieved from the nontargeted profiles by ANOVA. Eighteen of these metabolites were 10°C shift responsive in Col-0 (Supplemental Table S1). Sixteen of these metabolites were reported previously to also increase following a 4°C cold shock (Kaplan et al., 2004, 2007; Guy et al., 2008). Only myoinositol, which decreased after cold shock in the previous 4°C analyses, increased significantly in our 10°C shift experiments (Supplemental Table S1).
Figure 7.
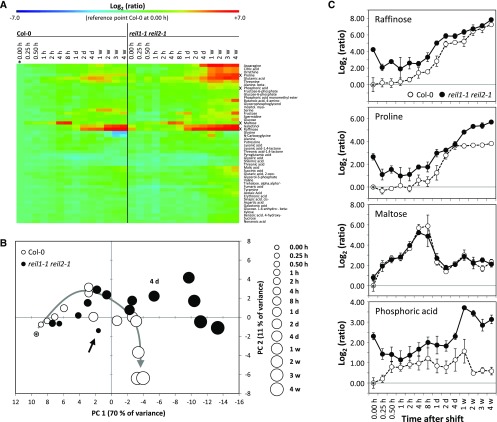
Relative changes of leaf primary metabolites in the nonacclimated and the 10°C cold-acclimating reil1-1 reil2-1 mutant compared with the Col-0 wild type. A, Heat map of relative changes compared with nonacclimated Col-0 (*). Mean ratios were log2 transformed and color coded according to the scale at top (n = 6). Metabolites presented in C are indicated (x). Metabolites were arranged by hierarchical clustering using Euclidian distance and complete linkage. B, Principal component analysis of the data set shown in A. Col-0 in the nonacclimated state is indicated by an asterisk within the 0.00-h circle to the left. The gray arrow indicates the progressing metabolic changes of Col-0 in the course of cold acclimation. The black arrow indicates nonacclimated reil1-1 reil2-1. Time after the cold shift is coded by circle size. The 4-d annotation indicates the time point of reil1-1 reil2-1 divergence from the wild type. C, Comparative time course of selected metabolite pools normalized to nonacclimated Col-0 indicated by an asterisk within the 0.00-h circle to the left (means ± se; n = 6). Plants were precultivated at 20°C and shifted to 10°C conditions at developmental stage ∼1.10 according to scheme C in Figure 1. Samples were harvested immediately before (0.00 h) and following the cold shift. Cold shift and sampling time points at full days (d) or weeks (w), except samplings at 0.25 to 8 h, were at 6 h (±5 min) after dawn of a 14-h/10-h day/night cycle. Two independent experiments with three replicate samples each per time point were performed. Samples were pools of mature leaves from at least two plants. Metabolism was profiled by multitargeted gas chromatography-mass spectrometry (GC-MS)-based technology. Metabolites with significant changes compared with the wild type, changes in the course of cold acclimation, and significant interactions of both effects were selected by two-way ANOVA (P < 0.001; Supplemental Table S1).
The responses of the reil1-1 reil2-1 mutant and Col-0 to the 10°C shift were highly similar up to 2 d after the cold shift (Fig. 7). However, the nonacclimated reil1-1 reil2-1 mutant already had a hidden metabolic phenotype compared with nonacclimated Col-0 (Fig. 7B, arrow). Many of the changed metabolite levels in the nonacclimated mutant were similar to the levels observed in cold-acclimating Col-0 (Fig. 7A). This observation included the well-characterized environmental stress metabolites of Arabidopsis, such as Pro and raffinose (Fig. 7C), but also other carbohydrates and amino acids that typically accumulate upon cold shock (Guy et al., 2008). Two days after the cold shift, mutant metabolism began to differ significantly from that of Col-0. Rather than deviating from the general trend of accumulating carbohydrates and amino acids, most cold-responsive metabolites accumulated further and beyond Col-0 levels in the mutant (Fig. 7, A and B; Supplemental Table S1). No metabolite pool had constitutively lower levels in the mutant, whereas phosphate, β-Ala, Pro, galactinol, and raffinose remained up-regulated throughout the complete time course (Fig. 7C; Supplemental Table S1).
To further characterize the metabolic phenotype of reil1-1 reil2-1 at standard cultivation temperature, we compared the nonacclimated mutant with the time course of cold-acclimating Col-0 (Supplemental Fig. S4). The metabolic profiles of nonacclimated reil1-1 reil2-1 were similar to those of Col-0, specifically the first 2 d after the cold shift. This similarity was revealed by both hierarchical clustering and principal component analyses (Supplemental Fig. S4A). Furthermore, the relative levels of metabolites in the nonacclimated reil1-1 reil2-1 correlated highly with the respective levels of Col-0 during the first 2 d after the cold shift and maintained correlation beyond these time points (Supplemental Fig. S4B). We hypothesized that the nonacclimated mutant may be temperature sensitized and was triggering premature metabolic cold responses at standard cultivation temperature.
Previous studies indicated that many cold-responsive primary metabolite pools were not highly cold specific but also may be modulated by heat or other stresses (Kaplan et al., 2004, 2007; Supplemental Table S1). Therefore, our temperature sensitization hypothesis required additional validation by transcriptome profiling.
reil1-1 reil2-1 Activates Transcriptional Cold Acclimation Responses at Standard Temperature
To validate our previous observations and to extend our insight into global systems responses of the mutant, we performed transcriptome profiles of mature leaves from the mutant and the Col-0 wild type at informative time points, such as the nonacclimated state (0 d of acclimation) and after 1 d, 1 week, and 3 weeks of 10°C cold acclimation (Supplemental Table S2). These time points were selected according to the preceding growth analyses (Fig. 2) and the better time-resolved metabolome analyses (Fig. 7; Supplemental Fig. S4). In the nonacclimated state, we aimed to further reveal the details of a hidden mutant phenotype. At 1 d and 1 week, we analyzed the short-term and long-term differential responses of the mutant that are not yet associated with differential growth. At 3 weeks after the cold shift, we investigated the long-term acclimation defect of the mutant, namely the effects that were associated with the inability of the mutant to resume growth.
The nonacclimated reil1-1 reil2-1 mutant differentially expressed 428 genes compared with nonacclimated Col-0 (Supplemental Table S3). These differentially expressed genes (DEGs) encoded 287 transcripts with increased abundance and 141 with decreased abundance (Table I), 42.2% (121) and 20.6% (29). In sum, 150 of these transcripts (Table I) were shared with the respectively changed 10°C cold-responsive transcripts of Col-0 at 1 d, 1 week, or 3 weeks after the cold shift, which also were calculated relative to the nonacclimated Col-0 wild type. Thirty-one of the 150 shared DEGs had constitutively increased transcript abundance in Col-0 even throughout the complete cold acclimation time course (Table I; Supplemental Table S3). In addition, relative expression levels of these shared 150 DEGs were highly correlated between the nonacclimated mutant and the respective transcript levels in Col-0 1 d, 1 week, or 3 weeks after acclimation to 10°C (Supplemental Fig. S5). However, the shared DEGs represented only a small fraction of the 10°C responsive genes of the Col-0 wild type (Table I).
Table I. Analysis of DEGs of nonacclimated reil1-1 reil2-1 relative to the nonacclimated Col-0 wild type (0 d) that also were cold-responsive transcripts of the Col-0 wild type at 1 day, 1 week, or 3 weeks after the shift to 10°C relative to nonacclimated Col-0.
Nonacclimated reil1-1 reil2-1 and Col-0 (0 d) were compared by heteroscedastic Student’s t testing at P < 0.01 (n = 5–6 independent pools of mature rosette leaves from two to three plants). 10°C cold-responsive transcripts were selected at P < 0.01 (n = 3 independent pools of mature rosette leaves from two to three plants). The set of 428 DEGs and the subset of 150 genes that also were 10°C shift responsive in Col-0 are reported in Supplemental Table S3. The column 1 d, 1 w, or 3 w lists the sum of genes that were differentially expressed during at least one time point; the column 1 d, 1 w, and 3 w lists the number of genes that were differentially expressed at all time points.
| Plant and Parameter | 0 d | 1 d | 1 Week | 3 Weeks | 1 d, 1 w, or 3 w | 1 d, 1 w, and 3 w |
|---|---|---|---|---|---|---|
| Col-0 | ||||||
| All | – | 3,301 | 2,711 | 3,275 | 6,537 | 594 |
| Up | – | 2,726 | 2,323 | 2,682 | 5,310 | 538 |
| Down | – | 575 | 388 | 593 | 1,227 | 56 |
| Number of DEGs that also were 10°C shift responsive in Col-0 | ||||||
| reil1-1 reil2-1 | ||||||
| Count | ||||||
| All | 428 | 80 | 77 | 100 | 150 | 31 |
| Up | 287 | 66 | 67 | 86 | 121 | 31 |
| Down | 141 | 14 | 10 | 14 | 29 | 0 |
| Percentage relative to day 0 | ||||||
| All | 100.0 | 18.7 | 18.0 | 23.4 | 35.0 | 7.2 |
| Up | 100.0 | 23.0 | 23.3 | 30.0 | 42.2 | 10.8 |
| Down | 100.0 | 9.9 | 7.1 | 9.9 | 20.6 | 0.0 |
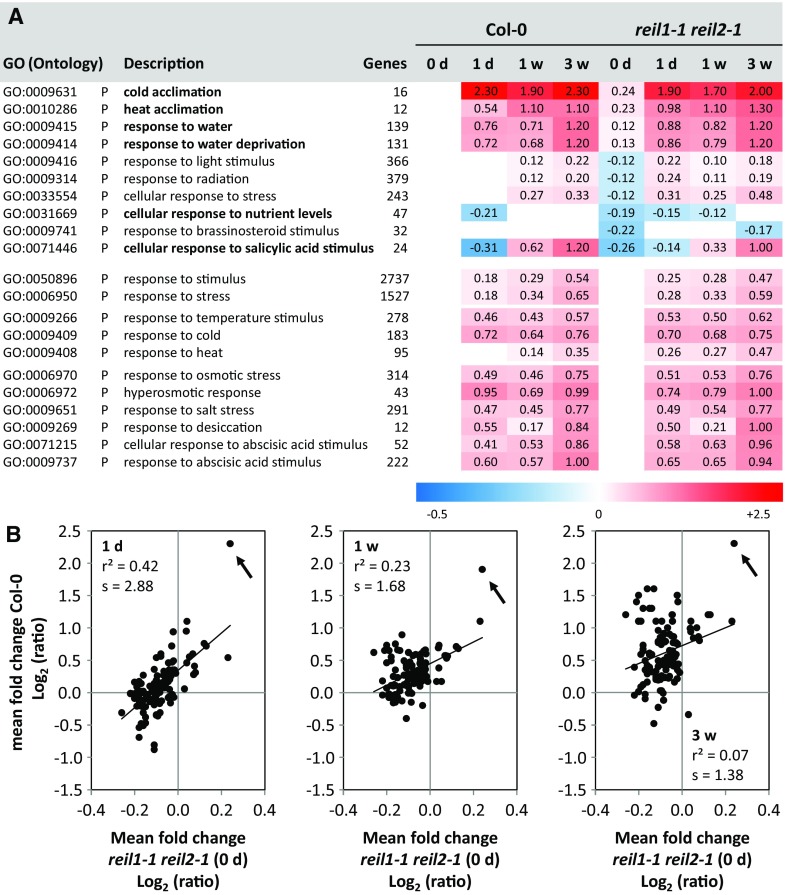
For this reason, we used functional enrichment analysis of differential gene expression by parametric analysis of gene set enrichment (Du et al., 2010) to test the full nonfiltered differential gene expression data set of nonacclimated reil1-1 reil2-1 relative to the nonacclimated Col-0 wild type. The method assessed the enrichment of up- or down-regulation among the gene sets of 1,940 Gene Ontology (GO) terms. These GO terms contained 135 that were linked to acclimation or responses to stimuli and stresses (Fig. 8A; Supplemental Table S4). Next to the differential gene expression enrichment analysis of nonacclimated reil1-1 reil2-1, we performed in parallel equivalent parametric analysis of gene set enrichment of the full nonfiltered differential gene expression data sets of 1-d, 1-week, or 3-week cold-acclimating Col-0, which also were calculated relative to nonacclimated Col-0. These analyses allowed comparison of the mean log2-FCs of the gene sets defined by the GO terms between the nonacclimated mutant and the three time points of 10°C acclimating Col-0 wild type. Pearson’s correlation analysis of the mean log2-FCs across the 135 stress-related GO terms revealed linear correlation between the mean log2-FCs of nonacclimated reil1-1 reil2-1 and Col-0 at 1 d after the shift to 10°C. The correlation attenuated over time, but the mean log2-FC of one GO term remained highly associated (Fig. 8B). To identify this and other GO terms that were relevant for insights into the physiological state of the nonacclimated mutant, we limited the analysis to GO terms with mean log2-FC > +0.1 or < −0.1. Furthermore, we ranked the resulting 59 from the initial 135 GO terms according to their significance of enrichment using z-scores (Du et al., 2010) and considered the GO terms with the top 10 z-score values to be most informative. All calculated mean log2-FCs, z-scores, and respective P values are reported in Supplemental Table S4.
