Abstract
Microenvironmental factors have a major impact on differentiation of embryonic stem cells (ESCs). Here, we report a novel phenomenon that size of ESC colonies has a significant regulatory role on stromal cells-induced differentiation of ESCs to neural cells. Using a robotic cell microprinting technology, we confine defined densities of ESCs within aqueous nanodrops over a layer of supporting stromal cells immersed in a second, immiscible aqueous phase to generate ESC colonies of defined sizes. We use temporal protein and gene expression studies and demonstrate that larger ESC colonies generate disproportionally more neural cells and longer neurite processes. Unlike previous studies that attribute neural differentiation of ESCs solely to interactions with stromal cells, we find that increased intercellular signaling of ESCs significantly enhances neural differentiation. This study offers an approach to generate neural cells with improved efficiency for potential use in translational research.
Keywords: colony size, neural differentiation, niche parameters, embryonic stem cells, stromal cells
1. Introduction
The potential to treat neurodegenerative diseases such as Parkinson’s disease and traumatic neural tissue injuries relies on external interventions including cell replacement therapies.[1,2] Embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) provide promising cell sources for neural tissue regeneration, modeling of degenerative diseases, and testing of neurotoxic effects of drugs.[3–6] In spite of the intense research over the past two decades, the use of stem cells for cell replacement therapies is hampered by the difficulty of mapping the roles of extrinsic and intrinsic factors and the ability to control the differentiation process to generate specific cell types. Spatial and temporal coordination of multiple physical and biochemical factors during differentiation of stem cells complicates this task.[7]
Differentiation of stem cells in embryoid body (EB) cultures mimics early stage embryonic development and results in precursor cells from all three germ layers.[8] Although addition of factors such as retinoic acid to cultures can promote neural differentiation, these chemicals may perturb the natural neural patterning and maturation of cells.[9,10] The use of an adherent monolayer of stem cells in chemically defined media containing a cocktail of specific factors provides a more efficient method to derive neural cells.[11] However, several reports show that the neural precursors obtained from the monolayer cultures do not develop neuronal functionality and networks.[11,12] Subsequent growth of differentiated cells followed by dissociation and re-plating on adherent surfaces is necessary to derive terminally differentiated cell types.[13,14] An alternative approach to generate pure populations of neural cells is co-culturing of stem cells with specific stromal cells.[15] Both physical contacts and paracrine signaling with the stromal cells induce stem cells to undergo neural differentiation. This approach resembles ESCs niche in vivo in terms of direct intercellular contacts and signaling, avoids using differentiation-inducing chemicals, and exclusively results in neural cells.
Biochemical and biophysical signaling cues in the local microenvironment dynamically modulate the fate of ESCs. A cohort of surface bound and soluble factors, interactions of ESCs with their neighboring cells and extracellular matrix proteins, and various epigenetic factors act synergistically to determine differentiation of ESCs to neural cell lineages.[16–19] While a majority of current research is centered on functionalizing specific biomolecules on scaffolds, or altering media compositions to gain a better control over the differentiation of ESCs, the role of niche mediated factors on regulating neural differentiation is less understood. The most studied factor is matrix stiffness that plays a critical role in fate determination of stem cells.[16,20–23]
We hypothesized that in addition to extrinsic paracrine signaling with stromal cells, intrinsic parameters such as the organization of ESCs and their autocrine factors determine the differentiation fate and efficiency of ESCs. For example, varying the size of ESC colonies can alter the concentration of endogenous differentiation-inducing soluble factors.[24,25] A few studies used EB cultures and investigated the effect of stem cell colony size on differentiation efficiency into three germ layers. Larger EBs yielded more cardiac cells while smaller EBs gave greater vascular differentiation.[26] A similar study showed enhanced ectodermal differentiation in smaller EBs, whereas larger EBs expressed more mesodermal and endodermal markers.[8] EB size-mediated cell fate was also observed in human ESCs where larger EBs showed greater propensity towards neural lineages, although a heterogeneous cell population resulted due to the use of EB cultures.[27]
To date, the effect of colony size on ESC differentiation in ESCs-stromal cells co-cultures remains unexplored. Our preliminary study showed that the expression of a neural lineage differentiation marker, beta-III tubulin, significantly increases in larger ESC colonies,[28] implying that in addition to the differentiation inducing signals from stromal cells, ESC colony size further regulates the neural differentiation process. To test this hypothesis, here we generate defined size ESC colonies on stromal cells and conduct a comprehensive gene and protein expression analysis to track the transition of ESCs to specific terminally-differentiated neural cells such as neurons, astrocytes, and oligodendrocytes. A major challenge to systematically study the effect of colony size in this co-culture environment is generating ESC colonies of defined sizes over a living layer of stromal cells to allow direct contacts between the two cell types. Methods to control the size of EBs using forced aggregation, encapsulating cells in hydrogels, and microfluidics are inadequate to address this need.[29–31] We address this issue using a cell microprinting technology based on a polymeric aqueous two-phase system (ATPS) with polyethylene glycol (PEG) and dextran (DEX) as phase-forming polymers. We robotically localize ESCs in an aqueous DEX phase nanodrop over a layer of supporting stromal cells immersed in the immiscible aqueous PEG phase. Importantly, the microprinting is non-contact and gentle to maintain full viability of both printed ESCs and stromal cells. Microprinted ESCs proliferate to form standalone colonies of defined sizes and differentiate into neural cells during culture. We study differentiation of ESCs in colonies by tracking temporal expression of neural genes and proteins over a two-week period and find that increasing the size of ESC colonies significantly and size-disproportionately enhances neural differentiation. Thus, this study elucidates the role of a niche parameter – colony size – on neural differentiation of ESCs in a controlled microenvironment and provides a potential approach to generate neural cells with improved efficiency.
2. Results and Discussion
2.1. Characterization of ATPS cell microprinting
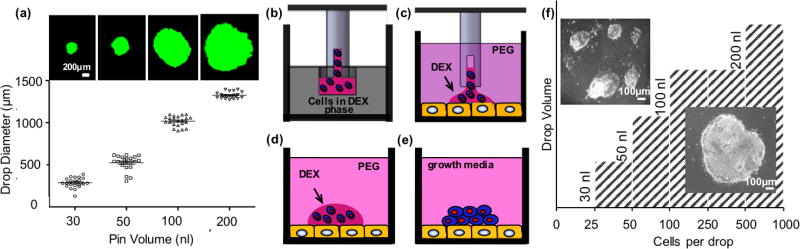
Evaluation of colony size effect on neural differentiation of ESCs requires generating individual colonies of defined sizes on stromal cells. We used a non-contact PEG-DEX ATPS cell microprinting technology to achieve this. Our first objective was to characterize changes in the DEX phase drop size by varying the volume of the drop dispensed onto a layer of stromal PA6 cells immersed in the PEG phase. We prepared an ATPS with 5.0%(w/v) PEG and 6.4%(w/v) DEX in the cell culture media. Hydrophobic slot pins were mounted on a robotic liquid handler pipetting head and dipped into a source vessel to load the FITC-labeled DEX solution. The pins were then lowered close to the surface of the PA6 cells monolayer in the PEG phase to allow the FITC-DEX phase drops to dispense. The dispensing of the DEX phase drops is autonomous because an ultralow interfacial tension between the two highly aqueous phases is insufficient to hold the DEX phase drops in the pins against the gravitational force.[32] Dispensing pins of volumes 30, 50, 100 and 200 nl generated drops ranging from 286 ± 59 µm to 1325 ± 37 µm in average diameter (Figure 1a). Printed drop diameter varied approximately linearly (R2 = 0.9) with the drop volume.
