Plant lipid droplets and their associated oleosin and other proteins have evolved and diversified in functions and cell species specificities and have been probed for commercial uses.
Abstract
Cytoplasmic lipid droplets (LDs) of neutral lipids (triacylglycerols [TAGs], sterylesters, etc.) are reserves of high-energy metabolites and other constituents for future needs. They are present in diverse cells of eukaryotes and prokaryotes. An LD has a core of neutral lipids enclosed with a monolayer of phospholipids and proteins, which play structural and/or metabolic roles. During the past 3 decades, studies of LDs in diverse organisms have blossomed after they were found to be involved in prevalent human diseases and industrial uses. LDs in plant seeds were studied before those in mammals and microbes, and the latter studies have since moved forward. Plant LDs carry a hallmark protein called oleosin, which has a long hydrophobic hairpin penetrating the TAG core and stabilizing the LD. The oleosin gene first appeared in green algae and has evolved in enhancing promoter strength, tandem repeats, and/or expression specificity, leading to the appearance of new LD organelles, such as tapetosomes in Brassicaceae. The synthesis of LDs occurs with TAG-synthesizing enzymes on the endoplasmic reticulum (ER), and nascent TAGs are sequestered in the acyl moiety region between the bilayers of phospholipids, which results in ER-LD swelling. Oleosin is synthesized on the cytosol side of the ER and extracts the LD from the ER-LD to cytosol. This extraction of LD to the cytosol is controlled solely by the innate properties of oleosin, and modified oleosin can redirect the LD to the ER lumen and then vacuoles. The breakdown of LDs requires lipase associating with core retromer and binding to peroxisomes, which then send the enzyme to LDs via tubular extensions. Two groups of LD-associated proteins, caleosin/dioxygenase/steroleosin and LD/oil body-associated proteins, participate in cellular stress defenses via enzymic activities and binding, respectively. The surface of LDs in all plant cells may be an inert refuge for these and other proteins, which exert functions on diverse cell components. Oleosin-LDs have been explored for commercial applications; successes in their uses will rely on overcoming conceptual and technical difficulties.
Cytoplasmic lipid droplets (LDs) of neutral lipids are present in diverse cells of eukaryotes and prokaryotes. An LD has a core of neutral lipids (usually triacylglycerols [TAGs] and steryl/wax esters) enclosed by a monolayer of phospholipids (PLs). Inserted into or attached to the surface of LDs are proteins, which, in general, play structural and/or metabolic roles. LDs usually represent reserves of high-energy metabolites and other constituents for future metabolic needs. Vegetative oils from plant LDs have been used for cooking, soaps, detergents, paints, and other products for centuries. During the past three decades, studies of LDs in diverse organisms have blossomed after they were found to be involved in prevalent human diseases and industrial utilizations. LDs are involved directly in obesity (Onal et al., 2017), genetic defects leading to disturbed LD homeostasis (Barbosa and Siniossoglou, 2017), storage of proteins for rapid development or specific metabolic needs (Welte and Gould, 2017), and as a food and assembly platform for pathogens (Nolan et al., 2017; Roingeard and Melo, 2017). Plant and microbe LDs can become warehouses for the potential industrial manufacture of pharmaceuticals and other high-value products (Garay et al., 2014; Matos, 2017) as well as renewable biodiesels (Taparia et al., 2016).
In recent years, the term LD has been adopted by most laboratories to replace the traditional names oil body, oil droplet, spherosome, oleosome, adiposome, etc. The number of publications on LDs has grown exponentially; 38, 1,125, and 5,650 in 1986, 1992, and 2016, respectively (Tirinato et al., 2017). LDs were studied extensively in plant seeds before mammals and microbes. Initial characterizations of the genes encoding LD proteins in plants, mammals, and bacteria were reported in 1987, 1993, and 1994, respectively (Vance and Huang, 1987; Greenberg et al., 1993; Pieper-Fürst et al., 1994).
LDs in diverse organisms have been reviewed, including those in nonplant organisms (Koch et al., 2014; Kory et al., 2016; Barbosa and Siniossoglou, 2017; Barneda and Christian, 2017; Chen and Goodman, 2017; Thiam and Beller, 2017; Walther et al., 2017; Welte and Gould, 2017) and plants (Chapman et al., 2012; Jolivet et al., 2013; Shimada and Hara-Nishimura, 2015; Pyc et al., 2017; Song et al., 2014). This article first provides succinct information on LDs in diverse organisms, with the intention of introducing to the plant community the latest advances as well as a generalization and comparison of LDs in all organisms. It will then emphasize plant LDs with oleosins and other proteins as well as their potential industrial applications. Relevant topics for future research will be suggested.
COMMON FEATURES OF LDs AND LD PROTEINS IN ALL ORGANISMS
LDs in all organisms share common features (see refs. in preceding section). Each LD has a neutral lipid core, which could be TAGs and/or steryl/wax esters, polyisoprene (rubber), or polyesters (in bacteria), etc. (For convenience, subsequent descriptions will use TAGs as a general term to represent some or all of these neutral lipids.) The core is enclosed by a monolayer of PLs (Fig. 1A). Inserted into the core or wrapped around the surface are proteins that serve to stabilize the LDs and exert metabolic functions. Only two LD proteins, oleosin and microalgae surface LD protein, have a hydrophobic polypeptide (72 and 40–60 residues, respectively) long enough to insert into the hydrophobic core (Table I). All other proteins have a short or no hydrophobic segment and may interact loosely or transiently with the surface of an LD or piggyback other LD proteins. LD proteins can be grouped according to their single, dual, or multiple functions of (1) structural, (2) enzymic (neutral lipid synthesis and degradation) and enzymic activity modulation, (3) membrane-protein trafficking (for detachment of LDs from the endoplasm reticulum [ER], fusion with other LDs, or interaction with the ER or other organelles), (4) signaling, and (5) using the LD surface as a refuge or platform for other cellular activities. Some of these proteins are ubiquitous in cells with and without LDs and are present on LDs and other subcellular compartments of the same cell.
Figure 1.
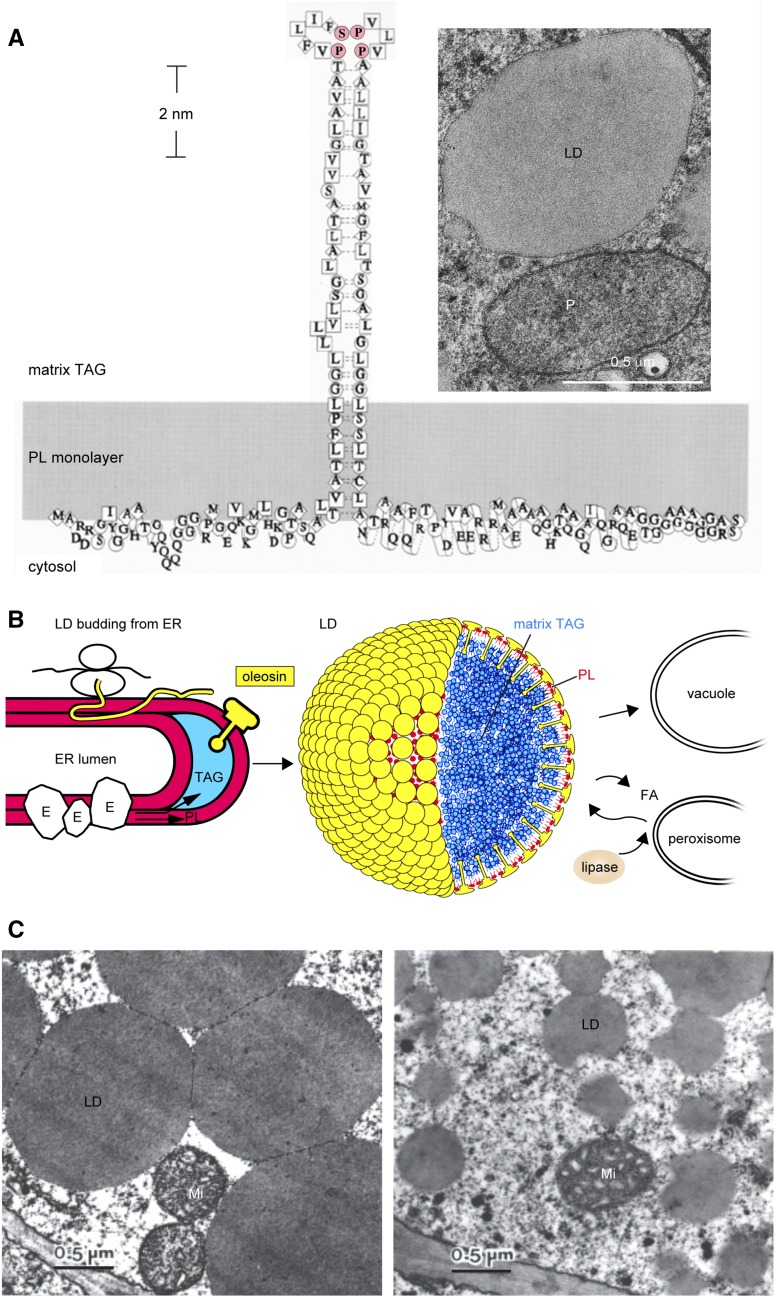
Seed LDs and oleosin. A, Model of an oleosin molecule on the surface of an LD (Tzen and Huang, 1992). The shaded area represents a monolayer of PLs with the head groups facing the cytosol. Symbols of amino acids and their hydropathy indices are as follows: square for very hydrophobic (4.5–3.8), diamond for hydrophobic (2.8–1.8), circle for amphipathic (−0.4 to −1.6), and no enclosure for hydrophilic (−3.2 to −4.5) residues. The N-terminal segment is shown without any well-defined structure. The central hydrophobic segment is depicted as a hairpin with a Pro knot at the loop (P and S in pink); its secondary structure is unknown. The C-terminal segment is an amphipathic α-helical structure interacting with the PL surface and a variable-length nonconserved peptide at the terminus. The monolayer of PL enclosing an LD is revealed in the inset electron micrograph of a shoot apex cell of a 1-d-old maize seedling (courtesy of Richard Trelease). LD, peroxisome (P), and plastid (at the top right corner; not labeled) are enclosed with one, two, and four electron-dense lines, representing one (one half-unit membrane), two (one unit membrane), and four (double membrane) PL layers, respectively. B, Model of LD synthesis during seed maturation and degradation during seed germination. Oleosin is shaped like a mushroom, which includes the mushroom head (N- and C-terminal amphipathic segments) and a stalk (the central hydrophobic hairpin; courtesy of Ariel Kuan [Tzen and Huang, 1992]). Left, Budding LD on rough ER during seed maturation. The overall structure includes the ER lumen, two PL layers (red), sequestered TAGs (blue), a ribosome with an mRNA synthesizing an oleosin polypeptide (thin yellow ribbon, of unknown configuration), and ER enzymes (irregular spheres [E]) for the synthesis of TAGs and PLs. All constituents are not drawn to scale. Middle, LD model, with oleosin (yellow) and PLs (red) enclosing the matrix TAGs (blue). All three types of molecules are drawn approximately to scale, but the diameter of the LD has been reduced 24 times to reveal clearly the surface structure. Right, LD degradation during germination. Lipase associated with core retromer binds to peroxisome, which transfers the lipase to LD for lipolysis; the enzymic product fatty acid (FA) is transferred to peroxisome for gluconeogenesis. A TAG-depleted ghost may join with the vacuole membrane. Alternatively, the whole LD is engulfed by a vacuole. All constituents are not drawn to scale. C, Electron micrographs of LDs in embryos of two maize lines with diverse TAG-oleosin ratios. LDs in Illinois High Oil line (with a high ratio of TAGs to oleosin) are larger and spherical. LDs in Illinois Low Oil line (with one-seventh the TAG-oleosin ratio) are smaller and have an irregularly contoured surface. Double-membraned mitochondria (Mi) are present. LDs isolated from the two lines are stable without coalescence or aggregation and maintain their respective sizes and shapes (Ting et al., 1996).
Table I. LD proteins in plants and photosynthetic microbes.
The main emphasis is on plants. Photosynthetic microbe proteins are shown for basic comparisons. The information is simplified for generalization; exceptions occur. Transient LD proteins, including lipase, phospholipase, and lipoxygenase, are not included but are described in the text. Genes encoding all LD proteins of Arabidopsis are listed in Table III. In the hydrophobic sequences, X represents a nonhydrophilic residue.
| Protein | Molecular Mass of Most Proteins; Phylogenic Distribution | No. of Amino Acids in Longest Hydrophobic Hairpin; Sequence; Conserved | Located on LDs | In Cell/Tissue without LD | Abundance on LD | Known Function, Binding Motif |
|---|---|---|---|---|---|---|
| Plants | ||||||
| Oleosina | 15–25 kD; green algae and plants | 72; X30PX5SPX3PX30; highly conserved | Yes | No | Cover LD completely | Structural, etc. |
| Caleosinb | 20–23 kD; dinoflagellate, fungi, green algae, and plants | 21; X8PX3PSPX2PX3; mildly conserved | Yes; likely also on ER, etc. | Yes; ubiquitous | <5% of oleosin in seed | Ca2+-binding motif, phosphorylation sites, peroxygenase activity |
| Steroleosinc | 40–41 kD; plants, unknown in other organisms | 24; X3PPX10PX8; mildly conserved | Yes; likely also on ER, etc. | Yes; mostly in seed | <5% of oleosin in seed | Sterol- and NADP-binding motifs, sterol-NADPH dehydrogenase |
| Dioxygenased | 73 kD; bacteria, fungi, plants, and animals | No long hydrophobic segment | Yes; likely more in cytosol | Yes; ubiquitous, highest in root | <1% of oleosin in seed | Heme-binding site, dioxygenase, induced by stresses, senescence |
| LD Associated Proteine | ∼25 kD; green algae and plants | No long hydrophobic segment | Yes; likely more in cytosol | Yes; ubiquitous, highest in seed | Unknown | Related to stresses |
| OB Associated Proteinf | ∼27 kD; bacteria, fungi, and plants | No long hydrophobic segment | Yes; LD < < cytosol | No; mostly in seed | Unknown | Related to stresses |
| Photosynthetic microbes | ||||||
| LD Surface Proteing | 40–60 kD; heterokont algae (diatoms, Nannochloropsis spp.) | 56; X7PX9PX10PX3PX23; highly conserved | Yes | |||
| Major LD Proteinh | 30 kD; green algae and some other algae | No long hydrophobic segment | Unknown | |||
| Oleosin | 15–25 kD; only green algae | See above | ||||
Mostly from Huang and Huang (2015).
Mostly from Naested et al. (2000) and Song et al. (2014).
Mostly from Pasaribu et al. (2016).
Shimada et al. (2014); searched by C.Y. Huang.
LDs are derived from the ER or the enclosing membrane in bacteria. TAG-synthesizing enzymes are present on diverse areas or subdomains of the ER or bacterial membrane or directly on the LD surface. Nascent TAGs are sequestered in the hydrophobic region between the bilayer of PLs or other membrane components, which results in a bud or swelling. The bud or swelling detaches from the ER or remains attached to the ER permanently. Diversification, enlargement, or degradation of LDs could occur via different mechanisms (Barneda and Christian, 2017; Thiam and Beller, 2017; Walther et al., 2017). LDs are degraded as needed under specific metabolic or physiological conditions. Two major mechanisms are recognized (Barbosa and Siniossoglou, 2017; Walther et al., 2017). First, preexisting and then activated lipase, or subsequently added active lipase on the LD surface catalyzes lipolysis. Second, LDs are engulfed via autophagy or by vacuoles.
LD ISOLATION AND THE BLURRED DEFINITION OF AN LD PROTEIN
The procedures for isolating LD fractions from different organisms are similar. After gentle cell breakage in a suitable buffer, the extract is centrifuged with an overlay to yield a floated LD fraction. The overlay acts as a wash that could include high salt, detergent, chaotic agent, organic solvent, etc. The overall objective is to obtain LDs without contaminants. The LD fraction is subjected to proteomics, lipidomics, and other studies.
