Abstract
The wide array of biological properties attributed to the CCN family of proteins (Perbal in Lancet 363(9402):62–64, 2004) led me to reconsider the possible relationship and roles that these proteins may play as a team, instead of acting on their own as individual regulators in various signaling pathways. The dynamic model which I present in this review stems from the contribution of the biological properties that we established for CCN3, one of the three founding members of the CCN family, which was identified by our group as the first CCN protein showing growth inhibitory properties (1992), expressed mainly in quiescent cells (1996), and showing anti-tumor activities in several cellular models both ex vivo and in vivo. At the present time CCN3 is the only member of the family that has been reported to negatively act on the progression of the cell cycle. The unique dual localisation of CCN3 in the nucleus and outside cells, either at the membrane or in the extracellular matrix, that I first established in 1999, and that now appears to be shared by several other CCN proteins, is a unique essential feature which can no longer be ignored. Based on the structural and functional properties of CCN3, shared by most of the CCN family members, I propose an « all in one » concept in which CCN proteins are team members with specific functions that are aimed at the same goal. This model accounts both for the functional specificity of the various CCN proteins, their sequential and opposite or complementary effects in various biological context, and for the biological consequences of their physical interaction and biological cross-regulation.
Keywords: Signaling coordination, Development, Angiogenesis, Differentiation, Inflammation, Growth control, Nuclear proteins, Matricellular proteins, Extracellular matrix (ECM), Tumorigenic potential, Regulation of cell cycle, Thrombospondin, Von Willebrand factor, Growth factors, IG Fbinding proteins (IGFBPs)
A short summary of CCN salient features
Even though the topic has already been discussed in several previous reviews dealing with different aspects of the biology of the CCN proteins, I believe that reconsidering structure-function aspects of these proteins from a different angle may help us to think in a new way about their place as members of a team aiming toward the same goal: that is to participate in the balance of signaling pathways contributing to the harmonious interactions of cells within the body and with their environment.
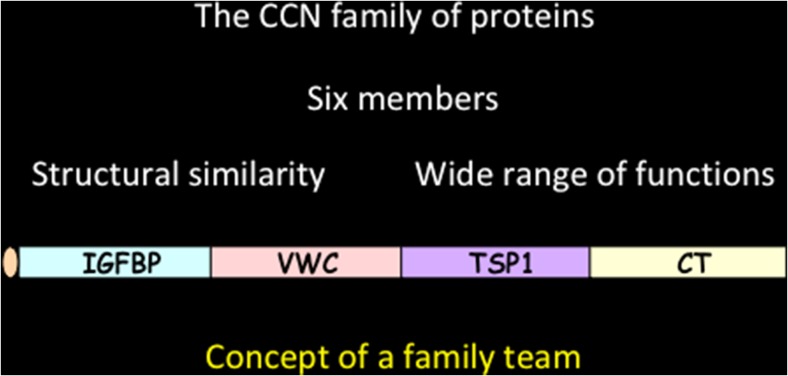
The most striking feature, which in my opinion has not attracted the attention that it deserves, is their structural tetramodular organization (Fig. 1). This tetramodular organization provides the basis for a very sophisticated array of biological properties for a unique group of regulatory proteins with acute roles in cell growth, differentiation, development, communication and response to microenvironmental signaling. As previously stated « The multimodular organisation of the CCN proteins raises interesting questions as to the participation of each module in the function of the full length protein. Either the activities of each module add up or they confer on the whole protein specific functions that might substitute or add to the function of the individual modules. » (Perbal 2001).
Fig. 1.
The structural organization of CCN proteins
Except for rCOP1, WIPS2 and CCN5 (orthologs encoded by the same gene, see below) which were reported lacking the carboxy-proximal CT module that is present in all other members, the CCN proteins are composed of four « construction bricks » assembled together as the result of evolutionary exon shuffling. The deep biological significance of this unique assembly has not been the matter of intense research.
To me, our ignorance of the biophysical properties of these molecules and the way that their constitutive modules interact to produce a tri-dimensional functional protein is the most important gap in the field.
This gap resulted, at least partly, from the inability of most investigators, those like us who were not « hard core » protein chemists, to deal with the great difficulties inherent in the physical characteristics of these proteins, and to obtain substantial amounts of full length native protein.1 Also, for many years, structuralists did not show a strong interest in these proteins whose wide and complex biological activities might have disuaded groups struggling to get « fundable projects ».2 Using techniques such as Circular Dichroism (CD), Nuclear Magnetic Resonance (NMR) spectroscopy, or Xray diffraction analysis, altogether are unique tools to establish and understand the roles of critical physical parameters governing both protein secondary and tertiary structure that underly conformational changes involved in interactions with ligands and with proteins including other CCN proteins.
Efforts that were initiated in the past (Holbourn et al. 2011) did not translate into a significant contribution to the understanding of CCN protein biology, but recently a publication reported the use of X-ray cristallography to identify the fine structure and binding properties of the VWC domain of CCN33 to the bone morphogenetic protein-2 (BMP2) (Xu et al. 2017).
As a reminder, the CCN proteins contain a VXC type C repeat that is present in other proteins involved in homeostasis, and is believed to participate in the late stages of oligomerisation. Together with the CT module, the presence of these two repeats in the CCN proteins is most likely responsible, at least in part, for the difficulty to purify large amounts of native biologically active CCN proteins in a stable form.
Another key feature of the CCN proteins is the considerable number of cystein residues (38) (except for CCN6 which lacks 4 residues in the VWC module) that are evolutionarily conserved, both in number and position in these 32–35 KDa CCN proteins from Frog to Human. Although anyone with a protein chemistry background would be struck by this feature, its biological significance reamains to be studied.
Even if one can easily predict that such a high number of potential disulfide bonds would be essential to the function of the CCN proteins, their individual contribution to any kind of function or physical interaction with a biological partner has not been fully investigated as yet. Although site specific mutagenesis of cystein residues has been proposed to be an easy way to approach their functions in the context of a whole protein,4 it seems that no wide range study of this type has been undertaken.
Recently, the mutation of cystein C241, which is localized within the TSP1 module of CCN3, proved to be essential for its secretion (Kim et al. 2018). The TSP1 repeat has been involved in cell cell interactions, inhibition of angiogenesis, and apoptosis (TSP1 Simple modular Architecture Research Tool (http://smart.embl.de/smart/do_annotation.pl? DOMAIN=SM00209).
