Abstract
Mycobacterium tuberculosis (Mtb) is a global health threat, compounded by the emergence of drug-resistant strains. A hallmark of pulmonary tuberculosis (TB) is the formation of hypoxic necrotic granulomas, which upon disintegration, release infectious Mtb. Furthermore, hypoxic necrotic granulomas are associated with increased disease severity and provide a niche for drug-resistant Mtb. However, the host immune responses that promote the development of hypoxic TB granulomas are not well described. Using a necrotic Mtb mouse model, we show that loss of Mtb virulence factors, such as phenolic glycolipids, decreases the production of the proinflammatory cytokine IL-17 (also referred to as IL-17A). IL-17 production negatively regulates the development of hypoxic TB granulomas by limiting the expression of the transcription factor hypoxia-inducible factor 1α (HIF1α). In human TB patients, HIF1α mRNA expression is increased. Through genotyping and association analyses in human samples, we identified a link between the single nucleotide polymorphism rs2275913 in the IL-17 promoter (–197G/G), which is associated with decreased IL-17 production upon stimulation with Mtb cell wall. Together, our data highlight a potentially novel role for IL-17 in limiting the development of hypoxic necrotic granulomas and reducing disease severity in TB.
Keywords: Infectious disease, Inflammation
Keywords: Cytokines, Tuberculosis, hypoxia
IL-17 negatively regulates the development of hypoxic necrotic granulomas and reduces disease severity in pulmonary tuberculosis.
Introduction
According to the World Health Organization (WHO), Mycobacterium tuberculosis (Mtb), the causative agent of pulmonary tuberculosis (TB), has surpassed human immunodeficiency virus (HIV) as the leading cause of death due to a single infectious agent. It is estimated that one-third of the global population is infected with Mtb. Although the majority of individuals are latently infected (LTBI), they have a 5% to 10% lifetime risk of reactivating TB, thus becoming an active reservoir of Mtb and facilitating TB transmission (1). This is further compounded by the increased prevalence of comorbidities, such as psoriasis and rheumatoid arthritis (2), as well as the emergence of drug-resistant Mtb, including both multidrug resistant (MDR) and extensively drug resistant (XDR) species. The hypoxic and necrotic microenvironment that develops within granulomas is thought to provide a niche for Mtb persistence and antibiotic resistance (3).
The TB granuloma, a hallmark of Mtb infection, is an organized conglomeration of immune cells consisting of lymphocytes, multinucleated giant cells, and macrophages, including foamy macrophages and epithelioid cells, and this structure functions to restrict mycobacterial growth and dissemination throughout the lung (4). During pulmonary TB disease in humans, the presence of caseum-filled hypoxic necrotic granulomas in the lungs is associated with severe disease, facilitating bacterial spread and compromising lung integrity. Human pulmonary TB lesions are hypoxic (5), and as a result, matrix metalloproteases such as MMP-1 associated with tissue remodeling are upregulated (5). Previous studies have shown that animal models of TB (e.g., rabbit, guinea pig, and nonhuman primate) develop heterogeneous granulomas, wherein hypoxia is closely associated with necrotic granulomas as opposed to solid granulomas (6). However, the host factors that regulate the development of such hypoxic necrotic granulomas during TB remain understudied.
In response to oxygen deprivation, the master transcriptional regulator hypoxia-inducible factor 1 (HIF1) is critical for metabolic adaptation within cells to promote survival (7, 8). Oxygen sensing of HIF1 is mediated through its regulatory subunit, HIF1α, which is degraded by the proteasome during normoxia and stabilized in the absence of oxygen (9). HIF1 facilitates the conversion of pyruvate from glucose, and shunts metabolism toward glycolysis, thereby promoting the final conversion to lactate. A similar shift toward glycolysis for ATP generation and away from mitochondrial oxidative phosphorylation, known as the Warburg effect, has been primarily studied in the context of cancer, in which HIF1 plays a key role in promoting the Warburg effect (10, 11).
While proinflammatory cytokines and Toll-like receptor (TLR) ligands have been shown to promote the metabolic switch towards glycolysis (12), less is known regarding how glycolysis is regulated during the formation of hypoxic granulomas during TB. Previous studies have reported increased IL-17–producing lymphocytes in human TB patients (13, 14). Moreover, a cooperative relationship between IL-17 (also referred to as IL-17A) and HIF1α promotes disease progression in rheumatoid arthritis (15) and stimulates glycolysis in cancer models (16). HIF1α also promotes IL-17 production through its major role as a transcriptional regulator of T helper 17 (Th17) polarization (17). In cancer, IL-17–producing Th17 cells also promote inflammation and tumor proliferation (18). Thus, we hypothesized that IL-17 may induce the development of hypoxic and necrotic granulomas during TB. Instead, we found that IL-17 plays a unique protective role in limiting hypoxia, through inhibition of HIF1α, and limiting myeloid-derived suppressor cell (MDSC) accumulation. In vivo neutralization of IL-17 in mice that develop necrotic TB granulomas induced hypoxia and increased HIF1α, resulting in severe susceptibility to Mtb HN878 (HN878) infection. Moreover, an HN878 mutant lacking phenolic glycolipids (PGLs) reduced IL-17 production in lung cells. This inverse relationship with IL-17 and TB disease translated to our findings in TB patients, in whom we identified a single nucleotide polymorphism (SNP), rs2275913, 197 bp upstream in the IL-17 promoter region where the G/G allele was linked with decreased IL-17 production and an increased association with TB. These studies together project an important, yet unrecognized, role for IL-17 in limiting hypoxia during TB. Considering the large population of LTBI patients that have existing comorbidities (2), our data caution the potential pathological consequences of the use of anti–IL-17 blocking agents for treatment of inflammatory conditions in TB patients.
Results
IL-17 limits HIF1α expression in vitro, while IL-17 neutralization exacerbates TB disease in vivo.
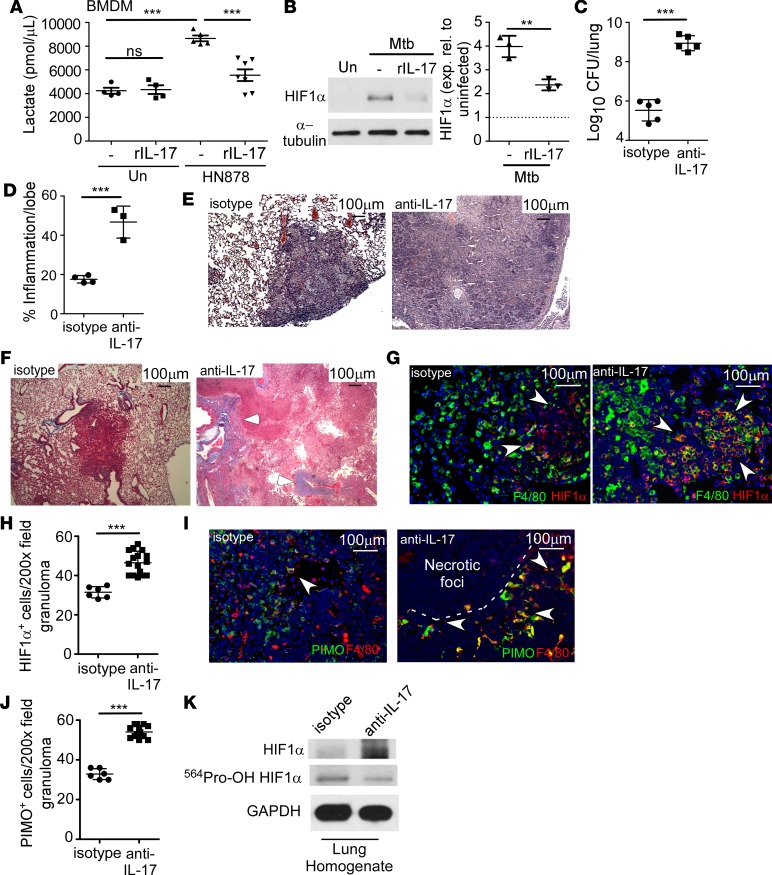
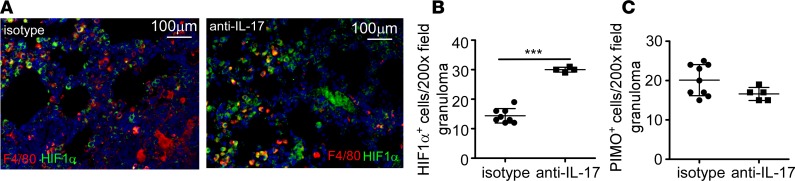
Lactate exits the cell via the monocarboxylate transport protein system, and accumulation of lactate reflects increases in the glycolytic pathway, a feature associated with hypoxia in multiple diseases (19) and Mtb-infected macrophages (20). Thus, we sought to determine whether IL-17 could modulate lactate production in Mtb-infected macrophages. C3HeB/FeJ (FeJ) mice develop necrotic, hypoxic, and caseous TB granulomas, similar to the necrotic granulomas found in human TB (21). Therefore, bone marrow–derived macrophages (BMDMs) isolated from FeJ mice were treated with recombinant IL-17 (rIL-17) and infected with HN878. Infection with HN878 induced production of lactate (Figure 1A) and HIF1α (Figure 1B), while treatment with rIL-17 inhibited lactate accumulation (Figure 1A) and decreased HIF1α expression (Figure 1B), indicating that IL-17 limits glycolysis and HIF1α expression in HN878-infected BMDMs. To determine the effects of IL-17 on the development of hypoxic necrotic granulomas during TB, HN878-infected mice received either isotype or IL-17–neutralizing antibody, and TB disease parameters were measured. Neutralization of IL-17 following HN878 infection resulted in substantially increased bacterial burden (Figure 1C and Supplemental Figure 1, A and C; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.92973DS1), severe pulmonary inflammation exhibiting necrotic lesions (Figure 1, D and E), and exacerbated fibrosis (Figure 1F), when compared with HN878-infected mice receiving isotype antibody. Abundant HN878 was observed to be diffusely localized, along with a large accumulation of Gr1+ (Ly6G/Ly6C) immune cells and inducible nitric oxide synthase–expressing (iNOS-expressing) cells within the granulomas of anti–IL-17–treated HN878-infected mice (Supplemental Figure 1, A–C). This was in sharp contrast to granulomas containing localized HN878 and few Gr1+ cells in lungs of isotype-treated mice (Supplemental Figure 1, A–C). The enhanced pathology and uncontrolled HN878 growth in anti–IL-17–treated HN878-infected mice coincided with increased expression of proinflammatory cytokines, including IL-1β, IL-6, tumor necrosis factor α (TNF-α), and granulocyte macrophage colony-stimulating factor (GM-CSF) in the lung (Supplemental Figure 1D), and increased weight loss at later time points (Supplemental Figure 1E). As our in vitro data suggested IL-17 might limit glycolysis, we determined whether HIF1α was stabilized in the absence of IL-17. HIF1α expression is a well-documented marker of hypoxia and necrosis in inflammatory diseases (19). While HIF1α-expressing macrophages were fewer and localized within the granulomas of isotype-treated HN878-infected lungs, HIF1α-expressing F4/80+ macrophages and Gr1+ myeloid cells were numerous and localized both inside large TB granulomas, as well as outside the necrotic zone in the lungs of anti–IL-17–treated HN878-infected mice (Figure 1, G and H, and Supplemental Figure 1F). Macrophages expressing the hypoxia marker, pimonidazole (PIMO), accumulated outside the necrotic zone within granulomas in anti–IL-17–treated HN878-infected FeJ lungs compared with non-necrotic granulomas in isotype-treated HN878-infected lungs (Figure 1, I and J). Importantly, mice infected with a lab-adapted strain, H37Rv, followed by IL-17 neutralization, did not exhibit increased Mtb burden or disease susceptibility (Supplemental Figure 1, G and H). Together, these data project an important role for IL-17 in limiting inflammation, hypoxia, and necrosis in FeJ mice infected with HN878.