An LD fraction so prepared would likely contain contaminating proteins that bind preferentially but fortuitously during the preparation. This contamination could be pinpointed and confirmed if the protein in question is known to be present solely in another subcellular compartment. In some cells of an organ or within the same cell, some or all LDs could still be associated with its original ER. When an LD preparation containing these ER-LD fragments is subjected to proteomics and lipidomics, some of the detected proteins and PLs could belong to the ER at the ER-LD junction and not those authentically associated with LDs. The desire to avoid contaminants in an LD fraction is understandable. However, some proteins in an LD fraction perceived to be contaminants may not be after all. For example, massive histones in Drosophila (Drosophila melanogaster) embryos are present authentically on LDs for temporary storage to be used for active nuclei production (Li et al., 2012). Or they may be virus core proteins using the LD surface as a platform for virus assembly (Roingeard and Melo, 2017). Some LD proteins are hydrophilic or amphipathic and do not bind to LDs tightly and actually are present in higher proportions in cytosol or other subcellular locations of the same cell or in cells containing no LDs. Whether these proteins are authentic LD proteins is debatable. If researchers opt for the cleanest possible LD fractions, they may lose opportunities to make new discoveries. It is a dilemma. In future studies, relatively clean and not-so-clean LD preparations could be obtained in parallel in the same experiment, their proteins and lipids compared, and the proteins in question examined by immunoconfocal microscopy.
LDs IN MAMMALS AND DROSOPHILA
Mammalian LDs have ∼50 LD proteins, although many are not present in the same cell or at the same developmental stage (Onal et al., 2017). LDs are synthesized on the ER and then detached, with the participation of Fat Storage Inducing Protein and SEIPIN protein (Walther et al., 2017). For LD degradation, lipase on LDs is activated with phosphorylation and interaction with a phosphorylated perilipin, the major LD protein in all vertebrates (Itabe et al., 2017). The activation also requires interaction with an additional protein called Comparative Gene Identification-58 (CGI-58; Oberer et al., 2011), which by itself may possess very mild enzyme activities of hydrolases and lysophosphatidate acyltransferase. These latter enzyme activities could produce signal molecules of phosphatidic acid and lysophosphatidic acid.
In a healthy person, LDs in cells act as centers to maintain lipid and energy homeostasis (Barbosa and Siniossoglou, 2017). When this homeostasis is upset by stresses or genetic defects, diseases occur (Onal et al., 2017). The diseases could include obesity (accumulation of LDs), fatty liver diseases (increase of LDs related or unrelated to excessive alcohol consumption), cholesterol ester storage disease (reduction of LDs and cholesterol ester degradation), atherosclerosis (increase of LDs possessing cholesterol esters), and lipodystrophies (reduction of LDs). In some forms of cancer, the cells increase in lipid synthesis, which results in LD accumulation of unknown physiological relevance (Tirinato et al., 2017). In neurodegeneration, mitochondrial dysfunction occurs due to the accumulation of reactive oxygen species, thereby resulting in LD accumulation in the adjacent glia cells, again of unknown physiological relevance (Welte, 2015).
LDs could be taken advantage of by intracellular pathogens (Nolan et al., 2017; Roingeard and Melo, 2017). After entering a cell, bacteria (e.g. Toxoplasma gondii) could use existing LDs as nutrients for growth and reproduction. Viruses (e.g. hepatitis C) may place their core protein on the LD surface for the assembly of new viruses. An antivirus mechanism in the host also exists. An innate antivirus protein (viperin) could be present on the LD surface and bind to the virus core protein and suppress viral replication.
The surface of LDs in Drosophila and some cultured mammalian cells can become a refuge for proteins for future use (Li et al., 2012; Welte and Gould, 2017). For example, in Drosophila embryos, special groups of histones accumulate on LDs and are used for the rapid synthesis of nuclei and cells during future development. These proteins can shuttle between the LD surface and the nuclei.
LDs IN MICROBES
When microbes are under environmental stress, such as nitrogen starvation in experimentation, they produce massive LDs, up to ∼50% of the cell weight, for future growth. These LDs possess two groups of surface proteins: enzymes for the metabolism of the core neutral lipids and proteins that play a structural role or act as enzyme activity modulators. Although some of these LD proteins are termed major, their abundance on LDs has not been demonstrated.
Bacteria
Most genera of bacteria produce LDs containing polyesters (poly 3-hydroxybutyrate and polyhydroxyalkanoates; Wältermann and Steinbüchel, 2005). These LDs contain an LD protein called phasin (Pieper-Fürst et al., 1994; Mezzina and Pettinari, 2016). Phasin does not have a long hydrophobic segment and is assumed to be associating with the LD surface rather than penetrating the LD core. Phasin somehow controls the number of LDs and activates LD-associated enzymes such as polyhydroxyalkanoate synthase and depolymerase. Bacteria of some other genera possess TAG-containing LDs and a surface protein called Major LD Small Protein (Ding et al., 2012; Zhang et al., 2017). The protein does not have a long hydrophobic segment and could bind to a specific regulator protein, which, in turn, binds to and activates the promoter of an operator of two genes encoding Major LD Small Protein and the regulator protein. This activation occurs in response to stresses and enhances the survival of the bacteria. Whether this direct gene activation also occurs with LDs in the nuclei of some mammalian cells (Welte, 2015) should be explored.
Yeast
Yeast LDs contain ∼25 proteins (Koch et al., 2014). Many of the proteins are enzymes for the synthesis and degradation of TAGs and sterylesters. They also include the regulatory protein CGI-58, which is present in all eukaryotes. Yeast LDs do not have structural proteins, but the LDs perform their functions properly. Oleosin or perilipin in transformed yeast targets LDs; the foreign protein replaces a full complement of the LD proteins and increases the number or size of LDs (Ting et al., 1997: Jacquier et al., 2013).
Microalgae
Neutral lipids of TAGs in LDs in microalgae have been actively studied because they are potential biodiesels. Three LD structural proteins in different microalgae have been identified (Table I). The first, LD Surface Protein, is present in Nannochloropsis spp. (Vieler et al., 2012), diatoms, and several other heterokont algae (Yoneda et al., 2016). This is the only LD protein other than oleosin in all organisms that has a long hydrophobic hairpin (∼56 residues with conserved Pro residues at the center; Table I). The second LD structural protein, termed Major LD Protein, is present in Chlamydomonas reinhardtii and some other microalgae (Moellering and Benning, 2010; Davidi et al., 2012). This protein does not possess a long hydrophobic segment and is assumed to be loosely bound to the LDs or ER subdomains at the ER-LD junction (Huang et al., 2013b). Whether these two LD proteins are abundant, covering the whole surface of an LD, has not been established. The third LD structural protein is oleosin in green algae. Oleosin is present minutely in C. reinhardtii and moderately in the more advanced Spirogyra grevilleana. Oleosin evolved in green algae, and its amount apparently is far from sufficient to cover the whole surface of algal LDs (Huang et al., 2013b). In addition to these three LD structural proteins, microalgae LDs (or ER, subdomains of ER, or ER-LD junctions) contain enzymes for the synthesis, degradation, acyl exchange, and lipid trafficking between LDs and the ER or plastids (Goold et al., 2015; Zienkiewicz et al., 2016). In applications, microalgae have been used for health food, fish feed, cosmetics, and others (Matos, 2017). The feasibility of using microalgae to produce biodiesels without government subsidies and public relations benefit remains unclear (Taparia et al., 2016).
LD PROTEINS IN PLANTS
Plant LD proteins can be divided into three groups (Table I): (1) oleosins playing structural and perhaps also docking site roles; (2) caleosin, dioxygenase, and steroleosin exerting enzyme activities to combat stresses; and (3) LD Associated Proteins and Oil Body (OB) Associated Protein with no hydrophobic segment or enzymic activity involved in responses to stresses. In addition, they may include TAG-synthesizing enzymes on LDs or ER-LD junctions and TAG-degrading enzymes (lipase) on LDs. The amount of oleosin is sufficient to cover the complete surface of a seed LD for a structural role, and the amounts of the other proteins could be relatively minute.
Oleosin Basic Characteristics
Oleosin is a small protein, mostly of 15 to 26 kD (Table I; Huang and Huang, 2015). On an LD, it has short amphipathic N- and C-terminal peptides orienting horizontally on or extending from the LD surface and a conserved central hydrophobic hairpin of ∼72 uninterrupted noncharged residues penetrating the LD core (Fig. 1A; Huang, 1992). The hairpin has two arms of ∼30 residues each linked with a loop of 12 most-conserved residues (X30PX5SPX3PX30, with X representing a large nonpolar residue in X5 and X3 and a nonhydrophilic residue in X30). The secondary structures of the hairpin have not been definitively determined or agreed upon (Alexander et al., 2002; Li et al., 2002; Gohon et al., 2011; Vindigni et al., 2013; Vargo et al., 2014; Huang and Huang, 2017). Technical and conceptual difficulties exist in the use of Fourier transform infrared and other physical means to determine the secondary structures of the hairpin on artificial LDs. These approaches could miss the native arrangements of the oleosins on an LD: (1) the N- and C-terminal peptides on an LD could hold the underlying hairpin to specific secondary structures; (2) the high density of oleosins on a native LD could dictate interactions of the hairpins of two adjacent oleosins with defined secondary structures; and (3) seed LDs may have two seed oleosin isoforms (SH oleosin and SL oleosin [see next paragraph]) existing in a dimer or multidimer arrangement. Regardless of its secondary structure, the hairpin is 5 to 6 nm long, penetrating the TAG core and stabilizing the whole LD (Fig. 1A).
Oleosin Lineages
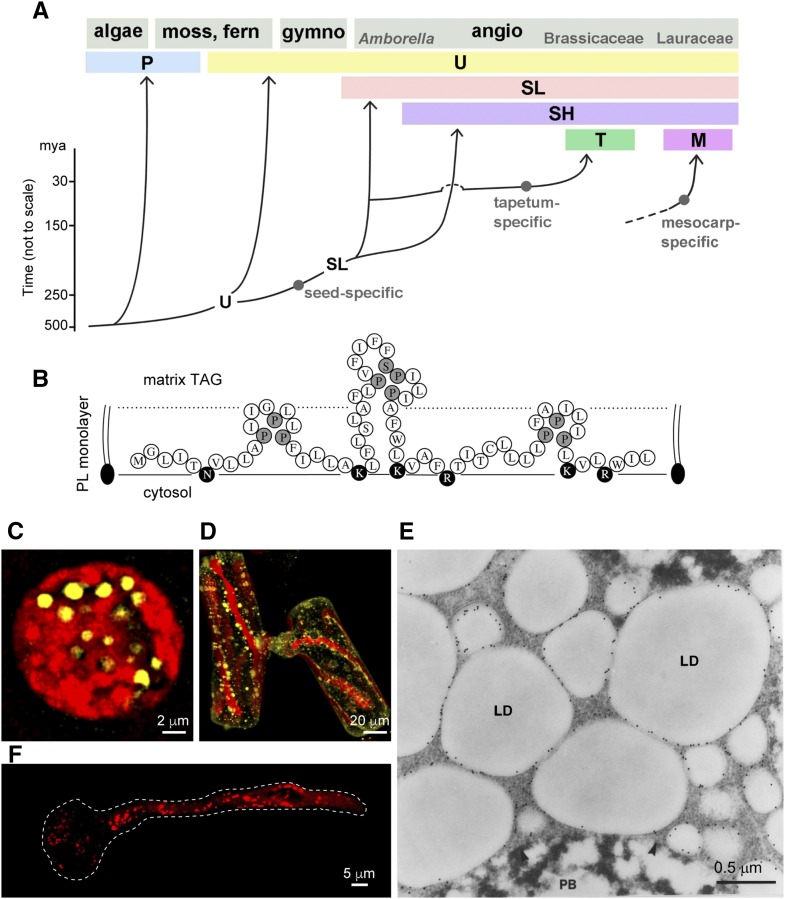
The hallmark of oleosin is its long hydrophobic hairpin. Bioinformatics analysis of 1,000 oleosins of diverse plants and green algae shows no exception to their hairpin sequence of approximately X30PX5SPX3PX30 (Huang and Huang, 2015). No pseudogene has been found, with the exception of dozens in Pinus and Picea spp., which have giant, highly redundant genomes (∼22,000 Mb versus 135 Mb in Arabidopsis [Arabidopsis thaliana]). Oleosin evolved in green algae, the ancestor of modern plants. The two modern, primitive green algae, C. reinhardtii and Volvox carteri, have a transcribed gene encoding an oleosin-like protein, which possesses one characteristic oleosin hairpin loop (PX5SPX3P) and two smaller loops, plus shorter hairpin arms (Fig. 2B; Huang et al., 2013b). This protein could be the precursor of oleosin or a remnant of a degenerating oleosin. In the two algae, the oleosin-like protein transcript is more abundant than the authentic oleosin transcript, even though both transcripts are at barely detectable levels. From primitive unicellular green algae, oleosin has evolved into new lineages associated with specialized LDs in plants of distinct phylogeny. At least six oleosin lineages (P, U, SL, SH, T, and M) are recognized (Fig. 2A). P oleosins evolved in green algae and are present in modern primitive species from green algae to ferns. They gave rise to U oleosins, whose genes are present universally in all species from mosses to advanced plants. U oleosins branched off to become specialized oleosins, which include the SL oleosins in all seed plants and the SH oleosins in angiosperms, the T oleosin in Brassicaceae, and the M oleosin in Lauraceae (Table II). Despite evolutionary diversifications, oleosins in all lineages maintain the conserved hairpin sequence, and their differences in the N- and C-terminal sequences show no appreciable correlation with the tissue or functional specifications of each lineage.
Figure 2.
Evolution of the six oleosin lineages and LDs housing some of these oleosins. A, Cartoon of the evolution of six oleosin lineages: P (primitive), U (universal), SL (seed low molecular weight), SH (seed high molecular weight), T (tapetum in Brassicaceae), and M (mesocarp in Lauraceae). The origin of M oleosin is unknown and is shown with a broken line. (Modified from Huang and Huang [2015].) B, Predicted polypeptide organization of an oleosin-like protein on the surface of an LD in the primitive green algae C. reinhardtii and V. carteri. The polypeptide has one hydrophobic hairpin with a loop identical to that of oleosin and two adjacent hairpins with less similar loops. Pivotal Pro and Ser residues are shaded, and hydrophilic residues are darkened. All three hairpins are shorter than the oleosin hairpin (for comparison, see Fig. 1A). The cytosol, the surface PL monolayer, and the matrix TAGs are labeled. This oleosin-like protein could be the precursor of oleosin or a remnant of degenerating oleosin (Huang et al., 2013b). C and D, P oleosin-containing LDs in C. reinhardtii zygote and conjugating S. grevilleana cells viewed by CLSM. The cells were stained for LDs with BODIPY (shown in yellow) in the presence of chloroplasts (autofluorescence [in red]; Huang et al., 2013b). E, SL and SH oleosin-containing LDs in maize embryo of seed (after 12 h of soaking) subjected to immunogold labeling with antibodies against the maize oleosin, viewed by electron microscopy. LD and protein body (PB) are visible (Fernandez et al., 1988). F, U oleosin-containing LDs in Arabidopsis pollen after germination. The pollen grain produces a tube that carries the two sperm nuclei to the ovary. LDs travel to the tube tip via streaming cytoskeleton. LDs were stained with Nile Red (shown in red). The boundary of the pollen grain and tube is shown with a dotted white line. (Courtesy of Ming-Der Huang.)
Table II. Types of LDs in plants.