Each of the constitutive modules is encoded by a sequence of DNA identical to an exon that can be found in the coding sequences of multidomain proteins known as Insulin-like growth factor binding proteins (IGFBPs), the Von Willebrand factor (VWF), and thrombospondin 1 (TSP1). The fourth construction brick is a module containing a cystine knot that is found in many different signaling proteins including growth factors, and is known to elicit dimerisation of the proteins in which it is present.
One can again wonder why no effort has apparently been put forward to examine the importance of this cystine knot motif for the ability to enter into the formation of homo and heterodimers especially in the light of the observation that CCN5 does not contain that motif.
The assembly of four structural modules, identical to parts of proteins involved in key regulatory functions, suggests that the CCN proteins are interacting with these functions. The shuffling of exons in extracellular proteins had already been suggested to supply « the molecular basis for a complex functional network allowing multiple regulation in and between nearly all tissues. It combines the most different functional complexes like, for example, the antibody framework, inflammation processes or the hematopietic system... » » (Bork 1991).
Altogether, the presence of these four modules in a single protein is not just a curiosity that is worth mentioning. It likely provides the cell with another « multisensing station » permitting coordinated interactions with key signaling pathways involved in the regulation of cell behavior,5 and « coordination of various functions within the world of extracellular proteins » (Bork 1991).
As in a soccer team, where center, forward, right and left wingers, and goal keeper have specialized roles, I see each of the CCN proteins as one team partner with specific functions that are functionally complementary to each other. Each member of the CCN family participates in the the same action and brings its own qualifications and specificity to the team.
The « all in one concept » part 1: A set of members showing opposite activities
The CCN family of proteins contains both negative and positive regulators of cell growth, differentiation and development.
The first evidence for members of the CCN family of proteins playing different functions stemmed from studies that we performed in the 1990s in the quest for a specific integration site for the Myeloblastosis Associated Virus type 1 in MAV1-induced avian nephroblastomas which had previously shown to represent a unique model of the human Wilm’s tumor (Perbal 1994). At that time, nearly all virologists involved in retroviral tumorigenesis studies were looking for insertion sites of viral sequences in the target host DNA.
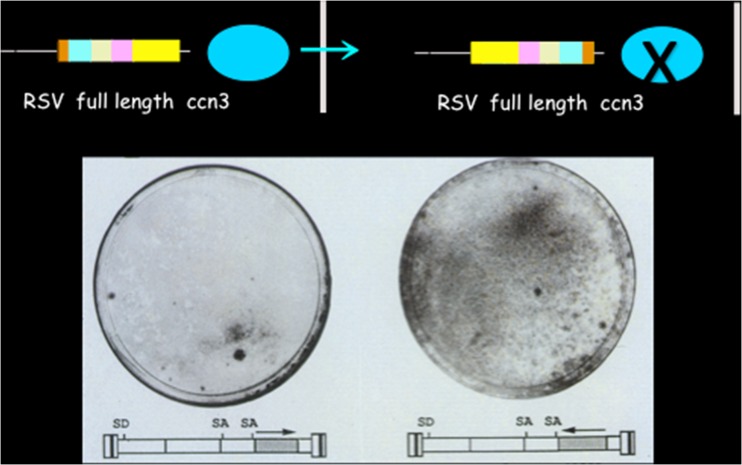
The molecular cloning of insertion sites for MAV1 in nephroblastoma led us to the discovery of CCN3 (originally designated NOV by our group), a protein showing antiproliferative activity when it is native and full length, and exhibiting transforming properties when it is amino-truncated (Fig. 2).6
Fig. 2.
Antiproliferative activity of ccn3
A few months before our manuscript was published, the group of L. Lau and G. Grotendorst had reported the identification of two secreted proteins, presently designated CCN1 and CCN2 that were pro-proliferative factors encoded by immediate early genes.
Based on the highly conserved primary sequence and multimodular structure of these proteins, that the three of us had reported, P. Bork (Bork 1993) proposed one year later that they were forming a new family of regulators that he designated CCN (for more details see Perbal 2013).
With our demonstration in 1992 that the secreted CCN3 protein was endowed with antiproliferative activity, while both CCN1 and CCN2 were growth stimulators, we had established for the first time that the family of proteins contained both negative and positive regulators. Subsequent experiments established that purified recombinant CCN3 inhibited cell growth.
The implications of this discovery were important and were clearly stated.7
As it is commonly illustrated by the functional organization of regulatory pathways in living organisms, from very simple enzymatic reactions up to complex arrays of intracellular and intercellular communication pathways in which positive and negative signals are required, the CCN family of proteins was therefore found to contain the necessary elements to balance their effects on cell proliferation, and most certainly optimize their role in other key biological functions in which they were likely involved.
Another important discovery within the CCN family of genes was the identification, a few years later, of rCOP1 as a negative regulator of cell growth, lost after transformation (Zhang et al. 1998).8 This finding thereby confirmed my previous suggestion that the CCN family of proteins was made of two types of counteracting factors.9 Interestingly the isolation of rCOP1 and the identification of three new CCN proteins in human tumors the same year (Pennica et al. 1998), brought the very first evidence of a CCN protein lacking the C-Terminal domain.
Considering that the C-terminal module of CCN3 is sufficient to induce a strong (80%) cellular growth inhibition (Bleau et al. 2007), and that the CCN5 protein (AKA rCOP1, WISP2) lacking the CT module also exhibits growth inhibitory effects in vascular cells (Lake et al. 2003),10 it appears that the negative effect of these two proteins involves different subdomains.
