A plant secondary metabolite modifies chromatin, primes defense, and controls plant disease in Arabidopsis.
Abstract
Modern crop production calls for agrochemicals that prime plants for enhanced defense. Reliable test systems for spotting priming-inducing chemistry, however, are rare. We developed an assay for the high-throughput search for compounds that prime microbial pattern-induced secretion of antimicrobial furanocoumarins (phytoalexins) in cultured parsley cells. The screen produced 1-isothiocyanato-4-methylsulfinylbutane (sulforaphane; SFN), a secondary metabolite in many crucifers, as a novel defense priming compound. While elucidating SFN’s mode of action in defense priming, we found that in Arabidopsis (Arabidopsis thaliana) the isothiocyanate provokes covalent modification (K4me3, K9ac) of histone H3 in the promoter and promoter-proximal region of defense genes WRKY6 and PDF1.2, but not PR1. SFN-triggered H3K4me3 and H3K9ac coincide with chromatin unpacking in the WRKY6 and PDF1.2 regulatory regions, primed WRKY6 expression, unprimed PDF1.2 activation, and reduced susceptibility to downy mildew disease (Hyaloperonospora arabidopsidis). Because SFN also directly inhibits H. arabidopsidis and other plant pathogens, the isothiocyanate is promising for the development of a plant protectant with a dual mode of action.
To supply the future world population with food, crop production needs to double by 2050 (UN Food and Agriculture Organization, 2009). The required boost in crop yield largely depends on effective plant protection, which is mostly achieved today with synthetic agrochemicals. Although safer than ever, chemical crop protection raises ecological and health concerns (Mascarelli, 2013; Lamberth et al., 2013). Therefore, safe and eco-friendly pest and disease control products are needed (Lamberth et al., 2013).
Phytochemicals that prime the plant immune system for enhanced defense are promising for sustainable crop protection (Beckers and Conrath, 2007; Conrath et al., 2015). When primed, plants respond to very low levels of a stimulus with earlier, sometimes faster and often more intense activation of defense than unprimed plants. This frequently reduces pest and disease susceptibility (Conrath et al., 2002; 2006; 2015; Beckers and Conrath, 2007; Frost et al., 2008). In Arabidopsis (Arabidopsis thaliana), priming is associated with an elevated level of microbial pattern receptors (Tateda et al., 2014), accumulation of dormant cellular signaling enzymes (Beckers et al., 2009), and covalent modification to chromatin (notably histone H3K4me3, H3K9ac, and DNA hypomethylation; Jaskiewicz et al., 2011a; Luna et al., 2012; López et al., 2011). Together, these events seem to provide the memory to the initial infection in that they poise defense genes for enhanced transcription upon reinfection (Conrath, 2011; Jaskiewicz et al., 2011a; Luna et al., 2012; Conrath et al., 2015). Other molecular mechanisms of priming remained unknown. Priming does not run up a major fitness bill and is hardly prone to pest or pathogen adaptation (van Hulten et al., 2006; Martinez-Medina et al., 2016). Thus, triggering priming by phytochemicals represents a promising means for sustainable pest and disease control.
Previous work with synthetic chemicals provided proof of this concept. Benzo(1,2,3)thiadiazole-7-carbothioic acid S-methyl ester (BTH; common name acibenzolar-S-methyl), a mimic of the phytochemical salicylate (SA), in addition to activating some defense genes directly (Ryals et al., 1996), primes plants for enhanced defense (Katz et al., 1998; Kohler et al., 2002) and protects multiple crops from disease (Ryals et al., 1996). In 1996, BTH was commercially launched as a plant immune activator (Ruess et al., 1996) with the trade name “Actigard.” However, BTH’s economic success was limited, mainly because of its strictly protective activity. To overcome the practical limitations of BTH, and take advantage of the low risk of pathogen adaptation, BTH is nowadays combined with conventional fungicides to achieve the best possible plant protection. For example, Bion M is a mixture of BTH with the fungicide mancozeb. The mixture performs particularly well on vegetables, even if mancozeb levels are much reduced (Leadbeater and Staub, 2014).
Another strategy for exploiting defense priming in practical pest and disease control is by identifying compounds combining insecticidal or antimicrobial activity with defense priming in a same molecule. The strobilurin fungicide pyraclostrobin (trade name “Headline”), in addition to its fungicide activity, primes crops and ornamental plants in the greenhouse and field for resistance to disease associated with enhanced yield (Herms et al., 2002; Koehle et al., 2003; 2006) even in abiotic stress conditions (Holmes and Rueber, 2007). Because of their broad spectrum of protection and the distinctive yield benefit, pyraclostrobin and other strobilurin fungicides became top-selling agrochemicals (Bartlett et al., 2002).
Today, the commercial success of crop protectants often relies on their ability to combine insecticidal or antimicrobial activity with defense priming in the treated crop (Beckers and Conrath, 2007; Conrath et al., 2015). However, identifying such chemistry is difficult because test systems for priming activity are rare. We developed a high-throughput screen for compounds that prime microbial pattern-induced furanocoumarin (phytoalexin) secretion in suspension-cultured parsley (Petroselinum crispum) cells. The screen produced 1-isothiocyanato-4-methylsulfinylbutane (sulforaphane [SFN]) as a novel defense-priming compound in plants. Because SFN is a natural compound with antimicrobial and insect deterrent activity (Tierens et al., 2001; Halkier and Gershenzon, 2006), the isothiocyanate may qualify for the development of a sustainable plant protectant.
RESULTS
Identification of SFN as a Novel Defense-Priming Compound
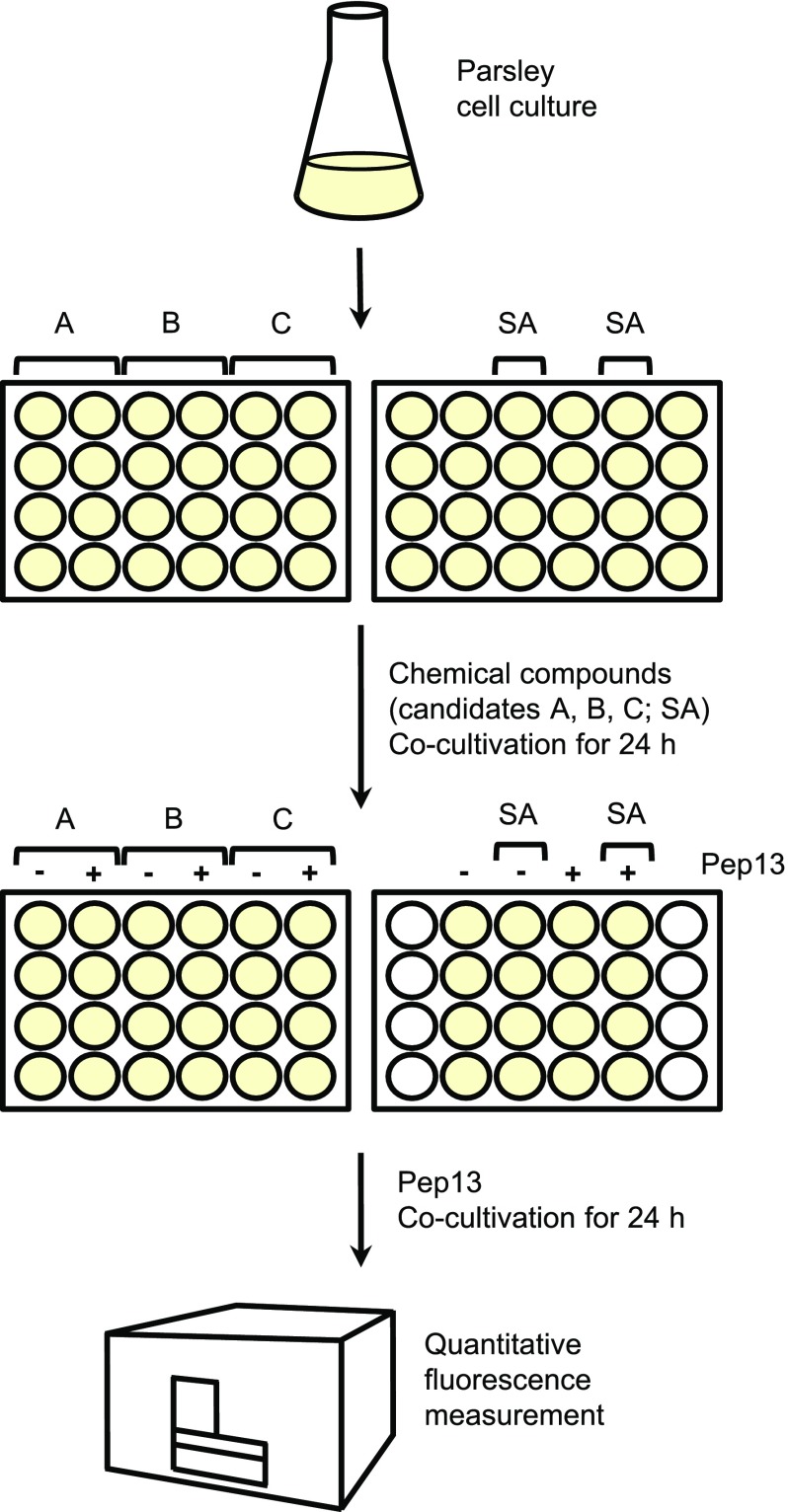
To spot novel defense-priming compounds, we optimized an assay that measures the enhancement by priming agents of furanocoumarin secretion provoked in cultured parsley cells by a moderate concentration of Pep13 (Kauss et al., 1992), a molecular pattern in the plant pathogen Phytophthora sojae (Brunner et al., 2002). Over the past approximately 25 years, the parsley-Pep13 interaction helped to disclose biochemical and molecular biological aspects of defense priming (Kauss et al., 1992; Katz et al., 1998), and identify novel priming-inducing chemistry (Katz et al., 1998; Siegrist et al., 1998). For enhanced throughput, we performed the test with 1-mL aliquots of cell culture in 24-well microtiter plates (Fig. 1). Cell culture aliquots were supplemented with the candidate compound (in DMSO), the natural priming compound salicylate (in DMSO; positive control), or DMSO (solvent control; Fig. 1). Upon shaking for 24 h in the dark, Pep13 (50 pm) was added to spur furanocoumarin synthesis and secretion. After another 24 h on the shaker, fluorescence of secreted furanocoumarins was quantified in a microtiter plate reader (Fig. 1). Compounds that significantly enhanced Pep13-induced furanocoumarin secretion were considered active at priming for enhanced defense.
