Abstract
Elotuzumab, targeting signaling lymphocytic activation molecule family 7 (SLAMF7), has been approved in combination with lenalidomide and dexamethasone (ELd) for relapsed/refractory multiple myeloma (MM) based on the findings of the phase III randomized trial ELOQUENT-2 (NCT01239797). Four-year follow-up analyses of ELOQUENT-2 have demonstrated that progression-free survival was 21% in ELd versus 14% in Ld. Elotuzumab binds a unique epitope on the membrane IgC2 domain of SLAMF7, exhibiting a dual mechanism of action: natural killer (NK) cell-mediated antibody-dependent cellular cytotoxicity (ADCC) and enhancement of NK cell activity. The ADCC is mediated through engagement between Fc portion of elotuzumab and FcgRIIIa/CD16 on NK cells. Enhanced NK cell cytotoxicity results from phosphorylation of the immunoreceptor tyrosine-based switch motif (ITSM) that is induced via elotuzumab binding and recruits the SLAM-associated adaptor protein EAT-2. The coupling of EAT-2 to the phospholipase Cg enzymes SH2 domain leads to enhanced Ca2+ influx and MAPK/Erk pathway activation, resulting in granule polarization and enhanced exocytosis in NK cells. Elotuzumab does not stimulate the proliferation of MM cells due to a lack of EAT-2. The inhibitory effects of elotuzumab on MM cell growth are not induced by the lack of CD45, even though SHP-2, SHP-1, SHIP-1, and Csk may be recruited to phosphorylated ITSM of SLAMF7. ELd improves PFS in patients with high-risk cytogenetics, i.e. t(4;14), del(17p), and 1q21 gain/amplification. Since the immune state is paralytic in advanced MM, the efficacy of ELd with minimal toxicity may bring forward for consideration of its use in the early stages of the disease.
Keywords: Elotuzumab, Multiple myeloma, SLAMF7, SLAM-associated protein (SAP), EAT-2
Introduction
Multiple myeloma (MM) is the second most common hematological malignancy in Western countries with 62 % of patients being older than 65 years at the time of diagnosis.1,2 According to the National Cancer Center in Japan, the number of patients with MM was 6697 in 2013, and that of deaths was 4129 in 2015; the five-year relative survival rate was 36.4% for MM patients diagnosed between 2000 to 2008.3 Regarding morbidity in 2015 based on age and gender, the proportions of patients older than 65 years were 90.1% for females and 87.9% for males, while those of patients older than 75 years were 69.1% for females and 60.9% for males.3 Due to its high incidence in the elderly and its incurability, there is an urgent need to develop effective and less toxic combination therapies for unfit or frail patients with MM.
The treatment outcomes of MM have significantly improved in the last decade or two due to the success of molecular targeting agents including thalidomide, lenalidomide, and bortezomib.4–7 According to the findings of a number of clinical trials, triplet induction therapy containing proteasome inhibitors (PIs) and immunomodulatory drugs (IMiDs) is the standard care for fit patients, whereas doublet induction therapy containing PIs or IMiD is administered to frail patients. In addition to the development of second- and third-generation PIs and IMiDs, monoclonal antibodies (mAb) will open a new era of MM treatments that selectively eliminate the malignant clone and reverse tumor-mediated immune paralysis.8–12 Elotuzumab is the first therapeutic mAb targeting SLAMF7 that has been approved for relapsed or refractory (RR) MM. It induces natural killer (NK) cell-mediated antibody-dependent cellular cytotoxicity (ADCC) and exerts stimulatory effects on immune cells, particularly NK cells, which are mediated by the engagement of elotuzumab with SLAMF7.13,14 Clinically, the combination of elotuzumab with lenalidomide and dexamethasone (ELd) is a promising treatment for frail patients regardless of the cytogenetic risk.8
In this review, we will focus on the efficacy and safety of elotuzumab for the treatment of RRMM. We will also discuss the biological characteristics of SLAMF7 and SLAM-associated protein (SAP), their expression and possible functions in normal cells and hematological malignancies, as well as the modes of action of elotuzumab. We will then propose optimal use and future directions for elotuzumab in the treatment of MM.
Elotuzumab for the Treatment of RRMM
Efficacy and safety of elotuzumab in combination with lenalidomide and dexamethasone
Elotuzumab was approved in combination with lenalidomide and dexamethasone (Ld) for patients with RRMM based on the findings of the phase III, randomized, open-label, multicenter trial, ELOQUENT-2 (NCT01239797).8 In ELOQUENT-2, the efficacy of elotuzumab combined with Ld (ELd) was evaluated for patients with RRMM who previously received one to three regimens. ELOQUENT-1 is still ongoing for patients with newly diagnosed MM (NDMM). ELOQUENT-2, in which 646 patients were randomized into ELd or Ld, demonstrated significant increase in overall response rate (ORR) and median PFS in ELd (Table 1).8 Progression-free survival (PFS) has significantly improved in patients older than 75 years, particularly those with refractory disease and high-risk cytogenetic abnormalities (CA), i.e. t(4;14), del(17p), and 1q21 gain/amplification. A subanalysis of the Japanese population from ELOQUENT-2 revealed similar outcomes to the global study as well as to the Japanese phase I study; ORR were 84% in ELd vs 86% in Ld, and PFS rates at two years were 48% in ELd vs 18% in Ld.15,16 A three-year follow-up and post-hoc analyses of ELOQUENT-2 recently confirmed that ELd provided a durable improvement in efficacy; ORR were 79% in ELd and 66% in Ld.17 ELd reduced the risk of disease progression/death by 27% versus Ld. Interim overall survival (OS) at 3 years was 60% with ELd versus 53% with Ld. Serum M-protein dynamic modelling showed slower tumor regrowth with ELd.17 An extended four-year follow-up of ELOQUENT-2 also demonstrated a sustained improvement in PFS in ELd versus Ld (21% vs 14%).18 Patients with ≥ very good partial response (VGPR) had the greatest reduction (35%) in risk of progression/death. Median OS was 48% in ELd versus 40% in Ld.18 These results further support the durable efficacy of ELd.
Table 1.
Antibody-containing novel combination regimens for RRMM.
| Regimen | Phase | N | ≧PR (%) | ≧VGPR (%) | ≧CR (%) | Median PFS (mo.) | References (Number) |
|---|---|---|---|---|---|---|---|
| Elotuzumab+Ld vs Ld | III | 321 vs 325 | 78.5 vs 65.5 | 31 vs 29 | 4 vs 7 | 19.4 vs 14.9 | Lonial S et al., 2015 (8) |
| Elotudumab+Bd vs Bd | II | 77 vs 75 | 65 vs 63 | 37 vs 27 | 4 vs 4 | 9.7 vs 6.9 | Jakubowiak A et al., 2016 (24) |
| Elotudumab+Td | II | 40 | 38 | 18 | 8 | 3.9 | Mateos MV et al., 2016 (25) |
| Daratumumab+Bd vs Bd | III | 251 vs 247 | 82.9 vs 63.2 | 59.2 vs 29.1 | 19.2 vs 9.0 | 60.7% vs 26.9% (at 12-mo) | Palumbo A et al., 2016 (9) |
| Daratumumab+Ld vs Ld | III | 286 vs 283 | 92.9 vs 76.4 | 75.8 vs 44.2 | 43.1 vs 19.2 | 83.2% vs 60.1% (at 12-mo) | Dimopoulos MA et al., 2016 (10) |
| Daratumumab+Pd | Ib | 103 | 60 | 42 | 17 | 8.8 (at a median follow-up of 13.1 mo.) | Chari A et al., 2017 (11) |
| Pembrolizumab+Pd | II | 48 | 60 | 27 | 8 | 17.4 (at a median follow-up of 15.8 mo.) | Badros A et al., 2017 (12) |
Rd, lenalidomide and dexamethazone; Bd, bortezomib and dexamethasone; Td, thalidomide and dexamethasone; Pd, pomalidomide and dexamethasone.
The safety, tolerability, and pharmacokinetics of intravenous elotuzumab have been assessed in a Phase I study of dose-escalation monotherapy at 10–20 mg/kg, demonstrating no maximum tolerated dose and modest activity with a best response of stable disease (SD).19 The other two Phase I or Phase I/II studies also reported that the safety and tolerability of elotuzumab in combination with bortezomib or lenalidomide were acceptable.20–22 Severe adverse events (AEs) in ELOQUENT-2 were 65% in ELd versus 57% in Ld; the most common grade 3/4 hematological AEs in ELd vs Ld were lymphocytopenia (77% vs 49%) followed by neutropenia (34% vs 44%), thrombocytopenia (19% vs 20%), and anemia (19% vs 21%).8 Grade 3/4 hematological AEs, except for lymphocytopenia, were less frequent with ELd than with Ld, which may be of particular benefit for frail elderly patients. Common non-hematological grade 3/4 AEs were fatigue (8% in both arms), diarrhea (5% in ELd vs 4% in Ld), and pyrexia (5% and 3% in both arms). Lymphocytopenia may develop as a result of the migration of peripheral lymphocytes including NK cells into the involved tissue sites.19 Infusion reactions (IRs) appearing as pyrexia, chills, and hypertension were very limited when compared with daratumumab, observed in 10% of ELd versus 45.3–50% of daratumumab-containing regimens.9–11 Premedication with antihistamines, acetaminophen, and dexamethasone have successfully prevented IRs, and now are standard of care as part of the treatment with this antibody treatment. A phase II study demonstrated that a 1-hour infusion of elotuzumab provided convenient alternative dosing.23
Elotuzumab in combination with bortezomib or thalidomide
The efficacy of elotuzumab combined with bortezomib or thalidomide was also evaluated (Table 1).24,25 A randomized Phase II study of elotuzumab combined with bortezomib and dexamethasone (EBd) versus bortezomib and dexamethasone (Bd), in which 152 patients with RRMM were randomized into EBd or Bd, has demonstrated slight increase in ORR and median PFS in EBd.24 Grade 3/4 AEs were reported in 53 patients (71%) with EBd versus 45 patients (69%) with Bd; the most common grade 3 or higher AEs of EBd vs Bd were infections (21% vs 13%) and thrombocytopenia (9% vs 17%).24 Grade 3/4 peripheral neuropathy (9% vs 12%), paresthesia (0% vs 5%), and thrombocytopenia were slightly less frequent in EBd than in Bd.24 Grade 1/2 IRs were observed in 5% of EBd; there were no grade 3 or higher IRs.
The efficacy of 10 mg/kg elotuzumab combined with 50–200 mg thalidomide and 40 mg dexamethasone (ETd) (with or without 50 mg cyclophosphamide), was also evaluated in a Phase II single-arm study with minimal additional toxicity.25 IRs were observed in 15% of ETd. This clinical trial showed ORR of 38% in 40 RRMM patients with a median of three prior regimens including bortezomib (98%) and lenalidomide (73%); median PFS and OS were 3.9 months and 16.3 months, respectively.25 These findings suggest that the combination of elotuzumab with bortezomib or thalidomide has potential as treatment option for patients with RRMM.
Biological Characteristics of SLAMF7 and its Adaptor Proteins
Biological characteristics of SLAMF receptors
SLAMF7 is one of the nine SLAMF receptors (SLAMF1-9) belonging to the CD2 subset of the immunoglobulin superfamily. It was originally identified as CS1 (CD2 subunit 1) by a subtractive hybridization between naïve B cell cDNA and that of memory B cells and plasma cells.13 Molecular cloning revealed that CS1 is a novel human NK cell receptor.26 SLAMF7 may also play a growth-promoting role and be involved in the autocrine expression of cytokines in normal B cells,27 whereas its function in normal plasma cells currently remains unknown.