Figure 8.
Functional enrichment analysis of stress- and stimuli-related differential gene expression in reil1-1 reil2-1 at standard cultivation temperature compared with cold-acclimating Col-0 and reil1-1 reil2-1 at 1 d (1d), 1 week (1w), or 3 weeks (3w) after the shift to 10°C. Differential gene expression was determined relative to nonacclimated Col-0 at standard cultivation temperature. Z-scores, P values, and mean log2 fold changes (log2-FCs) of genes belonging to 1,940 GO terms were calculated (Du et al., 2010; Supplemental Table S4). A, Heat map of mean log2-FC of the top 10 significantly enriched 135 GO terms that represented responses to stimuli or abiotic and biotic stresses. Ranking was according to top positive or negative z-score values with mean log2-FC > +0.1 or < −0.1. GO terms are listed with ontology, description, and number of constituting genes. Boldface in the GO description indicates shared enrichment of the nonacclimated mutant (0 d) with the acclimating Col-0 at 1 d, 1 week, or 3 weeks. Two general stimuli- and stress-response GOs as well as the temperature, cold, heat response, water, drought, and osmotic stress-related GOs were added for comparison. All top-scoring GO terms were biological processes (P). B, Mean log2-FC of GO terms from the nonacclimated mutant (0 d) and acclimating Col-0 at 1 d, 1 week, and 3 weeks (from left to right). All 135 acclimation- and response-related GO terms were included. The inset data show the Pearson’s correlation coefficient (r2) and the slope of a linear regression (s). The arrows indicate GO:0009631, cold acclimation.
The term with the highest mean log2-FC was GO:0009631, cold acclimation, and identical with the persistently correlated term of our correlation analysis (Fig. 8B). This GO term was followed by GO:0010286, heat acclimation, and two terms, GO:0009414 and GO:0009415, with highly overlapping gene sets that represented responses to water limitation (Fig. 8A). The triggering of temperature acclimation responses in the nonacclimated mutant was highly specific, because GOs that represented the general responses to temperature shifts, stresses, or stimuli were not activated prematurely. In the case of GO:0033554, cellular response to stress, the mean log2-FC was even significantly lower in the nonacclimated mutant relative to the wild type (Fig. 8A). All the mentioned general GO terms, including GO:0010286, heat acclimation, and to a minor extent also GO:0009408, response to heat, were part of the Col-0 wild-type response to the 10°C cold shift. Enrichment of drought-, salt-, osmotic stress-, and abscisic acid-related GO terms also was part of the Col-0 10°C response, but mean log2-FC did not exceed +0.1 in nonacclimated reil1-1 reil2-1 (Fig. 8A). The terms with the most decreased transcript abundance, which were shared between the nonacclimated mutant and 1-d cold-acclimating Col-0, were GO:0071446, cellular response to salicylic acid stimulus, and GO:0031669, cellular response to nutrient levels. The decreased responses to light or radiation and to brassinosteroid stimuli in the nonacclimated mutant were not part of the Col-0 cold response.
For further enrichment analysis, we applied the same selection criteria, mean log2-FC > +0.1 or < −0.1 and selection of the most significant enrichments according to z-score values, to all included 1,940 GO terms (Supplemental Fig. S6). The previously identified water limitation GOs and, in addition, two flavonoid pathway-related GO terms, GO:0009812 and GO:000983, ranked highest among the GO terms with increased transcript abundance. This observation was again shared between the nonacclimated mutant and acclimating Col-0. In contrast, the top-scoring down-regulated terms were mostly specific to the nonacclimated mutant and related to morphogenesis/development, the structural component of the chloroplast, nitrogen compound metabolism, and the cell wall (Supplemental Fig. S6).
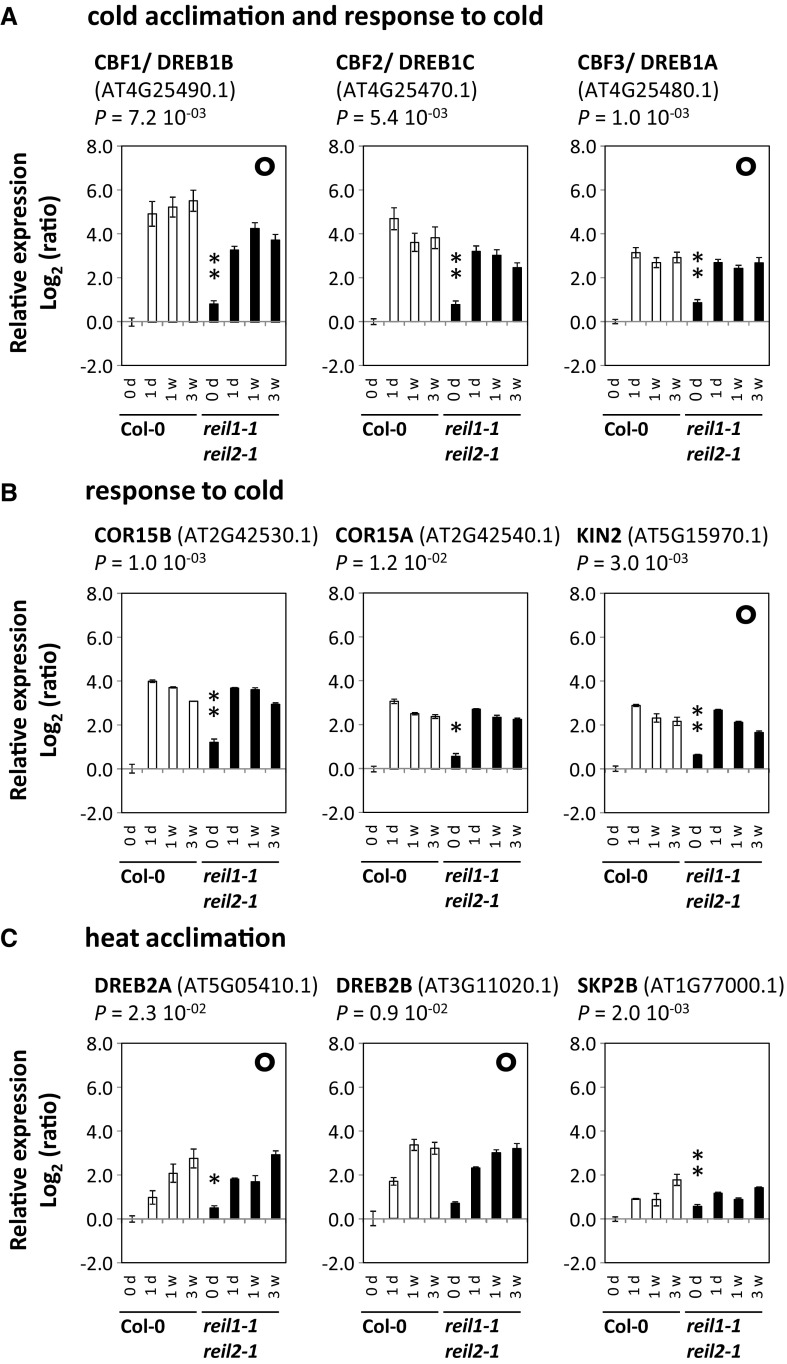
We subsequently analyzed the set of genes that were both changed in the nonacclimated mutant (Supplemental Table S3) and annotated by the GO terms GO:0009631, cold acclimation, or GO:0009409, response to cold. We found eight DEGs that belonged to these GO terms. The well-known regulators of cold acclimation (Thomashow, 1999; Shinozaki et al., 2003), C-REPEAT/DRE BINDING FACTOR1/DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN1B (CBF1/DREB1B, AT4G25490.1), CBF2/DREB1C (AT4G25470.1), and CBF3/DREB1A (AT4G25480.1), were part of this set (Fig. 9A). Together with COLD-REGULATED15A (COR15A, AT2G42540.1), COR15B (AT2G42530.1), and COLD-INDUCED2 (KIN2, AT5G15970.1; Fig. 9B), the three regulators also scored as high as robust cold-responsive genes compared with a previous meta-analysis of transcriptional responses to 4°C cold stress (Hannah et al., 2005).
Figure 9.
Differential expression of genes that were up-regulated in nonacclimated reil1-1 reil2-1 and changed significantly in 10°C cold-acclimating Col-0. A, DEGs (P < 0.01) in nonacclimated reil1-1 reil2-1 that belonged to GO term GO:0009631, cold acclimation. Note that these three genes also were part of GO:0009409, response to cold. B, Additional DEGs (P < 0.01 or P < 0.05) that belonged to GO term GO:0009409, response to cold. C, Selected genes that belonged to GO:0010286, heat acclimation. Note that DREB2A and DREB2B were increased slightly in the nonacclimated mutant and also part of the long-term Col-0 response to a 10°C shift (see Fig. 8). SKP2B was the only gene of GO:0010286, heat acclimation, that was significantly up-regulated at P < 0.01 in nonacclimated reil1-1 reil2-1. Relative expression values are log2-FCs (ratios) compared with nonacclimated Col-0 (means ± se). Headers show Arabidopsis gene names and gene codes in parentheses. Significance values of heteroscedastic Student’s t tests comparing nonacclimated mutant and nonacclimated Col-0 are indicated (*, 0.01 < P ≤ 0.05 and **, P ≤ 0.01). Circles in the top right corner of some graphs indicate that the respective genes also were part of the water limitation GO terms, GO:0009414 and GO:0009415.
We extended the same intersection analysis to the heat GO terms and found only one significant DEG in the nonacclimated mutant that belonged to GO:0010286, heat acclimation, namely S-PHASE KINASE-ASSOCIATED PROTEIN2B (SKP2B, AT1G77000.1). This protein is part of an E3 ubiquitin-ligase complex that degrades ARABIDOPSIS THALIANA HOMOLOG OF E2F C (AT1G47870.1), a likely regulator of cell division transition from skotomorphogenesis to photomorphogenesis (del Pozo et al., 2002). No significant DEG was found among the more general gene set of GO:0009408, response to heat. The enrichment result of the heat-related GO terms in nonacclimated reil1-1 reil2-1 was explained by slight increases of the mRNA abundance of many genes from the GO term, but none of the genes was changed significantly in expression. We subsequently investigated the expression of genes that encode DREB2 proteins in more detail (Fig. 9C). DREB2 proteins are transcription factors that are part of the DREB/CBF regulon and share the regulation of a subset of genes with DREB1/CBF factors (Yamaguchi-Shinozaki and Shinozaki, 2009; Lata and Prasad, 2011). Through this interaction, the DREB/CBF regulon integrates the control of gene expression following drought and cold stress. In contrast to DREB1/CBF gene expression that is rapidly activated upon cold stress, the expression of DREB2 genes is activated primarily upon heat and osmotic stresses. For this reason, DREB2 genes are assigned to the heat-, water limitation-, and osmotic stress-related GO terms. mRNA levels of both DREB2A and DREB2B were only slightly increased in nonacclimated reil1-1 reil2-1 (Fig. 9C) but were increased significantly as part of the long-term (1 week or longer) Col-0 response to 10°C (Figs. 8 and 9).
