SUMMARY
Structured mRNAs positioned downstream of the ribosomal decoding center alter gene expression by slowing protein synthesis. Here, we solved the cryo-EM structure of the bacterial ribosome bound to an mRNA containing a 3′ stem-loop that regulates translation. Unexpectedly, the E-site tRNA adopts two distinct orientations. In the first structure, normal interactions with the 50S and 30S E site are observed. However, in the second structure, although the E-site tRNA makes normal interactions with the 50S E site, its anticodon stem-loop moves ~54 Å away from the 30S E site to interact with the 30S head domain and 50S uL5. This position of the E-site tRNA causes the uL1 stalk to adopt a more open conformation that likely represents an intermediate state during E-site tRNA dissociation. These results suggest that structured mRNAs at the entrance channel restrict 30S subunit movement required during translation that slow E-site tRNA dissociation.
eTOC Blurb
Zhang, Hong et al. solved cryo-EM structures of the 70S ribosome interacting with mRNA containing a stem-loop at the mRNA entrance channel. The study provides insight into how the stem-loop interacts with uS3 and into the conformation of the E-site tRNA, suggesting how structured mRNAs impact translation.
INTRODUCTION
Ribosomes decode three-nucleotide mRNA codons into polypeptide sequences with a ~20 amino acids/second rate of amino acid incorporation into the growing nascent chain (Zaher and Green, 2009). This rate is influenced by both tRNA cellular abundance and the ability of mRNAs to adopt secondary structures that impede the helicase activity of the ribosome (Jacobson and Clark, 2016, Rodnina and Wintermeyer, 2016). In the latter case, these stable secondary structures are located downstream of the decoding center where they function as physical barriers to the mRNA transiting into the mRNA entrance channel and thus cause the ribosome to pause. Ribosomal pausing events are important recoding signals that can direct a subpopulation of ribosomes to decode a different mRNA reading frame (Farabaugh, 1996, Giedroc and Cornish, 2009, Dinman, 2012, Atkins et al., 2016) or allow the correct folding of the emerging peptide chain (Wachter, 2014, Jacobson and Clark, 2016, Rodnina and Wintermeyer, 2016).
Complementary base-pairing regions of mRNAs can interact to form more complex tertiary structures including stem-loops and pseudoknots. When located downstream of the decoding center, these stable mRNA structures slow translation by impeding the movement of tRNAs (translocation) through the ribosome and by decreasing the rate at which the exit(E)-site tRNA (E-tRNA) is ejected (Wen et al., 2008, Chen et al., 2013, Caliskan et al., 2014, Kim et al., 2014). Reducing the stability of structured mRNAs reverts translocation back to normal rates even though slow dissociation of E-tRNA persists (Chen et al., 2013). These studies indicate that structured mRNAs can decouple translocation and E-tRNA ejection but the molecular mechanism for this decoupling is unclear (Chen et al., 2013).
Here, we performed biochemical structure probing and determined a cryo-electron microscopy (cryo-EM) structure at near-atomic resolution of the 70S ribosome programmed with mRNA containing a downstream guanosine-cytosine (GC)-rich stem-loop. The stem-loop of this mRNA is from the E. coli dnaX mRNA which undergoes −1 frameshifting in the context of this stem-loop, an internal Shine-Dalgarno region and a polynucleotide sequence (Tsuchihashi and Kornberg, 1990, Tsuchihashi and Brown, 1992, Larsen et al., 1994, Larsen et al., 1997). The 3′-stem-loop of the dnaX mRNA induces a hyper-rotated state during ribosomal elongation (Qin et al., 2014) and impacts ribosome dynamics and tRNA translocation (Kim et al., 2014, Chen et al., 2014). We find that the stem-loop mainly forms interactions with ribosomal protein uS3, a helicase protein located at the surface of the mRNA entrance channel (“u” stands for universal according to the recently adopted ribosomal protein nomenclature (Ban et al., 2014)). Unexpectedly, the E-tRNA exists in two different positions providing insights into how structured mRNAs influence tRNA movement on the ribosome. These results suggest the mRNA stem-loop may restrict or interfere with dynamic 30S movements during ribosomal translocation and thus slow dissociation of E-tRNA.
RESULTS
The mRNA stem-loop forms in context of the 70S ribosome
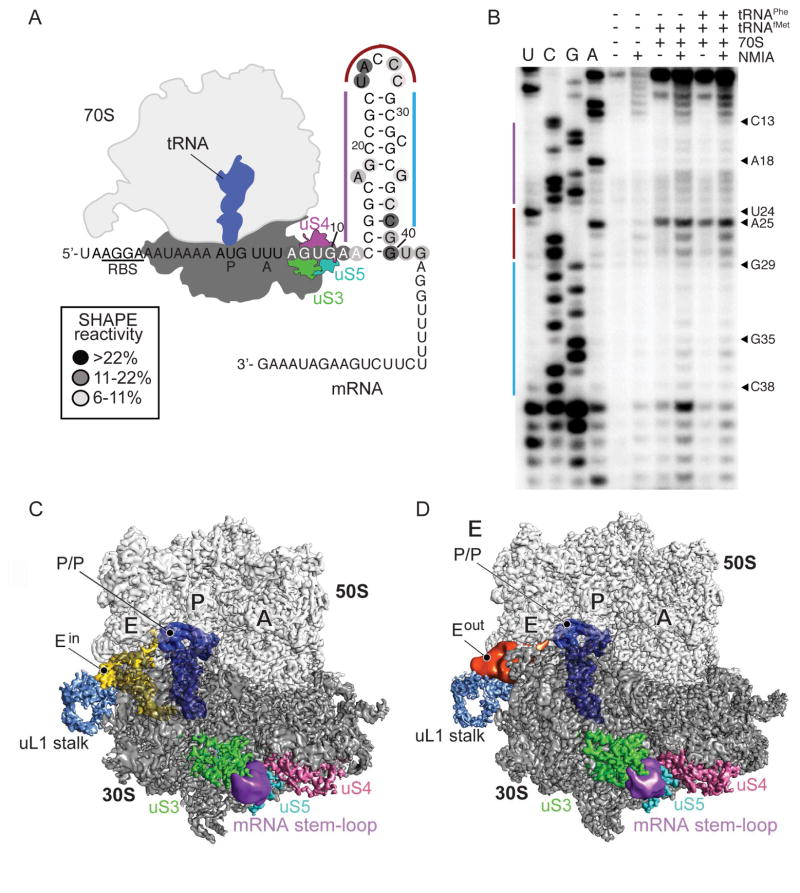
Previous single molecule FRET (smFRET) experiments of mRNA stem-loops and pseudoknots downstream of the ribosome decoding center or aminoacyl(A) site revealed that these mRNAs slow protein synthesis (Kim et al., 2014, Chen et al., 2014, Qin et al., 2014, Chen et al., 2013). In order to address whether such mRNAs adopt a partially or fully formed stem-loop, we formed 70S ribosome complexes with mRNA containing the 3′ stem-loop, P-site tRNAfMet and A-site tRNAPhe, and performed selective 2′-hydroxyl acylation analyzed by primer extension SHAPE analysis of the mRNA (Figure 1) (Smola et al., 2015). Our analysis revealed lower N-methylisatoic anhydride (NMIA) reactivity for nucleotides 13–17, 20–23 and 29–31 suggesting these nucleotides form a nine-nucleotide base-paired stem (Figures 1A, B; the first P-site codon nucleotide is numbered as +1). Nucleotide C13 demarcates the first base-paired nucleotide of the stem-loop and the nucleotides to the stem (nucleotides 13–17) form base-pairing interactions as assessed by the lack of NMIA reactivity. Residues predicted to form single-stranded or bulged regions (nucleotides A18 & G35) show increased levels of NMIA reactivity confirming that these nucleotides do not form base-pairing interactions. The five nucleotides in the loop (24–28) are also relatively flexible, with nucleotides U24 and A25 displaying high NMIA reactivity, suggesting highly dynamic and flexible nucleotides. Loop nucleotide C26 and predicted bulged nucleotide C32 are less dynamic, suggesting these residues may be forming tertiary interactions with either other mRNA regions or the ribosome itself. Nucleotides 29–31 and 33–34 located in the 3′ part of the stem-loop are involved in base-pairing interactions, while further towards the base of the stem, nucleotides 37–40 show increasing levels of NMIA reactivity. These data indicate the stem-loop is formed adjacent to the mRNA entrance channel.