Figure 1. IL-17 limits HIF1α and the metabolic shift to glycolysis, protecting against in vivo susceptibility to Mtb and development of hypoxic tuberculosis granulomas.
(A and B) FeJ bone marrow–derived macrophages (BMDMs) were infected with Mycobacterium tuberculosis (Mtb) clinical strain HN878 (MOI 1) and treated with rIL-17 (100 ng/ml) for 48 hours (n = 7 biological replicates) or left uninfected (n = 4 biological replicates) and treated with rIL-17 (n = 4 biological replicates), or untreated (n = 5 biological replicates), and (A) lactate accumulation was measured by lactate assay. (B) HIF1α expression was analyzed by Western blots (representative blot on left), which were quantified by densitometry (right, n = 3 biological replicates/group). Dashed line represents uninfected control. (C–K) FeJ mice were aerosol infected with approximately 100 CFU HN878 and received IL-17–blocking antibody (100 μg) between 10 and 30 (days postinfection [d.p.i.]), and lungs were harvested at 37 d.p.i. (C) Lung bacterial burden was assessed by plating from HN878-infected isotype (n = 5 mice) and anti–IL-17–treated mice (n = 5 mice). (D) Lung area per lobe covered by inflammation on H&E–stained formalin-fixed paraffin-embedded (FFPE) sections from HN878-infected isotype-antibody-treated (n = 4 mice) and anti–IL-17–treated mice (n = 3 mice) was quantified using the morphometric tool of the Zeiss Axioplan microscope. (E) Representative H&E images shown (×50 magnification). (F) Fibrosis was assessed using Masson’s trichrome staining (×50 magnification, arrowheads depict collagen deposition). (G) Immunofluorescence staining using antibodies specific for HIF1α (red) and F4/80 (green) was carried out on FFPE sections (arrows indicate HIF1α-expressing macrophages) and (H) HIF1α staining was quantified (isotype-treated, n = 5 mice; anti–IL-17–treated, n = 10 mice). (I) Hypoxia was determined using pimonidazole (PIMO, red) and F4/80 (green) staining by immunofluorescence, ×200 magnification (arrowheads depict areas of hypoxia) and (J) PIMO staining was quantified (isotype-treated, n = 5 mice; anti–IL-17–treated, n = 10 mice). All data shown as mean ± SD. **P < 0.01, ***P < 0.001 by 1-way ANOVA (A) or Student’s t test (B–D, H, and J). ns, not significant.
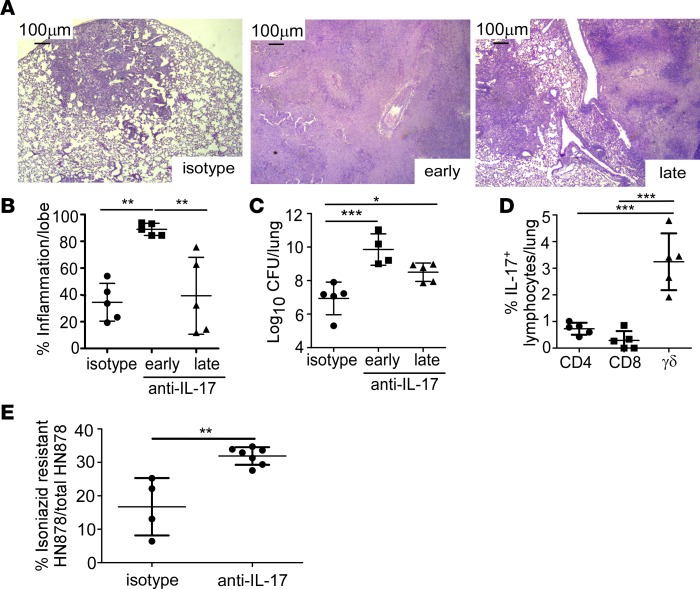
To determine whether early IL-17 produced by innate cells, or production of IL-17 by accumulating adaptive immune cells, was mediating the protective effect following HN878 infection, IL-17 was neutralized either early (10–24 days postinfection [d.p.i.]) or late (24–30 d.p.i.) following infection. Early neutralization of IL-17 was sufficient to induce extensive inflammation and necrosis at the time of terminal harvest (Figure 2, A and B), and correlated with substantially increased HN878 lung burden (Figure 2C). In contrast, later IL-17 neutralization did not result in severe disease progression and only slightly increased HN878 burden at the time of harvest (Figure 2, A–C). While Th17 cells are a major source of IL-17 in HN878 infection, γδ T cells can also produce IL-17 early after HN878 infection (22). Consistent with these findings, γδ T cells were the major lymphocytic source of IL-17 in HN878 lungs despite accumulation of CD4+ and CD8+ T cells producing IL-17 in the lungs of HN878-infected mice (Figure 2D).
Figure 2. Early IL-17 neutralization results in hypoxic granuloma formation and loss of Mtb control.
FeJ mice were aerosol infected with approximately 100 CFU Mycobacterium tuberculosis (Mtb) clinical strain HN878. IL-17 was neutralized (A–E) early (10–24 days postinfection [d.p.i]) or (A–D) late (24–31 d.p.i) and mice were harvested at 37 d.p.i. (A) Representative images from formalin-fixed, paraffin-embedded (FFPE) lung sections stained using H&E, ×50 magnification. (B) Lung inflammation in H&E–stained FFPE sections was quantified using the morphometric tool of the Zeiss Axioplan microscope (n = 5 mice/group). (C) Lung bacterial burden was assessed by plating lung homogenates of both isotype-antibody-treated (n = 5 mice) and IL-17–neutralized mice (n = 4 mice/early anti–IL-17; n = 5 mice/late anti–IL-17). (D) Lungs were processed to single-cell suspensions and flow cytometry was used to determine the lymphocyte populations producing IL-17 in the lungs of HN878-infected FeJ mice (n = 5 mice/group). (E) At 37 d.p.i., bacterial burden was plated and the percentage of isoniazid-resistant HN878 was measured by normalizing growth on isoniazid plates to total HN878 growth on non-isoniazid plates (n = 4 mice/isotype, n = 7 mice/anti–IL-17). All data shown as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 by 1-way ANOVA (B–D) or Student’s t test (E).
The W-Beijing lineage, of which HN878 is a member, is widely distributed geographically and emergence of this family may be associated with antibiotic resistance (23). Thus, we investigated the effects of neutralizing IL-17 on the development of antibiotic resistance in HN878 to isoniazid, a first-line antibiotic used against HN878. The frequency of isoniazid-resistant HN878 was 2-fold higher in anti–IL-17–treated mice compared with isotype-treated mice (Figure 2E), suggesting that neutralizing IL-17 supports a hypoxic environment that promotes the acquisition of antibiotic resistance. Together, these data provide potentially new evidence that IL-17 produced early upon HN878 infection, likely by γδ T cells, is required for limiting disease severity, limiting acquisition of antibiotic resistance, and conferring protective immunity against Mtb infection.
IL-17 neutralization leads to MDSC accumulation and decreased IFN-γ–producing CD4+ T cell responses during Mtb infection.
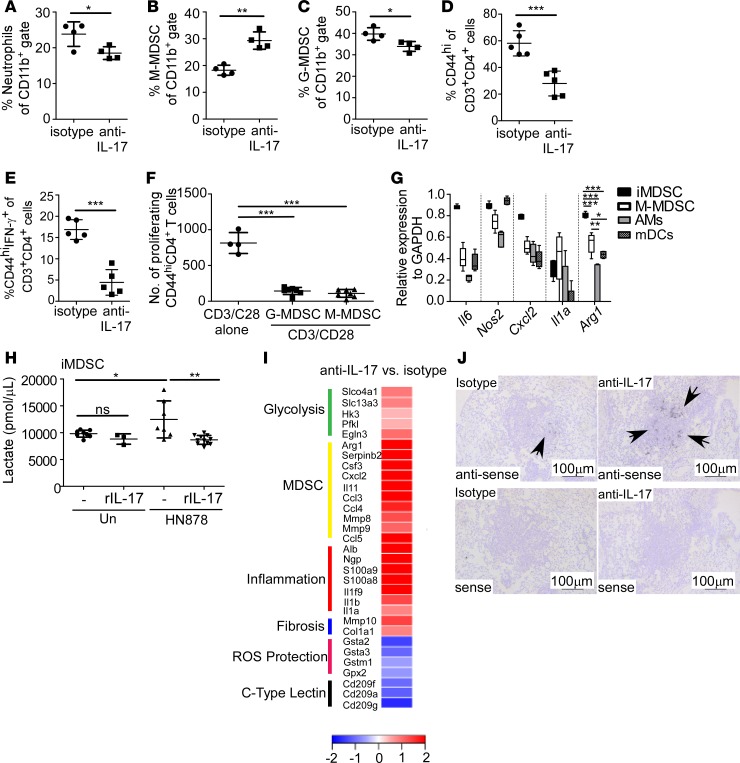
IL-17 has a well-documented role in the induction of chemokines that mediate neutrophil recruitment for pathogen control (22). Thus, we determined whether IL-17 neutralization affected neutrophil accumulation in HN878-infected lungs. While the frequency of lung neutrophils was lower, total numbers of infiltrating neutrophils were similar between isotype-treated and IL-17–neutralized HN878-infected mice (Figure 3A and Supplemental Figure 2A). In addition, rIL-17 treatment of HN878-infected macrophages did not alter their ability to control HN878 growth in vitro, suggesting that IL-17 was not directly activating macrophages for HN878 control (Supplemental Figure 2B). Recently, Knaul et al. showed that MDSCs accumulate in Mtb-infected lungs to modulate disease outcome (24). MDSCs are classified into either monocytic or granulocytic MDSCs. We found that the frequency and number of monocytic MDSCs (M-MDSCs; CD11b+Ly6GloLy6Chi; gating strategy in Supplemental Figure 2C) were higher in the lungs of HN878-infected mice following IL-17 neutralization, when compared with isotype-treated mice (Figure 3B and Supplemental Figure 2D). This was in contrast to the frequency of granulocytic MDSCs (G-MDSCs; CD11b+Ly6GhiLy6Clo; gating strategy in Supplemental Figure 2C), which had slightly lower frequencies but similar numbers in lungs of anti–IL-17–treated HN878-infected mice, when compared with isotype treatment (Figure 3C and Supplemental Figure 2E). Consistent with a suppressive role for MDSCs in T cell function in vivo, we observed a significant reduction in the frequency and numbers of activated CD4+ T cells, as well as activated CD44hiCD4+ IFN-γ–producing T cells in HN878-infected lungs following IL-17 neutralization (Figure 3, D and E, and Supplemental Figure 2, F and G). To determine the suppressive function of the MDSCs, we cocultured sorted M-MDSCs and G-MDSCs from anti–IL-17–treated animals with anti–CD3/CD28–stimulated, CFSE-stained CD4+ T cells. We found that proliferation of activated CD44+CD4+ T cells significantly decreased upon coculture with M-MDSCs or G-MDSCs (Figure 3F), suggesting that both M-MDSCs and G-MDSCs are functionally suppressive, and distinct from neutrophils, which do not exhibit suppressive function (24). These data suggest that IL-17 mediates protection through limiting M-MDSC accumulation in the lung, resulting in optimal IFN-γ–producing CD4+ T cell responses to mediate HN878 control.
Figure 3. IL-17 limits accumulation of monocytic MDSCs and shifts towards aerobic glycolysis.