Together with the activated expression of DREB2A and DREB2B, gene expression of the water limitation response terms, GO:0009414 and GO:0009415, was activated during Col-0 acclimation to 10°C (Fig. 8). Again, the enrichment result was based predominantly on a small overall increase of expression of many genes. The gene sets of the water limitation GO terms overlapped with the sets defined by heat stress GOs, such as DREB2A and DREAB2B (Fig. 9C), but also with the cold stress GO terms (Fig. 9, A and B). In detail, 28 of 183 cold response genes also belonged to the water limitation GOs, including CBF1/DREB1B (AT4G25490.1), CBF3/DREB1A (AT4G25480.1), and KIN2 (AT5G15970.1). Next to the overlapping genes, mRNAs of two water limitation-specific genes were changed significantly in the nonacclimated mutant, namely LIPID TRANSFER PROTEIN4 (AT5G59310.1) and HISTONE1-3 (HIS1-3, AT2G18050.1). HIS1-3 encodes a drought-inducible linker histone that is expressed preferentially in younger leaf tissue and root tips (Ascenzi and Gantt, 1999) and has a cell-to-cell mobile mRNA (Thieme et al., 2015).
reil1-1 reil2-1 Delays and Subsequently Hyperactivates Cold-Triggered Expression of Translation- and Ribosome-Related Genes
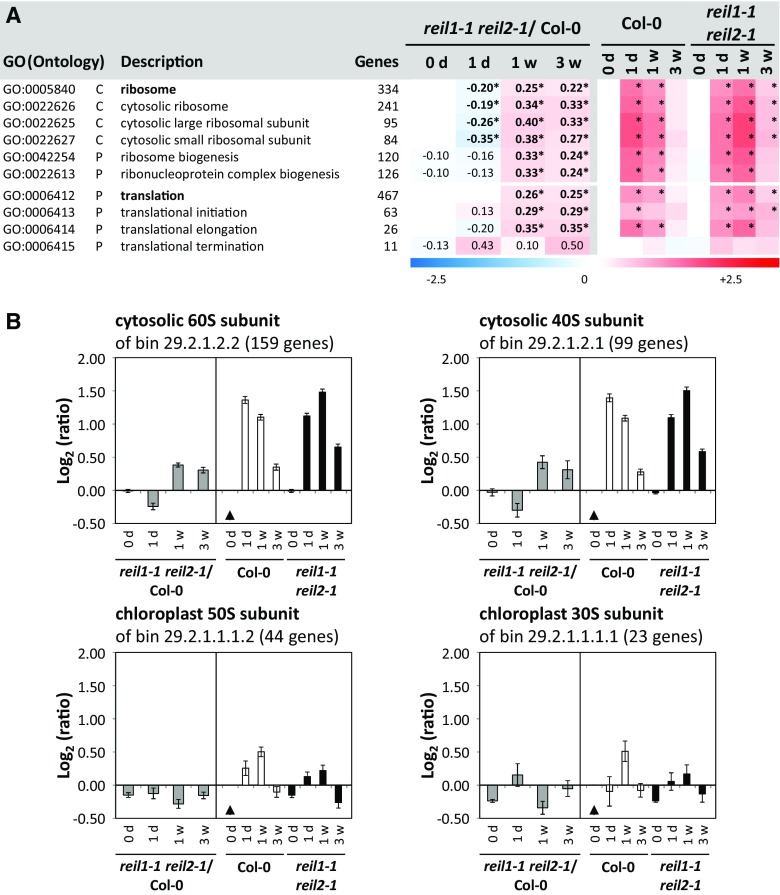
Because the absence of REIL proteins delayed accumulation of the eukaryotic 60S ribosomal subunit upon the shift to low temperature, we searched for transcriptional responses that were likely associated with this defect or that may partially compensate for REIL deficiency. For this purpose, we focused on ribosome- and translation-related genes, such as the members of the global GO terms GO:000584, ribosome, and GO:0006412, translation, and included more specified GO terms of cellular components and biological processes (Fig. 10). Col-0 highly activated the expression of cytosolic ribosome- and translation-related genes relative to the nonacclimated state at 1 d after the 10°C cold shift. This activation subsequently attenuated. In contrast, reil1-1 reil2-1 delayed these responses, as was revealed by an enrichment analysis of differential gene expression in the mutant compared with Col-0 at each time point (Fig. 10A). As a result of the delay, ribosome- and translation-related gene expression in reil1-1 reil2-1 was first reduced relative to Col-0 and then increased both at 1 and 3 weeks after the shift to 10°C. This pattern of differential gene expression was specific for the structural genes that constitute the cytosolic 60S and 40S ribosomal subunits (Barakat et al., 2001; Chang et al., 2005; Giavalisco et al., 2005) and did not extend to the structural genes of chloroplast ribosomes. These observations were confirmed (Fig. 10B) by additional analyses of the functional bins, which are equivalent to GO terms but were defined by the MapMan and PageMan software developers (Thimm et al., 2004; Usadel et al., 2006). Among the translation-related GO terms, translation elongation followed the general trend of structural cytosolic ribosome genes, whereas the terms translation initiation and termination were activated in the mutant throughout the 10°C cold acclimation (Fig. 10A).
Figure 10.
Functional enrichment analysis of ribosome- and translation-related gene expression in reil1-1 reil2-1 compared with Col-0 in the nonacclimated state and after the shift to 10°C. A, Heat map of mean log2-FCs from structural ribosome- and translation-related GO terms and selected more specified subterms. reil1-1 reil2-1/Col-0 columns represent mean log2-FCs > +0.1 or < −0.1 of mutant expression ratios compared with Col-0 in the nonacclimated state (0 d) and at the time points after the shift to 10°C, 1 d (1d), 1 week (1w), and 3 weeks (3w). Log2-FCs with significant positive or negative enrichment (i.e. z-score values > 2.5) are indicated by asterisks. Columns reil1-1 reil2-1 and Col-0 at 0d, 1d, 1w, and 3w represent mean log2-FC of expression ratios relative to nonacclimated Col-0. Information was extracted from mean log2-FCs, z-scores, and P values of a differential gene expression enrichment analysis covering genes belonging to 1,940 GO terms (Du et al., 2010; Supplemental Table S4). B, Mean log2-FC (±se) of cytosolic and chloroplast structural genes as defined by MapMan/PageMan functional bin annotations (Thimm et al., 2004; Usadel et al., 2006). Gray bars in each left subgraph are expression ratios of reil1-1 reil2-1/Col-0 at each given time point. Black and white bars in the right subgraphs represent the ratios relative to nonacclimated Col-0 (black triangles).
For a detailed analysis of relevant DEGs, we extracted 74 ribosome- or translation-related genes that were changed significantly at one or more of the four analyzed time points (Supplemental Table S5). Thirty-six of these DEGs were structural constituents of the cytosolic 60S and 40S subunits. Nine DEGs were part of translational initiation factors. One DEG was a translational elongation factor. Five DEGs were aminoacyl-tRNA synthetases, and nine genes were ribosome biogenesis factors. Furthermore, five of the 74 DEGs were previously annotated homologs of the yeast cytosolic 60S maturation machinery (Schmidt et al., 2013), namely the RLP24 homolog (AT2G44860), the RPL24A gene (AT2G36620), RPP0A (AT2G40010), the TIF6 homolog (AT3G55620), and a SSA1/2 homolog (AT3G09440.1). In addition, three previously studied plant biogenesis factors were present in this gene set, namely ESSENTIAL NUCLEAR PROTEIN1 (AT1G31660.1), HOMOLOG OF YEAST BRX1-1 (AT3G15460.1) and NUCLEOLIN2 (AT3G18610.1; Weis et al., 2015).
With few exceptions, such as RPL3B (AT1G61580) or the RPL13 homolog (AT3G48130), the differential gene expression patterns of the 74 extracted DEGs matched the general expression trends of their respective functional classes (Supplemental Table S5). One to 3 weeks after the shift to 10°C, most genes had increased mRNA abundance relative to Col-0, as exemplified by the 60S constituents RPP0A (AT2G40010) and RPL24A (AT2G36620) and the 40S constituent RPS8B (AT5G59240). Surprisingly, 12 DEGs belonged to the plastid 50S ribosomal subunit and two to the 30S ribosomal subunit. In contrast to the cytosolic RPs, the chloroplast RP components were down-regulated (Supplemental Table S5), such as the 50S component PRPL11 (AT1G32990) and the 30S component PRPS17 (AT1G79850).
Some ribosome- and translation-related genes deviated from the two general patterns of the up-regulated cytosolic and down-regulated chloroplast RPs. The deviating genes had mostly constitutively increased mRNA levels in the mutant relative to Col-0, for example the POLY(A)-BINDING PROTEIN PAB5 (AT1G71770) or the EUKARYOTIC TRANSLATION INITIATION FACTOR3 SUBUNIT C2 (EIF3C2, AT3G22860) of Arabidopsis eukaryotic translation initiation factor 3 (EIF3). The constitutive up-regulation of EIF3C2 mRNA relative to the wild type (Supplemental Table S5) was an exception compared with the other differentially expressed components of EIF3 (Burks et al., 2001). EIF3 subunit G2 (EIF3G2, AT5G06000) had first lower then higher mRNA levels relative to Col-0, while EIF3H1 (AT1G10840) had decreased transcript levels. Similarly, cytosolic aminoacyl-tRNA synthetase mRNAs (AT3G42723 and AT3G30770) were constitutively up-regulated or activated after the cold shift, such as the cytosolic threonyl-tRNA synthetase (AT1G17960).
reil1-1 reil2-1 Does Not Activate FLOWERING LOCUS T in Mature Leaves after the Cold Shift
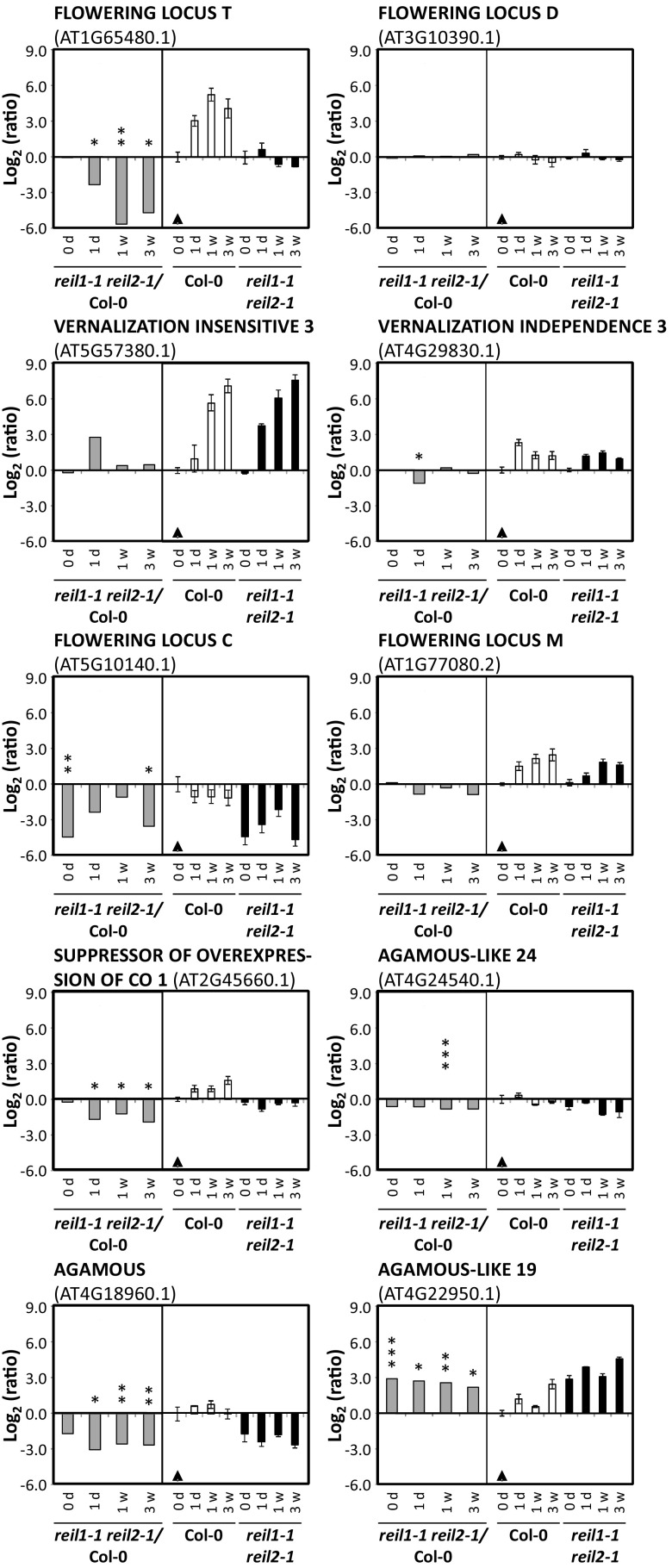
The transcripts of threonyl-tRNA synthetase (AT1G17960), aminoacyl-tRNA synthetase (AT3G42723), and EIF3C2 also were among a set of 64 genes with a greater than 4-fold change in expression in reil1-1 reil2-1 after 1 d, 1 week, and 3 weeks at 10°C. Almost two-thirds (41) of these 64 transcripts were already significantly different from the wild type in the nonacclimated state and may represent candidates of genes with REIL-dependent gene expression control or constitutive compensation responses to REIL deficiency (Supplemental Table S6). The 64 genes did not generate significant GO enrichment results when analyzed by singular enrichment analysis (Du et al., 2010), but FLOWERING LOCUS T (FT, AT1G65480.1) and genes encoding MADS domain transcription factors were prominent within this gene set (Fig. 11).