The information is simplified for generalization. Many exceptions are known. Less-studied LDs of special functions are not included. Sizes of LDs are greatly generalized to show differences among LD types. Comprehensive information on the distribution of nonoleosin LD proteins on various LDs is unavailable.
| Location/Name | Oleosin Lineages | Size (Generalized) | Morphology | Notable Biogenesis | Function (Projected or Proven) | Phylogeny Restriction | Additional Comments |
|---|---|---|---|---|---|---|---|
| μm | |||||||
| Seed LD | SH oleosin, SL oleosin U oleosin | 1 | Solitary, not aggregated | Released from the ER; degraded by cytosol lipase or engulfed by vacuoles | TAGs for gluconeogenesis during germination | All seed plants | Two oleosin isoforms could form a dimer |
| Pollen LD | U oleosin | 0.5 | Solitary, not aggregated | Transported along the cytoskeleton from pollen to tube tip | TAGs releasing FA for tube tip plasma membrane elongation | All seed plants | TAGs not for gluconeogenesis |
| Flower tapetosome | T oleosin | 5 | Cluster of alkane LDs and flavonoid vesicles | ER produces both oleosin-coated alkane LDs and flavonoid vesicles | Upon programmed cell death, LD alkane and flavonoid become pollen coat | Family Brassicaceae | In anther tapetum of only one family; with multitandem T oleosin genes |
| Aerial epidermis giant LD | U oleosin | 10 | Cluster of LDs and other vesicles, etc. | Have not been delineated | Hub of cuticle wax | Order Asparagales | In green aerial epidermis with thick cuticle of only one order |
| Fruit mesocarp large LD | M oleosin | 20 | Giant and small LDs, not aggregated | Small oleosin-coated LDs fuse with oleosinless giant LDs | Attract animals for seed dispersion | Many orders | Oleosin-coated small LDs found only in family Lauraceae (e.g. avocado) |
| Leaf, stem, root LD | Likely to have oleosin | 0.5 | Solitary, not aggregated | Not necessarily from ER budding | TAGs as refuge for toxic FFAs from damaged membranes upon stress or senescence; produce phytoalexin and brassinosteroids | All plants | Minimal number of LDs; induced by stresses and senescence |
| Rubber particle | No | 10 | Solitary, not aggregated | Released from cell upon wounding | Defense | Many orders | Polyisoprene instead of TAGs; two related proteins: SRPP and REF |
| Plastid plastoglobule | No | 2 | Solitary or associated with inner membrane | Released from plastid thylakoid membrane | Temporary refuge for toxic FFAs; fruit pigment to attract animals | All plants | Various neutral lipids: TAGs, carotenoids, etc. |
Caleosin (Peroxygenase), Steroleosin (Sterol Dehydrogenase), and Dioxygenase
Three proteins possess enzymic activities whose enzymic products are related to stress responses. Caleosin and steroleosin (Chen et al., 1999; Naested et al., 2000; Frandsen et al., 2001; Lin et al., 2002; Song et al., 2014) are likely partly associated with LDs and also the ER. Caleosin was first reported in microsomes (Frandsen et al., 1996), and their transcripts are ubiquitous in diverse tissues or organs with or without known LDs (Table I). Regardless, caleosin and steroleosin transcripts are more abundant in seeds than in any other organs. In seeds, their transcripts and proteins in isolated LDs represent 2% to 4% of those of oleosin. A seed LD is covered completely with oleosin (Fig. 1B, center model), and so the two nonoleosin proteins should occupy a small, diffused or concentrated, area of the LD surface. Caleosin and steroleosin each has a hydrophobic segment with conserved Pro residues (Table I), which could form a hairpin penetrating the surface monolayer of PLs of an LD. These hairpins would be analogous to the hairpin of oleosin. However, they are substantially shorter (the longest being 21 residues for caleosin and 24 residues for steroleosin, both one-third the length of the oleosin hairpin), and the hairpin residues are only mildly conserved among isoforms within the same species and among diverse species (Song et al., 2014; Pasaribu et al., 2016). Two of the Pro resides in the caleosin hairpin loop are not essential for LD binding (Müller et al., 2017). Theoretically, caleosin and steroleosin, with their short hydrophobic hairpins, would be equally stable on the monolayer of PLs of an LD and the bilayer of PLs of membranes. This prediction could explain the two proteins being associated with the ER or other membranes in cells with or without LDs. Each of the two proteins could bind competitively to the surface of LDs and membranes in all cells, and the maturing or mature seeds have more LD surface and, thus, more LD-bound caleosin and steroleosin.
Importantly, both caleosin and steroleosin possess enzyme activities. Caleosin has a Ca2+-binding motif and several phosphorylation sites and possesses peroxygenase activities that convert hydroperoxides of α-linolenic acid to various oxylipins as phytoalexins (Hanano et al., 2006; Blée et al., 2014; Charuchinda et al., 2015). Steroleosin has a sterol-binding site and an NADPH-binding site and is a member of a large family of hydroxysteroid dehydrogenases in diverse organisms (Lin et al., 2002; Lindemann, 2015; Zhang et al., 2016). Steroleosin can convert sterol substrates to brassinosteroids, which are plant hormones controlling various phases of growth and development. The origins of the substrates in the cell for the two enzymes are unknown; they could be the LD interior, LD surface PLs, and/or diverse membranes in the whole cell. Overall, caleosin and steroleosin may carry out functions important to cellular and physiological events related or unrelated to their association with LDs.
Another LD protein, dioxygenase, was found recently on LDs (Shimada et al., 2014). The enzyme should be present in cytosol also, because the protein does not have a long hydrophobic segment. In Arabidopsis, the dioxygenase transcript level is highest in roots (Table III). However, its transcript in leaves elevates substantially in response to senescence and biotic stresses. Earlier, the enzyme was studied extensively as a pathogen/senescence-induced enzyme for the synthesis of hydroperoxylinolenic acid during hypersensitive responses (Sanz et al., 1998; Marcos et al., 2015). This dioxygenase could collaborate with caleosin (peroxygenase) to produce stable oxylipins from α-linolenic acid as signaling molecules for defense against biotic and abiotic stresses (Shimada and Hara-Nishimura, 2015).
Table III. Genes encoding LD proteins in Arabidopsis thaliana.
Levels of transcripts (reads per kilobase million), on a relative basis, of genes within a lineage are simplified for generalization; they vary according to the organ, stage of development, etc. Levels of transcripts of genes encoding LD proteins of different lineages are, in general, SH = SL = T > > U and other nonoleosin proteins. T oleosin transcripts in flower buds are 1 order of magnitude lower than SL or SH oleosin transcripts in maturing seed; however, T oleosin transcripts are restricted to one tapetum cell layer in a flower bud, whereas SL and SH oleosin transcripts are present in most cells in a maturing seed. Transcript levels were obtained from the RNA sequencing databasea (http://ncbi.nlm.nih.gov/) and individual reports cited in the text, and the information was supplemented with TAIR-eFP database (https://www.arabidopsis.org/). Genes encoding oleosins, dioxygenase, LD Associated Proteins, and OB Associated Proteins are numbered according to those in reports cited in the text. Genes encoding the other LD proteins are listed and numbered according to their occurrence on chromosomes 1 to 5 and in the 5′ to 3′ direction.
| Protein Names | No. of Genes within the Lineage | Organs with Highest Transcript Level | Transcript Level, Relative within the Lineage | Gene Accession Numbers | Comments |
|---|---|---|---|---|---|
| Oleosin | |||||
| U oleosin | 3 | Seed, pollen | U1 = U2 = U3 | At1g48990 (U1) | |
| At3g18570 (U2) | |||||
| At2g25890 (U3) | |||||
| SL oleosin | 2 | Seed | SL2 > SL1 | At3g27660 (SL1) | |
| At5g40420 (SL2) | |||||
| SH oleosin | 3 | Seed | SH2 > SH1 > SH3 | At3g01570 (SH1) | |
| At4g25140 (SH2) | |||||
| At5g51210 (SH3) | |||||
| T oleosin | 9 | Anther tapetum | T5 > T3 > T6 | At5g07470 (T1) | Eight of the nine genes in tandem |
| At5g07520 (T2) | |||||
| At5g07530 (T3) | |||||
| At5g07540 (T4) | |||||
| At5g07550 (T5) | |||||
| At5g07560 (T6) | |||||
| At5g07571 (T9) | |||||
| At5g07600 (T7) | |||||
| At5g61610 (T8) | |||||
| Caleosin | 7 | C1 and C2 in anther; C3, C4, and C6 in seed; C5 in petal and sepal | C6 > C5 > C1 | At1g23240 (C1) | C1 and C2 in tandem; C3 and C4 in tandem |
| At1g23250 (C2) | |||||
| At1g70670 (C3) | |||||
| At1g70680 (C4) | |||||
| At2g33380 (C5) | |||||
| At4g26740 (C6) | |||||
| At5g29560 (C7) | |||||
| Steroleosin | 8 | S3, S5, S7, and S8 in seed; S4 and S6 in stem and embryo | S7 > S3 > S8 | At3g47350 (S1) | S1 and S2 in tandem; S4 to S8 in tandem |
| At3g47360 (S2) | |||||
| At4g10020 (S3) | |||||
| At5g50590 (S4) | |||||
| At5g50600 (S5) | |||||
| At5g50690 (S6) | |||||
| At5g50700 (S7) | |||||
| At5g50770 (S8) | |||||
| Dioxygenase | 2 | D1 in root; D2 ubiquitous; increased during senescence and biotic stresses | D1 > D2 | At3g01420 (D1) | |
| At1g73680 (D2) | |||||
| LD Associated Protein | 3 | Ubiquitous; increased during abiotic stresses | SR3 > > SR1 = SR2 | At1g67360 (SR1) | Also termed small rubber particle protein, SRPP |
| At2g47780 (SR2) | |||||
| At3g05500 (SR3) | |||||
| OB Associated Protein | 5 | Seed (late maturation) | OB1 > OB5 > OB2 | At1g05510 (OB1) | |
| At1g29680 (OB2) | |||||
| At2g31985 (OB3) | |||||
| At4g18920 (OB4) | |||||
| At5g45690 (OB5) | |||||
| Lipase | 4 | Ubiquitous | SDP1 = SDP2 = SGI | At5g04040 (SDP1) | CGI in peroxisome, not LD |
| At3g57140 (SDP1L) | |||||
| At1g33270 (SDP2) | |||||
| At4g24160 (CGI58L) | |||||
| PAP | 3 | Ubiquitous | PAP2 > PAP1 > PAP3 | At4g04020 (PAP1) | Plastid-lipid-associated protein; all in plastids, not LD |
| At4g22240 (PAP2) | |||||
| At2g35490 (PAP3) |
The RNA sequencing database from the National Center for Biotechnology Information included SRX424380 for 7- to 8-d seed, SRX424383 for 9- to 10-d seed, SRX424386 for 11- to 12-d seed, SRX463620 for leaf, SRX1425915 for root, SRX399566 for stage 6 flower bud, and SRX257730 for stage 12 flower bud.
LD Associated Protein and OB Associated Protein
LD Associated Protein (Horn et al., 2013; Gidda et al., 2016) and OB Associated Protein (López-Ribera et al., 2014) are two recently discovered LD proteins in plants (Table I). The two share fairly similar characteristics but do not have protein sequence similarities indicative of an evolutionary relationship. They are small proteins of 25 to 27 kD with no major recognized binding/functional motif. They do not have a long hydrophobic segment that would allow them to be tightly associated with LDs. LD Associated Protein is present mostly in cytosol in cells with minimal numbers of LDs, as illustrated by confocal laser scanning microscopy (CLSM), and when the cells are fed with FA to induce the formation of more LDs, a portion of the cytosolic protein is associated with LDs. In these latter cells, the protein-associated LD in vivo can be washed away with mild detergent. OB Associated Protein is present largely in cytosol, as shown with immunoblotting after gel electrophoresis of subcellular fractions. In seed cells, LD/OB Associated Protein transcripts are ∼1% of the oleosin transcript (Table III). Apparently, LD/OB Associated Proteins do not play an important structural role. There are differences between the two proteins. LD Associated Protein transcript is present ubiquitously in cells/tissues with and without known LDs and is more abundant in seed cells containing ample LDs. OB Associated Protein transcript in maize (Zea mays) and Arabidopsis seeds appears late in maturation after the oleosin transcript and drops off rapidly during germination; the transcript is largely absent in nonseed organs. LD Associated Protein was studied earlier as a protein on the surface of rubber particles and termed Small Rubber Particle Protein and Rubber Elongation Factor (Oh et al., 1999; Sando et al., 2009). These two rubber proteins are paralogs, the latter not having the C-terminal portion of the former. They do not have enzymatic activities. Rubber Elongation Factor was so termed because, in its presence, likely as an anchoring or stabilization center, the rubber polyisoprene molecule elongates faster. LD Associated Protein and rubber particle proteins belong to a large family of evolutionarily related proteins in plants and green algae; they are stress related and required for proper growth. Overall, LD/OB Associated Proteins could exert functions of resisting abiotic stresses and promoting normal growth related to or independent of their physical association with LDs.
LD/OB Associated Proteins and dioxygenase, as well as lipoxygenase, phospholipase, and monoacylglycerol hydrolase (to be described), possess no long hydrophobic or predicted membrane-insertion segment (López-Ribera et al., 2014; Shimada et al., 2014; Gidda et al., 2016). LD/OB Associated Proteins are present in cytosol and partly associated with LDs. The mode of association of the cytosolic nonoleosin proteins to the LDs in normal, stressed, and senescent cells is intriguing and indispensable to delineating their physiological functions. Experimentation with CLSM should be expanded to other techniques. In CLSM, a protein on the LD surface (concentrated in a small cell space) could be detected much easier than that in the cytosol (spread out in a huge volume). The assessment of the relative amount of the protein in cytosol and on LDs could be subjective. The promoter for expressing the gene encoding the studied protein for CLSM should be the native promoter to avoid overproducing or underproducing the protein (in more than one subcellular site) for localization studies. Immunoblotting of the protein in isolated LDs and cytosol should be performed with quantification techniques, taking into account the usually small volume of the LD fraction and the huge volume of the cytosolic fraction. The use of artificial LDs with and without oleosin instead of PL liposomes (Gidda et al., 2016) should be employed to assess if oleosin is the docking site.
LDs IN PLANTS
Oleosin evolved from green algae and has undergone diversification. Their associated LDs have undergone similar paths (Figs. 2, C–F, and 3). They are specialized in morphology, contents, functions, and presence among diverse or restricted species, organs, tissues, and cells. LDs in plants related to food and other uses, as well as in Arabidopsis, have been better studied. Plant LDs of less economic importance have not been adequately studied. This review will describe only the better-studied LDs (Table II).
Figure 3.
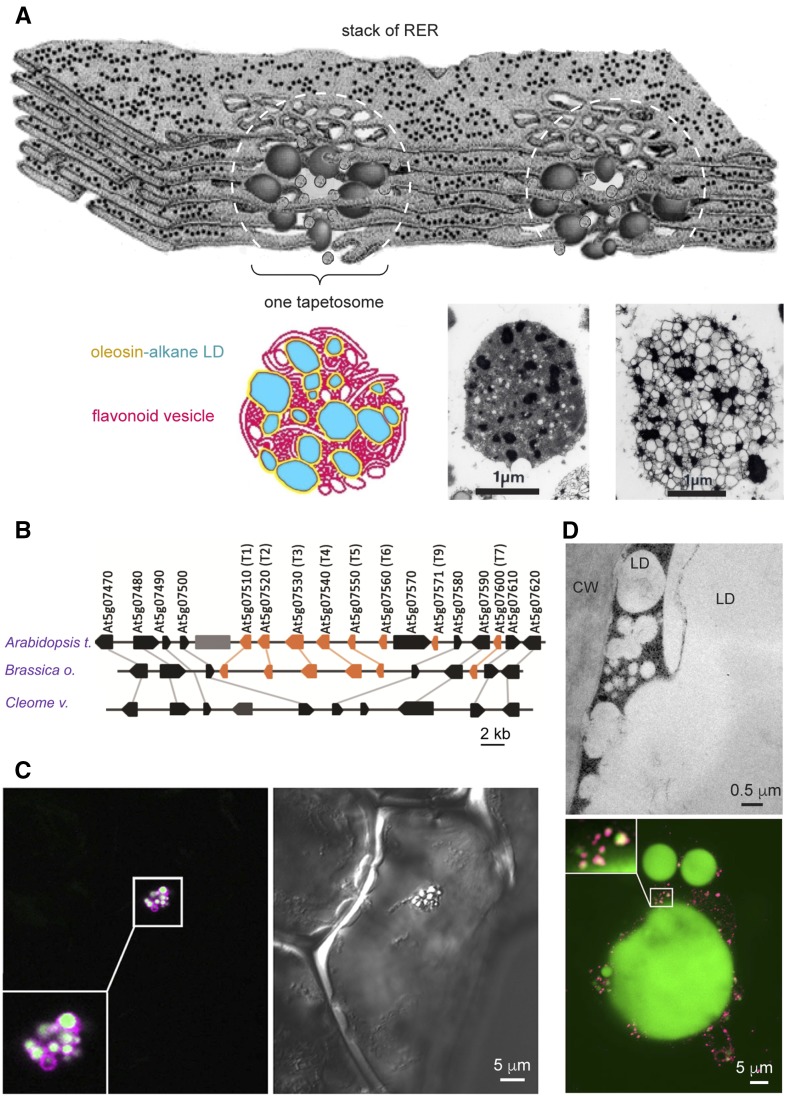
Specialized LDs in restricted tissues of specific plant phylogenies. A, T oleosin-containing LDs in tapetosomes being synthesized in rough ER in the tapetum of Brassica spp. In the model, a stack of rough ER produces alkane-containing, T oleosin- and PL-coated LDs and flavonoid-possessing vesicles at many locations. At each location (marked with a dotted white circle), numerous LDs and vesicles aggregate to form a tapetosome. The bottom shows a color model of a tapetosome having numerous alkane-containing (blue) LDs enclosed with a layer of oleosin-PL (yellow) associated with flavonoid-possessing vesicles (red). At right of the color model are electron micrographs of isolated tapetosomes without (middle) and with (right) osmotic swelling; after swelling of the tapetosome, internal LDs and vesicles can be seen. (A portion of the rough ER stack was traced from a drawing in a Trends in Cell Biology poster, 1998 [Hsieh and Huang, 2004, 2005]). B, T oleosin gene cluster in Brassicaceae and its absence in Cleomaceae in syntenic chromosome segments. Coding sequences of genes are indicated with colored arrows (orange for oleosin genes and black for nonoleosin genes) in genomic DNA from Arabidopsis, Brassica oleracea, and Cleome violacea. Orthologs among the genomes are linked with gray lines. The large gray box indicates a transposon (Huang et al., 2013a). C, U oleosin-containing LD cluster in vanilla leaf epidermis. Right, Differential interference contrast microscopy image shows the cell containing one LD cluster. Left, Immuno-CLSM image of the same cell reveals the LDs and oleosin. BODIPY-stained (in green) individual LDs in the cluster are visible. Antibodies against vanilla U oleosin reacted (in magenta) with the LDs. Oleosin appears more on the periphery of individual LDs, resulting in a magenta coat enclosing a white matrix (Huang and Huang, 2016). D, M oleosin-containing small LDs and oleosin-depleted large LD in avocado mesocarp. Top, Transmission electron microscopy image showing a portion of an avocado mesocarp cell containing large (10–50 μm) and small (less than 0.5 μm) LDs adjacent to the cell wall (CW). Bottom, Immuno-CLSM images show a large LD and numerous adjacent small LDs as well as a magnified junction between a large LD and numerous small LDs. BODIPY-stained (in green) large and small LDs are visible. Antibodies against avocado M oleosin reacted (in magenta) mostly with the small LDs. Oleosin appears more on the periphery of individual small LDs, resulting in a magenta coat enclosing a yellow matrix (Huang and Huang, 2016).