Figure 1.
Scheme of the high-throughput screen for identifying plant immune stimulants. A quantity of 1-mL aliquots of a 3-d-old parsley cell culture was transferred to individual wells of a 24-well microtiter plate containing a candidate compound for priming (A, B, or C) or the known priming activator SA (positive control). All compounds were dissolved in DMSO (<1%). Thus, DMSO (1%) treatment served as a negative control. Upon incubation for 24 h on a shaker, Pep13 (50 pm) was added to appropriate wells. After shaking for another 24 h, the fluorescence of secreted furanocoumarins was quantified in a microtiter plate reader.
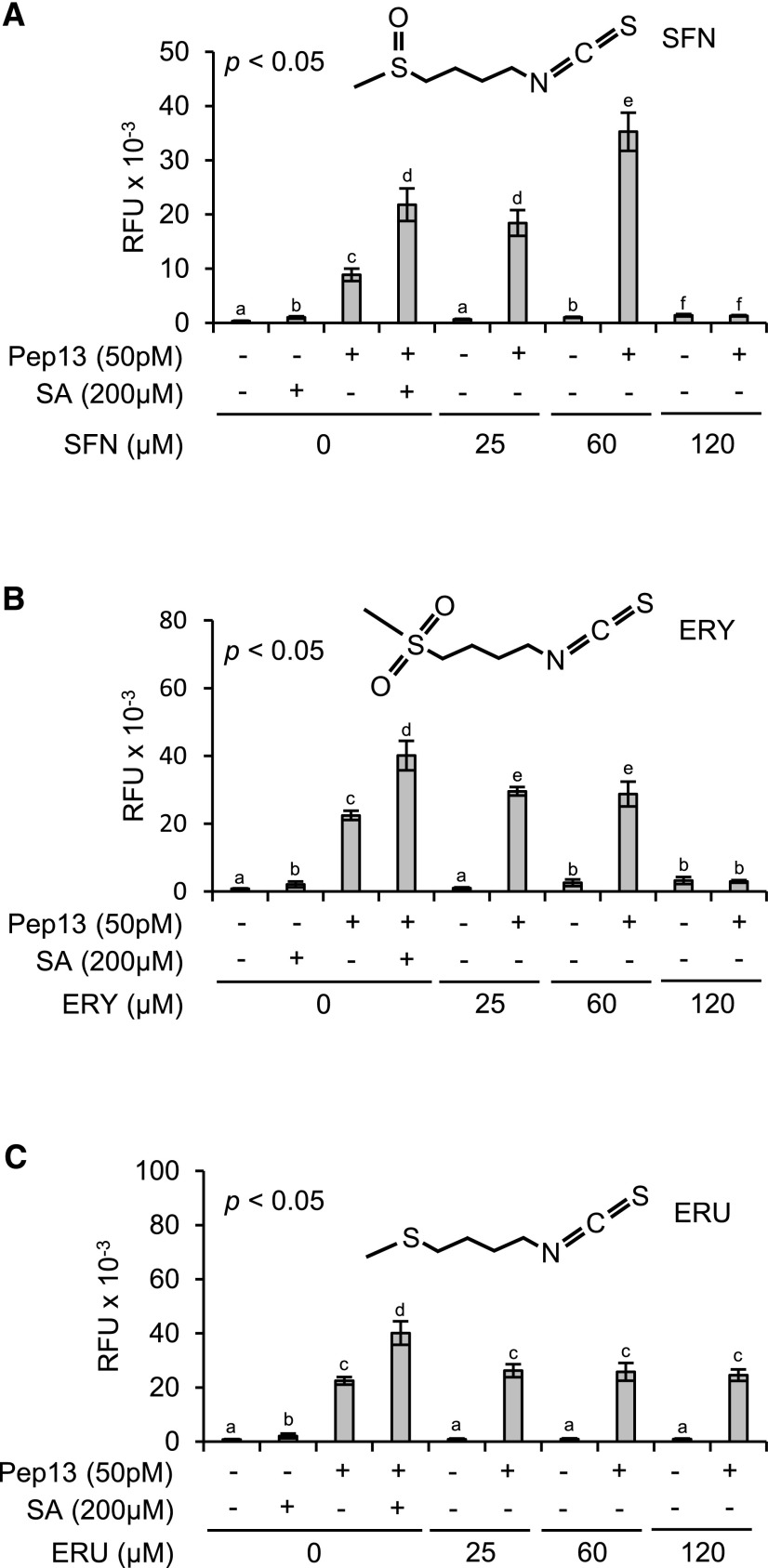
For unbiased screening, we randomly selected candidate compounds from commercial compound libraries. In three replications of the screening procedure, we identified SFN, an aliphatic isothiocyanate in many crucifers, as a novel defense-priming compound (Fig. 2A). Priming by SFN of Pep13-incuded furanocoumarin secretion was dose-dependent. At 25 μm, SFN was as active at priming as SA at 200 µm, whereas 60 μm SFN primed parsley cells better than 200 µm SA. No defense priming was seen when SFN was used at 120 µm (Fig. 2A). At this concentration, SFN noticeably harmed the cells, as made obvious by their mucilaginous appearance.
Figure 2.
Role of the –N=C=S group and the oxidation state of sulfur in the side chain. A quantity of 1-mL aliquots of a 3-d-old parsley cell culture in microtiter plates was treated with SFN (A), ERY (B), ERU (C), or SA (A–C, positive control). All compounds were dissolved in DMSO (0.25%). Therefore, treatment with 0.25% DMSO served as an additional control. After 24 h on a shaker, 50 pm Pep13 was added. After another 24 h, furanocoumarin fluorescence in the wells was determined. Data were analyzed by one-way ANOVA followed by posthoc Student’s t test. Different letters denote statistically significant differences with 95% confidence. Data presented are means ± sd (n > 6). RFU, Relative fluorescence units.