Figure 1. Interactions between the 70S and stem-loop of the mRNA.
(A) Schematic representation of 70S bound to mRNA containing a downstream stem-loop complex. Ribosomal binding site (RBS) denotes Shine-Dalgarno-like sequence. (B) RNA SHAPE probing of the mRNA with the NMIA reactivity mapped on the mRNA in panel A. (C) Cryo-EM maps of 70S-mRNA containing the E-site tRNAin (Ein) and (D) 70S-mRNA containing the E-site tRNAout (Eout) complex. The cryo-EM density for the E-site tRNA that adopts an “out” conformation (panel D) and the mRNA stem-loop is low-pass filtered to 10 Å and shown at the 2σ threshold level.
Structure of the mRNA stem-loop bound to the 70S
In an attempt to understand the structural basis for how the 3′ stem-loop regulates translation, we solved a high-resolution cryo-EM structure of the Thermus thermophilus 70S bound to the same mRNA as in the smFRET studies (Qin et al., 2014). The 70S was programmed with P- and A-site tRNAs, and mRNA containing the downstream GC-rich 3′ stem-loop (Figure 1A). Three-dimensional (3D) classification of cryo-EM particle projections showed three distinct ribosomal complexes: 70S bound to both P- and E-tRNAs, 70S bound to E-tRNA, and the large ribosomal subunit alone (Figure S1- related to Figure 1). Refinement of the structure corresponding to 70S bound to both P- and E-tRNAs produced a cryo-EM map with a global resolution of 3.1 Å and revealed the ribosome in a non-rotated conformation (Figure S1, S2- related to Figure 1). The map shows excellent map density for the P-tRNA, however, there is little to no density for the A-tRNA, the A-site mRNA codon (+4 – +6 nucleotides) and the nucleotides +7 to +12 that comprise the path from the 3′ edge of the A site to the start of the stem-loop (Figure 1A). While one reason for the absence of A-tRNA could be the lack of aminoacylation of the tRNA, we note many other structural studies of cognate tRNA-mRNA pairs where deacylated tRNAs are used, still bind the A site (Yusupov et al., 2001, Selmer et al., 2006, Korostelev et al., 2006, Jenner et al., 2010b). In support of an A site lacking tRNA, 16S rRNA decoding nucleotide A1492 is located inside helix h44 while A1493 is half-splayed out (Figure S3- related to Figure 1), similar to what has been observed when the A site lacks tRNAs (Jenner et al., 2010a). The E-tRNA is well-resolved in our maps although we did not program the 70S with tRNA (Figure S4- related to Figure 1). Non-specific, deacylated tRNA binding at the E site has been observed in a number of structural studies (Yusupov et al., 2001, Selmer et al., 2006, Korostelev et al., 2006). After employing local 3D classification and alignment on E-tRNA (discussed in detail below), we identified two different E-tRNA conformations. While the core of the ribosome in these two 70S conformers is resolved at near-atomic level (3.2 and 3.5 Å, respectively), the densities for the stem-loop and the 5′-Shine-Dalgarno region have much lower resolution (7–10 Å), suggesting conformational variability.
The interactions of the mRNA stem-loop with 30S ribosomal protein uS3
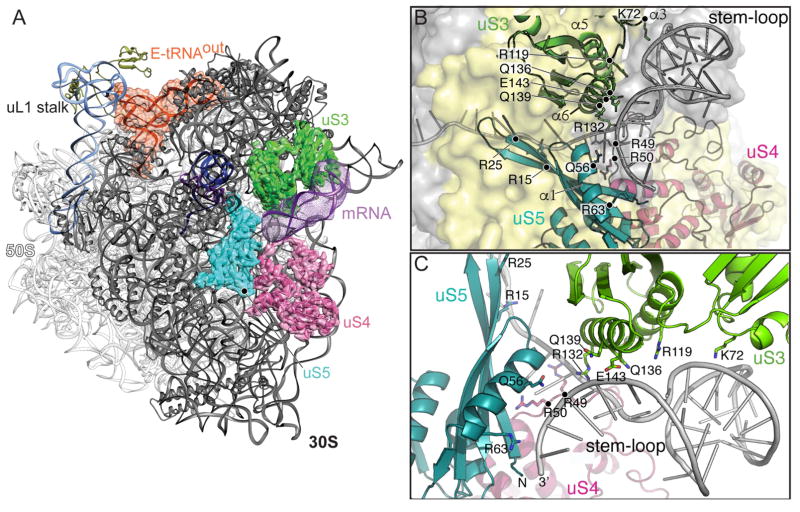
uS3, uS4 and uS5 are RNA helicase proteins located at the ribosome entrance channel on the 30S subunit (Takyar et al., 2005). A number of uS3 and uS4 arginine and lysine residues were predicted to contribute to this helicase activity; in these experiments, upon mutation to alanine, the ribosome was unable to unwind nucleic acid duplexex, likely by preventing electrostatic interactions with the mRNA phosphate backbone (Takyar et al., 2005). Considering that stable structured mRNAs can slow the rate of translation (Qu et al., 2011, Chen et al., 2013, Caliskan et al., 2014, Kim et al., 2014, Chen et al., 2014), a partial or full stem-loop is predicted to be adjacent to uS3, uS4 and uS5. To clearly visualize the mRNA density along its path, we applied a 10-Å low-pass filter to the difference map generated between the model and the cryo-EM maps. The map show that the stem-loop, which is reconstructed at low resolution (7–10 Å) likely due to its inherent positional flexibility, interacts with uS3, uS4 and uS5 at the entrance of the mRNA channel (Figure 2A). The densities for uS3, uS4 and uS5 are resolved at near-atomic resolution (3–4 Å).
Figure 2. Localization of the mRNA stem-loop and analysis of its interaction with the 30S subunit.
(A) Overview and details of the 70S-mRNA particle shown with the 30S in the forefront and cryo-EM densities for ribosomal proteins uS3 (green), uS4 (pink) and uS5 (cyan). (B) mRNA stem-loop in the same view as the panel A. uS3, uS4 and uS5 helices and residues potentially interacting with the stem-loop are shown. (C) A ~45° rotated view highlighting potential uS3 regions that interact with the stem-loop. The density for the mRNA stem-loop is low-pass filtered to 10 Å and shown at the 2σ threshold level.
