Abstract
Most vaccine adjuvants directly stimulate and activate antigen presenting cells but do not sustain immunostimulation of these cells. A high throughput screening (HTS) strategy was designed to identify compounds that would sustain NF-κB activation by a stimulus from the Toll-like receptor (TLR)4 ligand, lipopolysaccharide (LPS). Several pilot studies optimized the parameters and conditions for a cell based NF-κB reporter assay in human monocytic THP-1 cells. The final assay evaluated prolongation of LPS induced NF-κB activation at 12 h. The dynamic range of the assay was confirmed in a pilot screen of 14 631 compounds and subsequently in a main extensive screen with 166 304 compounds. Hit compounds were identified using an enrichment strategy based on unsupervised chemoinformatic clustering, and also by a naïve “Top X” approach. A total of 2011 compounds were then rescreened for levels of coactivation with LPS at 5 h and 12 h, which provided kinetic profiles. Of the 407 confirmed hits, compounds that showed correlation of the kinetic profiles with the structural similarities led to identification of four chemotypes: pyrimido[5,4-b]indoles, 4H-chromene-3-carbonitriles, benzo[d][1,3]dioxol-2-ylureas, and tetrahydrothieno[2,3-c]pyridines, which were segregated by 5 h and 12 h kinetic characteristics. Unlike the TLR4 agonistic pyrimidoindoles identified in previous studies, the revealed pyrimidoindoles in the present work did not intrinsically stimulate TLR4 nor induce NF-κB but rather prolonged NF-κB signaling induced by LPS. A 42-member combinatorial library was synthesized which led to identification of potent N3-alkyl substituted pyrimidoindoles that were not only active in vitro but also enhanced antibody responses in vivo when used as a coadjuvant. The novel HTS strategy led to identification of compounds that are intrinsically quiescent but functionally prolong stimulation by a TLR4 ligand and thereby potentiate vaccine efficacy.
Keywords: adjuvant, pyrimidoindole, NF-κB, LPS, TLR4
Graphical abstract
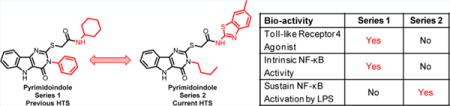
INTRODUCTION
The dearth of adjuvants in human vaccines has motivated the search for effective nontoxic immunomodulators that provide sustained immunoprotection particularly to high-risk populations such as the elderly.1–5 The use of Toll-like receptor (TLR) ligands has gained interest as sensors of innate immunity and stimulators of antigen-presenting cells (APCs) that prime protective immune responses.6 The use of TLR ligands as adjuvants can provide a potent activation stimulus to resident APCs and promote further APC recruitment to the site of antigen injection and subsequent delivery to the draining lymph nodes.2,3 In a prior high throughput screen (HTS) using the Förster resonance energy transfer (FRET) based CellSensor NF-κB-bla reporter in the human monocytic leukemic cell line, THP-1, we identified pyrimido[5,4-b]indole and 2-aminoquinazoline chemotypes that were characterized as TLR4/MD-2 ligands.7,8 These compounds were directly immunostimulatory and provided the basis for developing a lead compound shown to be an effective adjuvant in a murine model of influenza.9
Other Toll-like receptor ligands have also been developed as vaccine adjuvants as they are innate immune stimulators and can directly activate APCs.2,10–17 To initiate immune responses from vaccines, antigen-loaded activated APCs travel from the site of injection to regional lymph nodes and prime naïve T cells. During the time period that the APC travels from the vaccine injection site to a regional lymph node, the activation status is down-regulated (Figure 1A, red dashed line). Activation of innate immune responses is regulated through multiple negative feedback loops.18–20 Negative regulators of TLR signaling that have been reported to date include processing pathways (ubiquitination,21–23 degradation,24 stabilization25), molecular competition,20 promoter regulation,23 phosphatases,26,27 and epigenetic regulators.28–32 For example, interleukin-1 receptor-associated kinase (IRAK)-M,20 phosphatidylinositide (PI)3 kinase inhibitors,33 and anti-inflammatory cytokines (IL-1030 and transforming growth factor β34) have evolved to comodulate innate immune responses and inflammation.35 These negative feedback systems may adversely impact the ability of adjuvants to sustain APC activation and to induce optimal T lymphocyte memory responses.
Figure 1.
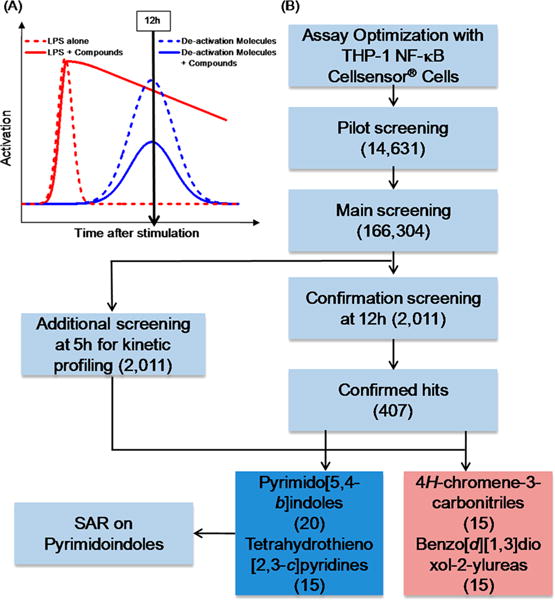
Theoretical kinetics of LPS activation and the HTS workflow strategy. (A) Theoretical kinetic profiles showing activation by LPS (dashed red line) and subsequent deactivation at 12 h due to an increase in deactivation molecules (dashed blue line). The addition of compounds that prolong the initial activation (solid red line) and also suppress the formation of deactivation molecules (solid blue line) were identified through HTS. (B) The cell based assay using THP-1 CellSensor NF-κB reporter cells was optimized for the identification of compounds that prolonged NF-κB activation when stimulated with LPS for 12 h. The assay was tested in a pilot screen of 14 631 compounds to validate the approach. In the main library screen, 166 304 compounds were screened in the presence of LPS for NF-κB activation at 12 h. Cluster based statistical analysis yielded 2011 compounds, which were retested in a confirmation screen at 12 h with LPS in duplicate. An additional screen was performed on these 2011 compounds for measuring NF-κB activity at 5 h to observe the activity kinetics. A total of 407 compounds that had confirmed activity at 12 h were structurally clustered using a Tanimoto index at 0.5. Four large families of chemotypes were identified as hits based on their kinetic profile of 5 h and 12 h that prolonged NF-κB activation with LPS. The pyrimidoindole class of compounds was selected for further SAR studies. The number in parentheses corresponds to the number of compounds.
Most vaccine development approaches have been to directly activate and stimulate APCs. However, prolonging the activation status of a cell by intercepting a negative regulatory pathway would be as valuable an approach to allow the impetus of the primary stimulus to be sustained during the transit time to the lymph node and to increase the number of T and B cell interactions the activated cell may have in the lymphoid environment. A compound disrupting one or more of the regulatory systems could sustain the activation status of a TLR stimulated APC. To date, HTS campaigns related to NF-κB have focused on identifying pathway inhibitors36–41 or intrinsic activators.7,8 However, no HTS campaign has focused on identifying compounds that sustain initial NF-κB stimulation by a TLR agonist. We hypothesized that identification of such small molecules could complement current TLR agonists as adjuvants and improve the efficacy of vaccines that are locally administered.
RESULTS AND DISCUSSION
Overall HTS Strategy and Design
Here, we aimed to identify compounds that manipulate negative feedback systems to sustain innate immune activation through a classic TLR4 ligand, lipopolysaccharide (LPS). We again used the CellSensor NF-κB-bla reporter in a human monocytic cell line, THP-1, as a model of functional monocyte-derived APCs for HTS. This cell-based assay was chosen as many of the cellular functions of APCs are retained and the dual fluorescence normalizes relative cell number and viability to NF-κB activity in each well. Typically, LPS stimulates NF-κB activation within the APC and then an increase in regulatory “deactivation” molecules (Figure 1A, blue dashed line) results in a decline in activation status. We sought to find small molecules that would prolong the LPS induced activation status by reducing or inhibiting these negative molecular regulators (Figure 1A, red and blue lines, respectively).
