Abstract
Background
The aim of this study is to find the potential miRNA expression signature capable of predicting survival time for cervical squamous cell carcinoma (CSCC) patients.
Methods
The expression of 332 miRNAs was measured in 131 (Training cohort) and 130 (Validation cohort) patients with CSCC in the Cancer Genome Atlas (TCGA) data portal. The miRNA expression signature was identified by Cox Proportion Hazard regression model to the Training data set, and subsequently validated in an independent Validation set. Kaplan-Meier curves and the receiver operating characteristic analyses of 5 years were used to access the overall survival of miRNA signature. MiRNA signature-gene target analysis was performed, followed by the construction of the regulatory network. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway analysis were used to explore the function of target genes of miRNA signature.
Results
A 2-miRNA expression signature of hsa-mir-642a and hsa-mir-378c associated with survivability was identified in CSCC. Both of them had a significant diagnostic and prognostic value of patients with CSCC. A total of 345 miRNA signature-target pairs were obtained in the miRNA signature-gene target regulatory network, in which 316 genes were targets of has-mir-378c and has-mir-642a. Functional analysis of target genes showed that MAPK signaling pathway, VEGF signaling pathway and endocytosis were the significantly enriched signal pathways that covered most genes.
Conclusions
The 2-miRNA signature adds to the prognostic value of CSCC. In-depth interrogation of the 2-miRNAs will provide important biological insights that finding and developing novel molecularly prediction to improve prognosis for CSCC patients.
Introduction
Cervical squamous cell carcinoma (CSCC), accounting for about 75–80% of all cervical cancers, is one of the most common gynecological malignancy and leads to the cancer death in women [1, 2]. Walboomers JM and Castellsague X et al found that CSCC was closely associated with high-risk human papillomavirus (HPV) infection [3, 4]. In addition, lymph node metastasis is one of diffusion routes that influence survival and prognosis of CSCC [5, 6]. Once lymph node metastasis occurs, the overall 5-year survival rate for early stage carcinoma of the uterine cervix is reduced to 53%, which lead to the high recurrence rate and poor prognosis of patients with CSCC [7–10]. As there are no valid diagnostic and therapeutic methods for CSCC, it is urgent to understand the pathological mechanism and find potential biological markers for diagnosis, therapy and prognosis of patients with CSCC. [11, 12]. Several genes have been identified as the diagnostic and prognostic biomarkers for CSCC. It has been demonstrated that the kinase family member 20a (KIF20A) protein is one potential biomarker for CSCC [13]. Additionally, Liu DQ et al suggested that receptor interacting serine/threonine kinase 4 (RIPK4) might act as a potential diagnostic and independent prognostic biomarker for patients with CSCC [14].
MicroRNAs (miRNAs) are small non-coding RNAs that are approximately 22 nt in size. They can modulate growth, proliferation, differentiation and apoptosis of cells by regulating target genes expression at the post-transcriptional level. As microRNAs stably present in almost all body fluids, they constitute a new class of non-invasive biomarkers [15–19]. It has been reported that the deregulation of miRNAs leads to the occurrence of a number of diseases, such as cancers in cervical [20]. MiR-23b/uPA is involved in the HPV-16 E6-associated cervical cancer development [21]. MiR-372 is down-regulated in cervical cancer tissues compared with normal cervical tissues [22]. The down-regulation of miR-143 is associated with lymph node metastasis and poor prognosis in cervical cancer [23, 24]. It is reported that 6 serum microRNAs including miR-1246, miR-20a, miR-2392, miR-3147, miR-3162-5p and miR-4484 has been identified in predicting lymph node metastasis of CSCC patients [25]. In addition, it is found that serum miR-206 is a powerful tool to predict chemoradiotherapy sensitivity in advanced-stage CSCC patients [26]. It is noteworthy that the identification of potential miRNAs that participate in survival prediction is essential for establishing novel prognosis strategies for CSCC. Recently, miRNA expression signatures related to prognosis have been found in number of malignancy [27]. Hence, we undertook to identify and validate a miRNA expression signature capable of predicting for survivability in CSCC patients.
Material and methods
TCGA data retrieval and analysis
The BCGSC__IlluminaHiSeq_miRNASeq data were acquired from Firebrowse (http://firebrowse.org/?cohort=LIHC&download_dialog=true, 2016-01-28). Level 3 (Reads-per-kilobase-million; RPKM) miRNA-Seq and Level 1 clinical data were downloaded from the TCGA data portal (http://tcga-data.nci.nih.gov/tcga) dataset. At the time of analysis, there were 307 clinical histories. Only those clinical histories with miRNA-seq values and sufficient follow-up data (261 cases) were used for further survival analysis. All these cases were randomly divided into Training cohort (131 cases) and Validation cohort (130 cases). There was no significant difference in gender, race, family history, tumor stage, vascular invasion, follow-up time and follow-up result between two cohorts. Clinical characteristics for two cohorts were shown in Table 1.
Table 1. The clinical characteristics of the patients in the two cohorts.
| Factor | All cohorts (n = 261) |
Training cohorts (n = 131) | Validation cohorts (n = 130) | p-value | |
|---|---|---|---|---|---|
| Age | Mean±SD | 48.77±13.49 | 48.52±13.12 | 49.01±13.91 | 0.7741 |
| Median | 47 | 47 | 47 | ||
| Race | Asian | 15 | 10 | 5 | 0.3705 |
| White | 185 | 88 | 97 | ||
| Black or African American | 24 | 13 | 11 | ||
| Native hawaiian or other pacific islander | 2 | 2 | 0 | ||
| American indian or Alaska native | 8 | 4 | 4 | ||
| Tumor grade | G1 | 15 | 7 | 8 | 0.7578 |
| G2 | 115 | 54 | 61 | ||
| G3 | 106 | 56 | 50 | ||
| G4 | 1 | 0 | 1 | ||
| Gx | 21 | 10 | 11 | ||
| Stage | I | 136 | 68 | 68 | 0.4456 |
| II | 61 | 34 | 27 | ||
| III | 40 | 21 | 19 | ||
| IV | 20 | 7 | 13 | ||
|
Lymphovascular Invasion |
present | 71 | 35 | 36 | 0.8577 |
| absent | 65 | 34 | 31 | ||
| Vital status | Alive | 201 | 101 | 100 | 0.4828 |
| Dead | 60 | 30 | 30 | ||
| Survival time | Mean±SD | 1032.85±1137.81 | 1054±1093.37 | 1011.53±1184.76 | 0.7637 |
| Median | 607 | 659 | 601.5 |
Identification and survival analysis of miRNA signature
In order to identify the survival time related miRNAs in CSCC, the single factor Cox proportional hazard (CoxPH) regression model was fitted to the Training cohort data. The statistical significance was set at p<0.05. After further adjustment, the multi-factor CoxPH regression model was used for identification of miRNA signature in the survival evaluation model of CSCC. A risk score (RS) was calculated using the coefficients from the model, and high vs. low risk patients were then compared in the Training cohort and Validation cohort using the log-rank test. Kaplan-Meier curves were used to plot overall survival with miRNA signature expression using Cutoff Finder (http://molpath.charite.de/cutoff). In addition, the receiver operating characteristic (ROC) analyses were performed to assess the 5 years’ survival rate of miRNA signature of CSCC by using pROC package in R language. The area under the curve (AUC) under binomial exact confidence interval was calculated and the ROC curve was generated.