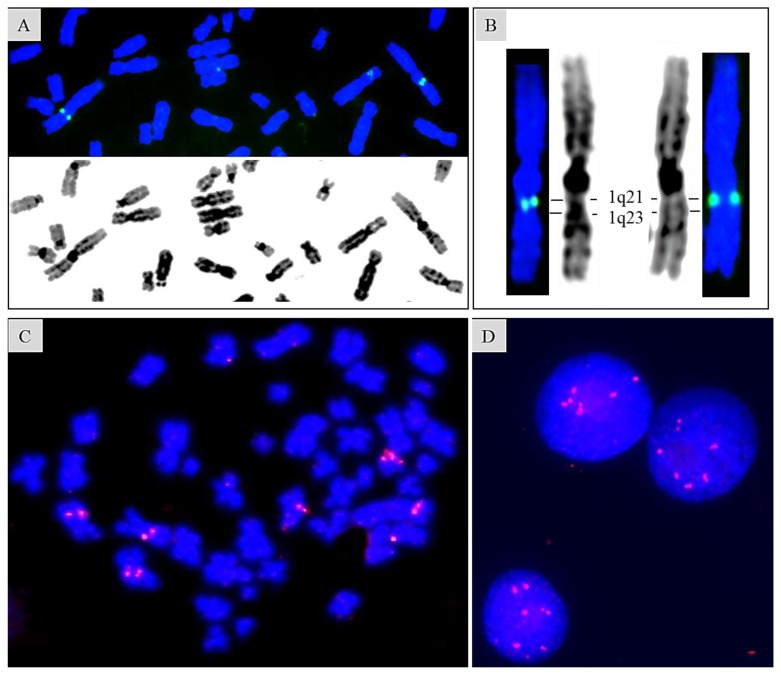
SLAMF receptors are type I transmembrane glycoproteins, except a glycosylphosphatidylinositol-anchored protein SLAMF2, which is widely expressed in hematopoietic cells but not in other tissues (Table 2). The genes encoding SLAMF receptors are reported to be clustered within an approximately 350-kb region at 1q23.3.28,29 Our fluorescence in situ hybridization (FISH) study assigned SLAMF7 to 1q21.3 using the BAC clone RP11-404F10 containing SLAMF2, SLAMF7, and SLAMF3 (Sakamoto N, Taniwaki M et al., unpublished) (Figures 1A and 1B). SLAMF7 is also included in the amplicon of chromosome 1q gain/amplification, which is a high-risk CA frequently detected in RRMM (Sakamoto N, Taniwaki M et al., unpublished) (Figures 1C and 1D).
Table 2.
Cytogenetic abnormalities valuable to predict prognosis of MM with candidate genes.
| Cytogenetic findings | Frequency (%) | Band | Candidate genes | Prognosis | |
|---|---|---|---|---|---|
| Primary changes | |||||
| IGH translocation | 45–55 | ||||
| t(11;14) | 15–20 | 11q13 | CCND1 | MYEOV | Good |
| t(4;14) | 15 | 4p16 | FGFR3 | MMSET | Poor |
| t(14;16) | 5–10 | 16q23 | MAF | WWOX | Poor |
| t(14;20) | 1–2 | 20q11 | MAFB | Poor | |
| t(8;14), t(8;22) | 1 | 8q24 | MYC | PVT1 | Poor |
| Hyperdiploidy | 45–55 | Good | |||
| Secondary changes | |||||
| 8q24 translocation | 13–22 | 8q24 | MYC | PVT1 | Poor |
| 8q24.1 gain | 15 | 8q24.1 | MYC | PVT1 | Unknown |
| del(13q)/-13 | 50 | 13q14 | RB | Poor by metaphase cytogenetics | |
| 13q13 | NBEA (BCL8B) | ||||
| del(17p) | 10–15 | 17p13 | TP53 | Poor | |
| 1q21 gain/amplification | 30–40 | 1q21–23 | PDZK1, CKS1B, BCL9, MUC1, RAB25, FCRL4 (IRTA1), FCRL5 (IRTA2), SLAMF7 | Poor | |
MYEOV, Myeloma-overexpressed; MMSET, multiple myeloma SET domain; WWOX, WW domain containing oxidoreductase; PVT1, plasmacytoma variant translocation 1; NBEA, neurobeachin; PDZK1, PDZ domain containing 1; CKS1B, cyclin-dependent kinases regulatory subunit 1; MUC1, mucin 1, cell surface associated; RAB25, RAB25, member RAS oncogene family; FCRL, Fc receptor-like protein gene; IRTA, immune receptor translocation-associated protein; SLAMF7, signaling lymphocyte activation molecule family 7.
Figure 1. Fluorescence in situ hybridization mapping of SLAMF7 gene on normal metaphase and MM cells.
(Sakamoto N, Taniwaki M et al., unpublished). FISH is performed as described as previously.98 (A) Representative mapping finding of SLAMF7 gene on a partial metaphase cell using BAC clone RP11-404F10 containing SLAMF2, SLAMF7, and SLAMF3. (B) An enlarged view of chromosomes 1 shown in (A). SLAMF7 gene is assigned to 1q21.3 in our FISH study, although reportedly to be at the chromosomal band 1q23.3. (C) (D) Amplification of SLAMF7 gene in a metaphase spread and interphase nuclei obtained from a MM patient harboring pseudodiploid karyotype with 1q gain.
SLAMF receptors are structurally characterized by distal Ig variable-like (IgV) and proximal C2-like (IgC2) domains within an extracellular portion and one or more immunoreceptor tyrosine-based switch motifs (ITSMs) within the cytoplasmic portion. The exception is that SLAMF3 has duplicated IgV-IgC2 sequences, and SLAMF8 and SLAMF9 lack tyrosine motifs.28,30,31 SLAMF receptors 1, 3, 5 to 7, and 9, are “self-ligands” that recognize the same receptor molecule on another cell as a ligand; SLAMF2 and SLAMF4 are “co-ligands” that recognize each other.27,28,32 Interactions between SLAMF receptors occur at their IgV domains between identical or different types of hematopoietic cells. The engagement of SLAMF receptors mediates regulatory effects on immune cells in the presence of the SLAM-associated protein (SAP) family of adaptors.26,33,34 Two SAP family adaptors have been identified in humans: SAP (SH2D1A) and EWS-Fli1-activated transcript-2 (EAT-2, SH2D1B), which are intracellular proteins containing the Src homology2 (SH2) domain devoid of enzymatic activity.28,31,35 Although most SLAMF receptors bind SAP and EAT-2, SLAMF7 is reported to be functionally controlled by EAT-2 only.33,36 However, RNA interference experiments have demonstrated that SLAMF7 may interact with SAP when the concentration of SAP is significantly higher than that of EAT-2 in cells.37 Hence, the selective binding of SLAMF7 to EAT-2 is due to its greater affinity to EAT-2 than SAP by nearly two orders of magnitude.37 Moreover, a recent study reported that SLAMF7 interacted with integrin Mac-1 instead of SAP adaptors utilizing signals involving immunoreceptor tyrosine-based activation motifs (ITAMs), which induced the promotion of phagocytosis.38 Further studies are needed in order to elucidate the exact role of SLAMF7 in myeloma cell pathophysiology.
SLAM-associated adaptor proteins and downstream signal transduction
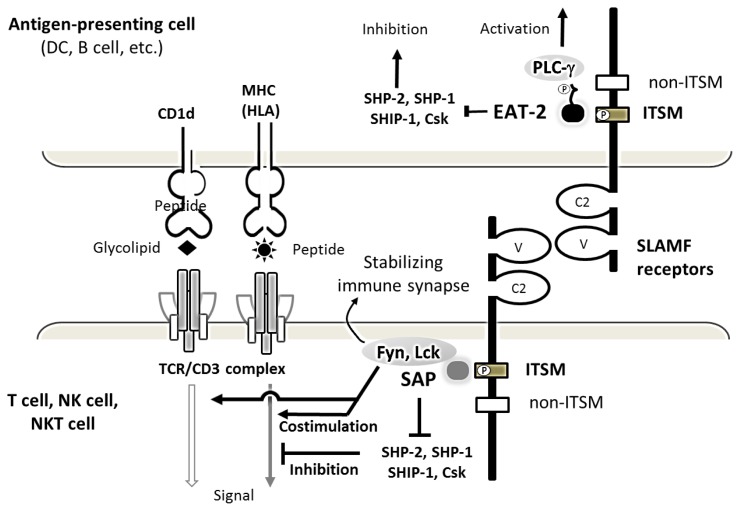
SLAMF functions as an either inhibitory or activating receptor depending on the availability of the SAP-related adaptor proteins, SAP and EAT-2. SAP is expressed in T, NK, NKT, and germinal center B cells. SAP expression has been reported in some Epstein-Barr virus (EBV)-transformed B cells, Hodgkin’s lymphoma, and angioimmunoblastic T-cell lymphoma.39–41 EAT-2 is expressed in NK cells and a range of antigen-presenting cells including monocytes.42,43 When the SLAMF receptor is engaged, tyrosine (Y) 281 located in ITSMs is phosphorylated, recruiting SAP or EAT-2.28,32 Through the SH2 domain, SAP or EAT-2 binds SLAMF at the phosphorylated ITSMs with overlapping specificities for activating and inhibitory binding partners. SAP contains an arginine-based motif in the SH domain, which mediates binding to the Src family protein Fyn, thereby stabilizing immune synapses (Figure 2).44 SAP also enhances adhesion between NK and target cells. On the other hand, EAT-2 controls NK cell function through the phospholipase Cg enzymes (PLC-g), Ca2+ fluxes, and the MAPK/Erk pathway, leading to granule polarization and the exocytosis of cytotoxic granules toward target cells (Figure 3).45 SAP and EAT-2 both prevent SLAMF receptors from interacting with inhibitory effectors such as SH2-domain-containing tyrosine phosphatase (SHP)-2, SHP-1, SH2 domain-containing 5′ inositol phosphatase (SHIP)-1, or C-terminal Src kinase (Csk).36,41 Hence, SLAMF receptors become inhibitory in the absence of SAP-related adaptors, suppressing the function of activating NK-cell receptors such as CD16, natural-killer group-2 member-D (NKG2D), and DNAX accessory molecule-1 (DNAM-1).32
Figure 2. Structure and function of SLAMF receptor in an immune synapse.
The SLAMF receptors are structurally characterized by IgV and IgC2 domains within an extracellular portion and one or more ITSMs, depicted as a closed rectangle, within the cytoplasmic portion. The mostly homophilic interactions between SLAMF receptors result in their costimulatory effects on TCR/CD3 complex signaling pathway. When the SLAMF receptor is engaged by its ligand, cytoplasmic domain ITSMs with tyrosine-based motifs undergo phosphorylation, recruiting adaptors proteins, SAP or EAT-2. SAP can then recruit the Src family protein tyrosine kinase Fyn or Lck, which is important for activation via SLAM family receptors. The coupling of EAT-2 carboxyl-terminal tail to the PLC-γ SH2 domains leads to an additional activation pathway. ITSM-like motif (non-ITSM) depicted as an unfilled rectangle does not bind SAP or EAT-2. SAP is mostly expressed in T cells, while EAT-2 is primarily expressed in antigen-presenting cells.