To identify compounds that prolonged the activation of APCs by LPS, we developed a tiered HTS strategy (Figure 1B). First, the conditions for the THP-1 CellSensor NF-κB-bla assay were optimized for a 12 h stimulation assay with LPS as described below. An initial pilot screen was performed with 14 631 representative compounds to evaluate the performance of the assay and quality control. The main HTS was then performed with a collection of 166 304 compounds that included the compounds used in the pilot screen to verify the reproducibility of the assay. We utilized our previously reported structure-based cluster analysis to select 2011 compounds for confirmation.42 These compounds were tested in duplicate in a confirmation screen for prolonged activation at 12 h, but we also included an additional 5 h screen to evaluate the kinetics of the stimulatory response. Approximately 20% (407 compounds) were confirmed as hits, and substructure-based clustering broadly classified some of these compounds into four chemotypes: pyrimido[5,4-b]indoles, 4H-chromene-3-carbonitriles, benzo[d][1,3]dioxol-2-ylureas, and tetrahydrothieno[2,3-c]pyridines. As we had prior success with developing a pyrimidoindole as a lead compound, this class was selected for further structure–activity relationship (SAR) studies.
Assay Optimization
For our LPS costimulation strategy, we optimized the THP-1 CellSensor NF-κB-bla assay for variables including (1) cell number, (2) preincubation time of thawed cells, (3) concentration of FBS, (4) incubation time, and (5) the dose and timing of a positive control compound, wortmannin, a PI3 kinase inhibitor that has been reported to augment initial NF-κB activity.43–45 In the first set of experiments, the tissue culture fetal bovine serum (FBS) concentration was kept constant at 10% while cell numbers were varied between 10 000 cells/well and 20 000 cells/well, and the preincubation time of thawed cells was either −16 h or 0 h relative to the test compound and LPS addition (Figure 2A). In addition, FRET response ratios were measured after 24 h for 100 ng/mL LPS coincubated with graded concentrations of wortmannin in the above-mentioned four different cell plating conditions. Higher FRET ratios were reliably demonstrated with 20 000 cells/well and a 16 h preincubation time of the thawed cells (Figure 2A). The potential for individual test compounds to bind to bovine albumin and other proteins in FBS could lead to a false negative output, so we probed the effect of lowering the FBS concentration (5%, 2%, or 1%) on prolonged LPS stimulation in the presence of graded concentrations of wortmannin (Figure 2B). Response ratios decreased with reduced concentrations of FBS and also at higher concentrations of wortmannin (Figure 2B). Thus, 5% FBS concentration was determined to be optimum for the assay.
Figure 2.
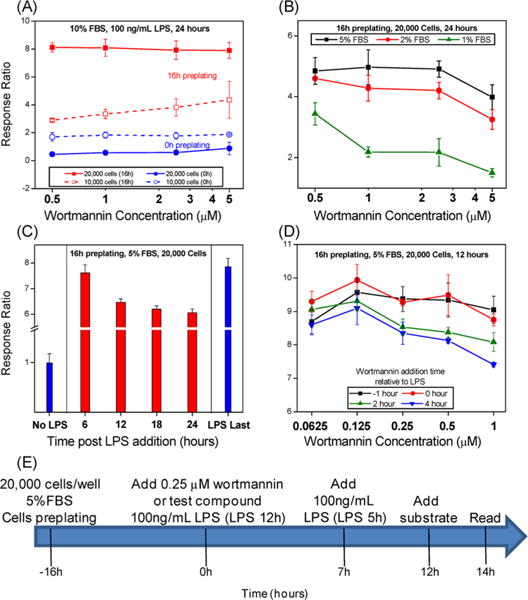
Optimization of assay conditions in THP-1 CellSensor assay. The assay conditions were optimized for the following parameters: (A) Cell numbers/well and cells preplating time, (B) FBS concentration, (C) incubation time, and (D) concentration of wortmannin and the time of its addition relative to LPS. (E) The schematic shows the optimum assay parameters and the assay protocol used for the HTS. Briefly, the cells were preplated at a density of 20 000 cells/well in 5% FBS 16 h prior to the addition of 100 ng/mL LPS alone or LPS with 0.25 μM wortmannin. An additional control (LPS 5 h) was added 5 h prior to the addition of FRET substrate at 12 h, and the plates were read at 14 h.
As the optimal incubation period of THP-1 Cellsensor NF-κB recommended by the manufacturer is 5 h to 6 h, a longer incubation time with THP-1 cells may lead to cellular stress responses as well as degradation of compounds due to metabolism. We were therefore interested to find the optimum incubation time to allow for natural decay of the LPS stimulation without losing the ability to distinguish an increase in the FRET signal due to aberrations in cellular metabolism or cell death. Using the previously optimized parameters (20 000 cells/well and 5% FBS), response ratios were measured at 6, 12, 18, and 24 h after stimulation with 100 ng/mL of LPS alone (Figure 2C). In addition, LPS was added after 18 h (LPS last) in untreated cells, and the response ratio was measured after 6 h to verify retention of the FRET activity in the cells after a long incubation time. LPS stimulation results in activation at 6 h; however, the response ratio shows a significant drop at 12 h which reached a plateau at later time points. The response ratio for LPS at 6 h and that for “LPS last” are almost identical, suggesting that THP-1 cells do not lose activity even after longer incubation time (Figure 2C). As there was no difference at 12 h and later time points, we chose 12 h for the HTS.
Next, we varied the dose and time of wortmannin (as a test compound) addition relative to LPS. Wortmannin at graded doses was added either 1 h before or at 0, 2, and 4 h after the addition of LPS. The response ratios were higher when wortmannin was added before or with LPS, but the signal decreased when wortmannin was added 2 h or 4 h after LPS (Figure 2D). Thus, the addition of 0.25 μM wortmannin concurrent with 100 ng/mL LPS was chosen as optimal for the assay. The final optimized conditions for the HTS to identify compounds that prolong initial activation of NF-κB by LPS are summarized in a schematic protocol (Figure 2E).
Pilot Screen and HTS
Using the optimized assay conditions, a pilot screen was first performed using 14 631 compounds selected to represent the diversity of the libraries in the chemical collection at University of California San Francisco (UCSF) Small Molecule Discovery Center (SMDC). The screen was performed using LPS alone (LPS 12 h) and LPS + 0.25 μM wortmannin as a positive control, 0.5% DMSO (no LPS) as a negative control, and an additional control included adding LPS at 5 h before adding substrate (LPS 5 h). The percent activation calculated using LPS 5 h as a reference is shown in Figure 3A. Statistical analysis showed good quality for the different controls, and we proceeded with the main screen, which included 166 304 test compounds on 527 plates. In addition, 27 plates containing 8627 compounds from the pilot screen were rescreened in triplicate within the main screen, in order to provide quality control information and to estimate confirmation rates. The percent activation plot for the main screen (Figure 3B) showed interplate variability for the LPS control (LPS 12 h). To mitigate for the variability, we calculated values based on the LPS control within each plate. Intraplate calculations for “distance to LPS” was calculated and used as the metric for compound selection. The main screen median Z′ was 0.74 (0.43–0.90). As in prior work, we used cluster enrichment methods including Murcko scaffold classes, Daylight functional clusters, and the Top X approach, to identify 2011 compounds for the confirmation and kinetic profiling screens (Supporting Information, Figure S1).42
Figure 3.
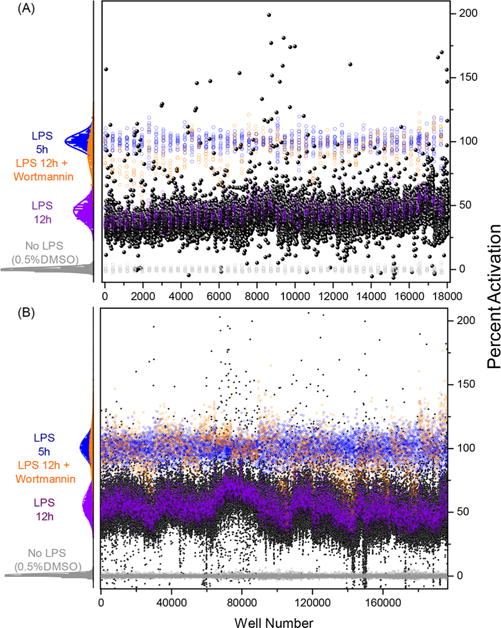
Pilot and HTS main screen. Percent activation values for the controls and test compounds from (A) the pilot screen and (B) the main screen. The histogram plot to the left for each screen shows the intra-assay statistics on the percent activation values for No LPS (0.5% DMSO, negative control), LPS 12 h (100 ng/mL LPS control added at time 0 h), LPS 12 h + wortmannin (positive control), and LPS 5 h (100 ng/mL LPS control added 5 h before the assay read out). The scatter plot to the right shows percent activation values for all the compounds and controls used in each screen. Each black circle represents an individual test compound, while controls are colored as in the histogram plot.
