Abstract
Potential relationship of vitamin D, vitamin D receptor (VDR), and vitamin D binding protein (DBP) have been suggested in the pathophysiology of hepatitis C virus (HCV) infection. The aim of this observational study is to determine vitamin D levels, and VDR and DBP genetic polymorphism according demographic and laboratory data in chronic HCV patients (CHC).
A total of 148 CHC patients gave serum samples for testing 25-hydroxyvitamin D (25 (OH)D) level by immunochemiluminometric assay (<20 ng/mL defined as deficient) and donated blood samples to allelic discrimination analysis using TaqMan assays. Analyzed single nucleotide polymorphisms (SNPs) were: VDR-rs7975232 (ApaI) C>A, rs731236 A>G (TaqI), rs1544410 C>T (BsmI), rs10735810 T>C (FokI) and carrier globulin/binding protein (GC)-rs4588 and rs7041 and the haplotype bAt [CCA]. Hepatic fibrosis was assessed using Fib-4 and Forns index.
Eighty-two (54.40%) patients demonstrated deficiency of vitamin D and this was associated to AST (P = .019 [CI: 1.003–1.034]), total cholesterol (P = .038 [CI: 1.004–1.164]), fibrosis grade (P < .001 [CI: 0.000–0.844]), and FokI (P = .028) allele T presence. Association was found between VDR polymorphism and fibrosis (BsmI andTaqI), triglycerides (TaqI), and HDL (FokI). DBP polymorphism was associated to HCV genotype (GC rs7041), previous HCV treatment, and GGT (GC rs4588).
In conclusion, low frequency of vitamin D deficiency was found, but VDR polymorphisms were frequently associated to fibrosis grade suggesting that they could be used as disease evaluation markers to understand the mechanisms underlying the virus–host interaction.
Keywords: fibrosis, hepatitis C, polymorphism, vitamin D
1. Introduction
Hepatitis C virus (HCV) is a major cause of chronic hepatitis responsible for more >185 million worldwide accounting a seroprevalence of 2.8%.[1–3] Although the incidence rate of HCV infection is decreasing in the developed countries, deaths from liver disease secondary to HCV infection will continue to increase over the next 20 years.[4]
In low- and middle-income countries, interferon and ribavirin (RBV)-based therapies are the first choices due to their availability and low cost. The combination of RBV and pegylated-interferon-alpha was considered the standard treatment for HCV infection in the last decade where those infected by genotypes 1 and 4 are treated for 48 weeks, and those infected by genotypes 2 and 3 are treated for 24 weeks.[5] Recently, the use of direct-acting antivirals has proven effective against HCV in developed countries, significantly decreasing morbidity and mortality. However, low- and middle-income countries, most of which are highly endemic areas, access to new antiviral drugs still is restricted due to high cost.[6]
Several predictors of successful treatment of chronic hepatitis C (CHC) using interferon and RBV have been identified,[7,8] such as factors related to the virus (infection by HCV genotype 2–3, basal HCV viral load, and HCV genetic variability) or to the host (liver inflammation, fibrosis, lower serum cholesterol values and interleukin 28B polymorphisms C/C, and vitamin D concentration).[9–11]
Calcitriol is an important modulator in the immune pathways and is also known to be involved in cell proliferation and differentiation.[12] Most of its biological effects are mediated through the vitamin D receptor (VDR), a member of the nuclear receptor superfamily of ligand-activated transcription factors[13–15] and directed to these cells connected with a binding protein. Carrier globulin/binding protein (GC)-globulin is the main serum vitamin D binding protein; it is polymorphic and characterized in Caucasian populations by a great variability at 2 loci (rs7041 and rs4588).[16]
The presence of the minor alleles in GC-globulin loci has been associated with lower circulating vitamin D levels,[17] instead VDR polymorphisms has been associated with rapid fibrosis progression (haplotype bAt[CCA]).[18] Vitamin D deficiency has been associated to low sustained virological response (SVR) among hepatitis C patients.[19,20] Moreover, vitamin D levels and single nucleotide polymorphisms (SNPs) involved in its metabolism have been related to anti-HCV therapy response.[21–23] In this way, our aim is to evaluate the influence of genetic determinants of 25-hydroxyvitamin D (25 (OH)D) serum levels and the association with demographic, metabolic, and virological data in chronic HCV patients living in South America.
2. Methods
2.1. Study design and population
This is an observational study that included 148 CHC patients resident in Rio de Janeiro recruited at Viral Hepatitis Ambulatory (Viral Hepatitis Laboratory, Oswaldo Cruz Institute, FIOCRUZ), Hepatology Unit (Clementino Fraga Filho University Hospital, UFRJ), General Medicine Department (Gaffrée Guinle University Hospital, UNIRIO), and Hepatology Unit (Antonio Pedro University Hospital, UFF). Nonprobabilistic recruitment was conducted from 2011 to 2012 year.
Patients were included if they had a virological diagnosis of CHC (anti-HCV and HCV-RNA reactive serum sample, with persistently abnormal alanine aminotransferase [ALT], at least 6 months) and more than 18 years of age. The infecting HCV genotypes were the following: 1, 2, 3, and 5. Patients were excluded if they were positive for serum hepatitis B surface antigen or antihuman immunodeficiency virus antibody, or exhibited other causes of hepatocellular injury (eg, any history of alcoholism, autoimmune hepatitis, advanced cirrhosis, or treatment with hepatotoxic drugs), and concomitant use of drugs known to affect serum vitamin D concentration and intravenous drug use. These factors were excluded due to potential source of bias.
