Abstract
The objective of this study was to purify, characterize, and phylogenetically and structurally analyze the dextranase produced by the fungus Pochonia chlamydosporia. Dextranase produced by the fungus P. chlamydosporia was purified to homogeneity in two steps, with a yield of 152%, purification factor of 6.84 and specific activity of 358.63 U/mg. Its molecular weight was estimated by SDS-PAGE at 64 kDa. The enzyme presented higher activity at 50 °C and pH 5.0, using 100 mM citrate–phosphate buffer, was inhibited by Ag1+, Hg2+, Cu2+, Mg2+, and presented KM of 23.60 µM. Mature dextranase is composed of 585 amino acids residues, with a predicted molecular weight of 64.38 kDa and pI 5.96. This dextranase showed a strong phylogenetic similarity when compared to Trichoderma harzianum dextranase. Its structure consists of two domains: the first composed by 15 β strands, and the second composed by a right-handed parallel β-helix.
Keywords: Enzyme, Verticillium chlamydosporium, Purification, 3D structure
Introduction
Dextranases (EC 3.2.1) are enzymes that hydrolyze α-(1,6), α-(1,2), α-(1,3) and α-(1,4) linkages in dextran polysaccharides. The main products from dextran hydrolysis by dextranase activity are glucose, isomaltose and isomalto-oligosaccharides and, thus, dextranases are commonly known as glucanases. Dextranases comprise a large and diverse group of enzymes which differ from one another due to the mode of action and resulted dextran hydrolysis products (Jaiswal and Kumar 2011; Picozzi et al. 2015).
Sugarcane, in the field, during transportation and in industry, is subject to microbial infections that lead to dextran production from sucrose. According to Boil and Wienese (2002), dextran presence in sugar processing results mainly from post-harvest delay and lack of hygiene in the factories.
Among the problems caused by dextran in the sugar industry we can mention sucrose concentration reduction as a result of its use in dextran production, lower sucrose recovery as a consequence of crystallization inhibition, and increase in the sugarcane juice viscosity (Khalikova et al. 2005). Moreover, dextran affects sugar quality, once syrup removal does not occur properly, forming residues in the sugar crystals (Batista 2014; Boil and Wienese 2002), as well as the formation of opaque, irregular, elongated and caramel-like crystals (Jiménez 2009). In addition to sucrose loss and worsening final product quality, the presence of dextran in the sugarcane juice also results in a higher equipment wear and compromise sucrose content quantification, which is the basis for the sugarcane payment (Singleton et al. 2002).
Physical separation methods, such as ultrafiltration, dialysis and reverse osmosis are useful methods for polysaccharide removal on a reduced scale, however, on a commercial scale, their use is not yet economically viable (Boil and Wienese 2002).
Therefore, in most cases, the methods used to remove dextran present in sugar solutions utilize enzymatic hydrolysis through dextranase use. However, for dextranase application to be economical, the availability of a suitable enzyme for the degree of dextran removal is required, which depends on time, temperature and dextran concentration (Boil and Wienese 2002; Picozzi et al. 2015).
Dextranase production was verified in some bacterial species, filamentous fungi and a small number of yeasts (Bhatia et al. 2010; Jiao et al. 2014; Zhang et al. 2016). Recently, our research group described the first report of dextranase production by the fungus Pochonia chlamydosoporia, a facultative parasite of eggs and female nematodes found in the soil in various regions of the world. This enzyme reduced the dextran content of sugarcane juice by 75%, with 12-h treatment (Sufiate et al. 2018).
Thus, the objective of this study was to purify, characterize, and phylogenetically and structurally analyze the dextranase produced by P. chlamydosporia.
Materials and methods
Dextranase production by P. chlamydosporia
P. chlamydosporia VC4 (syn. Verticillium chlamydosporium) was obtained from the soil of Viçosa city, Minas Gerais, southeast of Brazil (latitude 20°4502000S, longitude 42°5204000W), and was maintained on solid 2% Potato Dextrose Agar medium [2% PDA (w/v)] under refrigeration, at 4 °C. For inoculation in liquid culture medium, four equally sized disks were removed from petri dishes edges. These disks were inoculated into 250 mL flasks containing 100 mL of liquid medium previously autoclaved at 121 °C for 15 min. The liquid medium contained 10 g/L dextran, 5 g/L NaNO3, 4 g/L KH2PO4, 0.5 g/L MgSO4.H2O, 0.5 g/L KCl, 0.178 g/L ZnSO4.7H2O, 0.18 g/L FeSO4.7H2O and pH was adjusted to 5.5. After inoculation, the flasks were kept at 28 °C and 180 rpm for 7 days. After this period, the flasks content was filtered and centrifuged at 10,000×g for 20 min. The supernatant constituted the crude extract.
Enzyme activity and protein quantification
Dextranase activity was measured by evaluating the amount of reducing sugars through 3.5-dinitrosalicylic acid (DNS) method (Miller 1959). The enzyme assay was performed using 370 μL of 100 mM citrate–phosphate buffer pH 6.0, 100 μL of 1% dextran solution and 30 μL of enzymatic sample. Reagents were incubated at 50 °C for 10 min, and the reaction was stopped by adding 500 μL of the DNS reagent. Samples were heated for 5 min in a boiling water bath, and then 1 mL of water was added to each sample. Absorbance readings were performed at 540 nm. In order to obtain the amount of reducing sugars, a standard curve was constructed with varying glucose concentrations. One dextranase unit (U) was defined as the amount of the enzyme that catalyzes the release of 1 μmol of reducing sugar per minute under the assay conditions. All enzymatic activity measurements were performed in triplicate.