Confirmation Screen and Kinetic Profiling
The selected hits from the main screen were then evaluated in duplicate in a confirmation screen at 12 h and an additional screen at 5 h for kinetic profiling. Figure 4A shows a scatter plot of the data (distance to LPS) for these compounds from the 12 h confirmation screen on the ordinate axis and from 5 h kinetic profiling screen on the abscissa axis. A total of 407 compounds were identified as confirmed hits from the 12 h screen using a data-driven method that fits a mixture of two linear models utilizing both the primary and confirmation screen data for these compounds. All of the confirmed hits enhanced the NF-κB induction activity of LPS at 12 h and were broadly classified into four groups showing four general kinetic patterns (Figure 4B). On the basis of the 5 h screen, these compounds were either early inhibitors (5 h distance to LPS < 0) or early enhancers (5 h distance to LPS > 0). Within the early inhibitors, the compounds were grouped as either showing moderate increase or high increase in NF-κB induction compared to LPS at 12 h (green and orange symbols, respectively, Figure 4A), where the cutoff used was mean + SD of the LPS 5 h values from all the confirmation screen plates. Similarly, within the enhancer groups, compounds showed either moderate or high increases in NF-κB induction compared to LPS at 12 h (red and blue symbols, respectively, Figure 4A). Theoretical activation profiles for these groups of compounds are shown in Figure 4B with the same color patterns. The structures of these compounds were then subjected to substructure based clustering using the server based ChemMine tools (University of California, Irvine; http://chemmine.ucr.edu/tools/launch_job/Clustering/) and binning clustering application with a similarity cutoff of 0.5. The application allowed us to group these compounds into several chemotypes, and correlation of kinetic profiles with structural similarities led us to identify four major structural classes, namely, pyrimido[5,4-b]indoles, 4H-chromene-3-carbonitriles, benzo[d] [1, 3] dioxol-2-ylureas, and tetrahydrothieno[2,3-c]pyridines (Figure 4C). Most of the compounds in pyrimido[5,4-b]indole and tetrahydrothieno-[2,3-c]pyridine series were early enhancers, while the bulk of the compounds in the 4H-chromene-3-carbonitriles and benzo[d][1,3]dioxol-2-ylureas series were early inhibitors, as described earlier. Figure 4C shows the distribution of the compounds in a chemotype connected to a calculated geometric centroid from the coordinates of the individual compounds. The centroid for these four different chemotypes clearly lies in the four different regions with distinct kinetic activation profiles as shown in Figure 4B.
Figure 4.
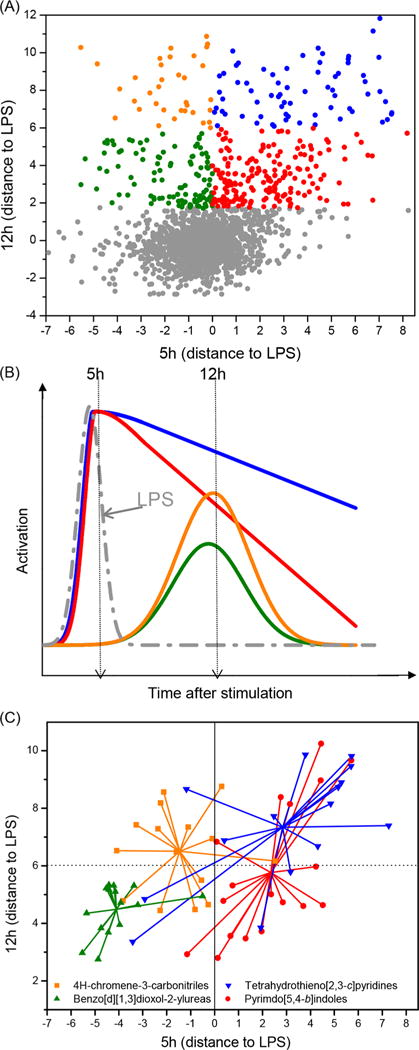
Confirmation and kinetic profile screens. (A) A scatter plot of distance to LPS values for 2011 compounds selected for confirmation and kinetic profile screens. The abscissa values correspond to the kinetic profile experiment while the ordinate values correspond to the confirmation screen experiment. (B) The compounds were divided into four categories based on their theoretical kinetic profiles for NF-κB activation with LPS. These compounds were either early inhibitors (5 h distance to LPS < 0) or early enhancers (5 h distance to LPS > 0). Within the early inhibitors, the compounds were grouped as either showing a moderate increase or high increase in NF-κB induction compared to LPS at 12 h (green and orange symbols and lines, respectively). Similarly, within the enhancer groups, compounds showed a moderate or high increase in NF-κB induction compared to LPS at 12 h (red and blue symbols and lines, respectively). (C) Cluster binning of confirmed hits indicates segregation of chemotypes into distinct kinetic profiles. Large clusters were colored based on chemotype, namely pyrimido[5,4-b]indoles (red circles), tetrahydrothieno[2,3-c]pyridines(blue triangles), benzo-[d][1,3]dioxol-2-ylureas (green triangles), and 4H-chromene-3-carbonitriles (orange squares). The center of the chemotype cluster was calculated as the geometric mean of the coordinates within a cluster. The colors are consistent with the kinetic profiles shown in B.
Structure–Activity Relationship Studies on Pyrimidoindoles
Pyrimidoindoles were identified as TLR4/MD2 agonists in a previous HTS,7,42 which was designed to identify compounds that induced NF-κB activation. The identification of this chemotype here as prolonging NF-κB induction warranted additional investigation into the structural differences to understand functional characteristics. Overall, 536 pyrimidoindoles were evaluated in both of the HTS campaigns, the bulk of which consisted of 357 amide-linked compounds (66.6%) and a small fraction of 27 ester-linked compounds (5%). Within the amide-linked compounds, there was a relatively higher number of N3-aryl (297 compounds) versus N3-methyl (60 compounds) substituted pyrimidoindoles pointing toward a bias for N3-aryl derivatives. A plot of potency signatures for the current HTS (distance to LPS) on the y-axis against previous HTS42 (percent activation) on the x-axis showed a clear demarcation for pyrimidoindoles designated as “hits” in the two different but related assays (Figure 5). The amide-linked hits identified in the previous HTS showed high dominance for N3-aryl substitution (73%) nearly matching the ratio present in the HTS library; however the amide-linked hits identified in the current HTS had a very high dominance for N3-methyl substitution (62%), despite being low in number (Figure 5). In addition, the current HTS also identified two active ester-linked compounds. Here, it is important to distinguish between the two HTS campaigns relative to the pyrimidoindoles chemotype. The first campaign was designed to identify compounds that alone had intrinsic ability to activate NF-κB, and as mentioned above, pyrimidoindoles bearing N3-aryl substitutions were found in this category, whereas those compounds bearing the N3-methyl substituent were inactive in the prior screen. In contrast, the present campaign was designed to identify compounds that could enhance or prolong an already-present TLR4 innate immune stimulus (LPS), and in this category, several pyrimidoindoles bearing the N3-methyl substituent were found to be active. With this in mind, we decided to pursue a combinatorial library of 42 compounds to explore a homologous series of N3-alkyl substitutions including N3-methyl, -ethyl, -propyl, -butyl, -isopropyl, and N3-aryl substitutions including N3-p-fluorophenyl and -p-methoxyphenyl. The other variation in the combinatorial library consisted of three esters (methyl, ethyl, and tert-butyl) and three amides linked to a 2-amino substituent of thiazole, benzothiazole, and 6-methylbenzothiazole at the C2 position as these substituents were identified as most potent in both the main and the 12 h confirmation screens.
Figure 5.
