Abstract
Studies of human erythropoiesis have relied, for the most part, on the in vitro differentiation of hematopoietic stem and progenitor cells (HSPC) from different sources. Here, we report that despite the common core erythroid program that exists between cord blood (CB)- and peripheral blood (PB)-HSPC induced towards erythroid differentiation in vitro, significant functional differences exist. We undertook a comparative analysis of human erythropoiesis using these two different sources of HSPC. Upon in vitro erythroid differentiation, CB-derived cells proliferated 4-fold more than PB-derived cells. However, CB-derived cells exhibited a delayed kinetics of differentiation, resulting in an increased number of progenitors, notably CFU-E. The phenotypes of early erythroid differentiation stages also differed between the two sources with a significantly higher percentage of IL3R−GPA−CD34+CD36+ cells generated from PB- than CB-HSPCs. This subset was found to generate both BFU-E and CFU-E colonies in colony-forming assays. To further understand the differences between CB- and PB-HSPC, cells at eight stages of erythroid differentiation were sorted from each of the two sources and their transcriptional profiles were compared. We document differences at the CD34, BFU-E, poly- and orthochromatic stages. Genes exhibiting the most significant differences in expression between HSPC sources clustered into cell cycle- and autophagy-related pathways. Altogether, our studies provide a qualitative and quantitative comparative analysis of human erythropoiesis, highlighting the impact of the developmental origin of HSPCs on erythroid differentiation.
Keywords: Erythropoiesis, experimental models, developmental biology, hematopoietic stem and progenitor cells
INTRODUCTION
Erythropoiesis is the biological process that regulates the formation of red blood cells. This process is highly coordinated and involves a series of differentiation stages that are initiated in hematopoietic stem and progenitor cells (HSPCs), cell types that possess self-renewing and multi-potential differentiation potential, respectively. HSPCs give rise to more committed hematopoietic progenitors including myeloid progenitors, megakaryocytes and erythroid progenitors[1]. Recent data have demonstrated that, in humans, red cells can originate directly from HSPCs or from a more committed progenitor[2]. The erythroid burst-forming unit (BFU-E) is the most primitive single-lineage committed erythroid progenitor that can be functionally defined. This BFU-E then differentiates into a more committed erythroid colony-forming unit (CFU-E) that ultimately differentiates into proerythroblasts, the first morphologically recognizable erythroid precursor in the bone marrow[3]. Four to five divisions, occurring over the course of four to five days, lead to the orthochromatic stage, followed by enucleation and subsequent formation of the nascent reticulocyte. Reticulocyte maturation is the final stage of erythropoiesis and is characterized by extensive membrane remodeling and the loss of all internal organelles[4, 5].
Studies of erythropoiesis have historically relied, in large part, on mouse models. However, these models have failed to recapitulate various physiologic and pathological aspects of human erythroid differentiation. Indeed, recent studies from our groups and others have highlighted divergent epigenetic, transcriptional, and proteomic landscapes between murine and human erythropoiesis[6–13]. To better study human erythropoiesis, in vitro culture systems have been developed. Since the original method reported by Fibach[14], these culture systems have been continually adapted and modified[15–19] and we are now able to use CD34+ cells to recapitulate all stages of erythropoiesis, from the HSPC through the reticulocyte stage. These culture systems have promoted the study of normal and disordered erythropoiesis, providing important insights into the mechanisms regulating human erythroid differentiation. However, the majority of these studies used a single source of CD34+ progenitor cells with the broad assumption that differentiation data obtained from fetal liver (FL), cord blood (CB), bone marrow (BM) and peripheral blood (PB) are roughly equivalent.[6, 8, 20–24]. In addition, comparisons from previous studies were complicated by the inability to isolate pure populations of cells at distinct erythroid stages, a bottleneck that has recently been alleviated by the immunophenotypic characterization of each distinct stage of erythropoiesis[18, 21].
Here, we undertook a comparative analysis of the erythroid differentiation potential of human CB and PB CD34+ cells, from the BFU-E to the orthochromatic erythroblast. We found that CB-derived CD34+ cells differentiate at a slower rate than their adult counterparts but exhibit an increased reticulocyte differentiation potential. Furthermore, our studies reveal the existence and maintenance of a population of progenitors that is specific to PB, giving rise to both BFU-E and CFU-E. To better understand the functional differences between CB- and PB-HSPCs, we isolated individual stages of erythropoiesis and performed global gene expression profiles. These data significantly contribute to our understanding of the impact of the developmental origin of HSPCs on erythroid differentiation and provide an important resource for future studies of normal and disordered human erythropoiesis.
MATERIAL AND METHODS
CD34+ cell culture, flow cytometry analysis and fluorescence-activated cell sorting of erythroid progenitors and precursors
CB was obtained from the New York Blood Center and PB from the New York Blood Center and the Stanford Blood Center. The present study was conducted in accordance with the declaration of Helsinki and was exempt from institutional review board approval since none of the human samples had identifiable personal information. CD34+ cells were purified from CB or non-mobilized PB leukoreduction filters by magnetic positive selection (Miltenyi Biotec). Erythroid differentiation of isolated CD34+ cells (Table 1) was monitored as a function of surface marker expression on a BD LSR Fortessa (Becton Dickinson) and data subsequently analyzed using FCS express 6 software (De Novo, Inc) as previously described[18, 21]. Pure populations of erythroid cells at distinct stages of differentiation were sorted using a MoFlo high-speed cell sorter (Beckman-Coulter) as previously described[18, 21].