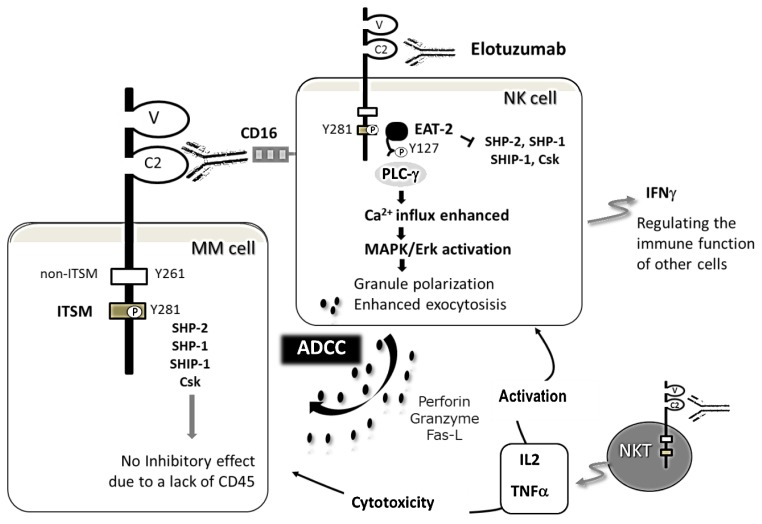
Figure 3. Effect of elotuzumab to NK, NKT, and MM cells.
The primary mechanism of action of elotuzumab is NK cell-mediated ADCC against MM cells. Elotuzumab also directly activates NK and NKT cells, but not MM cells, by its engagement with SLAMF7. This effect results in phosphorylation of tyrosine 281 (Y281) located in ITSMs, thereby recruiting a SLAM-associated adaptor EAT-2. EAT-2 binds to the SH2 domains of PLC-g, and leads to enhanced Ca2+ influx and MAPK/Erk pathway activation, finally resulting in granule polarization and enhanced exocytosis in NK cells. Tyrosine 261 (Y261), needed for the inhibitory function of mouse SLAMF7, is conserved in human SLAMF7.31 NKT cells are also activated via elotuzumab binding, resulting in the accelerated secretion of IL2 and TNFa, which induces the cytotoxicity of NK cells against MM cells.64 Elotuzumab binds to the proximal IgC2 domain of SLAMF 7.
The SAP gene located at Xq25 was identified as the causative gene altered in X-linked lymphoproliferative syndrome (XLP).46,47 Germline mutations or deletions in SAP have been implicated in XLP, resulting in aberrant functions of SLAMF1.48,49 Aberrant functions of SLAMF1, 2, and 6 caused by SAP mutations result in extreme sensitivity to EBV infection in patients with XLP. EBV-specific cytotoxic CD8+ T cells in XLP exhibit defects in the cytolysis of EBV-infected B cells. They escape an apoptotic death, which results in the uncontrolled proliferation of B cells and T cells, thereby causing fulminant infectious mononucleosis (60%), lymphomas (30%), and dysgammaglobulinemia (30%).48,50
Expression of SLAMF7 in Normal Cells, MM, and other Hematological Malignancies
Expression of SLAMF7 in normal cells and MM cells
SLAMF7 is expressed on NK cells, NKT cells, a subset of cytotoxic T-lymphocytes (CTLs) including CD8+ and CD4+ cells, mature dendritic cells (DCs), and activated B cells, regulating T- and B-cell functions. (Table 2).27,31–33,51,52 Normal plasma cells also highly express SLAMF7 at the mRNA and protein levels.13,14 SLAMF7 is not expressed in resting B cells, monocytes, granulocytes, or hematopoietic stem cells.13,14,36 On the other hand, SLAMF7 is highly expressed in neoplastic plasma cells from more than 95% of patients with MM, plasmacytoma,13,14 and plasma cell leukemia (PCL). It is also expressed in CD138 purified plasma cells from patients with monoclonal gammopathy of undetermined significance (MGUS) and smoldering MM (SMM).14 There have been no studies describing the higher expression of SLAMF7 in MM than in normal plasma cells. Soluble SLAMF7 (sSLMF7) lacking transmembrane and cytoplasmic domains was detected in patients with MM, particularly at advanced stages, but not in those with MGUS or healthy individuals.14 The role of sSLMF7 in myeloma cell pathophysiology remains to be elucidated.
Although SLAMF7 expression level in MM cells were independent of the cytogenetic subtypes of MM, one of the highest expression levels was found in t(4;14)-positive MM.13 A recent study demonstrated that the knockdown of SLAMF7 induced cell cycle G1 arrest or apoptosis, and also reduced colony formation in t(4;14) MM cells.53 Overexpressed SLAMF7 in t(4;14)-positive MM cell lines was down-regulated by MMSET shRNAs.53 These findings suggest a direct effect on the transcription of SLAMF7 by the MMSET protein. Although the mechanisms underlying the upregulation in plasma cells and MM cells currently remain unclear, a recent study demonstrated that SLAMF7 transcription was positively regulated by Blimp-1 (B lymphocyte-induced maturation protein-1) in NK cells and B cells.54 Blimp-1 is a known transcriptional repressor in macrophages, NK cells, B cells, T cells, and skin epithelial cells. Plasma cell function is controlled by Blimp-1 through the regulation of immunoglobulin secretion and the unfolded protein response.55
Expression of SLAMF7 in other hematological malignancies
Most of B-cell lymphomas including various histological subtypes and Hodgkin lymphoma do not express SLAMF7, as assessed by immunohistochemistry (IHC). Neither acute myeloid leukemias nor lymphoblastic leukemias express SLAMF7.13 The SLAMF7 protein was detected in 25% of peripheral T-cell lymphomas (PTCL) at a modest level using IHC. PTCL is a heterogeneous disease, but generally shows the CD4-positive phenotype. Using IHC, we identified various CD4+ Th subsets (Th1, Th2, Th17, and Treg) as possible normal counterparts of PTCL based on the expression of master regulators such as T-bet, GATA3, BCL6, RORγt, and FOXP3.56 These findings suggest that some functional subsets of CD4+ T cells expressing SLAMF7 exist. Recent studies demonstrated the clonal expansion of CD4+ CTLs expressing SLAMF7, granzyme A, IL-1β, and TGF-β1, at inflamed tissue sites of IgG4-related disease.52 Although CD4+ CTLs may develop from naïve T (Th0) and various Th subsets, Th1 cells regulated by T-bet represent the majority of CD4+ CTLs secreting IFN-g.57 CD4+ CTLs have been detected among peripheral blood lymphocytes under conditions of chronic viral infections and during antitumor responses.58,59
Dual Immunotherapeutic Mechanism of Elotuzumab
Elotuzumab induces NK cell-mediated ADCC
Elotuzumab is a humanized immunoglobulin G1 kappa (IgG1k) monoclonal antibody, that binds a unique epitope on the IgC2 domain of SLAMF7.13,14 Human IgG1 elicits ADCC and complement-dependent cytotoxicity (CDC) activities. However, elotuzumab and the novel anti-SLAMF7 mAb PDL241 did not mediate CDC.60,61 Elotuzumab-induced ADCC is mediated through the engagement of its Fc portion with FcgRIIIa/CD16 on NK cells.14,61 On the other hand, elotuzumab is unable to directly suppress the growth of MM cells. In MM cells lacking EAT-2, inhibitory molecules including SHP-2, SHP-1, SHIP-1, and Csk are recruited to the phosphorylated ITSMs of SLAMF7.62 However, inhibitory effects are not induced in MM cells, partly due to a lack of CD45. Elotuzumab also does not induce the proliferation of myeloma cells (Figure 3).45,62
Preclinical studies demonstrated that elotuzumab strongly induced cytotoxicity in established MM cell lines and primary samples including bortezomib-resistant MM cells when incubated with peripheral blood mononuclear cells (PBMCs) or purified NK cells.63 This anti-myeloma effect of elotuzumab was prevented when CD16 was inhibited.64 Elotuzumab alone does not affect the viability of MM cells without PBMCs or purified NK cells in vitro. SLAMF7 may also potentiate interactions between NK and target MM cells through its homotypic engagement recognizing the distal epitope IgV.65 NK cells activated by elotuzumab do not show cytotoxicity against autologous NK cells.14 In mice, the interaction between NK cells by the SLAMF7 engagement may enhance their function.36
Elotuzumab directly stimulates NK cells
Elotuzumab directly enhances the cytotoxic activity of NK cells in addition to primarily inducing ADCC against MM cells, giving rise to a dual immunotherapeutic mechanism of action.13,14,63 NK cell activation is mediated by the SLAMF adaptor proteins EAT-2 and SAP, the cooperated expression of which promotes the cytotoxic activity of NK cells. NK cell cytotoxicity is also dependent on PLCg1 and PLCg2.66 SAP promotes and stabilizes adhesion between NK cells and target cells in a dual manner: one is by the coupling of SLAMF receptors to the protein tyrosine kinase Fyn, and the other is by preventing SLAMF receptors from coupling inhibitory signals involving SHIP and SHP-1.67,68 On the other hand, EAT-2 does not enhance adhesion between NK and target cells, but controls NK cell function through PLCg, Ca2+ fluxes, and the MAPK/Erk pathway, leading to granule polarization and the exocytosis of cytotoxic granules toward target cells (Figure 3).45 NKT cells are also activated via elotuzumab binding, resulting in the accelerated secretion of IL2 and TNFa, which induces the cytotoxicity of NK cells against MM cells (Figure 3).64 While most SLAMF receptors bind SAP and EAT-2,35 SLAMF7 is functionally controlled by EAT-2, not SAP.34,35
A previous study showed that lenalidomide augmented elotuzumab-induced ADCC against MM cells in vitro.14,64 The enhanced NK cell function was associated with the up-regulation of IL-2Rα expression, IL-2 production by CD3+CD56+ lymphocytes including NKT cells, and TNFα production.64 Augmentations in NK-cell cytotoxic activity were also demonstrated with pomalidomide.69,70 Low-dose bortezomib71 and carfilzomib72 also augmented NK-cell cytotoxic activity against MM cells. This effect was associated with the enhanced expression of the activating or co-activating molecules of NK cells including MHC class I polypeptide-related sequence A (MICA), NKG2D, and DNAM-1 ligands (PVR and Nectin-2). These findings may provide the rationale for combining these agents with elotuzumab. However, further studies are needed in order to delineate which and how immune cells other than NK cells are modulated in their function by elotuzumab.
Quantity and quality of NK cells in MM
The quantity and quality of effector cells including NK cells are essential for ADCC activity. Peripheral blood (PB) NK cell counts from MM patients increased or showed no changes in the earlier stages and decreased in the advanced stages.73–75 Patients with MGUS also showed no changes in PB NK cell counts form those of the controls.74,76,77 On the other hand, NK cell counts in bone marrow (BM) from MM patients were reported to increase.73,78 However, the functions of NK cells differ among their subsets. CD56brightCD16-/dim NK cells are mainly responsible for the production of cytokines, while CD56dimCD16+ NK cells are mainly responsible for cytotoxic activities.75 CD16+ subsets were decreased in MM patients.79
Regarding the quality of NK cells in MM patients, previous studies suggested that they were dysfunctional and showed decreased or no cytotoxicity in advanced MM, while they remained functional in MGUS.79–83 NK cell dysfunction is often associated with the down-regulated expression of activating molecules including natural cytotoxicity receptors, NKG2D, and SLAMF4 (2B4) in BM NK-cells.84 Other studies also demonstrated the down-regulated expression of SLAMF4 and DNAM-1 in NK cells, and this was associated with a reduction in NK cell cytotoxicity against MM.83,85 MM cells escape NK cell cytotoxicity due to the lack of a HLA Class I loss, the shedding of surface MICA, and circulating MICA, which result in the down-regulation of NKG2D. NK cells from MM patients also express programmed death protein 1 (PD-1), which results in escape from immune surveillance.86,87 In mouse tumor models, an anti-PD-1 antibody enhances elotuzumab efficacy due to the production of tumor-infiltrating NK and CD8+ T cell activity.88 These findings may provide the rationale for combination therapy of elotuzumab and PD-1 blockade.