LDs in Seeds
General Characteristics
Seeds store TAGs in LDs as metabolite reserves for germination and seedling growth. LDs are present in various parts of a seed, including the embryo axis, cotyledon, endosperm and its aleurone layer, and megagametophyte, depending on the plant species. The cells of a mature oil seed are packed with LDs and storage protein bodies. Each spherical LD is ∼0.5 to 1.5 μm in diameter. It has a core of TAGs enclosed by a monolayer of PLs and oleosin (Figs. 1, A and B, and 2E). LDs with their surface completely covered with oleosins are highly stable in mature seed during prolonged storage, desiccation, and temperature stresses as well as in isolated preparations. The small size of LDs and their stability provide a large surface area per unit of TAG and facilitate binding to lipase or other subcellular structures for rapid TAG mobilization during germination. Indirect evidence from genetics, protein contents, and transcript levels (Lee et al., 1995; Huang and Huang, 2015) has suggested that the two seed oleosin isoforms SL and SH form a dimer or multidimer, which could provide further stability or functional attributes. Caleosin, dioxygenase, and steroleosin as well as LD Associated Protein and OB Associated Protein also are present on seed LDs, but in substantially lesser amounts (Table I). Whether oleosin is the docking site of these nonoleosin proteins with or without a short hairpin structure (Table I) remains to be explored.
Synthesis
LDs in seeds are synthesized on the ER. TAG-synthesizing enzymes are associated with extended regions or subdomains of the ER (Cao and Huang, 1986; Loer and Herman, 1993; Lacey et al., 1999; Shockey et al., 2006). TAGs synthesized on the ER are sequestered in the nonpolar acyl region of the bilayer of PLs, which results in ER-LD swelling (Fig. 1B). Oleosin is synthesized on the cytosolic side of the ER cotranslationally (Qu et al., 1986; Loer and Herman, 1993; Thoyts et al., 1995; van Rooijen and Moloney, 1995) via signal-recognition particle-guided mRNAs (Beaudoin et al., 2000) and extracts ER-budding LDs to the cytosol. Reducing the proportions of oleosin to TAGs in vitro via chemical reconstitution (Tzen and Huang, 1992) or in vivo via breeding (Ting et al., 1996; Fig. 1C), T-DNA insertion mutation (Siloto et al., 2006; Shimada et al., 2008), or gene knockdown (Siloto et al., 2006; Schmidt and Herman, 2008) generates larger LDs. LDs could detach from the ER to be the final LDs of defined sizes, or nascent small LDs could undergo fusion to form large LDs (Schmidt and Herman, 2008; Huang and Huang, 2016). These two possibilities may not be mutually exclusive and also could depend on the species and growth environment. They deserve to be tested. The mechanisms of proteins on the ER or at the ER-LD junction controlling the formation and detachment of budding LDs in plants should be explored. These proteins, including Fat Storage Inducing Protein and SEIPINS, have been studied extensively, albeit with their exact roles still undefined, in yeast and mammals (Chen and Goodman, 2017; Walther et al., 2017) and explored recently in plants (Cai et al., 2015b, 2017).
For the oleosin molecule per se, the C- and N-terminal peptides are not required for oleosin targeting ER-LDs (Abell et al., 2002, 2004; Beaudoin and Napier, 2002). However, the whole hairpin, including the N-portion residues, the full length of the hairpin arms, as well as the three Pro and one Ser residues at the hairpin turn, are necessary for proper oleosin targeting ER-LDs and extraction of budding LDs to the cytosol (Huang and Huang, 2017). Oleosin molecules modified with (1) the addition of an N-terminal ER target signal peptide to the oleosin, (2) retention of the hairpin N-portion residues, and (3) reduction of the hairpin length can enter the ER lumen and extract ER-budding LDs to the ER lumen. Further addition of a 12-residue vacuole-targeting propeptide to the modified oleosin allows the LDs in the ER lumen to move to the vacuoles. Thus, oleosin is the sole molecule responsible in vivo for the budding LDs entering the cytosol instead of the ER lumen (Fig. 1B).
Degradation
In young seedlings, TAGs of LDs are hydrolyzed to FAs for gluconeogenesis in peroxisomes and other subcellular compartments. Peroxisomes are newly synthesized and are adjacent to or in physical contact with existing LDs. This contact could reflect the cytoplasm being tightly packed with organelles, including the abundant peroxisomes and LDs, or the need for metabolite or material transport between the two organelles. Studies with Arabidopsis mutants have revealed a role for the physical contact. TAG lipase (Sugar-Dependent lipase [SDP1]; Eastmond, 2006) in association with core retromer (a protein complex for subcellular cargo trafficking) binds to peroxisomes, which then produce tubular extensions to transport the lipase to LDs (Thazar-Poulot et al., 2015). Physical contact between LDs and peroxisomes is negatively correlated with the presence of sucrose, reflecting a mechanism of end product feedback inhibition of TAG gluconeogenesis (Cui et al., 2016). CGI-58 protein, a lipase activity modulator on mammalian LDs, is present on peroxisomes in seed (Pyc et al., 2017). Thus, the junction between LDs and peroxisomes is highly active in lipolysis and the initiation of β-oxidation. Further study could investigate the mechanisms of the transport of lipase and possibly other proteins from peroxisomes to LDs as well as FAs from LDs to peroxisomes and the activation of FAs to acyl-CoA for peroxisomal β-oxidation. The study could follow and expand from the findings with yeast and mammals. The lipase transcript level peaks during seed maturation and is 10% to 30% (Eastmond, 2006; Kelly et al., 2013b) of the peak level in seedlings at the height of lipolysis. Whether in maturing seed the transcript is dormant or active in synthesizing inactive/active lipase and where this lipase is inside the cell that possesses abundant LDs but no peroxisomes should be explored. Lipase activity in isolated LDs is absent in mature seed and present in germinated seed. The activity of the lipase in Arabidopsis seed throughout maturation and germination has not been reported, likely due to the minute size of the seed and the difficulty in assaying authentic TAG lipase activity in total seed extracts.
The SDP1 lipase can hydrolyze TAGs, diacylglycerols, and monoacylglycerols, even though a different monoacylglycerol hydrolase also is present on LDs (and other subcellular sites; Kim et al., 2016). The transcript level of this latter hydrolase is higher in roots and leaves than in seeds (https://www.arabidopsis.org/). Some species contain a lipoxygenase on LD (and likely other subcellular sites), which could convert polyunsaturated acyl moieties in TAGs to hydroperoxy FAs before the FAs are acylated for β-oxidation in peroxisomes (Feussner et al., 2001; Rudolph et al., 2011). The monoacylglycerol hydrolase and lipoxygenase, as well as a phospholipase (to be described), do not have a long hydrophobic segment in their sequences and are assumed to be loosely associated with LDs and likely present in cytosol and other subcellular compartments.
In Arabidopsis, some LDs could be engulfed by vacuoles during germination (Poxleitner et al., 2006). This engulfment is in apparent discrepancy with the dogma that LDs work closely with peroxisomes for efficient LD gluconeogenesis.
Seed LDs are enclosed completely with monolayers of PLs and oleosin. Whether these layers need to be removed before lipase can act on the core TAGs is unknown. Isolated seedling LDs (with acquired lipase and after washing) of diverse species undergo autolysis, releasing most of the TAGs as free fatty acids (FFAs), apparently without the need for protease or phospholipase activities. Regardless, oleosin proteolysis via ubiquitination occurs during seed germination (Hsiao and Tzen, 2011; Deruyffelaere et al., 2015). This proteolysis could facilitate the lipolysis of LDs or represent a general mobilization of all proteins, including oleosins, for seedling growth. These uncertainties apply also to a phospholipase that occurs transiently on LDs (and likely other subcellular sites; Rudolph et al., 2011).
Among LD-TAG-hydrolyzing lipases in seedlings of all plant species, only the lipase SDP1 and its related SDP1-like (SDP1-L) lipase in Arabidopsis have their genes identified and proven to be the enzymes responsible for hydrolyzing LD TAGs (Eastmond, 2006). Early studies show that lipase hydrolysis of seed LDs varies substantially among species (Lin and Huang, 1983; Huang, 1987). In species of Brassica (a genus closely related to the Arabidopsis genus in the family Brassicaceae) and many other distant phylogenic species, lipase activity is absent in mature seed and appears after germination, concomitant with the loss of storage TAGs. The Brassica lipase is present on LDs and, unlike the maize LD lipase, can be washed away easily with dilute buffer. It has highest activities toward trierucin, triolein, and trilinolein, whose erucic, oleic, and linoleic moieties are the major acyl constituents of Brassicaceae seed TAGs; this diverse substrate specificity also is exhibited by the Arabidopsis SDP1 lipase (Eastmond, 2006). Future research should expand the findings with Arabidopsis to Brassica (for biochemical studies) to dissect the mechanism of LD hydrolysis in relation to the lipase activity throughout seed maturation and germination. In Arabidopsis, SDP1 transcript is present in diverse organs (Kelly et al., 2013b; Table III); its encoded lipase should be examined for possible involvement in the turnover of the nonabundant LDs in these organs. Whether the information acquired with Brassicaceae species could be extended to diverse plant species should be explored. A precedent is that the flower LD-tapetosome system is present only in Brassicaceae and not the closest family Cleomaceae (to be described). In maize scutellum during germination, cytosolic translatable lipase mRNA is absent in mature seed and peaks in level, along with the activity of de novo-synthesized lipase, at the highest stage of lipolysis (several days after imbibition). At this stage, the lipase is tightly bound to LDs and cannot be washed away with high salt or ethanol. After the depletion of TAGs, the lipase still binds to LD ghost-vacuolar membranes and remains in the cell (Wang and Huang, 1987).
LDs in Pollen
Pollen of all plants contains metabolite reserves, which are usually LDs of TAGs and/or starch grains (Fig. 2F). A pollen grain germinates on the female flower stigma and produces an extending tube to penetrate the style to reach the ovary. The two sperm nuclei move from the pollen grain proper along the tube to the ovary to fertilize the eggs. During tube extension, numerous cytoskeleton structures stream rapidly from the pollen proper to the tube tip, carrying with them cytoplasmic organelles, vesicles, and LDs (Cai et al., 2015a; Müller et al., 2017). The tube tip continues to grow toward the ovary. This movement requires the tip plasma membrane to reshuffle old constituents and likely acquire new constituents. Pollen LD TAGs apparently do not function in gluconeogenesis, because no or minimal peroxisomes or transcripts of β-oxidation and glyoxylate cycle enzymes are present in pollen before or after germination. Accordingly, the TAGs in LDs likely function to supply new FA for the de novo synthesis or partial reshuffling of the plasma membrane at the tube tip. Pollen LDs are coated with oleosin, caleosin, and steroleosin. Whether these LD proteins possess binding sites for the cytoskeleton and movement molecules for the rapid movement of LDs along the cytoskeleton from the pollen proper to the tube tip should be tested.
LD Clusters Termed Tapetosomes in the Flower Tapetum
Anthers are the male component in a flower. Each elongated anther consists of four cell layers enclosing a pollen sac, where haploid cells produced from meiosis mature to become pollen grains. The innermost cell layer, termed the tapetum, is most active and possesses dense cytoplasm. At a peak stage of tapetum development in Brassicaceae species, the tapetum cell is packed with two equally abundant spherical organelles, each ∼5 μm in diameter, called tapetosomes and elaioplasts (Wu et al., 1997; Hsieh and Huang, 2007; Huang et al., 2013a; Lévesque-Lemay et al., 2016). Each tapetosome is a cluster of dozens of oleosin-coated LDs (∼0.2–0.5 μm) of alkanes instead of TAGs and flavonoid-containing vesicles (∼0.2–0.5 μm; Fig. 3A). Each elaioplast is a nongreen lipid-storing plastid and includes numerous spherical lipid globules (plastoglobules) of sterylesters enclosed with Plastid-lipid Associated Protein. Subsequently, the tapetum cell undergoes programmed cell death, releasing alkanes, oleosins, and sterylesters to the adjacent maturing pollen grains, forming a pollen coat. The coat provides waterproofing and UV protection to the pollen grain. Inside the tapetum cell during the active stage of tapetosome formation, massive rough ER produces via budding oleosin-coated LDs (analogous to the synthesis of seed LDs) and flavonoid-containing vesicles (Fig. 3A). Dozens of adjacent LDs and flavonoid vesicles cluster to form a tapetosome. At the peak stage of tapetosome synthesis in the tapetum, massive oleosin is required, and ∼10% of the mRNA in the tissue encodes oleosin. For this massive oleosin production, the genome has evolved to have multitandem genes encoding tapetosome-specific oleosin (T oleosin). The Arabidopsis genome has eight T oleosin genes in tandem within a 26-kb segment (Fig. 3B). Tapetosomes evolved only recently in the family Brassicaceae, whose closest family Cleomaceae and other families of the order Brassicales do not have T oleosin genes or tapetosomes. The tapetosome system confers an advantage in that Brassicaceae pollen is more resistant to dehydration and possibly other stresses than Cleomaceae pollen. Cleome transformed with an Arabidopsis T-oleosin gene possesses T-oleosin and primitive tapetosomes in the tapetum and more alkanes and flavonoids in the pollen coat, resulting in the transformed Cleome pollen being more resistant to dehydration. A system similar to Brassicaceae tapetosomes that evolved via convergent evolution in distant phylogenic plant groups remains for investigation.
LD Clusters in Epidermis of Green Aerial Organs
The order Asparagales includes food and ornamental species, such as onion (Allium cepa), aloe (Aloe vera), and vanilla (Vanilla planifolia). Their green aerial organs of leaves, flowers, and ovaries are juicy and have a thick cuticle. Each epidermal cell of these organs contains LDs in solitary entities (∼0.5 μm diameter) and/or more conspicuously one to several large LD clusters (each of ∼10 μm in diameter with ∼10 LDs [Fig. 3C]; Kwiatkowska et al., 2010, 2015; Huang and Huang, 2016). Each LD cluster also appears to be associated with other subcellular structures such as cytoskeleton, as observed by electron microscopy. In leaves, the clusters are present in both the upper and lower epidermis but absent in the mesophyll. These and other observations have led to the proposal that the clusters are the warehouses of epidermal cuticle lipids to be exported to the epidermal surface. Each of the LDs in the cluster is coated with oleosin (Fig. 3C), and the clusters in isolated preparations do not fuse. The oleosin in the clusters belongs to the U oleosin lineage similar to the U and S oleosins on seed LDs. This lack of a unique oleosin lineage contrasts with the T oleosin in tapetosomes (preceding paragraph) and M oleosin in avocado (Persea americana) small LDs (next paragraph). More research is needed to advance our knowledge of the LD clusters in Asparagales.