Antibodies
APC-conjugated CD235a (GPA), PE-conjugated-CD34, FITC-conjugated-CD36 and cell viability marker 7-AAD were purchased from BD Biosciences, PE-conjugated α4 integrin from Miltenyi Biotec, PE-Cyanine7-conjugated-IL-3R (CD123) from eBioscience, and a mouse mAb against human Band3 was used as previously described[18].
Colony assay
Cells were diluted to a density of 200 cells in 1ml of MethoCult® H4434 classic medium for BFU-E colony assay with SCF, IL-3, GM-CSF and Epo and 1ml of MethoCult® H4330 medium for CFU-E colony assay with Epo only (Stem Cell Technologies) and incubated at 37°C in a humidified atmosphere with 5% CO2.. BFU-E and CFU-E colonies were defined according to the criteria described by Dover et al[25]. CFU-E colonies were counted on day 7 and BFU-E colonies were counted on day 15 by investigators blinded to the experimental conditions.
Cytospin preparation
Cytospins were prepared on slides (5x104 cells in 200 μl of PBS), using the Thermo Scientific Shandon 4 Cytospin. Slides were stained with May-Grünwald solution (Sigma) for 5 minutes, rinsed in 40 mM Tris buffer for 90 seconds, and subsequently stained with Giemsa solution (Sigma) for 15 minutes. Cells were imaged using a Leica DM2000 inverted microscope under 100× objective magnifications.
RNA-Seq and bioinformatics analysis
RNA was extracted from FACS-sorted cells at 8 distinct stages of erythropoiesis, derived from CB- and PB-CD34+ cells. cDNA libraries were prepared using the Illumina TruSeq kit and sequenced on the Illumina HiSeq 2500 (Epigenomics Core of Weill Cornell Medical College, New York). This sequencing strategy produced ~15–80 million 50bp single end reads per sample. For each distinct stage of erythropoiesis, 3 biological replicates were obtained from independently cultured and sorted samples from different donations. Quality control of reads was performed and low-quality reads removed. Reads were aligned to the hg19 reference genome using HISAT2[26]. Raw read counts were extracted from the aligned reads using the featureCounts program[27]. Differential expression analysis was assessed at each stage by comparing the two original sources of CD34+ cells using the DESeq2 bioconductor package[28]. Using gene expression data from each stage, the subset of genes differentially expressed as a function of HSPC source was split into 10 clusters using divisive hierarchical clustering. Gene ontology enrichment analysis of the gene sets was then performed on each cluster using the cluster Profiler R package[29] with the gene background of all genes expressed across all stages in both sources.
Statistical analysis
Statistical evaluations between different experimental groups were performed using GraphPad Prism 7 (unpaired t-test) and p<0.05 was considered to indicate statistical significance.
RESULTS
CB-HSPCs lead to more erythroid cells than PB-HSPCs
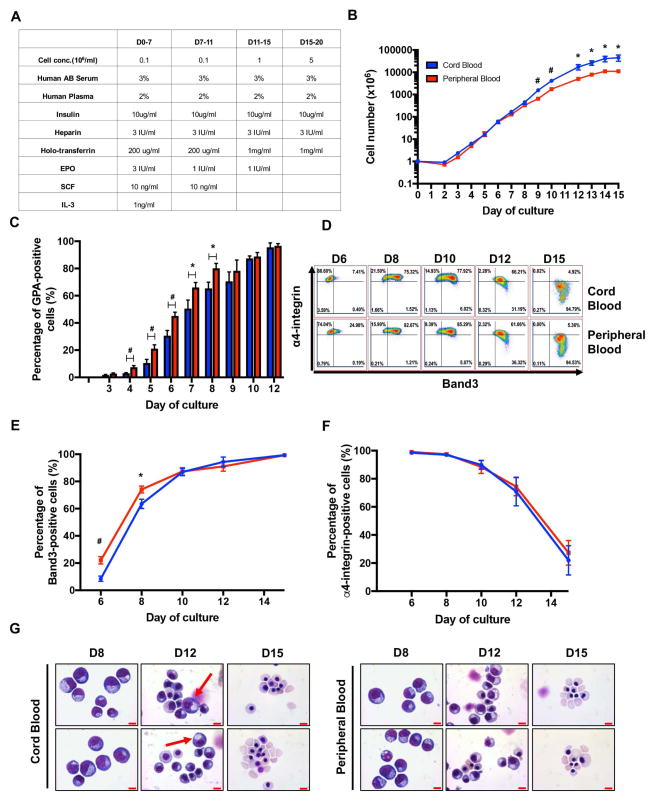
We first monitored the proliferative capacities of the two different sources of CD34+ cells during in vitro erythroid differentiation. CD34+ cells were isolated from CB or adult PB and then cultured for 15 days as described in the Material and Methods to induce their differentiation towards reticulocytes (Figure 1A). While a decrease in cell numbers was observed at Day 2 of culture of both CB and PB CD34+ cells, the cells recovered thereafter and started growing exponentially. From Day 3 to Day 6, there was no noticeable difference in the growth rates between the two sources of cells. However, while cells from CB continued to grow exponentially from Day 6 until Day 12, the growth of PB-derived cells was slower with significantly fewer cells by Day 9. By the end of culture, cell growth in both cultures plateaued, suggesting a terminal erythroid differentiation with decreased division (Figure 1G). CB-HSPCs yielded 4-fold more erythroid cells than PB-HSPCs under our culture conditions (Figure 1B, 45,085±14,103 vs. 10,988±1,218; p<0.05), suggesting that the former have an increased proliferation potential under conditions of erythroid differentiation.