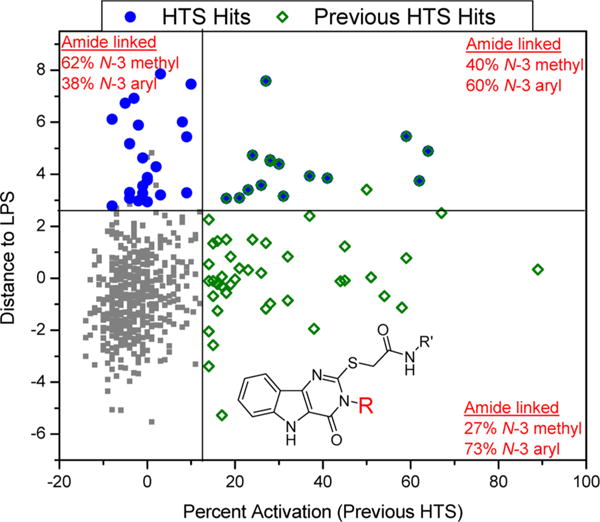
NF-κB activation data for pyrimidoindoles from two different HTS campaigns. A scatter plot of “distance to LPS” values from the current HTS on the Y axis and “percent activation” values from the previous HTS on the X axis for all the pyrimidoindoles shows clearly the distinct structural attribute for the C2–S-methylene acetamide linked molecules. The blue circles represent the hits identified in the current HTS campaign, while the green squares represent the hits identified in the previous HTS. The N3-aryl substituted pyrimidoindoles show intrinsic NF-κB activity while N3-methyl substituted pyrimidoindoles tend to prolong initial NF-κB activation by LPS without any intrinsic NF-κB activation.
The proposed compounds were synthesized as shown in Scheme 1. The common precursor ethyl 3-amino-1H-indole-2-carboxylate (1) was reacted with isothiocyanate chemset 2{1–7} to obtain the intermediate thiourea chemset 3{1–7}, followed by intramolecular condensation to yield the fused pyrimidine ring using acetyl chloride and ethanol to obtain chemset 4{1–7}. These compounds were then reacted with three different 2-chloroacetates 5{1–3} and 2-chloroacetamides 5{4–6} to obtain the 42 compound combinatorial library of differently N3- and C2-substituted pyrimidoindoles 6{1–7,1–6}.
Scheme 1.
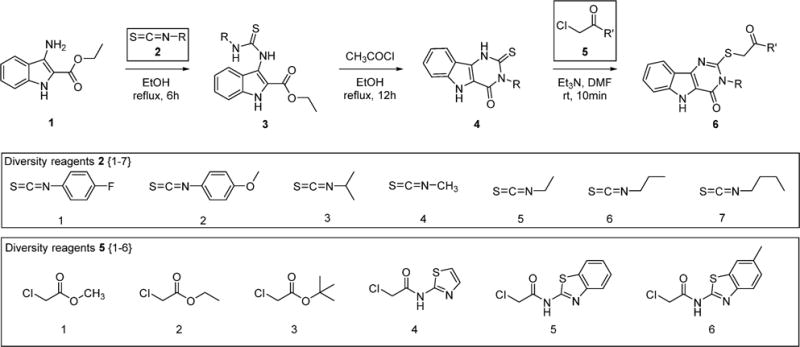
Syntheses of Differently N3- and C2-Substituted Pyrimidoindoles
These compounds were tested in the Cellsensor THP-1 cells at 5 μM concentration for the enhancement of NF-κB upregulation with LPS at 5 h and 12 h (Figure 6). The activity data were calculated as percent activation (%act), which was measured as FRET response ratios relative to the LPS alone control (Supporting Information, Table S1). None of the synthesized compounds significantly enhanced NF-κB induction at 5 h, while all the ester linked derivatives and thiazole amide linked compounds 6{1–7,1–4} showed negligible increases in NF-κB induction at 12 h compared to LPS alone. However, some of the amide-linked compounds with the 2-aminobenzothiazole and 2-amino-6-methylbenzothiazole substituents (6{1–7,5–6}) were very active and showed a distinct relationship between the N3-alkyl chain length and the NF-κB inducing activities in the presence of LPS at 12 h (Figure 6). In N3-alkyl substituted benzothiazole derivatives 6{3–7, 5} the percent activation value increases moderately for N3-methyl (6{4,5}, %act = 100), N3-ethyl (6{5,5}, %act = 120), N3-propyl (6{6,5}, %act = 119) derivatives; however, the N3-butyl derivative (6{4,5}, %act = 180) was found to be the most potent, and surprisingly the N3-isopropyl derivative (6{3,5}, % act = 75) showed NF-κB inhibitory effects with values lower than that of LPS alone. A similar trend was observed with the N3-alkyl substituted methylbenzothiazole derivatives 6{3–7,6} with both the N3-propyl and N3-butyl substituted derivatives being very potent (6{6,6}, %act = 190, 6{7,6}, %act = 192, respectively). The N3-isopropyl derivative (6{3,6}, %act = 85) with LPS showed reduction in NF-κB activation compared to the LPS alone, suggesting that a branched alkyl chain is not tolerated. In the N3-aryl substituted derivatives, the 4-fluorophenyl substituted compounds (6{1,5–6}) exhibited better potency than the corresponding 3-methoxyphenyl substituted analogs (6{2,5–6}).
Figure 6.
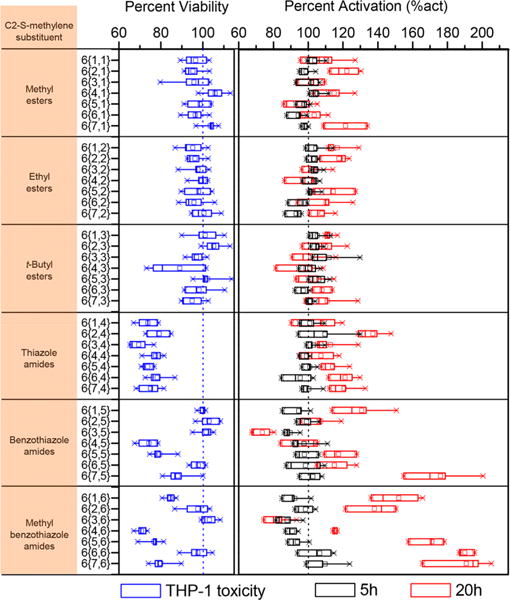
NF-κB induction assay and MTT toxicity in THP-1 cells. The viability (blue boxes) measured by MTT assay in THP-1 cells relative to DMSO (blue dotted line) and NF-κB percent activation for synthesized pyrimidoindole library compounds in the presence of LPS calculated relative to LPS alone (black dotted line) values at both 5 h (black boxes) and 20 h (red boxes). The values were statistically computed from two and four independent experiments for MTT cell viability and NF-κB activation assays, respectively in THP-1 cells. The C2–S-methylene substituent on the pyrimidoindole for the synthesized compounds is shown in the individual blocks on the left. Relative viability was calculated as a percentage of OD at 630 nm compared to DMSO. The OD at 630 nm for DMSO was 0.92 ± 0.1 (mean ± SD). Percent activation (%act) was calculated as a response ratio relative to LPS. The response ratio for LPS was 8.00 ± 1.2 and 2.58 ± 0.48 at 5 h and 20 h (mean ± SD), respectively.
Next, we assayed the toxicity of these compounds in THP-1 cells using an (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) MTT assay. Toxicity was measured as percent viability relative to the vehicle (0.5% DMSO). All of the weakly potent ester-linked compounds were found to be nontoxic (% viability >90) at 5 μM, while all of the thiazole substituted amide-linked compounds were found to be relatively toxic, suggesting an off-target effect related to the presence of the thiazole group. Among the N3-alkyl substituted benzothiazole or 6-methylbenzothiazole derivatives, all except the N3-propyl and -isopropyl derivatives were relatively more toxic.
In order to identify an optimum N3-alkyl chain length for maximal potency, we synthesized N3-pentyl (9a and 10a) and hexyl (9b and 10b) derivatives for the benzothiazole and 6-methylbenzothiazole substituted compounds as shown in Scheme 2. These higher chain analogs showed declining potency with increasing chain length (9a, %act = 154; 9b, %act = 143; 10a, %act = 139; 10b, %act = 142) and also were found to be relatively more toxic. We had earlier reported that N5-methylation of pyrimidoindoles reduces their toxicity without abrogating the activity at TLR4.7 Thus, we synthesized N5-methyl derivatives of the potent N3-butyl substituted derivatives 6{7,5–6}. These compounds, however, did not eliminate toxicity but lost their ability to sustain an LPS induced NF-κB response (11, %act = 94; 12, %act = 91).
Scheme 2.