Figure 11.
Selected differentially expressed flowering-related genes of nonacclimated (0 d) and 10°C cold-acclimating reil1-1 reil2-1 at 1 d (1d), 1 week (1w), or 3 weeks (3w) after the shift to 10°C. Relative expression values are mean log2-FC (±se). Gray bars in each left subgraph are expression ratios of reil1-1 reil2-1/Col-0 at the given time points. Black and white bars in the right subgraph represent the ratios relative to nonacclimated Col-0 (black triangles). *, 0.01 < P ≤ 0.05; **, 0.001 < P ≤ 0.01; and ***, P ≤ 0.001, by heteroscedastic Student’s t tests.
In contrast to Col-0, FT expression was not activated in reil1-1 reil2-1 after the cold shift (Fig. 11). FT is the key mobile flowering signal, the florigen that is produced in leaves, the tissue that we analyzed in this study, and acts at the shoot apex by triggering the transition from vegetative to reproductive growth. mRNAs of downstream elements of FT-mediated flower induction were detectable in mature leaves. In agreement with the lack of FT activation, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1/AGL20, AT2G45660.1) and AGAMOUS-LIKE24 (AGL24, AT4G24540.1) were down-regulated in reil1-1 reil2-1 leaves (Fig. 11). Transcripts of the FT partner protein FLOWERING LOCUS D (FD, AT3G10390.1), however, remained unaffected, as was expected of FD, which is expressed mainly in the shoot apex (Kobayashi and Weigel, 2007). Transcripts of the FT/FD-activated meristem identity genes, LEAFY (AT5G61850.1) and APETALA1 (AP1/AGL7, AT1G69120.1), also were detectable in our mRNA preparations from mature leaves but were not differentially expressed.
To further dissect the lack of FT induction in reil1-1 reil2-1, we analyzed the expression of vernalization factors and of FLOWERING LOCUS C (FLC, AT5G10140.1). FLC is one of the floral repressors that functions in leaves by repressing FT expression and, in the shoot apical meristem, by repressing FD expression (Kobayashi and Weigel, 2007). FLC is a central component of the vernalization response pathway and has decreased mRNAs levels in vernalized leaves (Wellmer and Riechmann, 2010). Thus, vernalization releases FLC-mediated inhibition of FT expression in the leaf and FD in the shoot apical meristem as prerequisites to the initiation of reproductive growth. If this pathway was responsible for the lack of FT induction in reil1-1 reil2-1 leaves, vernalization responses would be absent in reil1-1 reil2-1 and FLC expression would be up-regulated. We found that the opposite of our expectations occurred. Vernalization responses were not activated prematurely in nonacclimated reil1-1 reil2-1 and, as in Col-0, were strongly activated after the 10°C cold shift (Fig. 11; Supplemental Table S6). For example, transcripts of VERNALIZATION INSENSITIVE3 (AT5G57380.1), a cold-specific component of the polycomb-repressive complex 2 (Lee et al., 2015), and VERNALIZATION INDEPENDENCE3 (AT4G29830.1) were increased in both the mutant and Col-0 (Fig. 11). Apparently, reil1-1 reil2-1 has no premature activation of vernalization and wild-type-like vernalization responses (Song et al., 2012; Sun et al., 2013). Also contrary to our expectations, FLC expression decreased much more in cold-acclimating reil1-1 reil2-1 compared with Col-0, and surprisingly, FLC expression also was much lower prior to vernalization in the nonacclimated mutant (Fig. 11).
Obviously, the vernalization pathway did not explain the lack of FT activation in reil1-1 reil2-1. As a consequence, we investigated alternative FT effectors in the leaf (Wellmer and Riechmann, 2010). Neither the transcripts of the activator CONSTANS (CO, AT5G15840.1) nor the mRNAs of the FT repressors TEMPRANILLO1 (TEM1, AT1G25560.1), TEM2 (AT1G68840.1) SHORT VEGETATIVE PHASE (SVP/AGL22, AT2G22540.1), SCHLAFMUTZE (AT3G54990.1), SCHNARCHZAPFEN (AT2G39250.1), TARGET OF EARLY ACTIVATION TAGGED1 (TOE1) to TOE3 (AT2G28550.1, AT5G60120.1, and AT5G67180.1), nor the splice variant 2 mRNA of FLOWERING LOCUS M (FLM/AGL27, AT1G77080.2) that was part of our transcriptome analysis were differentially expressed (Fig. 11; Supplemental Table S6).
Among the remaining DEGs, other AGL transcription factors are part of our set of 64 top DEGs (Fig. 11; Supplemental Table S6). We found FLM-independent vernalization-induced AGL19 (AT4G22950.1; Schönrock et al., 2006; Liu et al., 2008) to have constitutively increased transcript levels in the mutant. Overexpression of AGL19 activates FT in an FLC-independent manner (Kim et al., 2013; Kang et al., 2015). Therefore, we concluded that the block of FT activation in reil1-1 reil2-1 is both downstream of AGL19-mediated FT activation and downstream of FLC-mediated release of FT repression.
In addition, eponymous AGAMOUS (AG, AT4G18960.1; Fig. 11) and AGL42 (also named FOREVER YOUNG FLOWER, AT5G62165.1; Supplemental Table S6) had constitutively decreased mRNA levels in reil1-1 reil2-1 leaves. The relevance of these observations remained unclear, because both genes are known for their roles in flower development rather than for a function in leaf differentiation. AG is the shoot apex-expressed floral organ identity gene responsible for stamen and carpel formation. AG controls flower development among other mechanisms by suppression of the leaf development program in emerging floral primordia (Ó’Maoiléidigh et al., 2013). In agreement with reduced AG expression in reil1-1 reil2-1 leaves, the transcript of CURLY LEAVES (AT2G23380.1) that suppresses the ectopic expression of AG in leaves (Kim et al., 1998) was not activated. AGL42 is highly expressed in young flowers prior to pollination rather than in leaves and controls floral senescence and abscission (Chen et al., 2011).
DISCUSSION
Cold Acclimation of the Col-0 Wild Type Is a Relevant Process for the Investigation of Cytosolic RBFs
Our first aim was to establish an experimental setup that enables the study of conserved or plant-specific functions of Arabidopsis REIL proteins and of their proposed role in cytosolic ribosome biogenesis (Schmidt et al., 2013, 2014). We previously discovered that the cold impact on growth of the Arabidopsis single and double reil mutants and respective mutants of yeast homologs was conserved (Schmidt et al., 2013). Also, the gene expression of one of the two reil isoforms in each species was activated after a shift to low temperatures, namely Arabidopsis reil2 (see Supplemental Fig. S5 of Schmidt et al., 2013) and yeast rei1 (Strassburg et al., 2010). These findings led us to focus this study on the cold acclimation process.
Our experiments establish cold acclimation of the Col-0 wild type to 10°C chilling temperature at the vegetative rosette stage, ∼1.10 (Boyes et al., 2001), as a physiologically relevant and experimentally feasible system for the study of cytosolic RBFs. We confirm the increase of reil2 mRNA under these conditions (Supplemental Table S2). Successful cold acclimation of Col-0 to 10°C starts with a growth arrest of approximately 1 week followed by resumed growth and development (Fig. 2), vegetative-to-regenerative phase transition, and flowering (Fig. 1). The growth arrest coincides with the proposed initial inhibition of protein translation after a cold shift that appears to be mediated by GENERAL CONTROL NON-DEREPRESSIBLE1 (GCN1)/GCN2-dependent phosphorylation of the α-subunit of eIF2α and the TARGET OF RAPAMYCIN pathway (Wang et al., 2017). Our data clearly indicate that Arabidopsis physiology is far from inactive during the initial lag phase, as is expected according to reports that show a requirement of de novo protein synthesis at low temperature. The requirement of active translation after a cold shift was demonstrated, for example, by mutants of translation elongation factor2-like protein, LOW EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 (Guo et al., 2002), or the Arabidopsis F-BOX PROTEIN7 (Calderón-Villalobos et al., 2007). Besides the induction of reil2 expression, Col-0 specifically activates the expression of genes that encode cytosolic RPs in contrast to the mostly nonresponsive chloroplast RP genes (Fig. 10; Supplemental Table S5). Apparently, the cytosolic translational apparatus is remodeled at the level of structural RP, already characterized RBFs, and both translation initiation and elongation factors (Fig. 10). The remodeling of the translational apparatus was confirmed with an expected delay between gene expression maximum and subsequent protein accumulation at the level of ribosome subunit and polysome composition (Figs. 5D and 10B). Reduced polysome abundance may still support the remodeling process and agrees with the growth arrest. In addition, the subunit composition changes dynamically, specifically the relative abundance of the 60S LSU (Fig. 5). The cause of the subunit dynamics cannot be determined unambiguously because our methods here do not distinguish between subunit recycling from polysomes and subunit accumulation caused by de novo biosynthesis. Nevertheless, the strong and specific activation of cytosolic RP gene expression and the subsequent relative increase of cytosolic subunits and cytosolic rRNA (Fig. 5) support the conclusion that a 10°C acclimation of Col-0 involves the activation of cytosolic ribosome biogenesis.
Cold-Acclimating reil1-1 reil2-1 Remains Viable and Maintains Wild-Type Cold Acclimation Responses
We selected the reil1-1 reil2-1 double mutant as the object of our systems analyses of Arabidopsis REIL function. Consequently, our systems analyses here cannot differentiate paralog-specific functions. However, we expected that experimental results will be clearer than those of the single mutants, which may have compensatory reactions of the remaining intact paralog. For example, REIL1 deficiency causes no obvious morphological phenotype in reil1-1. REIL1 deficiency can be almost completely compensated by REIL2 (Schmidt et al., 2013). In addition, the cold sensitivity of reil2 mutant growth is restricted only to the vegetative phase (Schmidt et al., 2013). reil1-1 reil2-1 was the first double homozygous mutant that we obtained and focused on, because reil2-1 has a stronger phenotype compared with reil2-2 (Schmidt et al., 2013). Importantly, the double homozygous reil1-1 reil2-2 that was not yet available when we performed the functional complementation and systems analyses of this study also is strongly growth impaired in the cold.
The strong response of reil1-1 reil2-1 to cold exposure (Fig. 1, B–D) raised our concern that reil1-1 reil2-1 may exhibit pleiotropic effects that obscure the primary effects of REIL deficiency. Because the reil1-1 reil2-1 mutant is morphologically similar to Col-0 under standard conditions (Figs. 1A and 2), we tried to minimize pleiotropic effects by performing shift experiments from standard temperature to cold conditions. We ruled out inverse temperature shifts as an option (Fig. 1E). Inverse temperature-shift experiments are not only materially limited due to the dwarf phenotype but also are biased by the activation of a rapid flowering program that was reminiscent of stress-induced early flowering (Xu et al., 2014).
The next concern was the possibility that reil1-1 reil2-1 cells may be destabilized after the cold shift, even though successful temperature-rescue experiments demonstrated that the reil1-1 reil2-1 mutation is nonlethal in the cold (Fig. 1B). Our results indicate that the reil1-1 reil2-1 phenotype is an arrest or rather a strong retardation of leaf expansion growth and of the development of new leaves at low temperature (Figs. 1 and 2). This interpretation is supported by unchanged electrolyte leakage, which, unlike the los1-1 mutant (Guo et al., 2002), indicates unaltered stability of the plasma membrane of rosette leaves both under nonacclimated conditions and after cold acclimation (Fig. 2C). In addition to the maintained cellular integrity and obviously unchanged acquisition of freezing tolerance, reil1-1 reil2-1 also remains physiologically capable of wild-type cold acclimation responses, both at metabolic (Fig. 7) and at transcriptomic (Fig. 9) levels. For example, transient maltose accumulation is a characteristic acclimation response caused by starch degradation following a 4°C cold shock (Kaplan et al., 2004, 2007; Maruyama et al., 2009). This hallmark response also occurred after a shift to 10°C and was virtually indistinguishable in the mutant and Col-0 (Fig. 7C). Similarly, transcriptional cold acclimation responses remained largely unaffected. For example, activation of the DREB/CBF regulon (Yamaguchi-Shinozaki and Shinozaki, 2009; Lata and Prasad, 2011), which integrates cold responses and the concurrent water limitation and osmotic responses (Supplemental Fig. S6), was highly similar to that of the wild type (Fig. 9). Even selected vernalization responses were unaffected (Fig. 11).