FeJ mice were aerosol infected with approximately 100 CFU Mycobacterium tuberculosis clinical strain HN878 and treated with isotype or anti–IL-17 antibody as described in Figure 1. At 30 days postinfection (d.p.i.), lungs were processed to a single-cell suspension, and flow cytometry was used to assess frequencies of (A) neutrophils (n = 4 mice/group), (B) monocytic myeloid-derived suppressor cells (M-MDSCs; n = 4 mice/group), (C) granulocytic MDSCs (G-MDSCs n = 4 mice/group), (D) activated CD44hiCD4+ T cells (n = 5 mice/group), and (E) activated CD44hiCD4+ IFN-γ–producing T cells (n = 5 mice/group) in the lungs. (F) FeJ mice were infected with approximately 100 CFU HN878 and treated with anti–IL-17. At 37 d.p.i., lung G-MDSCs and M-MDSCs were sorted based on the gating strategy shown in Supplemental Figure 2C and cocultured with anti–CD3/CD28–stimulated, CFSE-stained CD4+ T cells for 3 days, and the number of proliferating CD44hiCD4+ T cells normalized to 10,000 cells are shown (n = 4 biological replicates/anti–CD3/CD28 alone, n = 7 mice/G-MDSC or M-MDSC + anti–CD3/CD28). (G) qRT-PCR was used to determine expression of Nos2, Il6, Il1a, Arg1, and Cxcl2 in FACS-sorted M-MDSCs, alveolar macrophages (AMs), and myeloid DCs (mDCs) from HN878-infected mice (n = 5 mice) at 31 d.p.i., or HN878-infected in vivo–generated MDSCs (iMDSCs; n = 5 biological replicates) from FeJ bone marrow. (H) Lactate accumulation in culture supernatants of uninfected (n = 8 biological replicates), uninfected and rIL-17–treated (100 ng/ml, n = 3 biological replicates), untreated HN878-infected (MOI 0.1) (n = 8 biological replicates), and HN878-infected rIL-17–treated (n = 11 biological replicates) iMDSCs was assessed using a lactate assay. (I) RNA-Seq analysis depicting fold change of specific gene expression in lungs of anti–IL-17–treated HN878-infected FeJ mice (n = 5 mice) over the expression in isotype-treated HN878-infected mice (n = 3 mice) at 30 d.p.i. (J) Cxcl2 mRNA localization in lung was determined by in situ hybridization. Representative images are shown, ×100 magnification; arrows depict Cxcl2 expression within granulomas. All data shown as mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001 by Student’s t test (A–E), or 1-way ANOVA (F–H). ns, not significant.
To further address the mechanism by which IL-17 impacts M-MDSCs during HN878 infection, we found that sorted M-MDSCs from lungs of IL-17–depleted HN878-infected mice expressed mRNA for inflammatory molecules such as IL-1α, IL-6, arginase, iNOS, as well as the chemokine CXCL2, previously shown to be expressed by MDSCs (Figure 3G). Additionally, upon infection with HN878, in vivo–generated MDSCs (iMDSCs) also expressed mRNA for these proinflammatory molecules (Figure 3G). Notably, both M-MDSCs and iMDSCs expressed increased arginase 1 mRNA, an enzyme associated with the immunosuppressive activity of MDSCs (24), compared with alveolar macrophages (AMs) and myeloid dendritic cells (mDCs) (Figure 3G). To determine whether the metabolic state of iMDSCs shifts during HN878 infection, iMDSCs were differentiated from bone marrow and infected with HN878. Lactate levels increased upon in vitro infection, suggesting a shift toward glycolysis. However, rIL-17 treatment completely reversed lactate accumulation in culture supernatants (Figure 3H). Interestingly, adenosine triphosphatase (ATPase), a marker of glycolysis, colocalized with iNOS and F4/80 within granulomas of isotype-treated mice, while in anti–IL-17–treated mice, colocalization was increased and observed mostly within necrotic granulomas (Supplemental Figure 3A). Furthermore, stabilization of HIF1α upregulates lactate dehydrogenase A (LDH-A), an enzyme necessary in the final step of anaerobic glycolysis whereby L-lactate and NAD are converted to pyruvate and NADH, respectively (19, 25). Similar to ATPase, the expression of LDH-A was increased within F4/80+ macrophages in the necrotic lesions of anti–IL-17–treated HN878-infected mice (Supplemental Figure 3B). As both ATPase and LDH-A are markers of glycolysis, these data support a shift towards glycolysis following HN878 infection and anti–IL-17 treatment. Consistent with the findings above, mRNA expression of genes regulating glycolysis such as Slc2a1, Slca3, hexokinases (i.e., Hk1), and members of the phosphofructokinase family (i.e., Pfk1) (26), was significantly higher in lungs of anti–IL-17–treated HN878-infected mice when compared with levels in isotype-treated HN878-infected mice (Figure 3I and Supplemental Table 1). In addition, mRNA expression of genes associated with regulation, recruitment, and function of MDSCs such as Cxcl2, S100a8, S100a9, Arg1, Csf3, and Il11 was also higher in lungs of anti–IL-17–treated HN878-infected mice (Figure 3I and Supplemental Table 1). mRNA gene expression associated with inflammation and lung damage such as albumin, Il1a, Il1b, serum amyloid proteins (SAA), matrix metalloproteases (MMPs), Ccl-chemokines, and collagenase (27) were also highly expressed in lungs of anti–IL-17–treated HN878-infected mice when compared with levels in isotype-treated HN878-infected mice. Furthermore, mRNA for Cxcl2, a key MDSC-recruiting chemokine (24), was highly expressed and localized within lungs of HN878-infected mice undergoing IL-17 neutralization, when compared with isotype-treated HN878-infected lungs (Figure 3J). These data together suggest that inflammatory MDSCs and macrophages are present within hypoxic TB granulomas and induced to express HIF1α, resulting in increased glycolysis; IL-17 restricts this shift by limiting HIF1α expression and impairing accumulation of MDSCs in TB granulomas.
HN878 PGLs are prime inducers of IL-1β production in lung cells.
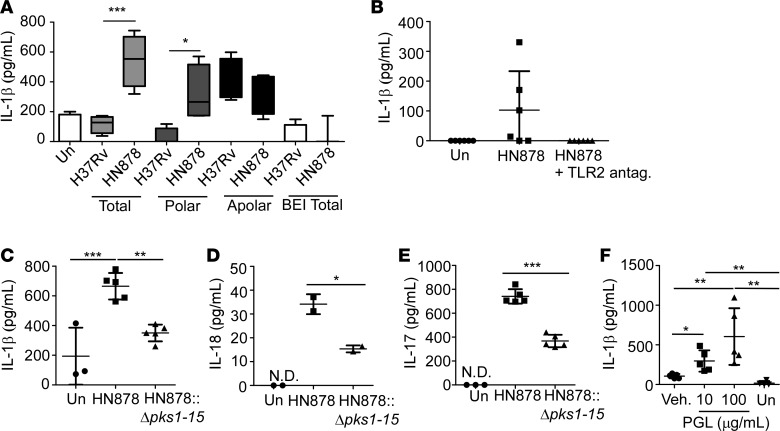
Cell wall components isolated from HN878, an emerging W-Beijing lineage strain, are potent inducers of IL-1β responses in C57BL/6J (B6) mouse lung DCs, thus mediating increased production of IL-17 in T cells (28). To delineate the molecular mechanism by which HN878 increases IL-1β production, we carried out MALDI-TOF analyses to compare the differential biomolecule expression in cell wall components between the clinical strain used in this study, HN878, and the lab-adapted strain, H37Rv, which does not effectively induce IL-17 (28). We identified 95 proteins that were increased in HN878 cell wall preparations, when compared with cell wall preparations from H37Rv (Supplemental Table 2). Consistent with previous studies (29), the protein product of the pks1 gene, necessary for PGL production, was by far the most elevated with 34-fold increased expression in cell wall preparation from HN878, when compared with H37Rv (Supplemental Table 2). To identify the major cell wall component of Mtb that mediates increased IL-1β induction in DCs, we utilized purified fractions of H37Rv- and HN878-derived acetone-soluble (apolar) and -insoluble (polar) lipids, mannose-capped lipoarabinomannan (ManLAM), and phosphatidylinositol mannoside (PIM) to stimulate bone marrow–derived DCs (BMDCs). While neither PIM nor ManLAM induced differential production of IL-1β (data not shown), HN878-derived total lipids and polar lipids, but not apolar lipids, induced higher protein levels of IL-1β in BMDCs, when compared with IL-1β production in BMDCs stimulated with similarly isolated derivatives from H37Rv (Figure 4A). As previously published, total lipids obtained from BEI Resources did not induce IL-1β production in DCs (28). Furthermore, loss of IL-1β production was observed in HN878 lipid–stimulated lung cells treated with purified anti–mouse CD282 (TLR2-blocking antibody), supporting a role for TLR2 in Mtb induction of IL-1β production (Figure 4B).
Figure 4. HN878 phenolic glycolipids (PGLs) drive inflammasome activation.
(A) Bone marrow–derived DCs (BMDCs) were stimulated for 48 hours with 10 μg/ml of either total lipids (n = 4 biological replicates/group), polar or apolar lipids, derived from either Mycobacterium tuberculosis lab-adapted strain H37Rv or clinical strain HN878 (n = 4 biological replicates), total lipids from BEI Resources (n = 4 biological replicates/H37Rv, n = 3 biological replicates/HN878), or left untreated (Un; n = 7 biological replicates). IL-1β in culture supernatants was measured by ELISA. (B) Lung cells were infected for 6 days with HN878 (MOI 0.1) following preincubation with a TLR2-blocking antibody (30 μg/ml) (n = 6 biological replicates/group). Supernatant IL-1β was measured by ELISA. (C–E) Lung cells isolated from FeJ mice were infected for 6 days with HN878 (MOI 0.1), HN878::Δpks1-15 (MOI 0.1), or left uninfected (Un). ELISA was used to measure (C) IL-1β (n = 3 biological replicates/Un, n = 5 biological replicates/infected group), (D) IL-18 (n = 2 biological replicates/group), and (E) IL-17 in culture supernatants (n = 3 biological replicates/Un, n = 5 biological replicates/infected group). (F) Lung cells isolated from FeJ mice were treated for 6 days with 10 μg/ml or 100 μg/ml PGL derived from HN878, vehicle (Veh), or left untreated (Un), and IL-1β was measured by ELISA (n = 5 biological replicates/PGL treatment, n = 4 biological replicates/untreated or Veh) (Student’s t test performed between vehicle and 10 μg/ml PGL treatment, 1-way ANOVA performed between all groups). All data shown as mean ± SD. N.D., not detectable. *P < 0.05, **P < 0.01, ***P < 0.001 by Student’s t test (D–F) or by 1-way ANOVA (A–C and F).
PGLs, produced by the polyketide synthase encoded by the pks1-15 gene, are a likely component of the acetone-insoluble polar lipids from HN878 (30), but are absent in preparations from H37Rv cell wall preparations, due to the absence of a functional pks1-15 gene (Supplemental Figure 4A). To determine the functional importance of PGLs in driving increased production of Th17 pathway cytokines, mouse total lung cells were infected with either HN878 or an Mtb mutant lacking the pks1-15 gene (HN878::Δpks1-15) (29). Indeed, HN878::Δpks1-15–infected lung cells produced lower amounts of IL-1β, when compared with infection with wild-type HN878, suggesting PGLs are the primary mediators of increased IL-1β responses in lung cells (Figure 4C). Furthermore, increased IL-18 production by lung cells following HN878 infection, and the corresponding decrease in IL-18 following infection with HN878::Δpks1-15 suggests that PGLs trigger activation of the inflammasome for production of IL-1β (Figure 4D). Importantly, the decreased induction of both IL-1β and IL-18 coincided with decreased IL-17 production in lung cells infected with HN878::Δpks1-15, when compared with HN878 (Figure 4E). However, HN878::Δpks1-15 infection induced some IL-17 production, suggesting that although PGLs play a dominant role in driving IL-17 production, other HN878-specific factors may still participate in the induction of IL-17 responses. Treatment of lung cells with purified PGLs alone was sufficient to induce IL-1β in a dose-dependent manner (Figure 4F). Furthermore, upon HN878 cell wall stimulation, glycolysis, measured via real-time quantification of extracellular acidification rate (ECAR), was enhanced in macrophages when compared with unstimulated macrophages (Supplemental Figure 4B). Together, these data support a critical role for PGL interaction with the host immune cells in the induction of glycolysis. Indeed, PGL-mediated induction of IL-17 may be one way to control pathogen-mediated hypoxic glycolysis.