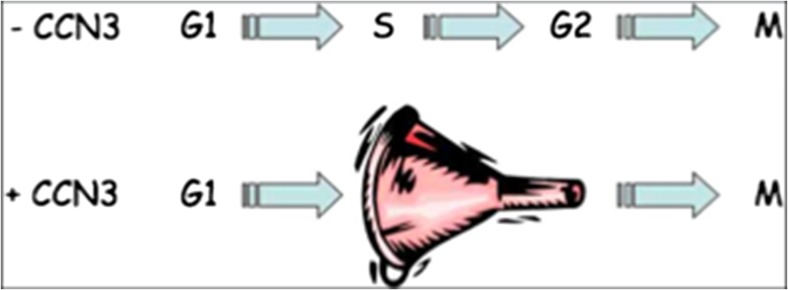
Mechanistically, the inhibitory effects of CCN3 is due to a brake on the cell cycle that results in an accumulation of cells induced by CCN3 at the S phase of the cell cycle (Fig. 3) (Bleau et al. 2007). It is quite unsettling that recent manuscripts do not take into account our observation and still cite the work of Liu et al. (1999) in which the authors claimed that CCN3 induced cell proliferation based on 3-H Thymidine incorporation in 3 T3 cells. The accumulation of cells at the S-phase of the cell cycle, which is induced by exogenous CCN3 fully accounts for the increased thymidine incorporation mentioned by Liu et al. without any need for CCN3-induced cell proliferation. Recent experimental evidence presented at the 9th International Workshop on the CCN family of genes also indicated that CCN2 is involved in regulation of cell cycle progression in primary adult human dermal fibroblasts (G. Fisher personal communication). This observation is in agreement with the opposite functions attributed to CCN3 and CCN2 in several different contexts (see below and « Bits and bytes » by A Leask 2009).
Fig. 3.
CCN3 interferes with cell cycle progression
The basis for the « all in one concept » was born: the members of the CCN family participate in a team playing the same game. Each member of the team has a specific set of functions and roles in the different phases of the play, that all concur with the same goal.
There is a great deal of evidence that the game is played at least in the extracellular matrix compartment where several other families of factors have been reported to play (Murphy-Ullrich and Sage 2014). However, the CCN family of proteins should not just be considered as another set of matricellular proteins with which they share common features. The CCN team appears as a potential « connector » between signaling pathways involving other matricellular proteins.
How these different actors cooperate and/or antagonize functionally is a very exciting and open question.
The « all in one concept » part 2: sequential expression and yin yang cross-regulation
Soon after the isolation of the first three founding members of the CCN family of proteins, their tetramodular structure suggested that the different structural modules might correspond to functional domains that can interact either sequentially or simultaneously with other partners, and that the final biological properties of the CCN proteins might be dependent upon different combinatorial effects » (Perbal 2001).
The sequential expression of CCN proteins correlated with cellular proliferation and differentiation programs was nicely uncovered in studies performed to understand the roles of CCN proteins in glomerular cell proliferation. In our early report on the characterization of CCN3 expression sites in developing human embryo (Chevalier et al. 1998), we had established that normal mesangial cells in human glomeruli expressed low levels of CCN3 RNA but were heavily loaded with CCN3 protein, an observation that suggested that the accumulation of CCN3 reflected a functional involvement of this protein in mature glomeruli. The stimulation of mesangial cell growth by PDGF (platelet derived growth factor) was accompanied by a down regulation of CCN3 expression, an observation in agreement with the antiproliferative effects of CCN3 that had been established ex vivo. By measuring the expression levels of CCN2 and CCN3 during anti-Thy1 induced glomerulonephritis, we could show that glomerular cell proliferation negatively correlated with CCN3 whereas CCN1 and CCN2 expression increased with cell numbers (Van Roeyen et al. 2008) The increase in cell number that occurs first in this system matched the increase in CCN1/CCN2 levels; in the second phase, the levels of CCN3 were found to follow the decrease in cell number that was accompanied with a reduction in both CCN1 ad CCN2 levels.
These results indicated for the first time that CCN proteins with opposite effects on proliferation were expressed sequentially in the anti-Thy1 model for glomerulonephritis, suggesting that a similar time dependent type of expression was a general feature of CCN proteins acting along other regulatory pathways.
Indeed, the same sequential pattern of expression involving the proliferative CCN2 protein and antiproliferative CCN3 was uncovered during the osteoblastic differentiation of pre-osteoblast, during which the expression of CCN2 was found to increase, whereas the expression of CCN3 correlatively decreased (Kawaki et al. 2008).
The use of this system also permitted establishing for the first time that the expression of both CCN2 and CCN3 were inversely correlated and were subject to cross regulatory signaling, the base for a yin yang type of cross regulation for the expression of CCN2 and CCN3. Indeed, knockout mice were also shown to overexpress CCN3.
The functions of the various team players were found to be dependent on each other and to be regulated in a context-dependent way.
The « all in one concept » part 3 physical interactions
In my opinion, the power of protein interactions had been underestimated in the CCN field as compared to the intense and productive attention that has been paid during the last century to the benefits resulting from combining in the same molecular scafolding both the catalytic and regulatory moities of many key enzymatic complexes. One of the most elegant systems is exemplified by the E. coli Aspartate transcarbamylase (ATCase), the first enzyme in the pyrimidine anabolic pathway, which is composed of two trimers of identical catalytic polypeptidic chains and three dimers of regulatory polypeptidic chains, altogether cross talking to ensure a very efficient allosteric response to the needs of the cell for pyrimidines. Upon evolutionary pressures the three first enzymatic activities of the biosynthestic pyrimidine pathway have been combined within a 240 KDa multifunctinal hexameric complex in which covalently linked catalytic domains of ATCase, carbamylphosphate synthetase (CPSase) and dihydroorotase (DHOase) catalyze each reaction in a coordinated manner.
The physical interactions that are holding together the four modules of CCN proteins and those which were reported to occur between CCN proteins are key features that helps regulate, both positively or negatively the degree of CCN protein interactions with potential targets.
Just like ice cream for which some associations of flavors often seem to work better to enhance both taste and pleasure, the physical interactions between CCN proteins is most certainly an essential factor allowing an increase and fine tuning of their biological potential, thereby ensuring that the combined actions of team members will be efficient and successful.
The presence of a CT module harboring a cystine knot that was shown to elicit dimerization of several growth factors including PDGF, nerve growth factor (NGF), transforming growth factor beta (TGF beta), and human chorionic gonadotropin, together with the VWC module that is involved in protein multimerization, strongly suggested that homo and heterodimers of CCN proteins would form under biological conditions.
Indeed, the physical interactions that were reported by several goups for various CCN proteins, indicated that the modular structure of the CCN proteins provided the basis for physical interactions taking place not only between CCN proteins themselves, but also with other proteins in which similar modules were represented.
The homodimerization and multimerization of CCN proteins with other regulatory factors involved in the control of various other signaling pathways offers a tremendous way of increasing the flexibility and power of regulatory hypercomplexes that are most probably responsible for the high level supervision and control of signaling pathways that are playing essential biological functions.