This study was approved by the Ethics Committee of FIOCRUZ (CAAE 41269015.3.0000.5248). Informed consent was obtained from each patient who was included in the study, and the study protocol was followed according to the ethical guidelines of the 1975 Declaration of Helsinki.
2.2. Clinical and laboratorial data
All serum samples were tested for anti-HCV using commercial EIA kit HCV Ab (Radim, Pomezia, Italy), where all anti-HCV-reactive samples were retested in duplicate.
HCV RNA viral load was determined using CobasTaqMan HCV Test (Roche Molecular Systems, Inc, Basel, Switzerland) or Abbott Real Time HCV m2000sp (Abbott Laboratories, IL) and expressed as IU/mL. HCV genotyping was performed using INNO-LIPA HCV II kit (Innogenetics, Zwijnaarden, Belgium) or Abbott Real Time HCV m2000sp (Abbott Laboratories) according to manufacturer's instruction.
Clinical and demographic data were collected simultaneously from all patients. A 12-hour overnight fasting blood sample was draw to determine serum levels of ALT, gamma-glutamyltransferase (GGT), total cholesterol (Tc), high-density lipoprotein (HDL) and low-density lipoprotein (LDL) cholesterol, triglycerides, plasma glucose concentration, and platelet count.
Baseline serum vitamin D level was assessed by measuring serum 25 (OH)D levels using automated immunochemiluminometric assay (ICMA) (Liason 25 (OH) Vitamin D, Diasorin, Varceli, Italy) with 20 ng/mL being considered the threshold value. Serum insulin was also determined using ICMA (Liason Insulin Assay, Diasorin, Varceli, Italy). Insulin resistance was determined using homeostasis model assessment method (HOMA ≥2).[24]
Hepatic fibrosis was assessed using Fib-4 and Forns index as the same as described previously.[25] When Fib-4 was less than 1.45, individual was considered as low fibrosis, and if Fib-4 was ≥1.45, Forns index was considered. Forns index below 4.2 was considered as low fibrosis, and index above or equal to 4.2 was considered high fibrosis.[25]
Genomic DNA was obtained from whole blood samples using QIamp DNA Mini Kit (Quiagen, Valencia, CA). The purified and elute DNA was quantified using a Nanodrop Spectrophotometer ND-1000 (Thermo Scientific, Rockford, IL). Thereafter, DNA was diluted in UltraPure DNase/RNase-Free water (Invitrogen Corporation, Camarillo, CA) to a concentration of 2 ng. The allelic discrimination analysis was performed using the TaqMan assays (Applied Biosystems, Foster City, CA) at ViiA7 Real-Time PCR System 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA). TaqMan Master mix (2.5 μL), TaqMan assay (0.25 μL), and H2O (1.25 μL) were added to a final volume of 4 μL per sample, and 1 μL of DNA was added for a total reaction volume of 5 μL. Reaction conditions for SNPs ApaI, FokI, TaqI, and rs4588 were: 10 minutes at 95 °C after 50 cycles of denaturation at 95 °C (15 seconds) and annealing at 60 °C (1 minute). To genotyping SNPs BsmI and rs7041 TaqMan Master mix (5.0 μL), TaqMan assay (0.50 μL), and H2O (3.5 μL) were added to a final volume of 9 μL per sample, and 1 μLof DNA was added for a total reaction volume of 10 μL. Reaction conditions for these polymorphisms were: 10 minutes at 95 °C after 50 cycles of denaturation at 95 °C (15 seconds) and annealing at 60 °C (1 minute) to BsmI and 61 °C (1 minute) to rs7041.[26]
We selected tag-SNPs from vitamin D metabolism-related genes, VDR and GC, and the haplotype bAt [CCA]. These SNPs were chosen using previous literature information.[18,19,26–28] Public databases were used to collect additional information about SNPs and genes: NCBI http://www.ncbi.nlm.nih.gov.
The analyzed SNPs were: VDR-rs7975232 (ApaI) C>A, rs731236 A>G (TaqI), rs1544410 C>T (BsmI), rs10735810 T>C (FokI) and GC-rs4588 and rs7041. According to Pubmed Database, the frequencies were: GC (rs7041): A = 0.4841/58741 (ExAC) C = 0.3816/1911 (1000 Genomes); GC (rs4588): T = 0.2528/30670 (ExAC) T = 0.2079/1041 (1000 Genomes); BsmI rs1544410: T = 0.2959/1482 (1000 Genomes); ApaI (rs7975232): C = 0.4813/58108 (ExAC) C = 0.4846/2427 (1000 Genomes); TaqI (rs731236): G = 0.3338/40509 (ExAC) G = 0.2766/1385 (1000 Genomes); and FokI (rs10735810): A = 0.3285/1645.
Haplotypes were constructed from the combination of the VDR polymorphisms BsmI, ApaI, and TaqI {bAt [CCA] [“b” (BsmI allele C), “A” (ApaI allele C), and “t” (TaqI allele A)]} and their frequencies were inferred using the PHASE 2.1.1 program (Matthew Stephens Lab, University of Chicago). To predict both allele frequencies will remain constant because they are in equilibrium, we calculated the Hardy–Weinberg equilibrium by X2 (P < .05).
2.3. Statistical analysis
Continuous data are expressed as mean ± standard deviation, and the categorical data are expressed as number (percentage). Comparisons of differences in categorical date between groups were performed using the Chi-square test. Distributions of continuous variables were analyzed by the Student t test or Mann–Whitney U test for the 2 groups where appropriate. Missing data were presented in tables when appropriate. Independent factors possibly affecting 25 (OH)D concentration (cut off value of 20 ng/mL) were determined by stepwise multiple logistic regression analysis.
