Abstract
Objective
To develop and validate a radiomics prediction model for individualized prediction of perineural invasion (PNI) in colorectal cancer (CRC).
Methods
After computed tomography (CT) radiomics features extraction, a radiomics signature was constructed in derivation cohort (346 CRC patients). A prediction model was developed to integrate the radiomics signature and clinical candidate predictors [age, sex, tumor location, and carcinoembryonic antigen (CEA) level]. Apparent prediction performance was assessed. After internal validation, independent temporal validation (separate from the cohort used to build the model) was then conducted in 217 CRC patients. The final model was converted to an easy-to-use nomogram.
Results
The developed radiomics nomogram that integrated the radiomics signature and CEA level showed good calibration and discrimination performance [Harrell’s concordance index (c-index): 0.817; 95% confidence interval (95% CI): 0.811–0.823]. Application of the nomogram in validation cohort gave a comparable calibration and discrimination (c-index: 0.803; 95% CI: 0.794–0.812).
Conclusions
Integrating the radiomics signature and CEA level into a radiomics prediction model enables easy and effective risk assessment of PNI in CRC. This stratification of patients according to their PNI status may provide a basis for individualized auxiliary treatment.
Keywords: Colorectal cancer, perineural invasion, prediction model, radiomics, nomogram
Introduction
Colorectal cancer (CRC) is the third most common cancer globally (1). Characterized by neoplastic invasion of nervous structures and spread along nerve sheaths, perineural invasion (PNI) has been recognized as an ominous pathologic feature with prevalence around 20%–30% at the time of resection in CRC patients (2,3). The presence of PNI at the time of resection has been reported as an independent prognostic factor indicating a more aggressive tumor phenotype with higher recurrent rate and reduced survival (3,4). The identification of PNI status has been suggested to assist in selecting patients who could potentially benefit from adjuvant therapy (5,6).
As a crucial part of preoperative workup for CRC patients in clinical practice, computed tomography (CT) facilitates noninvasive assessment compared with other modalities such as preoperative biopsy and serum test (7,8). However, conventional CT interpretation fails in the detection of PNI. Other than the commonly used qualitative imaging descriptors, recent advance in “radiomics” has enabled the extraction of quantitative descriptors from routinely acquired CT (9-11). In the last few years, several prognostic radiomics signatures have been proposed to quantify and monitor tumor phenotypic characteristics, with various prediction performances reported in the field of oncology (9). In CRC, several studies have been conducted on CT-based texture analysis (12-17). Preliminary evidence has indicated that texture analysis on CT images is potentially predictive of survival for CRC patients (15,16). In addition, CT texture analysis has been reported to be useful for prediction of therapeutic response after cytotoxic chemotherapy in CRC patients with liver metastasis (12,17). It is of interest that whether there is an association between the prognostic pathologic feature of PNI and radiomics features, which to the best of our knowledge has not been studied. Therefore, the aim of this study was to develop and validate a radiomics prediction model for individualized prediction of PNI in CRC.
Materials and methods
Patients
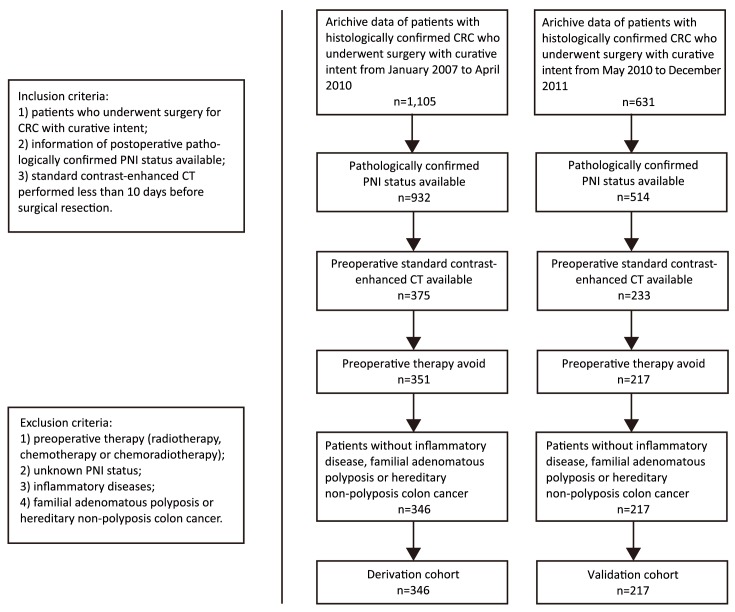
This retrospective study was approved by the institutional review board of Guangdong General Hospital, Guangdong Academy of Medical Sciences, and written informed consent was waived. The records of consecutive patients with colorectal adenocarcinomas who underwent surgical resection of their tumors with curative intent between January 2007 and April 2010 were reviewed to form the derivation cohort of this study, and 346 cases were enrolled. The records of consecutive patients between May 2010 and December 2011 were evaluated to form an independent temporal validation cohort, and 217 cases were enrolled. The patient recruitment process is presented in Figure 1.