Protein concentration was estimated at each fraction from the chromatographic columns by absorbance measured at wavelength of 280 nm. To determine the protein concentration in the crude extract and in the pools obtained from the chromatographic columns, Bradford method (Bradford 1976) was used.
Ion-exchange chromatography on DEAE-Sepharose and CM-Sepharose
A 5 mL crude extract volume was subjected to DEAE-Sepharose™ Fast Flow (Amershan Biosciences®) ion-exchange column previously equilibrated in 25 mM citrate–phosphate buffer pH 6.0, connected to an automatic collector and a peristaltic pump. Flow rate was kept constant at 0.5 mL/min. After sample application, 25 mL of 25 mM citrate–phosphate buffer pH 6.0 was applied. Resin-bound proteins were eluted with 25 mL of the same buffer solution with increasing linear NaCl concentrations up to 1 M. The collected fractions contained 3 mL each and those containing high dextranase activity were selected and assembled to form a pool.
This pool was subjected to a CM-Sepharose ™ Fast Flow (Amershan Biosciences®) ion exchange column previously equilibrated in 25 mM citrate–phosphate buffer pH 6.0, connected to an automatic collector and a peristaltic pump. The flow rate was kept constant at 0.5 mL/min. After sample application, 18 mL of 25 mM citrate–phosphate buffer pH 6.0 was applied. Resin-bound proteins were eluted with 18 mL of the same buffer solution with increasing linear NaCl concentrations up to 1 M. The collected fractions contained 1.5 mL each and those containing high dextranase activity were selected and assemble to form a pool, which consisted of the purified enzyme.
Page
Samples were subjected to 10% (w/v) polyacrylamide gel electrophoresis (SDS-PAGE) (Laemmli 1970). Electrophoresis was performed at 80 V, and the gel was stained with silver nitrate to allow protein visualization.
An in-gel activity assay was carried out in order to confirm the dextranase activity of the purified dextranase from P. chlamydosporia. Samples were applied to 10% native PAGE gel containing 1% blue dextran, and a voltage of 80 V was used. The gel was incubated in 100 mM citrate–phosphate buffer pH 6.0 for 1 h at 50 °C. Dextranase activity was detected as a clear band on blue background. Subsequently, the gel was stained with silver nitrate to confirm the enzyme presence.
Enzymatic characterization
pH and temperature effect on enzyme activity
An assay was conducted using 100 mM citrate–phosphate buffer with different pH values: 2.2; 3.0; 4.0; 5.0; 6.0; 7.0 and 8.0. Assays were performed under the conditions described above, using the buffer solution in such pH values. Dextranase activity was measured at different reaction incubation temperatures: 30, 40, 50 and 60 °C, keeping all the previously described conditions.
Thermal stability of the purified dextranase was evaluated by incubating enzymatic samples at 40 and 50 °C for 0 to 300 min, before being subjected to the enzymatic activity measurement.
Effect of metal ions and other reagents on enzymatic activity
The presence effect of the following ions, initially in saline form, on dextranase enzymatic activity was evaluated: ZnCl2, CaCl2, MgSO4, MnSO4, CuSO4, FeSO4, AgNO3, HgSO4, and EDTA (Ethylenediamine tetraacetic acid) and SDS (sodium dodecyl sulfate) reagents. Compounds were prepared at 10 mM concentration in 100 mM citrate–phosphate buffer pH 6.0. Enzymatic activity in the absence of any reagent was considered to be 100%. The other reaction conditions followed as described in Sect. 2.2.
Kinetic constants
Kinetic parameters KM (Michaelis–Menten constant) and Vmax (maximum velocity) were calculated by means of the velocity curve as a function of the substrate concentration, Michaelis–Menten model. For this, Curve Expert program, version 1.4 for Windows, was used.
In silico predictions, molecular modeling and phylogeny
FASTA sequence of dextranase precursor from P. chlamydosporia was retrieved from UNIPROTKB (ID A0A179FQV9). The target sequence was searched for similar sequences using BLASTp (protein–protein Basic Local Alignment Search Tool) (http://www.ncbi.nlm.nih.gov/BLAST/). Signal dextranase sequence was predicted using SignalP 4.0 server. Theoretical protein isoelectric point (pI) and molecular weight were calculated using the pI/MW tool (http://web.expasy.org/compute_pi/).
Dextranase sequences were selected from CAZy (http://www.cazy.org/GH49_characterized.html) and from National Center for Biotechnology Information (NCBI). The selected sequences were: P. chlamydosporia (XP_018144597.1), Aspergillus niger (BAA18971.1), Lipomyces starkeyi (AAS90631.1), Talaromyces minioluteus (AAB47720.1), Trichoderma harzianum (KKO97501.1), Talaromyces cellulolyticus (GAM43713.1), Sporothrix schenckii (ERS97553.1), Sporothrix brasiliensis (KIH89944.1) and Penicillium subrubescens (OKP15192.1). Dextranase amino acid sequences were aligned with ClustalX, and MEGA 7 was used to construct a neighbor joining (NJ) tree. The 3-dimensional (3D) structure for dextranase from P. chlamydosporia was predicted by protein modeling with the SWISS-MODEL server (http://swissmodel.expasy.org/), using a Talaromyces minioluteus (syn. Penicillium minioluteum) (UniProt Acession no. P48845) dextranase template.