Figure 1. Distinct kinetics and erythroid maturation potential of CB- and PB-derived CD34+ cells.
(A) Description of the erythroid differentiation culture system. (B) Growth curves of CB- and PB-derived CD34+ cells in erythroid culture medium (N=6). (C) Quantification of GPA expression, monitored by flow cytometry, on the indicated days of culture. Means±SEM are presented (N=9). (D) Terminal erythroid differentiation was monitored on the indicated days by flow cytometry and representative plots of α4-integrin versus Band3 within the GPA+ subset are shown (N=6). (E–F) Quantitative analysis of Band3 (E) and α4-integrin (F) expression on indicated days of culture (N=6). (G) Representative cytospin images of differentiated erythroblasts derived from CB and PB CD34+ cells. Arrows indicate the presence of more immature precursors in CB cultures; scale bar=10μm. *P<0.05, **P<0.01, #P<0.005.
CB-HSPCs exhibit a slower kinetics of EPO-induced differentiation than PB-HSPCs
The finding that CB-HSPCs proliferated to a significantly greater extent than their adult counterpart led us to investigate the differentiation capacities of the two sources of CD34+ cells. Using our recently published methods of immuno-phenotyping which allow for the characterization of each stage of human erythroid differentiation, from BFU-E to orthochromatic erythroblasts, we first monitored terminal erythroid differentiation using the expression of three surface markers, namely Glycophorin A (GPA), Band3 and α4-integrin[18]. Low, but detectable GPA expression was first observed in both cultures at Day 3. From Day 4 to Day 8, the levels of GPA increased in both cultures; however, the percentages of GPA-positive cells in adult PB-derived culture were significantly higher than that of their CB counterpart (Figure 1C). At Day 6, a mean of 42.4±3.2% of PB precursors were already GPA-positive as compared to 27.2±5.8% for CB precursors (p<0.01). Similar differences were detected at Day 8, with percentages of GPA-positive cells of 80.1±3.6% vs 65.4±4.6% for PB and CB precursors, respectively (p<0.05). This difference in GPA expression was associated with a delayed terminal erythroid differentiation in the CB-derived cultures, as documented by lower levels of Band3 expression at Day 6 and Day 8 (Figure 1D, E). After Day 10, differentiation was more similar for the two progenitors as reflected by expression patterns of GPA, Band3 and α4-integrin with completion of terminal erythroid differentiation (Figure 1C–F). Furthermore, Giemsa staining reflected the differentiation delay in CB-derived cultures; pro- and basophilic erythroblasts were still detected at Day 12 (Figure 1G, arrows). These data further suggest a slower kinetics of erythroid differentiation of CB-derived CD34+ HSPCs as compared to their adult PB-derived counterpart.
Increased persistence of CD34 expression in EPO-differentiated PB-HSPCs as compared to CB-HSPCs
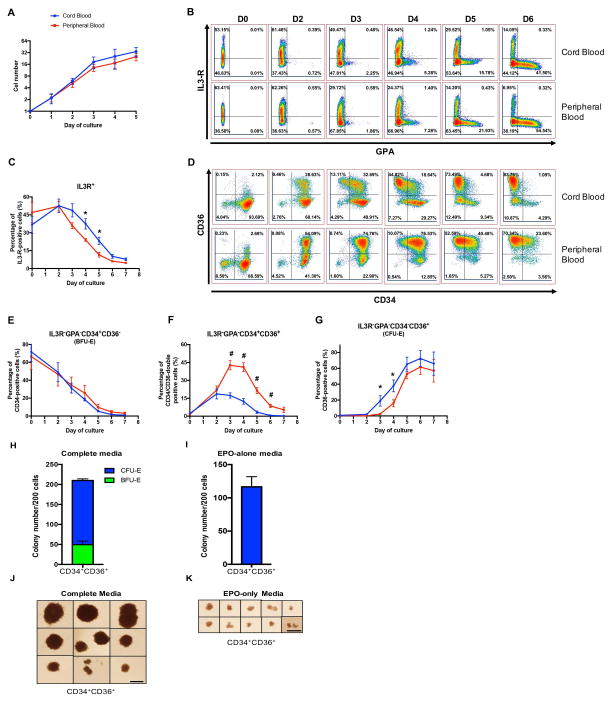
Since major differences were observed between Day 5 and Day 8 of culture, at a stage corresponding to the transition from CFU-E to proerythroblast, we further investigated the differentiation of erythroid progenitors and precursors based on the initial source of CD34+ cells. Proerythroblasts were sorted on the basis of their expression levels of GPA, Band3 and α4-integrin and their proliferation and differentiation towards orthochromatic erythroblasts were monitored. Although it is well documented that proerythroblasts undergo 4 to 5 cell divisions to generate orthochromatic erythroblasts, no comparative studies using highly pure populations of proerythroblasts have been performed. Interestingly, the growth patterns of proerythroblasts from both CB and adult PB were very similar (Figure 2A), suggesting that the growth differences observed between CB- and PB-HSPC sources are the result of a program initiated at the earlier progenitor levels.