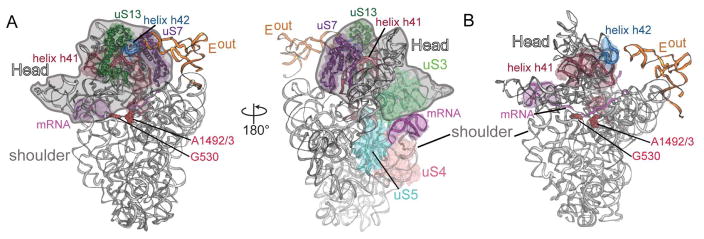
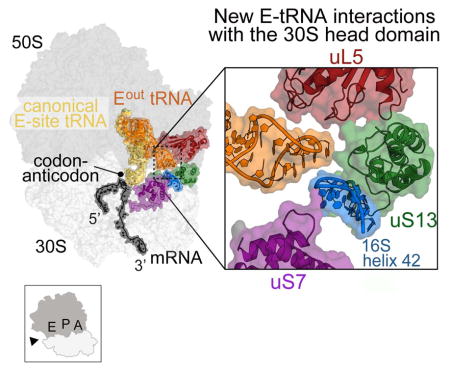
At the entrance of the mRNA channel, residues Arg49 and Arg50 of uS4 and Arg15 and Arg25 of uS5 are proximal to the mRNA (Figures 2B, C). In addition to pointing towards the mRNA path, uS4 Arg50 also forms interactions with the region of mRNA 5′ to the stem-loop. Arg44 and Arg47 of uS4 in E. coli were previously shown to be critical for ribosomal helicase activity, consistent with our structure in which the corresponding region within the uS4 α1-α2 linker (containing E. coli Arg47 or T. thermophilus Arg49) is proximal to the mRNA path (Figure 2; Figure S5B- related to Figure 2). uS5 variants R19A/R28A (T. thermophilus R15/R24) had little effect on activity (Takyar et al., 2005), and our structure suggests this is likely because each side chain faces the opposite direction of the mRNA path (Figures 2; Figure S5B- related to Figure 2). Instead, the side chains of uS5 residues Gln56 and Arg63 face towards the mRNA path (Figures 2B, C). Although the density surrounding the stem-loop is not of sufficient resolution to discern electrostatic interactions, the base of the stem-loop primarily interacts with uS3 α5 and α6, while the apical stem nucleotides and possibly the loop region interact with α3 (Figures 2B, C). Consistent with this interpretation, mutation of uS3 residues Arg131, Arg132 and Lys135 results in defective helicase activity (Takyar et al., 2005). The side chains of Arg131 and Lys135 face away from the path while residues Arg132 and Gln136 side chains are directed towards the mRNA (Figures 2B, C). uS3 residues located adjacent to the mRNA path include Lys72, Arg119, Gln139 and Glu143, which have not been previously proposed to interacting with mRNA (Figure 2C). The close interaction between the stem-loop and uS3 is functionally important because uS3 is associated with the 30S head domain while uS4 and uS5 are associated with the 30S body domain. The 30S head domain moves substantially during translocation of the tRNAs (Ratje et al., 2010, Tourigny et al., 2013, Zhou et al., 2014, Wasserman et al., 2016, Belardinelli et al., 2016), and the stem-loop associating closely with uS3 may interfere with this movement (Figure 6B).
Figure 6. Association of 30S head domain with Eout tRNA and the mRNA stem-loop.
(A) The anticodon stem-loop of Eout tRNA prods the 30S head domain via uS7, uS13 and helix h42, while the mRNA stem loop stacks against the opposite side of the 30S head domain via uS3. The head domain is shown in outline and 16S rRNA decoding nucleotides A1492/3 and G530 are shown to emphasize the position of the 30S A site. (B) 16S rRNA shown only to emphasize the connection between the mRNA stem-loop (magenta), the A-site decoding center (demarcated by G530 and A1492/3), helix h41 (red) and finally to helix h42 (blue) that interacts with Eout tRNA.
The anticodon stem-loop of the Eout-tRNA forms interactions with the 30S head domain
tRNAs bind both the small 30S subunit and large 50S of the ribosome: the anticodon stem-loop interacts with mRNA on the 30S while the tRNA acceptor arm interacts with the 50S. In the E site, 30S ribosomal proteins uS7 and uS11 contact the anticodon stem region and 23S rRNA Helix H88 and the uL1 protein interact with the T and D loops, respectively (Yusupov et al., 2001, Selmer et al., 2006, Korostelev et al., 2006) (Figure 3A). A further round of 3D classification of cryo-EM projections with the alignment focusing on the E-tRNA revealed the existence of two ribosome conformers. In the first E-tRNA conformation that we call the Ein conformation, the global resolution is 3.5 Å (Figures 1D & 3; Figure S1- related to Figure 1). The Ein tRNA maintains all canonical interactions with both the 30S and 50S as previously observed (Yusupov et al., 2001, Selmer et al., 2006, Korostelev et al., 2006) (Figure 3A). The second conformation has a global resolution of 3.2 Å and adopts a very different conformation than the Ein tRNA (Figures 1D & 3; Figure S1- related to Figure 1). In this E-tRNA conformation that we call Eout, nearly all interactions of the E-tRNA acceptor arm with Helix H88 are maintained with the 50S E site (Figure 3). However, the anticodon stem-loop pivots at the acceptor stem, moving the anticodon stem-loop ~54 Å away from 30S E site and thus, from the mRNA path (Figure 3B). In this orientation, the anticodon stem-loop of the Eout tRNA forms new interactions with the 30S head domain including uS7 (loop residues 112-115), uS13 (residues 10-13), and 16S rRNA helix h42 (nucleotides 1295–1304) as well as the 50S protein uL5 (loop residues 145-150) (Figure 3A).
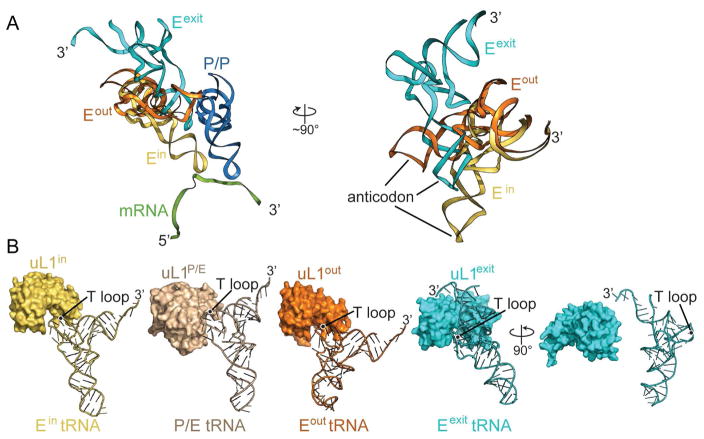
Figure 3. The anticodon stem-loop of Eout tRNA adopts a new position on the 70S.