Figure 1.
Aqueous Two-Phase System (ATPS) mediated niche microprinting. (a) The diameter of FITC-DEX drops printed on PA6 stromal cells monolayer immersed in PEG phase is varied using dispensing pins volume. (b) Dispensing pins are loaded with mESCs (blue) in the DEX phase from the source plate and (c) lowered into the culture plate containing PA6 cells (yellow) immersed in the PEG phase. (d) This results in autonomous dispensing of the pins content to form isolated drops confining mESCs. (e) Printed mESCs attach to the stromal layer and proliferate. (f) An experimental diagram showing the minimum number of cells in the DEX drops of different volumes to result in formation of a single colony on PA6 cells.
Next, we established an experimental phase diagram to determine a minimum ESC density for a given volume of DEX phase drops to result in a single, compact ESC colony on the stromal cells during incubation. A 6.4%(w/v) DEX phase solution containing mESCs was loaded into a 384-well plate (source plate). This suspension was loaded from the source plate into slot pins (Figure 1b) and dispensed onto a monolayer of PA6 cells immersed in the PEG phase solution (Figure 1c). The mESCs remained confined within the DEX phase drop (Figure 1d), adhered to the PA6 cells and proliferated over time (Figure 1e). DEX phase drops with four different volumes of 30, 50, 100, and 200 nl containing 25 – 1000 cells per drop were dispensed on a stromal cells layer. Each condition had at least 18 replicates. After six days of incubation, we evaluated whether the printed ESCs form a single colony or multiple interspaced colonies. The results are summarized in the phase diagram of Figure 1f. The hatch area to the right of each volume indicates cell densities that generated a single colony, whereas the blank area to the left of each volume gave multiple colonies. For a given ESC density, decreasing the drop volume reduces the dispersion of cells within the drop due to the smaller drop size (Figure 1a). Based on this set of data, we selected a DEX drop volume of 50 nl containing more than 50 ESCs to form single colonies for the following studies.
2.2. Generation of size-controlled colonies and neural differentiation
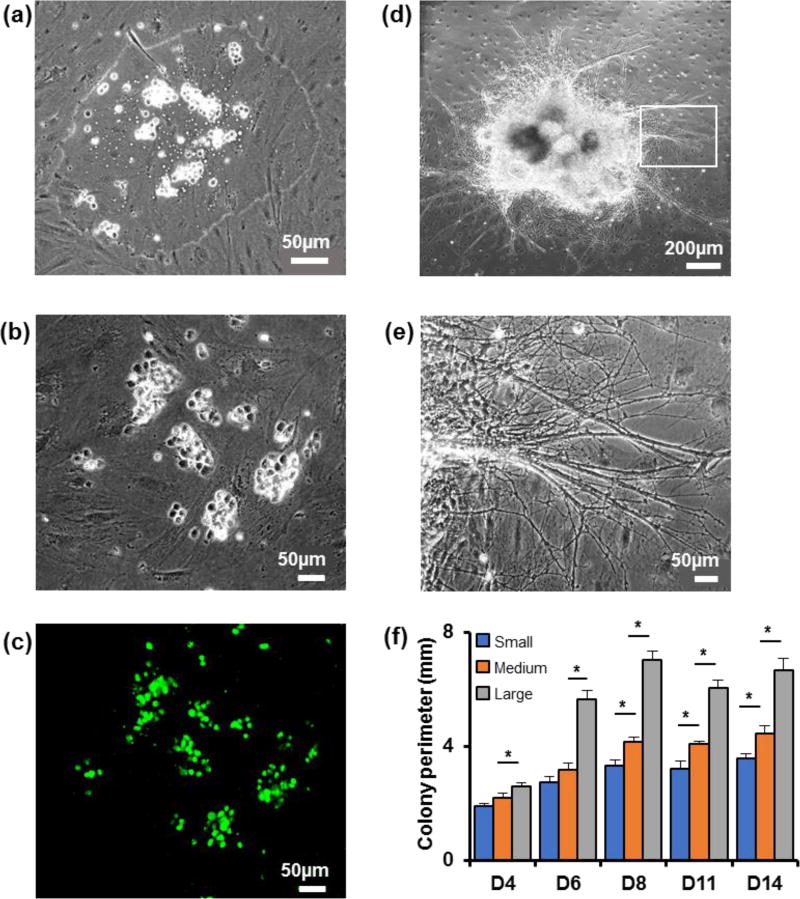
Microprinted mESCs remained partitioned to the DEX phase drops (Figure 2a) and adhered to the PA6 cells layer within three hours of printing (Figure 2b). Unlike other cell printing methods such as inkjet printing or membrane- or gel-embedded cell printing,[33–35] the ATPS printing approach does not exert any thermal, mechanical, or chemical stresses on cells.[28] Positive staining of printed mESCs for a pluripotency marker, Oct4, validated this point (Figure 2c). mESCs rapidly proliferated to generate a single colony and the layer of mitotically-arrested PA6 cells remained viable and intact (Figure 2d). Differentiating cells migrated toward the colony periphery and extended out dense and thick neurite processes (Figure 2e). The size of mESC colonies was controlled through the density of printed cells in the 50 nl DEX phase drops. Densities of 2×103, 5×103, and 1×104 cells/µl yielded individual mESC colonies with average diameters of 1.00±0.05 mm, 1.40±0.04 mm, and 2.00±0.10 mm. These colonies are labeled small, medium, and large. Regression analysis showed a linear correlation between cell count per colony with colony perimeter and colony area, with R2 values of 0.86 and 0.88, respectively (Figure S2). mESCs showed rapid proliferation over the first week of culture but the size of colonies did not significantly change thereafter (Figure 2f). This pattern suggests that cells proliferate and transition into a multipotent neural stem and progenitor cell stage during the first week of culture, subsequently acquire neuronal or glial progenitors stage, or terminally differentiate to neurons, astrocytes, or oligodendrocytes.
Figure 2.
Generation of controlled size colonies using ATPS. (a) mESCs microprinted on PA6 cells are confined within the DEX phase drop. (b) mESCs adhered to the underlying PA6 cells. (c) Microprinted mESCs are pluripotent and express Oct4 (green). This fluorescent image corresponds to the phase image of panel (b). (d) mESCs proliferate and form a single colony. The image shows a colony on day 8 of culture. (e) Neurite processes extend out from differentiating cells in mESC colonies. (f) Temporal changes in the perimeter of colonies developed from 100, 250, and 500 mESCs printed on PA6 cells. One-way ANOVA was used for statistical analysis. * p < 0.01. n=18. Error bars represent mean±S.E.M.