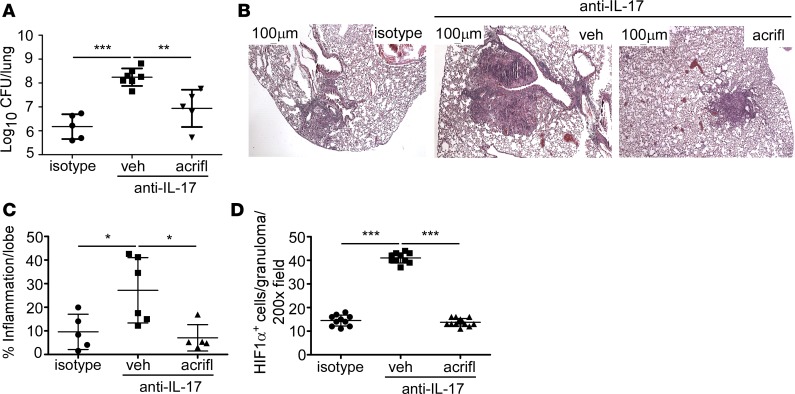
Inhibition of HIF1α can reverse disease susceptibility in IL-17–neutralized Mtb-infected FeJ mice.
Our in vitro data show that IL-17 treatment of macrophages and MDSCs limits glycolysis, while IL-17 neutralization in vivo resulted in increased HIF1α expression and hypoxia following HN878 infection (Figure 1). However, the kinetics of HIF1α expression in relation to the progression to a hypoxic lung environment in response to HN878 infection was unknown. Therefore, we measured PIMO and HIF1α expression and found that HIF1α, but not PIMO, expression is increased early in anti–IL-17–treated mice compared with isotype (Figure 5, A–C). As PIMO levels increase at later time points (Figure 1J) but not earlier (Figure 5C), HIF1α was increased prior to detectable changes in hypoxia. We next addressed whether increased TB disease and necrosis observed in IL-17–neutralized HN878-infected mice could be reversed by blocking HIF1α activity. Acriflavine is a compound that inhibits HIF1 dimerization and transcriptional activity by binding to the PAS-B domain of HIF1α, thereby reducing the HIF1-mediated hypoxic response (31). We treated HN878-infected mice with IL-17–neutralizing antibody followed by acriflavine to determine whether, in the absence of IL-17, dysregulated HIF1α drives increased disease and susceptibility to HN878. Our results show that HIF1α inhibition by acriflavine treatment completely reversed both the increased bacterial burden and the exacerbated inflammation in HN878-infected mice undergoing IL-17 neutralization (Figure 6, A–C). The effect of HIF1α inhibition was confirmed in vivo in mice receiving anti–IL-17, where acriflavine treatment resulted in reduced HIF1α expression within TB granulomas, when compared with HN878-infected mice receiving IL-17–neutralizing antibody alone (Figure 6D). Previously, acriflavine was shown to harbor bactericidal characteristics (32). The minimum inhibitory concentration (MIC) of acriflavine against HN878 was 25 μg/ml (Supplemental Table 3). In vivo, we detected an acriflavine concentration of 1.5 μg/ml within the lung 24 hours following acriflavine delivery. Therefore, the in vivo effect of acriflavine in the lung is independent of its direct inhibitory effect on HN878 at higher concentrations. Thus, our data demonstrate that IL-17 limits HIF1α expression to downregulate hypoxia, necrosis, and TB disease during HN878 infection. In contrast, upon IL-17 neutralization, HIF1α expression within macrophages is increased, resulting in increased hypoxia, necrosis, and profound susceptibility to infection.
Figure 5. HIF1α expression precedes exacerbated hypoxic granulomas in anti–IL-17–treated mice.
(A–C) FeJ mice were infected with approximately 100 CFU Mycobacterium tuberculosis clinical strain HN878, and received isotype or IL-17–neutralizing antibody (anti–IL-17) via i.p. from 10 to 24 days postinfection (d.p.i.) and sacrificed at 30 d.p.i. Lung immunofluorescence images of (A) HIF1α (green), F4/80 (red), and quantification of (B) HIF1α (n = 5 mice/group) and (C) pimonidazole (PIMO; n = 5 mice/group) staining by immunofluorescence at 30 d.p.i. are shown. All data shown as mean ± SD, ***P < 0.001 by Student’s t test.
Figure 6. HIF1α inhibition in vivo reverses IL-17 neutralization–mediated susceptibility to Mtb.
(A–D) Following infection with approximately 100 CFU Mycobacterium tuberculosis (Mtb) clinical strain HN878, FeJ mice received isotype or IL-17–neutralizing antibody via i.p. from 10 to 24 days postinfection (d.p.i.). At 24 d.p.i., anti–IL-17–treated mice received 2 mg/kg HIF1α inhibitor (acriflavine [acrifl]) or vehicle (veh) from 24 to 36 d.p.i. Mice were harvested at 37 d.p.i. and bacterial burden (n = 5 mice/isotype, n = 7 mice/anti–IL-17, n = 5 mice/anti–IL-17 + acrifl) (A), H&E–stained lungs (B), and inflammation (n = 5 mice/isotype, n = 6 mice/anti–IL-17, n = 7 mice/anti–IL-17 + acrifl) (C) were quantified. (D) HIF1α-expressing cells in representative granulomas in the lungs were measured by immunofluorescence microscopy (n = 5 mice/isotype, n = 8 mice/anti–IL-17, n = 7 mice/anti–IL-17 + acrifl). All data shown as mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001 by 1-way ANOVA (A, C, and D).
The IL-17 SNP rs2275913 is associated with susceptibility to TB.
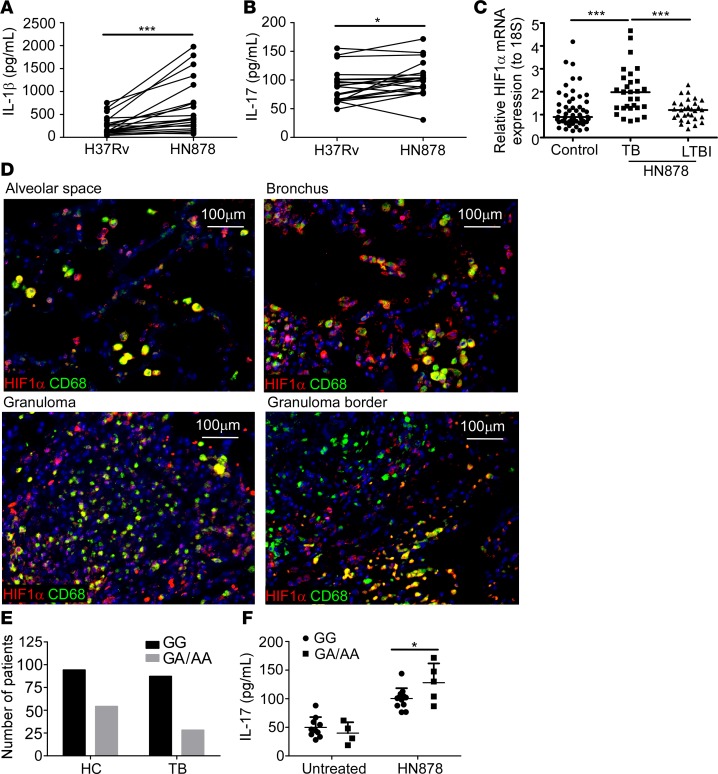
To further determine relevance to human TB, we first addressed if, when compared with H37Rv, the clinical isolate HN878 could differentially regulate the expression of IL-1β and IL-17 responses in peripheral blood mononuclear cells (PBMCs) from human TB patients. HN878-stimulated PBMCs from TB patients produced significantly higher amounts of IL-1β and IL-17 compared with H37Rv stimulation (Figure 7, A and B). The induction of cytokines was limited to Th17 pathway cytokines, as neither IFN-γ nor TNF-α levels were increased in PBMCs stimulated with HN878 cell wall preparation when compared with H37Rv stimulation (Supplemental Figure 5, A and B). Importantly, we also found increased expression of HIF1A mRNA in HN878-stimulated PBMCs of TB patients, when compared with HIF1A mRNA expression in PBMCs from healthy control individuals, or LTBI patients (Figure 7C). In addition, similar to lungs of HN878-infected mice, HIF1α protein expression colocalized mainly with CD68-expressing macrophages residing within and around the borders of granulomas from TB patients (Figure 7D). These data show that similar to murine cells, human PBMCs from TB patients respond to Mtb HN878 exposure by enhancing IL-1β and IL-17 production. Increased HIF1A mRNA expression was detected in PBMCs from TB patients and increased HIF1α protein within macrophages in TB granulomas, suggesting that hypoxia pathways are activated upon exposure to HN878.
Figure 7. IL-17 SNP rs2275913 differentially regulates IL-17 expression and is associated with TB.
Peripheral blood mononuclear cells (PBMCs) were isolated from tuberculosis (TB) patients and stimulated with 10 μg/ml of either Mycobacterium tuberculosis lab-adapted strain H37Rv or clinical strain HN878 cell wall extract. Levels of (A) IL-1β (n = 19 patients) and (B) IL-17 (n = 20 patents) in culture supernatants were measured by ELISA. (C) Expression of HIF1A mRNA following stimulation with 10 μg/ml HN878 cell wall extract was determined by RT-PCR in PBMCs from TB patients (n = 30), latently infected TB (LTBI) patients (n = 30), or household controls (n = 55). (D) Formalin-fixed, paraffin-embedded lung sections from TB patients were stained for CD68 (green) and HIF1α (red) by immunofluorescence. Representative images shown (×200 magnification). (E) TB patients or household controls (HC) were genotyped for G/A, G/G, or A/A alleles of the rs2275913 SNP using the Sequenom assay as described in the Methods (nTotal = 263 patients; n = 94 HC with G/G, n = 54 HC with G/A or A/A, n = 87 TB patients with G/G, and n = 28 TB patients with G/A or A/A). (F) IL-17 production in culture supernatants of HN878 cell wall extract–stimulated (10 μg/ml) PBMCs isolated from either G/A, A/A, or G/G genotype TB patients was measured by multiplex assay (n = 12 patients for G/G allele, n = 4 patients for untreated G/A or A/A alleles, and n = 5 patients for HN878-treated G/A or A/A alleles). *P < 0.05, **P < 0.01, ***P < 0.001 by 2-way ANOVA (A and B), 1-way ANOVA (C), or repeated-measures ANOVA (F).