As previously predicted (Perbal 2001), the existence in the CCN team, of a protein lacking the CT module can considerably increase the plasticity of the system, by freeezing the complexes in dead-end or constitutively active states, avoiding or favoring the complexation of ligands to the CT module of a CCN protein combined with a CT negative partner.
Considering that the CT module is also represented in a number of proteins sharing no other structural relationship with CCN proteins, the possibility of pairing a whole set of CT containing factors with CCN proteins would considerably increase the number of combinations between apparently unrelated functional pathways and make them work together (Perbal 2016).
The multimodular structure of the CCN proteins which has been maintained throughout evolutionary selective pressures provides the basis for extremely flexible and potentially complex type of hyperstructures that might have been required to serve the developmental needs and adaptative responses to environmental changes that have led to the emergence of mankind.
The « all in one concept » part 4: the multiple localisation of CCN protein isoforms
For the team members to play a concerted action, they need to be located on the same playgrounds.
The detection of CCN proteins in different cellular compartments raised interesting questions as to the biological significance of these localisations, some of them being quite unexpected (reviewed in Perbal 2001).
While some of the team players appear to be kept as a reserve to be used in complex games, involving a long run multitask strategy, others are placed at critical points in order to keep the system running at the basal pace, enabling the system to react more efficiently and rapidly in case of a need, by adding or changing the team members at play.
The way to make the best use of a fire in a fireplace and build it up rapidly when needed is to keep a small amount of embers that can be used to restart almost instantly a new fire upon addition of new kindling.
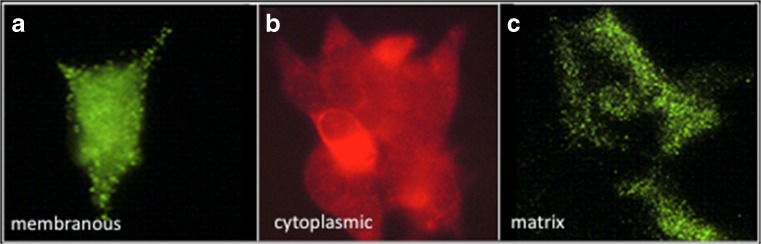
For example, we have established that CCN3 isoforms are present in the extracellular matrix, at the cellular membrane, within the cytoplasm, and within the nucleus of cells (Perbal 1999; Kyurkchiev et al. 2004; Perbal 2006) (Fig. 4). If the extracellular detection of CCN proteins containing signal peptides for secretion was not a real surprise, their nuclear localisation suggested that there might be different specialized isoforms playing distinct roles. The localization of several different regulatory factors both outside and within the nucleus of cells has been heavily documented.
Fig. 4.
Immunological detection of the CCN3 protein in human cultured cells
Interference of CCN3 with cell cycle progression is actually in good agreement with CCN3 being detected in the nucleus of several cell types (Perbal 1999, 2006) and interacting with the rpb7 subunit of the RNA polymerase, a subunit known to interact with a transcriptional mediator (Poss et al. 2013).
For a long time, the concept of nuclear CCN proteins was not accepted by many members of the CCN community who reluctantly considered the possibility that extracellular matrix proteins, participating in the regulation of key signaling pathways and interacting with membrane receptors (Lau 2016), might physiologically be acting in the nuclear cell compartment.
As discussed elsewhere (Perbal 2013), there is increasing evidence in favor of all CCN proteins showing a dual extracellular and nuclear localization, that seem to be dependent upon physiological conditions.
In the case of CCN3, the nuclear localization of amino-truncated forms was shown to be frequently associated with tumorigenicity although the production of short forms (whose localization was not identified) were also produced predominantly in myoblasts where differentiated myotubes contained only full length forms (B. Perbal unpublished).
The nuclear functions of CCN proteins needs to be carefully evaluated, as I feel that the detection of nuclear CCN proteins that is described in scattered reports that succeed in making their way to publication, is the « tree hiding the forest ».
Regretably the localization of CCN family members within the nucleus is the common feature that has been the least studied over the past 20 years.
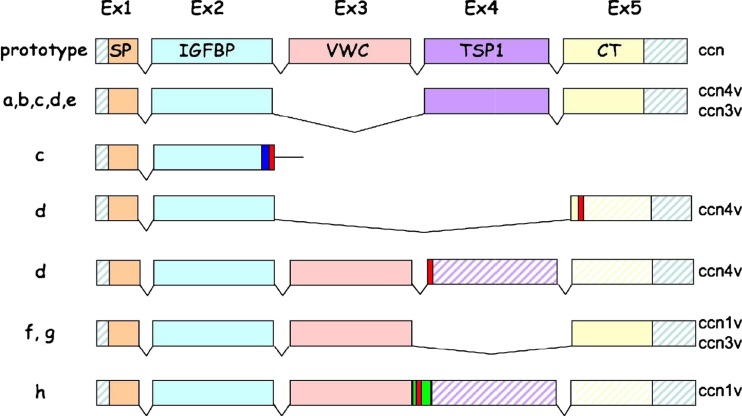
The production of isoforms in both normal and pathological conditions (Fig. 5) is another critical feature of the CCN proteins that has unfortunately not been fully exploited. Absence of an individual domains in some of these isoforms might be extremely informative about their biological function(s). Also some of them seem to be expressed in specific conditions and are likely playing specific roles in these contexts. In addition, the production of these isoforms and their transport to their site of action might provide us with precious information regarding the physical parameters that are requested for specific adressing of the CCN proteins.
Fig. 5.
Variants CCN3 proteins generated by alternative splicing
We know for instance for a long time that CCN3 is post-translationally modified by glycosylation that is not alone sufficient to account for the increase of apparent molecular weight observed between the unmodifed CCN3 protein and the deglycosylated CCN3 species (Joliot et al. 1992). We had previously established that secretion of CCN3 occurs rapidly following its intracellular production and that increased levels of intracellular CCN3 did not alter its secretion (Chevalier et al. 1998; Bleau et al. 2007) A recent study indicated that the TSP1 domain of CCN3 plays a critical role for both its secretion and intracellular function(s) (Kim et al. 2018).
The « all in one concept » part 5: the spatio-temporal regulation of CCN proteins expression
Considering the wide of physiological functions attributed to CCN proteins, it is no surprise that their expression may be very tightly regulated.