2.3. Colony size effect on neural differentiation of mESCs: Protein expression analysis
Direct contacts between mESCs and stromal PA6 cells was sufficient to induce neural cell differentiation, without a need for additional chemicals such as retinoic acid. Neural cell-specific markers were detectable in colonies of mESCs starting from the fourth day of culture and neurite processes emerged from differentiating cells. Expression of neural cell proteins was detected using immunocytochemistry and quantified using image processing techniques.
A. Neural stem and progenitor cell markers
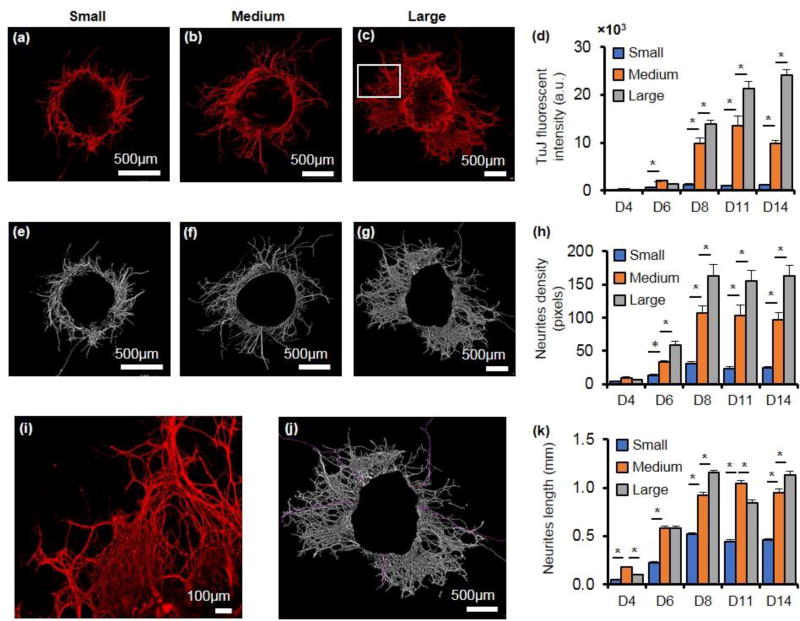
Fluorescent intensity measurements of colonies immunostained with neural stem and progenitor cell markers TUJ, Nestin, and NCAM over the two-week period of co-culture showed a rapid rise in the expression of all three proteins. Figure 3a–c shows TUJ-stained colonies of three different sizes on day 8. Evidently, the large colony has longer and denser neural processes extending out from its periphery compared to the medium and small colonies. The magnified image shows the high density of the neurite bundles of the large colony (Figure 3i). Since TUJ expression was primarily concentrated at the periphery of the colonies, the net fluorescent intensity measured with each colony was normalized against its perimeter to compare the protein expression among colonies of different sizes (Figure 3d). TUJ levels were consistently and significantly higher in the medium sized colonies than the small colonies starting from day six and at each subsequent measurement day. The large colonies also showed significantly greater TUJ expression than the medium sized colonies. Normalizing the data with respect to colony area or the number of cells per colony also gave similar results (data not shown). In addition, using an adaptive thresholding method, TUJ-positive colonies were analyzed for their total neurites density that accounted for both the length and thickness of the neural processes. Unlike fluorescent intensity measurements, the adaptive thresholding method is not sensitive to the intensity variations within an image and thus, is a more objective method of quantification of fluorescent images. Consistent with the fluorescent intensity data, neurites density of each colony significantly increased up to day 8 but remained unaltered in the second week of culture (Figure 3e–g). Comparing the three colony sizes also showed that normalized neurites density significantly increased by increase in the colony size for the duration of culture (Figure 3h). Moreover, our measurements of neurites length by manual tracing of the processes (shown in purple in Figure 3j) showed that larger colonies on average had longer neurite extensions (Figure 3k).
Figure 3.
TuJ expression varies with colony size. (a–c) Immunostained images of TUJ-positive colonies of 3 different sizes on day 8 of culture. Note the difference in the sizes of scale bars among the panels. (d) TUJ fluorescent intensity normalized against the perimeter of each colony shows a significant increase with increase in the colony size. (e–g) The immunostained images of panels (a–c) subjected to adaptive thresholding to determine density of neurites. (h) Total neurites density normalized with colony perimeter significantly increases in larger colonies. (i) Magnified view of the boxed portion of the panel (c) image showing neural processes. (j) Automated tracing of neural extensions (purple lines) in the image of panel (c) to measure the neurites length. (k) Longest neurites emerging from three different colony sizes. One-way ANOVA was used for all statistical analysis. * p < 0.01. n=18. Error bars represents mean±S.E.M.
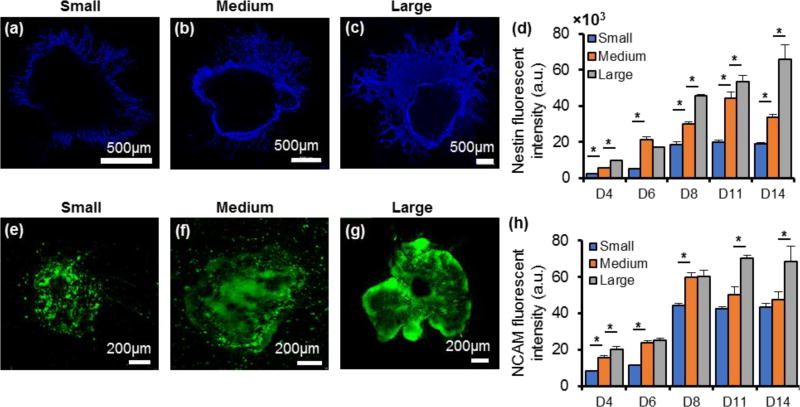
Fluorescently labeled Nestin-positive differentiated cells were more abundant with increase in the size of colonies (Figure 4a–c). These cells were mainly distributed at the periphery of the colonies and therefore, the measured fluorescent intensities were normalized against the perimeter of the colonies to make quantitative comparison of Nestin expression among the three colony sizes. We note that unlike TUJ staining that marked neural processes, Nestin marked the cell bodies (Figure S3a). Thus, we limited our analysis to the measured fluorescent signal intensity. Compared to the small colonies, Nestin expression was significantly higher in the medium size colonies throughout the experimental period. The expression of Nestin further enhanced in the large colonies and except for day six, the large colonies showed significantly greater Nestin expression than the medium size colonies throughout the two-week culture (Figure 4d). Normalizing the data with respect to colony area or the number of cells per colony also showed significant increase in Nestin expression with increase in colony size (data not shown). The fluorescent signal from NCAM-positive cells was mainly generated from the cell bodies within the colonies (Figure 4e–g). To compare NCAM expression among the three colony sizes, the measured fluorescent intensity was normalized against the area of each respective colony. Overall, increase in the colony size led to greater NCAM expression (Figure 4h).
Figure 4.