Role of the –N=C=S Group and the Oxidation State of Sulfur in the Side Chain
The many biological activities of SFN have been assigned to its –N=C=S moiety that has various cellular nucleophilic targets (Zhang et al., 1992). To investigate whether the –N=C=S group in SFN is sufficient for priming and if oxidation of sulfur in the side chain would affect SFN’s priming capacity, 1-isothiocyanato-4-methylsulfonylbutane (erysolin [ERY]) and 1-isothiocyanato-4-methylsulfanylbutane (erucin [ERU]) were tested for priming activity (Fig. 2, B and C). In ERU, the sulfur atom is not oxidized at all. In SFN, it bears one, and in its sulfonyl analog ERY, two oxygen atoms. With reference to SA, ERY displayed weaker priming activity than SFN at 25 and 60 µm, and this activity was gone at 120 µm ERY (Fig. 2B). ERU at 25, 60, and 120 µm did not much, if at all, prime parsley cells for enhanced Pep13-provoked furanocoumarin secretion (Fig. 2C). Therefore, the oxidation state of sulfur in the side chain seems to be critical, and the –N=C=S moiety insufficient, for SFN’s priming activity.
SFN Induces Covalent Modification to Histone H3
In eukaryotes, trimethylation of Lys residue 4 in histone H3 (H3K4me3), acetylation of Lys-9 in histone H3 (H3K9ac), and some other histone modifications in the promoter or body of gene accompany gene activity (Li et al., 2007). After transcription, they provide a memory for enhanced future gene expression (Badeaux and Shi, 2013). In Arabidopsis, H3K4me3 and H3K9ac in the promoter and promoter-proximal region associate with the primed state of enhanced WRKY6/29/53 defense gene readiness, before reactivation of WRKY6/29/53 transcription (Jaskiewicz et al., 2011a). H3K9ac on the WRKY6/53 promoter region was also linked to transgenerational defense priming in this plant (Luna et al., 2012). Because of the assumed critical role of H3K4me3 and H3K9ac in defense gene priming and transcription, we wondered whether SFN would modify histone H3 in the promoter/promoter-proximal region of WRKY6, PLANT DEFENSIN1.2 (PDF1.2), and PATHOGENESIS-RELATED1 (PR1). These loci were selected because WRKY6 is a reliable reporter gene for defense priming in Arabidopsis (Jaskiewicz et al., 2011a), and PDF1.2 and PR1 serve as marker genes for the jasmonate (JA)/ethylene (ET) and SA signaling pathways, respectively (Uknes et al., 1992; Penninckx et al., 1996; Koornneef and Pieterse, 2008). The drought-responsive RAB18 (Lång and Palva, 1992) and Rubisco genes served as control loci.
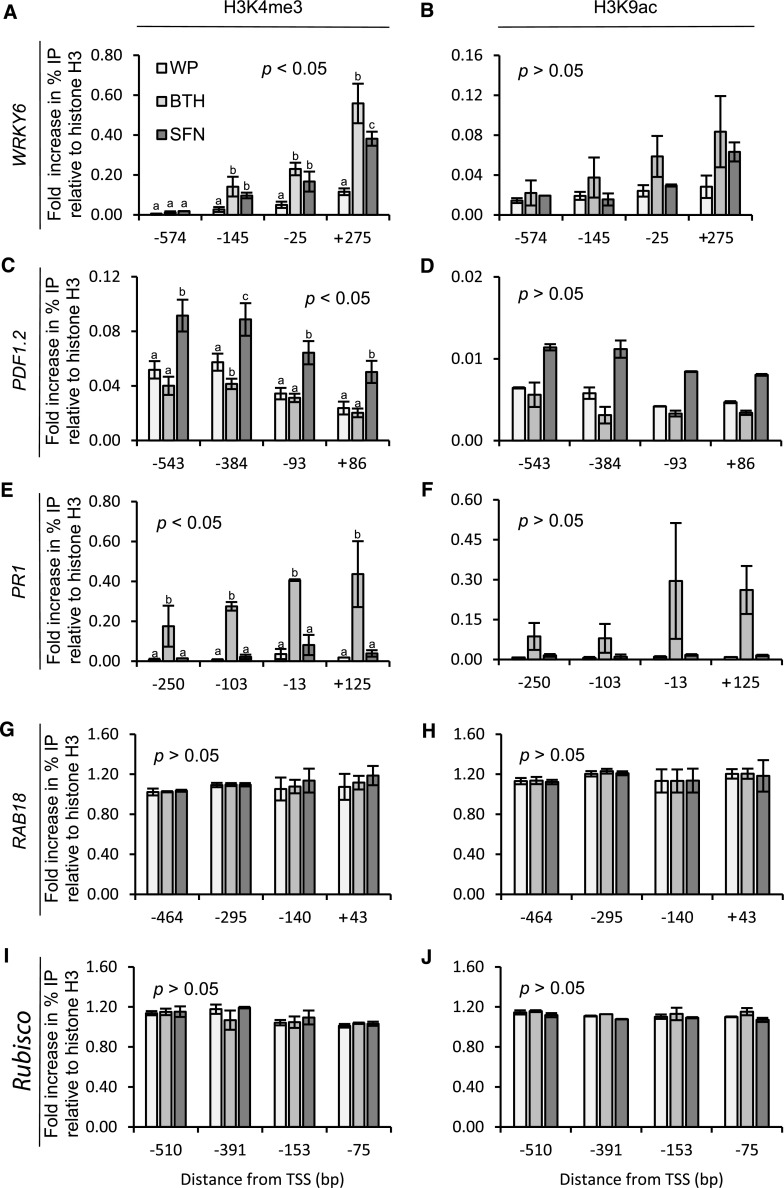
Our chromatin immunoprecipitation (ChIP) experiments disclosed that a wettable powder (WP) formulation of SFN or BTH (which served as a positive control for defense priming) enhanced H3K4me3 and, with less confidence, H3K9ac in the promoter and promoter-proximal region of WRKY6 (Fig. 3, A and B). SFN induced both these histone modifications also in the promoter region of PDF1.2, whereas BTH seemed to reduce the two epi-marks on histone H3 in the PDF1.2 promoter (Fig. 3, C and D), although this was not significant. In contrast, BTH, but not SFN, significantly enhanced H3K4me3 and seemingly also H3K9ac in the promoter/promoter-proximal region of PR1 (Fig. 3, E and F). SFN and BTH either did not, or did only marginally, change H3K4me3 and H3K9ac in the promoter/promoter-proximal region of RAB18 and Rubisco (Fig. 3, G–J). These findings demonstrated that in the regulatory region of defense genes, SFN and BTH induce covalent modification of histone H3 that accompanies primed or unprimed defense gene transcription (Fig. 5, A–C).
Figure 3.
Histone H3 modification in the promoter of Arabidopsis defense genes upon plant treatment with SFN or BTH. Plants were sprayed with WP formulations of SFN (450 µm) or BTH (100 µm). Application of WP served as a control for these treatments. At 24 h after treatment, leaves were harvested and subjected to chromatin extraction, isolation, and immunoprecipitation with antibodies to the H3K4me3 (A, C, E, G, and I) or H3K9ac (B, D, F, H, and J) epitopes. DNA abundance in the precipitate was measured by qPCR with primers specific to WRKY6 (A and B), PDF1.2 (C and D), PR1 (E and F), RAB18 (G and H), or Rubisco (I and J). Data give the fold increase in amplicon abundance compared to a sample from untreated control plants. We analyzed the data for every given position of gene by one-way ANOVA followed by posthoc Student’s t test. Different letters denote statistically significant differences with 95% confidence. Data are means ± sd (n > 3).
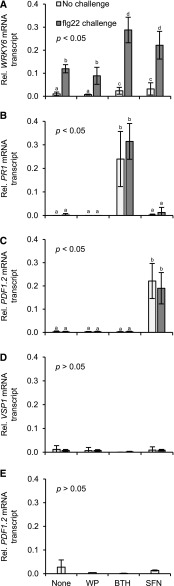
Figure 5.
Unprimed or primed defense gene activation in Arabidopsis genotypes treated with SFN or BTH. Wild type (A–D) and ein2-1 (E) plants were left untreated, sprayed with a WP formulation of SFN (450 µm) or BTH (100 µm), or WP only. 24 h later, three leaves of half of the plants for each treatment were challenged by infiltrating flg22 (200 nm; A–D) or left unchallenged (A–E). After 45 min, we extracted RNA from leaves and subjected it to RT-qPCR analysis using gene-specific primers (Supplemental Table S1). Data were analyzed by one-way ANOVA followed by posthoc Student’s t test. Different letters denote statistically significant differences with 95% confidence. Data presented are means ± sd (n > 6).