Results and discussion
Purification of dextranase produced by P. chlamydosporia
Dextranase produced by P. chlamydosporia was purified to homogeneity in two steps, the first step being anion-exchange resin chromatography (DEAE-Sepharose), and the second step a cation-exchange resin chromatography (CM-Sepharose). The enzyme was purified with a yield of 152%, purification factor of 6.84 and specific activity of 358.63 U/mg (Table 1). In the first purification stage, increased total dextranase activity was observed, which suggests inhibitor elimination during this step. This, consequently, can explain the high yield obtained for the process.
Table 1.
Purification steps of dextranase from Pochonia chlamydosporia
| Step | Total enzyme activity (U) | Total protein (mg) | Specific activity (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Crude extract | 1397.50 | 26.65 | 52.45 | 100 | 1 |
| DEAE Sepharose | 2224.09 | 16.02 | 138.81 | 159 | 2.65 |
| CM Sepharose | 2120.49 | 5.91 | 358.63 | 152 | 6.84 |
One dextranase unit (U) was defined as the amount of the enzyme that catalyzes the release of 1 μmol of reducing sugar per minute under the assay conditions
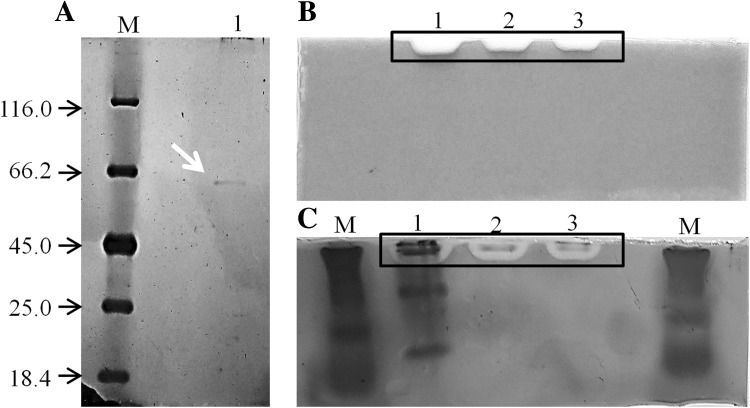
In SDS-PAGE (Fig. 1a), there is only a single band, corresponding to the purified dextranase molecular weight estimated at 64 kDa by mobility comparison with marker proteins. In the native PAGE prior to staining by silver nitrate (Fig. 1b) dextranase activity was confirmed in samples from the crude extract, DEAE-Sepharose chromatography and CM-Sepharose chromatography. Dextranase activity was observed as clear bands in the gel. This same native PAGE was stained by silver nitrate (Fig. 1c) in order to confirm dextranase presence and to observe other proteins occurrence in the samples. After staining with silver nitrate of native PAGE (Fig. 1c), it was possible to observe the presence of other protein in the crude extract besides dextranase, and that dextranase activity band corresponds to the only protein band in the sample from the second chromatography, confirming its purification to homogeneity.
Fig. 1.
Purified dextranase from Pochonia chlamydosporia on SDS-PAGE stained with silver nitrate. Lane M: Protein molecular weight markers (Thermo Scientific); Lane 1: purified dextranase (a). Clear bands, indicated by black rectangles, showing dextranase activity on a 10% native PAGE gel containing 1% (w/v) blue dextran, prior to staining with silver nitrate (b), and stained with silver nitrate (c). Lane M: Protein molecular weight markers (Thermo Scientific); Lane 1: crude extract; Lane 2: pool from DEAE-Sepharose; Lane 3: purified dextranase
This estimated molecular weight is close to the observed molecular weight values for purified dextranases from Chaetomium erraticum, Paecilomyces marquandii, Penicilillium aculeatum, Penicillium funiculosum and two dextranases from Chaetomium gracile, whose molecular weights were estimated at, respectively, 59, 73, 66.2, 67, 71 and 77 kDa (Abdel-Aziz et al. 2007; Hattori et al. 1981; Machado 2009; Mahmoud et al. 2014; Virgen-Ortíz et al. 2015).