Network construction of miRNA signature-targets
Identifying target genes is an important step in studying the function of miRNA in tissues. In this study, target genes of miRNA signature were obtained by miRWalk (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/). According to the miRNA-target pairs, miRNA signature-targets interaction network was established by Cytoscape software (http://www.cytoscape.org/).)
Functional annotation of miRNA signature targets
In order to study the biological function of target genes of miRNA signature, the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed by using the online software GeneCodis3 (http://genecodis.cnb.csic.es/analysis). The threshold of false discovery rate (FDR) < 0.05 was set as the criteria of statistical significance.
Results
Generation and validation of miRNA signature
The single factor CoxPH regression model fitted to the Training cohort yielded 47 miRNAs (Table 2). A group of miRNA signatures including hsa-mir-642a and hsa-mir-378c (Table 3) was identified that were most strongly associated with survival after multi-factor CoxPH regression model analysis. The miRNA signature was combined with their coefficients within the penalized model to yield the following equation:
Table 2. The single factor CoxPH regression model fitted to the Training cohort.
| miRNA | Coefficient | HR | 95%CI lower |
95%CI upper |
p-value |
|---|---|---|---|---|---|
| hsa-mir-361 | -0.741962146 | 0.476178666 | 0.337831316 | 0.671181479 | 2.27E-05 |
| hsa-mir-150 | -0.399833202 | 0.670431863 | 0.544302388 | 0.825788924 | 1.70E-04 |
| hsa-mir-642a | -0.933407714 | 0.393211473 | 0.241114727 | 0.64125184 | 1.84E-04 |
| hsa-mir-142 | -0.459467464 | 0.631619916 | 0.493836414 | 0.807845893 | 2.53E-04 |
| hsa-mir-378c | -0.704272826 | 0.494468008 | 0.330219908 | 0.74041148 | 6.28E-04 |
| hsa-mir-148b | -0.940559471 | 0.390409352 | 0.218272292 | 0.698299636 | 1.52E-03 |
| hsa-mir-502 | -0.823303663 | 0.438979018 | 0.259043417 | 0.743900693 | 2.22E-03 |
| hsa-mir-532 | -0.689571509 | 0.501791036 | 0.32109448 | 0.784174938 | 2.47E-03 |
| hsa-mir-548o | -1.431560505 | 0.238935771 | 0.093023518 | 0.613719019 | 2.94E-03 |
| hsa-mir-629 | -0.582879033 | 0.558288719 | 0.378627906 | 0.823199476 | 3.26E-03 |
| hsa-mir-3607 | -0.455590425 | 0.634073485 | 0.465711706 | 0.863300575 | 3.81E-03 |
| hsa-mir-140 | -0.831986343 | 0.435184003 | 0.245859481 | 0.770298201 | 4.29E-03 |
| hsa-mir-653 | -0.559648681 | 0.571409776 | 0.384603247 | 0.848950534 | 5.59E-03 |
| hsa-mir-3940 | -0.798489446 | 0.450008213 | 0.255437141 | 0.792787577 | 5.72E-03 |
| hsa-mir-204 | -0.503949703 | 0.604139769 | 0.417830953 | 0.87352279 | 7.39E-03 |
| hsa-mir-500b | -0.689845567 | 0.501653535 | 0.302748834 | 0.831237782 | 7.42E-03 |
| hsa-mir-659 | -0.984396955 | 0.373664495 | 0.177043262 | 0.788649926 | 9.80E-03 |
| hsa-mir-33b | -0.49164916 | 0.611616908 | 0.421114439 | 0.888298305 | 9.82E-03 |
| hsa-mir-331 | -0.661490245 | 0.516081673 | 0.311061687 | 0.856229823 | 1.04E-02 |
| hsa-mir-550a-2 | -0.713332169 | 0.490008682 | 0.282973877 | 0.848518284 | 1.09E-02 |
| hsa-mir-3074 | -0.533161438 | 0.58674707 | 0.388980125 | 0.885063534 | 1.10E-02 |
| hsa-mir-155 | -0.319675972 | 0.726384368 | 0.566546989 | 0.931315954 | 1.17E-02 |
| hsa-mir-34a | -0.601640189 | 0.547912219 | 0.34001366 | 0.882928646 | 1.35E-02 |
| hsa-mir-942 | -0.501600699 | 0.605560563 | 0.406007929 | 0.903193187 | 1.39E-02 |
| hsa-mir-101-1 | -0.572617152 | 0.564047308 | 0.356914365 | 0.891388514 | 1.42E-02 |
| hsa-mir-1306 | -0.582090351 | 0.558729205 | 0.349290949 | 0.893748679 | 1.52E-02 |
| hsa-mir-146a | -0.298734498 | 0.741756321 | 0.579025164 | 0.950221983 | 1.81E-02 |
| hsa-mir-3130-1 | -0.335178726 | 0.715210248 | 0.538178207 | 0.950476427 | 2.09E-02 |
| hsa-mir-766 | -0.453584546 | 0.635346636 | 0.432031166 | 0.93434312 | 2.12E-02 |
| hsa-mir-651 | -0.444810245 | 0.640945887 | 0.435193351 | 0.94397497 | 2.43E-02 |
| hsa-mir-589 | -0.602353886 | 0.547521315 | 0.323793065 | 0.925836971 | 2.46E-02 |
| hsa-mir-580 | -0.907437169 | 0.403557148 | 0.18125611 | 0.898498661 | 2.63E-02 |
| hsa-mir-128-2 | -0.488785221 | 0.613371052 | 0.397361642 | 0.946805146 | 2.73E-02 |
| hsa-mir-153-2 | -0.33037904 | 0.718651284 | 0.535409284 | 0.964607232 | 2.78E-02 |
| hsa-mir-550a-1 | -0.577582055 | 0.561253808 | 0.334287794 | 0.94231929 | 2.89E-02 |
| hsa-mir-186 | -0.704267306 | 0.494470737 | 0.262595738 | 0.931093978 | 2.92E-02 |
| hsa-mir-16-2 | -0.435460059 | 0.646966956 | 0.43718272 | 0.957417167 | 2.94E-02 |
| hsa-mir-423 | -0.630852633 | 0.532137889 | 0.29790113 | 0.950552733 | 3.31E-02 |
| hsa-mir-188 | -0.536683133 | 0.58468436 | 0.353621916 | 0.966726849 | 3.65E-02 |
| hsa-mir-3199-2 | 0.733244039 | 2.081823182 | 1.043660499 | 4.152679692 | 3.74E-02 |
| hsa-mir-128-1 | -0.506262412 | 0.602744184 | 0.373877643 | 0.971709751 | 3.77E-02 |
| hsa-mir-34c | -0.247813788 | 0.780505269 | 0.617220838 | 0.986986242 | 3.85E-02 |
| hsa-mir-335 | 0.328925144 | 1.389473842 | 1.017173297 | 1.898041918 | 3.87E-02 |
| hsa-mir-660 | -0.439107775 | 0.644611303 | 0.421343551 | 0.986187474 | 4.30E-02 |
| hsa-mir-145 | -0.347777277 | 0.706256158 | 0.502311525 | 0.993004812 | 4.55E-02 |
| hsa-mir-1468 | -0.518087872 | 0.595658435 | 0.358192455 | 0.990554007 | 4.59E-02 |
| hsa-mir-99a | -0.209225673 | 0.811212146 | 0.659987923 | 0.997086648 | 4.68E-02 |
HR: hazard ratio; CI: confidence interval
Table 3. miRNA signature in CSCC.