1.
Patient recruitment process.
Baseline data pertaining to demographics, pathology, carcinoembryonic antigen (CEA) value and staging were reviewed and recorded. Staging was performed according to the 7th edition of the American Joint Committee on Cancer (AJCC) TNM staging system (18). Histopathologic analysis for the determination of PNI status is listed in Appendix A1.
CT imaging acquisition and retrieval procedure
Acquisition parameters and retrieval procedure of abdominal CT are listed in Appendix A2.
Radiomics feature extraction
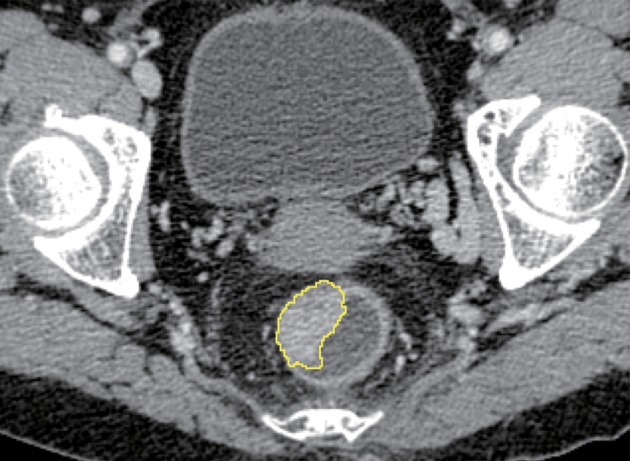
Portal venous phase contrast-enhanced CT data were loaded into in-house radiomics features extraction software with algorithms implemented in Matlab 2010a (Mathworks, Natick, USA). A region of interest (ROI) was delineated around the outline of the tumor for the largest cross-sectional area (Appendix Figure A1).
A total of 150 gray-level histogram and gray-level co-occurrence matrix (GLCM) radiomics features were extracted from the CT image, with the extraction algorithm listed in Appendix Table A1.
Inter- and intra-observer radiomics feature extraction reproducibility
Eighty randomly chosen cases were initially analyzed to assess inter-observer and intra-observer reproducibility of ROI-based radiomics feature extraction. Two radiologists with 10 (Reader 1) and 12 years (Reader 2) of experience in abdominal CT interpretation performed the ROI-based texture feature extraction procedure, in a blind fashion. Reader 1 repeated the procedure after one week, and subsequently performed the workflow for the remaining cases. Inter- and intra-class correlation coefficients (ICCs) were used to measure the intra- and inter-observer agreement of the radiomics features extraction. An ICC greater than 0.75 indicates good agreement. The similar workflow has been described in several published radiomics studies (13,19).
The inter-observer ICCs calculated on the basis of Reader 1’s first-extracted features and Reader 2’s ranged from 0.751 to 0.903. The intra-observer ICC calculated based on Reader 1’s second round of feature extraction ranged from 0.794 to 0.911. Therefore, all outcomes were based on the features extracted by Reader 1.
Statistical analysis
Patient characteristics
The Mann-Whitney U test was used to assess difference in patients’ age, whereas χ2 tests were used to assess the differences in other patient characteristics (sex, CEA level, and tumor location) between the derivation and validation cohort, and within the PNI-positive group and PNI-negative group. Differences in PNI positivity between patients with different characteristics (sex, CEA level, tumor location, and stage I–II vs. III–IV) were also assessed using χ2 tests.
Radiomics signature building and discrimination performance assessment
Radiomics signature building was performed in the derivation cohort. Using the least absolute shrinkage and selection operator (lasso) method (20), the most useful prognostic features were selected and a radiomics signature was built through a linear combination of the selected features weighted by their coefficients, with a radiomics score (Rad-score) calculated for each patient.
Because of the non-normal distribution of the data, the potential association of the radiomics signature with the presence of PNI was assessed in the derivation and validation cohort using the Mann-Whitney U test.