Response to elotuzumab and the polymorphism of FcgRIIIa/CD16
The FcgRIIIa/CD16 genotype may provide some guidance for the administration of elotuzumab to patients who are expected to have a favorable response. Since the allelic variation affects the affinity of FcgRIIIa for IgG1 antibodies, differential responses to mAb have been reported to correlate with specific polymorphisms.89,90 The presence of a valine (V) at position 158 of FcgRIIIa is associated with high-affinity to the Fc portion of IgG1 mAb, in contrast to phenylalanine (F) with low affinity. The high-affinity “VV” genotype of FcgRIIIa has been associated with enhanced ADCC in rituximab treatments for patients with follicular lymphoma.91,92
In a randomized phase II study of EBd versus Bd for RRMM, patients homozygous for the high-affinity FcgRIIIa V allele (VV) showed longer survival than those who were homozygous for the low-affinity FcgRIIIa F allele (FF).24 A subanalysis of PFS by the CD16a genotype showed no significant difference between VV and FF in ELOQUENT-2. A difference was noted between VV/VF and FF in the study of elotuzumab monotherapy, although the interpretation of this finding is limited by the small number of patients with each genotype.93 The incidence of the high-affinity VV allele is 59% in the Japanese population versus 17% in the populations of Western countries.24,94 In Japanese patients, the genetic FcgRIIIa-V158F polymorphism may have a significant impact on myeloma cell killing by ADCC.
Optimal use of ELd for the Treatment of RRMM
Three factors need to be considered in order to achieve better outcomes using ELd: risk of the disease, frailty of the patients, and the quantity and quality of effector cells. Prior to introducing elotuzumab, many patients were treated with lenalidomide-based regimens until disease progression as first-line therapy, and were lenalidomide refractory at the time of first relapse. Since elotuzumab is approved in combination with Ld for the treatment of RRMM, there are two possible conditions under which to administer elotuzumab: starting ELd as second-line later treatment or adding elotuzumab to Ld ongoing as first-line or later treatment. In the case of second-line or later treatment, patients with PR, VGPR, or CR using Ld may be the ideal candidates for the addition of elotuzumab. This is because PFS by tumor responses between the ELd and Ld groups was significantly better in patients who achieved PR or better than in patients with a minor response or SD in ELOQUENT-2.8
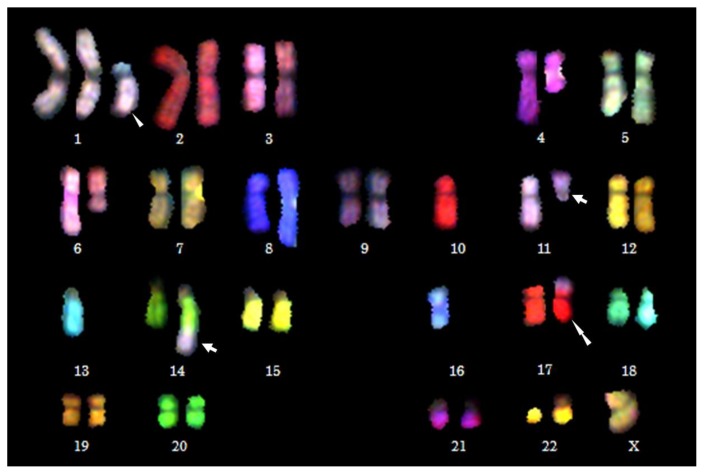
According to the ELOQUENT-2 study, elotuzumab is beneficial for patients with high-risk CA including del(17p), 1q21 gain/amplification, and particularly t(4;14). A direct effect on SLAMF7 transcription by the MMSET protein has provided the rationale to use elotuzumab for t(4;14)-positive MM patients.53 Secondary CA may impact adversely on treatment outcomes and survival in both NDMM and RRMM regardless of the primary high-risk CA (Table 3). For example, t(11;14) is not necessarily associated with a good, but with a poor prognosis when identified concomitantly with a high-risk secondary CA, such as 1q21 gain/amplification and del(17p) (Figure 4).95 In the novel agent era, chromosomal rearrangements at 8q24 is also high-risk CA.96,97 We previously detected 8q24 rearrangements involving MYC or PVT1 (plasmacytoma variant translocation 1) loci in 24% of patients with MM.98
Table 3.
SLAM family receptors: their expression and interaction with adaptor proteins.
| SLAMF Receptors | Ligands | Expression | Interaction with | Effectors | ||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Normal | MM | SAP | EAT-2 | |||
| SLAMF1 | SLAMF1 measles virus | T, B, DCs, Mϕ, Platelets | + | + | + | Fyn, Lck, SHIP1, Dok1, PKCθ, Akt |
|
| ||||||
| SLAMF2 | SLAMF4 CD2 | Lymphocytes, Immune cells, DCs, Endothelial cells | ++ | − | − | Fyn, Lck |
|
| ||||||
| SLAMF3 | SLAMF3 | T, B, NK, DCs, Mϕ | +++ | + | + | Fyn, Lck, ERK, AP2, Grb2 |
|
| ||||||
| SLAMF4 | SLAMF2 | NK, NKT, D8+ T, DCs, Mϕ, Eosinophils | + | + | + | Fyn, Lck, LAT, PI3K, Vav1, SHIP1, cCbl, ERK, p38, SHP1, SHP2 |
|
| ||||||
| SLAMF5 | SLAMF5 | T, B, NK, DCs, Mϕ, Granulocytes, Platelets, Mast cells, Eosinophils | + | + | + | Fyn, Lck |
|
| ||||||
| SLAMF6 | SLAMF6 | T, B, NK, DCs, Neutrophils | ++ | + | + | Fyn, Lck, PLC-γ, PI3K, SHP1, cCbl, Vav1 |
|
| ||||||
| SLAMF7 | SLAMF7 | NK, NKT, T, Plasma cells, B, DCs, Mϕ | +++ | − | + | Fyn, Lck, PLC-γ, Vav1, PI3K |
|
| ||||||
| SLAMF8 | SLAMF8 | Granulocytes, Mϕ, Monocytes, DCs | unknown | − | − | unknown |
|
| ||||||
| SLAMF9 | unknown | Immune cells | unknown | − | − | unknown |
SAP, SLAM-associated protein; EAT-2, EWS-Fli1-activated transcript-2; T, T cells; B, B cells; NKT, natural killer-T cells; NK, natural killer cells; DCs, dendritic cells; Mϕ, macrophages
Figure 4. Chromosomal abnormalities in a patient with t(11;14)-positive primary refractory PCL detected by multicolor spectral karyotyping (SKY).
(Goto M, Taniwaki M, et al. unpublished). SKY is performed as described as previously.98 Arrows indicate a balanced translocation t(11;14)(q13;q32). Three secondary CA are detected in this patient: der(1)t(1;16)(q10;p10) indicated by an arrowhead, monsomy 13 (−13), and der(17)t(4;17)(?;p13) indicated by a double arrowhead. Unbalanced translocations, der(1)t(1;16)(q10;p10) and der(17)t(4;17)(?;p13), result in 1q gain and 17p deletion, respectively, which are high-risk secondary CA in MM (Goto M, Taniwaki M, et al. unpublished).
Taking the modes of action of elotuzumab into consideration, the counts and functions of immune cells, particularly NK cells are crucial as already mentioned. In this regard, the findings of Phase II and III trails in patients with SMM have been encouraging. Elotuzumab monotherapy may delay progression to MM in patients with SMM, resulting in favorable PFS, because most patients achieved the best overall response of SD or MR, with ≥MR in 29% including PR in 10%.99 Early treatments with Ld in patients with high-risk SMM provided a significant benefit over observations in terms of time to progression.100 Since elotuzumab is well tolerated with minimal toxicity, elderly or frail patients who are ineligible for PI/MiD-based triplet therapy or transplantation are suitable candidates for ELd treatment. Moreover, the addition of elotuzumab to bortezomib, lenalidomide, and dexamethasone (LBd) is feasible without major additive AEs beyond what is already known about LBd, as demonstrated in SWOGS1211 trial.101 However, the efficacy of elotuzumab in combination with LBd needs to be studied.
Conclusions
A number of molecular targeting agents are currently available for MM; therefore, risk stratification and frailty assessments are critical for their optimal combination. Secondary CA are effective biomarkers, and more than 50% of patients are unfit because they are older than 75 years. However, even with the use of novel agents, MM remains incurable with recurrence and refractoriness to treatment, and frequently develops extramedullary disease and secondary plasma cell leukemia (sPCL) at the end stages of the disease. Although a number of clinical trials have attempted to achieve high tumor responses in RRMM using novel triplet therapy with second- and third-generation PIs and IMiDs, difficulties are associated with successfully treating extramedullary lesions and sPCL. Therefore, it is important not only to develop treatments with high tumor responses, but also to have early therapeutic interventions for MM. Moreover, 30–50% of MM patients are transplant-ineligible or unable to receive PI/IMiDs based triplets therapy.102,103 Hence, elotuzumab is promising and beneficial for the treatment of frail patients with MM.
The mechanisms of action of elotuzumab and the functional role of SLAMF7 in relation to pathophysiology of MM remain unclear. For example, what the signal transduction pathway of engaged SLAMF7 in MM cells is involved in is unknown, and which or how immune cells other than NK cells are implicated in killing MM cells has yet to be elucidated by elotuzumab. It will be beneficial for patients with RRMM to clarify whether elotuzumab has a marked impact on the recovery of immune paralysis in combination with other novel molecular targeting agents such as carfilzomib and pomalidomide. In order to address these questions, basic research is conducted to investigate the molecular mechanisms involving SLAMF receptors and SAP-related adaptors with their downstream molecules in the signal transduction pathway.
The efficacy of ELd with minimal toxicity and the paralytic immune state in advanced MM may bring forward for consideration of early therapeutic intervention in patients with SMM. However, studies are needed in order to clarify whether ELd is effective for patients with SMM.
Acknowledgments
This work was supported in part by a Grant-in-Aid for Scientific Research from The Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT KAKENHI 16K09856) (MT); by the National Cancer Center Research and Development Fund (29-A-3); by a grant (Practical Research for Innovative Cancer Control) from the Japan Agency for Medical Research and Development (AMED) (17ck0106348h0001); and by the Takeda Science Foundation and Astra Zeneca (JK).
Footnotes
Competing interests: The authors have declared that no competing interests exist.
References
- 1.Cancer Stat Facts: Myeloma. National Cancer Institute, Surveillance, Epidemiology, and End Results Program; [Accessed 25 December 2017]. https://seer.cancer.gov/statfacts/html/mulmy.html. [Google Scholar]
- 2.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D, Bray F. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–403. doi: 10.1016/j.ejca.2012.12.027. https://doi.org/10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 3.Center for Cancer Control and Information Service, National Cancer Center, Japan. Dec 25, 2017. https://ganjoho.jp/reg_stat/statistics/stat/summary.html.