Enzymatic characterization
pH and temperature effect on enzymatic activity
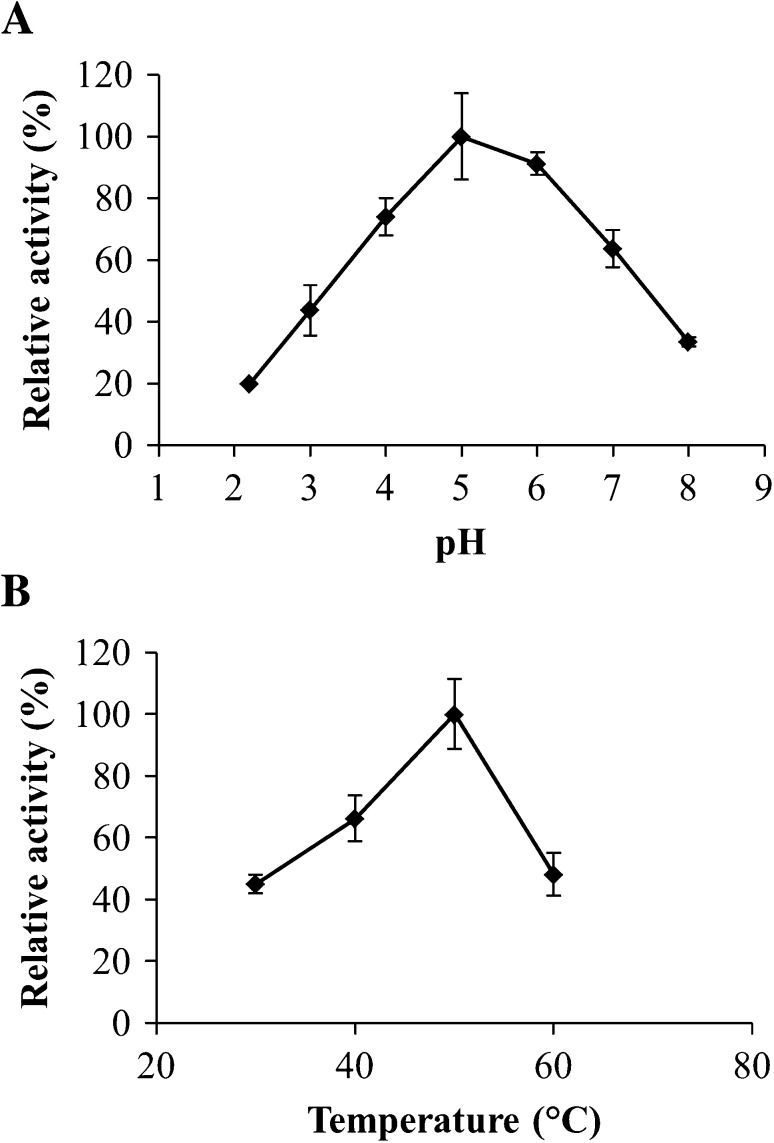
Purified P. chlamydosporia dextranase showed higher activity at pH 5.0 (Fig. 2a). In addition, it was observed that this dextranase has a high hydrolysis capacity also at pH 6.0, with relative activity above 90%, and at pH 4.0, with a 74% relative activity. Similarly, Machado (2009) determined that pH 6.0 provides higher activity of dextranase produced by P. marquandii, and that at pH 4.0, it has 90% relative activity. Virgen-Ortiz et al. (2015) reported that pH 5.2 results in increased activity of the dextranase produced by C. erraticum, and observed relative activity above 85% in the pH ranging from 4.0 to 6.0. However, when studying P. aculeatum dextranase, Mahmoud et al. (2014) observed that the highest activity pH was 4.5, and that from this pH value, dextranase activity decreased abruptly.
Fig. 2.
pH (a) and temperature (b) effect on the enzymatic activity of purified dextranase from Pochonia chlamydosporia. 2.43 and 2.44 U/mL were defined as 100% relative activity for pH and temperature effect, respectively
The highest activity temperature of purified P. chlamydosporia dextranase was 50 °C (Fig. 2b). At 60 °C the enzyme demonstrated only 48% activity, indicating that at this temperature protein denaturation may occur. These results are close to the results found by Mahmoud et al. (2014), which determined that dextranase produced by P. aculeatum exhibited higher activity at 45 °C, with relative activity of approximately 75% at 40 and at 50 °C. On the other hand, Virgen-Ortiz et al. (2015) observed that C. erraticum dextranase exhibited maximum activity at 60 °C and more than 85% of its maximum activity in the temperatures ranging from 55 to 65 °C, and Machado (2009) reported that P. marquandii dextranase presented higher activity temperature at 55, and at 60 °C it retained 80% of its activity. These results demonstrate that the dextranase produced by the fungus P. chlamydosporia presents similar properties when compared to those already described in the literature.
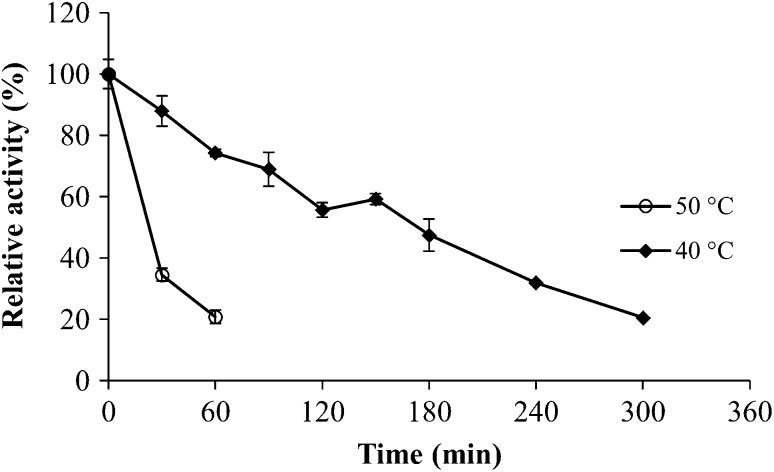
P. chlamydosporia dextranase thermal stability was tested at 40 and at 50 °C (Fig. 3), which corresponds to the reaction temperature which resulted in higher enzymatic activity. Dextranase showed low thermal stability at 50 °C, exhibiting 35% relative activity when previously incubated for 30 min prior enzymatic activity measurement. However, the enzyme was stable at 40 °C, with 48% relative activity when previously incubated for 180 min. Aspergillus penicillioides dextranase showed little thermal stability, with relative activity close to zero when incubated at 45 °C for 60 min (El-Shamy and Atalla 2014). Machado (2009) observed that dextranase produced by P. marquandii was much more stable at 37 °C, maintaining 85% of its initial activity after has been incubated for 24 h. On the other hand, when evaluating thermal stability at 55 °C, reaction temperature that resulted in higher activity, Machado (2009) also observed low stability, with relative activity, after 30 min of incubation, of approximately 20%. Although there are dextranases produced by bacteria much more stable at higher temperatures, the industrial application of these enzymes becomes impracticable due to the extremely low amount of dextranase produced (Wynter et al. 1995). Thus, despite having intermediate stability, P. chlamydosporia dextranase has the advantage of being produced in higher quantity, with 4.82 U/mL activity in the crude extract.