Nestin and NCAM expressions vary with colony size. (a–c) Immunostained images of Nestin-positive cells in colonies of different sizes on day 8. (d) Measured and normalized fluorescent intensities of Nestin-positive colonies during 14 days of culture. (e–g) Immunostained images of NCAM-positive cells in colonies of different sizes on day 8. and (h) Measured and normalized fluorescent intensities of NCAM-positive colonies during 14 days of culture. One-way ANOVA was used for both statistical analysis. * p < 0.01. n=18. Error bars represent mean±S.E.M.
Key conclusions of the above protein expression study are as follows: (i) Continuous increase in the expression of prominent neural stem/progenitor cell proteins, neurites density, and average neurites length indicates axonal development and neurites extension by differentiating mESCs; (ii) the plateau in the expression of the protein markers of neural cells after day eight suggests that differentiating cells transition past their progenitor stage toward specific neuronal or glial cells;[36] and importantly, (iii) larger mESC colonies show size-disproportionately enhanced expression of the neural proteins.
B. Specific neural cell markers
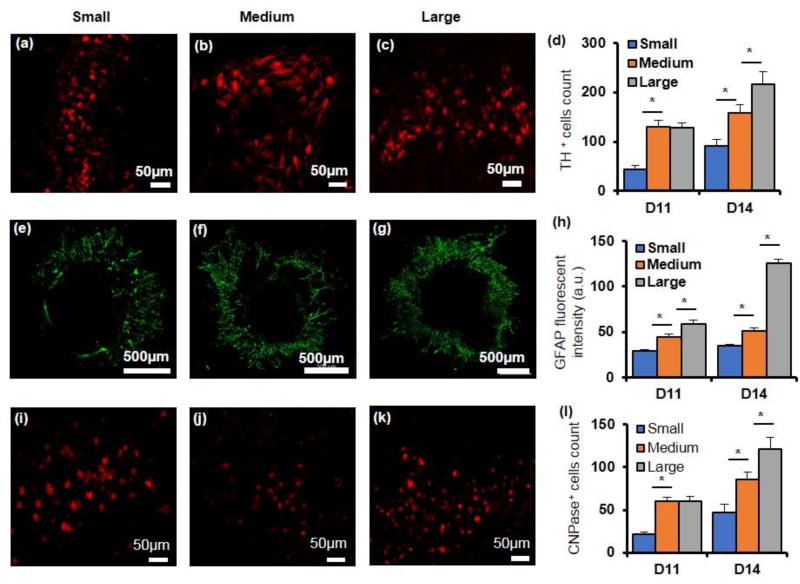
We immunostained the colonies for specific cells of the central nervous system dopaminergic neurons, astrocytes, and oligodendrocytes using TH, GFAP, and CNPase, respectively. This analysis was done only during the second week of culture to allow the differentiating cells to acquire the specific traits. Expression of TH was quantified by counting the number of positively stained cells and showed a significant increase in all colonies over time with the large colonies containing the highest number of TH-positive cells (Figure 5a–d). Expression of GFAP was quantified through fluorescent intensity measurements. Although cells in all three colony sizes showed positive staining, only the large colony had a significant rise in GFAP expression over time. Additionally, the colony size effect on GFAP expression became more pronounced on day 14 with the large colony containing 2.5 and 3.7 folds greater GFAP-expressing cells than the medium and small colonies, respectively (Figure 5e–h and Figure S3b). CNPase expression was also quantified by counting the number of cells stained positive in each colony. All three colony sizes showed greater number of oligodendrocytes over time, with the large colony containing markedly higher number of CNPase-positive cells (Figure 5i–l). We note that TH- and CNPase-expressing cells were mainly detected in the peripheral regions of colonies and the relatively small number of counted cells was in part due to the difficulty with visualizing positively-stained cells within the colonies that comprised of multiple layers of cells. Nevertheless, these data indicate that differentiation of mESCs into specific neural lineages can be reproducibly induced due to signaling with the stromal cells in a process that closely mimics embryonic neural differentiation, but importantly, the size of mESC colonies further modulates this process.[37]
Figure 5.
Expressions of specific neuronal and glial marker proteins vary with colony size. (a–c) Immunostained images of TH-positive cells at the periphery of colonies of different sizes on day 14. (d) Larger colonies contain greater number of TH-positive cells. (e–g) Immunostained images of GFAP-positive colonies of different sizes on day 14. (h) Normalized GFAP fluorescent intensity increases significantly with the colony size. (i–k) Immunostained images of CNPase-positive cells at the periphery of colonies of different sizes on day 14. (d) The number of CNPase-positive cells increases with colony size. One-way ANOVA was used for all statistical analysis. * p < 0.01. n=18. Error bars represent mean±S.E.M.
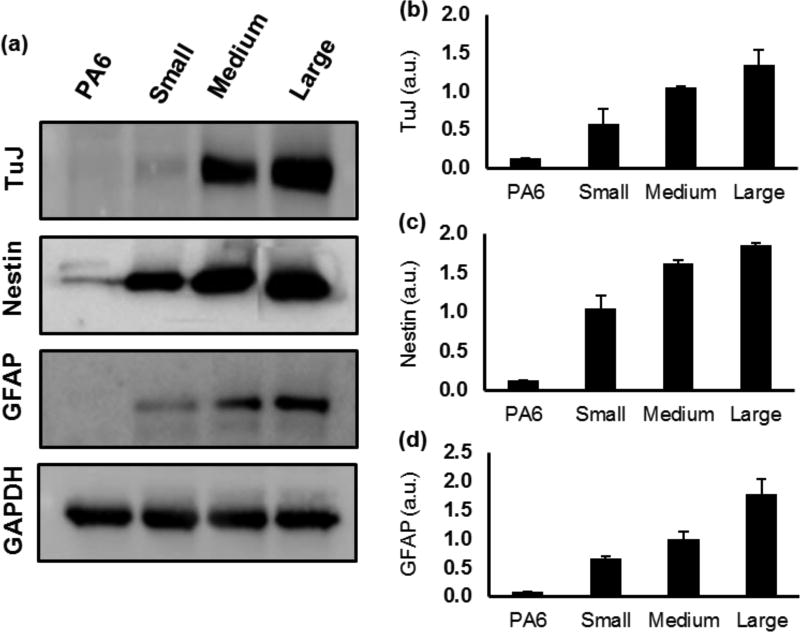
We recognize that quantification of protein expression based on fluorescent intensity measurements of immunostained samples is a semi-quantitative method; however, our approach to normalize the measured fluorescent intensity with respect to imaging magnification and exposure time (Supplementary Information and Figure S1), and validation of results with additional image processing techniques such as adaptive thresholding and neurites length measurements, ensured reliable quantitative comparisons of protein expression among different colony sizes. To further substantiate the validity of the protein quantification based on the analysis fluorescence images, we performed western blot analysis of select neural and glial marker proteins TUJ, Nestin, and GFAP. We used stromal PA6 cells as a negative control to ensure that these proteins are expressed by differentiating mESCs in the co-culture niche, and not by PA6 cells. Our western blot results corroborated well the fluorescence-based data quantification and showed greater expression of the proteins with increase in the colony size (Figure 6a–d). Collectively, our immunofluorescence-based quantification of protein expression and the western blot analysis establish that the size of stem cell colonies in co-culture with stromal cells is a major niche parameter that regulates the neural differentiation process.