SFN and BTH Unpack Chromatin at Defense Gene Promoters
Histone acetylation slacks the interaction of nucleosome neighbors, loosens the ionic DNA-histone interaction, and provides docking sites for regulatory proteins (Kanno et al., 2004). H3K4me3 is also known to recruit transcription coactivators to chromatin (Aasland et al., 1995). As the interaction of DNA with regulatory proteins in chromatin is less intense than the DNA-nucleosome interplay (Giresi and Lieb, 2009), nucleosome-depleted DNA elements interacting with regulatory proteins can experimentally be detected as open chromatin (Gaulton et al., 2010). To investigate whether modification of histone H3 by SFN and BTH (Fig. 3) would open chromatin in the promoter/promoter-proximal region of target genes, we performed formaldehyde-assisted isolation of regulatory DNA elements (FAIRE) associated with quantitative amplification of DNA using locus-selective PCR (FAIRE-qPCR; Giresi and Lieb, 2009). FAIRE is exceptionally powerful for identifying unpacked regulatory chromatin (Simon et al., 2012).
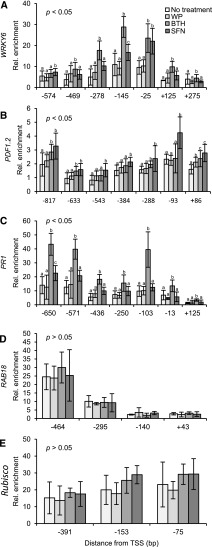
Figure 4A shows that treatment of Arabidopsis plants with SFN or BTH, both in WP, causes chromatin to open in the WRKY6 promoter/promoter-proximal region. Chromatin unpacking was detected within the −574- to +275-bp region relative to the transcription start site (TSS) and was most pronounced at −25 bp (SFN) and −145 bp (BTH) of all sites tested (Fig. 4A). In contrast to WRKY6, the PDF1.2 promoter opened in the −817- to +86-bp region after plant treatment with SFN, but less so upon treatment with BTH (Fig. 4B). Chromatin unpacking by SFN in the PDF1.2 promoter was maximal at −93 bp relative to the TSS (Fig. 4B). The PR1 promoter opened in the −650- to +125-bp region in response to BTH treatment with prominent peaks at −650-, −571-, and −103 bp, but did not open after SFN application (Fig. 4C). SFN and BTH did not open chromatin in the RAB18 promoter/promoter-proximal region (Fig. 4D). The overall chromatin unpacking recorded in the −295- to −464-bp region of RAB18 is likely due to the promoter start of the preceding, constitutively active, and antisense-oriented PLP3.b gene with a role in microtubule assembly. In the Rubisco promoter, chromatin seemed to be generally loosened after treatment with SFN and BTH, but typical peaks of chromatin opening were not detected (Fig. 4E).
Figure 4.
Detection of open chromatin in defense gene promoters/promoter-proximal regions. Arabidopsis plants were left untreated or sprayed with a WP formulation of SFN (450 µm) or BTH (100 µm). WP treatment served as a control for these treatments. After 24 h, we harvested leaves and subjected them to FAIRE. We performed qPCR with primers specific for different sites in the promoter/promoter-proximal region of WRKY6 (A), PDF1.2 (B), PR1 (C), RAB18 (D), and Rubisco (E). Data for every given position of gene were analyzed by one-way ANOVA followed by posthoc Student’s t test. Different letters denote statistically significant differences with 95% confidence. Data presented are means ± sd (n > 6).
A Dual Role for SFN in Defense Gene Activation
Because SFN caused covalent modification of histone H3 and chromatin opening in the WRKY6 and PDF1.2 but not PR1 regulatory regions (Figs. 3, A–F, and 4, A–C), we wondered whether the isothiocyanate affected the transcriptional response of WRKY6, PDF1.2, or PR1. To answer this question, we sprayed Arabidopsis plants with the WP formulation of SFN or BTH. Plants that were left untreated or sprayed with WP served as controls. At 24 h later, three leaves of an Arabidopsis plant were infiltrated with an aqueous solution of the microbial pattern flg22 to activate defense. After another 45 min, leaves were harvested and analyzed for accumulation of mRNA transcript of the WRKY6, PR1, and PDF1.2 defense genes (Fig. 5, A–C).
Although flg22 at 200 nm triggered the accumulation of WRKY6 mRNA transcript, this accumulation was somewhat, but not significantly, reduced upon preexposure to WP (Fig. 5A). Treatment with SFN or BTH only faintly caused WRKY6 expression (Fig. 5A). However, treating Arabidopsis with either of the two compounds enhanced the later WRKY6 activation by flg22 (Fig. 5A). PR1 expression was not activated by SFN, but was induced upon treatment with BTH and this response was somewhat, but not significantly, augmented upon flg22 challenge (Fig. 5B). By contrast, SFN, but not BTH, activated PDF1.2 expression, with no further enhancement by subsequent flg22 challenge (Fig. 5C).
The activation of PDF1.2 by SFN (Fig. 5C) let us conclude that the isothiocyanate stimulates JA and/or ET signaling. In Arabidopsis, the two hormone signals are transduced via two distinct but interconnected pathways, which both lead to PDF1.2 expression (Zarate et al., 2007). However, only the JA signaling pathway also activates expression of the VEGETATIVE STORAGE PROTEIN1 (VSP1) gene (Zarate et al., 2007). To disclose the pathway by which SFN activates PDF1.2 in Arabidopsis (Fig. 5C), we thus examined the accumulation of VSP1 mRNA transcript in samples from appropriately treated plants. As shown in Figure 5D, neither WP, nor BTH or SFN, activated Arabidopsis VSP1 before or after flg22 treatment. This suggests involvement of ET rather than JA in SFN-induced PDF1.2 expression. Consistent with this assumption, SFN did not activate PDF1.2 expression in the Arabidopsis ETHYLENE INSENSITIVE2-1 (ein2-1) mutant (Fig. 5E), which is blocked in ET signaling (Alonso et al., 1999).
SFN and Plant Protection
Because SFN seems to stimulate ET signaling in Arabidopsis, the isothiocyanate is likely to reduce the susceptibility to nectrotroph pathogens in this plant. However, SFN directly inhibits the growth of many infectious fungi (e.g. Plectosphaerella cucumerina) and bacteria (e.g. Pseudomonas syringae) already in the absence of plant (Tierens et al., 2001). Therefore, we will not be able to distinguish whether a reduction of disease susceptibility to such pathogens in SFN-treated plants is due to defense priming, direct inhibition of the pathogen, or both these possibilities.