LDs in Fruit Mesocarp
Some fruits of phylogenically diverse species possess voluminous, high-lipid, juicy mesocarp, which attracts animals to consume and disperse seeds. Those of economic importance include avocado, olive (Olea europaea), oil palm fruit (Elaeis guineensis), tung tree fruit (Vernicia fordii), and some sweet tropical fruits. In these fruits, the mesocarp cells contain several giant LDs (e.g. 10 μm versus 1 μm of seed LDs) of TAGs. Mesocarp TAGs are usually of higher saturation than seed TAGs. The giant mesocarp LDs do not have oleosin on the surface, and in isolated preparations, they fuse to become much larger, in contrast to the small, oleosin-coated stable seed LDs. A variation of these generalized descriptions occurs in the avocado mesocarp (Horn et al., 2013; Huang and Huang, 2015, 2016; Kilaru et al., 2015). Within the same mesocarp cell, the large LDs do not have coated oleosin, whereas the small LDs adjacent to them do (Fig. 3D). The small oleosin-coated LDs may be newly synthesized from ER on their way to fuse with the enlarging LDs; after fusion, the oleosin would not be retained on the large LDs. The transcript level is slightly higher for oleosin than the detected LD Associated Protein, whose subcellular locations in avocado are unknown. In avocado and other species of the family Lauraceae, the mesocarp oleosin, termed M oleosin, is distinct from the oleosins in seed and pollen and apparently evolved only in the family Lauraceae. The occurrence of oleosin on small LDs in the mesocarp of Lauraceae species apparently cannot be extended to the less-studied mesocarp of fruits of other plant families. No oleosin transcript has been reported in fruit mesocarp of oil palm (Bourgis et al., 2011) and tung tree (Zhi et al., 2017), and tung fruit mesocarp possesses LD Associated Protein transcript. Apparently, the coating of LDs with oleosin and possibly other proteins as well as the mechanism of LD biogenesis in mesocarp are species/family specific.
LDs in Leaves and Other Vegetative Organs
LDs occur minimally in nonstorage vegetative organs such as leaves, stems, and roots. If they are present, they could be considered less as storage warehouses and more as detoxification refuges. LD TAGs are derived from scavenged toxic FFAs from damaged membranes during stresses or senescence. The mode of synthesis of these LDs is unknown and could be dissimilar to the ER-budding model in seeds. More studies have involved leaf LDs (Sridhar et al., 1973; Wahlroos et al., 2003; Lersten et al., 2006; López-Ribera et al., 2014; Shimada et al., 2014; Gidda et al., 2016; Pyc et al., 2017). In healthy leaves, the number of LDs is very low. The number increases substantially when the leaves or plants are subjected to abiotic or biotic stresses, fed with FAs or sterols (a form of stress per se if the toxic FAs or sterols are unaltered and taken into the cytoplasm), during senescence, or at the end of the dark period in a diurnal cycle (could be running out of housekeeping bioenergy obtained during the light period). Studies of leaf LDs rely largely on confocal microscopy, because isolating the scarce LDs cleanly for biochemical studies could be difficult. Suspected LD proteins have been localized to the LDs in leaves transformed with a gene encoding the protein linked to a fluorescence protein. They include oleosin, caleosin, dioxygenase, LD Associated Protein, and OB Associated Protein. Localization of the external LD protein (attached to a fluorescent protein) on LDs does not necessarily mean that the native LD protein exists on LDs. Nevertheless, transcripts of these proteins in leaves can be detected at low levels, and their levels increase with stress. Thus, the presence of the encoded LD proteins is expected. Overall, the protein complement on leaf LDs should differ from that on seed LDs, in part because of the transient presence of these LDs. The pathogen- or senescence-induced LD dioxygenase can collaborate with caleosin (peroxygenase) to catalyze sequential reactions converting α-linolenic acid to stable oxylipins such as 2-hydroxyoctadecatrienoic acid. Oxylipins are signal molecules that could initiate a hypersensitive response to protect the whole plant (Marcos et al., 2015; Shimada and Hara-Nishimura, 2015). This is a novel potential function of LDs in leaves, seeds, and other organs for defense against abiotic and biotic stresses. The enhancement of dioxygenase transcript and protein (for phytoalexin synthesis) in the infected area clearly signifies a defense response to biotic invasion. Whether the increase in LDs in the same infected area represents a defense response also or a pathological consequence should be explored. LD Associated Protein is present on leaf LDs (OB Associated Protein present mostly in seed) and exert unspecified functions related to stress resistance. The two proteins do not have enzyme activities and, presumably, carry out functions via binding. LDs containing TAGs in senescing leaves also can be viewed as temporary warehouses of toxic FFAs from decaying membranes, and the TAGs would be subjected to gluconeogenesis with lipase and peroxisomes, etc., with the generated sugar transported to other parts of the plants. Whether LDs produced from various forms of stresses are subjected to similar gluconeogenesis awaits exploration. The only functional lipase (with its gene identified) for seed TAG mobilization is that from Arabidopsis (SDP1 and the related SDP1-L). Transcript of this lipase is ubiquitous in diverse organs, and its encoded lipase could be for LD breakdown in leaves and other nonseed organs (Kelly et al., 2013b).
Rubber Particles
More than 20,000 species of diverse plant orders produce abundant subcellular rubber particles (LDs containing polyisoprene) in their vegetative organs. In many species upon organ/tissue damage, the rubber particles together with other cellular materials are released as latex for defense against pathogens. The best known is the rubber tree (Hevea brasiliensis), whose latex rubber has been the major source of industrial rubber for a century (Berthelot et al., 2014). Inside the tree trunk cells, the rubber particles enlarge as the cells mature (Sando et al., 2009). Each rubber particle contains a core of polyisoprene enclosed by a monolayer of PLs embedded with two major proteins, Small Rubber Particle Protein and Rubber Elongation Factor (see preceding section on LD proteins). The two proteins would make the particles stable from fusion or aggregation. In the secreted latex, the rubber particles are fairly stable and aggregate or coalesce slowly. The two proteins are members of a large plant protein family involved in stress responses and proper plant growth, and their presence on rubber particles also may be related to these effects.
LDs (Plastoglobules) Inside Plastids
Plastoglobules are elongated or spherical LDs of 0.2 to 5 μm diameter associated with or detached from the internal thylakoid of a plastid (Deruère et al., 1994; Bréhélin and Kessler, 2008; Zeng et al., 2015). They are present in all plants and green algae. Each plastoglobule has a core of neutral lipids enclosed by a monolayer of amphipathic lipids (PLs and/or glycolipids) embedded with proteins. The neutral lipids vary greatly, depending on the plant species, tissues, growth stages, and environmental conditions. TAGs serve as a temporary refuge for toxic FFAs from damaged membranes. Yellowish carotenoids in small amounts in a healthy leaf, overshadowed by green chlorophyll until senescence, are for light reception in photosynthesis and quenching of excess energy in photoprotection. Carotenoids of diverse colors (yellow, gold, red, etc.) in petal, sepal, fruit coat, etc., in the absence of green chlorophyll, are for attracting animals. Other neutral lipids could be prenylquinones, plastoquinones, and FA phythyl esters. The major surface protein is called Plastid-lipid Associated Protein, fibrillin, or chromoplast-specific protein. The protein of ∼30 kD does not have a long hydrophobic segment and is assumed to interact with the surface of rather than penetrating the plastoglobule and play a structural role. Minor proteins could include lipid metabolic enzymes and signal proteins, depending on whether the plastoglobule is attached to or detached from the thylakoid.
SURFACE OF LDs BEING A REFUGE FOR DIFFERENT PROTEINS
Plant nonoleosin LD proteins, including caleosin, steroleosin, dioxygenase, and lipoxygenase, as well as LD/OB Associated Proteins, but excluding lipase, may or may not have a direct functional relationship with LDs. Knockout, knockdown, or overproduction mutants of these proteins may have the LD numbers or sizes altered as downstream responses or consequences. Some of these proteins may use the LD surface as a refuge for future activities at diverse subcellular locations under specific environmental cues. This hypothesis is based on the following considerations (evidence described and referenced earlier).
(1) The proteins are not associated tightly or uniquely with LDs. Caleosin and steroleosin have a hydrophobic hairpin substantially shorter (approximately one-third) than the oleosin hairpin, and the hairpin sequences are only mildly conserved (Table I). They would be equally stable on LDs and other membranous structures in the cell. Dioxygenase, lipoxygenase, and LD/OB Associated Proteins do not have a long hydrophobic segment and are predicted to be in cytosol (lipoxygenase) or shown to be in cytosol and on LDs (dioxygenase and LD/OB Associated Proteins). Transcripts of some of these proteins are present ubiquitously in organs with and without LDs (Tables I and III).
(2) Some of the proteins are in excessive amounts for enzyme activities. On seed LDs, caleosin and steroleosin represent 2% to 4% of oleosin. These two proteins have enzymic activities. Some of the enzymic products are signaling molecules functioning in minute quantities, whereas others, such as phytoalexin, may be in need of higher amounts for defense. Having the enzymes in high amounts may be unnecessary but could be beneficial in storage forms for quick actions upon stresses. Dioxygenase is present in a minute quantity, as judged from its transcript level.
(3) The enzyme substrates may be from various subcellular membranes. The origins of α-linolenic acid and sterols for the enzymic activities have not been defined and could come from various membranes upon stress-induced damage (Hong et al., 2016). They could come from the surface and core of LDs also, but seed LDs are comparatively stable in vivo, much more so than the membranous structures in the whole cell, upon desiccation and temperature stresses.
(4) The surface of LDs is the most inert structure among all physical or membranous structures inside a cell and, thus, is suitable for use as a refuge for proteins. LD surface activities are limited to lipolysis and physical interaction with peroxisomes at restricted locations and developmental stages. If the proteins, such as LD/OB Associated Proteins and dioxygenase, were in cytosol instead of on LDs, they would be more susceptible to unintended proteolysis.
(5) Plant LDs as a refuge for proteins unrelated to LDs directly would be analogous to animal LDs (Welte and Gould, 2017). For example, Drosophila LDs are a refuge for histones to be used for rapid nuclei and cell synthesis at a future developmental stage. The LD histones and other refuge proteins can shuffle between LDs and other subcellular compartments for different metabolism or activation purposes. The inertness of the LD surface as a refuge has been taken advantage of by some viruses, which assemble virus particles on LDs with minimized disturbance to the host cells (Roingeard and Melo, 2017).
Plant nonoleosin LD proteins, at least some of them, could use the LD surface as a refuge. Upon a signal induction, they could, with or without activation, shuffle among the LD surface, cytosol, and other membranous structures. The activation or reshuffling energy would come from protein phosphorylation (analogous to the activation of some mammalian LD proteins), palmitoylation, or other means. Alternatively, they could move with the whole LDs to different membranous structures along the cytoskeleton or via other mechanisms. These two possibilities are not mutually exclusive and could depend on the cell type, plant species, and environmental cue. In addition, LDs in seeds and vegetative organs such as leaves play different functional roles and may operate the LD refuges differently. It is unlikely that the toxic and amphipathic FFAs and sterols diffuse from the damaged membranous structures to the LD surface for enzymic reactions. The overall hypothesis, regardless of its validity, should be tested to delineate the structure and functions of the nonoleosin LD proteins and the LDs per se. The subcellular partition, the mode of the partition, and the possible reshuffling among subcellular structures of each of the proteins and their individual isoforms in cells with and without LDs upon stresses, pathogenicity, and senescence should be delineated. Examination also should be made of the possible movement of whole LDs to membranous structures or the reverse movement of membranous structures to LDs upon stresses and senescence.
EVOLUTION OF PLANT LDs AND THEIR ASSOCIATED PROTEINS
Primitive unicellular organisms evolved to possess neutral lipid-containing LDs, which conferred two survival advantages. First, the cells would convert toxic FFAs from damaged or aged membranes to nontoxic TAGs and/or other neutral lipids in LDs. Second, LDs would become storage warehouses for future growth when the organisms encountered stresses. For storage, neutral lipids instead of polysaccharides would be advantageous because they contain twice as much bioenergy. Enzymes for the synthesis and degradation of neutral lipids would evolve from existing membrane-associated acyltransferase and non-membrane-associated lipase, respectively, originally meant for membrane metabolism. Initially, the nascent neutral lipids were present between the acyl moieties of the di-PL or diglycolipid layer in membranes. This bulky presence would interfere with membrane functioning and lead to the release of the neutral lipids to cytosol as LDs. The released LDs would be partially stabilized with a monolayer of PLs or glycolipids from the original bilayer membrane, and perhaps also with some of the original membrane proteins. Further stability occurred with the evolutionary appearance of LD structural proteins. Also, proteins modulating the whole storage and breakdown process evolved, resulting in refinement of metabolic regulation. The structural and modulating roles of some of these proteins would cross over. The early-evolved LD protein in a very primitive organism would be retained by all organisms, and its mode of action would evolve and differentiate in a phylogeny-specific manner. An example is the different modes of action of CGI-58 in modern plants, animals, and fungi (James et al., 2010; Oberer et al., 2011; Pyc et al., 2017). The late-evolved proteins would be retained in specific phylogenies. They include phasin in many bacteria, Major LD Protein in some bacteria, LD Surface Protein in heterokont algae, Major LD Protein in some green algae and other algae (Table I), perilipin in vertebrates (Itabe et al., 2017), and oleosin in green algae and plants.
Oleosin evolved in green algae, the ancestor of modern plants. Oleosin might have evolved from a transmembrane or peripheral membrane protein via consecutive mutations of hydrophilic/polar residues to nonpolar residues, leading to the formation of the long hydrophobic hairpin of oleosin. The gene encoding the oleosin precursor did not have a strong promoter, and its duplication throughout evolution was modest (one gene in C. reinhardtii, three in S. grevilleana, and five to six in a true diploid advanced plant; Huang and Huang, 2015). Initially, the oleosin amount produced was too minute to even cover a small fraction of the surface of LDs, and its amount would increase gradually through evolution. Among modern organisms, the amount of oleosin on each LD is minimal in C. reinhardtii, detectable in S. grevilleana, moderate in Physcomitrella patens, and abundant in plants (Huang et al., 2009, 2013b). In Arabidopsis, two to three most-active S or T oleosin genes are expressed strongly in seed and tapetum, respectively, producing oleosin transcripts representing 10% of all cell transcripts (Huang et al., 2013a). Along the way, specific oleosins would evolve together with LDs, forming particular LD structures, such as tapetosomes and leaf epidermal LD clusters in restricted phylogenies. Currently, six oleosin lineages are known, some ubiquitous (e.g. U oleosin and seed SL and SH oleosins in seed plants) and others highly restricted. In view of this latter restriction of T oleosin and M oleosin each to only one plant family, we should expect the existence of more oleosin lineages not yet discovered.
Table III lists genes encoding all the LD-associated proteins in Arabidopsis. Tandem genes encoding T oleosin, caleosin, and steroleosin exist. For the four oleosin lineages, each lineage has two to nine gene members, whose transcript levels vary drastically but still of identical tissue specificities (Kim et al., 2002). For genes encoding each type of nonoleosin LD proteins, their transcripts vary significantly not only in levels but also in organ/tissue specificities. Some genes have highest transcript levels in organs not known to possess abundant LDs (e.g. dioxygenase D1, lipase CGI-58L, and monoacylglycerol hydrolase transcript levels being highest in roots). These diversifications of nonoleosin LD proteins of the same type suggest that the proteins carry out different organ/tissue-specific functions related or unrelated to LDs and/or that their levels, tissue specificity, and subcellular locations could be enhanced or modified upon stresses or senescence.
OLEOSIN-LDs FOR POTENTIAL INDUSTRIAL APPLICATIONS
Among all LD proteins, oleosin has the longest hydrophobic hairpin (Table I) and is tightly associated with LDs. Oleosin has been probed for industrial applications. Oleosin-LDs can be used as a natural and safe emulsifying agent in food and cosmetics adjuvants (Bhatla et al., 2010). Oleosin-LDs in foods could be allergenic (Jappe and Schwager, 2017), and removing or denaturing oleosin in these foods could be advantageous. The seed oleosin gene promoter is restricted to maturing seed and related to an array of oil synthesis genes. It is as strong if not stronger than the seed storage protein gene promoter. Thus, the seed oleosin promoter has been used extensively to express oil synthesis and modification genes in maturing seeds (see below). Several transcription regulatory genes also could be used to boost seed maturation signaling to induce oil synthesis (Weselake et al., 2009) in seeds and leaves. Major studies to probe the commercialization of oleosin-LDs are as follows.