Our investigations of reil1-1 reil2-1 obviously show surprisingly limited effects that are largely not confounded by cell death during cold acclimation. Therefore, we concluded that reil1-1 reil2-1 represents a valid and feasible system for the functional analysis of Arabidopsis REIL proteins. To demonstrate phenotype rescue, we reintroduced REIL1 and REIL2 into the mutant background. Phenotype rescue was achieved using the constitutive UBQ10 promoter and N-terminal FP fusion proteins (Fig. 3). The expression of N-terminal RFP and GFP fusion proteins in reil1-1 reil2-1 reconstituted the rosette morphologies of the reil1 and reil2 single mutants (Fig. 3, A and B) and their respective rate of seedling establishment (Fig. 3, C and D). Apparently, REIL function has little demand for exact timing and tissue localization of gene expression. Ectopic REIL expression by the UBQ10 promoter caused no observable detrimental effects except for a small delay in the appearance of first rosette leaves at 20°C (Supplemental Fig. S2).
Arabidopsis REIL Proteins Reveal Conserved and New Functional Aspects
Rescue of the reil1-1 reil2-1 phenotype was a prerequisite of studies that indicate conserved cytosolic localization of Arabidopsis REIL proteins (Fig. 4). REIL1 shows strictly conserved localization and is excluded from nuclei, vacuoles, and chloroplasts (Fig. 4). In contrast, REIL2 localization was more divergent and extended into the nucleus of guard cells or the root tip (Fig. 4, D–F). These observations and the previously reported rescue of the yeast ∆rei1 mutant by REIL1 but not by REIL2 (Schmidt et al., 2013) indicate that REIL1 may be the more conserved paralog, whereas REIL2 may be a neofunctionalized paralog. Localization of Arabidopsis REIL2 to the nucleus suggests that REIL2 may adopt a conditional function as a pre-60S nuclear assembly and export factor similar to yeast Nog1 (Wu et al., 2016). This suggestion, however, requires further validation, because we currently cannot rule out tissue- or condition-specific activation of a paralog selective protease. Protease action may cause local cleavage of RF fusion proteins so that the resulting free RFP may relocalize to the nucleus.
The conservation of REIL function is further evident by the requirement of REIL proteins for the rapid accumulation of cytosolic ribosome subunits after the cold shift (Fig. 5) and, in addition, by the association of the REIL1 protein with 60S LSU-containing ribosome fractions (Fig. 6). In contrast to yeast Rei1 and Reh1 (Hung and Johnson, 2006; Lebreton et al., 2006; Meyer et al., 2007), we also find an association of the Arabidopsis REIL1 protein with translating ribosome fractions, such as the 80S and polysome fractions (Fig. 6). This observation may point toward an additional role of Arabidopsis REIL for the cytosolic translation process or for subunit recycling. Again, further analyses are required, not least because our western-blot analyses indicate a weak and possibly more transient interaction of the REIL proteins with the Arabidopsis 60S LSU than the strong interactions that allow the capture of yeast Rei1/Reh1-60S complexes (Greber et al., 2012, 2016; Ma et al., 2017). The case of REIL1 is complicated by proteolysis after density gradient preparation. Currently, we cannot assign functional significance to REIL1 proteolysis. Arabidopsis REIL proteins and their complexes with the 60S LSU clearly need to be stabilized during preparation as a prerequisite for future functional analyses.
REIL Function Is Linked to Arabidopsis Temperature Perception and Development
Our analyses clearly indicate plant-specific functions of Arabidopsis REIL proteins. Metabolomic and transcriptomic evidence indicates premature triggering of cold acclimation responses in the mutant under standard growth conditions. Nonacclimated reil1-1 reil2-1 mimics the metabolic phenotype of 10°C cold-acclimating Col-0 (Fig. 7, A and B; Supplemental Fig. S4) and prematurely triggers the DREB/CBF regulon with a preference for the cold acclimation factors, CBF1/DREB1B, CBF2/DREB1C, and CBF3/DREB1A, over the heat and osmotic components DREB2A and DREB2B (Figs. 8 and 9). Indeed, CBF3/DREB1A activation explains the cold-induced carbohydrate metabolism and starch degradation. These metabolic processes apparently are under the control of CBF3/DREB1A but are not influenced by DREB2A overexpression in transgenic plants (Maruyama et al., 2009).
We conclude that either direct REIL function or REIL-dependent availability of translationally competent cytosolic 60S LSUs feedback on Arabidopsis temperature perception. In this feedback loop, REIL either directly or indirectly modifies temperature perception by suppressing premature low-temperature acclimation responses. REIL may modulate the energy- and resource-demanding ribosome biogenesis after the cold shift. Furthermore, REIL clearly does not operate a ribosome biogenesis switch but rather enhances the kinetics of cytosolic 60S LSU accumulation after the cold shift (Fig. 5) and the transcriptional activation of cytosolic RPs, RBFs, and translation-related genes (Fig. 10).
Besides the likely role of Arabidopsis REILs as kinetic modulators of cold-triggered cytosolic ribosome biogenesis, we discovered a link between REILs and the vegetative-to-regenerative phase transition in the cold. REIL proteins are required to trigger the transcription of FT mRNA in mature leaves at low-temperature conditions (Fig. 11). In this context, we suggest that REIL function overrides both FLC-mediated release of inhibition of FT expression and AGL19-dependent activation of FT expression. Altered gene expression of both factors apparently is part of the premature triggering of limited cold responses in nonacclimated reil1-1 reil2-1 (Fig. 11). REIL action also is apparently required upstream of FLC and AGL19 expression and is likely vernalization independent (Fig. 11). We suggest that REIL proteins execute a specific, yet either direct or indirect, input into the signaling chain that is required for FT mRNA activation in cold-acclimating mature leaves and, subsequently, for the vegetative-to-reproductive phase transition of the shoot apical meristem.
CONCLUSION
This work further establishes REIL proteins as likely RBFs with roles as kinetic modulators of cytosolic ribosome biogenesis or recycling in the cold that may help overcome the initial cold-induced inhibition of translation (Wang et al., 2017). We also reveal the possibility of functional changes of Arabidopsis REIL proteins during plant evolution and the integration of REIL function into plant-specific physiological processes, such as cold acclimation and flower induction in the cold. We summarize our conclusions on the role of REIL in Arabidopsis by a functional interaction model (Fig. 12).
Figure 12.
Functional interaction model that summarizes our insights into the role of REIL proteins in mature leaves before (20°/18°C, day/night) and after the shift to low-temperature conditions (10°/8°C). Bars = 1 cm.
Our investigations focused so far on mature leaf tissue as a first source of amply available leaf material for systems analyses. REILs clearly play a role in cold-acclimating mature leaves that were already preformed at standard temperature. Our results, however, indicate that REIL function also may be important for young leaf or root tissue that grows and develops in the cold. Therefore, future functional investigations may be specifically fruitful when focused on tissues that develop in the cold and on mechanisms that integrate ribosome biogenesis, cell propagation, and extension growth.
MATERIALS AND METHODS
Plant Material
The double homozygous reil1-1 reil2-1 mutant was created by Schmidt et al. (2013) by crossing the T-DNA insertion mutant SALK_090487 (reil1-1) that was obtained through the Nottingham Arabidopsis Stock Centre (Scholl et al., 2000) and GK_166C10 (reil2-1) of the GABI-Kat program (Rosso et al., 2003). T-DNA insertions had been characterized previously by sequencing of the left border insertion sites (Schmidt et al., 2013). Homozygosity of the insertion sites was verified by PCR amplification of genomic DNA with previously described T-DNA- and reil1- or reil2-specific oligonucleotides (Schmidt et al., 2013). According to the reported genetic background of the initial mutants, the ecotype Col-0 was used as the wild-type control throughout this study.
Growth Conditions
Generation of seed batches, storage, sterilization, and in vitro cultivation were as described previously (Schmidt et al., 2013). Arabidopsis (Arabidopsis thaliana) seeds were germinated under sterile conditions in long days with a 16-h/8-h day/night cycle and temperature settings as given below using one-half-strength Murashige and Skoog agar plates with 0.8% (w/v) agar and 2% (w/v) Suc (Murashige and Skoog, 1962). Seedlings at stage 1.02-1.03 (Boyes et al., 2001) were transferred, if not mentioned otherwise, to 10-cm-diameter pots with Arabidopsis soil (Stender) after a period of in vitro cultivation that was adjusted to the temperature-dependent rate of development. Pots with mutant and wild-type plants (n ≥ 10) were arrayed on growth chamber shelves in Latin square designs.
Constant Standard Temperature Conditions
Seedlings were transferred to soil after 10 d of in vitro cultivation at 20°C. Plants were acclimated to soil in short days with an 8-h/16-h day/night cycle at 20°C/18°C, 100 to 150 µmol m−2 s−1 light intensity, and 60% to 70% relative humidity using a controlled-environment chamber. After a total of 4 weeks, namely 10 d of in vitro cultivation and 18 d on soil, the wild type reached stage ∼1.10 (Fig. 1A).
Constant Low-Temperature Conditions
Seedlings were transferred to soil after 4 weeks of in vitro cultivation at 10°C. Plants were acclimated to soil using a controlled-environment chamber set to long days with a 16-h/8-h day/night cycle at 10°C/8°C, 100 to 150 µmol m−2 s−1 light intensity, and 60% to 70% relative humidity. After acclimation, plant cultivation was continued under identical conditions. Wild-type plants reached stage ∼1.10 after a total of 6 weeks, namely 28 d of in vitro cultivation and 2 weeks on soil. Care was taken throughout cultivation to maintain plants at 10°C without accidental transient shifts to elevated temperatures (Fig. 1B).
Temperature Shift from 20°C to 10°C or 4°C
At stage ∼1.10, plants were transferred from standard conditions to a controlled-environment chamber that was set to long days with a 16-h/8-h day/night cycle at 10°C/8°C, 100 to 150 µmol m−2 s−1 light intensity, and 60% to 70% relative humidity. The alternative settings were 4°C during both day and night at ∼90 µmol m−2 s−1 light intensity without humidity control. Control plants for temperature-shift experiments were moved in parallel to 16 h of light of 100 to 150 µmol m−2 s−1 light intensity at 20°C and 8 h of night at 18°C with 60% to 70% relative humidity (Fig. 1, C and D).
Temperature Shift from 10°C to Standard Temperature Conditions
Wild-type plants at stage ∼1.10 and simultaneously cocultivated mutant plants that had stage 1.02-1.03 were transferred from the 10°C/8°C settings to the 20°C/18°C conditions described above (Fig. 1C).
In Vitro Germination and Morphometric Growth Assays
In vitro germination assays of Col-0, reil1-1 reil2-1, and complemented reil1-1 reil2-1 were performed and the appearance of first juvenile rosette leaves was scored exactly as described previously (Schmidt et al., 2013). A 16-h/8-h day/night cycle was applied with either constant 20°C or constant 10°C. Strict maintenance of cold conditions was controlled to ensure the full suppression of reil1-1 reil2-1 rosette leaf emergence (Fig. 3, C and D; Supplemental Fig. S2).
Morphometric growth assays, namely measurements of leaf numbers and planar rosette leaf areas (Fig. 2, A and B), were performed as described previously (Schmidt et al., 2013). Plants were cultivated in single 10-cm-diameter pots on soil. Pots were arranged in a Latin square array. Three to 10 plants of each genotype, condition, and time point were analyzed after temperature shifts from 20°C standard conditions to either 10°C or 4°C as described above (Fig. 1, C and D). Photographs were taken at 4,928 × 3,264 pixel resolution in weekly intervals starting at the time of temperature transfer (i.e. week 0) with an SLR Nikon D5100 camera (Nikon), an AF-S DX Nikkor 18- to 55-mm, F/3.5-5.6 VR lens, and a 1-mm resolution scale on each photograph. Images were normalized to scale and analyzed with manual supervision using Adobe Photo Shop CS5 Extended (Adobe Photoshop version 11.0; Adobe Systems).