- 4.Kumar SK, Rajkumar SV, Dispenzieri A, Lacy MQ, Hayman SR, Buadi FK, Zeldenrust SR, Dingli D, Russell SJ, Lust JA, Greipp PR, Kyle RA, Gertz MA. Improved survival in multiple myeloma and the impact of novel therapies. Blood. 2008;111:2516–20. doi: 10.1182/blood-2007-10-116129. https://doi.org/10.1182/blood-2007-10-116129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar SK, Dispenzieri A, Lacy MQ, Gertz MA, Buadi FK, Pandey S, Kapoor P, Dingli D, Hayman SR, Leung N, Lust J, McCurdy A, Russell SJ, Zeldenrust SR, Kyle RA, Rajkumar SV. Continued improvement in survival in multiple myeloma: changes in early mortality and outcomes in older patients. Leukemia. 2014;28:1122–28. doi: 10.1038/leu.2013.313. https://doi.org/10.1038/leu.2013.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benboubker L, Dimopoulos MA, Dispenzieri A, Catalano J, Belch AR, Cavo M, Pinto A, Weisel K, Ludwig H, Bahlis N, Banos A, Tiab M, Delforge M, Cavenagh J, Geraldes C, Lee JJ, Chen C, Oriol A, de la Rubia J, Qiu L, White DJ, Binder D, Anderson K, Fermand JP, Moreau P, Attal M, Knight R, Chen G, Van Oostendorp J, Jacques C, Ervin-Haynes A, Avet-Loiseau H, Hulin C, Facon T FIRST Trial Team. Lenalidomide and dexamethasone in transplant-ineligible patients with myeloma. N Engl J Med. 2014;371:906–17. doi: 10.1056/NEJMoa1402551. https://doi.org/10.1056/NEJMoa1402551. [DOI] [PubMed] [Google Scholar]
- 7.Palumbo A, Gay F, Cavallo F, Di Raimondo F, Larocca A, Hardan I, Nagler A, Petrucci MT, Hajek R, Pezzatti S, Delforge M, Patriarca F, Donato F, Cerrato C, Nozzoli C, Yu Z, Boccadifuoco L, Caravita T, Benevolo G, Guglielmelli T, Vincelli D, Jacques C, Dimopoulos MA, Ciccone G, Musto P, Corradini P, Cavo M, Boccadoro M. Continuous therapy versus fixed duration of therapy in patients with newly diagnosed multiple myeloma. J Clin Oncol. 2015;33:3459–66. doi: 10.1200/JCO.2014.60.2466. [DOI] [PubMed] [Google Scholar]
- 8.Lonial S, Dimopoulos M, Palumbo A, White D, Grosicki S, Spicka I, Walter-Croneck A, Moreau P, Mateos MV, Magen H, Belch A, Reece D, Beksac M, Spencer A, Oakervee H, Orlowski RZ, Taniwaki M, Röllig C, Einsele H, Wu KL, Singhal A, San-Miguel J, Matsumoto M, Katz J, Bleickardt E, Poulart V, Anderson KC, Richardson P ELOQUENT-2 Investigators. Elotuzumab Therapy for relapsed or refractory multiple myeloma. N Engl J Med. 2015;373:621–31. doi: 10.1056/NEJMoa1505654. https://doi.org/10.1056/NEJMoa1505654. [DOI] [PubMed] [Google Scholar]
- 9.Palumbo A, Chanan-Khan A, Weisel K, Nooka AK, Masszi T, Beksac M, Spicka I, Hungria V, Munder M, Mateos MV, Mark TM, Qi M, Schecter J, Amin H, Qin X, Deraedt W, Ahmadi T, Spencer A, Sonneveld P CASTOR Investigators. Daratumumab, bortezomib, and dexamethasone for multiple myeloma. N Engl J Med. 2016;375:754–66. doi: 10.1056/NEJMoa1606038. https://doi.org/10.1056/NEJMoa1606038. [DOI] [PubMed] [Google Scholar]
- 10.Dimopoulos MA, Oriol A, Nahi H, San-Miguel J, Bahlis NJ, Usmani SZ, Rabin N, Orlowski RZ, Komarnicki M, Suzuki K, Plesner T, Yoon SS, Ben Yehuda D, Richardson PG, Goldschmidt H, Reece D, Lisby S, Khokhar NZ, O’Rourke L, Chiu C, Qin X, Guckert M, Ahmadi T, Moreau P POLLUX Investigators. Daratumumab, lenalidomide, and dexamethasone for multiple myeloma. N Engl J Med. 2016;375:1319–31. doi: 10.1056/NEJMoa1607751. https://doi.org/10.1056/NEJMoa1607751. [DOI] [PubMed] [Google Scholar]
- 11.Chari A, Suvannasankha A, Fay JW, Arnulf B, Kaufman JL, Ifthikharuddin JJ, Weiss BM, Krishnan A, Lentzsch S, Comenzo R, Wang J, Nottage K, Chiu C, Khokhar NZ, Ahmadi T, Lonial S. Daratumumab plus pomalidomide and dexamethasone in relapsed and/or refractory multiple myeloma. Blood. 2017;130:974–81. doi: 10.1182/blood-2017-05-785246. https://doi.org/10.1182/blood-2017-05-785246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Badros A, Hyjek E, Ma N, Lesokhin A, Dogan A, Rapoport AP, Kocoglu M, Lederer E, Philip S, Milliron T, Dell C, Goloubeva O, Singh Z. Pembrolizumab, pomalidomide, and low-dose dexamethasone for relapsed/refractory multiple myeloma. Blood. 2017;130:1189–97. doi: 10.1182/blood-2017-03-775122. https://doi.org/10.1182/blood-2017-03-775122. [DOI] [PubMed] [Google Scholar]
- 13.Hsi ED, Steinle R, Balasa B, Szmania S, Draksharapu A, Shum BP, Huseni M, Powers D, Nanisetti A, Zhang Y, Rice AG, van Abbema A, Wong M, Liu G, Zhan F, Dillon M, Chen S, Rhodes S, Fuh F, Tsurushita N, Kumar S, Vexler V, Shaughnessy JD, Jr, Barlogie B, van Rhee F, Hussein M, Afar DE, Williams MB. CS1, a potential new therapeutic antibody target for the treatment of multiple myeloma. Clin Cancer Res. 2008;14:2775–84. doi: 10.1158/1078-0432.CCR-07-4246. https://doi.org/10.1158/1078-0432.CCR-07-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tai Y-T, Dillon M, Song W, Leiba M, Li XF, Burger P, Lee AI, Podar K, Hideshima T, Rice AG, van Abbema A, Jesaitis L, Caras I, Law D, Weller E, Xie W, Richardson P, Munshi NC, Mathiot C, Avet-Loiseau H, Afar DE, Anderson KC. Anti-CS1 humanized monoclonal antibody HuLuc63 inhibits myeloma cell adhesion and induces antibody-dependent cellular cytotoxicity in the bone marrow milieu. Blood. 2008;112:1329–37. doi: 10.1182/blood-2007-08-107292. https://doi.org/10.1182/blood-2007-08-107292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iida S, Tobinai K, Taniwaki M, Shumiya Y, Nakamura T, Chou T. Phase I dose escalation study of high dose carfilzomib monotherapy for Japanese patients with relapsed or refractory multiple myeloma. Int J Hematol. 2016;104:596–604. doi: 10.1007/s12185-016-2070-7. https://doi.org/10.1007/s12185-016-2070-7. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki K, Sunami K, Ohashi K, Iida S, Mori T, Handa H, Matsue K, Miyoshi M, Bleickardt E, Matsumoto M, Taniwaki M. Randomized phase 3 study of elotuzumab for relapsed or refractory multiple myeloma: ELOQUENT-2 Japanese patient subanalysis. Blood Cancer J. 2017;7(3):e540. doi: 10.1038/bcj.2017.18. https://doi.org/10.1038/bcj.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dimopoulos MA, Lonial S, White D, Moreau P, Palumbo A, San-Miguel J, Shpilberg O, Anderson K, Grosicki S, Spicka I, Walter-Croneck A, Magen H, Mateos MV, Belch A, Reece D, Beksac M, Bleickardt E, Poulart V, Sheng J, Sy O, Katz J, Singhal A, Richardson P. Elotuzumab plus lenalidomide/dexamethasone for relapsed or refractory multiple myeloma: ELOQUENT-2 follow-up and post-hoc analyses on progression-free survival and tumour growth. Br J Haematol. 2017;178:896–905. doi: 10.1111/bjh.14787. https://doi.org/10.1111/bjh.14787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dimopoulos MA, Lonial S, White D, Moreau P, Mateos MV, San-Miguel J, Anderson KC, Shpilberg O, Grosicki S, Spicka I, Walter-Croneck A, Magen H, Belch A, Reece DE, Beksac M, Mekan S, Sy O, Anil K, Singhal AK, Richardson PG, Weisel K. In: Phase 3 ELOQUENT-2 Study: Extended 4-year follow-up of elotuzumab plus lenalidomide/dexamethasone vs lenalidomide/dexamethasone in relapsed/refractory multiple myeloma. Dimopoulos M, editor. EHA Learning Center; Jun 24, 2017. p. 181743. (Abstract release date: May 18, 2017) https://learningcenter.ehaweb.org/eha/2017/22nd/181743/meletios.a.dimopoulos.phase.3.eloquent-2.study.extended.4-year.follow-up.of.html. [Google Scholar]
- 19.Zonder JA, Mohrbacher AF, Singhal S, van Rhee F, Bensinger WI, Ding H, Fry J, Afar DE, Singhal AK. A phase 1, multicenter, open-label, dose escalation study of elotuzumab in patients with advanced multiple myeloma. Blood. 2012;120:552–9. doi: 10.1182/blood-2011-06-360552. https://doi.org/10.1182/blood-2011-06-360552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakubowiak AJ, Benson DM, Bensinger W, Siegel DS, Zimmerman TM, Mohrbacher A, Richardson PG, Afar DE, Singhal AK, Anderson KC. Phase I trial of anti-CS1 monoclonal antibody elotuzumab in combination with bortezomib in the treatment of relapsed/refractory multiple myeloma. J Clin Oncol. 2012;30:1960–5. doi: 10.1200/JCO.2011.37.7069. https://doi.org/10.1200/JCO.2011.37.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lonial S, Vij R, Harousseau JL, Facon T, Moreau P, Mazumder A, Kaufman JL, Leleu X, Tsao LC, Westland C, Singhal AK, Jagannath S. Elotuzumab in combination with lenalidomide and low-dose dexamethasone in relapsed or refractory multiple myeloma. J Clin Oncol. 2012;30:1953–9. doi: 10.1200/JCO.2011.37.2649. https://doi.org/10.1200/JCO.2011.37.2649. [DOI] [PubMed] [Google Scholar]
- 22.Richardson PG, Jagannath S, Moreau P, Jakubowiak AJ, Raab MS, Facon T, Vij R, White D, Reece DE, Benboubker L, Zonder J, Tsao LC, Anderson KC, Bleickardt E, Singhal AK, Lonial S 1703 study investigators. Elotuzumab in combination with lenalidomide and dexamethasone in patients with relapsed multiple myeloma: final phase 2 results from the randomised, open-label, phase 1b-2 dose-escalation study. Lancet Haematol. 2015;2(12):e516–e27. doi: 10.1016/S2352-3026(15)00197-0. https://doi.org/10.1016/S2352-3026(15)00197-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berenson J, Manges R, Badarinath S, Cartmell A, McIntyre K, Lyons R, Harb W, Mohamed H, Nourbakhsh A, Rifkin R. A phase 2 safety study of accelerated elotuzumab infusion, over less than 1 h, in combination with lenalidomide and dexamethasone, in patients with multiple myeloma. Am J Hematol. 2017;92:460–6. doi: 10.1002/ajh.24687. https://doi.org/10.1002/ajh.24687. [DOI] [PubMed] [Google Scholar]