Figure 2. CB-HSPCs exhibit a significantly higher CFU-E differentiation potential than PB-HSPCs.
(A) Growth curves of sorted proerythroblasts derived from CB (blue) and PB (red) CD34+ cells at the indicated time points (N=4). (B) Representative dot plots of GPA versus IL-3R expression at the indicated time points of erythroid differentiation. (C) Quantification of IL-3R expression at the indicated days of differentiation (N=6). (D) Representative dot plots of CD36 versus CD34 expression within the gated IL-3R−/GPA− population. BFU-E and CFU-E are identified as IL-3R−GPA−CD34+CD36− and IL-3R−GPA−CD34−CD36+, respectively. (E–G) Quantification of the percentages of IL-3R−GPA−CD34+CD36− (E), IL-3R−GPA−CD34+CD36+ (F), and IL-3R−GPA−CD34−CD36+ (G) cells at the indicated days of in vitro erythroid differentiation (N=6). (H–K) The erythroid colony forming ability of sorted IL-3R−GPA−CD34+CD36+ cells derived from PB was assessed in a colony-forming assay in complete medium supporting multiple lineage colony formation (H) and in the presence of EPO-alone (I), (N=3) and representative images of erythroid colonies generated from PB-derived IL-3R−GPA−CD34+CD36+ cells in complete media (J) or EPO-alone (K). Scale bar=1mm. *P<0.05, **P<0.01.
We therefore elected to focus on erythroid progenitors, and used the surface markers IL-3R, GPA, CD34 and CD36, to monitor differentiation of CB- or PB-HSPCs from Day 0 to Day 6. A similar pattern of expression of IL-3R and GPA was detected for both CB and PB-derived HSPCs, with a progressive loss of IL-3R and the acquisition of GPA (Figure 2B). Nevertheless, quantitatively, there were major differences between the two sources. Cells from both sources expressed comparative levels of IL-3R in the first two days of culture, but following that time point, cells derived from PB lost IL-3R at a faster rate than CB-derived progenitors (Figure 2B, C). Since erythroid colony-forming cells are enriched in the IL-3R-negative population, we further compared the differentiation potential of gated IL-3R−GPA− cells derived from both CB and PB (Figure 2D–F).
We previously showed that erythroid differentiation is characterized by an increased expression of CD36 and a loss of CD34. While the IL-3R−GPA−CD34+CD36− population predominantly generates more primitive BFU-E colonies, the IL-3R−GPA−CD34−CD36+ population contains CFU-E colony-forming cells. Using this gating strategy, we found that there was no difference in the relative percentages of immunophenotypically defined BFU-E (IL-3R−GPA−CD34+CD36−) between CB and PB throughout the culture process (Figure 2E). However, there were significantly more immunophenotypically defined CFU-E (IL-3R−GPA−CD34−CD36+) generated in CB in comparison to PB. Indeed, CFU-E represented 20% of the total cell population in CB-derived cultures at Day 3 while no CFU-E were found in PB. CFU-E appeared rapidly in PB-derived cultures thereafter and by Day 5, the percentages of CFU-E in CB- and PB-derived cultures were not statistically different (Figure 2G). However, it is important to note that a subset of CD34+CD36+ cells, phenotypically located in the transition between BFU-E and CFU-E, were present at significant numbers in PB- but not CB-derived cultures, from Days 3 to 6 of culture (Figures 2D, 2F).
These surprising findings led us to investigate the status and overall potential of the PB-derived CD34+CD36+ population. Indeed, these cells, potentially sharing characteristics with either BFU-E and/or CFU-E, have not been previously described. We sorted cells from this population and plated 200 cells to perform functional colony-forming assays. As one would expect, these cells respond to complete medium with SCF, IL-3, GM-CSF and EPO supporting multiple lineage colony formation (Figure 2H, J). In addition, we observed that these cells already have the ability to respond to EPO alone (Figure 2I, K), suggesting that they are already engaged in erythroid differentiation beyond the BFU-E stage. Taken together this data suggests that the CD34+CD36+ population, specific to PB-HSPCs, contains cells transitioning from a BFU-E to a CFU-E stage, with the ability to form both colonies.
Differences in the transcriptomes of CB-derived and PB-derived HSPC
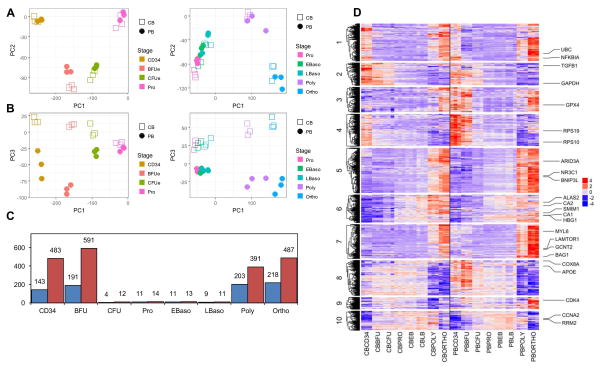
Using our defined surface markers, cells at each stage of erythroid differentiation were sorted to >95% purity and transcriptomes were compared as a function of the CD34+ cell source. The principle component analysis is shown in Figure 3A and demonstrates that cells at each distinct stage of development formed distinct clusters in the first two principle components (PC1 and PC2). However, all stages from CB and PB completely separated from each other into their own clusters when viewed using the 3rd principle component (PC1 versus PC3, Figure 3B). From this analysis, we can conclude that the differences between the two sources are smaller than the differences between stages.