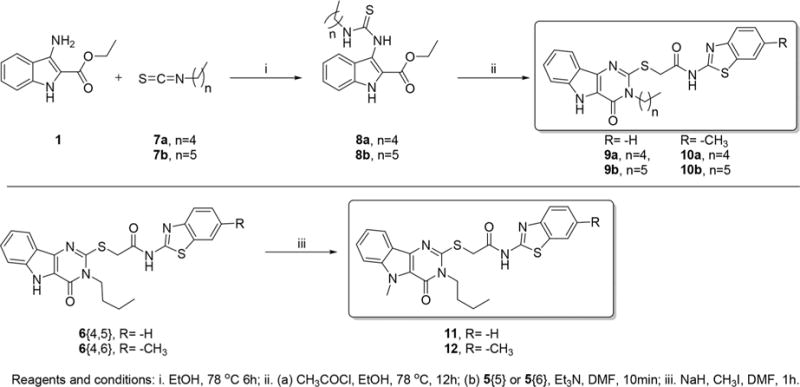
Syntheses of N3-Pentyl, -Hexyl, and N5-Methyl Substituted Derivatives of Select Pyrimidoindoles
Moreover, while these active N3-alkyl derivatives enhance the LPS induced NF-κB activation, they appear to do so by a mechanism that is independent of TLR4 binding, unlike the derivatives identified in the previous HTS. We evaluated the abilities these compounds have to activate the NF-κB and TLR4 signaling pathways using THP-1 NF-κB Cellsensor and human TLR4 HEK reporter cell lines, respectively. We found that these compounds lack activity in either of these assays as compared to the potent pyrimidoindoles identified in the previous HTS (Supporting Information, Figure S2).
For all the above reasons, we were interested to pursue vaccination studies in mice with these novel compounds in the presence of a TLR4 stimulus to evaluate their potential coadjuvant effects. We chose MPLA (monophosphoryl lipid A), a known TLR4 agonist and Food and Drug Administration (FDA) approved adjuvant, and ovalbumin (OVA) as an antigen for the vaccination studies. Examination of antigen-specific antibodies showed that coimmunization of MPLA with compounds 6{7,5}, 6{6,6}, or 6{7,6} induced statistically significant increases in antigen specific IgG titers when compared to mice immunized with MPLA alone (p < 0.05 compared to MPLA plus antigen, Figure 7).
Figure 7.
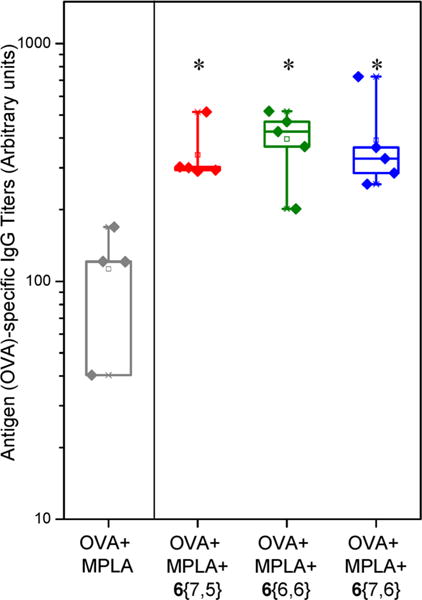
Coadjuvanticity of potent pyrimidoindole compounds with MPLA. Mice (n = 4–5 per group) were immunized via intramuscular route on day 0 and day 14 with antigen (ovalbumin, 20 μg/animal), MPLA (10 μg/animal), and compound 6{7,5} or 6{6,6} or 6{7,6} (100 nmol/animal). The immunized mice were bled on day 21, and OVA-specific IgG titers were measured using ELISA. Note that the potent compounds augmented the production of antigen-specific IgG by approximately 4 fold when coadjuvanted with MPLA compared to MPLA alone. *p < 0.05 compared to OVA + MPLA group using one-way ANOVA followed by Dunn’s post hoc testing.
CONCLUSIONS
In conclusion, a novel HTS assay using the THP-1 Cellsensor reporter cells has been standardized and used to identify compounds that prolong NF-κB activation when coadministered with a TLR4 stimulus (LPS). Hit compounds were identified using an enrichment strategy based on unsupervised chemoinformatic clustering of the compounds within the screen library, and also by a naïve “Top X” approach. Confirmation screening at 12 h and kinetic profile screening at 5 h yielded 407 confirmed hits. The correlation of the kinetic profiles with the structural similarities led to identification of four chemotypes including pyrimido[5,4-b]indoles. A 42-member combinatorial library of pyrimidoindoles was synthesized that led to identification of potent N3-alkyl substituted pyrimidoindole analogs. Although these potent compounds were intrinsically inactive toward NF-κB induction or TLR4 activation, when coadjuvanted with MPLA, they augmented the level of antigen-specific IgG production in mice. Many of the negative regulatory pathways involve kinase; hence future studies to elucidate the mechanisms of action of these derivatives will include kinase inhibitory assays among other pathways.
EXPERIMENTAL METHODS
Compound Library
A library of compounds was acquired from the UCSF Small Molecule Discovery Center consisting of 166 304 chemical entities from eight suppliers (Supporting Information, Figure S3).
High Throughput Screening and Statistical Analysis
HTS was performed by Thermo Fisher Scientific at their commercial facility (Madison, WI) in 384 well plates using Cellsensor THP-1 cells. The screen was performed using LPS alone (LPS 12 h) and LPS + 0.25 μM wortmannin as positive controls, 0.5% DMSO as a negative control, and an additional control included adding LPS at 5 h before substrate (LPS 5 h). There were 527 plates in the main screen and 18 plates each in the 12 h confirmation screen and in the 5 h kinetic screen.
Emission ratios were computed as the ratio of fluorescence density values at two wavelengths, fluorescein versus coumarin emission, after background subtraction using the mean of the cell-free wells. Response ratios (RR) were computed as the emission ratio divided by the mean emission ratio from no-lipid wells within the same plate. To optimize assay parameters, box plots and bar graphs were used to describe data. For compound selection, percent activation by plate was computed as the difference between RR from a particular well and the mean RR from no-LPS (0.5% DMSO) wells, divided by the difference of the mean RRs between LPS 5 h wells and no LPS (0.5% DMSO) wells. Quality checks in the pilot screen included making dot plots of percent activation values versus well number by well type, calculating Z′ for each plate, making scatter plots for duplicated compounds and outlier detection. In both the main screen and the confirmation screen, to account for the variability from the LPS 12 h wells (where LPS was added at 0 h), distance to LPS (standardized percent activation values against LPS 12 h, calculated as the difference between percent activation values for the test compound and mean of LPS 12 h control wells divided by the standard deviation for the LPS 12 h control wells within each plate) was used as the metric for compound selection. In the main screen, a cluster enrichment method42 was used to identify hits where Murcko scaffold classes, Daylight functional clusters, and the Top X approach (whether the distance to LPS first > 3) were used together to select compounds to carry forward. In the confirmation screen, compounds were identified as confirmed hits based on a mixture of two linear models using data from both confirmation and primary screens.
R (www.r-project.org, version 3.3.1) and Prism 6 (GraphPad Software, San Diego, CA) statistical software were used to carry out these analyses.46
Chemistry
Reagents and solvents used were obtained as at least reagent grade from commercial sources and used without further purification unless noted otherwise. Moisture or air-sensitive reactions were conducted under an argon atmosphere in an oven-dried (120 °C) glass apparatus. The solvents were removed under reduced pressure using standard rotary evaporators. Flash chromatography was carried out on a Biotage Isolera One (Charlotte, NC); while analytical thin-layer chromatography (TLC) was performed using precoated TLC silica gel 60 F254 aluminum sheets purchased from EMD (Gibbstown, NJ) and visualized using UV light. Reaction monitoring, compound characterization, and purity analysis were performed using a 1260 Infinity/6420 Triple Quad (Agilent Technologies, Inc., Santa Clara, CA) with a Supelco Discovery HS C18 column (Sigma-Aldrich). All the compounds were identified to be at least 98% pure by UV. Compounds of interest were analyzed by high resolution MS (HRMS) using an Agilent 6230 ESI-TOFMS (Santa Clara, CA). 1H NMR spectra were obtained on a Varian Mercury 400 (Varian, Inc., Palo Alto, CA). 13C HNMR spectra were obtained on a Varian VX 500 equipped with a Varian XSens 2 channel NMR Cold Probe (Varian, Inc., Palo Alto, CA). The chemical shifts are expressed in parts per million (ppm) using suitable deuterated NMR solvents.
General Procedure for the Synthesis of Compound 3{1–7}
To a solution of compound 1 (1 equiv) in warm ethanol was added the appropriate isothiocyanate 2{1–7} (1.1 equiv) dropwise with stirring. The reaction mixture was refluxed for 6 h and then allowed to cool at 4 °C overnight. Precipitated solids were filtered, washed with ethanol, and dried overnight under a vacuum to yield compounds 3{1–7} as white solids.