- 24.Jakubowiak A, Offidani M, Pégourie B, De La Rubia J, Garderet L, Laribi K, Bosi A, Marasca R, Laubach J, Mohrbacher A, Carella AM, Singhal AK, Tsao LC, Lynch M, Bleickardt E, Jou YM, Robbins M, Palumbo A. Randomized phase 2 study: elotuzumab plus bortezomib/dexamethasone vs bortezomib/dexamethasone for relapsed/refractory MM. Blood. 2016;127:2833–40. doi: 10.1182/blood-2016-01-694604. https://doi.org/10.1182/blood-2016-01-694604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mateos MV, Granell M, Oriol A, Martinez-Lopez J, Blade J, Hernandez MT, Martín J, Gironella M, Lynch M, Bleickardt E, Paliwal P, Singhal A, San-Miguel J. Elotuzumab in combination with thalidomide and low-dose dexamethasone: a phase 2 single-arm safety study in patients with relapsed/refractory multiple myeloma. Br J Haematol. 2016;175:448–56. doi: 10.1111/bjh.14263. https://doi.org/10.1111/bjh.14263. [DOI] [PubMed] [Google Scholar]
- 26.Boles KS, Mathew PA. Molecular cloning of CS1, a novel human natural killer cell receptor belonging to the CD2 subset of the immunoglobulin superfamily. Immunogenetics. 2001;52:302–7. doi: 10.1007/s002510000274. https://doi.org/10.1007/s002510000274. [DOI] [PubMed] [Google Scholar]
- 27.Lee JK, Mathew SO, Vaidya SV, Kumaresan PR, Mathew PA. CS1 (CRACC, CD319) induces proliferation and autocrine cytokine expression on human B lymphocytes. J Immunol. 2007;179:4672–8. doi: 10.4049/jimmunol.179.7.4672. https://doi.org/10.4049/jimmunol.179.7.4672. [DOI] [PubMed] [Google Scholar]
- 28.Cannons JL, Tangye SG, Schwartzberg PL. SLAM family receptors and SAP adaptors in immunity. Annu Rev Immunol. 2011;29:665–705. doi: 10.1146/annurev-immunol-030409-101302. https://doi.org/10.1146/annurev-immunol-030409-101302. [DOI] [PubMed] [Google Scholar]
- 29.Boles KS, Stepp SE, Bennett M, Kumar V, Mathew PA. 2B4 (CD244) and CS1: novel members of the CD2 subset of the immunoglobulin superfamily molecules expressed on natural killer cells and other leukocytes. Immunol Rev. 2001;181:234–49. doi: 10.1034/j.1600-065x.2001.1810120.x. https://doi.org/10.1034/j.1600-065X.2001.1810120.x. [DOI] [PubMed] [Google Scholar]
- 30.Schwartzberg PL, Mueller KL, Qi H, Cannons JL. SLAM receptors and SAP influence lymphocyte interactions, development and function. Nat Rev Immunol. 2009;9:39–46. doi: 10.1038/nri2456. https://doi.org/10.1038/nri2456. [DOI] [PubMed] [Google Scholar]
- 31.Veillette A, Guo H. CS1, a SLAM family receptor involved in immune regulation, is a therapeutic target in multiple myeloma. Crit Rev Oncol Hematol. 2013;88:168–7. doi: 10.1016/j.critrevonc.2013.04.003. https://doi.org/10.1016/j.critrevonc.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Veillette A. SLAM-family receptors: immune regulators with or without SAP-family adaptors. Cold Spring Harb Perspect Biol. 2010;2(3):a002469. doi: 10.1101/cshperspect.a002469. https://doi.org/10.1101/cshperspect.a002469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin M, Romero X, de la Fuente MA, Tovar V, Zapater N, Esplugues E, Pizcueta P, Bosch J, Engel P. CD84 functions as a homophilic adhesion molecule and enhances IFN-gamma secretion: adhesion is mediated by Ig-like domain 1. J Immunol. 2001;167:3668–76. doi: 10.4049/jimmunol.167.7.3668. https://doi.org/10.4049/jimmunol.167.7.3668. [DOI] [PubMed] [Google Scholar]
- 34.Bouchon A, Cella M, Grierson HL, Cohen JI, Colonna M. Activation of NK cell-mediated cytotoxicity by a SAP-independent receptor of the CD2 family. J Immunol. 2001;167:5517–21. doi: 10.4049/jimmunol.167.10.5517. https://doi.org/10.4049/jimmunol.167.10.5517. [DOI] [PubMed] [Google Scholar]
- 35.Detre C, Keszei M, Romero X, Tsokos GC, Terhorst C. SLAM family receptors and the SLAM-associated protein (SAP) modulate T cell functions. Semin Immunopathol. 2010;32:157–71. doi: 10.1007/s00281-009-0193-0. https://doi.org/10.1007/s00281-009-0193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cruz-Munoz ME, Dong Z, Shi X, Zhang S, Veillette A. Influence of CRACC, a SLAM family receptor coupled to the adaptor EAT-2, on natural killer cell function. Nat Immunol. 2009;10:297–305. doi: 10.1038/ni.1693. https://doi.org/10.1038/ni.1693. [DOI] [PubMed] [Google Scholar]
- 37.Wilson TJ, Garner LI, Metcalfe C, King E, Margraf S, Brown MH. Fine specificity and molecular competition in SLAM family receptor signaling. PLoS One. 2014;9:e92184. doi: 10.1371/journal.pone.0092184. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J, Zhong MC, Guo H, Davidson D, Mishel S, Lu Y, Rhee I, Pérez-Quintero LA, Zhang S, Cruz-Munoz ME, Wu N, Vinh DC, Sinha M, Calderon V, Lowell CA, Danska JS, Veillette A. SLAMF7 is critical for phagocytosis of haematopoietic tumour cells via Mac-1 integrin. Nature. 2017;544:493–7. doi: 10.1038/nature22076. https://doi.org/10.1038/nature22076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al-Alem U, Li C, Forey N, Relouzat F, Fondanèche MC, Tavtigian SV, Wang ZQ, Latour S, Yin L. Impaired Ig class switch in mice deficient for the X-linked lymphoproliferative disease gene Sap. Blood. 2005;106:2069–75. doi: 10.1182/blood-2004-07-2731. https://doi.org/10.1182/blood-2004-07-2731. [DOI] [PubMed] [Google Scholar]
- 40.Kis LL, Nagy N, Klein G, Klein E. Expression of SH2D1A in five classical Hodgkin’s disease-derived cell lines. Int J Cancer. 2003;104:658–61. doi: 10.1002/ijc.10986. https://doi.org/10.1002/ijc.10986. [DOI] [PubMed] [Google Scholar]
- 41.Roncador G, García Verdes-Montenegro JF, Tedoldi S, Paterson JC, Klapper W, Ballabio E, Maestre L, Pileri S, Hansmann ML, Piris MA, Mason DY, Marafioti T. Expression of two markers of germinal center T cells (SAP and PD-1) in angioimmunoblastic T-cell lymphoma. Haematologica. 2007;92:1059–66. doi: 10.3324/haematol.10864. https://doi.org/10.3324/haematol.10864. [DOI] [PubMed] [Google Scholar]
- 42.Morra M, Lu J, Poy F, Martin M, Sayos J, Calpe S, Gullo C, Howie D, Rietdijk S, Thompson A, Coyle AJ, Denny C, Yaffe MB, Engel P, Eck MJ, Terhorst C. Structural basis for the interaction of the free SH2 domain EAT-2 with SLAM receptors in hematopoietic cells. EMBO J. 2001;20:5840–52. doi: 10.1093/emboj/20.21.5840. https://doi.org/10.1093/emboj/20.21.5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calpe S, Erdos E, Liao G, Wang N, Rietdijk S, Simarro M, Scholtz B, Mooney J, Lee CH, Shin MS, Rajnavölgyi E, Schatzle J, Morse HC, 3rd, Terhorst C, Lanyi A. Identification and characterization of two related murine genes, Eat2a and Eat2b, encoding single SH2-domain adapters. Immunogenetics. 2006;58:15–25. doi: 10.1007/s00251-005-0056-3. https://doi.org/10.1007/s00251-005-0056-3. [DOI] [PubMed] [Google Scholar]
- 44.Chan B, Lanyi A, Song HK, Griesbach J, Simarro-Grande M, Poy F, Howie D, Sumegi J, Terhorst C, Eck MJ. SAP couples Fyn to SLAM immune receptors. Nat Cell Biol. 2003;5:155–60. doi: 10.1038/ncb920. https://doi.org/10.1038/ncb920. [DOI] [PubMed] [Google Scholar]
- 45.Pérez-Quintero LA, Roncagalli R, Guo H, Latour S, Davidson D, Veillette A. EAT-2, a SAP-like adaptor, controls NK cell activation through phospholipase C?, Ca++, and Erk, leading to granule polarization. J Exp Med. 2014;211:727–42. doi: 10.1084/jem.20132038. https://doi.org/10.1084/jem.20132038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coffey AJ, Brooksbank RA, Brandau O, Oohashi T, Howell GR, Bye JM, Cahn AP, Durham J, Heath P, Wray P, Pavitt R, Wilkinson J, Leversha M, Huckle E, Shaw-Smith CJ, Dunham A, Rhodes S, Schuster V, Porta G, Yin L, Serafini P, Sylla B, Zollo M, Franco B, Bolino A, Seri M, Lanyi A, Davis JR, Webster D, Harris A, Lenoir G, de St Basile G, Jones A, Behloradsky BH, Achatz H, Murken J, Fassler R, Sumegi J, Romeo G, Vaudin M, Ross MT, Meindl A, Bentley DR. Host response to EBV infection in X-linked lymphoproliferative disease results from mutations in an SH2-domain encoding gene. Nat Genet. 1998;20:129–35. doi: 10.1038/2424. https://doi.org/10.1038/2424. [DOI] [PubMed] [Google Scholar]
- 47.Sayos J, Wu C, Morra M, Wang N, Zhang X, Allen D, van Schaik S, Notarangelo L, Geha R, Roncarolo MG, Oettgen H, De Vries JE, Aversa G, Terhorst C. The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM. Nature. 1998;395:462–9. doi: 10.1038/26683. https://doi.org/10.1038/26683. [DOI] [PubMed] [Google Scholar]
- 48.Wu C, Sayos J, Wang N, Howie D, Coyle A, Terhorst C. Genomic organization and characterization of mouse SAP, the gene that is altered in X-linked lymphoproliferative disease. Immunogenetics. 2000;51:805–15. doi: 10.1007/s002510000215. https://doi.org/10.1007/s002510000215. [DOI] [PubMed] [Google Scholar]
- 49.Calpe S, Wang N, Romero X, Berger SB, Lanyi A, Engel P, Terhorst C. The SLAM and SAP gene families control innate and adaptive immune responses. Adv Immunol. 2008;97:177–250. doi: 10.1016/S0065-2776(08)00004-7. https://doi.org/10.1016/S0065-2776(08)00004-7. [DOI] [PubMed] [Google Scholar]
- 50.Nelson DL, Terhorst C. X–linked lymphoproliferative syndrome. Clin Exp Immunol. 2000;122:291–5. doi: 10.1046/j.1365-2249.2000.01400.x. https://doi.org/10.1046/j.1365-2249.2000.01400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Veillette A. NK cell regulation by SLAM family receptors and SAP-related adapters. Immunol Rev. 2006;214:22–34. doi: 10.1111/j.1600-065X.2006.00453.x. https://doi.org/10.1111/j.1600-065X.2006.00453.x. [DOI] [PubMed] [Google Scholar]