Figure 6.
Neural protein expressions vary with colony size. (a) Western blot images of the neural proteins Nestin and TUJ, and the glial protein GFAP from day 8 and day 14 samples, respectively. (b–d) Quantified data for Nestin, TUJ, and GFAP show increased expression of the proteins with larger colonies. n=2. Error bars represent mean±S.E.M.
2.4. Colony size effect on neural differentiation of mESCs: Gene expression analysis
Next, we investigated gene level regulation of colony size effect on neural differentiation of mESCs using a temporal gene expression study. mRNA samples were obtained daily from small, medium, and large colonies for a duration of two weeks to measure the expression levels of select gene markers of pluripotency (Oct4), neural stem cells (Nestin), neural progenitor cells (TuJ), and dopaminergic neuronal cells (TH). On each day, qPCR was performed on three experimental replicates for each colony size. And for each experiment, 14 samples were obtained daily over the two-week culture period. Using GAPDH and β-Actin as reference genes and undifferentiated mESCs as the negative control, ΔΔCt values were calculated and fold change values were represented as 2−ΔΔCt.[38]
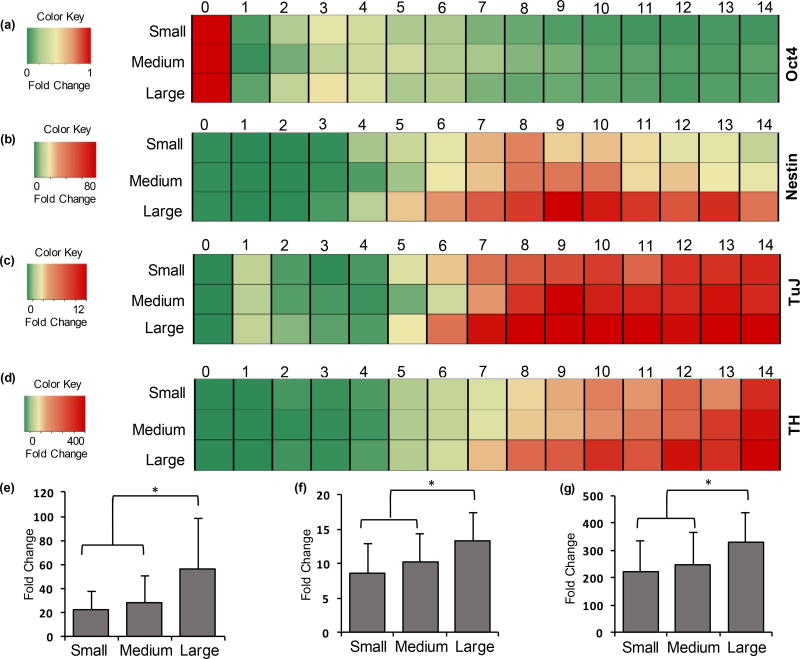
Consistent with the protein expression study, the neural genes showed the highest mRNA fold change in the large colonies, followed by the medium and small colonies. Figure 7 represents the temporal mRNA fold change data for Oct4 (Figure 7a), Nestin (Figure 7b), TuJ (Figure 7c), and TH (Figure 7d). It is evident from these heatmaps that the larger colonies showed a greater fold change for Nestin, TuJ, and TH as depicted by the darker shades of red in the corresponding heatmaps. Another key conclusion from Figure 7 is that the gene expression trajectory was similar in all three colony sizes. Oct4 expression steadily decreased over time, Nestin expression rose to a peak level around days 6–10 and declined thereafter, whereas the gene expression of markers of neural progenitor and specific neural cell lineages, TuJ and TH, continuously increased throughout the culture time. These results are consistent with the role of these markers in terms of loss of pluripotency (Oct4), commitment to a neural type (Nestin and TuJ), followed by differentiation into specific neural cell lineages (TH).
Figure 7.
Expression of neural genes varies with colony size. Heatmaps representing temporal mRNA fold change over 14 days of culture for (a) Oct4, (b) Nestin, (c) TuJ, and (d) TH in mESC colonies of three different sizes. One-way ANOVA on mRNA expressions of (e) Nestin (days 6–14), (f) TuJ (days 7–14), and (g) TH (days 7–14) show significantly higher expressions in large colonies compared to the medium and small colonies. * p<0.01. (e) n= 27, (f,g) n= 24. Error bars represent mean±S.D.
Next, we performed a statistical analysis on the gene expression data. As Oct4 expression quickly downregulated with minimal differences among the three colony sizes (Figure 7a), it was not considered for statistical analysis. Nestin showed minimal expression during the first five days of culture but showed transient elevation afterwards, whereas TuJ and TH showed a steady and significant expression increase during the second week of culture. This is consistent with our previous study that showed high Nestin gene expression precedes the expression of TuJ and TH genes.[39] To enable a direct statistical comparison between different colony sizes, the temporal fold change data for Nestin was averaged from days 6–14 and the temporal fold change data for TuJ and TH were averaged from days 7–14. The results were then subjected to one-way ANOVA. This analysis showed that the expression levels of Nestin, TuJ and TH were all significantly higher in large colonies compared to the medium and small colonies (Figure 7e–g). Although a clear increase was also seen in temporal gene expressions levels in the medium sized colonies compared to the small colonies (Figure 7b–d), the difference was not statistically significant. We observed significant experiment-to-experiment variations in the expression of selected genes during differentiation of mESCs (Figure S4), potentially masking statistically significant differences between the expression of these genes when comparing the small and medium colony sizes.
Unlike the gene expression data, all neural proteins showed disproportionately higher protein expression levels with increase in the colony size (Figure 3–5), indicating that mRNA levels cannot be used as surrogates for corresponding protein levels. Apart from transcription, key processes such as mRNA decay, translational efficiency, and protein degradation determine the protein expression levels in cells, thereby limiting the proportionality between gene and protein levels.[40] A study by Tian et al. on EML cell lines found that 35% of the studied factors showed significant changes at a protein level but not at the mRNA level.[41] Schwanhausser et al. found a correlation factor of only 0.41 between mRNA and protein level.[42] The significant increase of Nestin and TuJ expression at a protein level with increasing colony size indicates high translational efficiency and the stability of these structural proteins in differentiating cells.[43] Higher expression of TH implies greater number of neurons were generated in the large colony configuration. At a protein level, greater number of TH-positive neuronal cells were counted in the medium sized colonies compared to the small colonies (Figure 5d), potentially due to a richer neural stem cells content in the former configuration that leads to greater levels of specific neuronal cells.