| miRNA | Coefficient | 95%CI lower |
95%CI Upper |
p-value |
|---|---|---|---|---|
| hsa-mir-642a | -1.318 | 0.142266 | 0.5038 | 4.40E-05 |
| hsa-mir-378c | -1.006 | 0.205868 | 0.6491 | 5.93E-04 |
Risk Score = -1.318×log2 (RPKM of hsa-mir-642a)−1.006×log2 (RPKM of hsa-mir-378c).
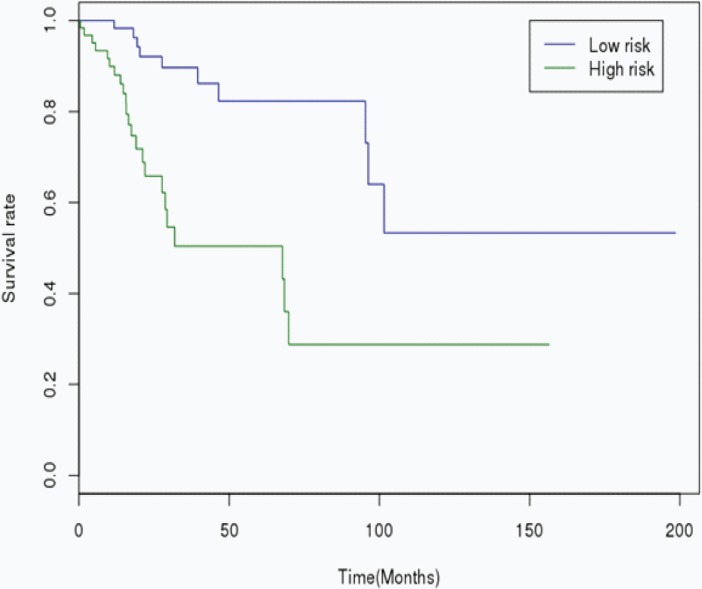
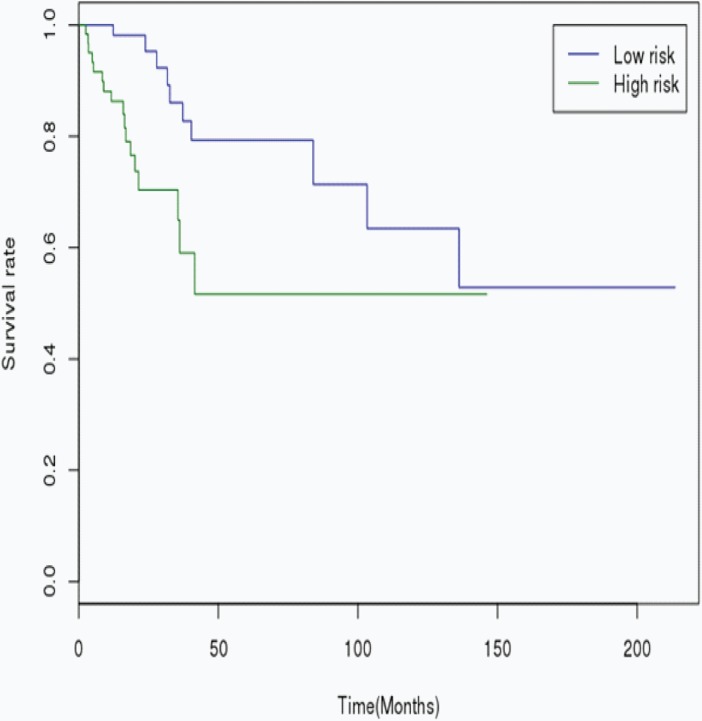
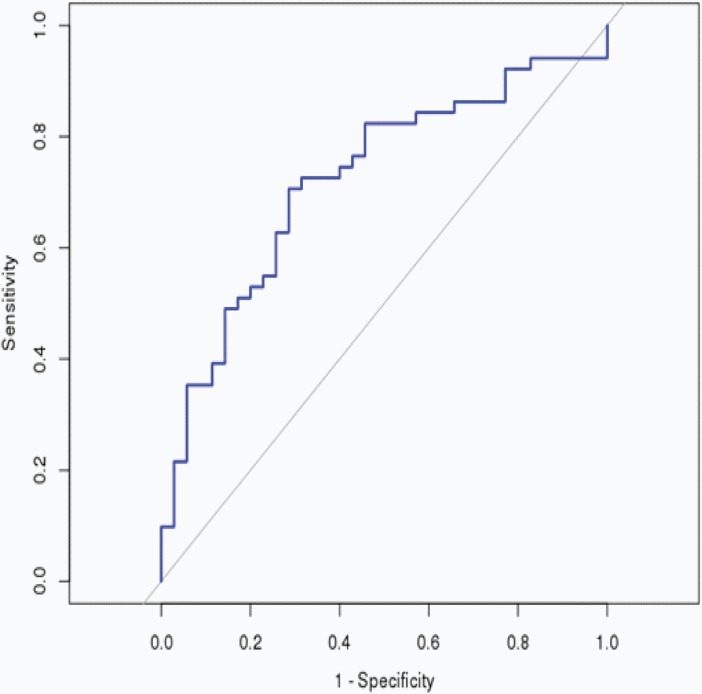
The RS was calculated for each patient in the Training cohort, in which the patients were dichotomized into either the “low risk” (< median), or the “high risk” (≥ median) group. A highly significant difference was observed between the high risk and the low risk group (p < 0.001), that was shown in Fig 1. When the same miRNA signature equation was applied to the Validation cohort, a similar significant difference was also observed between the high risk and the low risk group (p = 0.007), that was shown in Fig 2. Additionally, we performed 5 years’ survival analysis of miRNA signature by ROC and calculated the AUC to assess the discriminatory ability of miRNA signature (Fig 3). The AUC of the miRNA signature was 0.7221. Our result suggested that the miRNA signature could be the prognosis model for predicting the survival situation of CSCC.