- 52.Mattoo H, Mahajan VS, Maehara T, Deshpande V, Della-Torre E, Wallace ZS, Kulikova M, Drijvers JM, Daccache J, Carruthers MN, Castelino FV, Stone JR, Stone JH, Pillai S. Clonal expansion of CD4(+) cytotoxic T lymphocytes in patients with IgG4-related disease. J Allergy Clin Immunol. 2016;138:825–38. doi: 10.1016/j.jaci.2015.12.1330. https://doi.org/10.1016/j.jaci.2015.12.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xie Z, Gunaratne J, Cheong LL, Liu SC, Koh TL, Huang G, Blackstock WP, Chng WJ. Plasma membrane proteomics identifies biomarkers associated with MMSET overexpression in T(4;14) multiple myeloma. Oncotarget. 2013;4:1008–18. doi: 10.18632/oncotarget.1049. https://doi.org/10.18632/oncotarget.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim JR, Mathew SO, Mathew PA. Blimp-1/PRDM1 regulates the transcription of human CS1 (SLAMF7) gene in NK and B cells. Immunobiology. 2016;221:31–9. doi: 10.1016/j.imbio.2015.08.005. https://doi.org/10.1016/j.imbio.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 55.Tellier J, Shi W, Minnich M, Liao Y, Crawford S, Smyth GK, Kallies A, Busslinger M, Nutt SL. Blimp-1 controls plasma cell function through the regulation of immunoglobulin secretion and the unfolded protein response. Nat Immunol. 2016;17:323–30. doi: 10.1038/ni.3348. https://doi.org/10.1038/ni.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matsumoto Y, Horiike S, Ohshiro M, Yamamoto M, Sasaki N, Tsutsumi Y, Kobayashi T, Shimizu D, Uchiyama H, Kuroda J, Nomura K, Shimazaki C, Taniwaki M. Expression of master regulators of helper T-cell differentiation in peripheral T-cell lymphoma, not otherwise specified, by immunohistochemical analysis. Am J Clin Pathol. 2010;133:281–90. doi: 10.1309/AJCP0SBHYVLY5EML. https://doi.org/10.1309/AJCP0SBHYVLY5EML. [DOI] [PubMed] [Google Scholar]
- 57.Takeuchi A, Saito T. CD4 CTL, a Cytotoxic Subset of CD4+ T Cells, Their Differentiation and Function. Front Immunol. 2017;8:194. doi: 10.3389/fimmu.2017.00194. eCollection 2017. https://doi.org/10.3389/fimmu.2017.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xie Y, Akpinarli A, Maris C, Hipkiss EL, Lane M, Kwon EK, Muranski P, Restifo NP, Antony PA. Naive tumor-specific CD4(+) T cells differentiated in vivo eradicate established melanoma. J Exp Med. 2010;207:651–67. doi: 10.1084/jem.20091921. https://doi.org/10.1084/jem.20091921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quezada SA, Simpson TR, Peggs KS, Merghoub T, Vider J, Fan X, Blasberg R, Yagita H, Muranski P, Antony PA, Restifo NP, Allison JP. Tumor-reactive CD4(+) T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. J Exp Med. 2010;207:637–50. doi: 10.1084/jem.20091918. https://doi.org/10.1084/jem.20091918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Woo J, Vierboom MP, Kwon H, Chao D, Ye S, Li J, Lin K, Tang I, Belmar NA, Hartman T, Breedveld E, Vexler V, ‘t Hart BA, Law DA, Starling GC. PDL241, a novel humanized monoclonal antibody, reveals CD319 as a therapeutic target for rheumatoid arthritis. Arthritis Res Ther. 2013;15:R207. doi: 10.1186/ar4400. https://doi.org/10.1186/ar4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Collins SM, Bakan CE, Swartzel GD, Hofmeister CC, Efebera YA, Kwon H, Starling GC, Ciarlariello D, Bhaskar S, Briercheck EL, Hughes T, Yu J, Rice A, Benson DM., Jr Elotuzumab directly enhances NK cell cytotoxicity against myeloma via CS1 ligation: evidence for augmented NK cell function complementing ADCC. Cancer Immunol Immunother. 2013;62:1841–9. doi: 10.1007/s00262-013-1493-8. https://doi.org/10.1007/s00262-013-1493-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo H, Cruz-Munoz M-E, Wu N, Robbins M, Veillette A. Immune cell inhibition by SLAMF7 is mediated by a mechanism requiring src kinases, CD45, and SHIP-1 that is defective in multiple myeloma cells. Mol Cell Biol. 2015;35:41–51. doi: 10.1128/MCB.01107-14. https://doi.org/10.1128/MCB.01107-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Rhee F, Szmania SM, Dillon M, van Abbema AM, Li X, Stone MK, Garg TK, Shi J, Moreno-Bost AM, Yun R, Balasa B, Ganguly B, Chao D, Rice AG, Zhan F, Shaughnessy JD, Jr, Barlogie B, Yaccoby S, Afar DE. Combinatorial efficacy of anti-CS1 monoclonal antibody elotuzumab (HuLuc63) and bortezomib against multiple myeloma. Molecular Cancer Therapeutics. 2009;8:2616–24. doi: 10.1158/1535-7163.MCT-09-0483. https://doi.org/10.1158/1535-7163.MCT-09-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Balasa B, Yun R, Belmar NA, Fox M, Chao DT, Robbins MD, Starling GC, Rice AG. Elotuzumab enhances natural killer cell activation and myeloma cell killing through interleukin-2 and TNF-a pathways. Cancer Immunol Immunother. 2015;64:61–73. doi: 10.1007/s00262-014-1610-3. https://doi.org/10.1007/s00262-014-1610-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dong Z, Cruz-Munoz ME, Zhong MC, Chen R, Latour S, Veillette A. Essential function for SAP family adaptors in the surveillance of hematopoietic cells by natural killer cells. Nat Immunol. 2009;10:973–80. doi: 10.1038/ni.1763. https://doi.org/10.1038/ni.1763. [DOI] [PubMed] [Google Scholar]
- 66.Caraux A, Kim N, Bell SE, Zompi S, Ranson T, Lesjean-Pottier S, Garcia-Ojeda ME, Turner M, Colucci F. Phospholipase C-gamma2 is essential for NK cell cytotoxicity and innate immunity to malignant and virally infected cells. Blood. 2006;107:994–1002. doi: 10.1182/blood-2005-06-2428. https://doi.org/10.1182/blood-2005-06-2428. [DOI] [PubMed] [Google Scholar]
- 67.Kageyama R, Cannons JL, Zhao F, Yusuf I, Lao C, Locci M, Schwartzberg PL, Crotty S. The receptor Ly108 functions as a SAP adaptor-dependent on-off switch for T cell help to B cells and NKT cell development. Immunity. 2012;36:986–1002. doi: 10.1016/j.immuni.2012.05.016. https://doi.org/10.1016/j.immuni.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao F, Cannons JL, Dutta M, Griffiths GM, Schwartzberg PL. Positive and negative signaling through SLAM receptors regulate synapse organization and thresholds of cytolysis. Immunity. 2012;36:1003–16. doi: 10.1016/j.immuni.2012.05.017. https://doi.org/10.1016/j.immuni.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lagrue K, Carisey A, Morgan DJ, Chopra R, Davis DM. Lenalidomide augments actin remodeling and lowers NK-cell activation thresholds. Blood. 2015;126:50–60. doi: 10.1182/blood-2015-01-625004. https://doi.org/10.1182/blood-2015-01-625004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sehgal K, Das R, Zhang L, Verma R, Deng Y, Kocoglu M, Vasquez J, Koduru S, Ren Y, Wang M, Couto S, Breider M, Hansel D, Seropian S, Cooper D, Thakurta A, Yao X, Dhodapkar KM, Dhodapkar MV. Clinical and pharmacodynamic analysis of pomalidomide dosing strategies in myeloma: impact of immune activation and cereblon targets. Blood. 2015;125:4042–51. doi: 10.1182/blood-2014-11-611426. https://doi.org/10.1182/blood-2014-11-611426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Niu C, Jin H, Li M, Zhu S, Zhou L, Jin F, Zhou Y, Xu D, Xu J, Zhao L, Hao S, Li W, Cui J. Low-dose bortezomib increases the expression of NKG2D and DNAM-1 ligands and enhances induced NK and ?d T cell-mediated lysis in multiple myeloma. Oncotarget. 2017;18:5954–64. doi: 10.18632/oncotarget.13979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang G, Gao M, Zhang Y, Kong Y, Gao L, Tao Y, Han Y, Wu H, Meng X, Xu H, Zhan F, Wu X, Shi J. Carfilzomib enhances natural killer cell-mediated lysis of myeloma linked with decreasing expression of HLA class I. Oncotarget. 2015;6:26982–94. doi: 10.18632/oncotarget.4831. https://doi.org/10.18632/oncotarget.4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.García-Sanz R, González M, Orfão A, Moro MJ, Hernández JM, Borrego D, Carnero M, Casanova F, Bárez A, Jiménez R, Portero JA, San Miguel JF. Analysis of natural killer-associated antigens in peripheral blood and bone marrow of multiple myeloma patients and prognostic implications. Br J Haematol. 1996;93:81–8. doi: 10.1046/j.1365-2141.1996.4651006.x. https://doi.org/10.1046/j.1365-2141.1996.4651006.x. [DOI] [PubMed] [Google Scholar]
- 74.Omedé P, Boccadoro M, Gallone G, Frieri R, Battaglio S, Redoglia V, Pileri A. Multiple myeloma: increased circulating lymphocytes carrying plasma cell-associated antigens as an indicator of poor survival. Blood. 1990;76:1375–9. [PubMed] [Google Scholar]
- 75.Lopez-Verges S, Milush JM, Pandey S, York VA, Arakawa-Hoyt J, Pircher H, Norris PJ, Nixon DF, Lanier LL. CD57 defines a functionally distinct population of mature NK cells in the human CD56dimCD16+ NK-cell subset. Blood. 2010;116:3865–74. doi: 10.1182/blood-2010-04-282301. https://doi.org/10.1182/blood-2010-04-282301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tienhaara A, Pelliniemi TT. Peripheral blood lymphocyte subsets in multiple myeloma and monoclonal gammopathy of undetermined significance. Clin Lab Haematol. 1994;16:213–23. doi: 10.1111/j.1365-2257.1994.tb00414.x. https://doi.org/10.1111/j.1365-2257.1994.tb00414.x. [DOI] [PubMed] [Google Scholar]
- 77.Pessoa de Magalhães RJ, Vidriales MB, Paiva B, Fernandez-Gimenez C, García-Sanz R, Mateos MV, Gutierrez NC, Lecrevisse Q, Blanco JF, Hernández J, de las Heras N, Martinez-Lopez J, Roig M, Costa ES, Ocio EM, Perez-Andres M, Maiolino A, Nucci M, De La Rubia J, Lahuerta JJ, San-Miguel JF, Orfao A Spanish Myeloma Group (GEM); Grupo Castellano-Leones de Gammapatias Monoclonales, cooperative study groups. Analysis of the immune system of multiple myeloma patients achieving long-term disease control by multidimensional flow cytometry. Haematologica. 2013;98:79–86. doi: 10.3324/haematol.2012.067272. https://doi.org/10.3324/haematol.2012.067272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pérez-Andres M, Almeida J, Martin-Ayuso M, Moro MJ, Martin-Nu-ez G, Galende J, Hernandez J, Mateo G, San Miguel JF, Orfao A Spanish Network on Multiple Myeloma; Spanish Network of Cancer Research Centers. Characterization of bone marrow T cells in monoclonal gammopathy of undetermined significance, multiple myeloma, and plasma cell leukemia demonstrates increased infiltration by cytotoxic/Th1 T cells demonstrating a squed TCR-Vbeta repertoire. Cancer. 2006;106:1296–305. doi: 10.1002/cncr.21746. https://doi.org/10.1002/cncr.21746. [DOI] [PubMed] [Google Scholar]