Our experimental data support a critical role for IL-17 in limiting disease upon infection with HN878. Thus, we explored the potential role for SNPs in the human IL-17 locus that may be involved in the immune control of HN878 in humans. For this purpose we performed a high-throughput screen of 203 SNPs in human-host immune genes including genes belonging to the Th1 cell pathway (IFNG, TBX21), T follicular helper cell pathway (CXCR5, CXCL13, BCL6, ICOS, IL21), and the Th17 cell pathway (IL1B, IL23R, IL23, IL22, IL17A, and IL17F) (Supplemental Table 4). In a cohort of Mexican Amerindian individuals, we found that 1 of 3 SNPs, allele G of the SNP rs2275913, located at position –197 of the promoter region of the IL17 gene (IL17 –197G>A), was significantly associated with increased susceptibility to TB (χ2 P value = 0.038) (Figure 7E). Additionally, the other 2 SNPs associated with increased susceptibility to TB were in rs2069837 (IL6 gene, χ2 P value = 0.030) and rs2397084 (IL17F gene, χ2 P value = 0.045). To understand the relevance of G/A or A/A versus G/G genotype with respect to HN878 infection and IL-17, PBMCs from G/A, A/A, and G/G allele–expressing TB patients were stimulated in vitro with HN878 cell wall proteins and IL-17 production was measured. PBMCs from IL17 –197A allele–positive TB patients (–197G/A or –197A/A) displayed a steeper increase in IL-17 production than TB patients with the IL17 –197G/G genotype (repeated-measures ANOVA [RM-ANOVA] P value = 0.04) (Figure 7F). Although IL-17 levels were not different at baseline (P value = 0.3992) between the –197 G/G, G/A, and A/A groups, exposure to HN878 cell wall products led to a significant change in IL-17 levels (Figure 7F). This is in sharp contrast to PBMCs stimulated with Mtb H37Rv cell wall, wherein G/A or A/A and G/G displayed a similar increase in IL-17 with no significant difference between alleles (P value = 0.8631, data not shown). Interestingly, the IL17 –197G/G genotype was significantly associated with infection by drug-resistant Mtb strains, when compared with patients with the IL17 –197G/A or A/A genotype (RM-ANOVA P value = 0.03) (Supplemental Table 5). However, there was no significant correlation between lung damage and drug resistance controlling for the rs2275913 SNP (Pearson partial correlation coefficient = 0.16, P value = 0.40) (Supplemental Table 5). These data together provide potentially new evidence that polymorphisms in the IL17 promoter region may influence the production of this cytokine in response to HN878 infection, supporting an important role for IL-17 in host immunity to TB.
Discussion
Hypoxic necrotic granulomas are associated with increased TB disease severity and promotion of antibiotic resistance (3, 6). However, the host factors that regulate the development of hypoxic granulomas during TB are not well described. Utilizing a mouse model of necrotic TB, we show that IL-17 plays a critical role in limiting hypoxia and necrosis in HN878 infection by negatively regulating HIF1α expression and the subsequent metabolic shift toward glycolysis. Thus, IL-17 neutralization results in increased HIF1α expression, the promotion of hypoxic TB granuloma, and more severe disease outcomes. Importantly, in vivo inhibition of HIF1α reversed the severe disease outcome seen upon IL-17 neutralization in the necrotic mouse model. Furthermore, association of rs2275913 (a SNP within the human IL-17 promoter) with TB indicates an important role for IL-17 in TB disease. Thus, our data have important implications for TB disease and therapy. Firstly, our data show a potentially novel critical role for IL-17 in limiting HIF1α during TB. Secondly, our data caution the potential pathological consequences of the use of IL-17–blocking agents for treatment of inflammatory conditions in TB patients. Finally, as the development of HIF1α inhibitors for anticancer therapeutics are currently underway (33), our data propose that repurposing HIF1α inhibitors as an effective therapy for TB patients should be considered further.
We show here that IL-17 can regulate HIF1α and limit the downstream development of hypoxic TB granulomas. This is in complete contrast to the known role of IL-17 and Th17 cells in promoting inflammation and tumor proliferation during cancer (18, 34). It is possible that IL-17 can negatively regulate HIF1α by promoting its ubiquitination or inhibition of protein-protein interactions with HIF1α coactivators (35). More recently, microRNA-210 was shown to directly target HIF1α mRNA (17). Thus, early IL-17 production induced in response to Mtb infection may possibly utilize microRNAs to regulate HIF1α.
While IL-17 and HIF1α have been implicated in promoting cancer progression and shifting metabolism toward the Warburg effect, IL-17 may deviate from this previously characterized role in limiting HIF1α in the presence of pathogens such as Mtb. Furthermore, cancer studies have implicated IL-17 produced by Th17 cells in driving cancer progression (18). However, in our study, early IL-17 production, likely produced by γδ T cells, limits HIF1α and progression towards hypoxic necrotic granulomas. Thus, the kinetics and cellular source of IL-17 may promote divergent roles for IL-17 in either promoting or limiting hypoxia under conditions of inflammation and infection. Future studies delineating these 2 different functions of IL-17 in inflammation and infection will provide new insights into the role of IL-17 in hypoxia and host immunity to infection.
IL-17 itself is associated with increased MDSC accumulation in cancer models (36). MDSC-like cells have previously been described during TB disease in susceptible mouse models (24, 37) and early during disease in TB patients (38). Both in experimental models and humans, accumulation of MDSCs is associated with increased TB disease severity, and depletion of MDSCs reverses TB disease severity in Mtb-susceptible mice (24, 38). Consistent with these studies, our data show that IL-17 depletion results in increased HIF1α-expressing myeloid cells and accumulation of large numbers of MDSCs within TB granulomas, coinciding with increased disease severity and suppression of T cell responses. That MDSCs themselves have been implicated as IL-17 producers during Mtb infection (37) suggests that IL-17 made by MDSCs may be a necessary feedback loop to limit excessive HIF1α production in MDSCs. Interestingly, in a systemic Mycobacterium avium model of infection, mice deficient in HIF1α expression in myeloid cells demonstrated increased liver necrosis (39). A recent study also showed that in a high-dose model of Mtb Erdman infection, mice deficient in HIF1α in myeloid cells demonstrated increased susceptibility to infection (40). HIF1α was shown to induce IFN-γ–mediated killing by activating aerobic glycolysis (40), suggesting that the pathways that modulate host protection versus pathology are finely balanced. However, the above studies were conducted in B6 mice; further studies on the role of HIF1α in human or nonhuman primate Mtb infection models may provide additional insights into the role of HIF1α in inducing pathology, as shown here in necrotic mouse models, versus protection as shown in the non-necrotic B6 mouse model of Mtb infection.
PGLs are key virulence factors present in virulent emerging Mtb strains (29). Our data using HN878::Δpks1-15 as well as purified PGL from HN878, project PGLs as the primary inducers of IL-1β in BMDCs through TLR2, following subsequent induction of IL-17. Our data show no difference in disease progression between isotype- and anti–IL-17–treated H37Rv-infected mice, emphasizing an Mtb-strain-dependent protective role for IL-17 (28). These findings are consistent with the ability of PGLs to induce the chemokine CCL2 to promote the recruitment of permissive macrophages in the host (41). Incidentally, IL-17 can effectively induce CCL2 expression in the host under inflammatory conditions (42). Additionally, HN878 infection resulted in a metabolic shift towards glycolysis, while inducing IL-17 production. These results support a role for IL-17 as a critical host factor that counterbalances the pathogen-mediated glycolytic shift. Our studies show that innate T cells such as γδ T cells, along with CD4+ and CD8+ T cells, are all IL-17 producers following HN878 infection. That early depletion of IL-17 alone is sufficient to induce increased susceptibility in HN878-infected mice supports the idea that innate sources of IL-17 are the primary IL-17 producers mediating the protective effect seen following infection. Consistent with this idea, γδ T cells respond to IL-23 and TLR2 stimulation to induce IL-17 production (43). Thus, our studies suggest that early interactions between PGLs, likely through TLR2 interactions, mediate important immune events necessary for protection against TB and to delay progression of Mtb infection.
IL-17 is a primary target for alleviating inflammation such as psoriatic arthritis, rheumatoid arthritis, ankylosing spondylitis, and multiple sclerosis (44). Therefore, several candidates with targets along the IL-17/Th17 pathway are being developed for use in these inflammatory conditions. Monoclonal antibodies that selectively bind and neutralize IL-17 have been successful in phase III trials in psoriasis, showing superiority when compared with etanercept, a TNF-α blocker (44). Despite not having a clear role thus far for IL-17 in TB, as a precaution, the studies testing IL-17 pathway blockers in humans have excluded LTBI patients in the study design. However, our work described here clearly suggests a protective and indispensable role for IL-17 in TB. Thus, for the first time to our knowledge we provide experimental evidence to propose that TB patients should not be treated with IL-17 pathway blockers, as this may result in profound consequences for progression from latency to TB disease, or development of severe TB disease.
As we identified a prominent role for IL-17 as a HIF1α regulator, IL-17 host genetics may be important indicators of outcome such as drug resistance and progression of TB. Our studies show that the –197G/A and –197A/A alleles in rs2275913 in the IL-17 promoter are associated with increased IL-17 production in PBMCs from TB patients, and coincide with a decreased association with TB. Despite Mexico not being a geographical area where W-Beijing Mtb strains are highly prevalent (45, 46), our data are consistent with recent studies that have shown the rs2275913 –197G/G phenotype is associated with increased TB in Spanish and Brazilian cohorts of TB patients (47–49). Of the 203 SNPs studied, which spanned Th1, Th17, and Tfh pathways, the SNPs that were found to be significantly associated with TB were in the IL-17/Th17 pathway, supporting an important role for IL-17 in human TB immunity. Additionally, the –197A/A allele has been reported to display a higher affinity for nuclear factor of activated T cells (NFAT), a critical transcription factor involved in IL-17 regulation (50). Considering that W-Beijing Mtb strains are emerging worldwide, it is possible that PGL-carrying Mtb strains interact with host immune cells to differentially modulate IL-17 production in response to infection. Future work studying the role of IL-17 SNPs in geographical areas such as East Asia where PGL-carrying W-Beijing Mtb strains are highly prevalent would further clarify the functional role of IL-17 in Mtb pathogenesis.
In summary, we have identified IL-17 as a potentially novel negative regulator for Mtb-driven metabolic shifts toward glycolysis, primarily through the inhibition of HIF1α. These findings have significant implications in designing new therapeutics and therapies for treatment of TB.
Methods
Human TB study design
Human whole blood and serum samples were obtained from patients with TB and LTBI from the Tuberculosis Outpatient Clinic at the National Institute of Respiratory Diseases (INER) in Mexico City. Samples were taken prior to anti-Mtb treatment from patients that did not present comorbidities (i.e., type 2 diabetes, HIV infection, cancer, chronic obstructive pulmonary disease [COPD]). For SNP Sequenom array analysis, samples from 263 unrelated individuals (115 TB patients and 148 household contact controls) were used. TB diagnosis was based on WHO criteria with presence of clinical symptoms, detection of acid-fast bacilli in sputum smear samples, Mtb-positive cultures in Löwenstein-Jensen medium, and X-ray evidence of cavitary lesions in lung. Only patients older than 18 years were included in the study. Household contacts were unrelated individuals in close contact with TB patients, which remained asymptomatic with no evidence of positive Mtb cultures or detectable radiological lesions in the lung.
Stimulation of human PBMCs with Mtb cell wall
Human PBMCs were isolated by Ficoll Hypaque gradient (Lymphoprep; Axis-Shield POC AS). PBMCs (2.5 × 106 cells/ml) were plated in 24-well plates and incubated in the presence of Mtb cell wall extract (10 μg/ml, from H37Rv or HN878). After 4 days of incubation at 37°C and 7.5% CO2, cells were harvested, and cytokine levels in culture supernatants were determined using a Bio-Plex Luminex 200 instrument (Bio-Rad).
Human SNP genotyping
SNP genotyping was carried out using the i-PLEX Gold MassARRAY system (Sequenom, Inc.) in the Genomics and Proteomics Core Laboratories (GPCL) at the University of Pittsburgh.
Primer design.
Three primers were designed for each locus of interest using MassARRAY Assay Design 4.0. Assay Design software (Agena Bioscience) was used to determine primer set pooling to optimize multiplex reactions. Mass modifications were incorporated in the design of MassExtend primers, in order to maximize the mass differential between primers of different loci within a given multiplexed pool. Multiplexed pools were designed for up to 40 loci, depending on primer compatibility for the specific loci being assayed.