General Procedure for the Synthesis of Compound 4{1–7}
A solution of sodium ethoxide (10 equiv) was prepared in anhydrous ethanol and stirred for 5 min. Separately, compound 3{1–7} was dissolved in anhydrous ethanol and added to the sodium ethoxide solution. The reaction mixture was further refluxed for 12 h and then allowed to cool at 4 °C overnight. Precipitated solids were filtered and washed with cold ethanol to obtain compound 4{1–7} as white solids.
General Procedure for the Synthesis of Compound 6{1–7,1–6}
To a solution of compound 4{1–7} (1 equiv) in anhydrous DMF were added compound 5{1–6} (1.1 equiv) and triethylamine (2 equiv). The reaction mixture was heated to dissolve the components and then stirred at room temperature for 10 min or until the reaction was complete. The solvent was then removed under a vacuum to obtain the residue which was suspended in methanol with sonication. Precipitated solids were filtered and washed with excess methanol to give compound 6{1–7,1–6} as white solids.
Syntheses of Compounds 9 and 10
These compounds were synthesized in a manner similar to that of the combinatorial library using either pentyl isothiocyante (7a) for compound 8a or hexyl isothiocyanate (7b) for compound 8b and then cyclized and alkylated as described for syntheses of compounds 6{1–6,1–7} to give pentyl derivatives 9a and 9b and hexyl derivatives 10a and 10b, respectively.
Syntheses of Compounds 11 and 12
To a solution of compound 6{7,5} for compound 11 or compound 6{7,6} for compound 12 (1 equiv) in anhydrous DMF was added sodium hydride (1.1 equiv) at room temperature. The reaction mixture was stirred for 5 min followed by the addition of iodomethane (1.1 equiv) and stirred for an additional 2 h. The solvent was then removed under a vacuum, and the residue was dissolved in ethyl acetate, washed with water, dried over sodium sulfate, and concentrated to obtain the crude product which was purified using column chromatography to obtain compound 11 or 12.
Biology
Cell Lines and Reagents
The CellSensor NFκB-bla human monocytic THP-1 cell line was purchased from Thermo Fisher Scientific (Waltham, MA). This cell line contains a stably integrated beta-lactamase reporter gene under the control of the nuclear factor kappa B (NF-κB) response element (https://tools.thermofisher.com/content/sfs/manuals/CellSensor_NFkBbla_THP1_man.pdf). NF-κB activation results in beta-lactamase production, which shifts the fluorescence emission of the beta-lactamase substrate (LiveBLAzerTM-FRET B/G (CCF4-AM), Thermofisher) to favor coumarin (460 nm emission) over fluorescein (530 nm emission). Murine or human TLR4 HEK Blue cells were purchased from Invivogen (San Diego, CA). These cell lines were stably cotransfected with human or mouse TLR4, MD-2, and CD14 coreceptor genes and an inducible secreted embryonic alkaline phosphatase (SEAP) reporter gene. QuantiBlue was purchased from Invivogen. MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-dipheyl tetrazolium bromide) was purchased from Acros Organics, and ovalbumin was purchased from Worthington Biochemical Corporation. For the HTS, LPS E. coli 0111:B4 (Sigma-Aldrich) was used, and LPS-EB Ultrapure (cat# tlrl-3pelps, Invivogen) was used in the validation and subsequent SAR studies. Wortmannin,33 a nonspecific, covalent inhibitor of PI3K, and MPLA were purchased from Invivogen.
Measurement of NF-κB Activation Using CellSensor NF-κB-bla THP-1
CellSensor NF-κB-bla THP-1 cells were plated in 96-well plates at 80 μL (5 × 104 cells) per well in RPMI supplemented with 10% dialyzed fetal bovine serum (FBS, Omega Scientific, Inc., Tarzana, CA), 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 100 U/mL penicillin, and 100 μg/mL streptomycin (additives from Thermo Fisher Scientific). A total of 10 μL of 10× (50 μM) compound and 10 μL of 10× (100 ng/mL) LPS-EB in assay media were then added. Cells were incubated for 5 h and 20 h in 5% CO2 at 37 °C, after which 20 μL of 6× LiveBLAzer FRET B/G Substrate (CCF4-AM) mixture (prepared according to the manufacturer’s instructions) was added to each well. Plates were incubated at room temperature in the dark for 2 h. Fluorescence was measured on a Tecan Infinite M200 plate reader (Männedorf, Switzerland) at an excitation wavelength of 405 nm and emission wavelengths of 465 and 535 nm. Background values (cell free wells at the same fluorescence wavelength) were subtracted from the raw fluorescence intensity values, and the emission ratios were calculated as the ratio of background subtracted fluorescence intensities at 465 nm to background subtracted fluorescence intensities at 535 nm. The response ratio was then calculated using the formula [(emission ratio of a test well)/average emission ratio of wells with vehicle (0.5% DMSO)]. Compounds were ranked using “%act” metrics, which was calculated as [response ratio of the compound/response ratio of LPS]. In each assay, negative control wells were included that were cell-free and cells with vehicle (0.5% DMSO).
Cell Viability Assays
THP-1 cells were dispensed in 96-well plates (105 cells/well) and treated with 5 μM of each compound. After 18 h of incubation, a solution of MTT in assay media (0.5 mg/mL) was added to each well and further incubated for 4–6 h, followed by the addition of cell lysis buffer (15% w/v SDS and 0.12% v/v 12 N HCl aqueous solution), incubated overnight, and then absorbance measured at 570 nm using 650 nm as a reference with a plate reader.
In Vitro Assays Using TLR4 Reporter Cell Lines
Murine or human TLR4 HEK Blue cells (2.5 × 104 cells per well of a 96 well plate) were incubated with 5 μM of each test compound. The culture supernatants were harvested after a 20–24 h incubation period. SEAP activity in the supernatants was determined using QuantiBlue, and absorbance was read at 630 nm per the manufacturer’s instructions as previously reported.7
Animals
Seven to nine-week-old C57BL/6 (wild-type, WT) mice were purchased from The Jackson Laboratories (Bar Harbor, MA). All animal experiments received prior approval from the UCSD Institutional Animal Care and Use Committee.
In Vivo Adjuvant Activity Study
WT mice (n = 4–5 per group) were immunized in the gastronemius muscle with ovalbumin (20 μg/animal) mixed with MPLA (10 μg/animal) and compound 6{7,5} or 6{6,6} or 6{7,6} (100 nmol/animal) on days 0 and 14. On day 21, immunized mice were bled and OVA-specific IgG titers were measured by ELISA as previously described.47
Statistical Analysis for in Vitro and in Vivo Studies
Data for in vitro and in vivo studies are represented as mean ± standard error of the mean (SEM) or mean ± standard deviation (SD). Prism 6 (GraphPad Software, San Diego, CA) statistical software was used to obtain p-values for comparison between groups (p < 0.05 was considered significant) for in vivo study. One-way ANOVA followed by Dunn’s post hoc test was used to compare multiple groups.
Supplementary Material
Acknowledgments
We are grateful to Dr. Michael Harvey and his team at the Thermo Fisher Scientific, Madison, WI for performing the assay optimization, pilot, and HTS experiments as well as confirmation and kinetic screens at their facility.
Funding
We acknowledge the National Institutes of Health Adjuvant Discovery Program for funding (HHSN272200900034C and HHSN272201400051C, Principal Investigator, D.A.C.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
ABBREVIATIONS
- APC
antigen presenting cells
- DMF
N,N-dimethylformamide
- DMSO
dimethyl sulfoxide
- ELISA
enzyme-linked immunosorbent assay
- FDA
Food and Drug Administration
- FRET
Förster resonance energy transfer
- HTS
high-throughput screening
- Ig
immunoglobulin
- IL-10
interleukin-10
- IRAK
interleukin-1 receptor-associated kinase
- MPLA
monophosphoryl Lipid A
- MTT
(3-[4,5-dimethylthiazol-2-yl]-2,5-dipheyl tetrazolium bromide)
- NF-κB
nuclear factor kappa B
- OD
optical density
- OVA
ovalbumin
- PI3K
phosphatidylinositide-3 kinase
- RR
response ratio
- SDS
sodium dodecyl sulphate
- SEAP
secreted embryonic alkaline phosphatase
- TLC
thin layer chromatography
- WT
wild type
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscombsci.7b00080.