Fig 1. Kaplan-Meier curves showing CSCC patients dichotomized based on risk score in the Training cohort.
High risk is defined as a RS ≥ the median in the training cohort, and low risk is defined as a RS < the median in the Training cohort.
Fig 2. Kaplan-Meier curves showing CSCC patients dichotomized based on risk score in the Validation cohort.
High risk is defined as a RS ≥ the median in the training cohort, and low risk is defined as a RS < the median in the Validation cohort.
Fig 3. 5 years’ ROC curves of miRNA signature in CSCC.
The ROC curves were used to show the diagnostic ability of miRNA signature and miRNA signature with 1-Specificity (the proportion of false positive) and sensitivity (the proportion of true positive) and. The x-axis shows 1-specificity and y-axis shows sensitivity.
MiRNA signature-targets network
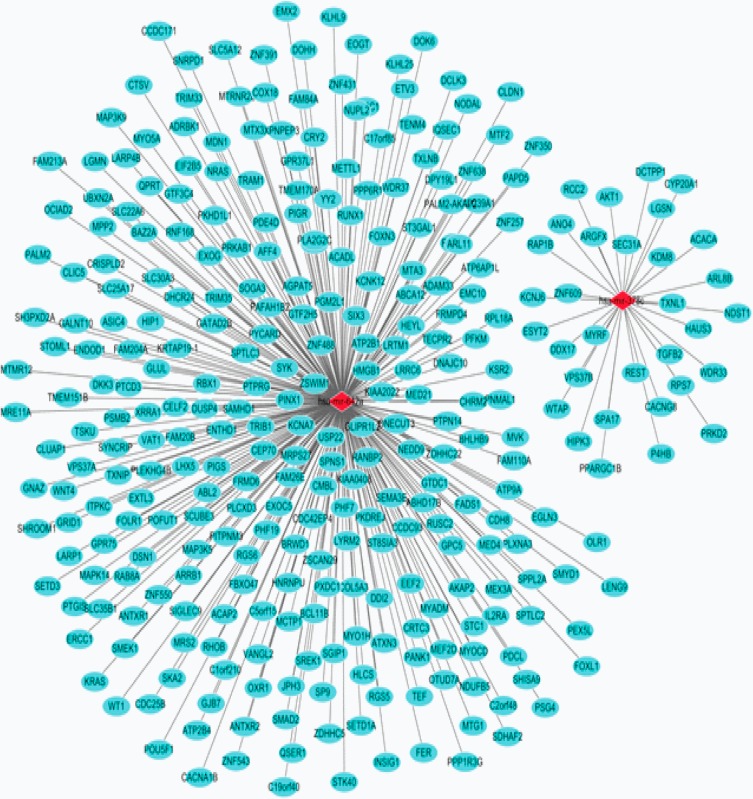
A total of 345 miRNA signature-target pairs were obtained by miRWalk, followed by the construction of the interaction network (Fig 4). In the network, 316 genes were targets of has-mir-642a and has-mir-378c. The red rhombus and blue-green ellipse represented the miRNA and target genes, respectively.
Fig 4. MiRNA signature-targets interaction network.
The red rhombus and blue-green ellipse represented the miRNA and target genes, respectively.
Functional annotation of miRNA signature targets
According to the GO enrichment analysis, intracellular signal transduction (FDR = 0.0001536), cell proliferation (FDR = 0.0001913) and blood coagulation (FDR = 0.0001913) were the most significantly enriched biological process; protein binding (FDR = 7.47E-12), nucleotide binding (FDR = 5.97E-09) and metal ion binding (FDR = 5.34E-08) were the most significantly enriched molecular function; nucleus (FDR = 5.68E-17), cytoplasm (FDR = 3.66E-15) and membrane (FDR = 7.10E-09) were the most significantly enriched cellular component. The top 15 GO terms were shown in Table 4. MAPK signaling pathway (FDR = 4.14E-05), VEGF signaling pathway (FDR = 8.89E-05) and endocytosis (FDR = 9.18E-05) were significantly enriched signal pathways that covered most genes. The top 15 KEGG terms were shown in Table 5.
Table 4. The enriched top 15 GO terms of miRNA signature targets.
| GO Items | Items Details | No. of genes | FDR |
|---|---|---|---|
| Biological process | |||
| GO:0035556 | intracellular signal transduction | 4 | 0.0001536 |
| GO:0008283 | cell proliferation | 3 | 0.0001913 |
| GO:0007596 | blood coagulation | 3 | 0.0001913 |
| GO:0030168 | platelet activation | 3 | 0.0001913 |
| GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | 3 | 0.0001913 |
| GO:0045087 | innate immune response | 3 | 0.0001913 |
| GO:0007165 | signal transduction | 3 | 0.0002376 |
| GO:0051146 | nerve growth factor receptor signaling pathway | 3 | 0.0002376 |
| GO:0048011 | striated muscle cell differentiation | 3 | 0.0002376 |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 18 | 0.0002497 |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | 6 | 0.0003527 |
| GO:0018105 | peptidyl-serine phosphorylation | 3 | 0.000354 |
| GO:0043066 | negative regulation of apoptotic process | 3 | 0.000354 |
| GO:0007281 | germ cell development | 3 | 0.000354 |
| GO:0006464 | protein modification process | 3 | 0.0007776 |
| Molecular function | |||
| GO:0005515 | protein binding | 89 | 7.47E-12 |
| GO:0000166 | nucleotide binding | 51 | 5.97E-09 |
| GO:0046872 | metal ion binding | 59 | 5.34E-08 |
| GO:0005524 | ATP binding | 33 | 5.95E-05 |
| GO:0046872 | metal ion binding | 37 | 6.92E-05 |
| GO:0008270 | zinc ion binding | 38 | 0.0001139 |
| GO:0003700 | sequence-specific DNA binding transcription factor activity | 7 | 0.0001349 |
| GO:0003677 | DNA binding | 7 | 0.0001768 |
| GO:0031625 | ubiquitin protein ligase binding | 4 | 0.0002863 |
| GO:0008134 | transcription factor binding | 11 | 0.0003429 |
| GO:0005525 | GTP binding | 4 | 0.0004299 |
| GO:0019003 | GDP binding | 4 | 0.0004299 |
| GO:0003924 | GTPase activity | 4 | 0.0004299 |
| GO:0003690 | double-stranded DNA binding | 6 | 0.0005831 |
| GO:0003697 | single-stranded DNA binding | 3 | 0.0009546 |
| Cellular component | |||
| GO:0005634 | nucleus | 112 | 5.68E-17 |
| GO:0005737 | cytoplasm | 106 | 3.66E-15 |
| GO:0016020 | membrane | 76 | 7.10E-09 |
| GO:0005829 | cytosol | 39 | 1.04E-07 |
| GO:0005730 | nucleolus | 38 | 1.10E-07 |
| GO:0043231 | intracellular membrane-bounded organelle | 11 | 1.23E-07 |
| GO:0016021 | integral to membrane | 53 | 8.89E-06 |
| GO:0043231 | intracellular membrane-bounded organelle | 13 | 1.73E-05 |
| GO:0016021 | integral to membrane | 68 | 4.08E-05 |
| GO:0005622 | intracellular | 26 | 0.0001472 |
| GO:0005654 | nucleoplasm | 14 | 0.0001523 |
| GO:0005625 | soluble fraction | 4 | 0.0001675 |
| GO:0005886 | plasma membrane | 4 | 0.0001675 |
| GO:0005624 | membrane fraction | 16 | 0.0002292 |
| GO:0005625 | soluble fraction | 8 | 0.000391 |
No.: number; FDR: false discovery rate.