Hardy–Weinberg equilibrium for each SNP was compared using the Pearson chi-square test (X2). 25 (OH)D concentration and the variance in each genotype was accessed in one-way ANOVA with Bonferroni correction. At univariate analyses, P-value were considered significant when P�<�.2 and in multivariate analyze P�<�.05. All the tests were performed with IBM SPSS Statistics 20.0 for Windows (Chicago, IL).
3. Results
3.1. Baseline characteristics
Demographic, virological, and clinical features of population studied are shown in Table 1. There were 56 men and 92 women, with mean age of 54.41 (±11.27). Elevated mean of ALT, AST, phosphatase, GGT, blood glucose, Tc, and LDL serum levels were observed in patients.
Table 1.
Demographic, laboratory, and metabolic data of 148 chronic hepatitis patients (CHC).
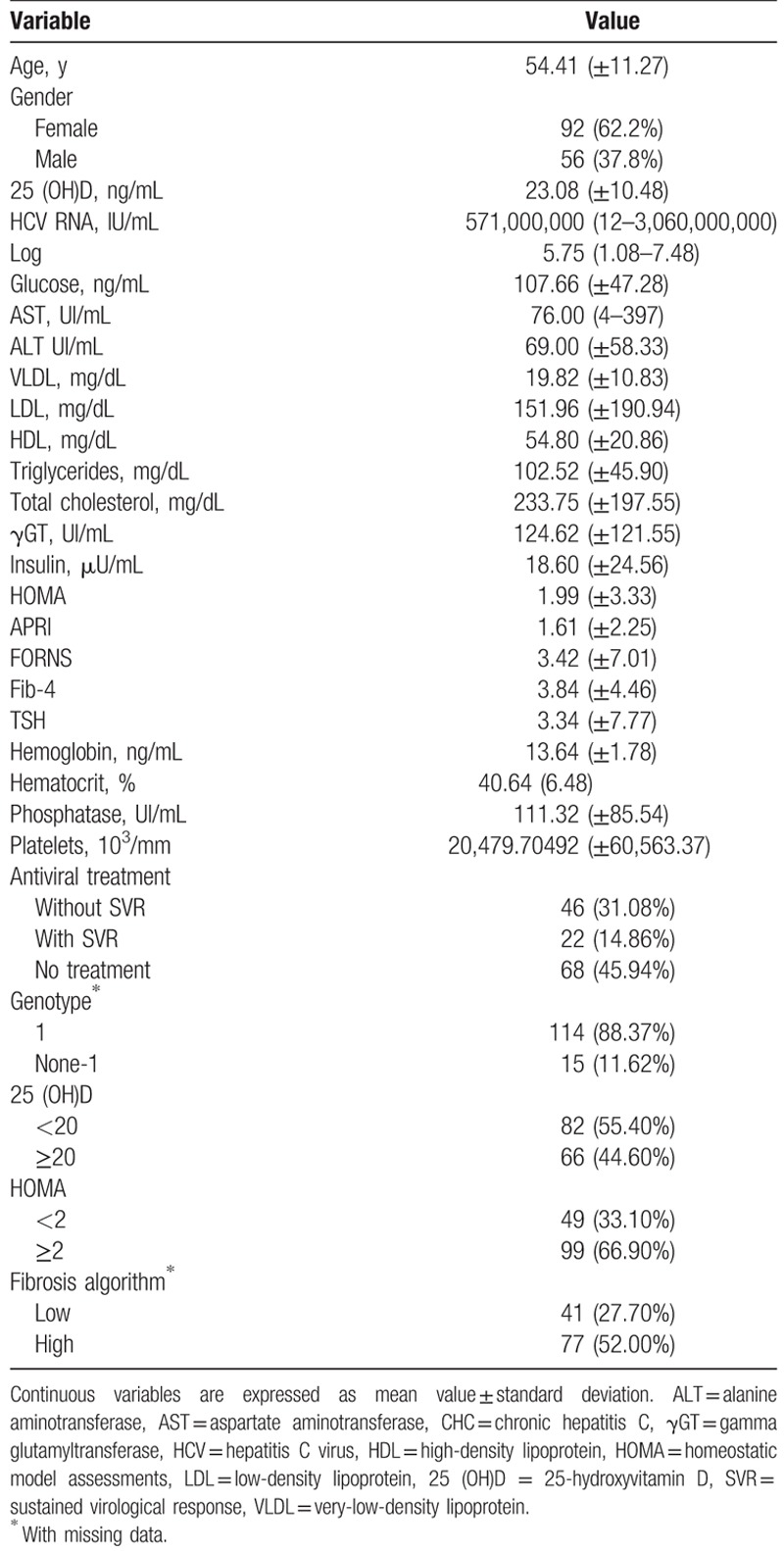
Normal median values were observed for: very-low-density lipoprotein (VLDL) cholesterol (19.82 ± 10.83 mg/dL), HDL (54.80 ± 20.86 mg/dL), platelets (20,479.70492 ± 60,563.37103/mm), hematocrit (40.64% ± 6.48%), and hemoglobin (13.64 ± 1.78 ng/mL) (anemic classification: hemoglobin values <12 ng/mL for women and <13 ng/mL for men). Using the cutoff value of 20 ng/mL, 82/148 (54.40%) patients demonstrated deficiency of vitamin D. HCV RNA viral load (log mean) was high and most prevalent genotype was 1 (88.37%). During this study, 68 CHC patients were treated for HCV using pegylated-interferon and RBV and in this subgroup, 46 (31.08%) did not achieve SVR. Sixty-eight patients (45.94%) were not treated for HCV. Using fibrosis algorithm, 52.00% (n = 77) presented high level. The HOMA index mean was also lower than the cutoff established in this work (1.99 ± 3.33), where 33.10% (n = 49) presented insulin resistance.