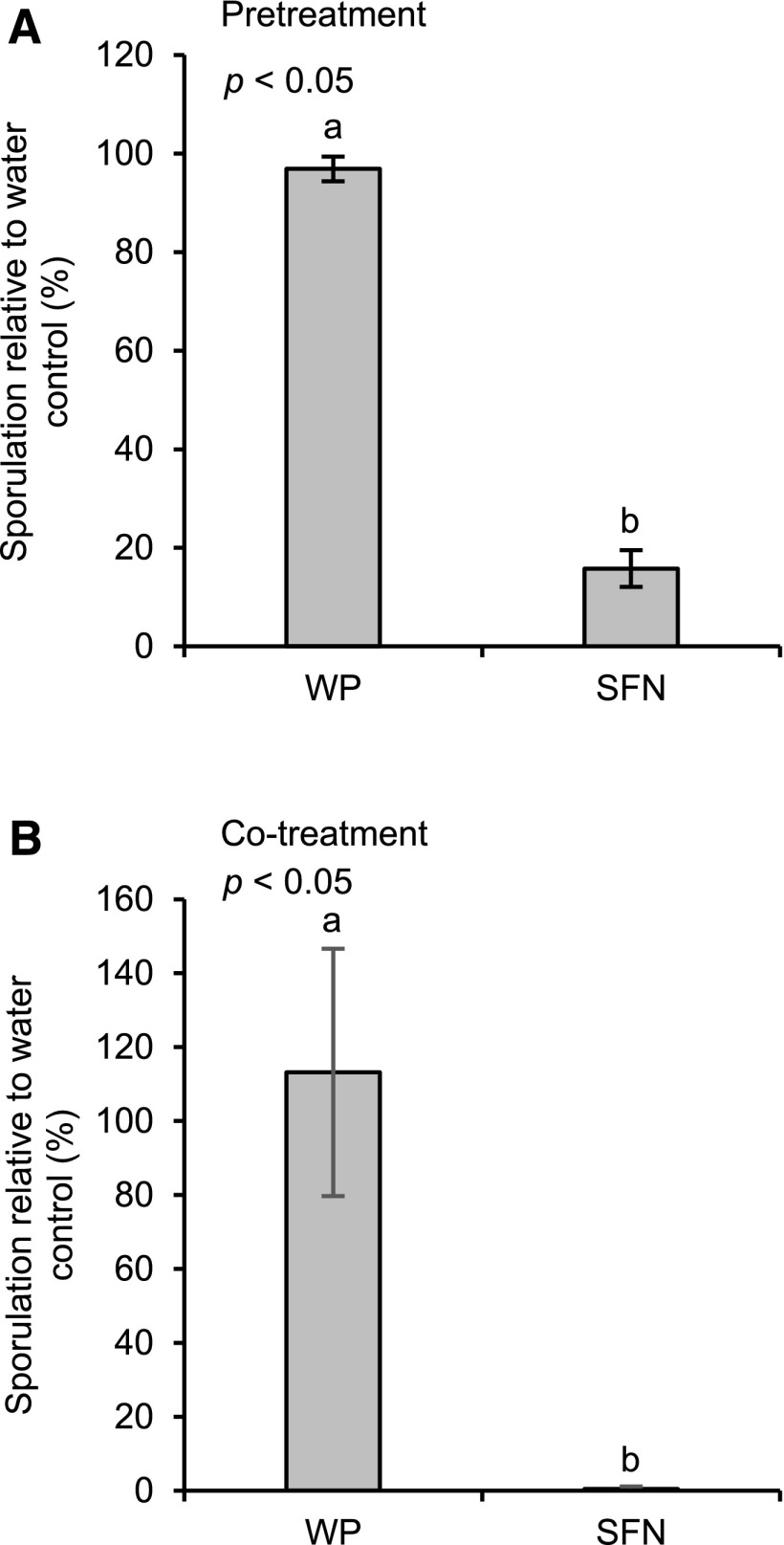
To investigate whether SFN affects the interaction of Arabidopsis with the infectious oomycete Hyaloperonospora arabidopsidis (Hpa), we sprayed Arabidopsis Col-0 plants with a WP formulation of SFN before we inoculated them with Hpa (race Noco; Uknes et al., 1992). The Noco race of Hpa causes downy mildew disease on Arabidopsis Col-0 (Coates and Beynon, 2010). Figure 6A discloses that pretreatment with SFN seems to reduce the susceptibility of Arabidopsis to downy mildew disease, as obvious by lower Hpa sporulation. This is suggesting that SFN activates defense priming, and by doing so, reduces the susceptibility to Hpa infection in Arabidopsis. However, because treatment of Arabidopsis leaves with Hpa conidiospores in a SFN solution causes an even stronger reduction of Hpa sporulation (Fig. 6B), the isothiocyanate seems to inhibit Hpa directly.
Figure 6.
SFN reduces downy mildew disease. A, Plants were treated with water (control), WP, or a WP formulation of SFN (450 µm). Twenty-four hours later, we spray-inoculated plants with a suspension of Hpa conidiospores (5 × 104 spores per mL of water). B, Conidiospores (5 × 104 spores per mL) were suspended in water (control), WP, or a WP formulation of SFN (450 µm). Spore suspensions were mixed and used for spray inoculation of plants. In A and B, inoculated plants were kept at high humidity in short day. After 7 d, we determined the number of spores released by Hpa. Data were analyzed by one-way ANOVA followed by posthoc Student’s t test. Different letters denote statistically significant differences with 95% confidence. Data presented are means ± sd. A, n > 7; B, n > 14.
DISCUSSION
We developed an assay for the high-throughput identification of chemical compounds with potential for plant-immunity-based, sustainable agriculture. The test measures the extent of furanocoumarin secretion by parsley culture cells challenged with Pep13 (Kauss et al., 1992; Katz et al., 1998; Siegrist et al., 1998). The advantage of the test over competitive assays is the high sensitivity of fluorescence detection and the need of only two treatments (activation of priming and Pep13 challenge; Fig. 1) before final analysis. A recently introduced competitive high-throughput assay for identifying priming-inducing chemistry evaluates the enhancement of P. syringae pv tomato avrRpm1-induced cell death in an Arabidopsis cell culture by priming agents (Noutoshi et al., 2012). The identification of several immune-priming compounds verified the suitability of the assay for discovering priming-inducing chemistry. However, because of requirement of bacterial challenge, cytochemical staining, washing, dye extraction, and absorbance measurement, the screen is rather elaborate. The same holds true for a more recently introduced respiratory activity-monitoring system for discovering priming-inducing chemistry (Schilling et al., 2015), which is innovative but suffers from low throughput.
Our screen identified the glucosinolate metabolite SFN as a novel defense-priming compound in plants. In addition to Pep13-induced furanocoumarin secretion in parsley (Fig. 2A), SFN primed WRKY6 for enhanced expression upon flg22 treatment (Fig. 5A) and directly activated PDF1.2 (Fig. 5C) in Arabidopsis. Thus, SFN, just like SA and BTH (Katz et al., 1998; Thulke and Conrath, 1998), seems to have a dual role in the activation of defense genes in plants.
Glucosinolate metabolites were known to be important to the Arabidopsis defense response (Clay et al., 2009), but their mode of action remained incompletely understood. Unfortunately, Arabidopsis mutants with impaired SFN perception or transduction are not yet available. Because SFN, although at higher concentrations than those applied here, induced cell death when infiltrated into Arabidopsis leaves (Andersson et al., 2015), we cannot completely exclude that the SFN-induced priming (Figs. 2A and 5A) and direct activation of defense genes (Fig. 5C) is mediated by incidental cell death. In addition, SFN directly inhibits the growth of many infectious bacteria (e.g. P. syringae) and fungi (e.g. Pl. cucumerina) in the absence of plant (Tierens et al., 2001). Therefore, we will not be able to distinguish whether a reduction of disease susceptibility to such pathogens in SFN-treated plants is due to defense priming in the plant, direct inhibition of the pathogen, or both these possibilities. The weaker toxicity of SFN to some other plant pathogens (e.g. Alternaria brassicicola and Botrytis cinerea) could be countered by its stimulating effects on ET-dependent immunity in the plant (Fig. 5, C–E).
Because ERY was less active, and ERU not active, at priming (Fig. 2), the oxidation state of sulfur in the side chain seems to be critical, and the –N=C=S moiety insufficient, for SFN’s priming activity. SFN and ERY (but not ERU) also induce activity of the phase-II detoxification enzymes quinone reductase and glutathione S-transferase in murine hepatoma cells (Zhang et al., 1992), suggesting a same or similar mode of action of these compounds in plants and mammals. Whether SFN activates quinone reductase and glutathione S-transferase in plants, and whether this possible activation is relevant to defense priming, remains to be seen. However, in seeming analogy to Arabidopsis, SFN inhibits histone deacetylase and increases histone acetylation while preventing prostate cancer (Myzak et al., 2004; 2006; Gibbs et al., 2009; Ho et al., 2009), further supporting histone modification as a key mode of action of SFN.
SFN enhanced H3K4me3 and H3K9ac in the promoter/promoter-proximal region of WRKY6 and the JA-responsive PDF1.2 gene (Fig. 3, A–D), whereas the SA mimic BTH reduced the two epi-marks in the PDF1.2 promoter (Fig. 3, C and D). By contrast, BTH, yet not SFN, enhanced H3K4me3 and H3K9ac in the promoter/promoter-proximal region of the SA-responsive PR1 gene (Fig. 3, E and F). Hence, the negative cross talk of the JA/ET and SA signaling pathways, which enables plants to fine-tune their immune response to pathogens with different lifestyles (Koornneef and Pieterse, 2008), seems to be under epigenetic control (Alvarez et al., 2010; Caarls et al., 2015).
SFN treatment caused unprimed PDF1.2 transcription before, and primed WRKY6 transcription after, flg22 challenge (Fig. 5, A and C). WRKY6 priming and PDF1.2 transcription by SFN coincide with H3K4me3 and H3K9ac (Fig. 3, A–D), chromatin unpacking in the promoter/promoter-proximal region of gene (Fig. 4, A and B), and reduced downy mildew disease (Fig. 6). These findings point to histone modification, enhanced DNA accessibility, and an antimicrobial effect as modes of SFN action in plant disease alleviation. H3K4me3, H3K9ac, and chromatin unpacking were found in the promoter of unprimed as well as primed genes (Figs. 3 and 4; Jaskiewicz et al., 2011a). Therefore, histone modifications other than H3K4me3 and H3K9ac, co-occurrence with other histone marks, regulatory RNAs, and/or nonhistone proteins (e.g. transcriptional coactivators, chromatin remodeling factors, histone variants) seem to determine whether transcription of a given gene will be unprimed or primed (Conrath, 2011; Badeaux and Shi, 2013; Weiner et al., 2016).