Table 5. The enriched top 15 KEGG terms of MiRNA signature targets.
| KEGG Items | Items_Details | No. of genes | FDR |
|---|---|---|---|
| hsa04010 | MAPK signaling pathway | 13 | 4.14E-05 |
| hsa04660 | T cell receptor signaling pathway | 4 | 7.86E-05 |
| hsa05160 | Hepatitis C | 4 | 7.86E-05 |
| hsa05211 | Renal cell carcinoma | 7 | 8.57E-05 |
| hsa04370 | VEGF signaling pathway | 5 | 8.89E-05 |
| hsa04144 | Endocytosis | 10 | 9.18E-05 |
| hsa04062 | Chemokine signaling pathway | 5 | 9.22E-05 |
| hsa04910 | Insulin signaling pathway | 3 | 9.85E-05 |
| hsa05223 | Non-small cell lung cancer | 3 | 9.85E-05 |
| hsa04012 | Endometrial cancer | 3 | 9.85E-05 |
| hsa04662 | B cell receptor signaling pathway | 3 | 9.85E-05 |
| hsa05215 | Prostate cancer | 3 | 9.85E-05 |
| hsa05218 | Melanoma | 3 | 9.85E-05 |
| hsa04530 | Tight junction | 3 | 9.85E-05 |
| hsa05200 | Pathways in cancer | 6 | 0.000105 |
No.: number; FDR: false discovery rate
Discussion
CSCC is one of the most common gynecological cancers that affect the health of women [1, 2]. In addition, the 5-year overall survival rate is about 80% [7, 28]. Even so, it is needed to understand the pathological mechanism and find potential survival related genes in the development of CSCC. In this study, we found a miRNA signature including hsa-mir-642a and hsa-mir-378c in CSCC, which could be a valuable tool in guiding treatment decisions for CSCC.
Hsa-mir-642a, a primate-specific miRNA, is a tumor suppressor. It is reported that hsa-mir-642a is differentially expressed in lung cancer cells [29]. Interaction of hsa-mir-642a-5p and Linc00974 can increase the expression of keratin 19 and activate Notch and TGF-β signaling pathways, which will increase the proliferation and invasion of hepatocellular carcinoma [30]. It is found that hsa-mir-642a is over expressed in the pediatric embryonal central nervous system neoplasm that is regarded as a prognostic parameter of patients [31]. In addition, the abnormal expression of hsa-mir-642a in myeloma cell lines significantly decreased protein levels of DEP domain containing MTOR interacting, which caused dedifferentiation of myeloma cells [32]. It is noteworthy that hsa-mir-642a is associated with cervical cancer prognosis [33]. Herein, we also found that hsa-mir-642a was related to survival time of patients with CSCC. Furthermore, cryptochrome circadian clock 2 (CRY2) was one of the target genes of hsa-mir-642a. Cryptochrome 2 is circadian clock gene and the hypermethylation of CRY2 is involved in DNA recombination and repair in long-term shift-workers [34]. It has been demonstrated that the genetic variation of CRY2 is related to metabolic characteristics of type 2 diabetes [35, 36]. In addition, CRY2 has been suggested to act as a modulator in the development of cancer [37]. The expression level of CRY2 in ovarian cancer is remarkably lower than those in normal ovary [38]. The polymorphism in CRY2 gene has been frequently found associated with increased risk or recurrence of breast and endometrial cancers [39, 40]. Our result showed that hsa-mir-642a was significantly associated with survival time of CSCC and could be a diagnostic and prognostic marker of CSCC.
It is reported that the expression of hsa-mir-378c may enhance cell survival and tumor growth [41]. It has been found that hsa-mir-378c is associated with Stage I and Stage II colon cancer compared with normal controls [42]. Additionally, the expression of hsa-mir-378c is significantly down-regulated in osteosarcoma, intrahepatic cholangiocarcinoma and advanced stage gastric cancer [43–45]. It is worth mentioning that hsa-mir-378c is the member of protective miRNA signatures and correlated with cervical cancer prognosis [33]. In this study, we found that hsa-mir-378c was one of the members of miRNA signatures in the CSCC survival analysis. Moreover, myelin regulatory factor (MYRF) was one of the target genes of hsa-mir-378c. MYRF is a myelin-associated gene and acts as a key transcription factor for oligodendrocyte differentiation and central nervous system myelination. [46–48]. In addition, it is the target gene of hsa-mir-423-5p and involved in the immune response or injury in the retina [49]. MYRF may be involved in the nervous and immune of CSCC. In a word, hsa-mir-378c played a crucial role in the CSCC and could be a diagnostic and prognostic marker in the development of CSCC.
According to the functional annotation analysis of miRNA signature targets, MAPK signaling pathway (FDR = 4.14E-05), VEGF signaling pathway (FDR = 8.89E-05) and endocytosis (FDR = 9.18E-05) were significantly enriched signal pathways that covered most genes. It has been shown that p38 MAPK is involved in a number of cellular processes, including cell survival and death [50, 51]. It is found that human papillomavirus (HPV) 16 E2 can induce apoptosis by inhibiting p38 MAPK/JNK signal pathway in CSCC, which is important for the in vitro growth and migration of cervical squamous carcinoma cells in response to HPV 16 E2 treatment [52]. VEGF has been identified as angiogenesis regulator and may be important to restrict tumor growth, progression and metastasis. Vascular proliferation is a characteristic of cervical cancer and high density of microvessels indicates a worse prognosis of the disease [53]. Tjalma W et al found that the expression level of VEGF was high in cervical cancers [54]. It is suggested that VEGF could stimulate tumor cell proliferation in the early stages and may be responsible for tumorigenesis of cervical cancer [55]. In addition, it has been demonstrated that VEGF could be the predictive biomarker for monitoring the recurrence of cervical cancer [56]. The endocytosis process is involved in regulating various biological process including cell cycle and apoptosis in cancer cells [57, 58]. It has been reported that endocytosis is associated with CSCC-specific alternative splicing events [59]. This suggested that MAPK, VEGF and endocytosis signal pathways may play an important role in CSCC. Inhibition of these signal pathways might be a useful therapeutic strategy for CSCC.