3.2. Association between serum 25 (OH)D levels, clinical and viral data in chronic HCV patients
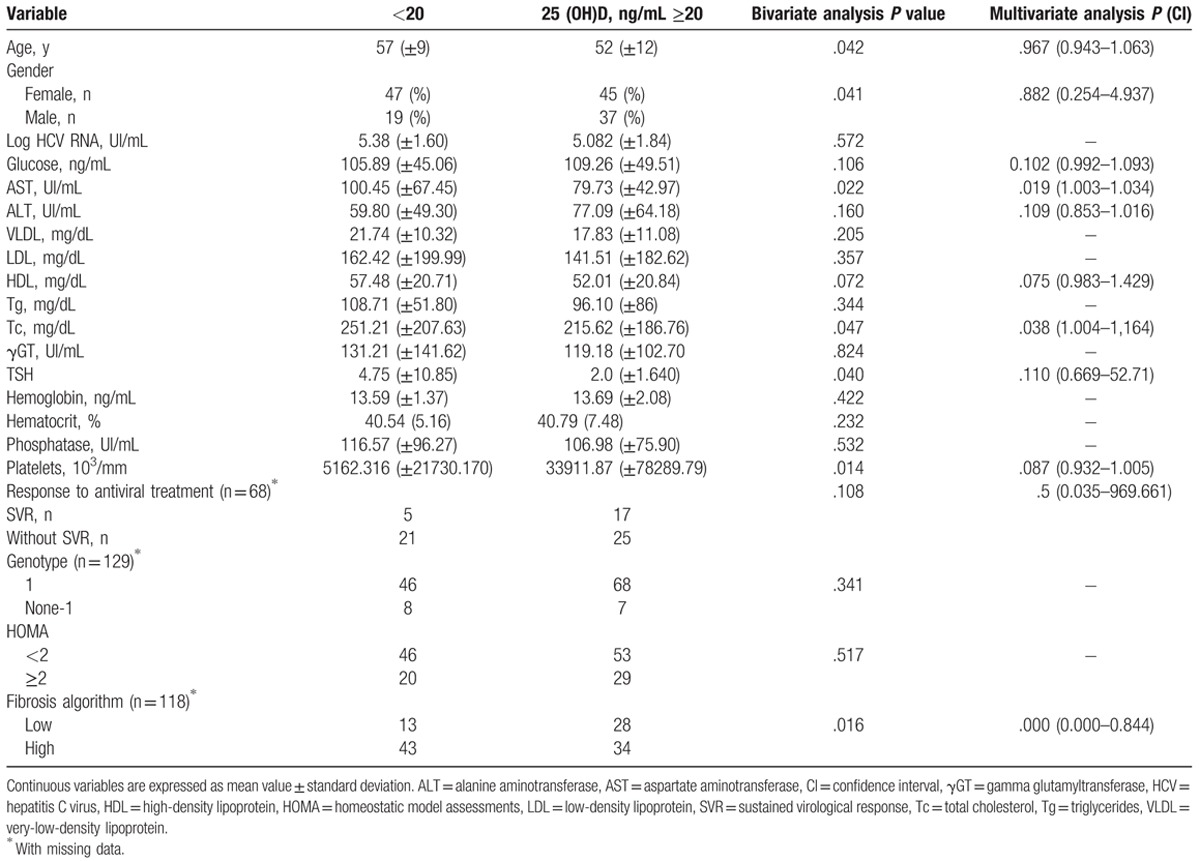
To assess the relationship among 25 (OH)D concentration and demographic and laboratory data in HCV patients, a cutoff value was established (deficiency <20 ng/mL). High mean values of age (P = .042), AST (P = .022), HDL (P = .072), triglycerides (0.047), SVR (P = .108), and presence of high fibrosis (P = .016) were associated to 25 (OH)D deficiency at univariate analysis.
Patients with high rates of platelets presented 25 (OH)D ≥20 ng/mL (P = .014), as also blood glucose (P = .106) and ALT (P = .160) were high in patients with 25 (OH)D ≥20 ng/mL (Table 2). At multiple linear regression analysis, AST (P = .019 [CI: 1.003–1.034]), Tc (P = .038 [CI: 1.004–1.164]), and fibrosis grade (P < .001 [CI: 0.000–0.844]) were significant.
Table 2.
Univariate and multivariate regression analysis of factors associated with 25 (OH) concentration.
3.3. Association between VDR and GC polymorphism according to 25 (OH)D level among HCV patients
To study the implication of these SNPs and 25 (OH)D concentration according to clinical and laboratory data of HCV patients, first our sample was categorized according to previously defined cutoff value (25 (OH)D deficiency < 20 ng/mL). Second, the alleles were categorized in risk allele/nonrisk allele and grouped, and presence/absence of haplotype bAt [CCA]. The genotype distribution of all investigated SNPs was in Hardy–Weinberg equilibrium. Only FokI was statistically significant (P = .028), where allele T were associated with 25 (OH)D ≥20 ng/mL.