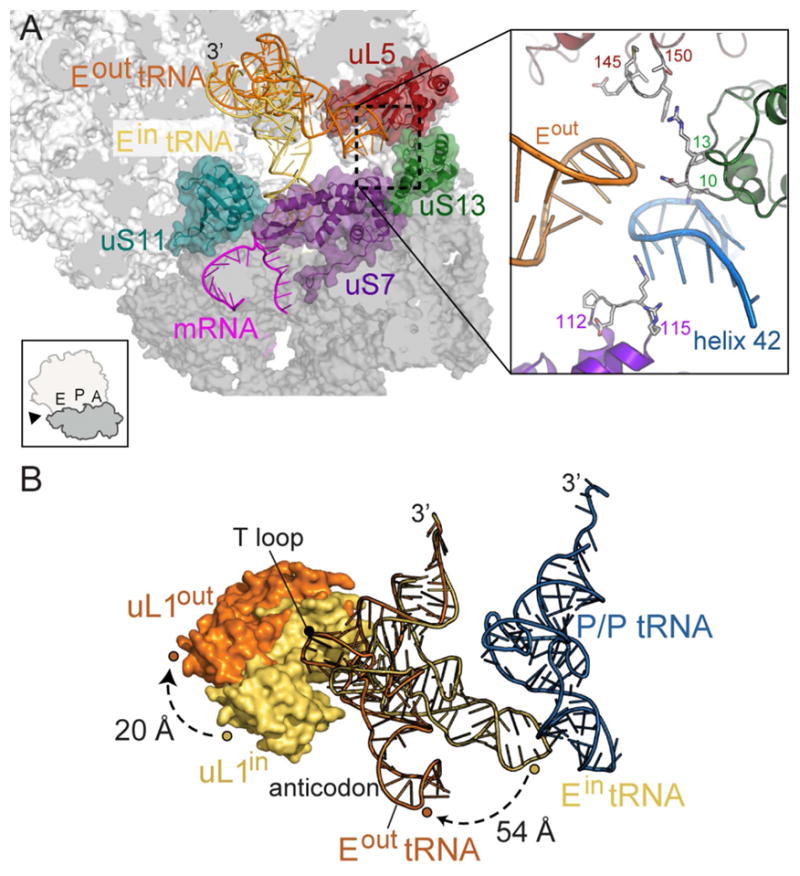
(A) Overview of the positions of the Eout and Ein tRNA on the 70S. The 3′ acceptor arms of both tRNAs superimpose on the 50S and the anticodon stem-loop of the Ein tRNA (yellow) is buried in the 30S E site where it interacts with mRNA. In the right inset panel, the Eout tRNA forms interactions with uS7, uS13 and 16S rRNA helix h42. Residues close to Eout tRNA are shown as sticks and the residue numbers are indicated. The left inset panel indicates the view of the ribosome in the main panel. (B) tRNA and uL1 protein conformations in Ein-uL1in and Eout-uL1out, both from this study. The uL1out protein moves ~20 Å, compared to uL1in. The anticodon stem-loop of the Eout tRNA is displaced ~54 Å compared to Ein tRNA, resulting in ~40° pivoting. The E-site tRNAs and uL1 proteins from the same structures are shaded similarly. The T-loop regions of Ein and Eout tRNAs interact directly with uL1.
The Eout tRNA causes uL1 to adopt an intermediate open conformation
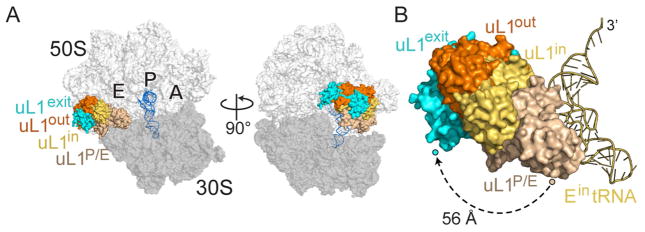
The solvent-exposed, flexible uL1 stalk adopts different conformations depending on the location of the E-tRNA (Fei et al., 2008, Cornish et al., 2009). During the elongation cycle, tRNA is decoded at the A site, transits to the P site after peptidyl transfer, and finally occupies the E site before leaving the ribosome. When the E site is empty during initiation, the uL1 stalk is flexible and can alternate between open and closed conformations (Fei et al., 2008, Munro et al., 2010, Cornish et al., 2009). After peptide bond formation, the two tRNAs move on the 50S to P/E and A/P hybrid states (P/E denotes the anticodon stem-loop location on the 30S P site and the acceptor arm of the tRNA on the 50S E site; A/P denotes the anticodon stem-loop location on the 30S A site and the acceptor arm of the tRNA on the 50S P site) and the 30S subunit rotates (Dunkle et al., 2011). In this hybrid state, the uL1 stalk assumes a fully closed conformation, blocking the E-tRNA from exiting (Tourigny et al., 2013, Bock et al., 2013) (Figure 4A; uL1P/E). After translocation of the tRNAs to E/E and P/P locations, the uL1 protuberance moves away from the 30S to assume a half-closed conformation (Figures 3B; uL1in; “in” refers to the corresponding Ein tRNA state) (Selmer et al., 2006). In the case of the Eout tRNA seen in our structure, the uL1 protein further moves by ~20 Å to a more open conformation (Figure 3B; compare uL1in to uL1out). Our structure shows that the Eout tRNA directly affects the location of uL1, consistent with other studies where the location of the E-tRNA impacts uL1 conformational dynamics (Fei et al., 2008, Cornish et al., 2009). The uL1out position we observe appears to be at a later stage in the E-tRNA dissociation pathway when compared to a fully closed state (P/E hybrid tRNA bound) and a half-closed state (E/E tRNA bound). These comparisons suggest we have trapped a conformational intermediate during E-tRNA dissociation (Figure 4B). In additional support of this, the uL1out conformation we find is less open than uL1exit (Figure 4B; compare uL1out and uL1exit), a state that appears to be further along the E-tRNA ejection pathway than what we observe (Figure 5; tRNAexit; see discussion below).
Figure 4. uL1 protein conformations.
(A) The uL1 protein adopts a fully closed state when bound to P/E tRNA (uL1P/E; PDB code 4V9H) and forms its most open state upon tRNA departing the ribosome (uL1exit; PBD code 4V79). The 30S is shown in dark grey, the 50S in light gray and the P-site tRNA in blue. The two E-tRNA states in this study are shown for comparison (Ein tRNA and Eout tRNA). (B) The same view as in the left panel A except Ein tRNA is included for reference. The displacement of uL1 protein from the uL1P/E position when the uL1 protuberance is in its most closed position to the most open conformation as seen with uL1exit is ~56 Å.
Figure 5. Comparison of Eexit tRNA with all E-tRNA states.
(A) The conformation of the Eexit tRNA (PBD code 4V79) compared to Ein tRNA and Eout tRNA (both from this study). The mRNA and P/P tRNA (PDB code 4V51) are shown for reference. In the right panel, a 90° rotation emphasizes the change in the position of the anticodon as indicated. (B) A comparison of how Ein tRNA (this study), P/E tRNA (PDB code 4V9H), Eout tRNA (this study), and Eexit tRNA (PBD code 4V79) interacts with uL1. The Ein, P/E and Eout tRNA interact with uL1 via their T-loops, while in the case of the Eexit tRNA (PBD code 4V79), the tRNA interacts with uL1 via its 3′ end and not through its T-loop. All four E-site tRNA-uL1 pairs are shown in the same orientations on the ribosome. The far right uL1-Eexit tRNA panel is rotated 90° to emphasize the interaction with the acceptor arm of the tRNA.
DISCUSSION
The mRNA bound to the 70S used in this study has a modified E. coli dnaX-like sequence containing a 3′-stem-loop which disrupts ribosome dynamics by either inducing a hyper-rotated state (Qin et al., 2014) or slows the movement of tRNAs through the ribosome (Kim et al., 2014, Chen et al., 2013, Chen et al., 2014). Our structures reveal a non-rotated ribosome with an unexpected E-tRNA conformation when the mRNA stem-loop is positioned at the entrance channel. This structure is consistent with prior studies that show complex mRNA secondary structures located downstream of the decoding center prevent hybrid state formation (Kim et al., 2014) and impact E-tRNA dissociation and translocation (Chen et al., 2013, Chen et al., 2014).