The first example of a spatio-temporal type of regulation governing the expression of a CCN protein within both a specific time frame and a specific cellular compartment, was reported in the case of CCN3 during chicken development (see below and Joliot et al. 1992).
From what our preliminary unpublished observations indicated, this situation does not seem to be restricted to CCN3 and it is predictible that gathering the information published regarding the specific times and places of production for other CCN proteins would certainly lead to meaningful comparisons and provide another informative picture of the team.
As a direct consequence of the spatio-temporal expression of CCN proteins, the combination between ligands and CCN targets will be dependent upon the bioavailability of both.
Among the data that paved the way to a better understanding of the regulatory processes involved in the control of CCN protein localisation in vivo, the studies aimed at deciphering the stability and turn over of CCN proteins highlighted the importance of their post-translational processing by the proteolytic machinery.
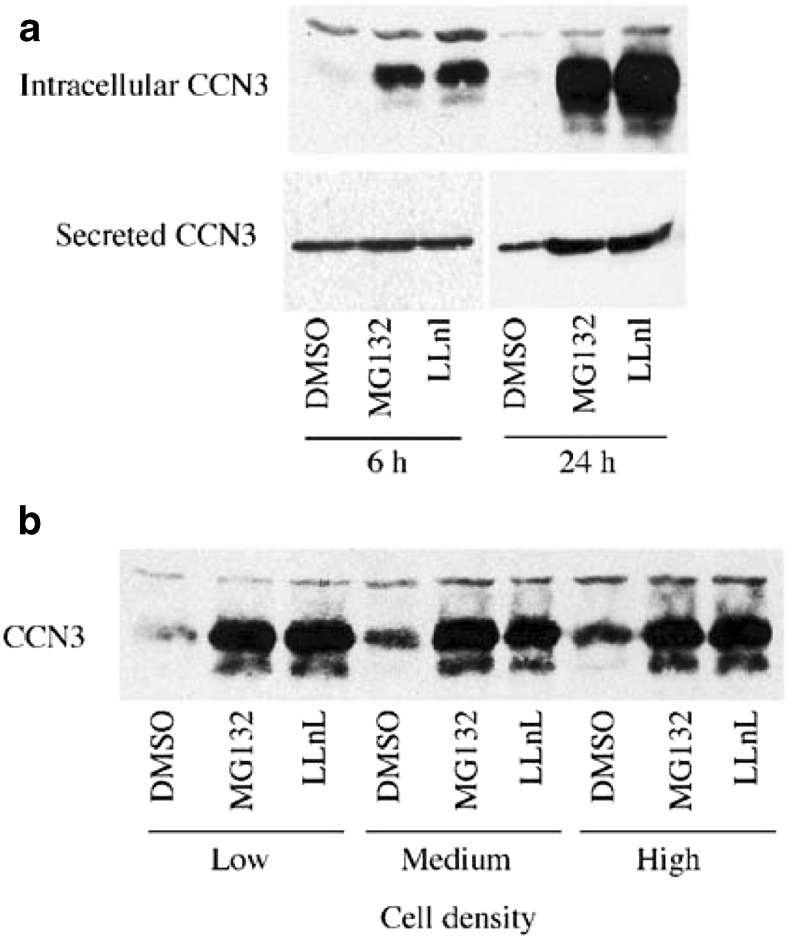
The quantities of CCN3 secreted into culture media is regulated by proteolysis involving the proteasome and « the amount of CCN3 expressed per cell was dependent upon cell density, the higher production being observed at high cell density » (Fig. 6) (Bleau et al. 2007).
Fig. 6.
Post-translational processing of CCN3. The amounts of intracellular and secreted CCN3 proteins are controlled by the proteasome
Although the details of prostranslational modifications of other CCN proteins is still unknown, one can easily predict that they likely constitute a key element dictating their fate and activities, since the intracellular production and sublocalization of CCN3 were also found to vary througout the cell cycle (Bleau et al. 2007).
A dynamic model for a CCN protein-based signaling coordination network
The various aspects of the « all in one concept » presented above:
the functional specificity and structural conservation of each family member
their sequential and cross-regulated expression
their opposite biological effects
the physical interactions of the team members with partners from within and outside the family
the post-translational processing and multiple localisations of the family members and
the spatio-temporal regulation of their local production;
all provide important clues for fine tuning of a model in which the CCN proteins, acting as team, are part of a coordination system permitting an integrated regulation of signaling.
The model on which I would like to elaborate stems from a previous prediction (Perbal 2001) that the local bioavailability of the CCN proteins and their partners is the key to the great variety of biological functions displayed by the family members.11 This model relies on the assumption that the CCN protein modules combine with regulatory elements that function as transmitters of incoming and outgoing signals from and to large families of factors involved in the regulation of key biological pathways controling normal and pathological conditions including cell growth and differentation, cellular homeostasis, production of growth factors, angiogenesis, inflammation, tumorigenesis, intra- and inter- cellular signaling, adaptation to microenvironmental modifications, etc.
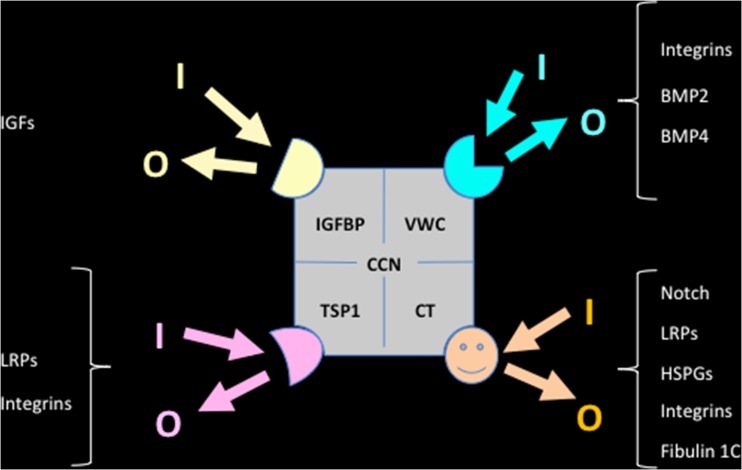
In this model, the four constitutive modules of CCN proteins bind different types of factors that allow specific connections to large families of cell biology regulators (Fig. 7). Furthermore, each particular module of a defined CCN protein can bind a series of different partners that will allow various types of connections with the pathways that cross talk to the CCN protein via this module .