In crucifers, including Arabidopsis, SFN and other isothiocyanates upon tissue disintegration are released from glucosinolates (1-thio-β-d-glucosides) by endogenous S-glycosyl hydrolases (myrosinases; Wittstock and Halkier, 2002). Whereas glucosinolates are considered biologically inactive isothiocyanate storage forms, SFN and other isothiocyanates serve as insect feeding and/or oviposition stimulants or deterrents (Halkier and Gershenzon, 2006). However, glucosinolates and their metabolites have also been shown to cause plant cell death (Andersson et al., 2015) and inhibit the growth of various infectious microbes (e.g. Pl. cucumerina, P. syringae, and Phakopsora pachyrhizi; Tierens et al., 2001; Supplemental Fig. S1) directly. Therefore, we cannot distinguish whether a reduction of disease susceptibility to pathogens in SFN-treated plants is due to defense priming, direct inhibition of the pathogen, or both these possibilities.
Here, we discovered a new role of SFN linking the glucosinolate pathway to defense priming. In the tritrophic interaction of crucifers, insects, and oomycetes, our work suggests that insect feeding on crucifers causes release of SFN from the glucosinolate glucoraphanin by myrosinase. SFN then imposes epigenetic modifications to histone H3 in the plant, associated with enhanced accessibility of DNA in defense gene regulatory regions. This specific chromatin environment seems to prepare defense genes for unprimed or primed transcription. By doing so, the plant may reduce the risk of microbial infection after insect feeding.
SFN combines insect deterrent (Halkier and Gershenzon, 2006), antimicrobial (Tierens et al., 2001; Supplemental Fig. S1), defense priming (Figs. 2A and 5A), and defense gene inducing (Fig. 5C) activity within a same molecule. In addition, SFN is a natural compound and is present in substantial quantities in human diet (Li and Zhang, 2013). Therefore, the isothiocyanate presents a promising candidate for the development of a novel, nonhazardous plant protectant with a dual mode of action.
MATERIALS AND METHODS
Parsley cell cultures and Arabidopsis (Arabidopsis thaliana) wild-type Col-0 and ein2-1 plants were grown, kept, and treated as described by Katz et al. (1998) and Beckers et al. (2009). BTH and WP were provided by Syngenta.
High-Throughput Screening for Defense Priming Compounds in Parsley Cell Cultures
At 3 d after subculture, 1 mL aliquots of parsley cell suspension were transferred to a 24-well CELLSTAR microtiter plate (Greiner). Cells were either left untreated (negative control), treated with 200 µm SA in DMSO (0.25%; positive priming control), or treated with the given concentration of test compound in DMSO. Cell suspensions were shaken at 100 rpm and at 25°C in the dark. After 24 h, 50 pm of custom-synthesized Pep13 (Thermo Fisher Scientific) was added to appropriate aliquots of cell suspension. Addition of a same volume of water served as a control for the Pep13 treatment. After another 24 h, relative furanocoumarin fluorescence in the supernatant of cell suspension was determined in a microtiter plate reader (Tecan) at 335 nm excitation and 398 nm emission.
Determination of Defense Priming in Arabidopsis
At 24 h after spraying plants with WP, BTH in WP, or SFN in WP, three leaves of half of the plants of each treatment were infiltrated with 200 nm custom-synthesized flg22 (Thermo Fisher Scientific) in water. The other half of plants was infiltrated with a same volume of water. Forty-five min after infiltration, leaves were harvested and snap-frozen in liquid nitrogen. RNA extraction and quantification of mRNA transcript accumulation by RT-qPCR was performed as described by Beckers et al. (2009) using gene-specific primers (Supplemental Table S1).
FAIRE-qPCR
For FAIRE, an existing protocol (Simon et al., 2012) was optimized for fully developed Arabidopsis leaves. Leaf tips of appropriately treated plants were harvested and subjected to three repetitive vacuum infiltrations (1, 1.5, and 1 min) in cross-linking buffer (0.4 m Suc, 10 mm HEPES, pH 7.8, 5 mm β-mercaptoethanol, 0.1 mm PMSF, and 3% formaldehyde). Cross-linking was quenched by three more vacuum infiltrations (1, 1.5, and 1 min) in Gly (final concentration 125 mm). Leaf tips were washed twice with tap water, dried in between paper towels and thoroughly ground in liquid nitrogen. The leaf powder was equally split into three microfuge tubes. A quantity of 850 μL of DNA extraction buffer (0.1 m Tris-HCl, pH 8.0, 0.1 m NaCl, and 0.05 m EDTA, pH 8.0) was added to each tube and samples were thoroughly mixed on a bench shaker. Chromatin was sheared at 4°C by sonication in 10 cycles (30 s on, 30 s off) in a Bioruptor (Diagenode). Samples were centrifuged (5 min, 16,100g, RT) and supernatants for the same treatments pooled in the same tube, mixed on a bench shaker, split into three aliquots of 700 μL (FAIRE samples), and stored at −20°C. Additionally, three 80-μL aliquots of each 700-μL sample were supplemented with 540 μL DNA extraction buffer, and incubated overnight at 65°C to reverse cross-linking (input samples). On the next day, samples were centrifuged (5 min, 16,100g, RT), and supernatants removed and transferred to a fresh microfuge tube. Supernatants were supplemented with an equal volume of phenol-chloroform (phenol/chloroform/isoamyl alcohol, 25:24:1). Tubes were thoroughly mixed for 20 s and centrifuged for 15 min at 20,800g at 4°C. Aqueous phase was transferred to a fresh microfuge tube and supplemented with an equal volume of chloroform. Samples were thoroughly mixed and centrifuged again. Aqueous phase was transferred to a fresh microfuge tube and two volumes of ice-cold ethanol (96%) added. DNA was precipitated for 30 min at −20°C and recovered by centrifugation in a microfuge (20 min, 20,800g, 4°C). Supernatant was removed, and the DNA pellet washed in 70% ethanol and centrifuged for 15 min at 20,800g and 4°C. Supernatant was removed, pellet-dried for 45 min on the bench, dissolved in 200 μL double-deionized water, and incubated for 15 min at 70°C. DNA samples were used as templates for qPCR using gene-specific primers (Supplemental Table S2). Relative enrichment of amplified DNA was calculated for different sites in the gene of interest and the ACTIN2 reference gene as described (Louwers et al., 2009). Briefly, the relative enrichment for each gene position (relative enrichmentgene position), and the ACTIN2 reference gene (relative enrichmentACTIN2), were calculated using the following formula:
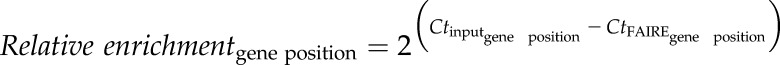 |
The relative enrichmentgene position was normalized to the relative enrichmentACTIN2 using the following formula:
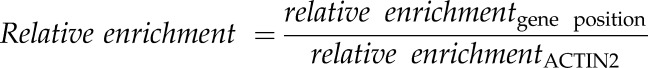 |
ChIP