Figures S1–S3, Table S1, experimental section with compound names, reaction yields, 1H NMR, MS characterization data for all the final compounds as well as 13C NMR and HRMS characterization data for key compounds, 1H NMR and LC-MS spectra for final compounds, 13C NMR and HRMS spectra for key compounds, and detailed synthetic protocol for reagents 5{4–6} (PDF)
ORCID
Nikunj M. Shukla: 0000-0002-5150-7827
Author Contributions
The manuscript was written through contributions by all the authors. All authors approved the final version of the manuscript. K.M. and M.P. performed statistical analysis of the HTS data. M.Ch., B.N., H.B.C., and N.M.S. synthesized compounds. T.H., A.A., S.Y., F.S.-K., and M.Co. performed biological assays. H.B.C. and N.M.S. performed chemotype clustering analysis. M.Ch., T.H., K.M., M.P., M.Co., D.A.C., H.B.C., and N.M.S. wrote the manuscript.
Notes
The authors declare no competing financial interest.
References
- 1.Van Buynder PG, Konrad S, Van Buynder JL, Brodkin E, Krajden M, Ramler G, Bigham M. The comparative effectiveness of adjuvanted and unadjuvanted trivalent inactivated influenza vaccine (TIV) in the elderly. Vaccine. 2013;31(51):6122–8. doi: 10.1016/j.vaccine.2013.07.059. [DOI] [PubMed] [Google Scholar]
- 2.Reed SG, Orr MT, Fox CB. Key roles of adjuvants in modern vaccines. Nat Med. 2013;19(12):1597–608. doi: 10.1038/nm.3409. [DOI] [PubMed] [Google Scholar]
- 3.O’Hagan DT, Rappuoli R, De Gregorio E, Tsai T, Del Giudice G. MF59 adjuvant: the best insurance against influenza strain diversity. Expert Rev Vaccines. 2011;10(4):447–62. doi: 10.1586/erv.11.23. [DOI] [PubMed] [Google Scholar]
- 4.Doherty M, Schmidt-Ott R, Santos JI, Stanberry LR, Hofstetter AM, Rosenthal SL, Cunningham AL. Vaccination of special populations: Protecting the vulnerable. Vaccine. 2016;34(52):6681–6690. doi: 10.1016/j.vaccine.2016.11.015. [DOI] [PubMed] [Google Scholar]
- 5.Camilloni B, Basileo M, Valente S, Nunzi E, Iorio AM. Immunogenicity of intramuscular MF59-adjuvanted and intradermal administered influenza enhanced vaccines in subjects aged over 60: A literature review. Hum Vaccines Immunother. 2015;11(3):553–63. doi: 10.1080/21645515.2015.1011562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammer GE, Ma A. Molecular control of steady-state dendritic cell maturation and immune homeostasis. Annu Rev Immunol. 2013;31:743–91. doi: 10.1146/annurev-immunol-020711-074929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan M, Hayashi T, Mathewson RD, Nour A, Hayashi Y, Yao S, Tawatao RI, Crain B, Tsigelny IF, Kouznetsova VL, Messer K, Pu M, Corr M, Carson DA, Cottam HB. Identification of Substituted Pyrimido[5,4-b]indoles as Selective Toll-Like Receptor 4 Ligands. J Med Chem. 2013;56(11):4206–4223. doi: 10.1021/jm301694x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nour A, Hayashi T, Chan M, Yao S, Tawatao RI, Crain B, Tsigelny IF, Kouznetsova VL, Ahmadiiveli A, Messer K, Pu M, Corr M, Carson DA, Cottam HB. Discovery of substituted 4-aminoquinazolines as selective Toll-like receptor 4 ligands. Bioorg Med Chem Lett. 2014;24(21):4931–4938. doi: 10.1016/j.bmcl.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goff PH, Hayashi T, Martinez-Gil L, Corr M, Crain B, Yao S, Cottam HB, Chan M, Ramos I, Eggink D, Heshmati M, Krammer F, Messer K, Pu M, Fernandez-Sesma A, Palese P, Carson DA. Synthetic Toll-like receptor 4 (TLR4) and TLR7 ligands as influenza virus vaccine adjuvants induce rapid, tained, and broadly protective responses. J Virol. 2015;89(6):3221–3235. doi: 10.1128/JVI.03337-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shukla NM, Salunke DB, Balakrishna R, Mutz CA, Malladi SS, David SA. Potent adjuvanticity of a pure TLR7-agonistic imidazoquinoline dendrimer. PLoS One. 2012;7(8):e43612. doi: 10.1371/journal.pone.0043612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salunke DB, Connelly SW, Shukla NM, Hermanson AR, Fox LM, David SA. Design and development of stable, water-soluble, human Toll-like receptor 2 specific monoacyl lipopeptides as candidate vaccine adjuvants. J Med Chem. 2013;56(14):5885–900. doi: 10.1021/jm400620g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasilakos JP, Tomai MA. The use of Toll-like receptor 7/8 agonists as vaccine adjuvants. Expert Rev Vaccines. 2013;12(7):809–19. doi: 10.1586/14760584.2013.811208. [DOI] [PubMed] [Google Scholar]
- 13.Maisonneuve C, Bertholet S, Philpott DJ, De Gregorio E. Unleashing the potential of NOD- and Toll-like agonists as vaccine adjuvants. Proc Natl Acad Sci USA. 2014;111(34):12294–9. doi: 10.1073/pnas.1400478111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Basto AP, Leitao A. Targeting TLR2 for vaccine development. J Immunol Res. 2014;2014:619410. doi: 10.1155/2014/619410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wheeler CM, Skinner SR, Del Rosario-Raymundo MR, Garland SM, Chatterjee A, Lazcano-Ponce E, Salmeron J, McNeil S, Stapleton JT, Bouchard C, Martens MG, Money DM, Quek SC, Romanowski B, Vallejos CS, Ter Harmsel B, Prilepskaya V, Fong KL, Kitchener H, Minkina G, Lim YK, Stoney T, Chakhtoura N, Cruickshank ME, Savicheva A, da Silva DP, Ferguson M, Molijn AC, Quint WG, Hardt K, Descamps D, Suryakiran PV, Karkada N, Geeraerts B, Dubin G, Struyf F. Efficacy, safety, and immunogenicity of the human papillomavirus 16/18 AS04-adjuvanted vaccine in women older than 25 years: 7-year follow-up of the phase 3, double-blind, randomised controlled VIVIANE study. Lancet Infect Dis. 2016;16(10):1154–68. doi: 10.1016/S1473-3099(16)30120-7. [DOI] [PubMed] [Google Scholar]
- 16.Aoshi T, Koyama S, Kobiyama K, Akira S, Ishii KJ. Innate and adaptive immune responses to viral infection and vaccination. Curr Opin Virol. 2011;1(4):226–32. doi: 10.1016/j.coviro.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Akira S. Innate immunity and adjuvants. Philos Trans R Soc, B. 2011;366(1579):2748–55. doi: 10.1098/rstb.2011.0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Hu Y, Deng WW, Sun B. Negative regulation of Toll-like receptor signaling pathway. Microbes Infect. 2009;11(3):321–7. doi: 10.1016/j.micinf.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 19.Qian C, Cao X. Regulation of Toll-like receptor signaling pathways in innate immune responses. Ann N Y Acad Sci. 2013;1283:67–74. doi: 10.1111/j.1749-6632.2012.06786.x. [DOI] [PubMed] [Google Scholar]
- 20.Turnis ME, Song XT, Bear A, Foster AE, Gottschalk S, Brenner MK, Chen SY, Rooney CM. IRAK-M removal counteracts dendritic cell vaccine deficits in migration and longevity. J Immunol. 2010;185(7):4223–32. doi: 10.4049/jimmunol.0903507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shembade N, Ma A, Harhaj EW. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science (Washington, DC, U S) 2010;327(5969):1135–9. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuk JM, Shin DM, Lee HM, Kim JJ, Kim SW, Jin HS, Yang CS, Park KA, Chanda D, Kim DK, Huang SM, Lee SK, Lee CH, Kim JM, Song CH, Lee SY, Hur GM, Moore DD, Choi HS, Jo EK. The orphan nuclear receptor SHP acts as a negative regulator in inflammatory signaling triggered by Toll-like receptors. Nat Immunol. 2011;12(8):742–51. doi: 10.1038/ni.2064. [DOI] [PubMed] [Google Scholar]