Figure 3. Principal Component Analysis (PCA), frequency and cluster analysis of differentially expressed genes as a function of erythroid stage and HSPC source.
(A–B) PCA plot of 3 biological replicates of CB- and PB-derived cells at 8 different stages of erythroid differentiation. (A) Cells were sorted according to the PC1 and PC2 principle components; CD34+ progenitors to proerythroblasts (Pro) are presented in the left panel and proerythroblasts to orthochromatic erythroblasts (Ortho) are presented in the right panel. (B) The same samples were sorted according to PC1 and PC3 principle components with progenitors and terminal differentiation stages presented in the left and right panels, respectively. (C) The number of differentially expressed genes between CB and PB at each stage of erythroid differentiation are presented. Blue bars indicate genes that are downregulated in PB as compared to CB while red bars indicate upregulated genes. (D) Expression values of differentially expressed genes between CB and PB, at each of 8 stages of erythroid differentiation, are shown as a heat map. In each row, the color code is used to distinguish expression levels of a particular gene as indicated in the legend. Rows are organized by divisive hierarchical clustering and split into 10 clusters for CB (left panel) and PB (right panel). Enriched GO terms for Cluster 1: unfolded proteins, Cluster 2: leukocyte migration and activation, Cluster 3: oxidative phosphorylation, Cluster 4: ribosomal proteins, Cluster 5: autophagy, Cluster 6: gas transport-related, Cluster 7: autophagy, Cluster 8: mitochondrial respiratory chain complexes and assembly, Cluster 9: RNA splicing and mitochondrial translation, and Cluster 10: cell cycle.
The principle component analysis also showed that the gene expression pattern significantly differed during the very early development stages (i.e. at the CD34 and BFU-E stages) and at the very late differentiation stages (i.e. Polychromatophilic and Orthochromatic erythroblasts). This is in agreement with the significant differences in differentially expressed genes between the two sources at each stage (Figure 3C).
We then undertook an in depth comparative analysis of the transcriptome of human erythropoiesis. Many of the differentially expressed genes corresponded to well characterized differences between fetal and adult erythropoiesis, and were used as internal controls for our analysis[30]. These included those responsible for hemoglobin synthesis with fetal globin genes (HBG1, HBG2, and HBE1) significantly up-regulated in CB while adult globin genes (HBB, and HBD) were up-regulated in PB. Additionally, the gene encoding the I-branching enzyme, responsible for the conversion of fetal i antigen to adult I blood group antigen GCNT2[31–33], as well as the CA1 and CA2 genes, encoding carbonic anhydrases 1 and 2[34, 35], were up-regulated in PB as compared to CB (Supplemental Fig. 1).
To gain further insights into the overall observed changes in the biological processes between CB and PB, the differentially expressed genes at all stages were clustered. Divisive hierarchical clustering was used on the gene expression profile across all stages for both sources and the dendrogram partitioned into 10 clusters as shown graphically in Figure 3D. Gene ontology (GO) enrichment analysis of the biological process gene sets was performed on each cluster (Supplementary table 1). Clusters 2,3,4 and 8 had gene expression patterns showing differences in expression at the CD34 and BFU-E stages between CB and PB. Of note, Cluster 2 genes showed higher expression at the CD34 and BFU-E stages in CB over PB and enriched GO terms (P.adj<10−6) corresponding to leukocyte migration and activation, consistent with CB being less committed to the erythroid lineage than PB which is in agreement with our earlier findings (Figure 2). Clusters 1,5,7 and 9 exhibited patterns with differences in expression at the late stages, Polychromatophilic and Orthochromatic erythroblasts. Among those, Clusters 5 and 7 presented genes with increased expression in PB compared to CB, and these genes are associated with autophagy (P.adj<10−6). Cluster 10 genes exhibited a general increase in expression across many stages from BFU-E to Polychromatophilic erythroblasts in CB-derived cultures and was enriched in GO terms related to cell cycle, mitosis and DNA replication (P.adj<10−6).
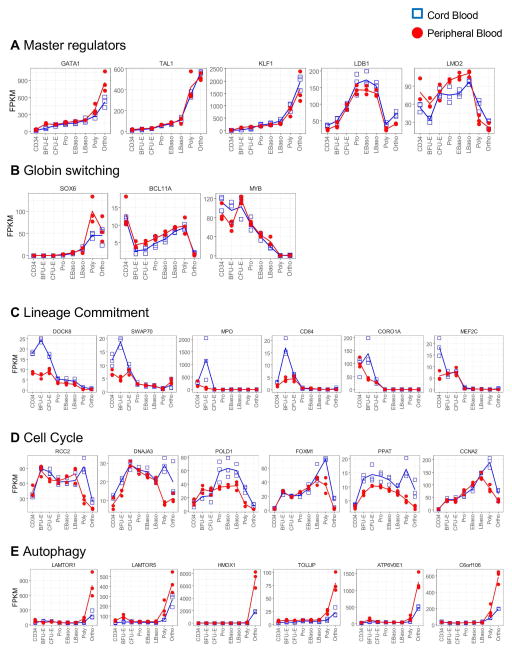
Within these clusters, we analyzed our transcriptome data on the basis of several genes critical for erythropoiesis or its regulation. The master regulators of erythropoiesis, defined as members of the core erythroid network[36], did not differ between CB and PB (Figure 4A), highlighting the similarities between the core differentiating programs. Recently, others have suggested that regulators of globin switching differ between CB and PB. In our study however, these did not significantly change (Figure 4B), except for MYB, which was described as unchanged in the Merryweather-Clarke study[23]. Consistent with the flow cytometry data, genes involved in lineage commitment were up-regulated in CB, at the CD34 and BFU-E stages (Figure 4C), supporting our data that CB-HSPCs have increased proliferative potential as compared to adult PB-HSPCs. This up-regulation was however transient and expression levels rapidly decreased as erythroid differentiation progresses to the CFU-E stage. We also observed that genes involved in cell cycle regulation were expressed at higher levels in CB than in PB (Figure 4D). Finally, we document that genes involved in the autophagy pathway were up-regulated in PB, specifically at the late stages (Figure 4E), consistent with a role for autophagy in late stages of erythropoiesis[37, 38]. Altogether, these data highlight the specificity in the developmental regulation of erythropoiesis and the differences between neonatal and adult erythropoiesis.