Fig. 3.
Thermal stability of purified dextranase from Pochonia chlamydosporia at 40 and 50 °C. 2.44 U/mL was defined as 100% relative activity
Effect of metal ions and other reagents on enzymatic activity
Ag1+ and Hg2+ ions strongly inhibited purified dextranase, both reducing dextranase activity to 17%. Cu2+, Mg2+, Fe2+, Ca2+ and Zn2+ ions reduced enzyme activity to, respectively, 51, 60, 71, 77 and 88%. The SDS and EDTA compounds had little influence on dextranase activity, reducing it to 79 and 81%, respectively (Table 2).
Table 2.
Effect of metal ions and other reagents on the enzymatic activity of purified dextranase from Pochonia chlamydosporia
| Compound | Relative activity (%) |
|---|---|
| Control | 100.00 ± 10.85 |
| Zn2+ | 87.81 ± 0.77 |
| Ca2+ | 77.00 ± 8.48 |
| Mg2+ | 59.67 ± 2.66 |
| Cu2+ | 51.05 ± 7.29 |
| Mn2+ | 88.96 ± 15.11 |
| Fe2+ | 70.76 ± 9.75 |
| Ag1+ | 17.22 ± 0.02 |
| Hg2+ | 17.40 ± 0.87 |
| EDTA | 80.52 ± 6.92 |
| SDS | 79.03 ± 6.68 |
P. marquandii dextranase activity increased by 20 and 15% in the presence of, respectively, Fe2+ and Mn2+ at 10 mM concentration and decreased by approximately 50% of its activity in the presence of Cu2+ and Pb2+ and 40% in the presence of Ag+, also at 10 mM concentration (Machado 2009). C. erraticum dextranase suffered total inhibition in the presence of 1 mM Ag+ and 77% inhibition in the presence of 1 mM Cu2+ and had its activity increased by 18 and 25% when incubated with, respectively, 1 mM Ca2+ and Co2+ (Virgen-Ortíz et al. 2015). Cu2+ and Hg2+ completely inhibited C. gracile dextranase, and Fe3+ inhibited its activity by 40% at 1 mM concentration (Hattori et al. 1981). P. funiculosum dextranase was totally inhibited by Hg2+, had its activity reduced by approximately 70% when incubated with Ag+, and was activated by 2 mM Co2+, Mn2+ and Cu2+ in 42, 39 and 21%, respectively (Sugiura et al. 1973).
Kinetic constants
The maximum velocity values and the Michaelis–Menten constant were determined by non-linear data adjustment to the Michaelis–Menten equation, with correlation coefficient (r2) of 0.9935. P. chlamydosporia dextranase presented KM and Vmax values of, respectively, 1.77 g/L and 2.09 mM glucose/min at 50 °C and pH 6.0 for dextran with molecular weight ranging from 60,000 to 90,000. Considering the average dextran molecular weight as 75,000, the KM value resulted in 23.60 μM. This value has the same order of magnitude of observed KM values for other dextranases. Abdel-Naby et al. (1999) found that P. funiculosum dextranase showed KM of 25 μM for dextran with molecular weight 260,000, and Machado (2009) reported KM of 7.86 μM for P. marquandii dextranase.
In silico predictions, molecular modeling and phylogeny
The P. chlamydosporia dextranase precursor sequence, deposited in UNIPROTKB (ID A0A179FQV9), has 601 amino acid residues, with a peptide signal of 16 residues. Mature protein is composed of 585 amino acid residues, exhibiting molecular weight and pI of, respectively, 64.38 kDa and 5.96. This predicted molecular weight value coincides with the molecular weight value of 64 kDa estimated empirically by SDS-PAGE in the present study.
In addition, the dextranase precursor sequence of P. chlamydosporia (XP_018144597.1) shares a high degree of identity with other fungi dextranases, with 69, 79, 76, 70, 70 and 67% identity with the dextranases of Lipomyces starkeyi (AAS90631.1), Trichoderma harzianum (KKO97501.1), Talaromyces cellulolyticus (GAM43713.1), Sporothrix schenckii (ERS97553.1), Sporothrix brasiliensis (KIH89944.1) and Penicillium subrubescens (OKP15192.1), respectively.