(1) Use Foreign Oleosin (as a Protectant or Metabolic Sink) and Enzymes (to Overcome Limiting Steps or Synthesize New Oils) to Produce More or New Oils in Seeds or Leaves
The desire to produce renewable biofuels to avert global warming has led to the exploration of biodiesel production in oilseeds, vegetative organs such as leaves and underground storage organs, and microalgae. The rationale for this exploration also could embrace the production of higher valued oil seed crops and health oils. The engineering approaches have included transforming plants or microalgae with foreign genes. These genes could be (1) a regulatory gene producing a transcription factor that activates the complete array of genes encoding oil-synthesizing enzymes, (2) a gene encoding a rate-limiting enzyme (e.g. diacylglycerol acyltransferase or a transmembrane protein), (3) a gene encoding a foreign desaturase or exerting a silencing effect on an internal desaturase gene for a desired desaturated acyl composition or new health oils, (4) an RNA interference (RNAi) construct to suppress lipase activity and, thus, TAG turnover in desiccating seed, and/or (5) a gene encoding oleosin to protect or act as a metabolic sink to house the new oils. Major advances have included the following (not a complete list). Seeds of several Brassica spp. have ∼40% oils and were engineered to produce more oils (Taylor et al., 2009; Savadi et al., 2016). Corn kernels have 1% to 4% oils largely restricted to the embryos and were engineered to enhance oil contents (Shen et al., 2010; Alameldin et al., 2017). Rice seed has 1% to 4% oil and was engineered to produce more oils (Liu et al., 2013). Leaves of diverse species (Winichayakul et al., 2013; Kim et al., 2015; El Tahchy et al., 2017; Yurchenko et al., 2017; and several others), sugarcane (Saccharum officinarum) organs (Zale et al., 2016), and underground starch-storing stems/roots (Liu et al., 2017) were engineered to produce new or more oils. A diatom was engineered to produce more oils (Zulu et al., 2017). Use of oleosin alone for the engineering could result in higher oil contents and more LDs in seed (Liu et al., 2013) and yeast cells (Jacquier et al., 2013).
Further advances in some of these successes of having more oils and LDs in seeds and leaves of transformed plants will need to overcome potential problems. Oils have twice as much bioenergy as storage starch or proteins, and a plant or organ will have to obtain extra photosynthetic energy to synthesize more oils or reshuffle bioenergy from other constituents, such as storage starch. Otherwise, a growth penalty will set in. Whether TAGs per se or oleosin (in addition to protecting the accumulated TAGs in LDs) can act as a metabolic sink to overcome the energy barrier remains to be seen. Existing crop plants such as canola (Brassica napus) and maize engineered to have increases in oils (or use of a low-oil variety as the starting experimental material) have been tested positively in the field on a short-term basis; whether larger increases will cause a growth penalty remains to be evaluated. High-oil (and diverse unsaturation) varieties of these oilseed crops exist, with oil contents usually negatively correlated with starch or other seed storage materials; most of these varieties do not perform as well as the established commercial varieties. The high-oil varieties, which do not carry the genetically modified label, also could be exploited. An increase in oils in leaves has been successful. Leaves are not a major metabolic sink organ, unlike maturing seed or underground storage organs. They have to be active and efficient in photosynthesis to support the growth of the whole plant. The diversion of leaves to storage organs, even partially, will affect photosynthesis and plant growth. In addition, LDs accumulated abundantly in the leaf cells will be bulky and will physically affect photosynthesis and other cellular activities. The argument for increasing oils in leftover leaves, stems, and roots in crops after seed harvest for biodiesel extraction is puzzling. These vegetative components were active and were constructed to serve their major physiological functions before seed harvest, and diversion of these functions would hurt plant growth. For centuries, agricultural efforts, intentionally or not, have increased the harvest index (bioenergy ratio of seed yield to vegetative components) to approach the theoretical maximum of 50%. Notwithstanding, the successes in the transformation studies (described in the preceding paragraph) have provided new and important knowledge of lipid metabolism in terms of its regulation and plasticity. Niches with sound conceptual foundations can be developed and explored for potential applications. The production of high-valued new oils could be pursued. The use of RNAi to suppress lipase transcript in maturing seeds could substantially reduce the ∼10% loss of TAGs during desiccation (Kelly et al., 2013a). CRISPR/Cas-targeted genome-editing technology should be used whenever possible to avoid the label of genetically modified. The current assessment is based on scientific deliberations and public perception of genetic modification, and proprietary considerations are beyond the scope of this article.
(2) Produce High-Value Protein via Its Engineered Attachment to Oleosin on LDs
Yeast and bacteria have been used to produce high-value proteins for medicinal and other purposes. This traditional approach could encounter a technical problem (in addition to the well-known lack of proper protein glycosylation in microbes). In laboratories, engineered microbes produce the foreign protein, which is then purified with chemical methods such as affinity chromatography. In scaling up production in factories, this purification process works less effectively and generates more contaminants. Attempts to overcome this purification problem and to take advantage of the low-cost agricultural production have led to the use of oleosin-LD technology (Bhatla et al., 2010; Nykiforuk et al., 2011; Huang et al., 2017). A high-value foreign protein is linked to oleosin on seed LDs via genetic engineering, and the foreign protein is released from floated LDs via a protease with substrate specificity tailored to a peptide linking the protein to oleosin. However, the scaled-up production with protein-oleosin-LDs could encounter the same scale-up problem in purity as the traditional microbe production. No industrial application with this approach has been reported 2 decades after the initiation of the research and development.
A modified and simpler approach for manufacturing industrial proteins requiring less purity is to attach the foreign enzyme to oleosin-LDs in seeds via genetic engineering and use the floated enzyme-oleosin-LDs without further purification or the crude extract directly (Yi et al., 2015; Leng et al., 2016; Liu et al., 2016; Montesinos et al., 2016; and other reports). Some of the engineered enzyme-oleosin-LDs have been shown to possess the expected enzymic or biological activities. This system offers an advantage in that the enzyme associated with oleosin-LD is less susceptible to proteolysis in vivo or in extracts. Its commercial feasibility remains to be seen.
(3) Enhance or Modify the Effects of Oleosin-LDs in Soybean Foods in Terms of Processing, Product Texture, and Nutrients
Thousands of soybean (Glycine max) products have been used as staple foods by different cultures in East and Southeast Asia for centuries. During the past 2 decades, their uses have spread globally. Many soybean products have gained a reputation for being health foods. The better-known items in the United States are soybean milk, tofu, soy yogurt, soya sauce, and various vegetarian foods. For most soybean foods, the bean is processed to products with different textures and available nutrients. In various soybean extracts, the water-soluble and abundant oleosin-LDs (including oleosin-LD ghosts after oil extraction with hexane) have tremendous effects on food stability (e.g. soymilk), textures (e.g. tofu and many other processed soybean products), and preservation of nutrients. Recent studies in Japan and China have probed the molecular structures and properties of oleosins on LDs, their interaction with storage proteins and soybean allergens as well as added non-soybean ingredients, and their modifications during and after different processing; the objective is to enhance soybean food qualities for different consumer preferences (Toda et al., 2008; Fujii, 2017; Matsumura et al., 2017; Peng et al., 2017; Zhao et al., 2017).
(4) Use Oleosin to Produce Micelles for Drug Delivery and Other Industrial Purposes
Oleosin is an emulsifying agent per se and has unique properties. It has been used as a platform protein on micelles, or even the sole component of micelles, for drug delivery and other purposes. For micelle production, oleosin and its modified variants are used to constitute micelles of different desired properties (Vargo et al., 2014). The size and surface properties of the micelles depend on the modified oleosin structures, the concentrations, and the media, etc. The micelles could have the oleosin C terminus attached to a cell receptor-binding peptide, so that the micelles can carry the drug to target specific human cells. The oleosin micelles also can be used as contrast-enhancing agents with ultrasonography and theranostic agents for diagnosis and therapy (Vargo et al., 2015). Oleosin has been used as a scaffold on LDs in yeast to assemble multiple enzymes for metabolic channeling in lipid synthesis (Lin et al., 2017).
PERSPECTIVE
LDs in all organisms are now recognized as a dynamic metabolic center for lipid homeostasis and other functions. Studies of LDs were initiated with plant seeds, and since then, LD studies with mammals and microbes have moved forward. Exchanges of research concepts and findings from diverse organisms have been ongoing and fruitful. LDs in all organisms share common features, and several LD proteins are evolutionarily related. This indicates the early evolution of LDs and some LD proteins in primitive organisms. Research with nonplant organisms has delineated the mechanism of LDs budding from the ER and the involvement of special proteins, such as Fat Storage Inducing Protein and SIEPIN. It also has defined the mode of LD catabolism whereby regulatory proteins and lipase bind transiently to or detach from LDs via phosphorylation and dephosphorylation and peroxisomes interact physically with LDs. Plant research on LDs should follow this progress and make advances and pinpoint diversifications. Diversifications are exemplified in the lipase modulator CGI-58, which transiently locates on LDs in humans but is present on peroxisomes in plants.
Stress-related proteins found on plant LDs during the past several years could function in combating abiotic and biotic stresses. The surface of LDs in plant cells could act as an inert refuge for nonoleosin proteins to perform major functions in other parts of the cells. This hypothesis should be explored actively. Regardless of any validation of the hypothesis, the research findings will delineate the functions of these proteins and the advantage of their association with LDs. All the nonoleosin LD proteins are not tightly associated with LDs (Table I). Caleosin and steroleosin have a short hydrophobic hairpin that would allow the protein to bind to various membranes in the cell and on the LD surface. Dioxygenase, lipoxygenase, and LD/OB Associated Proteins do not have a long hydrophobic segment and could be present in cytosol and on LDs. LD/OB Associated Proteins have been shown to be present in cytosol, and a portion is loosely associated with the LD surface. The mechanism of the association of these proteins with the LD surface and the changes of the association upon a physiological or pathological condition are indispensable information for delineating the functions of the proteins. LD Associated Protein has been tested for its in vitro binding to liposomes composed of various PLs, and no binding specificity has been identified (Gidda et al., 2016). The binding of the nonoleosin proteins to oleosin and its truncated forms on LDs or liposomes should be explored. Seed LDs are covered completely with oleosins, and the nonoleosin proteins would likely bind to the oleosin, at least partly. Nonseed LDs, such as those in leaves, may have their surface covered at least partly with oleosin. Deep sequencing of transcripts of various organs has revealed the presence of very low levels of oleosin transcripts in the leaves of Coffea, Pinus, Picea, Ginkgo, Linum, and Vernicia spp. (Simkin et al., 2006; Hyun et al., 2013; Cao et al., 2014; Huang and Huang, 2015). The low levels could simply reflect the very minimal numbers of LDs in these tissues. The oleosin transcript may increase, in parallel with the enhanced LD number and nonoleosin protein transcript, upon stresses or senescence. If oleosin is not involved in the specific binding of the nonoleosin proteins to LDs, other mechanism (e.g. phosphorylation and palmitoylation) should be explored.
The emphasis on oil synthesis and modification, as well as lipid metabolism related to stress responses, have minimized studies of LD breakdown in seedlings, which are perceived to be less related to applicability. The classic argument that good basic research will lead to applicability may be valid in studying LD breakdown. Studies with Arabidopsis reveal that the lipase transcript peaks during seed maturation, that it is 10% to 30% of the peak in seedlings at the height of lipolysis, and that it is present ubiquitously in diverse organs (Table III). In Arabidopsis and Brassica spp., lipase activity is absent in mature seed and appears in seedlings. Some activity of this lipase could be present in maturing seed and be responsible for the loss of ∼10% of TAGs during seed desiccation; this loss is common among many oilseed crops, including Brassica spp. RNAi of the lipase gene applied to Brassica spp. maturing seeds generates less lipase transcript and reduces the TAG loss during seed desiccation (Kelly et al., 2013a). Further research could investigate whether, in maturing seed and nonseed organs, the transcript is active in producing inactive or active lipase or the transcript or lipase becomes active only in seedlings. The mechanism of the possible storage of inactive transcript and/or lipase and their subsequent activation is intriguing.
Many intriguing scientific questions on LDs should be addressed. They have been mentioned throughout the text of this article. Some are also listed in Outstanding Questions.
Acknowledgments
I sincerely thank members of my laboratory for their excellent contributions to plant LD studies during the past decades.
Footnotes
The author’s research on lipid droplets was supported by grants from the USDA-National Research Initiative Grant Program, the National Science Foundation, and Hatch-AES.
Articles can be viewed without a subscription.