After this, the one-way ANOVA test with Bonferroni multiple comparison posttest (P < .05) was done. There was no statistically difference between SNPs genotypes and 25 (OH)D concentration in 148 Brazilian CHC patients (Fig. 1). ApaI: (CC vs CA [t = 0.172], CC vs AA [t = 0.199], CA vs AA [t = 0.430]); TaqI: (AA vs AG [t = 0.806], AA vs GG [t = 0.990], AG vs GG [t = 0.497]); BsmI: (CC vs CT [t = 1.281], CC vs TT [t = 0.564], CT vs TT [t = 0.278]); FokI: (CC vs CT [t = 1.786], CC vs TT [t = 0.961], CT vs TT [t = 2.040]); GC rs4588: (AA vs AC [t = 0.927], AA vs CC [t = 0.101], AC vs CC [t = 1.515]); GC rs7041: (TT vs TG [t = 0.610], TT vs GG [t = 2.001], TG vs GG [t = 1.565]).
Figure 1.
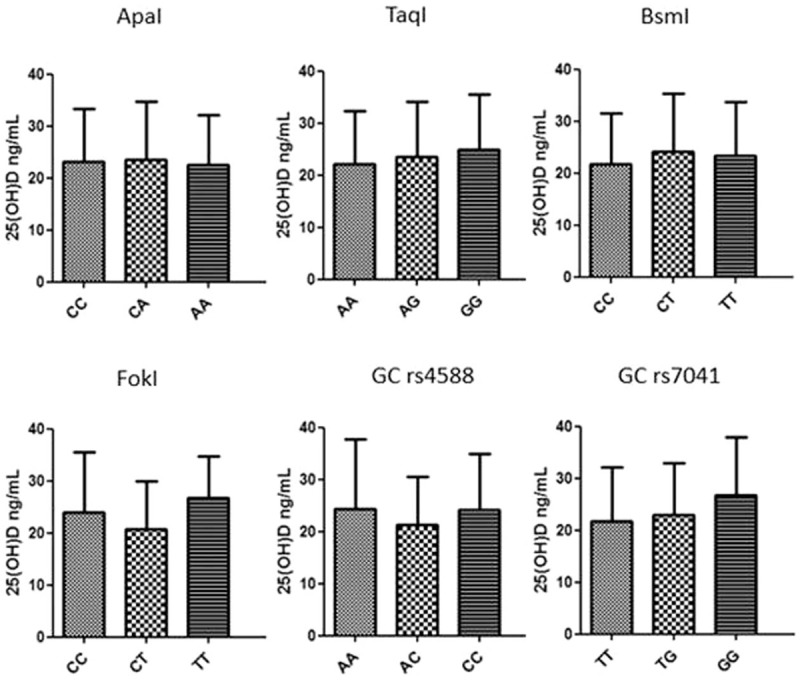
Association of VDR and GC gene polymorphism with 25 (OH)D concentration in Brazilian chronic HCV patients. No significant variance was found using ANOVA I test with Bonferroni correction (P < .05). GC = carrier globulin/binding protein, HCV = hepatitis C virus, 25 (OH)D = 25-hydroxyvitamin D, VDR = vitamin D receptor.
Besides that we carried out a bivariate and a linear regression analysis by comparing favorable allele group with demographic, biochemical, and viral data. The significant results are given in Table 3. In the multivariate analysis, using SPNs as dependent variable, we found association between BsmIrs1544410 (fibrosis algorithm), TaqI rs731236 (triglycerides and fibrosis algorithm), FokI rs10735810 (HDL), GC rs4588 (treated and none-treated, and GGT), and GC rs7041 (genotype 1 and none-1).
Table 3.
Univariate and multivariate regression analysis of factors associated with VDR-ApaI rs7975232, TaqI rs731236, BsmI rs1544410, FokIrs10735810, GC-rs4588, and GC-rs7041 SNPs.

4. Discussion
In vitamin D pathway, a series of studies had reported on interaction between vitamin D-related genes and 25 (OH)D levels. Among them, VDR and vitamin D-binding protein (DBP) genes played an important role in vitamin D pathway. Previous studies had shown that genetic variations in these genes had a significant effect on vitamin D levels in many populations including the Chinese,[29,30] Japanese,[31] Caucasian,[16,17,19,20,27] Brazilian healthy girls,[26] and in this study, FokI C allele in Brazilian CHC patients were associated to 25 (OH)D <20 ng/mL.
This way, we hypothesized that genetic determinants of 25 (OH)D serum levels may be associated with demographic, metabolic, and virological data in chronic HCV Brazilian patients. To our knowledge, this is the first study that describes the interaction between vitamin D-related genes and 25 (OH)D levels among HCV-infected patients from South America giving some new insight about these parameters in tropical regions.
In this study, the prevalence of vitamin D deficiency was 55.04% that is lower than previously observed in similar population in Brazil (63%)[25] and among Italians HCV patients (73%).[20,22,23,32] Moreover, severe fibrosis was found in patients with lower vitamin D concentration (Table 2). Lange et al[19] also found that the prevalence of vitamin D deficiency was greater in HCV patients with more advanced fibrosis.