To quantify the discrimination performance of the radiomics signature, the Harrell’s concordance index (c-index) was measured in both the derivation and independent validation cohort, with a bootstrapping technique used for internal validation. The c-index can be interpreted as the probability that a subject with a particular outcome is given a higher probability of that outcome by the predictor or model than a randomly chosen subject without the outcome (20). A value of 0.5 implies that the predictor or model has no discriminatory ability, and a value of 1.0 implies perfect discrimination (20).
Radiomics prediction model development
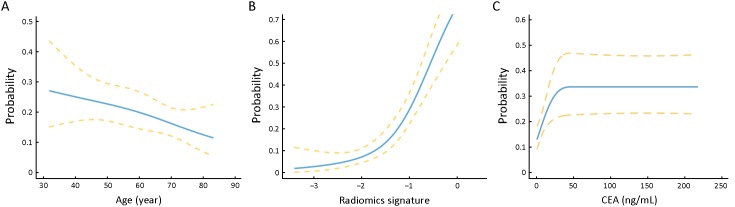
Multivariable logistic regression analysis was used to develop a prediction model in the derivation cohort. Candidate predictors included the developed radiomics signature, age, sex, tumor location, and CEA level. For age and the radiomics signature, a linear relationship with the PNI status was found to be a good approximation (Appendix A3), so these two variables were coded as continuous variables. As a nonlinear relationship was found for the CEA values with PNI status (Appendix A3), thus it was handled as a categorical variable with a threshold set at 5 ng/mL based on clinical consensus. Sex was considered as a binary variable, whereas tumor location was considered as a multi-categorical variable. Backward stepwise selection was applied, using the likelihood ratio test with the stopping rule being Akaike’s information criterion (significance level α was set to 0.157) (21). To facilitate easy calculation of individualized risk in clinical practice, the final model was converted to a radiomics nomogram.
Apparent prediction performance of the prediction model
In the derivation cohort, model performance was assessed in terms of calibration and discrimination. A calibration curve was plotted to compare the agreement between observed outcomes (Y-axis) and the predictions of the model (X-axis), where the perfect calibration should lie on or around a 45° line. The Hosmer-Lemeshow test was conducted, where a significant test statistic (P<0.05) implies a poor calibration (22). c-index was calculated to quantify the discrimination performance of the radiomics prediction model.
Validation of the prediction model
Model performance was validated in terms of calibration and discrimination, with calibration curve, Hosmer-Lemeshow test and c-index derived.
Regarding internal validity, the radiomics prediction model was subjected to a 1000-resampled bootstrapping technique.
For the independent validation cohort, the logistic regression formula of the model derived from the derivation cohort was applied to the results for all patients, with a total points calculated for each patient. Logistic regression in this cohort was then performed using the total points as a factor.
Stratification analysis
Previous studies have advocated that node-negative, PNI-positive stage II patients should be managed similarly to stage III patients and should receive currently available adjuvant therapy (3). Hence the need for taking PNI status into account when stratifying patients for adjuvant therapy has been underscored especially for stage II CRC patients (4,23,24). Therefore, in this study, stratification analysis of the nomogram prediction performance was done specifically in stage II CRC patients within the validation cohort.
R software (Version 3.0.1; R Foundation for Statistical Computing, Vienna, Austria) was used for the statistical analysis, with packages in use listed in Appendix A4. P<0.05 was considered statistically significant.
Results
Patient characteristics
The derivation cohort consisted of 346 cases (222 males and 124 females; mean age, 61.76±13.92 years), with 217 cases identified as the validation cohort (141 males and 76 females; mean age, 61.91±12.52 years). Other than the tumor location (P=0.030), no difference was found in clinical characteristics between the derivation and validation cohort within the PNI-positive group or PNI-negative group (P=0.065–0.872; Table 1). The PNI positivity was 18.8% and 15.7% in derivation and validation cohorts, respectively. In both the derivation cohort or validation cohort, there was significant difference in the PNI positivity between patients with abnormal CEA level and normal CEA level (P=0.001 for the derivation cohort, P=0.029 for validation cohort), as well as between stage I–II patients and stage III–IV patients (P<0.001, P=0.04); whereas there was no significant difference in the PNI positivity between patients with different sex or tumor location (P=0.071–0.722).
1.