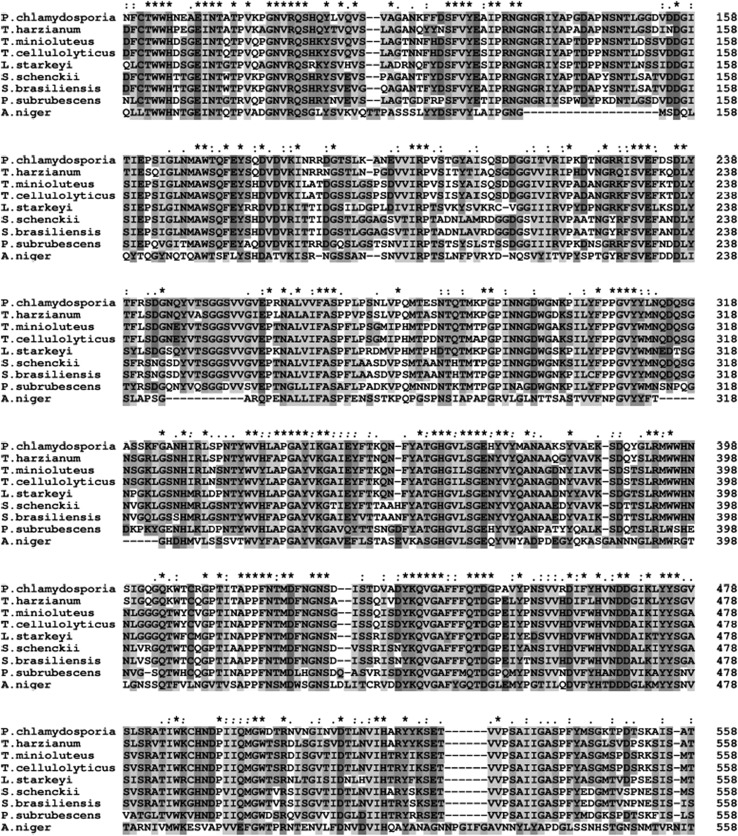
The selected fungi dextranases multiple sequence alignment revealed the occurrence of several conserved regions, occurring mostly among amino acid residues at positions ranging from 82 to 134, 324 to 377 and 403 to 478 (Fig. 4). Although sequence and structural homology methods are tools to determine global similarities between compared proteins, the enzyme molecular role is related to its active site. Thus protein comparisons focusing on global sequence and structural similarities may neglect proteins with conserved active sites but divergent sequences and structures (Morya et al. 2012).
Fig. 4.
Alignment of the dextranases from different fungi. Asterisk, colon and dot symbols indicate full conservation, and groups of strongly and weakly similar properties residues, respectively
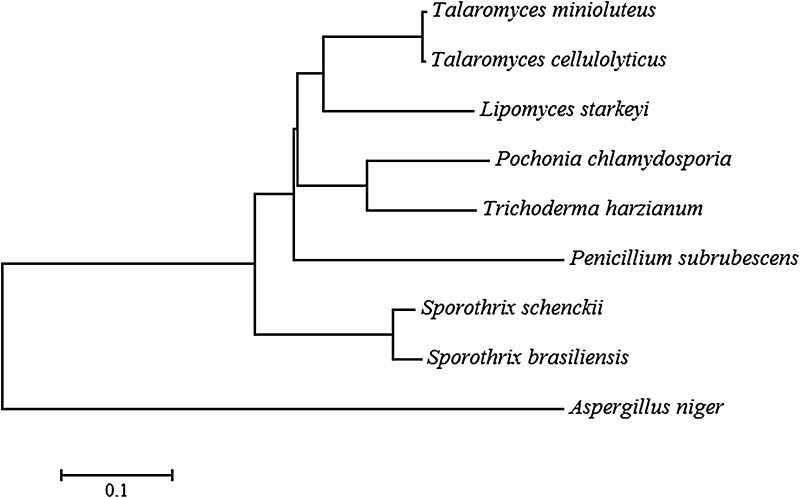
The phylogenetic tree constructed based on protein sequences using Neighbor Joining method suggested that dextranases from fungi had one common ancestor (Fig. 5). Phylogenetic analysis showed that these dextranases cluster into two subclades. One subclade contains dextranases from Sordariomycetes, including S. schenckii and S. brasiliensis. Another subclade consisted of six dextranases, and contains three clusters. The first cluster is composed by T. minoluteus and T. cellulolyticus, from Eurotiomycetes, and L. starkeyi from Saccharomycetes. The dextranase from P. chlamydosporia clusters with the dextranase from T. harzianum, both species belong to Sordariomycetes, constituting the second cluster. P. subrubescens, from Eurotiomycetes, was distinct from others and formed the third cluster. Thus, though the dextranase from P. chlamydosporia has a distant evolutionary relationship with dextranases from some fungi, it clusters with dextranases from others fungi belong to different orders.
Fig. 5.