References
- Abell BM, Hahn M, Holbrook LA, Moloney MM (2004) Membrane topology and sequence requirements for oil body targeting of oleosin. Plant J 37: 461–470 [DOI] [PubMed] [Google Scholar]
- Abell BM, High S, Moloney MM (2002) Membrane protein topology of oleosin is constrained by its long hydrophobic domain. J Biol Chem 277: 8602–8610 [DOI] [PubMed] [Google Scholar]
- Alameldin H, Izadi-Darbandi A, Smith SA, Balan V, Jonese AD, Orhun GE, Sticklen M (2017) Metabolic engineering to increase the corn seed storage lipid quantity and change its compositional quality. Crop Sci 57: 1854–1864 [Google Scholar]
- Alexander LG, Sessions RB, Clarke AR, Tatham AS, Shewry PR, Napier JA (2002) Characterization and modelling of the hydrophobic domain of a sunflower oleosin. Planta 214: 546–551 [DOI] [PubMed] [Google Scholar]
- Barbosa AD, Siniossoglou S (2017) Function of lipid droplet-organelle interactions in lipid homeostasis. Biochim Biophys Acta 1864: 1459–1468 [DOI] [PubMed] [Google Scholar]
- Barneda D, Christian M (2017) Lipid droplet growth: regulation of a dynamic organelle. Curr Opin Cell Biol 47: 9–15 [DOI] [PubMed] [Google Scholar]
- Beaudoin F, Napier JA (2002) Targeting and membrane-insertion of a sunflower oleosin in vitro and in Saccharomyces cerevisiae: the central hydrophobic domain contains more than one signal sequence, and directs oleosin insertion into the endoplasmic reticulum membrane using a signal anchor sequence mechanism. Planta 215: 293–303 [DOI] [PubMed] [Google Scholar]
- Beaudoin F, Wilkinson BM, Stirling CJ, Napier JA (2000) In vivo targeting of a sunflower oil body protein in yeast secretory (sec) mutants. Plant J 23: 159–170 [DOI] [PubMed] [Google Scholar]
- Berthelot K, Lecomte S, Estevez Y, Peruch F (2014) Hevea brasiliensis REF (Hev b 1) and SRPP (Hev b 3): an overview on rubber particle proteins. Biochimie 106: 1–9 [DOI] [PubMed] [Google Scholar]
- Bhatla SC, Kaushik V, Yadav MK (2010) Use of oil bodies and oleosins in recombinant protein production and other biotechnological applications. Biotechnol Adv 28: 293–300 [DOI] [PubMed] [Google Scholar]
- Blée E, Boachon B, Burcklen M, Le Guédard M, Hanano A, Heintz D, Ehlting J, Herrfurth C, Feussner I, Bessoule JJ (2014) The reductase activity of the Arabidopsis caleosin RESPONSIVE TO DESSICATION20 mediates gibberellin-dependent flowering time, abscisic acid sensitivity, and tolerance to oxidative stress. Plant Physiol 166: 109–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgis F, Kilaru A, Cao X, Ngando-Ebongue GF, Drira N, Ohlrogge JB, Arondel V (2011) Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proc Natl Acad Sci USA 108: 12527–12532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bréhélin C, Kessler F (2008) The plastoglobule: a bag full of lipid biochemistry tricks. Photochem Photobiol 84: 1388–1394 [DOI] [PubMed] [Google Scholar]
- Cai G, Parrotta L, Cresti M (2015a) Organelle trafficking, the cytoskeleton, and pollen tube growth. J Integr Plant Biol 57: 63–78 [DOI] [PubMed] [Google Scholar]
- Cai Y, Goodman JM, Pyc M, Mullen RT, Dyer JM, Chapman KD (2015b) Arabidopsis SEIPIN proteins modulate triacylglycerol accumulation and influence lipid droplet proliferation. Plant Cell 27: 2616–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, McClinchie E, Price A, Nguyen TN, Gidda SK, Watt SC, Yurchenko O, Park S, Sturtevant D, Mullen RT, et al. (2017) Mouse fat storage-inducing transmembrane protein 2 (FIT2) promotes lipid droplet accumulation in plants. Plant Biotechnol J 15: 824–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Zhang L, Tan X, Long H, Shockey JM (2014) Identification, classification and differential expression of oleosin genes in tung tree (Vernicia fordii). PLoS ONE 9: e88409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao YZ, Huang AHC (1986) Diacylglycerol acyltransferase in maturing oil seeds of maize and other species. Plant Physiol 82: 813–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KD, Dyer JM, Mullen RT (2012) Biogenesis and functions of lipid droplets in plants. J Lipid Res 53: 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charuchinda P, Waditee-Sirisattha R, Kageyama H, Yamada D, Sirisattha S, Tanaka Y, Mahakhant A, Takabe T (2015) Caleosin from Chlorella vulgaris TISTR 8580 is salt-induced and heme-containing protein. Biosci Biotechnol Biochem 79: 1119–1124 [DOI] [PubMed] [Google Scholar]
- Chen JCF, Tsai CCY, Tzen JTC (1999) Cloning and secondary structure analysis of caleosin, a unique calcium-binding protein in oil bodies of plant seeds. Plant Cell Physiol 40: 1079–1086 [DOI] [PubMed] [Google Scholar]
- Chen X, Goodman JM (2017) The collaborative work of droplet assembly. Biochim Biophys Acta 1862: 1205–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S, Hayashi Y, Otomo M, Mano S, Oikawa K, Hayashi M, Nishimura M (2016) Sucrose production mediated by lipid metabolism suppresses the physical interaction of peroxisomes and oil bodies during germination of Arabidopsis thaliana. J Biol Chem 291: 19734–19745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidi L, Katz A, Pick U (2012) Characterization of major lipid droplet proteins from Dunaliella. Planta 236: 19–33 [DOI] [PubMed] [Google Scholar]
- Deruère J, Römer S, d’Harlingue A, Backhaus RA, Kuntz M, Camara B (1994) Fibril assembly and carotenoid overaccumulation in chromoplasts: a model for supramolecular lipoprotein structures. Plant Cell 6: 119–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deruyffelaere C, Bouchez I, Morin H, Guillot A, Miquel M, Froissard M, Chardot T, D’Andrea S (2015) Ubiquitin-mediated proteasomal degradation of oleosin is involved in oil body mobilization during post-germinative seedling growth in Arabidopsis. Plant Cell Physiol 56: 1374–1387 [DOI] [PubMed] [Google Scholar]
- Ding Y, Yang L, Zhang S, Wang Y, Du Y, Pu J, Peng G, Chen Y, Zhang H, Yu J, et al. (2012) Identification of the major functional proteins of prokaryotic lipid droplets. J Lipid Res 53: 399–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ. (2006) SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell 18: 665–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Tahchy A, Reynolds KB, Petrie JR, Singh SP, Vanhercke T (2017) Thioesterase overexpression in Nicotiana benthamiana leaf increases the fatty acid flux into triacylgycerol. FEBS Lett 591: 448–456 [DOI] [PubMed] [Google Scholar]
- Fernandez DE, Qu R, Huang AHC, Staehelin LA (1988) Immunogold localization of the L3 protein of maize lipid bodies during germination and seedling growth. Plant Physiol 86: 270–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feussner I, Kühn H, Wasternack C (2001) Lipoxygenase-dependent degradation of storage lipids. Trends Plant Sci 6: 268–273 [DOI] [PubMed] [Google Scholar]
- Frandsen G, Müller-Uri F, Nielsen M, Mundy J, Skriver K (1996) Novel plant Ca2+-binding protein expressed in response to abscisic acid and osmotic stress. J Biol Chem 271: 343–348 [DOI] [PubMed] [Google Scholar]
- Frandsen GI, Mundy J, Tzen JTC (2001) Oil bodies and their associated proteins, oleosin and caleosin. Physiol Plant 112: 301–307 [DOI] [PubMed] [Google Scholar]
- Fujii T. (2017) Coagulation and rheological behaviors of soy milk colloidal dispersions. Biosci Biotechnol Biochem 81: 680–686 [DOI] [PubMed] [Google Scholar]
- Garay LA, Boundy-Mills KL, German JB (2014) Accumulation of high-value lipids in single-cell microorganisms: a mechanistic approach and future perspectives. J Agric Food Chem 62: 2709–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidda SK, Park S, Pyc M, Yurchenko O, Cai Y, Wu P, Andrews DW, Chapman KD, Dyer JM, Mullen RT (2016) Lipid droplet-associated proteins (LDAPs) are required for the dynamic regulation of neutral lipid compartmentation in plant cells. Plant Physiol 170: 2052–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohon Y, Vindigni JD, Pallier A, Wien F, Celia H, Giuliani A, Tribet C, Chardot T, Briozzo P (2011) High water solubility and fold in amphipols of proteins with large hydrophobic regions: oleosins and caleosin from seed lipid bodies. Biochim Biophys Acta 1808: 706–716 [DOI] [PubMed] [Google Scholar]
- Goold H, Beisson F, Peltier G, Li-Beisson Y (2015) Microalgal lipid droplets: composition, diversity, biogenesis and functions. Plant Cell Rep 34: 545–555 [DOI] [PubMed] [Google Scholar]
- Greenberg AS, Egan JJ, Wek SA, Moos MC Jr, Londos C, Kimmel AR (1993) Isolation of cDNAs for perilipins A and B: sequence and expression of lipid droplet-associated proteins of adipocytes. Proc Natl Acad Sci USA 90: 12035–12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanano A, Burcklen M, Flenet M, Ivancich A, Louwagie M, Garin J, Blée E (2006) Plant seed peroxygenase is an original heme-oxygenase with an EF-hand calcium binding motif. J Biol Chem 281: 33140–33151 [DOI] [PubMed] [Google Scholar]
- Hong Y, Zhao J, Guo L, Kim SC, Deng X, Wang G, Zhang G, Li M, Wang X (2016) Plant phospholipases D and C and their diverse functions in stress responses. Prog Lipid Res 62: 55–74 [DOI] [PubMed] [Google Scholar]
- Horn PJ, James CN, Gidda SK, Kilaru A, Dyer JM, Mullen RT, Ohlrogge JB, Chapman KD (2013) Identification of a new class of lipid droplet-associated proteins in plants. Plant Physiol 162: 1926–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao ES, Tzen JTC (2011) Ubiquitination of oleosin-H and caleosin in sesame oil bodies after seed germination. Plant Physiol Biochem 49: 77–81 [DOI] [PubMed] [Google Scholar]
- Hsieh K, Huang AHC (2004) Endoplasmic reticulum, oleosins, and oils in seeds and tapetum cells. Plant Physiol 136: 3427–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh K, Huang AHC (2005) Lipid-rich tapetosomes in Brassica tapetum are composed of oleosin-coated oil droplets and vesicles, both assembled in and then detached from the endoplasmic reticulum. Plant J 43: 889–899 [DOI] [PubMed] [Google Scholar]
- Hsieh K, Huang AHC (2007) Tapetosomes in Brassica tapetum accumulate endoplasmic reticulum-derived flavonoids and alkanes for delivery to the pollen surface. Plant Cell 19: 582–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AHC. (1987) Lipases. In Stumpf PK, Conn EE, eds, Lipids II. Academic Press, New York: pp 91–119 [Google Scholar]
- Huang AHC. (1992) Oil bodies and oleosins in seeds. Annu Rev Plant Physiol Plant Mol Biol 43: 177–200 [Google Scholar]
- Huang CY, Chen PY, Huang MD, Tsou CH, Jane WN, Huang AHC (2013a) Tandem oleosin genes in a cluster acquired in Brassicaceae created tapetosomes and conferred additive benefit of pollen vigor. Proc Natl Acad Sci USA 110: 14480–14485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Chung CI, Lin YC, Hsing YIC, Huang AHC (2009) Oil bodies and oleosins in Physcomitrella possess characteristics representative of early trends in evolution. Plant Physiol 150: 1192–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Huang AHC (2017) Unique motifs and length of hairpin in oleosin targets the cytosolic side of endoplasmic reticulum and budding lipid droplets. Plant Physiol 174: 2248–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Yang J, Guan L, Yi S, Du L, Tian H, Guo Y, Zhai F, Lu Z, Li H, et al. (2017) Expression of bioactive recombinant human fibroblast growth factor 10 in Carthamus tinctorius L. seeds. Protein Expr Purif 138: 7–12 [DOI] [PubMed] [Google Scholar]
- Huang MD, Huang AHC (2015) Bioinformatics reveal five lineages of oleosins and the mechanism of lineage evolution related to structure/function from green algae to seed plants. Plant Physiol 169: 453–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MD, Huang AHC (2016) Subcellular lipid droplets in vanilla leaf epidermis and avocado mesocarp are coated with oleosins of existing and novel phylogenic lineages. Plant Physiol 171: 1867–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang NL, Huang MD, Chen TLL, Huang AHC (2013b) Oleosin of subcellular lipid droplets evolved in green algae. Plant Physiol 161: 1862–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun TK, Kumar D, Cho YY, Hyun HN, Kim JS (2013) Computational identification and phylogenetic analysis of the oil-body structural proteins, oleosin and caleosin, in castor bean and flax. Gene 515: 454–460 [DOI] [PubMed] [Google Scholar]
- Itabe H, Yamaguchi T, Nimura S, Sasabe N (2017) Perilipins: a diversity of intracellular lipid droplet proteins. Lipids Health Dis 16: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier N, Mishra S, Choudhary V, Schneiter R (2013) Expression of oleosin and perilipins in yeast promotes formation of lipid droplets from the endoplasmic reticulum. J Cell Sci 126: 5198–5209 [DOI] [PubMed] [Google Scholar]
- James CN, Horn PJ, Case CR, Gidda SK, Zhang D, Mullen RT, Dyer JM, Anderson RGW, Chapman KD (2010) Disruption of the Arabidopsis CGI-58 homologue produces Chanarin-Dorfman-like lipid droplet accumulation in plants. Proc Natl Acad Sci USA 107: 17833–17838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jappe U, Schwager C (2017) Relevance of lipophilic allergens in food allergy diagnosis. Curr Allergy Asthma Rep 17: 61. [DOI] [PubMed] [Google Scholar]
- Jolivet P, Acevedo F, Boulard C, d’Andréa S, Faure JD, Kohli A, Nesi N, Valot B, Chardot T (2013) Crop seed oil bodies: from challenges in protein identification to an emerging picture of the oil body proteome. Proteomics 13: 1836–1849 [DOI] [PubMed] [Google Scholar]
- Kelly AA, Shaw E, Powers SJ, Kurup S, Eastmond PJ (2013a) Suppression of the SUGAR-DEPENDENT1 triacylglycerol lipase family during seed development enhances oil yield in oilseed rape (Brassica napus L.). Plant Biotechnol J 11: 355–361 [DOI] [PubMed] [Google Scholar]
- Kelly AA, van Erp H, Quettier AL, Shaw E, Menard G, Kurup S, Eastmond PJ (2013b) The sugar-dependent1 lipase limits triacylglycerol accumulation in vegetative tissues of Arabidopsis. Plant Physiol 162: 1282–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilaru A, Cao X, Dabbs PB, Sung HJ, Rahman MM, Thrower N, Zynda G, Podicheti R, Ibarra-Laclette E, Herrera-Estrella L, et al. (2015) Oil biosynthesis in a basal angiosperm: transcriptome analysis of Persea americana mesocarp. BMC Plant Biol 15: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HU, Hsieh K, Ratnayake C, Huang AHC (2002) A novel group of oleosins is present inside the pollen of Arabidopsis. J Biol Chem 277: 22677–22684 [DOI] [PubMed] [Google Scholar]
- Kim HU, Lee KR, Jung SJ, Shin HA, Go YS, Suh MC, Kim JB (2015) Senescence-inducible LEC2 enhances triacylglycerol accumulation in leaves without negatively affecting plant growth. Plant Biotechnol J 13: 1346–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim RJ, Kim HJ, Shim D, Suh MC (2016) Molecular and biochemical characterizations of the monoacylglycerol lipase gene family of Arabidopsis thaliana. Plant J 85: 758–771 [DOI] [PubMed] [Google Scholar]
- Koch B, Schmidt C, Daum G (2014) Storage lipids of yeasts: a survey of nonpolar lipid metabolism in Saccharomyces cerevisiae, Pichia pastoris, and Yarrowia lipolytica. FEMS Microbiol Rev 38: 892–915 [DOI] [PubMed] [Google Scholar]
- Kory N, Farese RV Jr, Walther TC (2016) Targeting fat: mechanisms of protein localization to lipid droplets. Trends Cell Biol 26: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowska M, Polit JT, Stępiński D, Popłońska K, Wojtczak A, Domίnguez E, Heredia A (2015) Lipotubuloids in ovary epidermis of Ornithogalum umbellatum act as metabolons: suggestion of the name ‘lipotubuloid metabolon.’ J Exp Bot 66: 1157–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowska M, Stepinski D, Poplonska K, Wojtczak A, Polit JT (2010) “Elaioplasts” of Haemanthus albiflos are true lipotubuloids: cytoplasmic domains rich in lipid bodies entwined by microtubules. Acta Physiol Plant 32: 1189–1196 [Google Scholar]
- Lacey DJ, Beaudoin F, Dempsey CE, Shewry PR, Napier JA (1999) The accumulation of triacylglycerols within the endoplasmic reticulum of developing seeds of Helianthus annuus. Plant J 17: 397–405 [Google Scholar]
- Lee K, Ratnayake C, Huang AHC (1995) Genetic dissection of the co-expression of genes encoding the two isoforms of oleosins in the oil bodies of maize kernel. Plant J 7: 603–611 [DOI] [PubMed] [Google Scholar]
- Leng SH, Yang CE, Tsai SL (2016) Designer oleosomes as efficient biocatalysts for enhanced degradation of organophosphate nerve agents. Chem Eng J 287: 568–574 [Google Scholar]
- Lersten NR, Czlapinski AR, Curtis JD, Freckmann R, Horner HT (2006) Oil bodies in leaf mesophyll cells of angiosperms: overview and a selected survey. Am J Bot 93: 1731–1739 [DOI] [PubMed] [Google Scholar]
- Lévesque-Lemay M, Chabot D, Hubbard K, Chan JK, Miller S, Robert LS (2016) Tapetal oleosins play an essential role in tapetosome formation and protein relocation to the pollen coat. New Phytol 209: 691–704 [DOI] [PubMed] [Google Scholar]
- Li M, Murphy DJ, Lee KHK, Wilson R, Smith LJ, Clark DC, Sung JY (2002) Purification and structural characterization of the central hydrophobic domain of oleosin. J Biol Chem 277: 37888–37895 [DOI] [PubMed] [Google Scholar]
- Li Z, Thiel K, Thul PJ, Beller M, Kühnlein RP, Welte MA (2012) Lipid droplets control the maternal histone supply of Drosophila embryos. Curr Biol 22: 2104–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JL, Zhu J, Wheeldon I (2017) Synthetic protein scaffolds for biosynthetic pathway colocalization on lipid droplet membranes. ACS Synth Biol 6: 1534–1544 [DOI] [PubMed] [Google Scholar]