In the present study, Fib-4 and Forns index were used to determine fibrosis level since they are useful for predicting fibrosis grade in the absence of biopsy.[33,34] Forns index can be used to differentiate patients with mild (F0–F1) fibrosis from those with severe (F2–F4) fibrosis demonstrating diagnostic performance of AUROC 0.76 to 0.79 for the detection of significant fibrosis. FIB-4 index correctly identified patients with severe fibrosis (F3–F4) and cirrhosis with an AUROC of 0.85 and 0.91, respectively. An FIB-4 index <1.45 had a negative predictive value of 94.7% to exclude severe fibrosis and a sensitivity of 74.3%. An FIB-4 index higher than 3.25 had a positive predictive value of 82.1% for the confirmation of significant fibrosis (F3–F4), with a specificity of 98.2%.[35,36]
Vitamin D deficiency is common among patients with liver diseases. Whether vitamin D status can also affect liver function is poorly understood. In this study, low 25 (OH)D concentration was associated with high AST and cholesterol total value. Skaaby et al[37] investigate the association between vitamin D status, liver enzymes, and incident liver disease in 2649 individuals and found that the risk of having a high level of ALT, AST, or GGT tended to be higher for lower vitamin D levels, in accordance with our results.
There is conflicting information when it comes to the link between cholesterol and vitamin D levels. Population studies show that people with lower vitamin D levels are more likely to have high cholesterol, although this does not prove a cause and effect relationship despite being according our findings.[38] To the National Institutes of Health, there is insufficient evidence to determine any relationship between vitamin D intake and cholesterol levels.
It was not found an association between vitamin D levels and HCV RNA mean values. Conversely, Gerova et al[39] found an inverse relationship between 25 (OH)D and HCV-RNA levels in Bulgarian HCV patients. These differences could be due to variation in the 25 (OH)D assay used, vitamin D cutoff, and the different ethnicities and geographic latitude of populations studied, and clinical trials.
A strong association of vitamin D sufficiency to TT genotype in FokI was found. This way, FokI C>T polymorphisms may be used as a molecular marker to predict the risk and to evaluate the disease severity of HCC in those infected with HCV since low concentrations of vitamin D is directly associated with increased liver injury in Brazilian population. Differently, Shehab et al[40] observed that VDR genetic polymorphisms are significantly associated with the occurrence of HCV-related HCC especially T allele carriers which could be considered as a risk factor of hepatocellular carcinoma in Egyptian patients.
Regarding VDR and DBP genes polymorphisms in Brazilian population, the BsmI, ApaI, TaqI, and the rs4588 and rs7041 polymorphisms were most consistently associated with lower 25 (OH)D levels.[26,40] However, in the present study both SNPs were not associated with lower levels of 25 (OH)D after applying Bonferroni correction. Nevertheless, Falleti et al[16] demonstrated that normal serum vitamin D levels and the carriage of GC-globulin WT isoform strongly predict the achievement of SVR after PEG-interferon plus RBV antiviral therapy. Some studies found that the carriage of ApaI CC genotype and TaqI AA genotype had significant higher viral load as compared to those with ApaI CA/AA type and TaqI AG type, respectively.[28] Besides that, VDR gene variants can modulate biological effects of vitamin D without influencing serum vitamin D levels.[15,41]
Some variables could explain these differences. It is well established that there are racial differences in the relations between the 25 (OH)D concentrations, parathyroid hormone concentration, and calcium homeostasis.[14] Racial differences in vitamin D physiology or race-specific factors that modify the effects of vitamin D may affect the immune response to HCV.[42]
We did not find a significant association between VDR bAt [CCA] haplotype and clinical and laboratory data in HCV patients as the same as Hung et al[28] who also did not find any relationship between this haplotype and response to peginterferon plus RBV therapy. These genetic variations have been described as important modulators of several chronic liver diseases such as primary biliary cirrhosis and autoimmune hepatitis.[43]
In the present study, fibrosis was associated to BsmI rs1544410 and TaqI rs731236 what was not observed among Chinese population with chronic HCV infection.[28] To explain the relation between high serum triglycerides and the effect of TaqI rs731236, a hypothetical explanation is related to the mechanism in which vitamin D induces the expression of VLDL-cholesterol receptors in some types of cell.[44] This situation is associated with an elevation of the levels of VLDL and triglycerides and is common in CHC patients.[45] The relationship between HCV genotype and GC remains unknown.
This study presents some limitations, like the absence of race information or ancestry profile, sun exposure, and season of collection that could create some bias regarding vitamin D measures.
In conclusion, the present study suggests a significant association of VDR FokI rs10735810 and the 25 (OH)D concentration in CHC patients, strong association between fibrosis algorithm, 25 (OH)D, BsmI rs15444, and TaqI rs731236, and finally the association between GC rs7041 (Genotype 1/ None-1). These data could provide helpful information for understanding the complex mechanisms underlying the virus–host interaction and the variations observed in antiviral therapy responses.
Acknowledgment
The authors thank Adilson José de Almeida for clinical assessment, Juliana Custódio Miguel and Elisangela Ferreira da Silva for technical assistance in serological tests, and Vanessa Alves Marques for technical assistance in molecular assays.
Footnotes
Abbreviations: ALT = alanine aminotransferase, CHC = chronic hepatitis C, GC = carrier globulin/binding protein, GGT = gamma-glutamyltransferase, HCV = hepatitis C virus, HDL = high-density lipoprotein, LDL = low-density lipoprotein, 25 (OH)D = 25-hydroxyvitamin D, RBV = ribavirin, SNP = single nucleotide polymorphism, SVR = sustained virological response, Tc = total cholesterol, VDR = vitamin D receptor, VLDL = very-low-density lipoprotein.