Electrolyte Leakage Assays
Electrolyte leakage measurements of cellular integrity and assessment of acquired freezing tolerance (Fig. 2C) were according to previously published standardized protocols (Rohde et al., 2004; Thalhammer et al., 2014). Tubes with three stacked mature leaves or up to half of a rosette to match equal leaf amounts were placed in test tubes with a bottom layer of double distilled water and kept at 4°C on ice. Tubes with nonfrozen control samples were maintained on ice to determine the basal electrolyte content. Test tubes for the freezing assays were transferred to a cooling bath, equilibrated 30 min at −1°C, and ice nucleation initiated for a subsequent 30-min period (Thalhammer et al., 2014). Test tubes were then cooled at a rate of 4°C h−1. Four to eight samples per 1°C temperature interval were removed from the bath and immediately placed on ice. All frozen and nonfrozen control samples were kept together overnight on ice in a 4°C chamber and then diluted by double distilled water. Paired electrical conductivities of leaked electrolytes from initially nonboiled samples and total electrolyte content from subsequently boiled samples were determined. The percentage of electrolyte leakage was calculated and normalized to the electrolyte content of nonfrozen control samples. The temperature of 50% normalized electrolyte leakage was calculated by the LOGEC50 value of sigmoidal fitted curves using GraphPad Prism 3.0 software (Thalhammer et al., 2014).
SDS-PAGE and Western-Blot Analysis
One hundred milligrams of liquid nitrogen flash-frozen and ground leaf material was extracted in denaturing SDS loading buffer that contained 50 mm Tris-HCl at pH 6.8, 4% (w/v) SDS, 30% (w/v) glycerol, 0.1 m DTT, 0.05% (w/v) Bromophenol Blue, and cOmplete protease inhibitor (Sigma-Aldrich) at a dose of one tablet per 1.5 mL of extraction buffer. Western-blot analysis was conducted as described previously (Grefen et al., 2009) after the separation of 18 µL of total protein in loading buffer at a 0.2 mg µL−1 concentration by 12% (w/v) acrylamide SDS-PAGE.
REIL-specific, 14-amino acid polyclonal peptide antibodies with an additional N-terminal Cys leader for carrier protein conjugation were epitope selected by the OptimumAntigen design tool (GenScript) and obtained from the same company. The anti-atREIL1.1 antigen peptide was (C)MSKRGETMRTKIGV, representing amino acid positions 377 to 390 of REIL1, C-terminal positions −28 to −15. Anti-atREIL2.1 was directed against (C)KRTDNKVTSRTLGS, equivalent to amino acid positions 305 to 318 of REIL2, C-terminal positions −91 to −78. The RFP-specific polyclonal antibody (anti-RFP) was obtained from Abcam. Polyclonal anti-RPL13B antibodies (AS13 2650/anti-L13-1) directed against the ∼23.8-kD RPL13B 60S subunit protein encoded by At3g49010 (Barakat et al., 2001; Sormani et al., 2011) were obtained from Agrisera. Anti-RFP, anti-RPL13B, and anti-atREIL1.1 were used at 1:5,000 dilution, and anti-atREIL2.1 was used at 1:3,000 dilution. The primary antibodies were detected by an anti-rabbit IgG-horseradish peroxidase secondary antibody that was diluted 1:10,000. Protein-antibody complexes were visualized by ECL reagents (ThermoFisher Scientific Life Technologies). Western blots were analyzed by the G:BOX F3 automated gel-imaging system (Syngene) and processed with the Molecular Analyst image-analysis software (Bio-Rad Laboratories).
Construction of FP-REIL Reporter Lines
All expression vectors were cloned and amplified by Gateway recombination cloning technology (Earley et al., 2006; Grefen et al., 2010). Clones containing reil genes were constructed after PCR amplification of cDNA from reverse-transcribed mRNA using the SalI and NotI restriction sites of the pENTR 1A vector and transformed into TOP10 competent Escherichia coli cells (Invitrogen Life Technologies). Successful cloning events were verified by colony PCR and sequencing of prepared vector DNA (LGC Genomics). Gateway vectors were cloned and amplified by E. coli ccdB survival cells (Invitrogen Life Technologies) and selected by 50 µg mL−1 spectinomycin and 25 mg mL−1 chloramphenicol. Vectors with the expression cassettes UBQ10::GFP-REIL1, UBQ10::RFP-REIL1, UBQ10::GFP-REIL2, UBQ10::RFP-REIL2, and UBQ10::REIL2 were placed under the control of the Arabidopsis UBQ10 promoter (AT4G05320) using the site-specific recombination sites attB1 and attB2. The LR recombination reaction included a mixture of 150 ng of pUBN-GFP-DEST or pUBN-RFP-DEST vector (Grefen et al., 2010; http://www.zmbp.uni-tuebingen.de/dev-genetics/grefen/resources/ubiquitin10-vectors.html), 150 ng of the pENTR clones with the respective reil genes, and 0.5 μL of LR clonase II (Invitrogen Life Technologies).
Vectors were transformed into reil1-1 reil2-1 mutant plants (Schmidt et al., 2013) by Arabidopsis in planta Agrobacterium tumefaciens transformation (Bent, 2000). UBQ10::GFP(RFP)-REIL reporter lines in the reil1-1 reil2-1 mutant background were screened at the T1 generation by 50 mg mL−1 BASTA (glufosinate) herbicide Murashige and Skoog plates. Resistant seedlings were selected at the cotyledon stage for generation of the T2 generation. Segregation of the T2 generation was checked after 30 d of in vitro cultivation at 10°C. Homozygous individuals were selected at stage 1.02-1.04, tested positively for the expression of protein fluorescence, and transferred to soil for seed-batch propagation under standard greenhouse conditions.
PCR
PCR was performed with 36 cycles, 15 s of denaturation at 94°C followed by 30 s of annealing at 55°C and 2 min of elongation at 72°C, using total RNA that was extracted using the RNeasy Plant Mini Kit (Qiagen). cDNA was synthesized from total RNA by SuperScript II Reverse Transcriptase (Invitrogen Life Technologies). The reil1-1 reil2-1 mutant background was verified by T-DNA-specific forward oligonucleotide primers, LBb1 (SALK) and GKo8409 (GABI-Kat), and reil gene-specific reverse primers (see Supplemental Table S5 of Schmidt et al., 2013). The forward and reverse oligonucleotide primers for the verification of the presence of the expression cassettes were nGFPfor 5′-AAGGGCGAGGAGCTGTT-3′ (UBQ10::GFP-REIL1), nRFPfor 5′-GAGGACGTCATCAAGGAGT-3′ (UBQ10::RFP-REIL1) combined with 3′-noncoding REIL1_at4g31420rev2 5′-GTGCGGCCGCAAGTGGCACATACACAG-3′ or nGFPfor 5′-AAGGGCGAGGAGCTGTT-3′ (UBQ10::GFP-REIL2), nRFPfor 5′-GAGGACGTCATCAAGGAGT-3′ (UBQ10::RFP-REIL2), and REIL2_at2g24500for2 5′-GCGTCGACGATGTCAGGTTTAGCTTGT-3′ (UBQ10::REIL2) combined with REIL2_at2g24500rev2 5′-GTGCGGCCGCGAAAATCGAGGAGCAAGA-3′, respectively.
Fluorescence Microscopy for FP-REIL Protein Localization
Seeds of the functionally complemented FP-REIL reporter lines UBQ10::RFP-REIL1 (line 49) and UBQ10::RFP-REIL2 (line 109) were germinated and grown in vitro as described above. Fluorescence microscopy of biological replicates of root tips and cotyledons from three to five individual plants was performed after 7 to 10 d of growth at 20°C or 7 d after the subsequent shift to 10°C. To visualize the subcellular localization of RFP fluorescence, a Leica TCS SP5 confocal laser scanning microscope was used with a 63× objective. Projection stacks of multiple confocal sections, overlays, and false-color renderings were generated using the manufacturer’s LSM software (Leica Microsystems). RFP excitation was achieved using a 561-nm laser. Emission was monitored between 572 and 634 nm. Chlorophyll fluorescence was obtained using the same excitation settings, but emission spectra were recorded in the range 652 to 727 nm.
Microfluidic Electrophoresis-Based Analysis of Relative rRNA Abundance
The relative abundance of cytosolic and chloroplast rRNA species was determined by microfluidic electrophoresis using an Agilent 2100 Bioanalyzer and the Agilent RNA 6000 Nano Kit according to the manufacturer’s instructions (Agilent Technologies). Samples of total RNA were extracted from 50 mg fresh weight of mature rosette leaves by the TRIzol reagent method (Ambion). rRNA ratios were determined as described previously (Walter et al., 2010; Salvo-Chirnside et al., 2011). Four biological replicates that represent preparations from independent pools of two or more plants, with two technical repetitions each, were analyzed per genotype and temperature condition.
Density Gradient Analysis of Arabidopsis Ribosomes
Ribosome fractions were isolated from three independent pools of mature Arabidopsis leaves from two or more plants that were harvested at approximately 6 ± 0.5 h after dawn according to Kawaguchi et al. (2004) and Piques et al. (2009) with minor modifications. Briefly, 100 mg of frozen leaf tissue was homogenized 30 min on ice in 0.5 mL of polysome extraction buffer (i.e. 200 mm Tris-HCl, pH 9, 200 mm KCl, 25 mm EGTA, 36 mm MgCl2, 5 mm DTT, 50 μg mL−1 cycloheximide, 50 μg mL−1 chloramphenicol, 0.5 mg mL−1 heparin, 1% (v/v) Triton X-100, 1% (v/v) Tween 20, 1% (w/v) Brij-35, 1% (v/v) IGEPAL CA-630, 2% (v/v) polyoxyethylene, and 1% (w/v) deoxycholic acid; Sigma-Aldrich). All procedures were carried out at 4°C. After clarification by centrifugation at 14,000g for 10 min, hydrated tissue was placed on a QIAshredder spin column of the RNeasy Plant Mini Kit (Qiagen) and centrifuged at 14,000g for 1 min to remove cell debris. Volumes of 0.5 mL of extract were loaded onto 9 mL of a 15% (w/v) to 60% (w/v) Suc gradient cushion. The loaded gradient was ultracentrifuged at 33,000g for 18 h at 4°C in a six-bore SW 41 Ti rotor (Beckman).
One analysis run comprised a set of six samples, 2 × 2 or 4 × 1 independently extracted replicates of the investigated conditions and two fixed control samples, namely a nonsample control with empty extraction buffer and a positive control prepared from a previously harvested and long-term stored homogenous reference batch of Col-0 rosette leaves. This reference batch was generated at developmental stage ∼1.10 from plants that were cultivated at standard temperature conditions. The Suc gradients were separated into 40 fractions of approximately 250 µL by a programmable density gradient fractionation system (Teledyne Isco), which recorded a continuous absorbance profile at 254 nm. After quality control of the Col-0 reference sample profile, absorbance profiles were exported and background subtracted by the nonsample control profile. Areas of ribosome fractions and total absorbance were determined by Cromas Lite 2.1 software (http://chromas-lite.software.informer.com/2.1/). To account for variations of loaded sample amounts, ribosome areas were normalized through division by total peak area.
Identification and Western-Blot Analysis of Ribosome Fractions
Ribosomal fractions were located within the profiles by screening of the whole Suc density profile range. All 40 fractions were analyzed by 1.2% (w/v) agarose gel electrophoresis of TRIzol total RNA preparations. rRNA-containing fractions were chosen for microfluidic electrophoresis (Salvo-Chirnside et al., 2011) and qualitative analysis of rRNA species (Fig. 5; Supplemental Fig. S3), with cytoplasmic 25S and 18S rRNA and chloroplast 23S and 16S rRNA annotations according to Nishimura et al. (2010) and Tiller et al. (2012).
For western blotting, complete 250-µL Suc density gradient fractions were precipitated by 600 µL of 100% (v/v) methanol, 150 µL of chloroform, and 400 µL of water. After a short vortex step and 10 min of centrifugation at 14,000g, the upper layer was carefully removed and a second washing step with 650 µL of methanol was performed. The final centrifugation was 20 min at 14,000g. The collected pellet was suspended in 24 μL of SDS-PAGE loading buffer, and western-blot analysis continued as described above.