ChIP analysis was performed as described by Haring et al. (2007) and Jaskiewicz et al. (2011b). Briefly, leaf tissue of appropriately treated plants was harvested and subjected to three repeated vacuum infiltrations (1.5 min, 1 min, 1.5 min) in cross-linking buffer (0.4 m Suc, 10 mm HEPES, pH 7.8, 5 mm β-Mercaptoethanol, 0.1 mm PMSF, and 1% formaldehyde). Cross-linking was quenched by three more vacuum infiltrations (1 min each) of Gly (final concentration 125 mm). Leaves were washed with tap water, dried in between paper towels and thoroughly ground in liquid nitrogen. Chromatin was extracted from ground leaf tissue as described by Bowler et al. (2004). Extracted chromatin was sonicated to fragments of approximately 400 bp. For minimizing unspecific background signals, 200 μL of sonicated chromatin was transferred to a 1.8-mL preclearing solution (50 mm Tris-HCl (pH 8.0), 1 mm EDTA-NaOH (pH 8.0), 150 mm NaCl, 0.1% Triton X-100, and 50 μL protein A agarose) in a 2-mL microfuge tube and incubated on an overhead shaker for 1 h at 4°C. For later normalization, 40 μL of the precleared chromatin solution was removed and stored at −20°C until further analysis (input sample). For immunoprecipitation of specific histone-DNA complexes, 400 μL of precleared chromatin solution was incubated in a 1.5-mL microfuge tube and in the presence of 40 μL of protein A agarose and the appropriate antibody (anti-H3K4me3 [Diagenode], anti-H3K9ac [Millipore; antibody sample]). After 2 h overhead rotation at 4°C, samples were centrifuged for 2 min at 2,000g and 4°C in a microfuge. Supernatants were discarded and the pellet subsequently washed for 10 min at 4°C with 900 μL of low salt buffer (20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.1% SDS, 1% Triton X-100, and 2 mm EDTA-NaOH, pH 8.0), 900 μL high salt buffer (20 mm Tris-HCl , pH 8.0, 500 mm NaCl, 0.1% SDS, 1% Triton X-100, and 2 mm EDTA-NaOH, pH 8.0), 900 μL LiCl buffer (20 mm Tris-HCl, pH 8.0, 1 mm EDTA-NaOH, pH 8.0, 250 mm lithium chloride, 1% Nonidet P-40, and 1% sodium deoxycholate), and twice with 900 μL TE buffer (10 mm EDTA-NaOH, pH 8.0, and 1 mm Tris-HCl, pH 8.0). Cross-linking was reversed by overnight incubation at 65°C in 100 μL of buffer G (62.5 mm Tris, pH 6.8, 200 mm NaCl, 2% SDS, and 10 mm DTT). DNA was isolated using the MSB Spin PCRapace kit (Stratec Molecular) and quantified by qPCR using gene-specific primers (Supplemental Table S2). The percentage (% IP) of the immunoprecipitated DNA sample (antibody sample) relative to the DNA sample without immunoprecipitation (input sample) was calculated for each gene position and antibody (anti-H3, anti-H3K4me3, and anti-H3K9ac) with the following formula:
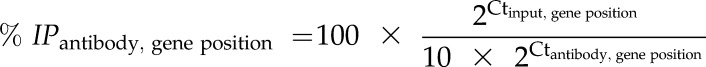 |
The factor 10 was used to normalize the different volumes used for input and antibody samples during ChIP (see the above text). For each gene position and antibody (anti-H3K4me3, anti-H3K9ac) the % IPantibody, gene position was normalized to the amount of histone H3 at the appropriate gene position (% IPH3, gene position) using the following formula:
 |
For a detailed written and visualized protocol of the ChIP procedure, see Jaskiewicz et al. (2011b).
Determining Arabidopsis Susceptibility to Hpa
Two-to-three-week-old Arabidopsis plants were left untreated or treated with WP or a WP formulation of SFN. At 24 h after treatment, plants were inoculated by spraying with a conidiospore suspension of Hpa (5 × 104 spores per mL of water). Alternatively, conidiospores (5 × 104 spores per mL) were suspended in water, WP, or a WP formulation of SFN. Spore suspensions were thoroughly mixed on a bench shaker and used for spray inoculation of 2- to 3-week-old Arabidopsis plants. In both cases (pretreatment and cotreatment), inoculated plants were covered with a transparent lid to ensure high humidity and kept in short day condition. At 7 d after inoculation, the number of spores released by Hpa was determined as described (Schmitz et al., 2010).
Accession Numbers
The Arabidopsis Genome Initiative accession number for ein2-1 is At5g03280.
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Gene-specific primers for measurement of mRNA transcript abundance by RT-qPCR.
Supplemental Table S2. Gene-specific primers used in FAIRE and ChIP.
Supplemental Figure S1. SFN inhibits P. pachyrhizi spore germination in vitro.
Acknowledgments
We thank Gerold J. M. Beckers, Laura Buglioni, Michal R. Jaskiewicz, and Caspar J. Langenbach for discussions. G. J. M. Beckers and C. Langenbach are thanked for advice, and Stefan Kusch and Anja Reinstädler for help with plant infections. We thank Eva Reimer-Michalski for composing the VTOC chart.
Footnotes
[OPEN]Articles can be viewed without a subscription.
References
- Aasland R, Gibson TJ, Stewart AF (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci 20: 56–59 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Alvarez ME, Nota F, Cambiagno DA (2010) Epigenetic control of plant immunity. Mol Plant Pathol 11: 563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson MX, Nilsson AK, Johansson ON, Boztaş G, Adolfsson LE, Pinosa F, Petit CG, Aronsson H, Mackey D, Tör M, Hamberg M, Ellerström M (2015) Involvement of the electrophilic isothiocyanate sulforaphane in Arabidopsis local defense responses. Plant Physiol 167: 251–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badeaux AI, Shi Y (2013) Emerging roles for chromatin as a signal integration and storage platform. Nat Rev Mol Cell Biol 14: 211–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett DW, Clough JM, Godwin JR, Hall AA, Hamer M, Parr-Dobrzanski B (2002) The strobilurin fungicides. Pest Manag Sci 58: 649–662 [DOI] [PubMed] [Google Scholar]
- Beckers GJM, Conrath U (2007) Priming for stress resistance: from the lab to the field. Curr Opin Plant Biol 10: 425–431 [DOI] [PubMed] [Google Scholar]
- Beckers GJM, Jaskiewicz M, Liu Y, Underwood WR, He SY, Zhang S, Conrath U (2009) Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21: 944–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J (2004) Chromatin techniques for plant cells. Plant J 39: 776–789 [DOI] [PubMed] [Google Scholar]
- Brunner F, Rosahl S, Lee J, Rudd JJ, Geiler C, Kauppinen S, Rasmussen G, Scheel D, Nürnberger T (2002) Pep-13, a plant defense-inducing pathogen-associated pattern from Phytophthora transglutaminases. EMBO J 21: 6681–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caarls L, Pieterse CMJ, Van Wees SCM (2015) How salicylic acid takes transcriptional control over jasmonic acid signaling. Front Plant Sci 6: 170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates ME, Beynon JL (2010) Hyaloperonospora arabidopsidis as a pathogen model. Annu Rev Phytopathol 48: 329–345 [DOI] [PubMed] [Google Scholar]
- Conrath U. (2011) Molecular aspects of defence priming. Trends Plant Sci 16: 524–531 [DOI] [PubMed] [Google Scholar]
- Conrath U, Beckers GJM, Flors V, García-Agustín P, Jakab G, Mauch F, Newman MA, Pieterse CM, Poinssot B, Pozo MJ, Pugin A, Schaffrath U, et al. ; Prime-A-Plant Group (2006) Priming: getting ready for battle. Mol Plant Microbe Interact 19: 1062–1071 [DOI] [PubMed] [Google Scholar]
- Conrath U, Beckers GJM, Langenbach CJG, Jaskiewicz MR (2015) Priming for enhanced defense. Annu Rev Phytopathol 53: 97–119 [DOI] [PubMed] [Google Scholar]
- Conrath U, Pieterse CMJ, Mauch-Mani B (2002) Priming in plant-pathogen interactions. Trends Plant Sci 7: 210–216 [DOI] [PubMed] [Google Scholar]
- Frost CJ, Mescher MC, Carlson JE, De Moraes CM (2008) Plant defense priming against herbivores: getting ready for a different battle. Plant Physiol 146: 818–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulton KJ, Nammo T, Pasquali L, Simon JM, Giresi PG, Fogarty MP, Panhuis TM, Mieczkowski P, Secchi A, Bosco D, Berney T, Montanya E, et al. (2010) A map of open chromatin in human pancreatic islets. Nat Genet 42: 255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs A, Schwartzman J, Deng V, Alumkal J (2009) Sulforaphane destabilizes the androgen receptor in prostate cancer cells by inactivating histone deacetylase 6. Proc Natl Acad Sci USA 106: 16663–16668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi PG, Lieb JD (2009) Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (formaldehyde assisted isolation of regulatory elements). Methods 48: 233–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkier BA, Gershenzon J (2006) Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 57: 303–333 [DOI] [PubMed] [Google Scholar]
- Haring M, Offermann S, Danker T, Horst I, Peterhänsel C, Stam M (2007) Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herms S, Seehaus K, Koehle H, Conrath U (2002) A strobilurin fungicide enhances the resistance of tobacco against tobacco mosaic virus and Pseudomonas syringae pv tabaci. Plant Physiol 130: 120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho E, Clarke JD, Dashwood RH (2009) Dietary sulforaphane, a histone deacetylase inhibitor for cancer prevention. J Nutr 139: 2393–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes J, Rueber D (2007) Soybean yield response to Headline fungicide application. Iowa State Research Farm Progress Reports. http://www.ag.iastate.edu/farms/05reports/n/SoybeanYield.pdf