Patient characteristics
| Characteristics | PNI-positive [n (%)] | P | PNI-negative [n (%)] | P | ||
| Derivation cohort | Validation cohort | Derivation cohort | Validation cohort | |||
| IQR, interquartile range; CEA, carcinoembryonic antigen; PNI, perineural invasion; P-value was derived from the comparison of patient characteristics between derivation cohort and validation cohort, within PNI-positive subgroup and PNI-negative subgroup respectively. | ||||||
| Age
[median (IQR)] (year) |
62 (50.0, 70.0) | 61 (51.5, 67.8) | 0.865 | 63 (54.0, 73.0) | 64 (53.0, 72.0) | 0.872 |
| Sex | 0.515 | 0.577 | ||||
| Male | 48 (73.8) | 23 (67.6) | 174 (61.9) | 118 (64.5) | ||
| Female | 17 (26.2) | 11 (32.4) | 107 (38.1) | 65 (35.5) | ||
| Tumor location | 0.555 | 0.030 | ||||
| Cecum-ascending colon | 20 (30.8) | 7 (20.6) | 49 (17.4) | 52 (28.4) | ||
| Transverse-descending colon | 5 (7.7) | 4 (11.8) | 20 (7.1) | 12 (6.6) | ||
| Sigmoid colon | 10 (15.4) | 8 (23.5) | 80 (28.5) | 38 (20.8) | ||
| Rectum | 30 (46.2) | 15 (44.1) | 132 (47.0) | 81 (44.3) | ||
| CEA level | 0.542 | 0.065 | ||||
| Normal | 29 (44.6) | 13 (38.2) | 188 (66.9) | 107 (58.5) | ||
| Abnormal | 36 (55.4) | 21 (61.8) | 93 (33.1) | 76 (41.5) | ||
| TNM staging | ||||||
| I | 1 (1.5) | 1 (2.9) | 0.307 | 57 (20.3) | 28 (15.3) | 0.186 |
| II | 9 (13.8) | 9 (26.5) | 103 (36.7) | 74 (40.4) | ||
| III | 43 (66.2) | 21 (61.8) | 93 (33.1) | 70 (38.3) | ||
| IV | 12 (18.5) | 3 (8.8) | 28 (10.0) | 11 (6.0) | ||
Radiomics signature building and discrimination performance assessment
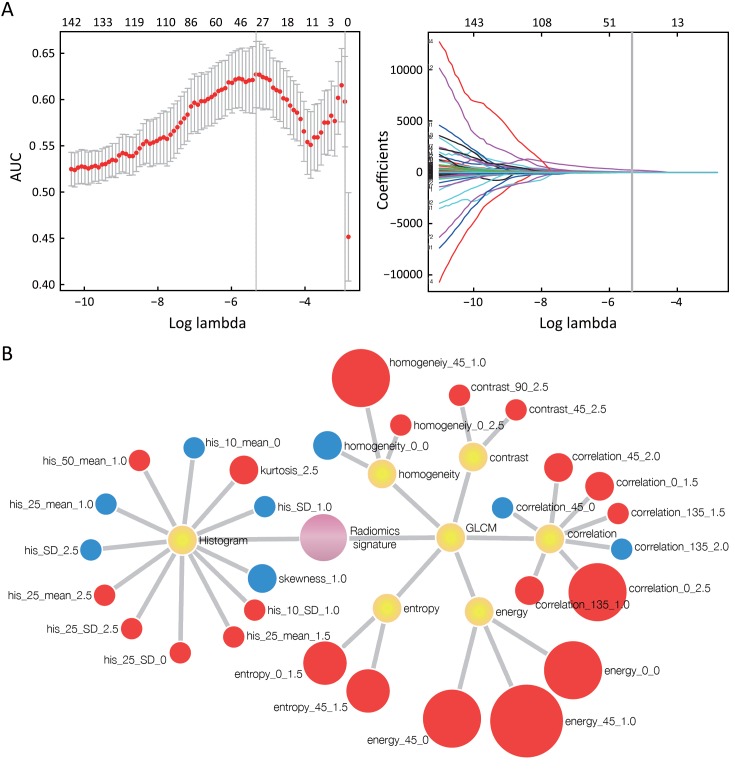
A total of 29 radiomics features with non-zero coefficients were selected (Figure 2A). A radiomics signature was built based on these features and corresponding coefficients. A Rad-score was calculated for each patient based on the radiomics signature (Appendix A5). Individual contribution of the 29 features to the radiomics signature building is described in Figure 2B.
2.