Fig. 7.
Schematic representation of a few interactions occuring between CCN protein modules and regulators of cell signaling. The resulting incoming (I) and outgoing (O) signaling cross-talk involve pathways that include FGF - VEGF - BMPs - LRPs - IGFs - Notch - Integrins - HSPGs – Fibronectin - Aggrecan – Vibronectin – Fubulin 1C – AKT – MAPKs - ROS - NF-kB - RANK - p53 and Ca2+
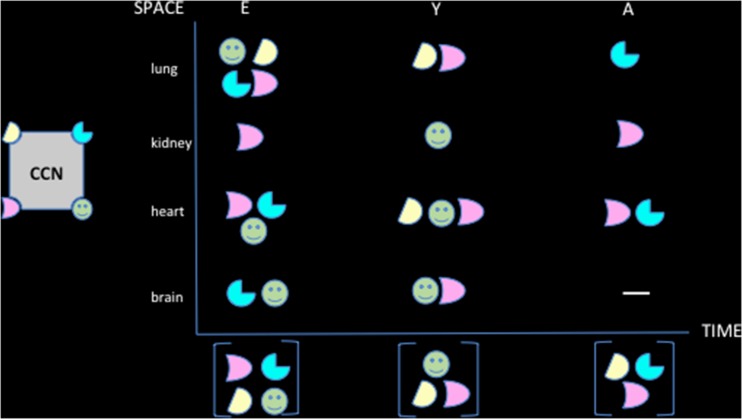
The bioavailability of its partners defines a range of possible interactions within the spatial environment in which each CCN protein is produced as a result of a temporal regulation of expression. The various combinations that occur as a result of combinatorial events that depend upon spatio-temporal production and bioavailability of the CCN protein partners are key to the wide variety of specific functions that are played by the team members (Fig. 8).
Fig. 8.
The combinatorial events depend upon the bioavailability of the partners
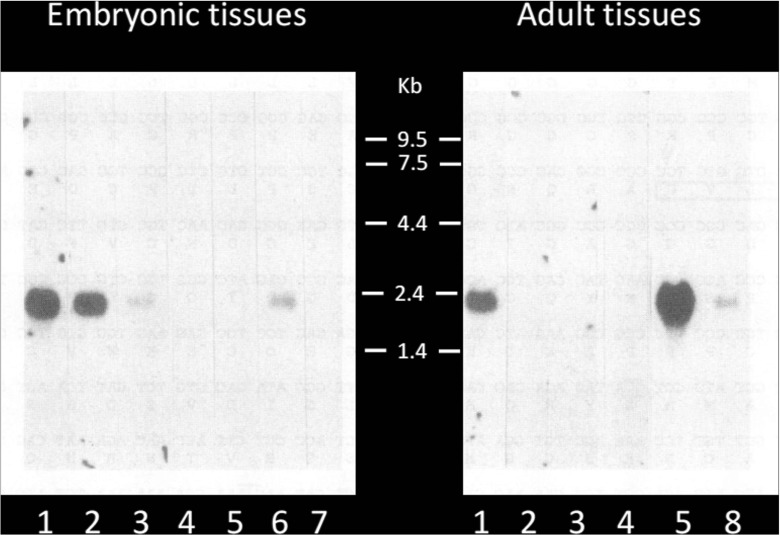
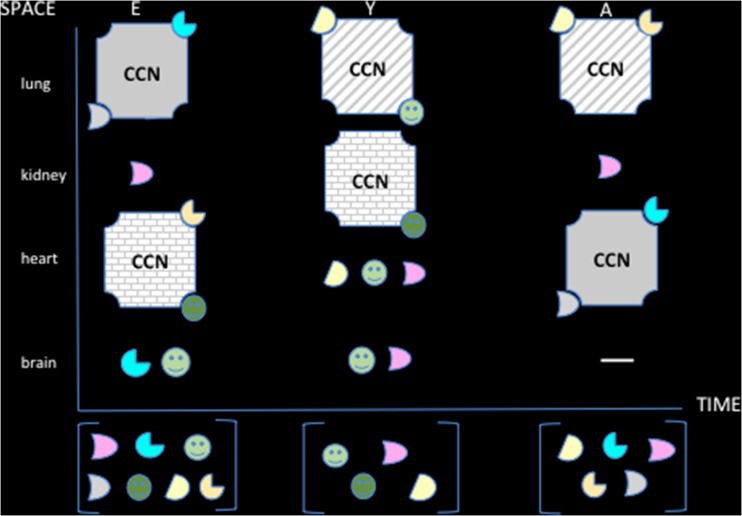
Another level of complexity and flexibility is generated by the temporal and spatial regulation of the CCN proteins as reported for CCN3 (Fig. 9). For example, potential partners will be expressed in a tissue that do not express the CCN with which it can interact (Fig. 10).
Fig. 9.
Spatio-temporal regulation of ccn3 RNA expression during chicken developement. The samples analyzed were purified form 1 : brain ; 2 : heart ; 3 muscle ; 4 liver ; 5 : lung ; 6 : intestine ; 7 : yolk sac ; 8 : spleen (Joliot et al. 1992)
Fig. 10.
The combinatorial events depend upon space, time, variety of ligands and proteins
The considerable variety of combinations offered by this model constitutes the basis for a scafolding system in which the CCN proteins act as connectors to permit the coordinated level of regulation that is mandatory to achieve a balanced series of differentiation, growth and developmental steps.
The Centralized Communication Network (CCN) concept could now be extended to a Centralized Coordination Network concept that would account for the multifunctional biological properties of the CCN proteins.
Acknowledgements
I am deeply grateful to Dr. Herman Yeger for his critical review of the manuscript and to Annick for her help and support.
Footnotes
Several groups, including mine, have in the past used recombinant proteins and individual domains produced in bacterial or baculoviral expression systems which proved to show biological activities exhibited by the native proteins (for CCN3 see Perbal et al. 1999 and Lazar et al. 2007). Unfortunately these proteins do not show the whole panel of post transcriptional modifications that have been suspected for a long time to be essential for native eucaryotic proteins to interact with their physiological partners.