The primary interactions with the downstream mRNA stem-loop are mediated by ribosomal helicase protein uS3, a component of the 30S head domain located at the mRNA entrance channel. In contrast, the other helicase proteins uS4 and uS5, which are part of the ribosome shoulder domain, make minimal interactions with the stem-loop (Figure 2). During decoding of tRNAs at the A site, both the head and shoulder domains move relative to one another to signal recognition of a cognate interaction with the mRNA (Ogle et al., 2002) (Figure 6A). Translocation requires large-scale ratcheting of the 30S subunit followed by head domain swiveling during which uS3 is displaced by 12–14 Å, while uS4 and uS5 remain largely unperturbed (Zhou et al., 2013, Zhou et al., 2014). Although the A and E sites are on opposite sides of the ribosome, these regions are connected by 16S rRNA (Figure 6B). The Eout tRNA interacts with helix h42, a 16S rRNA helix that is connected to the decoding center helix h44 via the long helix h43 (Figure 6B). One possibility is that the stem-loop may lock uS3 in a conformation that prevents 30S head domain movement. The inhibition of such dynamics would interfere with tRNA movement, in turn, slowing the dissociation of the E-tRNA consistent with smFRET experiments (Kim et al., 2014, Chen et al., 2013, Chen et al., 2014). Whether this signal is communicated to the E site via helices h42/h43/44 to slow Eout tRNA ejection is unknown.
The anticodon stem-loop of the Eout tRNA no longer interacts with mRNA in the 30S E site and instead, forms interactions with the 30S head domain including uS7, uS13, and 16S rRNA helix h42, and 50S protein uL5 (Figure 3A). Noncognate, deacylated tRNA is bound in the E site, and it is unclear whether it is tRNAPhe (anticodon 5′-GAA-3′) or tRNAfMet (anticodon 5′-CAU-3′). However, regardless of which tRNA is bound, the interaction is noncognate in nature (the E-site mRNA codon is AAA). Numerous ribosome structures have been determined in which deacylated tRNA is bound at the E site and forms a noncognate interaction; yet, in all these structures, the overall location of the E-tRNA conforms to a canonical Ein position (Yusupov et al., 2001, Selmer et al., 2006, Korostelev et al., 2006). The inability to observe the Eout tRNA position in previous studies could arise from a transient nature of this interaction which would therefore may not be observed in structural studies using X-ray crystallography. However, in the presence of the downstream structured mRNA, the dissociation of E-tRNA is slowed, allowing this state to be populated to a greater extent and thus its visualization in our cryo-EM analysis.
In two previous cryo-EM studies, the E-tRNA was also found to occupy multiple positions with the most relevant to this study being the “E2” (Agrawal et al., 2000) and “POST3” states (Fischer et al., 2010). Since they are both very similar, we refer to them collectively as Eexit tRNA (Figure 5). In these two structures, the Eexit tRNA also only interacts with the 50S subunit and this E-tRNA position has been proposed to be an intermediate state preceding tRNA ejection (Fischer et al., 2010) (Figure 5A). The Eexit tRNA does not interact with either normal 50S and 30S E sites but instead, interacts exclusively with the uL1 protein via its acceptor arm (Figure 5B). In contrast, in all other structures to date and in our structures presented in this study, uL1 interacts with the E-tRNA via its T-loop (Figure 5B). Therefore, given the Eexit tRNA has no interactions with the 50S or 30S E sites and has minimal interactions with uL1, the Eexit conformation likely represents a state at a late point in E-tRNA dissociation from the ribosome (Fischer et al., 2010).
The Eout tRNA position we observe is distinct from the Eexit tRNA (Figure 5, compare Eout to Eexit). We propose that the Eout tRNA in our structure represents an on-pathway state earlier in its dissociation from the ribosome given the manner in which the uL1 interacts with the Eout tRNA and the conformational trajectory of uL1in. As mentioned, uL1 interacts with Eout tRNA via its T-loop in a manner consistent with all previously solved structures including fully accommodated E-tRNA state (Ein) and a hybrid P/E tRNA state (Yusupov et al., 2001, Selmer et al., 2006, Korostelev et al., 2006, Dunkle et al., 2011)(Figure 5B). Additionally, uL1out also appears to be on the same trajectory as uL1in and uL1P/E (that contain Ein and P/E tRNAs, respectively), albeit at a later step (Figure 4). Taken together, these interactions suggest our Eout tRNA is an on-pathway intermediate.
In light of studies demonstrating that GC-rich stem-loops downstream of the decoding center slow translocation (Wen et al., 2008, Chen et al., 2013) and prevent hybrid state formation (Kim et al., 2014), the alternate location of the Eout tRNA in our structure provides insights into two possible ways that structured mRNAs may regulate this step of translation. First, our structure provides a rationale for how Eout tRNA interferes with and sterically blocks the movement of the P/P tRNA to a P/E hybrid state. Second, the interaction between the Eout tRNA and the 30S head domain may also restrict the intersubunit ratcheting that is required to move the tRNAs through the ribosome, consistent with studies showing dnaX mRNA impacts ribosome dynamics and results in −1 frameshifting (Qin et al., 2014, Kim et al., 2014, Chen et al., 2013, Chen et al., 2014). Thus, the inability to fully eject the E-tRNA in combination with the restriction of ribosome dynamics may lead to a programmed slowing of translocation (Chen et al., 2013) (Figure 6B).
STAR METHODS
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial strains | ||
| E. coli MRE 600 | N/A | |
| Thermus thermophilus HB8 | Bioexpression and Fermentation Facility, University of Georgia | N/A |
| Recombinant DNA | ||
| Plasmid: pdnaX-pUC19 | (Qin et al., 2014) | N/A |
| Oligonucleotides | ||
| 5′-CTTTATCTTCAGAAGAAAAACC-3′ | IDT | N/A |
| tRNAfMet | Chemical Block | N/A |
| tRNAPhe | Chemical Block | N/A |
| Deposited Data | ||
| T. thermophilus 70S ribosome for three-dimensional classifications (Cryo-EM image processing) | (Bai et al., 2013) | EMDB code 2277 |
| T. thermophilus 70S ribosome for model building by rigid docking | (Polikanov et al., 2015) | PDB code 4Y4O |
| T. thermophilus 70S ribosome for extracting uL1 coordinates | (Gao et al., 2009) | PDB code 4V5F |
| T. thermophilus 70S ribosome in translocational intermediate state containing P/E tRNA | (Tourigny et al., 2013) | PDB code 4V9H |
| E. coli 70S ribosome containing uL1exit and Eexit tRNA | (Bock et al., 2013) | PDB code 4V79 |
| T. thermophilus 70S ribosome for extracting P/P tRNA | (Selmer et al., 2006) | PDB code 4V51 |
| T. thermophilus 70S ribosome in translocational intermediate (chimeric) state | (Ratje et al., 2010) | PDB code 4W29 |
| 70S ribosome containing Ein | This study | PDB code 5UQ7 |
| 70S ribosome containing Eout | This study | PDB code 5UQ8 |
| Software and Algorithms | ||
| MotionCorr | (Li et al., 2015) | http://cryoem.ucsf.edu/software/driftcorr.html |
| CTFFIND3 | (Mindell and Grigorieff, 2003) | http://grigoriefflab.janelia.org/ctf |
| ResMap | (Kucukelbir et al., | http://resmap.sourceforge.net/ |
| RELION1.2 | (Scheres, 2012) | http://www2.mrc-lmb.cam.ac.uk/relion |
| UCSF Chimera | (Pettersen et al., 2004) | https://www.cgl.ucsf.edu/chimera/ |
| COOT | (Emsley et al., 2010) | https://www.phenix-online.org |
| PHENIX 1.11.1-3575 | (Adams et al., 2010) | https://www.phenix-online.org |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests may be directed to, and will be fulfilled by the Lead Author, Christine M. Dunham, Ph.D. (christine.m.dunham@emory.edu).