ESCs utilize paracrine and autocrine signaling to communicate with their surrounding cells, self-renew, and influence their own differentiation fate.[44,45] In this study, we modulated the intercellular interactions among the differentiating mESCs by controlling the size of mESC colonies and evaluated the resulting changes in the efficiency of neural differentiation. Metabolites of mESCs contain growth factors, transcription factors, morphogens, and activins with important roles in regulating neural differentiation.[25,45,46] Our previous study identified a set of soluble factors secreted by differentiating mESCs that significantly enhance the yield of neural cells.[25] Greater concentrations of these factors with increase in the stem cell colony size is potentially responsible for the resulting disproportionate enhancement in neural differentiation of mESCs. Parnas et al. also observed a similar increase in neuronal cell development and maturation in terms of axonal outgrowth, synapse formation, and neurotransmitters secretion by increase in the cell density.[47] It is important to note that mESC colonies of different sizes were exposed to similar levels of paracrine factors of PA6 cells because of identical number of the stromal cells used. Therefore, the role of stromal cells-mediated paracrine induction of neural differentiation is expected be similar for all colony sizes and that, colony size-mediated enhancement of neural differentiation is due to self-regulation of differentiating mESCs.
The mESCs-mediated self-regulation of neural differentiation is most likely due to surface bound and soluble secreted factors.[15] Several studies have reported that surface molecules as well as soluble signaling molecules mediate regulatory mechanisms to determine the fate of stem cells. For example, membrane-bound molecules such as E-cadherin, galectin, integrins, and proteoglycans as well as secreted factors such as epidermal growth factors (EGF) and basic fibroblast growth factor (bFGF) regulate proliferation and self-renewal of neural stem cells.[48–50] Interestingly, in an experiment to investigate the influence of cell density on the generation of neural stem cells, Tropepe et al. found that under high density plating conditions, EGF-mediated mitogenic effects were more dominant than bFGF-mediated mitogenic effects but low cell density plating reversed this observation.[50] Collectively, these data support our finding that increasing the size of colonies of mESCs microprinted on stromal cells has a compounding effect to enhance differentiation to neural cells.
3. Conclusions
We used a cell microprinting technology to generate standalone mESC colonies of defined sizes on a layer of stromal cells to elucidate the impact of colony size on neural cell differentiation. Increasing the size of mESC colony disproportionately enhanced differentiation to neural cells. Larger colonies displayed greater expression of protein markers of neural cells and extended longer neurite processes. Temporal gene expression analysis of neural gene markers showed differential expression levels in different colony sizes wherein large colonies showed higher expression levels compared to medium and small colony sizes throughout the two-week culture period. Thus, our study offers a mechanistic understanding of the unexplored role of a niche parameter of stem cells, i.e., colony size, on generation of neural cells. This study provides an approach to engineer an efficient ESC microenvironment to obtain neural precursor cells for potential use in translational research that may eventually lead to regenerative therapies of neurodegeneration.
4. Experimental Section
4.1. Maintenance of Cells and Preparation of Stromal Cells
Mouse embryonic stem cells (mESCs) (EB5, Riken) were maintained on 0.1% gelatin-coated dishes in GMEM (Life Technologies) supplemented with 1% fetal bovine serum (FBS, Sigma), 10% knockout serum replacement (KSR, Life Technologies), 2 mM glutaMAX (Life Technologies), 0.1 mM non-essential amino acids (NEAA, Life Technologies), 1 mM sodium pyruvate (Life Technologies), 0.1 mM 2-mercaptoethanol (Life Technologies), and 2000 U/ml leukemia inhibitory factor (Millipore). PA6 cells (Riken) –a stromal cell line derived from mouse skull bone marrow– were also maintained on gelatin coated dishes in αMEM (Life Technologies) supplemented with 10% FBS and 1% antibiotic (Life Technologies). To prepare feeder cells for mESCs, PA6 cells were grown to a confluent monolayer on a gelatin-coated 35 mm Petri dish and mitotically inactivated with 10 µg/ml mitomycin-c (Sigma) for 2 hrs. PA6 cells were washed three times with PBS and then incubated overnight at 37°C and 5% CO2 in a “differentiation” medium containing GMEM supplemented with 10% KSR, 2 mM glutaMAX, 0.1 mM NEAA, 1 mM sodium pyruvate, and 0.1 mM 2-mercaptoethanol before co-culturing with mESCs.
4.2. Microprinting of mESCs onto Stromal Cells
Polyethylene glycol (PEG, Mw: 35 kDa) and dextran (DEX, Mw: 500 kDa) were dissolved in the differentiation medium at concentrations of 5.0% (w/v) and 12.8% (w/v), respectively.[32] mESCs were suspended in an equal volume of the medium and the DEX phase solution to yield a desired cell density and reduce DEX concentration to 6.4% (w/v). Different slot pins with volumes of 30, 50, 100, and 200 nl and the cell densities of 25, 50, 100, 250, 500, and 1000 cells/droplet were used to generate mESC colonies through microprinting of mESCs on the feeder stromal cells, to generate the phase diagram (Figure 1f). Cell densities of 2×103, 5×103 and 1×104 cells/µl (i.e., 100, 250, and 500 cells per 50 nl) were used to generate mESC colonies of three distinct sizes for differentiation studies. The mESCs-DEX phase suspension was loaded into a 384-well plate (source plate). A liquid handler (SRT Bravo, Agilent) was used to load 50 nl of the suspension into slot hydrophobic pins (VP Scientific) (Figure 1b). The pins were then slowly lowered into a 35 mm Petri dish containing a monolayer of PA6 cells immersed in 1 ml of the PEG phase solution (Figure 1c). The DEX phase containing mESCs autonomously dispensed from each pin and formed a drop on the PA6 cell layer (Figure 1d). The mESCs adhered to the PA6 cells (Figure 1e). Polymeric solutions were replaced with fresh differentiation medium after 3 hours of incubation. Cultures were maintained in the differentiation medium for 8 days and with 1× N2 supplementation for an additional 6 days (Life Technologies). Nine mESC colonies of similar size with a center-to-center spacing of 9.0 mm were generated in 35 mm Petri dishes and cultured for 14 days.
4.3. Immunofluorescence and Imaging
For Immunocytochemistry, mESC colonies were fixed in 3.7% formaldehyde on days 4, 6, 8, 11, and 14 followed by blocking with 5% donkey serum for 1 hour. Both primary and secondary antibodies solutions were prepared in PBS containing 0.05% Tween 20 (Sigma). The following primary antibodies were used: affinity purified goat Oct4 (Neuromics) marked pluripotent stage of mESCs, affinity purified chicken Nestin (Neuromics) identified neural stem cells, mouse monoclonal NCAM (Santa Cruz), and rabbit monoclonal class III β-tubulin (Biolegend) marked neural progenitor stage. Similarly, rabbit monoclonal TH (Abcam) identified dopaminergic neurons, affinity purified chicken GFAP (Neuromics) identified astrocytes, and mouse CNPase (Covance) identified oligodendrocytes. All secondary antibodies were raised in donkey. Since each sample was double or triple stained for multiple marker proteins, multiple secondary antibodies tagged with aminomethylcoumarin (AMCA), fluorescein isothiocyanate (FITC), Rhodamine red, Alexa Fluor 488 and Alexa Fluor 592 (Jackson Immunoresearch) were used. Each colony was imaged in sections at 10× or 20× using an inverted fluorescent microscope (Axio Observer, Zeiss) equipped with a high-resolution camera (AxioCam MRm, Zeiss). Sections were merged using Photoshop (Adobe) and resulting images were used to quantify protein expression through image analysis.