Systems Analyses
Sampling
Samples for paired metabolome and transcriptome analyses were prepared from Arabidopsis Col-0 and reil1-1 reil2-1 mutant plants at stage ∼1.10 that were transferred 6 h after the start of the light period from the 20°C conditions to the 10°C regime described above. Mature leaves were harvested at 0 h (i.e. nonacclimated) before transfer and 0.25, 0.5, 1, 2, 4, 8, 24, 48, and 72 h after transfer as well as at 1, 2, 3, and 4 weeks after transfer. Daily and weekly samplings were all at approximately 6 h after dawn. Three replicate samples each were collected in two independent experiments and immediately frozen in liquid nitrogen. Each replicate sample was a pool of mature leaf material from at least two rosettes. Leaf tissue was frozen in liquid nitrogen immediately after harvest.
GC-MS-Based Metabolome Analysis
All collected leaf material was ground and weighed in the frozen state, and a polar metabolite fraction was extracted by standardized methanol/chloroform/water extraction and liquid phase separation as described previously (Roessner et al., 2000; Dethloff et al., 2014). The polar metabolite fraction was methoxyaminated and trimethylsilylated, and GC-MS profiling was performed by electron-impact ionization/time-of-flight mass spectrometry using an Agilent 6890N24 gas chromatograph (Agilent Technologies) and a Pegasus III time-of-flight mass spectrometer (LECO Instrumente). GC-MS peak abundances (responses) were processed into a standardized numerical data matrix that was normalized by sample fresh weight and internal standard, [U-13C]sorbitol. Metabolites were annotated using the TagFinder software (Luedemann et al., 2008) by mass spectral and retention time index matching to the reference collection of the Golm Metabolome Database (http://gmd.mpimp-golm.mpg.de/; Kopka et al., 2005; Hummel et al., 2010) and to the mass spectra of the NIST database (https://www.nist.gov/srd/nist-standard-reference-database-1a-v17). Normalized metabolite response ratios were calculated per metabolite relative to the measurement of nonacclimated Col-0 and log2 transformed.
Microarray Transcriptome Analysis
Total plant RNA of samples from the time points 0 (nonacclimated, i.e. before transfer), 1 d, 1 week, and 3 weeks was isolated from leaf tissue that was paired backup material of the metabolome study using the RNeasy Plant Mini Kit and RNase-free DNase (Qiagen). Three replicate samples, and in the case of nonacclimated plants, five to six replicate samples, that represented independent pools of mature rosette leaves from two or three plants were analyzed. RNA integrity number was calculated by microfluidic electrophoresis using an Agilent 2100 Bioanalyzer and 2100 Expert software (Agilent Technologies). RNA samples with RNA integrity number > 8 were sent to the 4×44K Agilent expression profiling service of ATLAS Biolabs. Gene expression data sets were quantile normalized using the normalize.quantile routine of the preprocessCore R statistical computing and graphics package (https://www.r-project.org/). The 4×44K Agilent expression profiling microarray contains variable numbers of redundant gene probes. Gene expression information was averaged if redundant probes were present to avoid bias of functional enrichment analyses. Gene expression information was normalized per gene to nonacclimated Col-0. Alternatively, the differential gene expression of mutant over Col-0 was calculated at each time point. The mode of normalization is indicated within legends and tables (Supplemental Table S2).
Statistical Analyses and Data Visualizations
Quantile-normalized log2-transformed ratios of DEGs or differentially accumulated metabolites were analyzed and visualized by Microsoft Excel software of the Office Professional Plus 2010 package (Microsoft), by the Multiple Experiment Viewer (version 4.9.0; http://www.mybiosoftware.com/mev-4-6-2-multiple-experiment-viewer.html), and by MetaGeneAlyse version 1.7.1 (http://metagenealyse.mpimp-golm.mpg.de/). Significance thresholds (P) of statistical analyses were as follows: one-way and two-way ANOVA, P < 0.001; Kruskal-Wallis test, P < 0.001; heteroscedastic Student’s t test, P < 0.01. All calculated P values are reported in Supplemental Tables S1 and S2. Details of the data reduction and visualization methods, namely hierarchical cluster analysis and principal component analysis, are reported in the legends to the figures and tables.
Functional Enrichment Analyses of Gene Expression Data
Functional enrichment analysis of differential gene expression by parametric analysis of gene set enrichment (Du et al., 2010), singular enrichment analysis (Du et al., 2010), and visualizations were performed at agriGO (http://bioinfo.cau.edu.cn/agriGO). Singular enrichment analysis investigates user-defined gene sets without consideration of numerical gene expression levels and provides information on the enrichment of GO categories among these genes (Du et al., 2010). The parametric analysis of gene set enrichment processes lists of genes with log2-transformed numerical differential gene expression values from one or more experimental conditions. Parametric analysis of gene set enrichment provides information on the enrichment of either expression increases or decreases across gene sets of GO terms. Additional and confirmatory enrichment analyses of gene expression were done by MapMan version 3.5.0 (http://mapman.gabipd.org/web/guest/pageman) using the functional bins (equivalent to GO terms) that were defined by the MapMan and PageMan software developers (Thimm et al., 2004; Usadel et al., 2006).
Accession Numbers
The expression data sets were uploaded to the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) and are available through accession number GSE101111.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Characterization of the UBQ10::GFP(RFP)-REIL reporter lines of the reil1-1 reil2-1 mutant.
Supplemental Figure S2. Comparison of early seedling development at 20°C to 10°C after constitutive expression of GFP-REIL and RFP-REIL fusion proteins under the control of the UBQ10 promoter in reil1-1 reil2-1.
Supplemental Figure S3. Suc density gradient separation optimized for the separation of the 50S and 60S ribosome fractions from Arabidopsis leaves.
Supplemental Figure S4. Comparison of leaf primary metabolite profiles from nonacclimated reil1-1 reil2-1 to the Col-0 time course of 10°C cold acclimation.
Supplemental Figure S5. Correlation analysis of gene expression in nonacclimated reil1-1 reil2-1 at standard cultivation temperature compared with cold-acclimating Col-0 at 1 d, 1 week, or 3 weeks after the shift to 10°C.
Supplemental Figure S6. Functional enrichment analysis of differential gene expression in reil1-1 reil2-1 at standard cultivation temperature compared with Col-0 and reil1-1 reil2-1 at 1 d, 1 week, or 3 weeks after the shift to 10°C.
Supplemental Table S1. Metabolite profiles and statistical data analyses of the nonacclimated and 10°C cold-acclimating reil1-1 reil2-1 mutant compared with the Col-0 wild type.
Supplemental Table S2. Gene expression profiles and statistical data analyses of mature leaves from the nonacclimated and 10°C cold-acclimating reil1-1 reil2-1 mutant compared with the Col-0 wild type.
Supplemental Table S3. Shared DEGs between the nonacclimated reil1-1 reil2-1 mutant and cold-acclimating Col-0 at 1 d, 1 week, or 3 weeks after the shift to 10°C.
Supplemental Table S4. Functional enrichment analysis of differential gene expression in the nonacclimated and cold-acclimating reil1-1 reil2-1 mutant at 1 d, 1 week, or 3 weeks after the shift to 10°C compared with nonacclimated and cold-acclimating Col-0.
Supplemental Table S5. Differential gene expression of ribosome- or translation-related genes in the nonacclimated and cold-acclimating reil1-1 reil2-1 mutant.
Supplemental Table S6. Differential expression of 64 genes that were significantly changed more than 4-fold at least once in the nonacclimated or cold-acclimating reil1-1 reil2-1 mutant and selected flowering- and vernalization-related genes.
Acknowledgments
We acknowledge the longstanding support by Dr. L. Willmitzer, Dr. M. Stitt, and Dr. R. Bock (Max-Planck-Institute of Molecular Plant Physiology).
Footnotes
We acknowledge funding by the Max Planck Society. We thank the National Council for the Improvement of Higher Education-CAPES of Brazil for the scholarship provided to A.A.P.F.
Articles can be viewed without a subscription.
References
- Ascenzi R, Gantt JS (1999) Molecular genetic analysis of the drought-inducible linker histone variant in Arabidopsis thaliana. Plant Mol Biol 41: 159–169 [DOI] [PubMed] [Google Scholar]
- Barakat A, Szick-Miranda K, Chang IF, Guyot R, Blanc G, Cooke R, Delseny M, Bailey-Serres J (2001) The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol 127: 398–415 [PMC free article] [PubMed] [Google Scholar]
- Basu U, Si K, Warner JR, Maitra U (2001) The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol Cell Biol 21: 1453–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent AF. (2000) Arabidopsis in planta transformation: uses, mechanisms, and prospects for transformation of other species. Plant Physiol 124: 1540–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning KS, Bailey-Serres J (2015) Mechanism of cytoplasmic mRNA translation. The Arabidopsis Book 13: e0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burks EA, Bezerra PP, Le H, Gallie DR, Browning KS (2001) Plant initiation factor 3 subunit composition resembles mammalian initiation factor 3 and has a novel subunit. J Biol Chem 276: 2122–2131 [DOI] [PubMed] [Google Scholar]
- Byrne ME. (2009) A role for the ribosome in development. Trends Plant Sci 14: 512–519 [DOI] [PubMed] [Google Scholar]
- Calderón-Villalobos LIA, Nill C, Marrocco K, Kretsch T, Schwechheimer C (2007) The evolutionarily conserved Arabidopsis thaliana F-box protein AtFBP7 is required for efficient translation during temperature stress. Gene 392: 106–116 [DOI] [PubMed] [Google Scholar]
- Chang IF, Szick-Miranda K, Pan S, Bailey-Serres J (2005) Proteomic characterization of evolutionarily conserved and variable proteins of Arabidopsis cytosolic ribosomes. Plant Physiol 137: 848–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MK, Hsu WH, Lee PF, Thiruvengadam M, Chen HI, Yang CH (2011) The MADS box gene, FOREVER YOUNG FLOWER, acts as a repressor controlling floral organ senescence and abscission in Arabidopsis. Plant J 68: 168–185 [DOI] [PubMed] [Google Scholar]
- Creff A, Sormani R, Desnos T (2010) The two Arabidopsis RPS6 genes, encoding for cytoplasmic ribosomal proteins S6, are functionally equivalent. Plant Mol Biol 73: 533–546 [DOI] [PubMed] [Google Scholar]
- Degenhardt RF, Bonham-Smith PC (2008) Arabidopsis ribosomal proteins RPL23aA and RPL23aB are differentially targeted to the nucleolus and are disparately required for normal development. Plant Physiol 147: 128–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Boniotti MB, Gutierrez C (2002) Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCFAtSKP2 pathway in response to light. Plant Cell 14: 3057–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demoinet E, Jacquier A, Lutfalla G, Fromont-Racine M (2007) The Hsp40 chaperone Jjj1 is required for the nucleo-cytoplasmic recycling of preribosomal factors in Saccharomyces cerevisiae. RNA 13: 1570–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethloff F, Erban A, Orf I, Alpers J, Fehrle I, Beine-Golovchuk O, Schmidt S, Schwachtje J, Kopka J (2014) Profiling methods to identify cold-regulated primary metabolites using gas chromatography coupled to mass spectrometry. Methods Mol Biol 1166: 171–197 [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Giavalisco P, Wilson D, Kreitler T, Lehrach H, Klose J, Gobom J, Fucini P (2005) High heterogeneity within the ribosomal proteins of the Arabidopsis thaliana 80S ribosome. Plant Mol Biol 57: 577–591 [DOI] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Montellese C, Ban N (2012) Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nat Struct Mol Biol 19: 1228–1233 [DOI] [PubMed] [Google Scholar]
- Greber BJ, Gerhardy S, Leitner A, Leibundgut M, Salem M, Boehringer D, Leulliot N, Aebersold R, Panse VG, Ban N (2016) Insertion of the biogenesis factor Rei1 probes the ribosomal tunnel during 60S maturation. Cell 164: 91–102 [DOI] [PubMed] [Google Scholar]
- Grefen C, Donald N, Hashimoto K, Kudla J, Schumacher K, Blatt MR (2010) A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J 64: 355–365 [DOI] [PubMed] [Google Scholar]
- Grefen C, Obrdlik P, Harter K (2009) The determination of protein-protein interactions by the mating-based split-ubiquitin system (mbSUS). Methods Mol Biol 479: 217–233 [DOI] [PubMed] [Google Scholar]