Figure 4. Expression profiles of specific genes that are critical for erythroid differentiation in CB- and PB-derived progenitors.
(A) Expression profiles of master regulators of erythropoiesis are presented as a function of differentiation stage and HSPC source (CB, blue; PB, red), with 3 samples per stage. (B) Expression profiles of genes involved in hemoglobin switching are presented as in (A). (C) Presentation of the top differentially expressed genes in cluster 2, enriched in GO terms related to lineage commitment. (D) Presentation of the top differentially expressed genes in cluster 10, enriched in GO terms related to cell cycle. (E) Presentation of the top differentially expressed genes in clusters 5 and 7, enriched in GO terms related to autophagy.
DISCUSSION
Erythropoiesis has been extensively studied with much research focusing on terminal erythroid differentiation. As such we have a better understanding of the molecular events leading to the formation of the reticulocyte from the pro-erythroblastic stage[39]. However, much less is known regarding the differentiation of human HSPCs to the precursor stages, and the events leading from the BFU-E to CFU-E progenitor stages have remained largely unexplored, mainly due to the difficulty of obtaining highly enriched populations of human erythroid progenitors. Different models have been used to study erythropoiesis, and all present their own unique advantages as well as challenges. Differences between species have been highlighted, notably with regards to changes in their transcriptomes during erythroid differentiation[36]. In the present study, we present evidence that even within the same species, the transcriptomes of erythroid progenitors differ as a function of the developmental source of HSPCs.
One of the main strengths of our study is that we used only one variable, the origin of the CD34+ cells. We induced differentiation of CD34+ cells from either CB or adult PB using the same culture conditions. We report that these cells exhibit distinct kinetics of differentiation and proliferation. CB-HSPCs lead to the generation of a significantly higher number of reticulocytes than adult PB-HSPC (4-fold) but their kinetics of differentiation is slower. Since the kinetics of growth from sorted pro-erythroblasts to orthochromatic erythroblasts are similar (Figure 2A), one can rule out terminal differentiation as potential mechanism for the differences observed. However, we document that CB-HSPCs retain IL-3R on their surface for a longer period time. In this context, and in accordance with our data, one may suggest that these cells can generate more immunophenotypically-defined CFU-E, which in turn will lead to a higher number of reticulocytes than PB-HSPCs.
In addition to the immunophenotypically defined IL3R−GPA−CD34−CD36+ that we previously characterized as a population leading to CFU-E in colony-forming assays, we observe the presence of a population specific to PB-HSPC, immunophenotypically defined as IL3R−GPA−CD34+CD36+. Using functional assays, we show that this population represents a transitory state during erythroid differentiation, able to give rise to both BFU-E and CFU-E in vitro. BFU-E generated from this population are heterogeneous in size, which, according to the empirical definition of a BFU-E[40], suggests that they represent both early and late BFU-E. It will be of interest to determine why this population is only very transient detected during the erythroid differentiation of CB-HSPCs.
This study is the first to compare the transcriptomes of sorted erythroid populations derived from PB and CB, allowing for a direct comparison of stage-specific gene expression during human erythropoiesis. One elegant study published in 2016 compared the transcriptome of human erythropoiesis using CD34+ cells from CB and adult PB and highlighted a similarity between the erythroid gene expression dynamics of adult and neonatal erythropoiesis[23]. In our study, we found similar data regarding the genes encoding the core erythroid program; however, we also found significant differences. In their study, Merryweather-Clarke et al. concluded that the transcriptomes of erythroid progenitors and precursors derived from CB- and adult PB-HSPCs were not different. The apparent discrepancies may be due to differences in the in vitro culture systems used in the two studies as well as differences in the analyzed subsets. To control for changes between progenitor populations, all our subsets were sorted, fostering high-resolution transcriptomics analyses.
The culture system utilized here together with the data sets for CB- and PB-derived erythroid progenitors will allow for a more robust evaluation of changes in pathological contexts. Our comparative analysis will undoubtedly foster research of multiple diseases where differentiation is being carried out on PB-HSPCs but the majority of comparative data have been obtained using CB-HSPCs.