EXPERIMENTAL MODELS
E. coli MRE 600 70S ribosomes were purified as previously described (Fagan et al., 2014) with a few modifications. Briefly, cells were grown in LB to an OD600 of ~0.7, cooled on ice for 20 min, and centrifuged for 10 min at 6,000 rpm at 4°C. The pellet was resuspended in buffer (100 mM NH4Cl, 10 mM MgCl2, 0.5 mM EDTA, 10 mM K-HEPES pH 7.6, 6 mM β-mercaptoethanol (β-Me), DNase I (10 units/mL lysate)), lysed in an EmulsiFlex cell disruptor and clarified by centrifugation (30 min at 70,000 x g). The supernatant was pelleted over a sucrose cushion (1.1 M sucrose, 500 mM NH4Cl, 10 mM MgCl2, 0.5 mM EDTA, 10 mM K-HEPES pH 7.6) for 17 hrs at 158,000 x g at 4°C, the ribosome pellet was resuspended in buffer (400 mM KCl, 10 mM MgOAc, 1.5 M (NH4)2SO4, 10 mM Tris-Cl pH 7.5) and applied to a Toyopearl butyl-650S column equilibrated in the same buffer. The ribosomes were purified using a reverse (NH4)2SO4 gradient and further separated by zonal ultracentrifugation in a Ti15 zonal rotor with a 10%–40% sucrose gradient in buffer (50 mM KCl, 10 mM NH4Cl, 10 mM MgCl2, 0.25 mM EDTA, 10 mM K-HEPES pH 7.5). Fractions were pooled, dialyzed overnight into storage buffer (50 mM KCl, 10 mM NH4Cl, 10 mM MgCl2, 5 mM K-HEPES pH 7.5, and 6 mM β-Me), flash frozen in liquid nitrogen and stored in aliquots at −80°C.
T. thermophilus 70S ribosomes were purified as previously described (Selmer et al., 2006). Briefly, frozen cell paste was resuspendend in buffer (100 mM NH4Cl, 10 mM MgOAc, 0.5 mM EDTA, 20 mM K-HEPES pH 7.5, 6 mM β-Me; 1.5 mL/g), lysed in an EmulsiFlex cell disruptor and clarified by centrifugation (30 min at 70,000 x g). The supernatant was layered over a 1.1 M sucrose gradient (1.1 M sucrose, 0.5 M KCl, 10 mM MgOAc, 0.5 mM EDTA, 20 mM K-HEPES, pH 7.5) and centrifuged at 40,000 rpm in a 45 Ti rotor for 17.5 hrs. The pellet was resuspended in buffer (400 mM KCl, 10 mM MgOAc, 1.5 M (NH4)2SO4, 20 mM Tris-Cl, pH 7.5) and then applied to Toyopearl butyl-650S column equilibrated in the same buffer. The peak containing ribosomes was diluted two-fold in buffer (10 mM NH4Cl, 10 mM MgOAc, 0.25 mM EDTA, 10 mM K-HEPES pH 7.5) and pelleted overnight by centrifugation at 40,000 rpms in a 45Ti rotor at 4°C. The ribosomes were separated by zonal ultracentrifugation in a Ti15 zonal rotor with a 10%–40% sucrose gradient in buffer (50 mM KCl, 10 mM NH4Cl, 10 mM MgCl2, 0.25 mM EDTA, 10 mM K-HEPES pH 7.5). The ribosomes were dialyzed into final storage buffer (50 mM KCl, 10 mM NH4Cl, 10 mM MgOAc, 5 mM K-HEPES pH 7.5), concentrated, flash frozen in liquid nitrogen and stored in aliquots at −80°C.
METHOD DETAILS
RNA structure probing assays
The mRNA containing a 3′ stem-loop was a kind gift from the Cornish lab (Qin et al., 2014). The pUC19 plasmid was overexpressed and purified, digested with BamHI, further purified, in vitro transcribed, purified by denaturing PAGE and refolded according to standard protocols (Linpinsel and Conn, 2012). The mRNA produced is 86 nucleotides and contains a Shine-Dalgarno sequence (underlined), an AUG start codon (bold) and a 3′ stem-loop sequence (italicized) (5′-GGUAAGGAAAUAAAAAUGUUUAGUGAACCGGCAGCC GCUACCCGCGCGCGGCCGGUGAGGUUUUUCUUCUGAAGAUAAAG AUAUCG-3′). E. coli 70S ribosomes (1.2 μM) were incubated with mRNA (1 μM) at 37°C for 6 mins followed by the addition of P-site tRNAfMet (2.4 μM) for 30 mins and A-site tRNAPhe (2.4 μM) for an additional 20 mins. Both tRNAfMet and tRNAPhe were purchased from Chemical Block. All the 70S ribosome complexes were formed in buffer containing 5 mM K-HEPES pH 7.5, 50 mM KCl, 10 mM NH4Cl, 10 mM MgOAc, and 6 mM β-Me. RNA reactivity was tested using N-methylisatoic anhydride (NMIA), a commonly used chemical in RNA structure probing experiments (Smola et al., 2015). NMIA (13 mM in DMSO) was incubated with either free mRNA (0.5 μM) or E. coli programmed 70S complexes (1.2 μM containing tRNAfMet (0.6 μM) and mRNA (0.5 μM)) were incubated with NMIA at 37°C for 90 mins. For negative controls, NMIA was replaced by DMSO alone. RNA was isolated by ethanol precipitation (350 mM NaCl, 0.04 mg/ml glycogen, 0.44 mM EDTA, pH 8.0, and 77% ethanol) at −80°C for 30 mins. A 5′-end labeled [γ32P] DNA oligo (5′-CTTTATCTTCAGAAGAAAAACC-3′; 0.36 μM) complementary to the 3′ end of isolated mRNA (0.12 μM) were annealed at 65°C for 5 mins, at 35°C for 5 mins, and on ice for 1 min essentially as previously published (Fredrick and Noller, 2002). Due to the placement of the DNA primer used in the extension, the structure of nucleotides 3′ to G42 are not visible by SHAPE analysis. Primer extension was performed with Protoscript®II Reverse Transcriptase (3 units/μl) (NEB) in the presence of 10 mM DTT and 0.5 mM dNTP mix (0.5 mM dATP, 0.5 mM dGTP, 0.5 mM dCTP, 0.5 mM dTTP) in the supplied buffer. The reactions (20 μl) were incubated at 25°C for 5 mins and at 42°C for 60 mins. Finally, 19 μl acid stop mix (138 mM Tris-HCl unbuffered, 73% formamide, 0.86X TBE, 43 mM EDTA pH 8.0, 0.02% bromophenol blue, 0.02% xylene cyanol) was added and the mixtures were incubated at 95°C for additional 5 mins. To make an RNA hydrolysis sample, 1 μl of 4 M NaOH was added to a reaction and incubated at 95°C for 5 mins. Samples were analyzed on a preheated, denaturing 7M urea, 1X TBE, 12% polyacrylamide sequencing gel for 1 hr, dried, and exposed to a phosphor storage screen. and the intensity of the bands were measured by Typhoon FLA 700 Imager and quantified using ImageQuant software (GE Healthcare). The background reactivity from no NMIA lanes were subtracted and reactivities of each mRNA nucleotide (nucleotides 7–42) was normalized and values from at least two replicates were averaged and classified as 6–11%, 11–22% and >22%.