Radiomics features selection. (A) Tuning parameter (λ) selection in the least absolute shrinkage and selection operator method (lasso) model used ten-fold cross-validation. Lasso coefficient profiles of the 150 texture features. A coefficient profile plot was produced against the log-lambda sequence. The vertical line was drawn at the value selected using ten-fold cross-validation, where the optimal λ resulted in 29 non-zero coefficients; (B) Individual contribution of the 29 features to the radiomics signature building. Nodes represent the 29 features of the radiomics signature. Size of each node represents the degree of contribution of individual feature to the signature building, according to its coefficient during the feature selection. Nodes marked in blue represent features with negative contribution to perineural invasion (PNI) (+); whereas those marked in red representing features with positive contribution to PNI (+). Nodes marked in yellow represent the feature subgroups. Among all the subgroups, subgroup of gray-level co-occurrence matrix (GLCM) features (Energy) achieves the highest contribution to the radiomics signature building.
There was a significant association between the radiomics signature and the presence of PNI (P<0.0001 for both the derivation and validation cohort). PNI-positive patients generally had higher Rad-scores compared with the PNI-negative patients {median [interquartile range (IQR)]: derivation cohort, –0.95 (–1.37, –0.30)vs. –1.80 (–2.36, –1.35); validation cohort, –1.11 (–1.48, –0.80) vs. –1.73 (–2.30, –1.23)}.
Regarding discrimination performance, the mean c-index of the radiomics signature was 0.809 [95% confidence interval (95% CI): 0.802–0.816] in the derivation cohort, 0.810 through the bootstrapping internal validation, and 0.777 (95% CI: 0.767–0.787) in the validation cohort.
Radiomics prediction model development
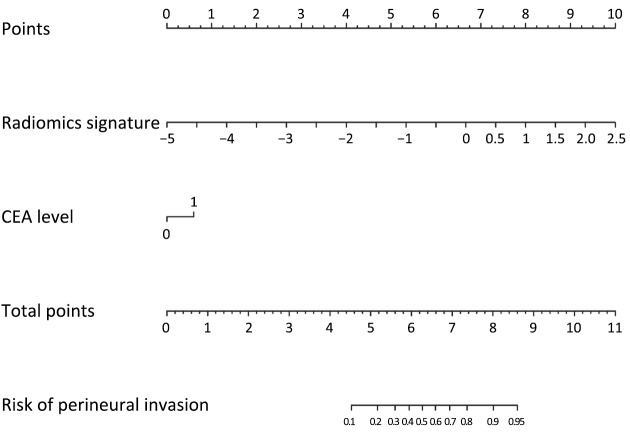
The final model consists of two predictors (radiomics signature and CEA level; Table 2). The converted nomogram is shown in Figure 3.
2.
Prediction model developed based on the derivation cohort
| Intercept and predictors | β | OR | 95% CI | P |
| CEA, carcinoembryonic antigen; OR, odds ratio; 95% CI, 95% confidence interval; The predicted probability of perineural invasion (PNI) can be calculated using the following formula: P (PNI) = 1/{1 + exp [– (0.504 + 1.683 × radiomics signature + 0.755 × CEA level)]}. Predictor value is one when PNI is positive and zero when negative. | ||||
| Intercept | 0.504 | 0.171 | ||
| Radiomics signature | 1.683 | 5.382 | 3.310, 8.747 | <0.0001 |
| CEA level | 0.755 | 2.128 | 1.136, 3.980 | 0.018 |
3.
The radiomics nomogram. The nomogram integrates two items: the radiomics signature and carcinoembryonic antigen (CEA) level. Locate the patient’s radiomics score (Rad-score) that calculated based on the radiomics signature on the “Radiomics signature” axis, followed by drawing a line straight upward to the “Points” axis to determine how many points toward the probability of perineural invasion (PNI) the patient receives for his Rad-score. After repeating the process for the CEA level, sum the points achieved for each of the two predictors. Finally we located the final sum on the “Total Points” axis and then drew a line straight down to derive the patient’s probability of PNI.
Apparent prediction performance of prediction model
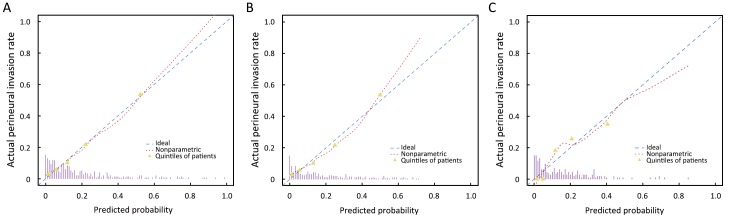
In derivation cohort, the calibration curve depicted good agreement between prediction and observation (Figure 4A; P=0.276). Good discrimination was also shown, with a c-index of 0.817 (95% CI: 0.811–0.823).