- 23.Zhou J, Wu R, High AA, Slaughter CA, Finkelstein D, Rehg JE, Redecke V, Hacker H. A20-binding inhibitor of NF-kappaB (ABIN1) controls Toll-like receptor-mediated CCAAT/enhancer-binding protein beta activation and protects from inflammatory disease. Proc Natl Acad Sci U S A. 2011;108(44):E998–1006. doi: 10.1073/pnas.1106232108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho PC, Tsui YC, Feng X, Greaves DR, Wei LN. NF-kappaB-mediated degradation of the coactivator RIP140 regulates inflammatory responses and contributes to endotoxin tolerance. Nat Immunol. 2012;13(4):379–86. doi: 10.1038/ni.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwasaki H, Takeuchi O, Teraguchi S, Matsushita K, Uehata T, Kuniyoshi K, Satoh T, Saitoh T, Matsushita M, Standley DM, Akira S. The IkappaB kinase complex regulates the stability of cytokine-encoding mRNA induced by TLR-IL-1R by controlling degradation of regnase-1. Nat Immunol. 2011;12(12):1167–75. doi: 10.1038/ni.2137. [DOI] [PubMed] [Google Scholar]
- 26.An H, Zhao W, Hou J, Zhang Y, Xie Y, Zheng Y, Xu H, Qian C, Zhou J, Yu Y, Liu S, Feng G, Cao X. SHP-2 phosphatase negatively regulates the TRIF adaptor protein-dependent type I interferon and proinflammatory cytokine production. Immunity. 2006;25(6):919–28. doi: 10.1016/j.immuni.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 27.An H, Hou J, Zhou J, Zhao W, Xu H, Zheng Y, Yu Y, Liu S, Cao X. Phosphatase SHP-1 promotes TLR- and RIG-I-activated production of type I interferon by inhibiting the kinase IRAK1. Nat Immunol. 2008;9(5):542–50. doi: 10.1038/ni.1604. [DOI] [PubMed] [Google Scholar]
- 28.O’Neill LA, Sheedy FJ, McCoy CE. MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat Rev Immunol. 2011;11(3):163–75. doi: 10.1038/nri2957. [DOI] [PubMed] [Google Scholar]
- 29.Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, Hua M, Li N, Yao H, Cao X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat Immunol. 2011;12(9):861–9. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 30.Ma F, Liu X, Li D, Wang P, Li N, Lu L, Cao X. MicroRNA-466l upregulates IL-10 expression in TLR-triggered macrophages by antagonizing RNA-binding protein tristetraprolin-mediated IL-10 mRNA degradation. J Immunol. 2010;184(11):6053–9. doi: 10.4049/jimmunol.0902308. [DOI] [PubMed] [Google Scholar]
- 31.Liu X, Zhan Z, Xu L, Ma F, Li D, Guo Z, Li N, Cao X. MicroRNA-148/152 impair innate response and antigen presentation of TLR-triggered dendritic cells by targeting CaMKIIalpha. J Immunol. 2010;185(12):7244–51. doi: 10.4049/jimmunol.1001573. [DOI] [PubMed] [Google Scholar]
- 32.El Gazzar M, Church A, Liu T, McCall CE. MicroRNA-146a regulates both transcription silencing and translation disruption of TNF-alpha during TLR4-induced gene reprogramming. J Leukocyte Biol. 2011;90(3):509–19. doi: 10.1189/jlb.0211074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arcaro A, Wymann MP. Wortmannin is a potent phosphatidylinositol 3-kinase inhibitor: the role of phosphatidylinositol 3,4,5-trisphosphate in neutrophil responses. Biochem J. 1993;296:297–301. doi: 10.1042/bj2960297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naiki Y, Michelsen KS, Zhang W, Chen S, Doherty TM, Arditi M. Transforming growth factor-beta differentially inhibits MyD88-dependent, but not TRAM- and TRIF-dependent, lipopolysaccharide-induced TLR4 signaling. J Biol Chem. 2005;280(7):5491–5. doi: 10.1074/jbc.C400503200. [DOI] [PubMed] [Google Scholar]
- 35.Kondo T, Kawai T, Akira S. Dissecting negative regulation of Toll-like receptor signaling. Trends Immunol. 2012;33(9):449–58. doi: 10.1016/j.it.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 36.Sakamoto H, Egashira S, Saito N, Kirisako T, Miller S, Sasaki Y, Matsumoto T, Shimonishi M, Komatsu T, Terai T, Ueno T, Hanaoka K, Kojima H, Okabe T, Wakatsuki S, Iwai K, Nagano T. Gliotoxin suppresses NF-kappaB activation by selectively inhibiting linear ubiquitin chain assembly complex (LUBAC) ACS Chem Biol. 2015;10(3):675–81. doi: 10.1021/cb500653y. [DOI] [PubMed] [Google Scholar]
- 37.Peng YM, Zheng JB, Zhou YB, Li J. Characterization of a novel curcumin analog P1 as potent inhibitor of the NF-kappaB signaling pathway with distinct mechanisms. Acta Pharmacol Sin. 2013;34(7):939–50. doi: 10.1038/aps.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma YM, Peng YM, Zhu QH, Gao AH, Chao B, He QJ, Li J, Hu YH, Zhou YB. Novel CHOP activator LGH00168 induces necroptosis in A549 human lung cancer cells via ROS-mediated ER stress and NF-kappaB inhibition. Acta Pharmacol Sin. 2016;37(10):1381–1390. doi: 10.1038/aps.2016.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li G, Diogo D, Wu D, Spoonamore J, Dancik V, Franke L, Kurreeman F, Rossin EJ, Duclos G, Hartland C, Zhou X, Li K, Liu J, De Jager PL, Siminovitch KA, Zhernakova A, Raychaudhuri S, Bowes J, Eyre S, Padyukov L, Gregersen PK, Worthington J, Gupta N, Clemons PA, Stahl E, Tolliday N, Plenge RM. Human genetics in rheumatoid arthritis guides a high-throughput drug screen of the CD40 signaling pathway. PLoS Genet. 2013;9(5):e1003487. doi: 10.1371/journal.pgen.1003487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang X, Lv B, Li P, Ma X, Wang T, Zhou Q, Wang X, Gao X. Bioactivity-integrated UPLC/Q-TOF-MS of Danhong injection to identify NF-kappaB inhibitors and anti-inflammatory targets based on endothelial cell culture and network pharmacology. J Ethnopharmacol. 2015;174:270–6. doi: 10.1016/j.jep.2015.08.026. [DOI] [PubMed] [Google Scholar]
- 41.Beck A, Vinik Y, Shatz-Azoulay H, Isaac R, Streim S, Jona G, Boura-Halfon S, Zick Y. Otubain 2 is a novel promoter of beta cell survival as revealed by siRNA high-throughput screens of human pancreatic islets. Diabetologia. 2013;56(6):1317–26. doi: 10.1007/s00125-013-2889-x. [DOI] [PubMed] [Google Scholar]
- 42.Pu M, Hayashi T, Cottam H, Mulvaney J, Arkin M, Corr M, Carson D, Messer K. Analysis of high-throughput screening assays using cluster enrichment. Stat Med. 2012;31(30):4175–89. doi: 10.1002/sim.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fukao T, Tanabe M, Terauchi Y, Ota T, Matsuda S, Asano T, Kadowaki T, Takeuchi T, Koyasu S. PI3K-mediated negative feedback regulation of IL-12 production in DCs. Nat Immunol. 2002;3(9):875–881. doi: 10.1038/ni825. [DOI] [PubMed] [Google Scholar]
- 44.Guha M, Mackman N. The Phosphatidylinositol 3-Kinase-Akt Pathway Limits Lipopolysaccharide Activation of Signaling Pathways and Expression of Inflammatory Mediators in Human Monocytic Cells. J Biol Chem. 2002;277(35):32124–32132. doi: 10.1074/jbc.M203298200. [DOI] [PubMed] [Google Scholar]
- 45.Hazeki K, Kinoshita S, Matsumura T, Nigorikawa K, Kubo H, Hazeki O. Opposite effects of wortmannin and 2-(4-morpholinyl)-8-phenyl-1(4H)-benzopyran-4-one hydrochloride on toll-like receptor-mediated nitric oxide production: negative regulation of nuclear factor-{kappa}B by phosphoinositide 3-kinase. Mol Pharmacol. 2006;69(5):1717–1724. doi: 10.1124/mol.105.021162. [DOI] [PubMed] [Google Scholar]
- 46.Grün B, Leisch F. FlexMix Version 2: Finite Mixtures with Concomitant Variables and Varying and Constant Parameters. J Stat Soft. 2008;28(4) doi: 10.18637/jss.v028.i04. [DOI] [Google Scholar]
- 47.Chan M, Hayashi T, Kuy CS, Gray CS, Wu CCN, Corr M, Wrasidlo W, Cottam HB, Carson DA. Synthesis and Immunological Characterization of Toll-Like Receptor 7 Agonistic Conjugates. Bioconjugate Chem. 2009;20(6):1194–1200. doi: 10.1021/bc900054q. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