Funding/support: This study was supported by Fundação de Amparo a Pesquisa do Estado do Rio de Janeiro (FAPERJ), Brazilian National Counsel of Technological and Scientific Development (CNPq), and Oswaldo Cruz Foundation (FIOCRUZ).
The authors have no conflicts of interest to disclose.
References
- [1].Georgel P, Schuster C, Zeisel MB, et al. Virus-host interactions in hepatitis C virus infection: implications for molecular pathogenesis and antiviral strategies. Trends Mol Med 2010;16:277–86. [DOI] [PubMed] [Google Scholar]
- [2].Lauer GM, Walker BD. Hepatitis C virus infection. N Engl J Med 2001;345:41–52. [DOI] [PubMed] [Google Scholar]
- [3].Mastro TD, Morrison CS, Hamilton CD. Determining the incidence of hepatitis C virus infection in populations: an important tool for epidemic control. J Infect Dis 2016;214:339–40. [DOI] [PubMed] [Google Scholar]
- [4].Razavi H, Elkhoury AC, Elbasha E, et al. Chronic hepatitis C virus (HCV) disease burden and cost in the United States. Hepatology 2013;57:2164–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jacobson IM, Pawlotsky J-M, Afdhal NH, et al. A practical guide for the use of boceprevir and telaprevir for the treatment of hepatitis C. J Viral Hepat 2012;19:1–26. [DOI] [PubMed] [Google Scholar]
- [6].Yilmaz H, Yilmaz EM, Leblebicioglu H. Barriers to access to hepatitis C treatment. J Infect Dev Ctries 2016;10:308–16. [DOI] [PubMed] [Google Scholar]
- [7].Asselah T, Estrabaud E, Bieche I, et al. Hepatitis C: viral and host factors associated with non-response to pegylated interferon plus ribavirin. Liver Int 2010;30:1259–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fattovich G, Covolo L, Bibert S, et al. IL28B polymorphisms, IP-10 and viral load predict virological response to therapy in chronic hepatitis C. Aliment Pharmacol Ther 2011;33:1162–72. [DOI] [PubMed] [Google Scholar]
- [9].Rauch A, Kutalik Z, Descombes P, et al. Genetic variation in IL28B is associated with chronic hepatitis c and treatment failure: a genome-wide association study. Gastroenterology 2010;138:1338–45. [DOI] [PubMed] [Google Scholar]
- [10].Villar LM, Del Campo JA, Ranchal I, et al. Association between vitamin D and hepatitis C virus infection: a meta-analysis. World J Gastroenterol 2013;19:5917–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Assone T, Paiva A, Fonseca LAM, et al. Genetic markers of the host in persons living with HTLV-1, HIV and HCV infections. Viruses 2016;8:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].von Essen MR, Kongsbak M, Schjerling P, et al. Vitamin D controls T cell antigen receptor signaling and activation of human T cells. Nat Immunol 2010;11:344–9. [DOI] [PubMed] [Google Scholar]
- [13].Rukin NJ, Strange RC. What are the frequency, distribution, and functional effects of vitamin D receptor polymorphisms as related to cancer risk? Nutr Rev 2007;65:96–101. [DOI] [PubMed] [Google Scholar]
- [14].Uitterlinden AG, Fang Y, Van Meurs JBJ, et al. Genetics and biology of vitamin D receptor polymorphisms. Gene 2004;338:143–56. [DOI] [PubMed] [Google Scholar]
- [15].Wang TJ, Zhang F, Richards JB, et al. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet 2010;376:180–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Falleti E, Bitetto D, Fabris C, et al. Vitamin D binding protein gene polymorphisms and baseline vitamin D levels as predictors of antiviral response in chronic hepatitis C. Hepatology 2012;56:1641–50. [DOI] [PubMed] [Google Scholar]
- [17].Zierold C, Mings JA, DeLuca HF. Regulation of 25-hydroxyvitamin D3-24-hydroxylase mRNA by 1,25-dihydroxyvitamin D3 and parathyroid hormone. J Cell Biochem 2003;88:234–7. [DOI] [PubMed] [Google Scholar]
- [18].Baur K, Mertens JC, Schmitt J, et al. The vitamin D receptor gene bAt (CCA) haplotype impairs the response to pegylated-interferon/ribavirin-based therapy in chronic hepatitis C patients. Antivir Ther 2012;17:541–7. [DOI] [PubMed] [Google Scholar]
- [19].Lange CM, Bojunga J, Ramos-Lopez E, et al. Vitamin D deficiency and a CYP27B1-1260 promoter polymorphism are associated with chronic hepatitis C and poor response to interferon-alfa based therapy. J Hepatol 2011;54:887–93. [DOI] [PubMed] [Google Scholar]
- [20].Ladero JM, Torrejón MJ, Sánchez-Pobre P, et al. Vitamin D deficiency and vitamin D therapy in chronic hepatitis C. Ann Hepatol;12:199–204. [PubMed] [Google Scholar]
- [21].D’Avolio A, Ciancio A, Siccardi M, et al. Negative predictive value of IL28B, SLC28A2, and CYP27B1 SNPs and low RBV plasma exposure for therapeutic response to PEG/IFN-RBV treatment. Ther Drug Monit 2012;34:722–8. [DOI] [PubMed] [Google Scholar]
- [22].Bitetto D, Fabris C, Falleti E, et al. Vitamin D deficiency and HCV chronic infection: what comes first? J Hepatol 2011;55:944–5. [DOI] [PubMed] [Google Scholar]
- [23].Bitetto D, Fabris C, Fornasiere E, et al. Vitamin D supplementation improves response to antiviral treatment for recurrent hepatitis C. Transpl Int 2011;24:43–50. [DOI] [PubMed] [Google Scholar]