4.
Calibration curves of the radiomics model prediction. (A) Calibration curve in the derivation cohort (Hosmer-Lemeshow test; P=0.276); (B) Calibration curve in the internal validation cohort (Hosmer-Lemeshow test; P=0.132); (C) Calibration curve in the independent validation cohort (Hosmer-Lemeshow test; P=0.132). Calibration curves depict the calibration of the radiomics prediction model in terms of the agreement between the predicted probability of perineural invasion (PNI) and observed rate of PNI. The Y-axis represents the actual observed PNI rate whereas the X-axis represents the model predicted PNI probability. The diagonal blue dash line represents a perfect prediction by an ideal model. The dashed smooth curve reflects the relation between observed rate of PNI and predicted probability of PNI using the radiomics prediction model. Triangles indicate the incidence of PNI in quintiles of patients with similar predicted probabilities. Spikes at the bottom represent distribution of predicted probabilities of PNI.
Validation of prediction model
Through the bootstrapping internal validation, good calibration was observed (Figure 4B; P=0.132). Good discrimination performance was internally validated, with a c-index of 0.812.
Regarding the independent validation cohort, good calibration (Figure 4C; P=0.132) and good discrimination performance were validated (c-index: 0.803; 95% CI: 0.794–0.812).
Stratification analysis
Good discrimination ability of the prediction model was demonstrated for stage II CRC patients in the validation cohort (c-index: 0.779; 95% CI: 0.753–0.805).
Discussion
In this study, a radiomics model integrating a constructed radiomics signature and CEA level was developed and validated for individualized prediction of PNI in CRC patients, the conversion of which into an easy-to-use nomogram may assist in effective risk stratification of patients who could potentially benefit from adjuvant therapy in clinical practice.
The developed radiomics signature in this study showed significant association with PNI, which successfully stratified patients according to their risk of PNI (P<0.0001). Compared to the prognostic studies conducted with individual CT-based texture features in CRC (15,16), the construction of a radiomics signature that combines a panel of features as a prognostic imaging marker may be a superior choice, which has nowadays been regarded as a more powerful method to affect clinical management, suggested by previous “-omics” studies (25-27). Prediction performance shown by the constructed radiomics signature (c-index: 0.777; 95% CI: 0.767–0.787) supports the concept that radiomics quantitative image-driven biomarkers have potential to extract in vivo biologic information and further expand the horizons of imaging interpretations toward greater precision to improve diagnostic and prognostic accuracy in decision support (10).
As the radiomics approach extracts high-dimension feature data with quite a large number of candidate predictors, the process of selecting relevant predictor variables to build a radiomics signature is a challenging endeavor (10). The problem of overfitting may occur and magnify as the ratio of the number of observations to the number of predictors decreases, especially with small sample sizes. Therefore, as an alternative to the competing model selection procedures (such as stepwise regression that magnifies problems associated with overfitting), the lasso method was used to select radiomics features in this study, which has the advantage of shrinking some estimates to exactly zero and regularizing regression coefficients to account for potential overfitting (20). Additionally, as overfitting is a central problem in “-omics” based prediction modelling, the validity of the developed model should be considered for new patients (28). Therefore, in our study, a bootstrapping re-sampling procedure and temporary independent validation were used to address the issue of model uncertainty. As shown by the results, the prediction model showed comparable performance after internal and independent validation.
Among the potential clinical candidate predictors, the CEA level was found to be an independent predictor of PNI, which is consistent with the published studies that have demonstrated an association between elevated serum CEA level and positive PNI (29). Although the CEA value was initially recorded as a continuous variable, given the priori clinical consensus that the threshold value for the level of CEA used in clinical practice was ≤5 ng/mL and >5 ng/mL, the linear relationship between the recorded continuous values of CEA and the PNI status was checked. As the restricted cubic spline showed that there was a nonlinear relationship between the CEA values and the PNI status, CEA was handled as a categorical variable with 5 ng/mL determined as the threshold based on clinical consensus. Although CEA was associated with the PNI status of CRC patients, its use as a single predictor alone fails to reliably estimate the absolute probability or risk of PNI, which is crucial to risk-stratify patients for different treatment strategies as required in precision medicine. In the past decade, emerging biomarkers with potential clinical value identified by advances in high-throughput biotechnologies have triggered calls for model building that combines multiple biomarkers ( 27,30). As demonstrated in our study, the developed prediction model which integrated the radiomics signature and independent clinical predictors showed satisfactory performance in assisting the estimation of risks of PNI, with good discrimination (c-index: 0.803; 95% CI: 0.794–0.812) and calibration (model-predicted risk of PNI agrees closely with the observed risk; P=0.132).