- Jaskiewicz M, Conrath U, Peterhänsel C (2011a) Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep 12: 50–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskiewicz M, Peterhänsel C, Conrath U (2011b) Detection of histone modifications in plant leaves. J Vis Exp 55: e3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Kanno Y, Siegel RM, Jang MK, Lenardo MJ, Ozato K (2004) Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol Cell 13: 33–43 [DOI] [PubMed] [Google Scholar]
- Katz VA, Thulke OU, Conrath U (1998) A benzothiadiazole primes parsley cells for augmented elicitation of defense responses. Plant Physiol 117: 1333–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauss H, Theisinger-Hinkel E, Mindermann R, Conrath U (1992) Dichloroisonicotinic and salicylic acid, inducers of systemic acquired resistance, enhance fungal elicitor responses in parsley cells. Plant J 2: 655–660 [Google Scholar]
- Koehle H, Conrath U, Herms S, Schlundt T, Johnson N, Stammler G (2003) Method for immunizing plants against bacterioses. Patent WO2003075663
- Koehle H, Conrath U, Seehaus K, Niedenbrueck M, Tavares-Rodrigues M-A, Sanchez W, Begliomini E, Oliveira C (2006) Method of inducing virus tolerance of plants. US Patent 20060172887
- Kohler A, Schwindling S, Conrath U (2002) Benzothiadiazole-induced priming for potentiated responses to pathogen infection, wounding, and infiltration of water into leaves requires the NPR1/NIM1 gene in Arabidopsis. Plant Physiol 128: 1046–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef A, Pieterse CMJ (2008) Cross talk in defense signaling. Plant Physiol 146: 839–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberth C, Jeanmart S, Luksch T, Plant A (2013) Current challenges and trends in the discovery of agrochemicals. Science 341: 742–746 [DOI] [PubMed] [Google Scholar]
- Lång V, Palva ET (1992) The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 20: 951–962 [DOI] [PubMed] [Google Scholar]
- Leadbeater A, Staub T (2014) Exploitation of induced resistance: a commercial perspective. In DR Walters, AC Newton, GF Lyon, eds, Induced Resistance for Plant Defense: A Sustainable Approach to Crop Protection, Ed 2 Wiley & Sons, Chichester, UK, pp 300–315 [Google Scholar]
- Li B, Carey M, Workman JL (2007) The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang T (2013) Targeting cancer stem cells with sulforaphane, a dietary component from broccoli and broccoli sprouts. Future Oncol 9: 1097–1103 [DOI] [PubMed] [Google Scholar]
- López A, Ramírez V, García-Andrade J, Flors V, Vera P (2011) The RNA silencing enzyme RNA polymerase v is required for plant immunity. PLoS Genet 7: e1002434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louwers M, Bader R, Haring M, van Driel R, de Laat W, Stam M (2009) Tissue- and expression level-specific chromatin looping at maize b1 epialleles. Plant Cell 21: 832–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna E, Bruce TJA, Roberts MR, Flors V, Ton J (2012) Next-generation systemic acquired resistance. Plant Physiol 158: 844–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Medina A, Flors V, Heil M, Mauch-Mani B, Pieterse CM, Pozo MJ, Ton J, van Dam NM, Conrath U (2016) Recognizing plant defense priming. Trends Plant Sci 21: 818–822 [DOI] [PubMed] [Google Scholar]
- Mascarelli A. (2013) Growing up with pesticides. Science 341: 740–741 [DOI] [PubMed] [Google Scholar]
- Myzak MC, Hardin K, Wang R, Dashwood RH, Ho E (2006) Sulforaphane inhibits histone deacetylase activity in BPH-1, LnCaP and PC-3 prostate epithelial cells. Carcinogenesis 27: 811–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myzak MC, Karplus PA, Chung FL, Dashwood RH (2004) A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer Res 64: 5767–5774 [DOI] [PubMed] [Google Scholar]
- Noutoshi Y, Okazaki M, Kida T, Nishina Y, Morishita Y, Ogawa T, Suzuki H, Shibata D, Jikumaru Y, Hanada A, Kamiya Y, Shirasu K (2012) Novel plant immune-priming compounds identified via high-throughput chemical screening target salicylic acid glucosyltransferases in Arabidopsis. Plant Cell 24: 3795–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IA, Eggermont K, Terras FR, Thomma BP, De Samblanx GW, Buchala A, Métraux JP, Manners JM, Broekaert WF (1996) Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8: 2309–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruess W, Mueller K, Knauf-Beiter G, Kunz W, Staub T (1996) Plant activator CGA 245704: an innovative approach for disease control in cereals and tobacco. In Proceedings, Brighton Crop Protection Conference, Pests and Diseases, Brighton, UK, Nov. 18–21. British Crop Protection Council, Farnham, UK, pp 53–60 [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner H-Y, Hunt MD (1996) Systemic acquired resistance. Plant Cell 8: 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling JV, Schillheim B, Mahr S, Reufer Y, Sanjoyo S, Conrath U, Büchs J (2015) Oxygen transfer rate identifies priming compounds in parsley cells. BMC Plant Biol 15: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz G, Reinhold T, Göbel C, Feussner I, Neuhaus HE, Conrath U (2010) Limitation of nocturnal ATP import into chloroplasts seems to affect hormonal crosstalk, prime defense, and enhance disease resistance in Arabidopsis thaliana. Mol Plant Microbe Interact 23: 1584–1591 [DOI] [PubMed] [Google Scholar]
- Siegrist J, Mühlenbeck S, Buchenauer H (1998) Cultured parsley cells, a model system for the rapid testing of abiotic and natural substances as inducers of systemic acquired resistance. Physiol Mol Plant Pathol 53: 223–238 [Google Scholar]
- Simon JM, Giresi PG, Davis IJ, Lieb JD (2012) Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat Protoc 7: 256–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateda C, Zhang Z, Shrestha J, Jelenska J, Chinchilla D, Greenberg JT (2014) Salicylic acid regulates Arabidopsis microbial pattern receptor kinase levels and signaling. Plant Cell 26: 4171–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thulke O, Conrath U (1998) Salicylic acid has a dual role in the activation of defence-related genes in parsley. Plant J 14: 35–42 [DOI] [PubMed] [Google Scholar]
- Tierens KFM-J, Thomma BPHJ, Brouwer M, Schmidt J, Kistner K, Porzel A, Mauch-Mani B, Cammue BPA, Broekaert WF (2001) Study of the role of antimicrobial glucosinolate-derived isothiocyanates in resistance of Arabidopsis to microbial pathogens. Plant Physiol 125: 1688–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992) Acquired resistance in Arabidopsis. Plant Cell 4: 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UN Food and Agriculture Organization (2009) Global agriculture towards 2050. Proceedings of the High-Level Expert Forum, Rome, October 12-13, 2009
- van Hulten M, Pelser M, van Loon LC, Pieterse CMJ, Ton J (2006) Costs and benefits of priming for defense in Arabidopsis. Proc Natl Acad Sci USA 103: 5602–5607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A, Lara-Astiaso D, Krupalnik V, Gafni O, David E, Winter DR, Hanna JH, Amit I (2016) Co-ChIP enables genome-wide mapping of histone mark co-occurrence at single-molecule resolution. Nat Biotechnol 34: 953–961 [DOI] [PubMed] [Google Scholar]
- Wittstock U, Halkier BA (2002) Glucosinolate research in the Arabidopsis era. Trends Plant Sci 7: 263–270 [DOI] [PubMed] [Google Scholar]
- Zarate SI, Kempema LA, Walling LL (2007) Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol 143: 866–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Talalay P, Cho CG, Posner GH (1992) A major inducer of anticarcinogenic protective enzymes from broccoli: isolation and elucidation of structure. Proc Natl Acad Sci USA 89: 2399–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]