4.4. Image Analysis
For each marker, net fluorescent intensity of the immunostained colony was measured from images in ImageJ after subtracting the background using the following relation:
The fluorescent intensity was normalized against the exposure time and imaging magnification to compare data among images taken from different sizes of mESCs colonies and on different days. A detailed description of the normalization process is included as supplementary information. Furthermore, the net fluorescent intensity of each marker protein from each colony was normalized against the circumference of the colony for unbiased comparison among colonies of different sizes.
Additionally, neural differentiation of mESCs colonies stained with TUJ was quantified using an adaptive thresholding plugin in ImageJ. Each image containing a colony was converted to a binary image. Neurites density resulting from the colony, defined as the total white pixels count considering both length and thickness of neurites, was computed. The colony was cropped out from each image after thresholding and then total neurites density was determined from the portion of the image containing only neural processes. Furthermore, an average neurite length was measured for each colony by tracing 10 random processes in images using a NeuronJ plugin in ImageJ.
4.5. Western Blot of Marker Proteins
Total protein was quantified for each sample using BCA quantification assay kit (Life Technologies). Then, equal amount of protein from each sample was load onto a gel (Biorad) for electrophoresis. After transferring the proteins onto nitrocellulose membranes, the membranes were blocked with 5% BSA (Sigma) and probed with the primary antibodies: Rabbit monoclonal class III β-tubulin (TuJ, Cell Signaling), Affinity purified chicken Nestin (Neuromics) and Affinity purified chicken GFAP IgY (Neuromics). Horseradish peroxidase (HRP)-conjugated secondary antibodies (Cell Signaling) and ECL chemiluminescence detection kit (GE Healthcare) were used to visualize the blots with FluorChem E imaging system (ProteinSimple). GAPDH (Cell Signaling) was used as a loading control and lysate from monolayer of PA6 cells was used as negative control to ensure detected proteins represent differentiating mESCs.
4.6. Gene Expression Analysis
Experimental samples were lysed every day for two weeks using a TRK lysis buffer (Omega Biotek) and homogenized by passing through homogenizer mini columns (Omega Biotek). Total RNA was isolated from the samples using an RNA isolation kit (Omega Biotek). DNase was removed using RNase-free DNase kit (Omega Biotek). Purity and concentration of isolated RNA was assessed using OD 260/280 spectrophotometry (Synergy H1M, Biotek instruments). cDNA was synthesized from 1 µg of total RNA using random hexamer primers (Roche).
Real time q-PCR was performed with a Lightcycler 480 II instrument using a SYBR Green Master Mix (Roche). Briefly, 50 ng of cDNA was combined with forward and reverse primers and the SYBR green Master Mix diluted to a final volume of 15 µl. The reactions were pre-incubated at 95°C for 5 min followed by 45 cycles of amplification, i.e., at 95°C for 10 sec, at 60°C for 10 sec, and at 72°C for 10 sec. Specific primer sequences for all the genes investigated are listed in Supplementary Table S1. Expression levels of mRNA for different marker genes were calculated relative to GAPDH and β-Actin using the ΔΔCt method and the fold change in mRNA expression was determined according to the 2−ΔΔCt method.
4.7. Statistical Analysis
All experiments were performed in duplicates for immunostaining and western blot and in triplicates for q-PCR. For protein expression analysis using image processing, at least 18 colonies were imaged and analyzed for each colony size. For gene expression analysis, daily expression levels from days 6 to 14 (for Nestin) and days 7 to 14 (for TuJ and TH) were averaged to increase the sample size. Statistical tests were performed using one-way ANOVA and Fisher’s post hoc test in Minitab 16 software. All error bars represent mean± S.E.M. unless stated otherwise. Statistical significance was defined at p<0.05.
Supplementary Material
Acknowledgments
This research is supported by grants 1264562 from National Science Foundation, CA182333 from National Institutes of Health, and TECG20140954 from Ohio Third Frontier.
Footnotes
Disclosure of Potential Conflict of Interest
The authors declare no potential conflicts of interest with respect to the research, authorship and/or publication of this article.
Contributor Information
Ramila Joshi, Department of Biomedical Engineering, The University of Akron, Akron, Ohio 44325, USA.
Pradip Shahi Thakuri, Department of Biomedical Engineering, The University of Akron, Akron, Ohio 44325, USA.
James C. Buchanan, Department of Biomedical Engineering, The University of Akron, Akron, Ohio 44325, USA
Dr. Jun Li, Department of Mathematical Sciences, Kent State University, Kent, Ohio 44242, USA
Dr. Hossein Tavana, Department of Biomedical Engineering, The University of Akron, 260 S. Forge St., Akron, OH 44325, Tel: (330) 972-6031, Fax: (330) 374-8834
References
- 1.Lindvall O, Kokaia Z, Martinez-Serrano A. Nat. Med. 2004;10(Suppl):S42. doi: 10.1038/nm1064. [DOI] [PubMed] [Google Scholar]
- 2.Matthys OB, Hookway TA, McDevitt TC. Curr. Stem Cell Reports. 2016;2:43. doi: 10.1007/s40778-016-0030-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolf Trumbower SL, Reier PJ, Behrman AL, Ross HH, Ambrosio F. 2016 doi: 10.2522/ptj.20150063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi Y, Inoue H, Wu JC, Yamanaka S. Nat. Rev. Drug Discov. 2017;16:115. doi: 10.1038/nrd.2016.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suri S, Singh A, Nguyen AH, Bratt-Leal AM, McDevitt TC, Lu H. Lab Chip. 2013;13:4617. doi: 10.1039/c3lc50663k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jakel RJ, Schneider BL, Svendsen CN. Nat. Rev. Genet. 2004;5:136. doi: 10.1038/nrg1268. [DOI] [PubMed] [Google Scholar]
- 7.Perrimon N, Pitsouli C, Shilo B. Cold Spring Harb. Perspect. Biol. 2012;4:1. doi: 10.1101/cshperspect.a005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park J, Cho CH, Parashurama N, Li Y, Berthiaume F, Toner M, Tilles AW, Yarmush ML. Lab Chip. 2007;7:1018. doi: 10.1039/b704739h. [DOI] [PubMed] [Google Scholar]