- Lin LJ, Tai SSK, Peng CC, Tzen JTC (2002) Steroleosin, a sterol-binding dehydrogenase in seed oil bodies. Plant Physiol 128: 1200–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YH, Huang AHC (1983) Lipase in lipid bodies of cotyledons of rape and mustard seedlings. Arch Biochem Biophys 225: 360–369 [DOI] [PubMed] [Google Scholar]
- Lindemann P. (2015) Steroidogenesis in plants: biosynthesis and conversions of progesterone and other pregnane derivatives. Steroids 103: 145–152 [DOI] [PubMed] [Google Scholar]
- Liu M, Chu S, Ai J, Li H, Chen Z, Huang S, Jiang C, Li X (2016) Application of oleosin-flanked keratinocyte growth factor-2 expressed from Arabidopsis thaliana promotes hair follicle growth in mice. Biotechnol Lett 38: 1611–1619 [DOI] [PubMed] [Google Scholar]
- Liu Q, Guo Q, Akbar S, Zhi Y, El Tahchy A, Mitchell M, Li Z, Shrestha P, Vanhercke T, Ral JP, et al. (2017) Genetic enhancement of oil content in potato tuber (Solanum tuberosum L.) through an integrated metabolic engineering strategy. Plant Biotechnol J 15: 56–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WX, Liu HL, Qu Q (2013) Embryo-specific expression of soybean oleosin altered oil body morphogenesis and increased lipid content in transgenic rice seeds. Theor Appl Genet 126: 2289–2297 [DOI] [PubMed] [Google Scholar]
- Loer DS, Herman EM (1993) Cotranslational integration of soybean (Glycine max) oil body membrane protein oleosin into microsomal membranes. Plant Physiol 101: 993–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Ribera I, La Paz JL, Repiso C, García N, Miquel M, Hernández ML, Martínez-Rivas JM, Vicient CM (2014) The evolutionary conserved oil body associated protein OBAP1 participates in the regulation of oil body size. Plant Physiol 164: 1237–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos R, Izquierdo Y, Vellosillo T, Kulasekaran S, Cascón T, Hamberg M, Castresana C (2015) 9-Lipoxygenase-derived oxylipins activate brassinosteroid signaling to promote cell wall-based defense and limit pathogen infection. Plant Physiol 169: 2324–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos AP. (2017) The impact of microalgae in food science and technology. J Am Oil Chem Soc 94: 1333–1350 [Google Scholar]
- Matsumura Y, Sirison J, Ishi T, Matsumiya K (2017) Soybean lipophilic proteins: origin and functional properties as affected by interaction with storage proteins. Curr Opin Colloid Interface Sci 28: 120–128 [Google Scholar]
- Mezzina MP, Pettinari MJ (2016) Phasins, multifaceted polyhydroxyalkanoate granule-associated proteins. Appl Environ Microbiol 82: 5060–5067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering ER, Benning C (2010) RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot Cell 9: 97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos L, Bundó M, Izquierdo E, Campo S, Badosa E, Rossignol M, Montesinos E, San Segundo B, Coca M (2016) Production of biologically active cecropin A peptide in rice seed oil bodies. PLoS ONE 11: e0146919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller AO, Blersch KF, Gippert AL, Ischebeck T (2017) Tobacco pollen tubes: a fast and easy tool for studying lipid droplet association of plant proteins. Plant J 89: 1055–1064 [DOI] [PubMed] [Google Scholar]
- Naested H, Frandsen GI, Jauh GY, Hernandez-Pinzon I, Nielsen HB, Murphy DJ, Rogers JC, Mundy J (2000) Caleosins: Ca2+-binding proteins associated with lipid bodies. Plant Mol Biol 44: 463–476 [DOI] [PubMed] [Google Scholar]
- Nolan SJ, Romano JD, Coppens I (2017) Host lipid droplets: an important source of lipids salvaged by the intracellular parasite Toxoplasma gondii. PLoS Pathog 13: e1006362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykiforuk CL, Shen Y, Murray EW, Boothe JG, Busseuil D, Rhéaume E, Tardif JC, Reid A, Moloney MM (2011) Expression and recovery of biologically active recombinant apolipoprotein AI(Milano) from transgenic safflower (Carthamus tinctorius) seeds. Plant Biotechnol J 9: 250–263 [DOI] [PubMed] [Google Scholar]
- Oberer M, Boeszoermenyi A, Nagy HM, Zechner R (2011) Recent insights into the structure and function of comparative gene identification-58. Curr Opin Lipidol 22: 149–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SK, Kang H, Shin DH, Yang J, Chow KS, Yeang HY, Wagner B, Breiteneder H, Han KH (1999) Isolation, characterization, and functional analysis of a novel cDNA clone encoding a small rubber particle protein from Hevea brasiliensis. J Biol Chem 274: 17132–17138 [DOI] [PubMed] [Google Scholar]
- Onal G, Kutlu O, Gozuacik D, Dokmeci Emre S (2017) Lipid droplets in health and disease. Lipids Health Dis 16: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasaribu B, Chung TY, Chen CS, Jiang PL, Tzen JTC (2016) Identification of steroleosin in oil bodies of pine megagametophytes. Plant Physiol Biochem 101: 173–181 [DOI] [PubMed] [Google Scholar]
- Peng X, Wang Y, Xing J, Wang R, Shi X, Guo S (2017) Characterization of particles in soymilks prepared by blanching soybean and traditional method: a comparative study focusing on lipid-protein interaction. Food Hydrocoll 63: 1–7 [Google Scholar]
- Pieper-Fürst U, Madkour MH, Mayer F, Steinbüchel A (1994) Purification and characterization of a 14-kilodalton protein that is bound to the surface of polyhydroxyalkanoic acid granules in Rhodococcus ruber. J Bacteriol 176: 4328–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poxleitner M, Rogers SW, Lacey Samuels A, Browse J, Rogers JC (2006) A role for caleosin in degradation of oil-body storage lipid during seed germination. Plant J 47: 917–933 [DOI] [PubMed] [Google Scholar]
- Pyc M, Cai Y, Greer MS, Yurchenko O, Chapman KD, Dyer JM, Mullen RT (2017) Turning over a new leaf in lipid droplet biology. Trends Plant Sci 22: 596–609 [DOI] [PubMed] [Google Scholar]
- Qu R, Wang SM, Lin YH, Vance VB, Huang AHC (1986) Characteristics and biosynthesis of membrane proteins of lipid bodies in the scutella of maize (Zea mays L.). Biochem J 235: 57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roingeard P, Melo RCN (2017) Lipid droplet hijacking by intracellular pathogens. Cell Microbiol 19: e12688. [DOI] [PubMed] [Google Scholar]
- Rudolph M, Schlereth A, Körner M, Feussner K, Berndt E, Melzer M, Hornung E, Feussner I (2011) The lipoxygenase-dependent oxygenation of lipid body membranes is promoted by a patatin-type phospholipase in cucumber cotyledons. J Exp Bot 62: 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sando T, Hayashi T, Takeda T, Akiyama Y, Nakazawa Y, Fukusaki E, Kobayashi A (2009) Histochemical study of detailed laticifer structure and rubber biosynthesis-related protein localization in Hevea brasiliensis using spectral confocal laser scanning microscopy. Planta 230: 215–225 [DOI] [PubMed] [Google Scholar]
- Sanz A, Moreno JI, Castresana C (1998) PIOX, a new pathogen-induced oxygenase with homology to animal cyclooxygenase. Plant Cell 10: 1523–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savadi S, Naresh V, Kumar V, Bhat SR (2016) Seed-specific overexpression of Arabidopsis DGAT1 in Indian mustard (Brassica juncea) increases seed oil content and seed weight. Botany 94: 177–184 [Google Scholar]
- Schmidt MA, Herman EM (2008) Suppression of soybean oleosin produces micro-oil bodies that aggregate into oil body/ER complexes. Mol Plant 1: 910–924 [DOI] [PubMed] [Google Scholar]
- Shen B, Allen WB, Zheng P, Li C, Glassman K, Ranch J, Nubel D, Tarczynski MC (2010) Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol 153: 980–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada TL, Hara-Nishimura I (2015) Leaf oil bodies are subcellular factories producing antifungal oxylipins. Curr Opin Plant Biol 25: 145–150 [DOI] [PubMed] [Google Scholar]
- Shimada TL, Shimada T, Takahashi H, Fukao Y, Hara-Nishimura I (2008) A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J 55: 798–809 [DOI] [PubMed] [Google Scholar]
- Shimada TL, Takano Y, Shimada T, Fujiwara M, Fukao Y, Mori M, Okazaki Y, Saito K, Sasaki R, Aoki K, et al. (2014) Leaf oil body functions as a subcellular factory for the production of a phytoalexin in Arabidopsis. Plant Physiol 164: 105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockey JM, Gidda SK, Chapital DC, Kuan JC, Dhanoa PK, Bland JM, Rothstein SJ, Mullen RT, Dyer JM (2006) Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18: 2294–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siloto RMP, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM (2006) The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin AJ, Qian T, Caillet V, Michoux F, Ben Amor M, Lin C, Tanksley S, McCarthy J (2006) Oleosin gene family of Coffea canephora: quantitative expression analysis of five oleosin genes in developing and germinating coffee grain. J Plant Physiol 163: 691–708 [DOI] [PubMed] [Google Scholar]
- Song W, Qin Y, Zhu Y, Yin G, Wu N, Li Y, Hu Y (2014) Delineation of plant caleosin residues critical for functional divergence, positive selection and coevolution. BMC Evol Biol 14: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar R, Buddenhagen TW, Ou SH (1973) Presence of lipid bodies in rice leaves and their discolouration during pathogenesis. Experimentia 19: 959–960 [Google Scholar]
- Taparia T, Mvss M, Mehrotra R, Shukla P, Mehrotra S (2016) Developments and challenges in biodiesel production from microalgae: a review. Biotechnol Appl Biochem 63: 715–726 [DOI] [PubMed] [Google Scholar]
- Taylor DC, Zhang Y, Kumar A, Francis T, Giblin EM, Baton DL, Ferrie JR, Laroche A, Shah S, Zhu W, et al. (2009) Molecular modification of triacylglycerol accumulation by over-expression of DGAT1 to produce canola with increased seed oil content under field conditions. Botany 87: 533–543 [Google Scholar]
- Thazar-Poulot N, Miquel M, Fobis-Loisy I, Gaude T (2015) Peroxisome extensions deliver the Arabidopsis SDP1 lipase to oil bodies. Proc Natl Acad Sci USA 112: 4158–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam AR, Beller M (2017) The why, when and how of lipid droplet diversity. J Cell Sci 130: 315–324 [DOI] [PubMed] [Google Scholar]
- Thoyts PJE, Millichip MI, Stobart AK, Griffiths WT, Shewry PR, Napier JA (1995) Expression and in vitro targeting of a sunflower oleosin. Plant Mol Biol 29: 403–410 [DOI] [PubMed] [Google Scholar]
- Ting JTL, Balsamo RA, Ratnayake C, Huang AHC (1997) Oleosin of plant seed oil bodies is correctly targeted to the lipid bodies in transformed yeast. J Biol Chem 272: 3699–3706 [DOI] [PubMed] [Google Scholar]
- Ting JTL, Lee K, Ratnayake C, Platt KA, Balsamo RA, Huang AHC (1996) Oleosin genes in maize kernels having diverse oil contents are constitutively expressed independent of oil contents: size and shape of intracellular oil bodies are determined by the oleosins/oils ratio. Planta 199: 158–165 [DOI] [PubMed] [Google Scholar]
- Tirinato L, Pagliari F, Limongi T, Marini M, Falqui A, Seco J, Candeloro P, Liberale C, Di Fabrizio E (2017) An overview of lipid droplets in cancer and cancer stem cells. Stem Cells International 2017: 1656053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda K, Yagasaki K, Takahashi K (2008) Relationship between protein composition and coagulation reactivity, particulate formation, and incorporation of lipids in soymilk. Biosci Biotechnol Biochem 72: 2824–2830 [DOI] [PubMed] [Google Scholar]
- Tzen JTC, Huang AHC (1992) Surface structure and properties of plant seed oil bodies. J Cell Biol 117: 327–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance VB, Huang AHC (1987) The major protein from lipid bodies of maize: characterization and structure based on cDNA cloning. J Biol Chem 262: 11275–11279 [PubMed] [Google Scholar]
- van Rooijen GJH, Moloney MM (1995) Structural requirements of oleosin domains for subcellular targeting to the oil body. Plant Physiol 109: 1353–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargo KB, Sood N, Moeller TD, Heiney PA, Hammer DA (2014) Spherical micelles assembled from variants of recombinant oleosin. Langmuir 30: 11292–11300 [DOI] [PubMed] [Google Scholar]
- Vargo KB, Zaki AA, Warden-Rothman R, Tsourkas A, Hammer DA (2015) Superparamagnetic iron oxide nanoparticle micelles stabilized by recombinant oleosin for targeted magnetic resonance imaging. Small 11: 1409–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieler A, Brubaker SB, Vick B, Benning C (2012) A lipid droplet protein of Nannochloropsis with functions partially analogous to plant oleosins. Plant Physiol 158: 1562–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vindigni JD, Wien F, Giuliani A, Erpapazoglou Z, Tache R, Jagic F, Chardot T, Gohon Y, Froissard M (2013) Fold of an oleosin targeted to cellular oil bodies. Biochim Biophys Acta 1828: 1881–1888 [DOI] [PubMed] [Google Scholar]
- Wahlroos T, Soukka J, Denesyuk A, Wahlroos R, Korpela T, Kilby NJ (2003) Oleosin expression and trafficking during oil body biogenesis in tobacco leaf cells. Genesis 35: 125–132 [DOI] [PubMed] [Google Scholar]
- Wältermann M, Steinbüchel A (2005) Neutral lipid bodies in prokaryotes: recent insights into structure, formation, and relationship to eukaryotic lipid depots. J Bacteriol 187: 3607–3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Chung J, Farese RV Jr (2017) Lipid droplet biogenesis. Annu Rev Cell Dev Biol 33: 491–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SM, Huang AHC (1987) Biosynthesis of lipase in the scutellum of maize kernel. J Biol Chem 262: 2270–2274 [PubMed] [Google Scholar]
- Welte MA. (2015) Expanding roles for lipid droplets. Curr Biol 25: R470–R481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte MA, Gould AP (2017) Lipid droplet functions beyond energy storage. Biochim Biophys Acta 1862: 1260–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weselake RJ, Taylor DC, Rahman MH, Shah S, Laroche A, McVetty PBE, Harwood JL (2009) Increasing the flow of carbon into seed oil. Biotechnol Adv 27: 866–878 [DOI] [PubMed] [Google Scholar]
- Winichayakul S, Scott RW, Roldan M, Hatier JHB, Livingston S, Cookson R, Curran AC, Roberts NJ (2013) In vivo packaging of triacylglycerols enhances Arabidopsis leaf biomass and energy density. Plant Physiol 162: 626–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SSH, Platt KA, Ratnayake C, Wang TW, Ting JTL, Huang AHC (1997) Isolation and characterization of neutral-lipid-containing organelles and globuli-filled plastids from Brassica napus tapetum. Proc Natl Acad Sci USA 94: 12711–12716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi S, Yang J, Huang J, Guan L, Du L, Guo Y, Zhai F, Wang Y, Lu Z, Wang L, et al. (2015) Expression of bioactive recombinant human fibroblast growth factor 9 in oil bodies of Arabidopsis thaliana. Protein Expr Purif 116: 127–132 [DOI] [PubMed] [Google Scholar]
- Yoneda K, Yoshida M, Suzuki I, Watanabe MM (2016) Identifications of a major lipid droplet protein in a marine diatom Phaeodactylum tricornutum. Plant Cell Physiol 57: 397–406 [DOI] [PubMed] [Google Scholar]
- Yurchenko O, Shockey JM, Gidda SK, Silver MI, Chapman KD, Mullen RT, Dyer JM (2017) Engineering the production of conjugated fatty acids in Arabidopsis thaliana leaves. Plant Biotechnol J 15: 1010–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zale J, Jung JH, Kim JY, Pathak B, Karan R, Liu H, Chen X, Wu H, Candreva J, Zhai Z, et al. (2016) Metabolic engineering of sugarcane to accumulate energy-dense triacylglycerols in vegetative biomass. Plant Biotechnol J 14: 661–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Du J, Wang L, Pan Z, Xu Q, Xiao S, Deng X (2015) A comparative analysis of chromoplast differentiation reveals complex protein changes associated with plastoglobule biogenesis and remodeling of protein systems in sweet orange flesh. Plant Physiol 168: 1648–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Yang L, Ding Y, Wang Y, Lan L, Ma Q, Chi X, Wei P, Zhao Y, Steinbach A, et al. (2017) Bacterial lipid droplets bind to DNA via an intermediary protein that enhances survival under stress. Nat Commun 8: 15979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Cheng ZJ, Gan L, Zhang H, Wu FQ, Lin QB, Wang JL, Wang J, Guo XP, Zhang X, et al. (2016) OsHSD1, a hydroxysteroid dehydrogenase, is involved in cuticle formation and lipid homeostasis in rice. Plant Sci 249: 35–45 [DOI] [PubMed] [Google Scholar]
- Zhao L, Kong X, Zhang C, Hua Y, Chen Y (2017) Soybean P34 probable thiol protease probably has proteolytic activity on oleosins. J Agric Food Chem 65: 5741–5750 [DOI] [PubMed] [Google Scholar]
- Zhi Y, Taylor MC, Campbell PM, Warden AC, Shrestha P, El Tahchy A, Rolland V, Vanhercke T, Petrie JR, White RG, et al. (2017) Comparative lipidomics and proteomics of lipid droplets in the mesocarp and seed tissues of Chinese tallow (Triadica sebifera). Front Plant Sci 8: 1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zienkiewicz K, Du ZY, Ma W, Vollheyde K, Benning C (2016) Stress-induced neutral lipid biosynthesis in microalgae: molecular, cellular and physiological insights. Biochim Biophys Acta 1861: 1269–1281 [DOI] [PubMed] [Google Scholar]
- Zulu NN, Popko J, Zienkiewicz K, Tarazona P, Herrfurth C, Feussner I (2017) Heterologous co-expression of a yeast diacylglycerol acyltransferase (ScDGA1) and a plant oleosin (AtOLEO3) as an efficient tool for enhancing triacylglycerol accumulation in the marine diatom Phaeodactylum tricornutum. Biotechnol Biofuels 10: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]