Phylogenetic tree constructed by Neighbor-joining tree method based on dextranases sequences from different fungi species. Pochonia chlamydosporia 170 [XP_018144597.1], Aspergillus niger ATCC 9642 [BAA18971.1], Lipomyces starkeyi KSM22M [AAS90631.1], Talaromyces minioluteus MUCL 38929 [AAB47720.1], Trichoderma harzianum T6776 [KKO97501.1], Talaromyces cellulolyticus Y-94 [GAM43713.1], Sporothrix schenckii ATCC 58251 [ERS97553.1], Sporothrix brasiliensis 5110 [KIH89944.1] and Penicillium subrubescens CBS 132785 [OKP15192.1]
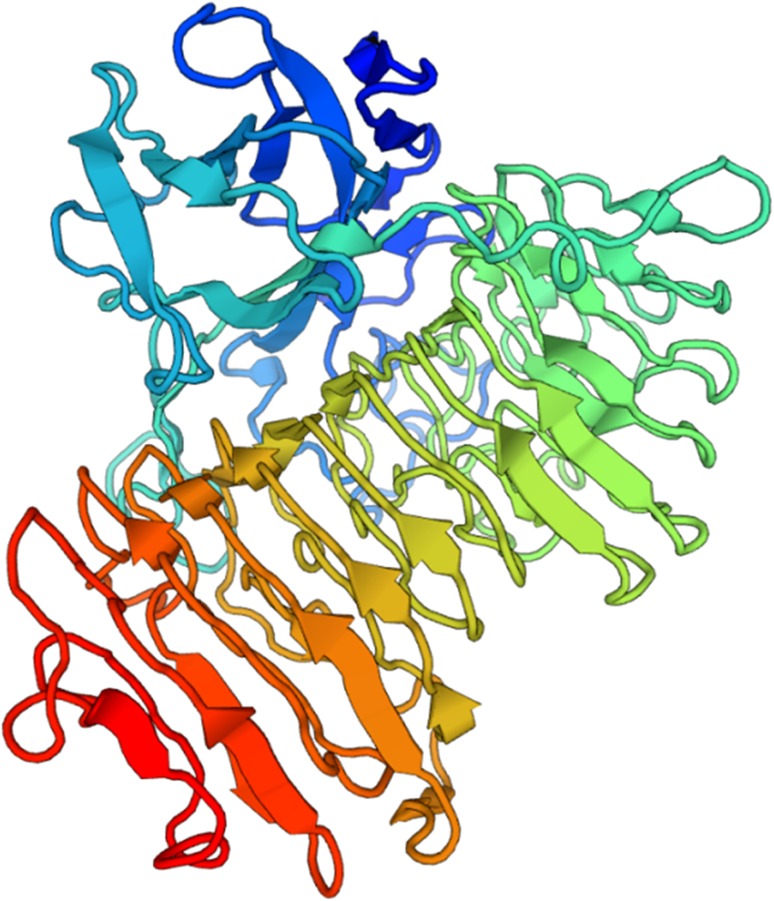
The 3D structure (Fig. 6) of the dextranase from P. chlamydosporia was modeled using SWISS-MODEL employing dextranase structure from Penicillium minioluteum (anamorph: T. minoluteum) (PDB code: 1ogm.1) as template, sharing 75.44% identity. Dextranase from P. chlamydosporia consists of two domains. The first domain is composed by 211 residues forming 15 β strands. The other domain consists in a right-handed parallel β-helix. The two domains are connected by several residues from the N-terminus of the β -helix. Asp395 is the catalytic amino acid residue from P. minioluteum dextranase, template of the dextranase structure of this present study (Larsson et al. 2003). This residue is shown conserved in the dextranases submitted to alignment, corresponding to the Asp422 residue in the P. chlamydosporia dextranase with the signal peptide (Fig. 4). This indicates that this residue may play catalytic role in the dextranase target of this study, since enzymes Asp residues are essential to enzyme activity involving glucans hydrolysis (Wang et al. 2014).
Fig. 6.
3D structure of dextranase from Pochonia chlamydosporia predicted using the dextranase from Talaromyces minioluteus as template
P. chlamydosporia dextranase predicted structure is similar to that found for dextranases of different species. The dextranase structure of Arthrobacter oxydans predicted by Wang et al. (2014) is also composed of two domains, the first domain consisting of 13 β-strands containing 200 residues and the second domain a right-handed β-helix. In addition, isopullulanase from A. niger, an enzyme belonging to GH 49, is also structurally similar to dextranases, having two domains: one composed of 13 β-strands and the other domain composed of a right-handed β-helix (Mizuno et al. 2008).
Acknowledgements
The authors would like to thank the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), the Fundação de Amparo à Pesquisa de Minas Gerais (FAPEMIG), and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for the financial support.
Compliance with ethical standards
Conflicts of interest
The authors declare that they have no conflict of interest.