- 79.Jurisic V, Srdic T, Konjevic G, Markovic O, Colovic M. Clinical stage-depending decrease of NK cell activity in multiple myeloma patients. Med Oncol. 2007;24:312–7. doi: 10.1007/s12032-007-0007-y. https://doi.org/10.1007/s12032-007-0007-y. [DOI] [PubMed] [Google Scholar]
- 80.Dosani T, Carlsten M, Maric I, Landgren O. The cellular immune system in myelomagenesis: NK cells and T cells in the development of MM and their uses in immunotherapies. Blood Cancer J. 2015;5:e321. doi: 10.1038/bcj.2015.49. https://doi.org/10.1038/bcj.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Famularo G, D’Ambrosio A, Quintieri F, Di Giovanni S, Parzanese I, Pizzuto F, Giacomelli R, Pugliese O, Tonietti G. Natural killer cell frequency and function in patients with monoclonal gammopathies. J Clin Lab Immunol. 1992;37:99–109. [PubMed] [Google Scholar]
- 82.De Rossi G, De Sanctis G, Bottari V, Tribalto M, Lopez M, Petrucci MT, Fontana L. Surface markers and cytotoxic activities of lymphocytes in monoclonal gammopathy of undetermined significance and untreated multiple myeloma. Increased phytohemagglutinin-induced cellular cytotoxicity and inverted helper/suppressor cell ratio are features common to both diseases. Cancer Immunol Immunother. 1987;25:133–6. doi: 10.1007/BF00199953. https://doi.org/10.1007/BF00199953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fauriat C, Mallet F, Olive D, Costello RT. Impaired activating receptor expression pattern in natural killer cells from patients with multiple myeloma. Leukemia. 2006;20:732–733. doi: 10.1038/sj.leu.2404096. https://doi.org/10.1038/sj.leu.2404096. [DOI] [PubMed] [Google Scholar]
- 84.Costello RT, Boehrer A, Sanchez C, Mercier D, Baier C, Le Treut T, Sébahoun G. Differential expression of natural killer cell activating receptors in blood versus bone marrow in patients with monoclonal gammopathy. Immunology. 2013;139:338–41. doi: 10.1111/imm.12082. https://doi.org/10.1111/imm.12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.El-Sherbiny YM, Meade JL, Holmes TD, McGonagle D, Mackie SL, Morgan AW, Cook G, Feyler S, Richards SJ, Davies FE, Morgan GJ, Cook GP. The requirement for DNAM-1, NKG2D, and NKp46 in the natural killer cell-mediated killing of myeloma cells. Cancer Res. 2007;67:8444–9. doi: 10.1158/0008-5472.CAN-06-4230. https://doi.org/10.1158/0008-5472.CAN-06-4230. [DOI] [PubMed] [Google Scholar]
- 86.Benson DM, Jr, Bakan CE, Mishra A, Hofmeister CC, Efebera Y, Becknell B, Baiocchi RA, Zhang J, Yu J, Smith MK, Greenfield CN, Porcu P, Devine SM, Rotem-Yehudar R, Lozanski G, Byrd JC, Caligiuri MA. The PD-1/PD-L1 axis modulates the natural killer cell versus multiple myeloma effect: a therapeutic target for CT-011, a novel monoclonal anti-PD-1 antibody. Blood. 2010;116:2286–94. doi: 10.1182/blood-2010-02-271874. https://doi.org/10.1182/blood-2010-02-271874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ray A, Das DS, Song Y, Richardson P, Munshi NC, Chauhan D, Anderson KC. Targeting PD1-PDL1 immune checkpoint in plasmacytoid dendritic cell interactions with T cells, natural killer cells and multiple myeloma cells. Leukemia. 2015;29:1441–4. doi: 10.1038/leu.2015.11. https://doi.org/10.1038/leu.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bezman NA, Jhatakia A, Kearney AY, Brender T, Maurer M, Henning K, Jenkins MR, Rogers AJ, Neeson PJ, Korman AJ, Robbins MD, Graziano RF. PD-1 blockade enhances elotuzumab efficacy in mouse tumor models. Blood Advances. 2017;1:753–65. doi: 10.1182/bloodadvances.2017004382. http://www.bloodadvances.org/content/bloodoa/1/12/753.full.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koene HR, Kleijer M, Algra J, Roos D, von dem Borne AE, de Haas M. Fc gammaRIIIa-158V/F polymorphism influences the binding of IgG by natural killer cell Fc gammaRIIIa, independently of the Fc gammaRIIIa-48L/R/H phenotype. Blood. 1997;90:1109–14. [PubMed] [Google Scholar]
- 90.Wu J, Edberg JC, Redecha PB, Bansal V, Guyre PM, Coleman K, Salmon JE, Kimberly RP. A novel polymorphism of Fc?RIIIa (CD16) alters receptor function and predisposes to autoimmune disease. J Clin Invest. 1997;100:1059–70. doi: 10.1172/JCI119616. https://doi.org/10.1172/JCI119616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Weng WK, Levy R. Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma. J Clin Oncol. 2003;21:3940–7. doi: 10.1200/JCO.2003.05.013. https://doi.org/10.1200/JCO.2003.05.013. [DOI] [PubMed] [Google Scholar]
- 92.Cartron G, Dacheux L, Salles G, Solal-Celigny P, Bardos P, Colombat P, Watier H. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood. 2002;99:754–8. doi: 10.1182/blood.v99.3.754. https://doi.org/10.1182/blood.V99.3.754. [DOI] [PubMed] [Google Scholar]
- 93.Poulart V, Jou Y-M, Delmonte T, Robbins M. Fc-gamma receptor polymorphisms and progression-free survival: Analysis of three clinical trials of elotuzumab in multiple myeloma. In: Poulart V, editor. EHA Learning Center; Jun 9, 2016. p. 132830. (Abstract release date: May 19, 2016) https://learningcenter.ehaweb.org/eha/2016/21st/132830/valerie.poulart.fc-gamma.receptor.polymorphisms.and.progression-free.survival.html?f=m3e968. [Google Scholar]
- 94.dbSNP Short Genetic Variations. NCBI; https://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?rs=396991. [Google Scholar]
- 95.Leiba M, Duek A, Amariglio N, Avigdor A, Benyamini N, Hardan I, Zilbershats I, Ganzel C, Shevetz O, Novikov I, Cohen Y, Ishoev G, Rozic G, Nagler A, Trakhtenbrot L. Translocation t(11;14) in newly diagnosed patients with multiple myeloma: Is it always favorable? Genes Chromosomes Cancer. 2016;55:710–8. doi: 10.1002/gcc.22372. https://doi.org/10.1002/gcc.22372. [DOI] [PubMed] [Google Scholar]
- 96.Walker BA, Wardell CP, Brioli A, Boyle E, Kaiser MF, Begum DB, Dahir NB, Johnson DC, Ross FM, Davies FE, Morgan GJ. Translocations at 8q24 juxtapose MYC with genes that harbor superenhancers resulting in overexpression and poor prognosis in myeloma patients. Blood Cancer J. 2014;4:e191. doi: 10.1038/bcj.2014.13. https://doi.org/10.1038/bcj.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Glitza IC, Lu G, Shah R, Bashir Q, Shah N, Champlin RE, Shah J, Orlowski RZ, Qazilbash MH. Chromosome 8q24.1/c-MYC abnormality: a marker for high-risk myeloma. Leuk Lymphoma. 2015;56:602–7. doi: 10.3109/10428194.2014.924116. https://doi.org/10.3109/10428194.2014.924116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nagoshi H, Taki T, Hanamura I, Nitta M, Otsuki T, Nishida K, Okuda K, Sakamoto N, Kobayashi S, Yamamoto-Sugitani M, Tsutsumi Y, Kobayashi T, Matsumoto Y, Horiike S, Kuroda J, Taniwaki M. Frequent PVT1 rearrangement and novel chimeric genes PVT1-NBEA and PVT1-WWOX occur in multiple myeloma with 8q24 abnormality. Cancer Res. 2012;72:4954–62. doi: 10.1158/0008-5472.CAN-12-0213. https://doi.org/10.1158/0008-5472.CAN-12-0213. [DOI] [PubMed] [Google Scholar]
- 99.Richardson P, Wong E, Stockerl-Goldstein K, Rosenbaum C, Dhodapkar M, Jou Y-M, Lynch M, Robbins M, Bleickardt E, Jagannath S. A phase 2 open-label, multicenter study of elotuzumab monotherapy in patients with high-risk smoldering multiple myeloma. In: Jagannath S, editor. EHA Learning Center; Jun 12, 2016. p. 135309. (Abstract release date: May 19, 2016) https://learningcenter.ehaweb.org/eha/2016/21st/135309/sundar.jagannath.a.phase.2.open-label.multicenter.study.of.elotuzumab.html?f=m3. [Google Scholar]
- 100.Mateos MV, Hernández MT, Giraldo P, de la Rubia J, de Arriba F, Corral LL, Rosi-ol L, Paiva B, Palomera L, Bargay J, Oriol A, Prosper F, López J, Argui-ano JM, Quintana N, García JL, Bladé J, Lahuerta JJ, Miguel JF. Lenalidomide plus dexamethasone versus observation in patients with high-risk smouldering multiple myeloma (QuiRedex): long-term follow-up of a randomised, controlled, phase 3 trial. Lancet Oncol. 2016;17:1127–36. doi: 10.1016/S1470-2045(16)30124-3. https://doi.org/10.1016/S1470-2045(16)30124-3. [DOI] [PubMed] [Google Scholar]
- 101.Usmani SZ, Sexton R, Ailawadhi S, Shah JJ, Valent J, Rosenzweig M, Lipe B, Zonder JA, Fredette S, Durie B, Hoering A, Bartlett B, Orlowski RZ. Phase I safety data of lenalidomide, bortezomib, dexamethasone, and elotuzumab as induction therapy for newly diagnosed symptomatic multiple myeloma: SWOG S1211. Blood Cancer J. 2015;5:e334. doi: 10.1038/bcj.2015.62. https://doi.org/10.1038/bcj.2015.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Palumbo A, Bringhen S, Mateos MV, Larocca A, Facon T, Kumar SK, Offidani M, McCarthy P, Evangelista A, Lonial S, Zweegman S, Musto P, Terpos E, Belch A, Hajek R, Ludwig H, Stewart AK, Moreau P, Anderson K, Einsele H, Durie BG, Dimopoulos MA, Landgren O, San Miguel JF, Richardson P, Sonneveld P, Rajkumar SV. Geriatric assessment predicts survival and toxicities in elderly myeloma patients: an International Myeloma Working Group report. Blood. 2015;125:2068–74. doi: 10.1182/blood-2014-12-615187. Correction in: Blood. 2016 Mar 3; 127(9): 1213. Correction in: Blood. 2016 Aug 18; 128(7): 1020 https://doi.org/10.1182/blood-2014-12-615187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ozaki S, Handa H, Saitoh T, Murakami H, Itagaki M, Asaoku H, Suzuki K, Isoda A, Matsumoto M, Sawamura M, Konishi J, Sunami K, Takezako N, Hagiwara S, Kuroda Y, Chou T, Nagura E, Shimizu K. Trends of survival in patients with multiple myeloma in Japan: A multicenter retrospective collaborative study of the Japanese Society of Myeloma. Blood Cancer J. 2015;5:e349. doi: 10.1038/bcj.2015.79. https://doi.org/10.1038/bcj.2015.79. [DOI] [PMC free article] [PubMed] [Google Scholar]