Sample amplification.
Target loci were amplified within the samples by multiplex PCR. After completion of PCR, dNTPs and primers were removed.
MassExtend.
Excess MassExtend primers corresponding to the loci represented by the amplification primers used were pooled. Higher-mass primers were added at a higher concentration to adjust for signal drop off during spectra acquisition. Single-base extension was carried out appropriate to the relative mass of the primer and amplification carried out. Clean resin and water were added to MassExtend reaction products and samples were incubated in clean resin at room temperature with mixing for 15 minutes and centrifuged at 3,200 g for 5 minutes.
NanoDispense, spectra acquisition, and analysis. Samples were dispensed to a SpectraChip using the MassARRAY Nanodispenser (Agena Bioscience) according to the manufacturer’s instructions. Spectrachips were loaded into the MassARRAY analyzer and spectra were acquired for each sample. MassARRAY Typer software uses the known mass of the MassExtend primers to identify each locus, and the increase caused by each distinct nucleotide to identify the alleles present in the sample.
Data analysis method.
Data analysis and interpretation were carried out by the Bioinformatics Analysis Core (BAC) at the GPCL, University of Pittsburgh. Statistical association analysis was performed using a χ2 test. SNP genotyping data corresponding to both control (household controls, n = 86 patients) and test (TB patients, n = 85 patients) groups from plexes 1–6 were pooled into 1 data file. As part of discovery, SNPs with P values less than or equal to 0.05 were considered as SNPs of interest (SOIs), putatively significant, and designated for follow-up validation. An additional set of control (n = 63) and TB (n = 30) patients were included for rs2275913 SNP genotyping.
IL-17 SNP genotyping by PCR
Genomic DNA (gDNA) was extracted from PBMCs isolated from TB patients using a Qiagen Blood Mini Kit (Qiagen). The IL17 −197G>A (rs2275913) SNP was analyzed. Genotyping was performed using a validated TaqMan 5′ nuclease assay (C__15879983_10, Life Technologies). Each PCR reaction used 15 ng of total gDNA, according to the manufacturer’s instructions. All PCRs were performed using 96-well plates in a Step One Plus Real-Time PCR system (Life Technologies).
RNA-Seq sample preparation and sequencing
Total RNA was isolated from lung tissue using an RNeasy RNA isolation kit (Qiagen). Each sample was assessed using the Qubit 2.0 fluorometer (Invitrogen, Thermo Fisher Scientific) and Agilent Tapestation 2200 (Agilent Technologies). Sequencing libraries were generated using a TruSeq RNA Access Library Prep Kit (Illumina) following the manufacturer’s protocol. Cluster generation and 75-bp paired-read dual-indexed sequencing was performed on the NextSeq 500 platform (Illumina). RNA-Seq analysis was performed using the Maverix Analytic Platform (Maverix Biomics).
Accession numbers
RNA-Seq data supporting the findings of this study have been deposited in BioProject with the accession code SRP113787. SNP data are available as part of the rs2275913 entry (batch ID 1062914) in the NCBI SNP data base (https://www.ncbi.nlm.nih.gov/snp/).
Mtb infection in mice
C3HeB/FeJ (FeJ) mice were obtained from The Jackson Laboratory and bred at Washington University in St. Louis. Experimental mice were age- and sex-matched and used between 6 and 8 weeks of age. Mtb wild type and mutant strains of HN878 (BEI Resources and Gilla Kaplan) were cultured to mid-log phase in Proskauer Beck medium containing 0.05% Tween 80 and frozen in 1-ml aliquots at –80°C. Mice were aerosol infected with approximately 100 CFU of bacteria using a Glas-Col airborne infection system (51). At given time points, organs were harvested, homogenized, and serial dilutions of tissue homogenates plated on 7H11 plates and CFU determined. IL-17–blocking antibody (BE0173, Bio X Cell) or corresponding isotype control was administered (100 μg intraperitoneally [i.p.]) every 2 days as denoted. As acriflavine is fluorescent, acriflavine in lung homogenates from 37 d.p.i. was determined to be 1.5 μg/ml with a spectrophotometer and the fluorescence compared against an acriflavine standard curve.
Preparation of mycobacterial extracts for LC-MS
HN878 strains were grown on 7H11 agar supplemented with OADC (oleic acid, albumin, dextrose, and catalase enrichment supplement) for 14 days. Live bacteria were harvested and serially extracted at 37°C with chloroform/methanol (2:1, v/v and 1:2 v/v) and chloroform/methanol/water (10:10:3, v/v/v) for 2 hours per extraction. Extracts were washed with sterile endotoxin-free water (Folch wash), and the resulting organic phase dried down under a flux of nitrogen. Dried pellets were extracted with cold acetone overnight at –20°C under stirring conditions. Acetone-soluble extracts were aliquoted, dried, and kept at –80°C. Acetone-insoluble extracts were suspended in chloroform/methanol (1:1, v/v), aliquoted, dried, and kept at –80°C. HN878 PGL was purified by preparative thin layer chromatography (TLC) using a solvent system of chloroform/methanol (95:5, v/v). PGL was visualized as being positive by UV light and by using 10% sulfuric acid in ethanol as the developer. BEI Resources total lipids were obtained from bacteria grown to late-log phase in glycerol-alanine-salts broth and inactivated by γ irradiation as described in the NIH contract AI-75320 (https://www.beiresources.org).
Cell wall preparation in phosphate-buffered saline (PBS) was lyophilized and dissolved in 80 μl of Tris-HCl buffer (pH 7.6, 100 mM) containing SDS (4%) and dithiothreitol (DTT, 100 mM) and heated (95°C for 5 minutes). Samples were diluted with 300 μl of Tris-HCl buffer (pH 8.5, 100 mM) containing 8 M urea and transferred to Microcon filtration units (YM-30). SDS was removed using the filter-aided-sample-preparation (FASP) method (52). After buffer exchange, 100 μl of buffer (ammonium bicarbonate, pH 7.8, 50 mM) was pipetted into the Microcon filtration unit and trypsin was added (1 μg/μl). The digest was incubated for 4 hours at 37°C, and then overnight in a humid chamber after the addition of another aliquot of trypsin. The digest was acidified (5 μl of neat formic acid) and peptides were recovered by centrifugation to the lower chamber. Acidified peptides were treated with ethyl acetate as previously described (53). Peptides were desalted by solid-phase extraction on a Beckman BioMek NxP robot with C4 and porous graphite carbon Nutips (Glygen) (54). Peptides that eluted with acetonitrile (60% in 1% formic acid) were combined, dried in a vacuum centrifuge, dissolved in acetonitrile/formic acid (1%/0.1%, 25 μl), and pipetted into autosampler vials (SUN-SRi) for LC-MS analysis.
LC-MS
Peptide mixtures from solid-phase extraction were separated using a nano-LC (2D Plus pump, Nanoflex module and AS1 auto-sampler) (Eksigent) in a dual column configuration. The nano-LC was coupled to a TripleTOF 5600+ mass spectrometer (AB SCIEX) with a Digital Picoview source and a silica PicoTip emitter (10 μm) (New Objectives). cHiPLC columns (ChromXP C18 (200 μm × 15 cm; particle size 3 μm, 120 Å) were equilibrated in acetonitrile (2%) containing 0.1% formic acid. Solvent A is water containing 0.1% formic acid. Organic gradients were produced by increasing the percentage of solvent B (0.1% formic acid in acetonitrile) using the gradient program (minutes:%B) 0:2, 5:0, 720:30, 760:80, 765:80, 770:2, and 790:2. Samples (10 μl) were loaded at a flow rate of 800 nl/min over 25 minutes. Data were acquired using an ion spray voltage of approximately 3 kV, curtain gas of 10 psi, nebulizer gas of 14 psi, and an interface heater temperature of 175°C. The mass spectrometer was operated with a resolution (full width at half maximum [FWHM]) of greater than 25,000FWHM and 15,000FWHM for TOFMS and MS2 scans, respectively. For data-dependent acquisition, survey scans were acquired in 250 ms from which 100 product ion scans were selected for MS2 acquisition with a dwell time of 100 ms. The Q1 was set to unit, which corresponds to 0.7 Th. MS2 spectra were recorded with a dwell time of 100 ms. Four time bins were summed for each scan at a pulser frequency value of 15.4 kHz through monitoring of the 40 GHz multichannel TDC detector with 4-anode/channel detection. A rolling collision energy was applied to all precursor ions for collision-induced dissociation using the equation CE = (slope × [m/z]) + intercept, where the slope for all charges above 2+ is 0.0625 and the intercept is –3,–5, and –6 for 2+, 3+, and 4+ respectively.
Protein identification
MS data were processed utilizing the AB SCIEX MS Data Converter v 1.3 (AB SCIEX) by converting the raw data files (*.wiff) to the Mascot generic format (*.mgf). Processed files were used for protein database searches using Mascot (Matrix Science; version 2.5.1). An Mtb database (Tuberculist_20140910, 4031 entries) was used to search the LC-MS data. The parent ion tolerance and MS2 fragment tolerance were set to 10 ppm and 0.05 Da, respectively. Carbamidomethyl of cysteine was specified as a fixed modification and oxidation of methionine was set as a variable modification. Protein identifications were performed using Scaffold, version 4.4.8 (Proteome Software Inc.) implementing the Protein and Peptide Prophet algorithms (55, 56). Peptide identifications were accepted with greater than 90.0% probability. Protein identifications were accepted if they could be established at greater than 95.0% probability and contained at least 2 peptides with unique sequences. Protein probabilities were assigned using the Protein Prophet algorithm.
Lung cell preparation and flow cytometry
Lung cell suspensions were prepared as previously described (51). Antibodies used for analyses were CD11b (clone M1/70, TonBo Biosciences), CD44 (clone IM7, eBioscience), and IFN-γ (clone XMG1.2, TonBo Biosciences). The following antibodies were from BD Biosciences: CD11c (clone HL3), Ly6G (clone 1A8), Ly6C (clone AL-21), CD3 (clone 500A2), CD4 (clone RM4-5), CD8 (clone 53-6.7), and IL-17A (clone TC11-18H10). Cells were acquired with the Becton Dickinson (BD) Fortessa flow cytometer using FACSDiva software, or the BD FACSJazz flow cytometer with FACSDiva software. Cells were gated based on forward-by-side scatter characteristics and the frequency of specific cell types was calculated using FlowJo (Tree Star Inc.).
In vitro cell culture
BMDMs, BMDCs, and MDSCs were generated from FeJ bone marrow as previously described (24, 51). Supernatants were collected 24 hours after infection for protein or metabolic analyses. M-MDSCs (CD11b+Ly6GloLy6Chi) were sorted using a BD FACSJazz flow cytometer and FACSDiva software. Where specified, cells were treated with the TLR2-blocking, purified anti–mouse CD282 (30 μg/ml, clone 6C2, Affymetrix eBioscience), or rIL-17 (100 ng/ml, R&D Systems) for 30 minutes or 24 hours prior to Mtb infection, respectively. BMDCs and total lung cells were treated with HN878 or H37Rv components, such as cell wall or lipid preparations (10 μg/ml each, or 20 μg/ml BEI Resources, obtained under NIH contract AI-75320, or prepared in house as described before).
In vitro Mtb infection
BMDMs, MDSCs, or lung cell preparations were infected with the specified MOI of HN878, HN878::Δpks1-15, or H37Rv, in antibiotic-free cDMEM. BMDMs and MDSCs were cultured for 24 hours, and total lung cells for 6 days. For Western blot analyses, BMDMs were infected for 48 hours. Supernatants were collected and cells were either stained for analysis by flow cytometry, or lysed with 0.05% SDS and plated for assessment of infection.