Conclusions
In summary, we have identified and successfully validated a 2-miRNA signature of hsa-mir-642a and hsa-mir-378c in patients with CSCC. The signature adds to the potential predictive role in the survival time of CSCC patients. Therefore, we can detect the expression of hsa-mir-642a and hsa-mir-378c in the blood to predict the survival time of patients with CSCC, which will improve the clinical outcome for patients with CSCC.
Data Availability
All relevant data are within the paper.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Intaraphet S, Kasatpibal N, Siriaunkgul S, Sogaard M, Patumanond J, Khunamornpong S, et al. Prognostic impact of histology in patients with cervical squamous cell carcinoma, adenocarcinoma and small cell neuroendocrine carcinoma. Asian Pacific journal of cancer prevention: APJCP. 2013;14(9):5355–60. Epub 2013/11/02. . [DOI] [PubMed] [Google Scholar]
- 2.Wang SS, Sherman ME, Hildesheim A, Lacey JV Jr., Devesa S. Cervical adenocarcinoma and squamous cell carcinoma incidence trends among white women and black women in the United States for 1976–2000. Cancer. 2004;100(5):1035–44. Epub 2004/02/26. doi: 10.1002/cncr.20064 . [DOI] [PubMed] [Google Scholar]
- 3.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. The Journal of pathology. 1999;189(1):12–9. Epub 1999/08/19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F . [DOI] [PubMed] [Google Scholar]
- 4.Castellsague X, Diaz M, de Sanjose S, Munoz N, Herrero R, Franceschi S, et al. Worldwide human papillomavirus etiology of cervical adenocarcinoma and its cofactors: implications for screening and prevention. Journal of the National Cancer Institute. 2006;98(5):303–15. Epub 2006/03/02. doi: 10.1093/jnci/djj067 . [DOI] [PubMed] [Google Scholar]
- 5.Trattner M, Graf AH, Lax S, Forstner R, Dandachi N, Haas J, et al. Prognostic factors in surgically treated stage ib-iib cervical carcinomas with special emphasis on the importance of tumor volume. Gynecologic oncology. 2001;82(1):11–6. Epub 2001/06/28. doi: 10.1006/gyno.2001.6252 . [DOI] [PubMed] [Google Scholar]
- 6.Suprasert P, Srisomboon J, Kasamatsu T. Radical hysterectomy for stage IIB cervical cancer: a review. International journal of gynecological cancer: official journal of the International Gynecological Cancer Society. 2005;15(6):995–1001. Epub 2005/12/14. doi: 10.1111/j.1525-1438.2005.00259.x . [DOI] [PubMed] [Google Scholar]
- 7.Yeh SA, Wan Leung S, Wang CJ, Chen HC. Postoperative radiotherapy in early stage carcinoma of the uterine cervix: treatment results and prognostic factors. Gynecologic oncology. 1999;72(1):10–5. Epub 1999/01/16. doi: 10.1006/gyno.1998.5217 . [DOI] [PubMed] [Google Scholar]
- 8.Hosaka M, Watari H, Mitamura T, Konno Y, Odagiri T, Kato T, et al. Survival and prognosticators of node-positive cervical cancer patients treated with radical hysterectomy and systematic lymphadenectomy. International journal of clinical oncology. 2011;16(1):33–8. Epub 2010/09/16. doi: 10.1007/s10147-010-0123-0 . [DOI] [PubMed] [Google Scholar]
- 9.Kasamatsu T, Onda T, Sawada M, Kato T, Ikeda S, Sasajima Y, et al. Radical hysterectomy for FIGO stage I-IIB adenocarcinoma of the uterine cervix. British journal of cancer. 2009;100(9):1400–5. Epub 2009/04/30. doi: 10.1038/sj.bjc.6605048 ; PubMed Central PMCID: PMCPmc2694432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma D, Cheng Y, Zhang Y, Guo Y, Li Z, Li G. Expression of CDC42 in cervical squamous cell carcinoma and its correlation with clinicopathologic characteristics. Chinese journal of cancer research = Chung-kuo yen cheng yen chiu. 2013;25(6):656–61. Epub 2014/01/05. doi: 10.3978/j.issn.1000-9604.2013.11.04 ; PubMed Central PMCID: PMCPmc3872540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao LJ, Guo SY, Cai YQ, Gu PQ, Su YJ, Gong H, et al. Cooperation of decay-accelerating factor and membrane cofactor protein in regulating survival of human cervical cancer cells. BMC cancer. 2009;9:384 Epub 2009/11/03. doi: 10.1186/1471-2407-9-384 ; PubMed Central PMCID: PMCPmc2774863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel S, Chiplunkar S. Host immune responses to cervical cancer. Current opinion in obstetrics & gynecology. 2009;21(1):54–9. Epub 2009/01/07. doi: 10.1097/GCO.0b013e32831a9890 . [DOI] [PubMed] [Google Scholar]
- 13.Zhang W, He W, Shi Y, Gu H, Li M, Liu Z, et al. High Expression of KIF20A Is Associated with Poor Overall Survival and Tumor Progression in Early-Stage Cervical Squamous Cell Carcinoma. PloS one. 2016;11(12):e0167449 Epub 2016/12/13. doi: 10.1371/journal.pone.0167449 ; PubMed Central PMCID: PMCPmc5152822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu DQ, Li FF, Zhang JB, Zhou TJ, Xue WQ, Zheng XH, et al. Increased RIPK4 expression is associated with progression and poor prognosis in cervical squamous cell carcinoma patients. Scientific reports. 2015;5:11955 Epub 2015/07/08. doi: 10.1038/srep11955 ; PubMed Central PMCID: PMCPmc4493702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang XW, Heegaard NH, Orum H. MicroRNAs in liver disease. Gastroenterology. 2012;142(7):1431–43. Epub 2012/04/17. doi: 10.1053/j.gastro.2012.04.007 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu R, Chen X, Du Y, Yao W, Shen L, Wang C, et al. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clinical chemistry. 2012;58(3):610–8. Epub 2011/12/24. doi: 10.1373/clinchem.2011.172767 . [DOI] [PubMed] [Google Scholar]
- 17.Jurmeister S, Baumann M, Balwierz A, Keklikoglou I, Ward A, Uhlmann S, et al. MicroRNA-200c represses migration and invasion of breast cancer cells by targeting actin-regulatory proteins FHOD1 and PPM1F. Molecular and cellular biology. 2012;32(3):633–51. Epub 2011/12/07. doi: 10.1128/MCB.06212-11 ; PubMed Central PMCID: PMCPmc3266604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hennessey PT, Sanford T, Choudhary A, Mydlarz WW, Brown D, Adai AT, et al. Serum microRNA biomarkers for detection of non-small cell lung cancer. PloS one. 2012;7(2):e32307 Epub 2012/03/06. doi: 10.1371/journal.pone.0032307 ; PubMed Central PMCID: PMCPmc3289652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Q, Lu Z, Li H, Lu J, Guo L, Ge Q. Next-generation sequencing of microRNAs for breast cancer detection. Journal of biomedicine & biotechnology. 2011;2011:597145 Epub 2011/07/01. doi: 10.1155/2011/597145 ; PubMed Central PMCID: PMCPmc3118289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez I, Gardiner AS, Board KF, Monzon FA, Edwards RP, Khan SA. Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene. 2008;27(18):2575–82. Epub 2007/11/14. doi: 10.1038/sj.onc.1210919 ; PubMed Central PMCID: PMCPmc2447163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Au Yeung CL, Tsang TY, Yau PL, Kwok TT. Human papillomavirus type 16 E6 induces cervical cancer cell migration through the p53/microRNA-23b/urokinase-type plasminogen activator pathway. Oncogene. 2011;30(21):2401–10. Epub 2011/01/19. doi: 10.1038/onc.2010.613 . [DOI] [PubMed] [Google Scholar]