The PNI-positive rate varies by AJCC stage in this study (positive rate: stage I–II vs. stage III–IV = 3.96% vs. 28.47%), which is consistent with the literature showing that PNI is associated with more advanced disease in CRC based on its correlation with higher stages. In stage IV CRC patients with resectable tumors who underwent curative operations, PNI was reported to be a prognostic factor of both survival and recurrence (31). Compared with stage IV CRC patients, prognosis is more variable for stage II and stage III. As reported in a previous study, the overall survival of patients deteriorated in the order of PNI-negative at stage II, PNI-positive at stage II/PNI-negative at stage III, and PNI-positive at stage III. It was thereby proposed that patients at stages II and III could be further divided according to their PNI status to provide a basis for individualized auxiliary treatment (32). It is worth noting that for stage II CRC patients, postoperative chemotherapy did not improve the 5-year disease-free survival (DFS) of those with PNI-negative tumors, whereas the PNI-positive chemotherapy group showed higher 5-year DFS. Overall, the particular interest in determining the potential role of PNI in therapy stratification is in node-negative stage II CRC patients who currently have the option to consider receiving adjuvant chemotherapy. Therefore, in this study, we performed a stratification analysis which showed that the radiomics prediction model performed well for stage II CRC patients (c-index: 0.779; 95% CI: 0.753–0.805).
In addition to the ability to predict the outcome of interest accurately, an optimal clinical prediction model should have the potential for ease of use in the clinical setting. Therefore, the prediction model proposed in this study was presented as an easy-to-use nomogram, which permits easy calculation of PNI risk tailored to each individual patient during clinical encounters, and assists clinical decision-making. Both the internal validation and prospective temporal validation of the model performance support the robustness of the developed radiomics nomogram in this study.
One major limitation within this work is that the semantic features that are commonly used in the radiology lexicon to describe ROI on CT images were not considered as potential model candidates. Although tumor size has shown prognostic value in several solid tumors (33,34), primary tumor of CRC is relatively flexible and grows with no preferred direction, which may result in various tumors shapes and potential interreader variability or even inaccuracy in the tumor diameter or size measuring. Regarding the candidate predictor selection involving radiological parameters, inter-observer variability caused by subjective interpretation is a specific concern, for which candidate predictors should be reliably measured and well defined by any observer (35). For N stage, size criteria are not sufficiently accurate as the majority of rectal cancer lymph node metastases occur in nodes less than 6 mm in size (36). Therefore, simple semantic features were not considered as candidate predictors in this study to prevent potential bias in the estimation of the association between predictors and PNI status.
A further limitation is that our study only focuses on one of the many site-specific independent prognostic factors, which include tumor deposits, circumferential resection margin, microsatellite instability, and KRAS gene analysis. These prognostic factors are well supported in the literature and generally used in patient management (18). Therefore, it is of interest to determine whether the radiomics approach could provide important information regarding these prognostic factors. In order to enable evidence-based clinical decision support, an optimal way to integrate the radiomics approach in conjunction with these prognostic factors to correlate with clinical outcomes data warrants further investigation.
Third, whole-tumor analysis was not performed in this study. Instead, radiomic features were extracted from the largest cross-sectional area of tumor. Theoretically, whole-tumor analysis may represent more diverse components of tumor heterogeneity by avoiding sampling errors that may result from single slice selection. Although there have been previous results supporting this idea, a study aiming to determine whether texture features of untreated hepatic metastatic CRC relate to pathologic features and clinical outcomes reported that a single slice-2D texture analysis was adequate (14). Moreover, a more recent study by Ahn et al. has reported that while skewness observed on 2D analysis and SD observed on 3D analysis were both independent predictors for the prediction of therapeutic response after cytotoxic chemotherapy in patients with liver metastasis from CRC, the odd ratio of skewness seen on 2D analysis was higher than that of the SD observed on 3D analysis (12). Therefore, considering that the 3D whole-tumor analysis is more computationally complex and time-consuming, we only delineated the largest cross-sectional area for features extraction. Further investigations are warranted to determine whether the features extracted from a single largest cross-sectional slice may adequately describe the characteristics of CRC.
Conclusions
Integrating the radiomics signature and CEA level into a radiomics prediction model enables easy and effective risk assessment of PNI in CRC. This stratification of patients according to their PNI status may provide a basis for individualized auxiliary treatment.