- [24].Eslam M, Kawaguchi T, Del Campo JA, et al. Use of HOMA-IR in hepatitis C. J Viral Hepat 2011;18:675–84. [DOI] [PubMed] [Google Scholar]
- [25].Melo-Villar L, Lampe E, de Almeida AJ, et al. Hypovitaminosis D and its relation to demographic and laboratory data among hepatitis C patients. Ann Hepatol;14:457–463. [PubMed] [Google Scholar]
- [26].Santos BR, Mascarenhas LPG, Boguszewski MCS, et al. Variations in the vitamin D-binding protein (DBP) gene are related to lower 25-hydroxyvitamin D levels in healthy girls: a cross-sectional study. Horm Res Paediatr 2013;79:162–8. [DOI] [PubMed] [Google Scholar]
- [27].Lange CM, Bibert S, Kutalik Z, et al. A genetic validation study reveals a role of vitamin D metabolism in the response to interferon-alfa-based therapy of chronic hepatitis C. PLoS One 2012;7:40159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hung C-H, Hu T-H, Lu S-N, et al. Association of vitamin D receptor gene polymorphisms with response to peginterferon plus ribavirin in Asian patients with chronic hepatitis C. J Formos Med Assoc 2016;115:278–83. [DOI] [PubMed] [Google Scholar]
- [29].Gong Y-G, Li Y-N, Zhang W-H, et al. Correlation between vitamin D receptor genetic polymorphism and 25-hydroxyvitamin D3 in vitamin D deficiency rickets. Zhongguo Dang Dai Er Ke Za Zhi 2010;12:544–6. [PubMed] [Google Scholar]
- [30].Ren Y, Liu M, Zhao J, et al. Serum vitamin D3 does not correlate with liver fibrosis in chronic hepatitis C. World J Gastroenterol 2015;21:11152–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Miki D, Ochi H, Hayes CN, et al. Variation in the DEPDC5 locus is associated with progression to hepatocellular carcinoma in chronic hepatitis C virus carriers. Nat Genet 2011;43:797–800. [DOI] [PubMed] [Google Scholar]
- [32].Petta S, Cammà C, Scazzone C, et al. Low vitamin D serum level is related to severe fibrosis and low responsiveness to interferon-based therapy in genotype 1 chronic hepatitis C. Hepatology 2010;51:1158–67. [DOI] [PubMed] [Google Scholar]
- [33].Sharma S, Khalili K, Nguyen GC. Non-invasive diagnosis of advanced fibrosis and cirrhosis. World J Gastroenterol 2014;20:16820–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Valva P, Ríos DA, De Matteo E, et al. Chronic hepatitis C virus infection: serum biomarkers in predicting liver damage. World J Gastroenterol 2016;22:1367–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Vallet-Pichard A, Mallet V, Nalpas B, et al. FIB-4: an inexpensive and accurate marker of fibrosis in HCV infection. Comparison with liver biopsy and fibrotest. Hepatology 2007;46:32–6. [DOI] [PubMed] [Google Scholar]
- [36].Holmberg SD, Lu M, Rupp LB, et al. Noninvasive serum fibrosis markers for screening and staging chronic hepatitis C virus patients in a large US cohort. Clin Infect Dis 2013;57:240–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Skaaby T, Husemoen LLN, Borglykke A, et al. Vitamin D status, liver enzymes, and incident liver disease and mortality: a general population study. Endocrine 2014;47:213–20. [DOI] [PubMed] [Google Scholar]
- [38].Hsia J, Heiss G, Ren H, et al. Calcium/vitamin D supplementation and cardiovascular events. Circulation 2007;115:846–54. [DOI] [PubMed] [Google Scholar]
- [39].Gerova DI, Galunska BT, Ivanova II, et al. Prevalence of vitamin D deficiency and insufficiency in Bulgarian patients with chronic hepatitis C viral infection. Scand J Clin Lab Invest 2014;74:665–72. [DOI] [PubMed] [Google Scholar]
- [40].Shehab H, Sabry D, Mukhtar M, et al. Vitamin D receptor foki gene polymorphism predicted poor response to treatment in chronic HCV genotype 4. Int J Biomed 2016;6:265–70. [Google Scholar]
- [41].Santos BR, Mascarenhas LPG, Satler F, et al. Vitamin D deficiency in girls from South Brazil: a cross-sectional study on prevalence and association with vitamin D receptor gene variants. BMC Pediatr 2012;12:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gutiérrez OM, Farwell WR, Kermah D, et al. Racial differences in the relationship between vitamin D, bone mineral density, and parathyroid hormone in the National Health and Nutrition Examination Survey. Osteoporos Int 2011;22:1745–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Fan L, Tu X, Zhu Y, et al. Genetic association of vitamin D receptor polymorphisms with autoimmune hepatitis and primary biliary cirrhosis in the Chinese. J Gastroenterol Hepatol 2005;20:249–55. [DOI] [PubMed] [Google Scholar]
- [44].Ochs-Balcom HM, Chennamaneni R, Millen AE, et al. Vitamin D receptor gene polymorphisms are associated with adiposity phenotypes. Am J Clin Nutr 2011;93:5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ginsberg HN, Zhang YL, Hernandez-Ono A. Regulation of plasma triglycerides in insulin resistance and diabetes. Arch Med Res 2005;36:232–40. [DOI] [PubMed] [Google Scholar]