- 9.Papalopulu N, Clarke JD, Bradley L, Wilkinson D, Krumlauf R, Holder N. Development. 1991;113:1145. doi: 10.1242/dev.113.4.1145. [DOI] [PubMed] [Google Scholar]
- 10.Lee LMY, Leung C-Y, Tang WWC, Choi H-L, Leung Y-C, McCaffery PJ, Wang C-C, Woolf aS, Shum aSW. Proc. Natl. Acad. Sci. 2012;109:13668. doi: 10.1073/pnas.1200872109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ying Q-L, Stavridis M, Griffiths D, Li M, Smith A. Nat. Biotechnol. 2003;21:183. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 12.Bouhon IA, Joannides A, Kato H, Chandran S, Allen ND. Stem Cells. 2006;24:1908. doi: 10.1634/stemcells.2006-0031. [DOI] [PubMed] [Google Scholar]
- 13.Banda E, Grabel L. Methods Mol. Biol. 2015;1307:289. doi: 10.1007/7651_2014_67. [DOI] [PubMed] [Google Scholar]
- 14.Gunther K, Appelt-Menzel A, Kwok CK, Walles H, Metzger M, Edenhofer F. J Stem Cell Res Ther. 2016;6 [Google Scholar]
- 15.Kawasaki H, Mizuseki K, Nishikawa S, Kaneko S, Kuwana Y, Nakanishi S, Nishikawa SI, Sasai Y. Neuron. 2000;28:31. doi: 10.1016/s0896-6273(00)00083-0. [DOI] [PubMed] [Google Scholar]
- 16.Lutolf MP, Gilbert PM, Blau HM. Nature. 2009;462:433. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juliandi B, Abematsu M, Nakashima K. Dev. Growth Differ. 2010;52:493. doi: 10.1111/j.1440-169X.2010.01175.x. [DOI] [PubMed] [Google Scholar]
- 18.M L, ffrench C, Kazani C Ilias, Lathia Justin. StemBook. 2008;1 [Google Scholar]
- 19.Tavana H, Mosadegh B, Zamankhan P, Grotberg JB, Takayama S. Biotechnol. Bioeng. 2011;108:2509. doi: 10.1002/bit.23190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 21.Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, Chen S, Engler AJ. Nat. Mater. 2014:1. doi: 10.1038/nmat4051. advance on. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu J, Wang YK, Yang MT, Desai RA, Yu X, Liu Z, Chen CS. Nat Methods. 2010;7:733. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guvendiren M, Burdick JA. Adv. Healthc. Mater. 2013;2:155. doi: 10.1002/adhm.201200105. [DOI] [PubMed] [Google Scholar]
- 24.Peerani R, Onishi K, Mahdavi A, Kumacheva E, Zandstra PW. PLoS One. 2009;4 doi: 10.1371/journal.pone.0006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joshi R, Buchanan JC, Tavana H. Integr. Biol. 2017 doi: 10.1039/c7ib00038c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hwang CM, Kim SK, Kim JH, Khademhosseini a, Lee SH. 2009 IEEE 35th Annu. Northeast Bioeng. Conf; 2009. p. 5. [Google Scholar]
- 27.Bauwens CL, Peerani R, Niebruegge S, Woodhouse Ka, Kumacheva E, Husain M, Zandstra PW. Stem Cells. 2008;26:2300. doi: 10.1634/stemcells.2008-0183. [DOI] [PubMed] [Google Scholar]
- 28.Tavana BH, Mosadegh B, Takayama S. Adv. Mater. 2010;22:2628. doi: 10.1002/adma.200904271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khademhosseini A, Suh KY, Jon S, Eng G, Yeh J, Chen G-J, Langer R. Annu. Rev. Biomed. Eng. Anal. Chem. Langmuir. 2001;3:335. [Google Scholar]
- 30.Cohen DL, Malone E, Lipson H, Bonassar LJ. Tissue Eng. 2006;12:1325. doi: 10.1089/ten.2006.12.1325. [DOI] [PubMed] [Google Scholar]
- 31.Ng ES, Davis RP, Azzola L, Stanley EG, Elefanty AG. Blood. 2005;106 doi: 10.1182/blood-2005-03-0987. [DOI] [PubMed] [Google Scholar]
- 32.Atefi E, Joshi R, Mann JA, Tavana H. ACS Appl. Mater. Interfaces. 2015;7:21305. doi: 10.1021/acsami.5b05757. [DOI] [PubMed] [Google Scholar]
- 33.Cui X, Dean D, Ruggeri ZM, Boland T. Biotechnol. Bioeng. 2010;106:963. doi: 10.1002/bit.22762. [DOI] [PubMed] [Google Scholar]
- 34.Du Y, Ghodousi M, Qi H, Haas N, Xiao W, Khademhosseini A. Biotechnol. Bioeng. 2011;108:1693. doi: 10.1002/bit.23102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faulkner-Jones A, Greenhough S, King JA, Gardner J, Courtney A, Shu W. Biofabrication. 2013;5:15013. doi: 10.1088/1758-5082/5/1/015013. [DOI] [PubMed] [Google Scholar]
- 36.Zhang S. Brain Pathol. 2006;16:132. doi: 10.1111/j.1750-3639.2006.00008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abranches E, Silva M, Pradier L, Schulz H, Hummel O, Henrique D, Bekman E. PLoS One. 2009;4 doi: 10.1371/journal.pone.0006286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. Clin. Chem. 2009;55:611. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 39.Joshi R, Buchanan JC, Paruchuri S, Morris N, Tavana H. PLoS One. 2016;11:e0166316. doi: 10.1371/journal.pone.0166316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Signatures E, Can C, Two E, Of T, Abundance P, Cell H, Christine L, Abreu S, Ko D, Le S-Y, Shapiro BA, Burns SC, Boutz DR, Sandhu D, Marcotte EM, Penalva LO, Biology S, Biology M, Health T, Antonio S, Science M, Program N. Mol. Syst. Biol. 2010;6:1. doi: 10.1038/msb.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian Q, Stepaniants SB, Mao M, Weng L, Feetham MC, Doyle MJ, Yi EC, Dai H, Thorsson V, Eng J, Goodlett D, Berger JP, Gunter B, Linseley PS, Stoughton RB, Aebersold R, Collins SJ, Hanlon WA, Hood LE. Mol. Cell. Proteomics. 2004;3:960. doi: 10.1074/mcp.M400055-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Nature. 2011;473:337. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 43.Vogel C, Marcotte EM. Nat. Rev. Genet. 2012;13:227. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peerani R, Rao BM, Bauwens C, Yin T, Wood GA, Nagy A, Kumacheva E, Zandstra PW. EMBO J. 2007;26:4744. doi: 10.1038/sj.emboj.7601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Discher DE, Mooney DJ, Zandstra PW. Science (80-.) 2009;324:1673. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J-Q, Yu X-B, Ma B-F, Yu W-H, Zhang A-X, Huang G, Mao FF, Zhang X-M, Wang Z-C, Li S-N, Lahn BT, Xiang AP. Neuroreport. 2006;17:981. doi: 10.1097/01.wnr.0000227977.60271.ca. [DOI] [PubMed] [Google Scholar]
- 47.Parnas D, Linial M. Dev. Brain Res. 1997;101:115. doi: 10.1016/s0165-3806(97)00057-6. [DOI] [PubMed] [Google Scholar]
- 48.Karpowicz P, Willaime-Morawek S, Balenci L, DeVeale B, Inoue T, van der Kooy D. J Neurosci. 2009;29:3885. doi: 10.1523/JNEUROSCI.0037-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anna Wade JJP, McKinney Andrew. Biochim Biophys Acta. 2014;1840:2520. doi: 10.1016/j.bbagen.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tropepe V, Sibilia M, Ciruna BG, Rossant J, Wagner EF, van der Kooy D. Dev. Biol. 1999;208:166. doi: 10.1006/dbio.1998.9192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.