In summary, our data demonstrate that the source of CD34+ cells is critical for the study of erythropoiesis, in normal and disordered conditions. It also provides a qualitative and quantitative comparative analysis of human erythropoiesis from CB and adult PB. All of the transcriptome data are deposited on Gene Expression Omnibus (GSE107218, password for review purpose only: qbshcykkfpozjcl). These data will provide a framework for studies of disordered human erythropoiesis and will foster the ex vivo evaluation of new therapeutic strategies for patients with bone marrow failure syndromes.
Supplementary Material
Expression profiles of α-, β-, γ- and δ-globin chains (top row), are presented as a function of erythroid differentiation stage and HSPCS source (CB, blue; PB, red; top row). Expression profiles of carbonic anhydrases genes 1 and 2 (CA1 and CA2) and the glucosaminyl (N-Acetyl) transferase 2, I-branching enzyme (I-blood group antigen) are presented (bottom row).
Supplemental Table 1. List of the GO-enriched terms related to Figure 3.
Acknowledgments
This research was supported in part by National Institutes of Health grants DK26263 (to N.M.) and DK32094 (to N.M. and N.T.), DK090145 (to A.N.), HL134043 (subcontract to L.B), and by the Pediatric Cancer Foundation (to L.B.). A.N is the recipient of an ASH Bridge award. L.B. is the recipient of a St. Baldrick’s Scholar award. N.T. and S.K are supported by a French national (ANR) research grant and the French Laboratory of excellence (Labex) GR-Ex.
Footnotes
CONFLICT OF INTEREST DISCLOSURE
The authors declare no competing financial interests.
AUTHORSHIP
H.Y. and J.H. designed and performed research, analyzed data, and edited the manuscript; J.J., J.L., Y.W., Y.H., and N.W. performed research, analyzed data, and edited the manuscript; X.A., and C.H. analyzed data and edited the manuscript; and S.K., N.T, N.M., A.N. and L.B designed research, analyzed data, and wrote the manuscript.
References
- 1.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Notta F, Zandi S, Takayama N, et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science. 2016;351:aab2116. doi: 10.1126/science.aab2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koury MJ, Haase VH. Anaemia in kidney disease: harnessing hypoxia responses for therapy. Nat Rev Nephrol. 2015;11:394–410. doi: 10.1038/nrneph.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanc L, Vidal M. Reticulocyte membrane remodeling: contribution of the exosome pathway. Curr Opin Hematol. 2010;17:177–183. doi: 10.1097/MOH.0b013e328337b4e3. [DOI] [PubMed] [Google Scholar]
- 5.Ney PA. Normal and disordered reticulocyte maturation. Curr Opin Hematol. 2011;18:152–157. doi: 10.1097/MOH.0b013e328345213e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.An X, Schulz VP, Li J, et al. Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood. 2014;123:3466–3477. doi: 10.1182/blood-2014-01-548305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.An X, Schulz VP, Mohandas N, et al. Human and murine erythropoiesis. Curr Opin Hematol. 2015;22:206–211. doi: 10.1097/MOH.0000000000000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pishesha N, Thiru P, Shi J, et al. Transcriptional divergence and conservation of human and mouse erythropoiesis. Proc Natl Acad Sci U S A. 2014;111:4103–4108. doi: 10.1073/pnas.1401598111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ulirsch JC, Lacy JN, An X, et al. Altered chromatin occupancy of master regulators underlies evolutionary divergence in the transcriptional landscape of erythroid differentiation. PLoS Genet. 2014;10:e1004890. doi: 10.1371/journal.pgen.1004890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brand M, Ranish JA, Kummer NT, et al. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat Struct Mol Biol. 2004;11:73–80. doi: 10.1038/nsmb713. [DOI] [PubMed] [Google Scholar]
- 11.Hricik T, Federici G, Zeuner A, et al. Transcriptomic and phospho-proteomic analyzes of erythroblasts expanded in vitro from normal donors and from patients with polycythemia vera. Am J Hematol. 2013;88:723–729. doi: 10.1002/ajh.23487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montel-Hagen A, Blanc L, Boyer-Clavel M, et al. The Glut1 and Glut4 glucose transporters are differentially expressed during perinatal and postnatal erythropoiesis. Blood. 2008;112:4729–4738. doi: 10.1182/blood-2008-05-159269. [DOI] [PubMed] [Google Scholar]
- 13.Richardson BM, Heesom KJ, Parsons SF, et al. Analysis of the differential proteome of human erythroblasts during in vitro erythropoiesis by 2-D DIGE. Proteomics Clin Appl. 2009;3:1123–1134. doi: 10.1002/prca.200900013. [DOI] [PubMed] [Google Scholar]
- 14.Fibach E, Manor D, Oppenheim A, et al. Proliferation and maturation of human erythroid progenitors in liquid culture. Blood. 1989;73:100–103. [PubMed] [Google Scholar]
- 15.Dulmovits BM, Appiah-Kubi AO, Papoin J, et al. Pomalidomide reverses gamma-globin silencing through the transcriptional reprogramming of adult hematopoietic progenitors. Blood. 2016;127:1481–1492. doi: 10.1182/blood-2015-09-667923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giarratana MC, Kobari L, Lapillonne H, et al. Ex vivo generation of fully mature human red blood cells from hematopoietic stem cells. Nat Biotechnol. 2005;23:69–74. doi: 10.1038/nbt1047. [DOI] [PubMed] [Google Scholar]