- Guo Y, Xiong L, Ishitani M, Zhu JK (2002) An Arabidopsis mutation in translation elongation factor 2 causes superinduction of CBF/DREB1 transcription factor genes but blocks the induction of their downstream targets under low temperatures. Proc Natl Acad Sci USA 99: 7786–7791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy C, Kaplan F, Kopka J, Selbig J, Hincha DK (2008) Metabolomics of temperature stress. Physiol Plant 132: 220–235 [DOI] [PubMed] [Google Scholar]
- Hannah MA, Heyer AG, Hincha DK (2005) A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet 1: e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Van Lijsebettens M, Candela H, Micol JL, Tsukaya H (2012) Ribosomes and translation in plant developmental control. Plant Sci 191-192: 24–34 [DOI] [PubMed] [Google Scholar]
- Hummel J, Strehmel N, Selbig J, Walther D, Kopka J (2010) Decision tree supported substructure prediction of metabolites from GC-MS profiles. Metabolomics 6: 322–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung NJ, Johnson AW (2006) Nuclear recycling of the pre-60S ribosomal subunit-associated factor Arx1 depends on Rei1 in Saccharomyces cerevisiae. Mol Cell Biol 26: 3718–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase M, Toh-e A (2004) Ybr267w is a new cytoplasmic protein belonging to the mitotic signaling network of Saccharomyces cerevisiae. Cell Struct Funct 29: 1–15 [DOI] [PubMed] [Google Scholar]
- Johnson AW, Lund E, Dahlberg J (2002) Nuclear export of ribosomal subunits. Trends Biochem Sci 27: 580–585 [DOI] [PubMed] [Google Scholar]
- Kang MJ, Jin HS, Noh YS, Noh B (2015) Repression of flowering under a noninductive photoperiod by the HDA9-AGL19-FT module in Arabidopsis. New Phytol 206: 281–294 [DOI] [PubMed] [Google Scholar]
- Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, Sung DY, Guy CL (2004) Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol 136: 4159–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Kopka J, Sung DY, Zhao W, Popp M, Porat R, Guy CL (2007) Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant J 50: 967–981 [DOI] [PubMed] [Google Scholar]
- Kawaguchi R, Girke T, Bray EA, Bailey-Serres J (2004) Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J 38: 823–839 [DOI] [PubMed] [Google Scholar]
- Kim GT, Tsukaya H, Uchimiya H (1998) The CURLY LEAF gene controls both division and elongation of cells during the expansion of the leaf blade in Arabidopsis thaliana. Planta 206: 175–183 [DOI] [PubMed] [Google Scholar]
- Kim W, Latrasse D, Servet C, Zhou DX (2013) Arabidopsis histone deacetylase HDA9 regulates flowering time through repression of AGL19. Biochem Biophys Res Commun 432: 394–398 [DOI] [PubMed] [Google Scholar]
- Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N (2011) Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 334: 941–948 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Weigel D (2007) Move on up, it’s time for change: mobile signals controlling photoperiod-dependent flowering. Genes Dev 21: 2371–2384 [DOI] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, et al. (2005) GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21: 1635–1638 [DOI] [PubMed] [Google Scholar]
- Lata C, Prasad M (2011) Role of DREBs in regulation of abiotic stress responses in plants. J Exp Bot 62: 4731–4748 [DOI] [PubMed] [Google Scholar]
- Lebreton A, Saveanu C, Decourty L, Rain JC, Jacquier A, Fromont-Racine M (2006) A functional network involved in the recycling of nucleocytoplasmic pre-60S factors. J Cell Biol 173: 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Yun JY, Zhao W, Shen WH, Amasino RM (2015) A methyltransferase required for proper timing of the vernalization response in Arabidopsis. Proc Natl Acad Sci USA 112: 2269–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Chen H, Er HL, Soo HM, Kumar PP, Han JH, Liou YC, Yu H (2008) Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135: 1481–1491 [DOI] [PubMed] [Google Scholar]
- Lo KY, Li Z, Bussiere C, Bresson S, Marcotte EM, Johnson AW (2010) Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol Cell 39: 196–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedemann A, Strassburg K, Erban A, Kopka J (2008) TagFinder for the quantitative analysis of gas chromatography-mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics 24: 732–737 [DOI] [PubMed] [Google Scholar]
- Ma C, Wu S, Li N, Chen Y, Yan K, Li Z, Zheng L, Lei J, Woolford JL Jr, Gao N (2017) Structural snapshot of cytoplasmic pre-60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1. Nat Struct Mol Biol 24: 214–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K, Takeda M, Kidokoro S, Yamada K, Sakuma Y, Urano K, Fujita M, Yoshiwara K, Matsukura S, Morishita Y, et al. (2009) Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol 150: 1972–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AE, Hoover LA, Craig EA (2010) The cytosolic J-protein, Jjj1, and Rei1 Function in the removal of the pre-60 S subunit factor Arx1. J Biol Chem 285: 961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AE, Hung NJ, Yang P, Johnson AW, Craig EA (2007) The specialized cytosolic J-protein, Jjj1, functions in 60S ribosomal subunit biogenesis. Proc Natl Acad Sci USA 104: 1558–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Mustroph A, Juntawong P, Bailey-Serres J (2009) Isolation of plant polysomal mRNA by differential centrifugation and ribosome immunopurification methods. Methods Mol Biol 553: 109–126 [DOI] [PubMed] [Google Scholar]
- Nishimura K, Ashida H, Ogawa T, Yokota A (2010) A DEAD box protein is required for formation of a hidden break in Arabidopsis chloroplast 23S rRNA. Plant J 63: 766–777 [DOI] [PubMed] [Google Scholar]
- Ó’Maoiléidigh DS, Wuest SE, Rae L, Raganelli A, Ryan PT, Kwasniewska K, Das P, Lohan AJ, Loftus B, Graciet E, et al. (2013) Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell 25: 2482–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm D, Simm S, Darm K, Weis BL, Ruprecht M, Schleiff E, Scharf C (2016) Proteome distribution between nucleoplasm and nucleolus and its relation to ribosome biogenesis in Arabidopsis thaliana. RNA Biol 13: 441–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse VG, Johnson AW (2010) Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem Sci 35: 260–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell KM, Bass BL (2009) Functional redundancy of yeast proteins Reh1 and Rei1 in cytoplasmic 60S subunit maturation. Mol Cell Biol 29: 4014–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendle AF, Clark GP, Boon R, Lewandowska D, Lam YW, Andersen J, Mann M, Lamond AI, Brown JWS, Shaw PJ (2005) Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol Biol Cell 16: 260–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piques M, Schulze WX, Höhne M, Usadel B, Gibon Y, Rohwer J, Stitt M (2009) Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Mol Syst Biol 5: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L (2000) Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J 23: 131–142 [DOI] [PubMed] [Google Scholar]
- Rohde P, Hincha DK, Heyer AG (2004) Heterosis in the freezing tolerance of crosses between two Arabidopsis thaliana accessions (Columbia-0 and C24) that show differences in non-acclimated and acclimated freezing tolerance. Plant J 38: 790–799 [DOI] [PubMed] [Google Scholar]
- Rosado A, Li R, van de Ven W, Hsu E, Raikhel NV (2012) Arabidopsis ribosomal proteins control developmental programs through translational regulation of auxin response factors. Proc Natl Acad Sci USA 109: 19537–19544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Salvo-Chirnside E, Kane S, Kerr LE (2011) High throughput silica-based purification of RNA from Arabidopsis seedlings in a 96-well format. Plant Methods 7: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, Dethloff F, Beine-Golovchuk O, Kopka J (2013) The REIL1 and REIL2 proteins of Arabidopsis thaliana are required for leaf growth in the cold. Plant Physiol 163: 1623–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, Dethloff F, Beine-Golovchuk O, Kopka J (2014) REIL proteins of Arabidopsis thaliana interact in yeast-2-hybrid assays with homologs of the yeast Rlp24, Rpl24A, Rlp24B, Arx1, and Jjj1 proteins. Plant Signal Behav 9: e28224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl RL, May ST, Ware DH (2000) Seed and molecular resources for Arabidopsis. Plant Physiol 124: 1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönrock N, Bouveret R, Leroy O, Borghi L, Köhler C, Gruissem W, Hennig L (2006) Polycomb-group proteins repress the floral activator AGL19 in the FLC-independent vernalization pathway. Genes Dev 20: 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz E, Tohge T, Zuther E, Fernie AR, Hincha DK (2015) Natural variation in flavonol and anthocyanin metabolism during cold acclimation in Arabidopsis thaliana accessions. Plant Cell Environ 38: 1658–1672 [DOI] [PubMed] [Google Scholar]
- Schulz E, Tohge T, Zuther E, Fernie AR, Hincha DK (2016) Flavonoids are determinants of freezing tolerance and cold acclimation in Arabidopsis thaliana. Sci Rep 6: 34027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 6: 410–417 [DOI] [PubMed] [Google Scholar]
- Song J, Angel A, Howard M, Dean C (2012) Vernalization: a cold-induced epigenetic switch. J Cell Sci 125: 3723–3731 [DOI] [PubMed] [Google Scholar]
- Sormani R, Masclaux-Daubresse C, Daniel-Vedele F, Chardon F (2011) Transcriptional regulation of ribosome components are determined by stress according to cellular compartments in Arabidopsis thaliana. PLoS ONE 6: e28070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassburg K, Walther D, Takahashi H, Kanaya S, Kopka J (2010) Dynamic transcriptional and metabolic responses in yeast adapting to temperature stress. OMICS 14: 249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C (2013) R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 340: 619–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalhammer A, Hincha DK, Zuther E (2014) Measuring freezing tolerance: electrolyte leakage and chlorophyll fluorescence assays. Methods Mol Biol 1166: 15–24 [DOI] [PubMed] [Google Scholar]
- Thieme CJ, Rojas-Triana M, Stecyk E, Schudoma C, Zhang W, Yang L, Miñambres M, Walther D, Schulze WX, Paz-Ares J, et al. (2015) Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat Plants 1: 15025. [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Thomashow MF. (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Tiller N, Weingartner M, Thiele W, Maximova E, Schöttler MA, Bock R (2012) The plastid-specific ribosomal proteins of Arabidopsis thaliana can be divided into non-essential proteins and genuine ribosomal proteins. Plant J 69: 302–316 [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Bläsing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, et al. (2006) PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lijsebettens M, Vanderhaeghen R, De Block M, Bauw G, Villarroel R, Van Montagu M (1994) An S18 ribosomal protein gene copy at the Arabidopsis PFL locus affects plant development by its specific expression in meristems. EMBO J 13: 3378–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Piepenburg K, Schöttler MA, Petersen K, Kahlau S, Tiller N, Drechsel O, Weingartner M, Kudla J, Bock R (2010) Knockout of the plastid RNase E leads to defective RNA processing and chloroplast ribosome deficiency. Plant J 64: 851–863 [DOI] [PubMed] [Google Scholar]
- Wang J, Lan P, Gao H, Zheng L, Li W, Schmidt W (2013) Expression changes of ribosomal proteins in phosphate- and iron-deficient Arabidopsis roots predict stress-specific alterations in ribosome composition. BMC Genomics 14: 783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Li H, Zhao C, Li S, Kong L, Wu W, Kong W, Liu Y, Wie Y, Zhu JK, et al. (2017) The inhibition of protein translation mediated by AtGCN1 is essential for cold tolerance in Arabidopsis thaliana. Plant Cell Environ 40: 56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis BL, Kovacevic J, Missbach S, Schleiff E (2015) Plant-specific features of ribosome biogenesis. Trends Plant Sci 20: 729–740 [DOI] [PubMed] [Google Scholar]
- Wellmer F, Riechmann JL (2010) Gene networks controlling the initiation of flower development. Trends Genet 26: 519–527 [DOI] [PubMed] [Google Scholar]
- Wu S, Tutuncuoglu B, Yan K, Brown H, Zhang Y, Tan D, Gamalinda M, Yuan Y, Li Z, Jakovljevic J, et al. (2016) Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature 534: 133–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu MY, Zhang L, Li WW, Hu XL, Wang MB, Fan YL, Zhang CY, Wang L (2014) Stress-induced early flowering is mediated by miR169 in Arabidopsis thaliana. J Exp Bot 65: 89–101 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2009) DREB regulons in abiotic-stress-responsive gene expression in plants. In Yamada T, Spangenberg G, eds, Molecular Breeding of Forage and Turf. Springer Science & Business Media, New York, pp 15–27 [Google Scholar]