On many occasions, since the early 1990s, efforts to attract colleagues specialized in physicochemical protein analysis to work on CCN proteins, proved unsuccessful and the feeling developed that proteins showing such a complex array of biological properties were not good candidates for this type of studies.
The authors quote CCN3 was amplified from cDNA (a kind gift from Dr. Paul Kemp). Hopefully this clone was not the one used in previous studies from Kemp’s group (Ellis et al., 2000) who claimed that their « data indicate that « NOV is not an effective inhibitor of proliferation, at least in vitro ». In our hands the human and chicken recombinant CCN3 proteins that we purified, and that were also used by others, showed both anti-proliferative and anti-tumor activities.
Perbal B, Lau L, Lyons K, Kubota S, Yeger H, Fisher G.(2016) Report on the 8th international workshop on the CCN family of genes - Nice November 3–8, 2015. J Cell Commun Signal. 10:77–86.
« CCN proteins might act as multipotent matchmakers, allowing the scaffolding of a complex network of regulatory proteins responsible for the regulation of several fundamental functions during normal life, development, and probably death. The identification of other proteins potentially interacting with NOV has established that each of the four constitutive domains of the native protein is required alone or in combination with others to promote the interactions » (Perbal 2001)
I wish to take this opportunity to point out an erroneous affirmation that was published regarding the biological properties of CCN3 and that conveyed wrong concepts. Although our results had clearly established that CCN3 inhibited cell growth on plastic, Ellis et al. (2000) quoted nine years later in « the data suggest that it [CCN3] is an inhibitor of cell proliferation, first demonstrated for chick embryo fibroblasts (CEFs) grown in soft agar » This affirmation is erroneous: i) CEF do not grow in agar unless they are transformed, and ii) our experimental conditions used primary CEF infected by retroviral expression vectors expressing CCN3 either in the sense or in the antisense orientations. When cells were plated, only those which do not expressed CCN3 could grow. Those which expressed CCN3 did not grow. The statement of Ellis et al. would imply that CCN3 avoids CEF-infected cells cloning without anchorage.
« The discovery of nov had three immediate implications: (1) The structural relation that had been reported between cyr61 and ctgf was extended to another gene, thereby leading to the concept of a new family of genes (a family is usually defined as a group of at least three subjects sharing common characteristics). (2) nov provided the first example of aberrant expression of a CCN gene being associated with tumour development, therefore indicating that the biological properties of this new class of growth regulators extended beyond the normal response to growth stimulation. (3) The growth inhibitory effect of nov on chicken embryonic fibroblasts (CEFs) also provided the first evidence that the CCN family of genes contains negative regulators and suggested that nov might counterbalance the stimulatory effects of the immediate early CCN proteins. » (Perbal 2001)
« The rCop-1 gene was found to be expressed only when primary REFs and MEFs began to age or became senescent during passage in culture » The authors also proposed « Since the rCOP-1 protein lacks the C-terminal module of the canonical CCN family members, one can expect it to act as a « dominant negative » effector in the homo and heterotropic multimerization of CCN proteins, either with other members of the CCN family or with proteins which do not belong to the CCN family but physically interact with CCN family members » (Zhang et al. 1998 )
In their manuscript, the authors claim that « rCOP-1 represents a new class of CCN proteins that have functions opposing those of previously indentified members »…. The authors did not seem to fully appreciate that the anti proliferative activity of NOV was sufficient enough to distinguish it from other CCN proteins (see Perbal 2013)
Surprisingly, 11 years after we had experimentally demonstrated that CCN3 was an antiproliferative factor in several different cell types, the authors of this manuscript falsely reported that « Although it is hypothesized that CCN3 may be a negative regulator of proliferation, there is no direct evidence to date supporting this function. » Furthermore, they also claimed that « recombinant CCN3 has been unable thus far to inhibit proliferation of cells in culture, including VSMCs » and added that their observations suggested « the intriguing possibility that CCN5 may represent a naturally occurring antagonist to the 4-domain members of the CCN family »!
« A clue to understanding the discrepancies observed for the expression of the CCN genes in cancer cells might come from the multimodular structure of the CCN proteins and their potential ability to interact sequentially or simultaneously with different partners, the bioavailability of which might be a key element in the manifestation of the positive or negative effects attributed to the CCN proteins »… « CCN proteins might act as multipotent matchmakers, allowing the scaffolding of a complex network of regulatory proteins responsible for the regulation of several fundamental functions during normal life, development, and probably death. The identification of other proteins potentially interacting with NOV has established that each of the four constitutive domains of the native protein is required alone or in combination with others to promote the interactions. ». B. Perbal (2001)
This review is based on the presentation that I delivered as an introduction to the 9th International Workshop on the CCN family of genes held in Saint-Malo France (Nov 2-7th, 2017)
References
- Bleau AM, Planque N, Lazar N, Zambelli D, Ori A, Quan T, Fisher G, Scotlandi K, Perbal B. Antiproliferative activity of CCN3: involvement of the C-terminal module and post-translational regulation. J Cell Biochem. 2007;101(6):1475–1491. doi: 10.1002/jcb.21262. [DOI] [PubMed] [Google Scholar]
- Bork P. Shuffled domains in extracellular proteins. FEBS Lett. 1991;286:47–54. doi: 10.1016/0014-5793(91)80937-X. [DOI] [PubMed] [Google Scholar]
- Bork P. The modular architecture of a new family of growth regulators related to connective tissue growth factor. FEBS Lett. 1993;327:125–130. doi: 10.1016/0014-5793(93)80155-N. [DOI] [PubMed] [Google Scholar]