Electron cryo-microscopy
3.5 μL aliquots of ~50 nM T. thermophilus 70S complexes were applied to a glow-discharged holey carbon grid (Quantifoil R2/2, 200 mesh) coated with continuous thin carbon film, and subsequently vitrified using a Vitrobot Mark IV (FEI Company. The specimen was visualized with a Titan Krios electron microscope (FEI) operating at 300 kV accelerating voltage, at a nominal magnification of 29,000X using a K2 Summit direct electron detector (Gatan, Inc.) in counting mode, corresponding to a pixel size of 1.0 Å on the specimen level. Images were recorded with defocus values in the range of 0.6 to 2.5 μm and a dose rate of ~5.0 electrons per Å2 per second. The total exposure time was set to 6.0 s and intermediate frames were recorded every 0.1 s resulting in an accumulated dose of ~ 30 electrons per Å2 and a total of 60 frames per micrograph.
QUANTIFICATION AND STATISTICAL ANALYSIS
Image processing
A total of 3,071 cryo-EM images were recorded during one microscope session. Each image movie stack was subjected to whole-frame motion correction using MotionCorr (Li et al., 2015), and then evaluated to remove micrographs with obvious astigmatism and Thon ring profiles showing lower than ~3 Å resolution. A sum of all frames in each image stack of 2,640 micrographs was used for further processing. CTF parameters for each micrograph were determined by CTFFIND3 (Mindell and Grigorieff, 2003).
Particle extraction, two-dimensional classification and three-dimensional classification were performed using RELION1.2 (Scheres, 2012) with a binned dataset at a pixel size of 2 Å and a box size of 192 X 192 pixels as previously described with minor modification (Shalev-Benami et al., 2016). In total, 294,286 particles were extracted using semi-automated particle picking. Reference-free two-dimensional classification was used to discard non-ribosomal or damaged particles. Unsupervised three-dimensional classifications with a 60-Å low-pass filtered cryo-EM map of T. thermophilus 70S ribosome (emdb 2277) as initial reference was performed to sort conformational and compositional heterogeneity. The first round of three-dimensional classification was run with 8 classes, resulting in 58,434 particles containing the intact 70S ribosome in presence of P- and E-tRNAs. A further round of three-dimensional classification with alignment focusing on E-tRNA was performed to differentiate conformers of Eout and Ein.
Statistical single-particle movie processing was performed in RELION 1.2 at a pixel size of 1 Å per pixel and a box size of 384 × 384 pixels, in which all the frames in image stack were used, running averages was set to 5 and standard deviation to one pixel for the translational alignment. In addition, a resolution-dependent radiation damage model was used to weight B-factor for each movie frame by calculating independent half-reconstructions for each frame separately. The resulting “polished” particles were used for the final refinement, resulting in the maps of Eout and Ein with a nominal global resolution of 3.2 Å and 3.5 Å, respectively.
Model building and refinement
For initial model building, the coordinates of T. thermophilus 70S ribosome (PDB code 4Y4O) were rigid-docked into cryo-EM maps using UCSF Chimera (Pettersen et al., 2004). tRNA coordinates for P and E sites were manually fit using COOT (Emsley et al., 2010). Because the original 70S model lacked the coordinates for the uL1 protein, the coordinates containing 23S rRNA and uL1 protein were extracted from a previously solved ribosome structure (PDB code 4V5F), aligned to the uL1 stalk region of the initially docked 70S ribosome, rebuilt in COOT. The assembled complex was then subjected to real space rigid body refinement in PHENIX (Adams et al., 2010) (Table S1- related to Figure 1). Model overfitting was evaluated through its refinement against one of two cryo-EM half maps. FSC curves were calculated between the resulting model and the half map used for refinement (red curve, Figure S2C- related to Figure 1), as well as between the resulting model and the other half map for cross-validation (green curve, Figure S2C- related to Figure 1). Coordinates for mRNA were then manually built into the refined model.
Reported resolutions are based on the gold-standard Fourier shell correlation (FSC) value of 0.143. High-resolution noise substitution was used to correct for the effects of soft masking on the FSC curves. All density maps were corrected for the modulation transfer function (MTF) of the K2 Summit direct detector and then sharpened by applying a negative B-factor estimated using automated procedures. Local resolution was determined using ResMap (Kucukelbir et al., 2014) with half-reconstructions as input maps.
DATA AND SOFTWARE AVAILABILITY
Please refer to the KEY RESOURCE TABLE section above.
ADDITIONAL RESOURCES
All deposited coordinates discussed here can be found in the RCSB Protein Data Bank (http://www.rcsb.org) or Protein Data Bank in Europe (http://www.ebi.ac.uk/pdbe/) under PDB codes 5UQ7 and 5UQ8.
Supplementary Material
Table 1.
Model building and refinement.
| Data collection | ||
|
| ||
| Complex | 70S-E/E in | 70S-E out |
| Particles | 22,993 | 36,361 |
| Pixel size (Å) | 1.0 | |
| Defocus range (μm) | 0.6–2.5 | |
| Voltage (kV) | 300 | |
| Electron dose (e−/Å2) | 30 | |
|
| ||
| Refinement | ||
|
| ||
| CCmap_model | 0.84 | 0.81 |
|
| ||
| Model quality | ||
|
| ||
| RMSD | ||
| Bond lengths (Å)/Bond angles (°) | 0.008/0.71 | 0.009/0.72 |
| Ramachandran plot statistics | ||
| Most favored (%) | 92.2 | 92.3 |
| Outliers (%) | 0.07 | 0.05 |
| Rotamer outliers (%) | 0.04 | 0 |
| C-beta deviations | 0 | 0 |
| Clashscore | 19.08 | 16.91 |
| B_isomean | 163.3 | 146.9 |
Highlights.
Cryo-EM structures of the 70S bound to an mRNA stem-loop reveal interactions with uS3
E-site tRNA shows a unique conformation in the presence of the mRNA stem-loop
E-site tRNA forms interactions with the 30S head domain
Structured mRNAs regulate the conformation of tRNAs to affect translation
Acknowledgments
We acknowledge P. Cornish for reagents, B. Calderon for technical assistance, and G. Conn, J. Dunkle, K. Fredrick, E. Hoffer and H. Zaher for critical reading of the manuscript. This work was supported in part by National Institutes of Health (NIH) GM093278 award and Emory University Department of Biochemistry funds (CMD).
Footnotes
AUTHOR CONTRIBUTIONS
YZ, acquisition of data, analysis and interpretation of data, revising the article. SH, analysis and interpretation of data, drafting and revising the article. AR, acquisition of data. GS, interpretation of data, revising the article. CMD, conception and design, interpretation of data, drafting and revising the article.