References
- Abdel-Aziz MS, Fatma Talkhan N, Janson J. Purification and characterization of dextranase from a new strain of Penicillium funiculosum. J Appl Sci Res. 2007;3:1509–1516. [Google Scholar]
- Abdel-Naby MA, Ismail A-MS, Abdel-Fattah AM, Abdel-Fattah AF. Preparation and some properties of immobilized Penicillium funiculosum 258 dextranase. Process Biochem. 1999;34:391–398. doi: 10.1016/S0032-9592(98)00127-7. [DOI] [Google Scholar]
- Batista MCT (2014) Produção de dextranases a partir de bagaço de malte: caracterização e avaliação do potencial de aplicaçao em indústria sucroalcooleira. Thesis, Universidade Federal do Paraná
- Bhatia S, Bhakri G, Arora M, Uppal SK, Batta SK. Dextranase production from Paecilomyces lilacinus and its application for dextran removal from sugarcane juice. Sugar Tech. 2010;12:133–138. doi: 10.1007/s12355-010-0026-4. [DOI] [Google Scholar]
- Boil PGMD, Wienese S (2002) Enzymic reduction of dextran in process-laboratory evaluation of dextranases. In: Proceedings of South African Sugar Technologist Association 76:435–443
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- CAZy—Carbohydrate Active Enzymes (2017) http://www.cazy.org/Glycoside-Hydrolases.html. Acessed 16 March 2017
- El-Shamy AR, Atalla SMM. Immobilization of dextranase by Aspergillus penicillioides NRC 39 and its properties. Afr J Microbiol Res. 2014;8:3893–3900. [Google Scholar]
- Hattori A, Ishibashi K, Minato S. The purification and characterization of the dextranase of Chaetomium gracile. Agric Biol Chem. 1981;45:2409–2416. [Google Scholar]
- Jaiswal P, Kumar S. Impact of media on isolation of dextranase producing fungal strains. J Sci Res. 2011;55:71–76. [Google Scholar]
- Jiao Y, Wang S, Lv M, Jiao B, Li W, Fang Y, Liu S. Characterization of a marine-derived dextranase and its application to the prevention of dental caries. J Ind Microbiol Biotechnol. 2014;41:17–26. doi: 10.1007/s10295-013-1369-0. [DOI] [PubMed] [Google Scholar]
- Jiménez ER. Dextranase in sugar industry: a review. Sugar Tech. 2009;11:124–134. doi: 10.1007/s12355-009-0019-3. [DOI] [Google Scholar]
- Khalikova E, Susi P, Korpela T. Microbial dextran-hydrolyzing enzymes: fundamentals and applications. Microbiol Mol Biol Rev. 2005;69:306–325. doi: 10.1128/MMBR.69.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larsson AM, Andersson R, Ståhlberg J, Kenne L, Jones TA. Dextranase from Penicillium minioluteum: reaction course, crystal structure, and product complex. Structure. 2003;11:1111–1121. doi: 10.1016/S0969-2126(03)00147-3. [DOI] [PubMed] [Google Scholar]
- Machado FPP (2009) Produção, purificação e caracterização bioquímica da dextranase de Paecilomyces marquandii. Thesis, Universidade Federal de Viçosa
- Mahmoud KF, Gibriel AY, Amin AA, Nessrien MN, Yassien NM, El Banna HA. Microbial production and characterization of dextranase. Int J Curr Microbiol Appl Sci. 2014;3:1095–1113. [Google Scholar]
- Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- Mizuno M, Koide A, Yamamura A, Akeboshi H, Yoshida H, Kamitori S, Sakano Y, Nishikawa A, Tonozuka T. Crystal structure of Aspergillus niger isopullulanase, a member of glycoside hydrolase family 49. J Mol Biol. 2008;376:210–220. doi: 10.1016/j.jmb.2007.11.098. [DOI] [PubMed] [Google Scholar]
- Morya VK, Yadav S, Kim EK, Yadav D. In silico characterization of alkaline proteases from different species of Aspergillus. Appl Biochem Biotechnol. 2012;166:243–257. doi: 10.1007/s12010-011-9420-y. [DOI] [PubMed] [Google Scholar]
- Picozzi C, Meissner D, Chierici M, Ehrmann MA, Vigentini I, Foschino R, Vogel RF. Phage-mediated transfer of a dextranase gene in Lactobacillus sanfranciscensis and characterization of the enzyme. Int J Food Microbiol. 2015;202:48–53. doi: 10.1016/j.ijfoodmicro.2015.02.018. [DOI] [PubMed] [Google Scholar]
- Singleton V, Horn J, Bucke C, Adlard M. A new polarimetric method for the analysis of dextran and sucrose. J Am Soc Sugarcane Technol. 2002;22:112–119. [Google Scholar]
- Sufiate BL, Soares FEF, Gouveia AS, Moreira SS, Cardoso EF, Tavares GP, Braga FR, Araujo JV, Queiroz JH (2018) Statistical tools application on dextranase production from Pochonia chlamydosporia (VC4) and its application on dextran removal from sugarcane juice. An Acad Bras Cienc (accepted) [DOI] [PubMed]
- Sugiura M, Ito A, Ogiso T, Kato K, Asano H. Studies on dextranase: purification of dextranase from Penicillium funiculosum and its enzymatic properties. Biochim Biophys Acta. 1973;309:357–362. doi: 10.1016/0005-2744(73)90034-X. [DOI] [PubMed] [Google Scholar]
- Virgen-Ortíz JJ, Ibarra-Junquera V, Escalante-Minakata P, Ornelas-Paz JJ, Osuna-Castro JA, González-Potes A. Kinetics and thermodynamic of the purified dextranase from Chaetomium erraticum. J Mol Catal B Enzym. 2015;122:80–86. doi: 10.1016/j.molcatb.2015.08.020. [DOI] [Google Scholar]
- Wang X, Lu M, Wang S, Fang Y, Wang D, Ren W, Zhao G. The atmospheric and room-temperature plasma (ARTP) method on the dextranase activity and structure. Int J Biol Macromol. 2014;70:284–291. doi: 10.1016/j.ijbiomac.2014.07.006. [DOI] [PubMed] [Google Scholar]
- Wynter CVA, Galeal CF, Cox LM, Dawsonl MW, Patel BK, Hamilton S, De Jersey J, Inkerman PA. Thermostable dextranases: screening, detection and preliminary characterization. J Appl Bacteriol. 1995;79:203–212. doi: 10.1111/j.1365-2672.1995.tb00936.x. [DOI] [Google Scholar]
- Zhang Y, Li R, Zhang H, Wu M, Hu X. Purification, characterization, and application of a thermostable dextranase from Talaromyces pinophilus. J Ind Microbiol Biotechnol. 2016;44:317–327. doi: 10.1007/s10295-016-1886-8. [DOI] [PubMed] [Google Scholar]