- Chevalier G, Yeger H, Martinerie C, Laurent M, Alami J, Schofield PN, Perbal B. NovH: differential expression in developing kidney and Wilm's tumors. Am J Pathol. 1998;152:1563–1575. [PMC free article] [PubMed] [Google Scholar]
- Ellis PD, Chen Q, Barker PJ, Metcalfe JC, Kemp PR. Nov gene encodes adhesion factor for vascular smooth muscle cells and is dynamically regulated in response to vascular injury. Arterioscler Thromb Vasc Biol. 2000;20:1912–1919. doi: 10.1161/01.ATV.20.8.1912. [DOI] [PubMed] [Google Scholar]
- Holbourn KP, Malfois M, Acharya KR. First structural glimpse of CCN3 and CCN5 multifunctional signaling regulators elucidated by small angle x-ray scattering. J Biol Chem. 2011;286:22243–22249. doi: 10.1074/jbc.M111.225755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot V, Martinerie C, Dambrine G, Plassiart G, Brisac M, Crochet J, Perbal B. Proviral rearrangements and overexpression of a new cellular gene (nov) in myeloblastosis-associated virus type 1- induced nephroblastomas. Mol Cell Biol. 1992;12:10–21. doi: 10.1128/MCB.12.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaki H, Kubota S, Suzuki A, Lazar N, Yamada T, Matsumura T, Ohgawara T, Maeda T, Perbal B, Lyons KM, Takigawa M. Cooperative regulation of chondrocyte differentiation by CCN2 and CCN3 shown by a comprehensive analysis of the CCN family proteins in cartilage. J Bone Miner Res. 2008;23:1751–1764. doi: 10.1359/jbmr.080615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Yang H, Min JK, Park YJ, Jeong SH, Jang SW, Shim S. CCN3 secretion is regulated by palmitoylation via ZDHHC22. Biochem Biophys Res Commun. 2018;495:2573–2578. doi: 10.1016/j.bbrc.2017.12.128. [DOI] [PubMed] [Google Scholar]
- Kyurkchiev S, Yeger H, Bleau AM, Perbal B. Potential cellular conformations of the CCN3(NOV) protein. Cell Commun Signal. 2004;2(1):9. doi: 10.1186/1478-811X-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake A, Bialik A, Walsh K, Castellot J., Jr CCN5 is a growth arrest-specific gene. Am J Pathol. 2003;162:219–231. doi: 10.1016/S0002-9440(10)63813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau LF. Cell surface receptors for CCN proteins. J Cell Commun Signal. 2016;10:121–127. doi: 10.1007/s12079-016-0324-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar N, Manara C, Navarro S, Bleau AM, Llombart-Bosch A, Scotlandi K, Planque N, Perbal B (2007) Domain-specific CCN3 antibodies as unique tools for structural and functional studies. J Cell Commun Signal 1:91–102 [DOI] [PMC free article] [PubMed]
- Leask A. Yin and Yang: CCN3 inhibits the pro-fibrotic effects of CCN2. J Cell Commun Signal. 2009;3:161–162. doi: 10.1007/s12079-009-0056-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Liu XJ, Crowe PD, Kelner GS, Fan J, Barry G, Manu F, Ling N, De Souza EB, Maki RA. Nephroblastoma overexpressed gene (NOV) codes for a growth factor that induces protein tyrosine phosphorylation. Gene. 1999;238:471–478. doi: 10.1016/S0378-1119(99)00364-9. [DOI] [PubMed] [Google Scholar]
- Murphy-Ullrich JE, Sage EH. Revisiting the matricellular concept. Matrix Biol. 2014;37:1–14. doi: 10.1016/j.matbio.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennica D, Swanson TA, Welsh JW, Roy M, Lawrence D, Lee J, Brush J, Taneyhill L, Deuel B, Lew M, Watanabe C, Cohen R, Melhem M, Finley G, Quirke P, Goddard A, Hillan K, Gurney A, Botstein D, Levine A. WISP genes are members of the connective tissue growth factor family that are up-regulated in Wnt-1- transformed cells and aberrantly expressed in human colon tumors. Proc Natl Acad Sci U S A. 1998;95:14717–14722. doi: 10.1073/pnas.95.25.14717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal B. Contribution of MAV-1-induced nephroblastoma to the study of genes involved in human Wilms’ tumor development. Crit Rev Oncog. 1994;5:589–613. [PubMed] [Google Scholar]
- Perbal B. Nuclear localization of NOV protein: a potential role for nov in the regulation of gene expression. Mol Pathol. 1999;52:84–91. doi: 10.1136/mp.52.2.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal B. NOV (nephroblastoma overexpressed) and the CCN family of genes: structural and functional issues. Mol Pathol. 2001;54:57–79. doi: 10.1136/mp.54.2.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal B. CCN proteins: multifunctional signalling regulators. Lancet. 2004;363(9402):62–64. doi: 10.1016/S0140-6736(03)15172-0. [DOI] [PubMed] [Google Scholar]
- Perbal B. New insight into CCN3 interactions–nuclear CCN3: fact or fantasy? Cell Commun Signal. 2006;4:6. doi: 10.1186/1478-811X-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal B. CCN proteins a centralized communication network. J Cell Commun Signal. 2013;7:169–177. doi: 10.1007/s12079-013-0193-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal B, Lau L, Lyons K, Kubota S, Yeger H, Fisher G (2016) Report on the 8th international workshop on the CCN family of genes - Nice November 3-8, 2015. J Cell Commun Signal 10:77–86 [DOI] [PMC free article] [PubMed]
- Perbal B, Martinerie C, Sainson R, Werner M, He B, Roizman B (1999) The C-terminal domain of the regulatory protein NOVH is sufficient to promote interaction with fibulin 1C: a clue for a role of NOVH in celladhesion signaling Proc Natl Acad Sci U S A 96:869–874 [DOI] [PMC free article] [PubMed]
- Poss ZC, Ebmeier CC, Taatjes DJ. The mediator complex and transcription regulation. Crit Rev Biochem Mol Biol. 2013;48:575–608. doi: 10.3109/10409238.2013.840259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roeyen CR, Eitner F, Scholl T, Boor P, Kunter U, Planque N, Gröne HJ, Bleau AM, Perbal B, Ostendorf T, Floege J. CCN3 is a novel endogenous PDGF-regulated inhibitor of glomerular cell proliferation. Kidney Int. 2008;73:86–94. doi: 10.1038/sj.ki.5002584. [DOI] [PubMed] [Google Scholar]
- Xu ER, Blythe EE, Fischer G, Hyvönen M. Structural analyses of von Willebrand factor C domains of collagen 2A and CCN3 reveal an alternative mode of binding to bone morphogenetic protein-2. J Biol Chem. 2017;292:12516–12527. doi: 10.1074/jbc.M117.788992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Averboukh L, Zhu W, Zhang H, Jo H, Dempsey PJ, Coffey RJ, Pardee AB, Liang P. Identification of rCop-1, a new member of the CCN protein family, as a negative regulator for cell transformation. Mol Cell Biol. 1998;18:6131–6141. doi: 10.1128/MCB.18.10.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]