RNA isolation and quantitative real-time PCR
Total RNA was extracted as specified by the manufacturer from lung tissue or cultured cells using the Qiagen RNeasy Mini kit (Qiagen). The primer and probe sequences targeting genes were purchased commercially (ABI Biosystems). Gene expression was calculated relative to GAPDH expression and log 10-fold induction over the control group was assessed using the ΔΔCT calculation.
Immunohistochemistry
Lung lobes were instilled with 10% neutral buffered formalin, embedded in paraffin, and processed as previously described (27). Endogenous biotin was neutralized by adding avidin, followed by incubation with biotin (both Sigma-Aldrich). Sections were probed with anti-mouse F4/80 (clone Cl:A3-1, AbD Serotec) or mouse anti-human CD68 (clone PG-M1, Thermo Fisher Scientific) to detect mouse or human macrophages. Rat specific antibodies against Gr-1 (Ly6G/Ly6C, clone RB6-8C5, BD-Pharmingen) were used to detect neutrophils and monocytes inside pulmonary granulomas. Rabbit antibodies against HIF1α (IHC-00460, Bethyl Laboratories Inc.) and FITC-conjugated mouse anti-PIMO antibodies (MAb 4.3.11.3, Hypoxyprobe-1 kit, Hypoxyprobe, Inc.) were used to define the hypoxic environments in TB granulomas. Goat antibodies specific for NOS2 (clone M-19, Santa Cruz Biotechnology) and rabbit anti-Mtb (MBS534825, MyBiosource.com) were used to visualize activated macrophages and mycobacteria in infected lungs. Rabbit anti–LDH-A (GTX101416, Gene Tex) and rabbit anti–V-ATPase (GTX119155, Gene Tex) were used to detect metabolic shifts toward glycolysis. Primary antibodies were detected with Alexa Fluor 568 donkey anti–goat IgG (A11057, Thermo Fisher Scientific), Alexa Fluor 488 donkey anti–rat IgG (A21208, Thermo Fisher Scientific), Alexa Fluor 488 rabbit anti–fluorescein/Oregon green (A11090, Thermo Fisher Scientific), FITC-conjugated donkey anti-mouse (715-095-150, Jackson Immunoresearch Laboratories), FITC-conjugated donkey anti-rabbit (711-096-152, Jackson Immunoresearch Laboratories), Cy3-conjugated donkey anti-rabbit (711-166-152, Jackson Immunoresearch Laboratories) and biotinylated donkey anti-rat (712-066-150, Jackson Immunoresearch Laboratories). Biotinylated antibodies were revealed with Cy5-Streptavidin (19-4317-82, eBiosciences) and Alexa Fluor 555-Streptavidin (S32355, Thermo Fisher Scientific). To prevent bleaching and counterstain nuclei, slides were mounted with Prolong Gold antifade with DAPI (P36931, Thermo Fisher Scientific). Representative pictures were taken with a Zeiss Axioplan microscope and recorded with a Hamamatsu camera. To measure fibrosis, paraffin-embedded lung slides were deparaffinized with xylene and processed as previously described with Masson’s trichrome staining (27).
PIMO
PIMO (Hypoxyprobe-1 kit) was dissolved in sterile PBS and 100 mg/kg was i.p. injected 2 hours prior to harvest. PIMO was detected on paraffin-embedded lungs.
Mycobacteria growth indicator tubes
Sterile water (4 ml) was added to an 8-ml sterile polystyrene tube with cap containing 8 to 10 glass beads. With a sterile loop, HN878 colonies (<14 days old) were removed and suspended in Middlebrook 7H9 broth. Tubes were vortexed (2–3 minutes) and the suspension held at room temperature for 20 minutes without perturbation. Supernatant was transferred to a new 8-ml sterile polystyrene tube with cap and left for another 15 minutes. Suspension was adjusted to a 0.5 McFarland standard using a spectrophotometer. Following a 1:5 dilution in sterile water, a 0.5-ml suspension was used to prepare growth control dilutions and to inoculate Mycobacteria growth indicator tubes containing acriflavine (dissolved in sterile endotoxin-free water at 1 mg/ml). Acriflavine concentrations tested were 0, 0.25, 0.75, 1.5, 3, and 25 mg/ml. Experiments were performed in triplicate except for the negative control (no drug) and positive control (100 mg/ml, toxic level), which were both done in 6 replicates. Tubes were placed in a BACTEC automated instrument to monitor for growth.
Lactate detection
Lactate was measured in the supernatants of variously treated cells using the Lactate Assay Kit (Sigma-Aldrich) according to the manufacturer’s protocol. Lactate was detected by absorbance at 570 nm (A570).
In situ hybridization
Paraffin-embedded tissue sections were deparaffinized and washed in ethanol. Stringent in situ hybridization (50°C with 0.1 M DTT in the hybridization mix) was performed with 35S-labeled riboprobes as previously described (57). Images were visualized using an Olympus BX41 microscope and captured using a SPOT RT3 digital camera (SPOT Imaging Solutions).
Protein determination by ELISA and multiplex assay
Protein levels for cytokines and chemokines in culture supernatants or lung homogenates were measured using a mouse Luminex assay (Millipore) or ELISA kits (R&D Systems, MBL International Corporation).
Protein determination by Western blot
Mouse lung homogenates were lysed in RIPA Lysis Buffer (Santa Cruz Biotechnology) and heat denatured in reducing sample buffer (Thermo Fisher Scientific). Proteins were separated in 4% to 20% polyacrylamide gradient gels (Bio-Rad) and transferred onto PVDF membranes. Nonspecific binding was blocked with 5% milk in TBST (Tris-buffered saline, 0.1% Tween 20), and membranes were probed with primary antibodies against HIF1α (1:500; NB100-449, Novus), and GAPDH (1:1,000; sc-25778, Santa Cruz Biotechnology) followed by incubation with HRP-conjugated anti–rabbit IgG secondary antibody (1:10,000; sc-2030, Santa Cruz Biotechnology) and developed by Clarity Western ECL Substrate (Bio-Rad).
Real-time extracellular flux assay
Cells (1 × 105 per sample) were stimulated with HN878 cell wall fraction (NR14830, BEI Resources) at 20 μg/ml for 24 hours. Real-time ECAR was measured using the XF-96 Extracellular Flux Analyzer (Seahorse Bioscience) (58). Three consecutive measurements were obtained under basal conditions.
Statistics
For murine studies, differences between the means of 2 groups were analyzed using the 2-tailed Student’s t test in Prism 5 (GraphPad). Differences between the means of 3 or more groups were analyzed using 1-way ANOVA with Tukey’s post-test in GraphPad Prism 5. A P value less than 0.05 was considered significant. In human studies, differences between baseline and Mtb-specific induction of IL-17 by PBMCs of the rs2275913 SNP, as well as association between the rs2275913 SNP and drug-resistant Mtb were compared by RM-ANOVA. Pearson correlation coefficient analysis was used to determine lung damage score association with the rs2275913 SNP.
Study approval — mouse and human study subjects/samples
All mice were used in accordance with the NIH guidelines for housing and care of laboratory animals and in accordance with Washington University in St. Louis Institutional Animal Care and Use Committee guidelines (approved under Protocol 20160129). All efforts were made to minimize suffering and pain as described in this approved protocol. TB patients and contacts were recruited from the collaborative programs of TB detection between the Tuberculosis Outpatient Clinic of the INER in Mexico City and the Oaxaca State Health Authorities. The collection of samples was performed after patients’ approval and consent letter signature obtained. The study was approved by the Institutional Ethics Review Board of the INER.
Author contributions
RDG, KLG, RG, SD, SAK, and JZ conceived the experiments. RDG, KLG, SD, M. Ahmed, RG, SG, TAR, MMT, MASL, ACL, LJA, MLGH, GRM, MAM, RES, MB, TS, JA, WH, JRM, JBT, JMBL, and JZ carried out experiments. RDG, KLG, RG, TAR, JA, JBT, WH, JRM, M.N. Artyomov, NAH, JLW, and SAK conducted analyses. TAR, GK, JKK, RT, and JZ provided resources and expertise. RDG, KLG, and SAK wrote the paper. SAK and JRM provided funding. SAK provided supervision, and overall project administration.
Supplementary Material
Acknowledgments
Version 1. 10/05/2017
Electronic publication
Footnotes
This work was supported by Washington University in St. Louis, NIH grant HL105427 to SAK, American Lung Association Senior Research Training Fellowship RT-30592 to KLG, and NIH/NHLBI T32 HL007317-37 to RDG. JRM was supported by funds of the Department of Medicine, University of Rochester, and NIH grant U19 AI91036. This work was also supported by the NIH through grants UL1 RR024153 (University of Pittsburgh) and UL1 TR000005, and through the ICTS UL TR000448 to Washington University in St. Louis. This project was funded, in part, by The Foundation for Barnes-Jewish Hospital and their generous donors, and the Competitive Medical Research Fund of the University of Pittsburgh Medical Center. The protein identifications and LC-MS analyses were generated at the Washington University Proteomics Shared Resource (WU-PSR). The WU-PSR is supported by the WU Institute of Clinical and Translational Sciences (NCATS UL1 TR000448), the WU Mass Spectrometry Research Resource (NIGMS P41 GM103422), and the Siteman Comprehensive Cancer Center (NCI P30 CA091842). The authors thank Sarah Squires for animal breeding, and Deborah Hollingshead (GPCL, University of Pittsburgh) and Yun Ju Sung (Division of Biostatistics at Washington University in St. Louis) for assistance with genomic analyses. We thank Kimberly Thomas for critical reading of the manuscript.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2017;2(19):e92973. https://doi.org/10.1172/jci.insight.92973.
This work was supported by Washington University in St. Louis, NIH grant HL105427 to SAK, American Lung Association Senior Research Training Fellowship RT-30592 to KLG, and NIH/NHLBI T32 HL007317-37 to RDG. JRM was supported by funds of the Department of Medicine, University of Rochester, and NIH grant U19 AI91036. This work was also supported by the NIH through grants UL1 RR024153 (University of Pittsburgh) and UL1 TR000005, and through the ICTS UL TR000448 to Washington University in St. Louis. This project was funded, in part, by The Foundation for Barnes-Jewish Hospital and their generous donors, and the Competitive Medical Research Fund of the University of Pittsburgh Medical Center. The protein identifications and LC-MS analyses were generated at the Washington University Proteomics Shared Resource (WU-PSR). The WU-PSR is supported by the WU Institute of Clinical and Translational Sciences (NCATS UL1 TR000448), the WU Mass Spectrometry Research Resource (NIGMS P41 GM103422), and the Siteman Comprehensive Cancer Center (NCI P30 CA091842). The authors thank Sarah Squires for animal breeding, and Deborah Hollingshead (GPCL, University of Pittsburgh) and Yun Ju Sung (Division of Biostatistics at Washington University in St. Louis) for assistance with genomic analyses. We thank Kimberly Thomas for critical reading of the manuscript.
Contributor Information
Racquel Domingo-Gonzalez, Email: rdomingo@wustl.edu.
Shibali Das, Email: shibalidas@wustl.edu.
Mushtaq Ahmed, Email: mahmed@wusm.wustl.edu.
Monika Bambouskova, Email: monika.bambouskova@wustl.edu.
Radha Gopal, Email: Radha.Gopal4@chp.edu.
Suhas Gondi, Email: shsg48@gmail.com.
Alfredo Cruz-Lagunas, Email: alfredocl@gmail.com.
Gustavo Ramirez-Martinez, Email: grmunam@yahoo.com.mx.
Ramón Espinosa-Soto, Email: skilyine@hotmail.com.
Tamanna Sultana, Email: tamanna.sultana@thermofisher.com.