- 22.Tian RQ, Wang XH, Hou LJ, Jia WH, Yang Q, Li YX, et al. MicroRNA-372 is down-regulated and targets cyclin-dependent kinase 2 (CDK2) and cyclin A1 in human cervical cancer, which may contribute to tumorigenesis. The Journal of biological chemistry. 2011;286(29):25556–63. Epub 2011/06/08. doi: 10.1074/jbc.M111.221564 ; PubMed Central PMCID: PMCPmc3138314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, Ma C, Zhang W, Chen Z, Ma L. Down regulation of miR-143 is related with tumor size, lymph node metastasis and HPV16 infection in cervical squamous cancer. Diagnostic pathology. 2014;9:88 Epub 2014/04/30. doi: 10.1186/1746-1596-9-88 ; PubMed Central PMCID: PMCPmc4039059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang L, Lin JX, Yu YH, Zhang MY, Wang HY, Zheng M. Downregulation of six microRNAs is associated with advanced stage, lymph node metastasis and poor prognosis in small cell carcinoma of the cervix. PloS one. 2012;7(3):e33762 Epub 2012/03/23. doi: 10.1371/journal.pone.0033762 ; PubMed Central PMCID: PMCPmc3306296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, Yao D, Li Y, Chen H, He C, Ding N, et al. Serum microRNA expression levels can predict lymph node metastasis in patients with early-stage cervical squamous cell carcinoma. International journal of molecular medicine. 2013;32(3):557–67. Epub 2013/06/27. doi: 10.3892/ijmm.2013.1424 ; PubMed Central PMCID: PMCPmc3782554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han Y, Liu M, Wang Z, Huang M, Xu N, Wu L. Serum MicroRNAs Related with Chemoradiotherapy Resistance in Advanced-Stage Cervical Squamous Cell Carcinoma. Translational oncology. 2017;10(3):378–84. Epub 2017/04/23. doi: 10.1016/j.tranon.2017.03.005 ; PubMed Central PMCID: PMCPmc5397578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews Cancer. 2006;6(11):857–66. Epub 2006/10/25. doi: 10.1038/nrc1997 . [DOI] [PubMed] [Google Scholar]
- 28.Xu L, Sun FQ, Wang ZH. Radical trachelectomy versus radical hysterectomy for the treatment of early cervical cancer: a systematic review. Acta obstetricia et gynecologica Scandinavica. 2011;90(11):1200–9. Epub 2011/07/02. doi: 10.1111/j.1600-0412.2011.01231.x . [DOI] [PubMed] [Google Scholar]
- 29.Qin X, Yu S, Zhou L, Shi M, Hu Y, Xu X, et al. Cisplatin-resistant lung cancer cell-derived exosomes increase cisplatin resistance of recipient cells in exosomal miR-100-5p-dependent manner. International journal of nanomedicine. 2017;12:3721–33. Epub 2017/05/30. doi: 10.2147/IJN.S131516 ; PubMed Central PMCID: PMCPmc5439933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang J, Zhuo H, Zhang X, Jiang R, Ji J, Deng L, et al. A novel biomarker Linc00974 interacting with KRT19 promotes proliferation and metastasis in hepatocellular carcinoma. Cell death & disease. 2014;5:e1549 Epub 2014/12/06. doi: 10.1038/cddis.2014.518 ; PubMed Central PMCID: PMCPmc4649834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braoudaki M, Lambrou GI, Giannikou K, Milionis V, Stefanaki K, Birks DK, et al. Microrna expression signatures predict patient progression and disease outcome in pediatric embryonal central nervous system neoplasms. Journal of hematology & oncology. 2014;7:96 Epub 2015/01/01. doi: 10.1186/s13045-014-0096-y ; PubMed Central PMCID: PMCPmc4342799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quwaider D, Corchete LA, Misiewicz-Krzeminska I, Sarasquete ME, Perez JJ, Krzeminski P, et al. DEPTOR maintains plasma cell differentiation and favorably affects prognosis in multiple myeloma. Journal of hematology & oncology. 2017;10(1):92 Epub 2017/04/20. doi: 10.1186/s13045-017-0461-8 ; PubMed Central PMCID: PMCPmc5395780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu B, Ding JF, Luo J, Lu L, Yang F, Tan XD. Seven protective miRNA signatures for prognosis of cervical cancer. Oncotarget. 2016;7(35):56690–8. Epub 2016/07/23. doi: 10.18632/oncotarget.10678 ; PubMed Central PMCID: PMCPmc5302945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Y, Stevens RG, Hoffman AE, Tjonneland A, Vogel UB, Zheng T, et al. Epigenetic impact of long-term shiftwork: pilot evidence from circadian genes and whole-genome methylation analysis. Chronobiology international. 2011;28(10):852–61. Epub 2011/11/15. doi: 10.3109/07420528.2011.618896 ; PubMed Central PMCID: PMCPmc3631105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nature genetics. 2010;42(2):105–16. Epub 2010/01/19. doi: 10.1038/ng.520 ; PubMed Central PMCID: PMCPmc3018764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ribel-Madsen R, Fraga MF, Jacobsen S, Bork-Jensen J, Lara E, Calvanese V, et al. Genome-wide analysis of DNA methylation differences in muscle and fat from monozygotic twins discordant for type 2 diabetes. PloS one. 2012;7(12):e51302 Epub 2012/12/20. doi: 10.1371/journal.pone.0051302 ; PubMed Central PMCID: PMCPmc3519577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L, Ptacek LJ, Fu YH. Diversity of human clock genotypes and consequences. Progress in molecular biology and translational science. 2013;119:51–81. Epub 2013/08/01. doi: 10.1016/B978-0-12-396971-2.00003-8 ; PubMed Central PMCID: PMCPmc4169291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sobia R, Saqib M. Circadian rhythm and its role in malignancy. Journal of Circadian Rhythms. 2010;8(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shih MC, Yeh KT, Tang KP, Chen JC, Chang JG. Promoter methylation in circadian genes of endometrial cancers detected by methylation-specific PCR. Molecular carcinogenesis. 2006;45(10):732–40. Epub 2006/05/10. doi: 10.1002/mc.20198 . [DOI] [PubMed] [Google Scholar]