DECLARATION OF INTERESTS
The authors declare no competing interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal RK, Spahn CM, Penczek P, Grassucci RA, Nierhaus KH, Frank J. Visualization of tRNA movements on the Escherichia coli 70S ribosome during the elongation cycle. J Cell Biol. 2000;150:447–60. doi: 10.1083/jcb.150.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins JF, Loughran G, Bhatt PR, Firth AE, Baranov PV. Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai XC, Fernandez IS, Mcmullan G, Scheres SH. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. Elife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Beckmann R, Cate JH, Dinman JD, Dragon F, Ellis SR, Lafontaine DL, Lindahl L, Liljas A, Lipton JM, et al. A new system for naming ribosomal proteins. Curr Opin Struct Biol. 2014;24:165–9. doi: 10.1016/j.sbi.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belardinelli R, Sharma H, Caliskan N, Cunha CE, Peske F, Wintermeyer W, Rodnina MV. Choreography of molecular movements during ribosome progression along mRNA. Nat Struct Mol Biol. 2016;23:342–8. doi: 10.1038/nsmb.3193. [DOI] [PubMed] [Google Scholar]
- Bock LV, Blau C, Schroder GF, Davydov Ii, Fischer N, Stark H, Rodnina MV, Vaiana AC, Grubmuller H. Energy barriers and driving forces in tRNA translocation through the ribosome. Nat Struct Mol Biol. 2013;20:1390–6. doi: 10.1038/nsmb.2690. [DOI] [PubMed] [Google Scholar]
- Caliskan N, Katunin VI, Belardinelli R, Peske F, Rodnina MV. Programmed −1 Frameshifting by Kinetic Partitioning during Impeded Translocation. Cell. 2014;157:1619–31. doi: 10.1016/j.cell.2014.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Zhang H, Broitman SL, Reiche M, Farrell I, Cooperman BS, Goldman YE. Dynamics of translation by single ribosomes through mRNA secondary structures. Nat Struct Mol Biol. 2013;20:582–8. doi: 10.1038/nsmb.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Petrov A, Johansson M, Tsai A, O’leary SE, Puglisi JD. Dynamic pathways of −1 translational frameshifting. Nature. 2014 doi: 10.1038/nature13428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Staple DW, Hoang L, Hickerson RP, Noller HF, Ha T. Following movement of the L1 stalk between three functional states in single ribosomes. Proc Natl Acad Sci U S A. 2009;106:2571–6. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman JD. Mechanisms and implications of programmed translational frameshifting. Wiley Interdiscip Rev RNA. 2012;3:661–73. doi: 10.1002/wrna.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–4. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan CE, Maehigashi T, Dunkle JA, Miles SJ, Dunham CM. Structural insights into translational recoding by frameshift suppressor tRNASufJ. RNA. 2014;20:1944–54. doi: 10.1261/rna.046953.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh PJ. Programmed translational frameshifting. Annu Rev Genet. 1996;30:507–28. doi: 10.1146/annurev.genet.30.1.507. [DOI] [PubMed] [Google Scholar]
- Fei J, Kosuri P, Macdougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30:348–59. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–33. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- Fredrick K, Noller HF. Accurate translocation of mRNA by the ribosome requires a peptidyl group or its analog on the tRNA moving into the 30S P site. Mol Cell. 2002;9:1125–31. doi: 10.1016/s1097-2765(02)00523-3. [DOI] [PubMed] [Google Scholar]
- Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–9. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedroc DP, Cornish PV. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res. 2009;139:193–208. doi: 10.1016/j.virusres.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson GN, Clark PL. Quality over quantity: optimizing co-translational protein folding with non-‘optimal’ synonymous codons. Curr Opin Struct Biol. 2016;38:102–10. doi: 10.1016/j.sbi.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner L, Demeshkina N, Yusupova G, Yusupov M. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat Struct Mol Biol. 2010a;17:1072–8. doi: 10.1038/nsmb.1880. [DOI] [PubMed] [Google Scholar]
- Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol. 2010b;17:555–60. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- Kim HK, Liu F, Fei J, Bustamante C, Gonzalez RL, Jr, Tinoco I., Jr A frameshifting stimulatory stem loop destabilizes the hybrid state and impedes ribosomal translocation. Proc Natl Acad Sci U S A. 2014;111:5538–43. doi: 10.1073/pnas.1403457111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–77. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–5. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen B, Gesteland RF, Atkins JF. Structural probing and mutagenic analysis of the stem-loop required for Escherichia coli dnaX ribosomal frameshifting: programmed efficiency of 50% J Mol Biol. 1997;271:47–60. doi: 10.1006/jmbi.1997.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen B, Wills NM, Gesteland RF, Atkins JF. rRNA-mRNA base pairing stimulates a programmed −1 ribosomal frameshift. J Bacteriol. 1994;176:6842–51. doi: 10.1128/jb.176.22.6842-6851.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zheng S, Agard DA, Cheng Y. Asynchronous data acquisition and on-the-fly analysis of dose fractionated cryoEM images by UCSFImage. J Struct Biol. 2015;192:174–8. doi: 10.1016/j.jsb.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linpinsel JL, Conn GL. General protocols for preparation of plasmid DNA template, RNA in vitro transcription, and RNA purification by denaturing PAGE. Methods Mol Biol. 2012;941:43–58. doi: 10.1007/978-1-62703-113-4_4. [DOI] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–47. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Munro JB, Altman RB, Tung CS, Cate JH, Sanbonmatsu KY, Blanchard SC. Spontaneous formation of the unlocked state of the ribosome is a multistep process. Proc Natl Acad Sci U S A. 2010;107:709–14. doi: 10.1073/pnas.0908597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–32. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Polikanov YS, Melnikov SV, Soll D, Steitz TA. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat Struct Mol Biol. 2015;22:342–344. doi: 10.1038/nsmb.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P, Yu D, Zuo X, Cornish PV. Structured mRNA induces the ribosome into a hyper-rotated state. EMBO Rep. 2014;15:185–90. doi: 10.1002/embr.201337762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Wen JD, Lancaster L, Noller HF, Bustamante C, Tinoco I., Jr The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature. 2011;475:118–21. doi: 10.1038/nature10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–6. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Wintermeyer W. Protein Elongation, Co-translational Folding and Targeting. J Mol Biol. 2016;428:2165–85. doi: 10.1016/j.jmb.2016.03.022. [DOI] [PubMed] [Google Scholar]
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–30. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FVT, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–42. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Shalev-Benami M, Zhang Y, Matzov D, Halfon Y, Zackay A, Rozenberg H, Zimmerman E, Bashan A, Jaffe CL, Yonath A, et al. 2.8-A Cryo-EM Structure of the Large Ribosomal Subunit from the Eukaryotic Parasite Leishmania. Cell Rep. 2016;16:288–94. doi: 10.1016/j.celrep.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola MJ, Rice GM, Busan S, Siegfried NA, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat Protoc. 2015;10:1643–69. doi: 10.1038/nprot.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- Tourigny DS, Fernandez IS, Kelley AC, Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science. 2013;340:1235490. doi: 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchihashi Z, Brown PO. Sequence requirements for efficient translational frameshifting in the Escherichia coli dnaX gene and the role of an unstable interaction between tRNA(Lys) and an AAG lysine codon. Genes Dev. 1992;6:511–9. doi: 10.1101/gad.6.3.511. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi Z, Kornberg A. Translational frameshifting generates the gamma subunit of DNA polymerase III holoenzyme. Proc Natl Acad Sci U S A. 1990;87:2516–20. doi: 10.1073/pnas.87.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter A. Gene regulation by structured mRNA elements. Trends Genet. 2014;30:172–81. doi: 10.1016/j.tig.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Wasserman MR, Alejo JL, Altman RB, Blanchard SC. Multiperspective smFRET reveals rate-determining late intermediates of ribosomal translocation. Nat Struct Mol Biol. 2016;23:333–41. doi: 10.1038/nsmb.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen JD, Lancaster L, Hodges C, Zeri AC, Yoshimura SH, Noller HF, Bustamante C, Tinoco I. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–96. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- Zaher HS, Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009;136:746–62. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lancaster L, Donohue JP, Noller HF. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science. 2013;340:1236086. doi: 10.1126/science.1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lancaster L, Donohue JP, Noller HF. How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation. Science. 2014;345:1188–91. doi: 10.1126/science.1255030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.