James Lyons-Weiler, Email: jim@ipaknowledge.org.
Jesus Arcos, Email: jesusarcos@gmail.com.
Maria de la Luz Garcia-Hernandez, Email: Luz_Garcia-hernandez@URMC.Rochester.edu.
Noor Al-Hammadi, Email: noor@wubios.wustl.edu.
Reid Townsend, Email: rtownsend@wustl.edu.
Joan-Miquel Balada-Llasat, Email: joan.balada@osumc.edu.
Gilla Kaplan, Email: Gilla.Kaplan@gatesfoundation.org.
William Horne, Email: william.horne@chp.edu.
Jay K. Kolls, Email: jay.kolls@chp.edu.
Javier Rangel-Moreno, Email: Javier_Rangel-Moreno@URMC.Rochester.edu.
References
- 1.Dye C, Glaziou P, Floyd K, Raviglione M. Prospects for tuberculosis elimination. Annu Rev Public Health. 2013;34:271–286. doi: 10.1146/annurev-publhealth-031912-114431. [DOI] [PubMed] [Google Scholar]
- 2.Cantini F, et al. Guidance for the management of patients with latent tuberculosis infection requiring biologic therapy in rheumatology and dermatology clinical practice. Autoimmun Rev. 2015;14(6):503–509. doi: 10.1016/j.autrev.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 3.Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64(6):2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guirado E, Schlesinger LS. Modeling the Mycobacterium tuberculosis granuloma — the critical battlefield in host immunity and disease. Front Immunol. 2013;4:98. doi: 10.3389/fimmu.2013.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belton M, et al. Hypoxia and tissue destruction in pulmonary TB. Thorax. 2016;71(12):1145–1153. doi: 10.1136/thoraxjnl-2015-207402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Via LE, et al. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect Immun. 2008;76(6):2333–2340. doi: 10.1128/IAI.01515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassavaugh J, Lounsbury KM. Hypoxia-mediated biological control. J Cell Biochem. 2011;112(3):735–744. doi: 10.1002/jcb.22956. [DOI] [PubMed] [Google Scholar]
- 8.Carmeliet P, et al. Role of HIF-1α in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394(6692):485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 9.Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci U S A. 1998;95(14):7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 11.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krawczyk CM, et al. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood. 2010;115(23):4742–4749. doi: 10.1182/blood-2009-10-249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jurado JO, et al. IL-17 and IFN-γ expression in lymphocytes from patients with active tuberculosis correlates with the severity of the disease. J Leukoc Biol. 2012;91(6):991–1002. doi: 10.1189/jlb.1211619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng MY, et al. Interleukin 17-producing γδ T cells increased in patients with active pulmonary tuberculosis. Cell Mol Immunol. 2008;5(3):203–208. doi: 10.1038/cmi.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hot A, Zrioual S, Lenief V, Miossec P. IL-17 and tumour necrosis factor α combination induces a HIF-1α-dependent invasive phenotype in synoviocytes. Ann Rheum Dis. 2012;71(8):1393–1401. doi: 10.1136/annrheumdis-2011-200867. [DOI] [PubMed] [Google Scholar]
- 16.Straus DS. TNFα and IL-17 cooperatively stimulate glucose metabolism and growth factor production in human colorectal cancer cells. Mol Cancer. 2013;12:78. doi: 10.1186/1476-4598-12-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, Flach H, Onizawa M, Wei L, McManus MT, Weiss A. Negative regulation of Hif1a expression and TH17 differentiation by the hypoxia-regulated microRNA miR-210. Nat Immunol. 2014;15(4):393–401. doi: 10.1038/ni.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang SH, et al. T helper 17 cells play a critical pathogenic role in lung cancer. Proc Natl Acad Sci U S A. 2014;111(15):5664–5669. doi: 10.1073/pnas.1319051111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Neill LA, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol. 2016;16(9):553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gleeson LE, et al. Cutting edge: Mycobacterium tuberculosis induces aerobic glycolysis in human alveolar macrophages that is required for control of intracellular bacillary replication. J Immunol. 2016;196(6):2444–2449. doi: 10.4049/jimmunol.1501612. [DOI] [PubMed] [Google Scholar]
- 21.Cooper AM. Mouse model of tuberculosis. Cold Spring Harb Perspect Med. 2015;5(2):a018556. doi: 10.1101/cshperspect.a018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das S, Khader S. Yin and yang of interleukin-17 in host immunity to infection. F1000Res. 2017;6:741. doi: 10.12688/f1000research.10862.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parwati I, van Crevel R, van Soolingen D. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect Dis. 2010;10(2):103–111. doi: 10.1016/S1473-3099(09)70330-5. [DOI] [PubMed] [Google Scholar]
- 24.Knaul JK, et al. Lung-residing myeloid-derived suppressors display dual functionality in murine pulmonary tuberculosis. Am J Respir Crit Care Med. 2014;190(9):1053–1066. doi: 10.1164/rccm.201405-0828OC. [DOI] [PubMed] [Google Scholar]
- 25.Corzo CA, et al. HIF-1α regulates function and differentiation of myeloid-derived suppressor cells in the tumor microenvironment. J Exp Med. 2010;207(11):2439–2453. doi: 10.1084/jem.20100587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi L, Eugenin EA, Subbian S. Immunometabolism in tuberculosis. Front Immunol. 2016;7:150. doi: 10.3389/fimmu.2016.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monin L, et al. Helminth-induced arginase-1 exacerbates lung inflammation and disease severity in tuberculosis. J Clin Invest. 2015;1(12):4699–4713. doi: 10.1172/JCI77378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gopal R, et al. Unexpected role for IL-17 in protective immunity against hypervirulent Mycobacterium tuberculosis HN878 infection. PLoS Pathog. 2014;10(5):e1004099. doi: 10.1371/journal.ppat.1004099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reed MB, et al. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature. 2004;431(7004):84–87. doi: 10.1038/nature02837. [DOI] [PubMed] [Google Scholar]
- 30.Daffé M, Lacave C, Lanéelle MA, Lanéelle G. Structure of the major triglycosyl phenol-phthiocerol of Mycobacterium tuberculosis (strain Canetti) Eur J Biochem. 1987;167(1):155–160. doi: 10.1111/j.1432-1033.1987.tb13317.x. [DOI] [PubMed] [Google Scholar]
- 31.Lee K, Zhang H, Qian DZ, Rey S, Liu JO, Semenza GL. Acriflavine inhibits HIF-1 dimerization, tumor growth, and vascularization. Proc Natl Acad Sci U S A. 2009;106(42):17910–17915. doi: 10.1073/pnas.0909353106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Rodrigues L, Villellas C, Bailo R, Viveiros M, Aínsa JA. Role of the Mmr efflux pump in drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2013;57(2):751–757. doi: 10.1128/AAC.01482-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3(10):721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 34.Welte T, Zhang XH. Interleukin-17 could promote breast cancer progression at several stages of the disease. Mediators Inflamm. 2015;2015:804347. doi: 10.1155/2015/804347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bárdos JI, Ashcroft M. Negative and positive regulation of HIF-1: a complex network. Biochim Biophys Acta. 2005;1755(2):107–120. doi: 10.1016/j.bbcan.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 36.Chen X, et al. IL-17 producing mast cells promote the expansion of myeloid-derived suppressor cells in a mouse allergy model of colorectal cancer. Oncotarget. 2015;6(32):32966–32979. doi: 10.18632/oncotarget.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Obregón-Henao A, Henao-Tamayo M, Orme IM, Ordway DJ. Gr1(int)CD11b+ myeloid-derived suppressor cells in Mycobacterium tuberculosis infection. PLoS One. 2013;8(11):e80669. doi: 10.1371/journal.pone.0080669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.du Plessis N, et al. Increased frequency of myeloid-derived suppressor cells during active tuberculosis and after recent Mycobacterium tuberculosis infection suppresses T-cell function. Am J Respir Crit Care Med. 2013;188(6):724–732. doi: 10.1164/rccm.201302-0249OC. [DOI] [PubMed] [Google Scholar]
- 39.Cardoso MS, Silva TM, Resende M, Appelberg R, Borges M. Lack of the transcription factor hypoxia-inducible factor 1α (HIF-1α) in macrophages accelerates the necrosis of Mycobacterium avium-induced granulomas. Infect Immun. 2015;83(9):3534–3544. doi: 10.1128/IAI.00144-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braverman J, Sogi KM, Benjamin D, Nomura DK, Stanley SA. HIF-1α is an essential mediator of IFN-γ-dependent immunity to Mycobacterium tuberculosis. J Immunol. 2016;197(4):1287–1297. doi: 10.4049/jimmunol.1600266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cambier CJ, et al. Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature. 2014;505(7482):218–222. doi: 10.1038/nature12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Onishi RM, Gaffen SL. Interleukin-17 and its target genes: mechanisms of interleukin-17 function in disease. Immunology. 2010;129(3):311–321. doi: 10.1111/j.1365-2567.2009.03240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khader SA, et al. In a murine tuberculosis model, the absence of homeostatic chemokines delays granuloma formation and protective immunity. J Immunol. 2009;183(12):8004–8014. doi: 10.4049/jimmunol.0901937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartlett HS, Million RP. Targeting the IL-17-T(H)17 pathway. Nat Rev Drug Discov. 2015;14(1):11–12. doi: 10.1038/nrd4518. [DOI] [PubMed] [Google Scholar]
- 45.Merker M, et al. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat Genet. 2015;47(3):242–249. doi: 10.1038/ng.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bocanegra-García V, et al. Molecular assessment, drug-resistant profile, and spacer oligonucleotide typing (spoligotyping) of Mycobacterium tuberculosis strains from Tamaulipas, México. J Clin Lab Anal. 2014;28(2):97–103. doi: 10.1002/jcla.21650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ocejo-Vinyals JG, et al. The IL-17 G-152A single nucleotide polymorphism is associated with pulmonary tuberculosis in northern Spain. Cytokine. 2013;64(1):58–61. doi: 10.1016/j.cyto.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 48.Shi GC, Zhang LG. Influence of interleukin-17 gene polymorphisms on the development of pulmonary tuberculosis. Genet Mol Res. 2015;14(3):8526–8531. doi: 10.4238/2015.July.28.22. [DOI] [PubMed] [Google Scholar]
- 49.Milano M, et al. Single Nucleotide polymorphisms in IL17A and IL6 are associated with decreased risk for pulmonary tuberculosis in Southern Brazilian population. PLoS One. 2016;11(2):e0147814. doi: 10.1371/journal.pone.0147814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Espinoza JL, et al. A genetic variant in the IL-17 promoter is functionally associated with acute graft-versus-host disease after unrelated bone marrow transplantation. PLoS One. 2011;6(10):e26229. doi: 10.1371/journal.pone.0026229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khader SA, et al. IL-23 and IL-17 in the establishment of protective pulmonary CD4+ T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat Immunol. 2007;8(4):369–377. doi: 10.1038/ni1449. [DOI] [PubMed] [Google Scholar]
- 52.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 53.Erde J, Loo RR, Loo JA. Enhanced FASP (eFASP) to increase proteome coverage and sample recovery for quantitative proteomic experiments. J Proteome Res. 2014;13(4):1885–1895. doi: 10.1021/pr4010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen ZW, Fuchs K, Sieghart W, Townsend RR, Evers AS. Deep amino acid sequencing of native brain GABAA receptors using high-resolution mass spectrometry. Mol Cell Proteomics. 2012;11(1):M111.011445. doi: 10.1074/mcp.M111.011445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 56.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 57.Slight SR, et al. CXCR5+ T helper cells mediate protective immunity against tuberculosis. J Clin Invest. 2013;123(2):712–726. doi: 10.1172/JCI65728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang SC, et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat Immunol. 2014;15(9):846–855. doi: 10.1038/ni.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.