- 40.Dai H, Zhang L, Cao M, Song F, Zheng H, Zhu X, et al. The role of polymorphisms in circadian pathway genes in breast tumorigenesis. Breast cancer research and treatment. 2011;127(2):531–40. Epub 2010/10/28. doi: 10.1007/s10549-010-1231-2 . [DOI] [PubMed] [Google Scholar]
- 41.Lee DY, Deng Z, Wang CH, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(51):20350–5. Epub 2007/12/14. doi: 10.1073/pnas.0706901104 ; PubMed Central PMCID: PMCPmc2154434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Yan F, Zhao Q, Zhan F, Wang R, Wang L, et al. Circulating exosomal miR-125a-3p as a novel biomarker for early-stage colon cancer. Scientific reports. 2017;7(1):4150 Epub 2017/06/25. doi: 10.1038/s41598-017-04386-1 ; PubMed Central PMCID: PMCPmc5482839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Z L, J S, MT C, WK W. The role of microRNAs in intrahepatic cholangiocarcinoma. Journal of Cellular\s&\smolecular Medicine. 2017;21(1):177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao H, Li M, Li L, Yang X, Lan G, Zhang Y. MiR-133b is down-regulated in human osteosarcoma and inhibits osteosarcoma cells proliferation, migration and invasion, and promotes apoptosis. PloS one. 2013;8(12):e83571 Epub 2014/01/07. doi: 10.1371/journal.pone.0083571 ; PubMed Central PMCID: PMCPmc3877051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bibi F, Naseer MI, Alvi SA, Yasir M, Jiman-Fatani AA, Sawan A, et al. microRNA analysis of gastric cancer patients from Saudi Arabian population. BMC genomics. 2016;17(Suppl 9):751 Epub 2016/10/22. doi: 10.1186/s12864-016-3090-7 ; PubMed Central PMCID: PMCPmc5073958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weng Q, Wang J, Wang J, Tan B, Wang J, Wang H, et al. Folate Metabolism Regulates Oligodendrocyte Survival and Differentiation by Modulating AMPKalpha Activity. Scientific reports. 2017;7(1):1705 Epub 2017/05/13. doi: 10.1038/s41598-017-01732-1 ; PubMed Central PMCID: PMCPmc5431811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emery B, Agalliu D, Cahoy JD, Watkins TA, Dugas JC, Mulinyawe SB, et al. Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell. 2009;138(1):172–85. Epub 2009/07/15. doi: 10.1016/j.cell.2009.04.031 ; PubMed Central PMCID: PMCPmc2757090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koenning M, Jackson S, Hay CM, Faux C, Kilpatrick TJ, Willingham M, et al. Myelin gene regulatory factor is required for maintenance of myelin and mature oligodendrocyte identity in the adult CNS. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2012;32(36):12528–42. Epub 2012/09/08. doi: 10.1523/jneurosci.1069-12.2012 ; PubMed Central PMCID: PMCPmc3752083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olivares AM, Jelcick AS, Reinecke J, Leehy B, Haider A, Morrison MA, et al. Multimodal Regulation Orchestrates Normal and Complex Disease States in the Retina. Scientific reports. 2017;7(1):690 Epub 2017/04/08. doi: 10.1038/s41598-017-00788-3 ; PubMed Central PMCID: PMCPmc5429617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wada T, Penninger JM. Mitogen-activated protein kinases in apoptosis regulation. Oncogene. 2004;23(16):2838–49. Epub 2004/04/13. doi: 10.1038/sj.onc.1207556 . [DOI] [PubMed] [Google Scholar]
- 51.Liu B, Fang M, Lu Y, Lu Y, Mills GB, Fan Z. Involvement of JNK-mediated pathway in EGF-mediated protection against paclitaxel-induced apoptosis in SiHa human cervical cancer cells. British journal of cancer. 2001;85(2):303–11. Epub 2001/07/20. doi: 10.1054/bjoc.2001.1910 ; PubMed Central PMCID: PMCPmc2364054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao LJ, Gu PQ, Zhao W, Ding WY, Zhao XQ, Guo SY, et al. The role of globular heads of the C1q receptor in HPV 16 E2-induced human cervical squamous carcinoma cell apoptosis is associated with p38 MAPK/JNK activation. Journal of translational medicine. 2013;11:118 Epub 2013/05/09. doi: 10.1186/1479-5876-11-118 ; PubMed Central PMCID: PMCPmc3651870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Obermair A, Bancher-Todesca D, Bilgi S, Kaider A, Kohlberger P, Mullauer-Ertl S, et al. Correlation of vascular endothelial growth factor expression and microvessel density in cervical intraepithelial neoplasia. Journal of the National Cancer Institute. 1997;89(16):1212–7. Epub 1997/08/20. . [DOI] [PubMed] [Google Scholar]
- 54.Tjalma W, Weyler J, Weyn B, Van Marck E, Van Daele A, Van Dam P, et al. The association between vascular endothelial growth factor, microvessel density and clinicopathological features in invasive cervical cancer. European journal of obstetrics, gynecology, and reproductive biology. 2000;92(2):251–7. Epub 2000/09/21. . [DOI] [PubMed] [Google Scholar]
- 55.Fujiwaki R, Hata K, Iida K, Maede Y, Miyazaki K. Vascular endothelial growth factor expression in progression of cervical cancer: correlation with thymidine phosphorylase expression, angiogenesis, tumor cell proliferation, and apoptosis. Anticancer research. 2000;20(2b):1317–22. Epub 2000/05/16. . [PubMed] [Google Scholar]
- 56.Bachtiary B, Selzer E, Knocke TH, Potter R, Obermair A. Serum VEGF levels in patients undergoing primary radiotherapy for cervical cancer: impact on progression-free survival. Cancer letters. 2002;179(2):197–203. Epub 2002/03/13. . [DOI] [PubMed] [Google Scholar]
- 57.Boucrot E, Kirchhausen T. Endosomal recycling controls plasma membrane area during mitosis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(19):7939–44. Epub 2007/05/08. doi: 10.1073/pnas.0702511104 ; PubMed Central PMCID: PMCPmc1876551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lehtonen S, Shah M, Nielsen R, Iino N, Ryan JJ, Zhou H, et al. The endocytic adaptor protein ARH associates with motor and centrosomal proteins and is involved in centrosome assembly and cytokinesis. Molecular biology of the cell. 2008;19(7):2949–61. Epub 2008/04/18. doi: 10.1091/mbc.E07-05-0521 ; PubMed Central PMCID: PMCPmc2441659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo P, Wang D, Wu J, Yang J, Ren T, Zhu B, et al. The landscape of alternative splicing in cervical squamous cell carcinoma. OncoTargets and therapy. 2015;8:73–9. Epub 2015/01/08. doi: 10.2147/OTT.S72832 ; PubMed Central PMCID: PMCPmc4278777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.