- 17.Giarratana MC, Rouard H, Dumont A, et al. Proof of principle for transfusion of in vitro-generated red blood cells. Blood. 2011;118:5071–5079. doi: 10.1182/blood-2011-06-362038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu J, Liu J, Xue F, et al. Isolation and functional characterization of human erythroblasts at distinct stages: implications for understanding of normal and disordered erythropoiesis in vivo. Blood. 2013;121:3246–3253. doi: 10.1182/blood-2013-01-476390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oburoglu L, Tardito S, Fritz V, et al. Glucose and glutamine metabolism regulate human hematopoietic stem cell lineage specification. Cell Stem Cell. 2014;15:169–184. doi: 10.1016/j.stem.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 20.Gautier EF, Ducamp S, Leduc M, et al. Comprehensive Proteomic Analysis of Human Erythropoiesis. Cell Rep. 2016;16:1470–1484. doi: 10.1016/j.celrep.2016.06.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Hale J, Bhagia P, et al. Isolation and transcriptome analyses of human erythroid progenitors: BFU-E and CFU-E. Blood. 2014;124:3636–3645. doi: 10.1182/blood-2014-07-588806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merryweather-Clarke AT, Atzberger A, Soneji S, et al. Global gene expression analysis of human erythroid progenitors. Blood. 2011;117:e96–108. doi: 10.1182/blood-2010-07-290825. [DOI] [PubMed] [Google Scholar]
- 23.Merryweather-Clarke AT, Tipping AJ, Lamikanra AA, et al. Distinct gene expression program dynamics during erythropoiesis from human induced pluripotent stem cells compared with adult and cord blood progenitors. BMC Genomics. 2016;17:817. doi: 10.1186/s12864-016-3134-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu J, Shao Z, Glass K, et al. Combinatorial assembly of developmental stage-specific enhancers controls gene expression programs during human erythropoiesis. Dev Cell. 2012;23:796–811. doi: 10.1016/j.devcel.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dover GJ, Chan T, Sieber F. Fetal hemoglobin production in cultures of primitive and mature human erythroid progenitors: differentiation affects the quantity of fetal hemoglobin produced per fetal-hemoglobin-containing cell. Blood. 1983;61:1242–1246. [PubMed] [Google Scholar]
- 26.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 28.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sankaran VG, Xu J, Orkin SH. Advances in the understanding of haemoglobin switching. Br J Haematol. 2010;149:181–194. doi: 10.1111/j.1365-2141.2010.08105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marsh WL. Anti-i: a cold antibody defining the Ii relationship in human red cells. Br J Haematol. 1961;7:200–209. doi: 10.1111/j.1365-2141.1961.tb00329.x. [DOI] [PubMed] [Google Scholar]
- 32.Marsh WL, Nichols ME, Reid ME. The definition of two I antigen components. Vox Sang. 1971;20:209–217. doi: 10.1111/j.1423-0410.1971.tb00432.x. [DOI] [PubMed] [Google Scholar]
- 33.Yu LC, Twu YC, Chang CY, et al. Molecular basis of the adult i phenotype and the gene responsible for the expression of the human blood group I antigen. Blood. 2001;98:3840–3845. doi: 10.1182/blood.v98.13.3840. [DOI] [PubMed] [Google Scholar]
- 34.Boyer SH, Siegel S, Noyes AN. Developmental changes in human erythrocyte carbonic anhydrase levels: coordinate expression with adult hemoglobin. Dev Biol. 1983;97:250–253. doi: 10.1016/0012-1606(83)90083-0. [DOI] [PubMed] [Google Scholar]
- 35.Weil SC, Walloch J, Frankel SR, et al. Expression of carbonic anhydrase and globin during erythropoiesis in vitro. Ann N Y Acad Sci. 1984;429:335–337. doi: 10.1111/j.1749-6632.1984.tb12358.x. [DOI] [PubMed] [Google Scholar]
- 36.Nandakumar SK, Ulirsch JC, Sankaran VG. Advances in understanding erythropoiesis: evolving perspectives. Br J Haematol. 2016;173:206–218. doi: 10.1111/bjh.13938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Betin VM, Singleton BK, Parsons SF, et al. Autophagy facilitates organelle clearance during differentiation of human erythroblasts: evidence for a role for ATG4 paralogs during autophagosome maturation. Autophagy. 2013;9:881–893. doi: 10.4161/auto.24172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Griffiths RE, Kupzig S, Cogan N, et al. The ins and outs of human reticulocyte maturation: autophagy and the endosome/exosome pathway. Autophagy. 2012;8:1150–1151. doi: 10.4161/auto.20648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ji P. New insights into the mechanisms of mammalian erythroid chromatin condensation and enucleation. Int Rev Cell Mol Biol. 2015;316:159–182. doi: 10.1016/bs.ircmb.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 40.Axelrad A, McLeod D, Shreeve M, Heath D. Properties of Cells that Produce Erythropoietic Colonies In Vitro. In: Robinson W, editor. Hemopoiesis in Culture. US Government Printing Office; Bethesda, MD: 1974. pp. 226–237. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression profiles of α-, β-, γ- and δ-globin chains (top row), are presented as a function of erythroid differentiation stage and HSPCS source (CB, blue; PB, red; top row). Expression profiles of carbonic anhydrases genes 1 and 2 (CA1 and CA2) and the glucosaminyl (N-Acetyl) transferase 2, I-branching enzyme (I-blood group antigen) are presented (bottom row).
Supplemental Table 1. List of the GO-enriched terms related to Figure 3.