Acknowledgements
All the authors have contributed significantly and have approved the manuscript. This work was supported by the National Key Research and Development Program of China (No. 2017YFC1309100) and the National Natural Scientific Foundation of China (No. 81771912, 81701782 and 81601469).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
A1.
Region of interest (ROI) delineation. An ROI was delineated around the outline of the tumor for the largest cross-sectional area.
A2.
Examination of the linear assumption of relationships between the continuous variables and perineural invasion (PNI) status. Linear relationships with the PNI status were found to be a good approximation for the age of patients (A) and the radiomics signature (B). While a nonlinear relationship was observed between the carcinoembryonic antigen (CEA) values and the PNI status (C).
A1.
Radiomics feature extraction algorithm
| Type | Description | Calculation formula | Feature |
| SD, standard deviation; GLCM, gray-level co-occurrence matrix; X(i), the intensity of gray level i; N, the sum of pixels in the image; σ, the Laplacian of Gaussian filter value applied, which could be 0, 1.0, 1.5, 2.0 and 2.5; M, the number of pixels in the histogram on the percentage of (1-β); x, y, the spatial coordinates of the pixel; P(i, j), the co-occurrence matrix by the δ=1 and θ(0°, 45°, 90°, 135°); Ng, the number of discrete intensity levels in the image; μ, the mean of P(i,j); μx (i), the mean of Px (i); μy(j), the mean of Py (j); σx (i), the standard deviation of Px (i); σy (j), the standard deviation of Py (j); α, the considered direction, which could be 0°, 45°, 90°, and 135°; β, the top percentage of the histogram curve, which could be 50%, 25%, and 10%. | |||
| Gray-level histogram features | |||
| Skewness | Degree of asymmetry around the mean value in the gray level histogram |
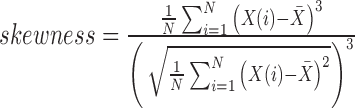
|
skewness_σ |
| Kurtosis | Sharpness of the gray level histogram |
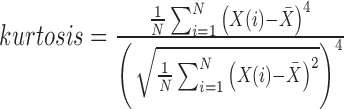
|
kurtosis_σ |
| Mean | Average value of the gray level histogram |
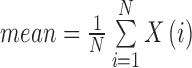
|
mean_σ |
| SD | Stability of the gray level histogram |
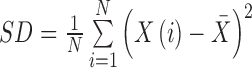
|
SD_σ |
| Percentile mean; percentile SD | Calculated from the top 50%, 25%, and 10% of the histogram curve |
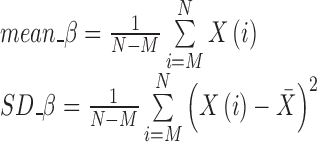
|
mean_β_σ SD_β_σ |
| GLCM features | |||
| Contrast | Measures local intensity variation, reflects the uniformity of image grayscale distribution and the degree of thickness in texture |
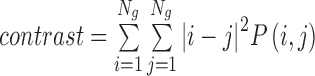
|
contrast_α_σ |
| Correlation | The gray level linear dependence between the pixels at the specified positions relative to each other |
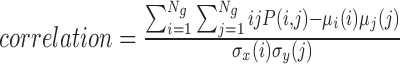
|
correlation_α_σ |
| Entropy | The inhomogeneity of an image |
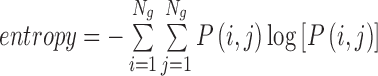
|
entropy_α_σ |
| Energy | The sum of squares of entries in the GLCM |
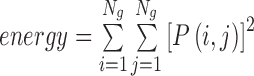
|
energy_α_σ |
| Homogeneity | The inverse of the Contrast weight |
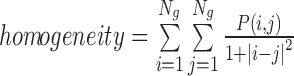
|
homogeneity_α_σ |
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. <DOI: 10.1002/ijc.29210> <PMID: 25220842>] [DOI] [PubMed] [Google Scholar]
- 2.Liebig C, Ayala G, Wilks JA, et al. Perineural invasion in cancer: a review of the literature. Cancer. 2009;115:3379–91. doi: 10.1002/cncr.24396. [Liebig C, Ayala G, Wilks JA, et al. Perineural invasion in cancer: a review of the literature. Cancer 2009;115:3379-91. <DOI: 10.1002/cncr.24396> <PMID: 19484787>] [DOI] [PubMed] [Google Scholar]
- 3.Liebig C, Ayala G, Wilks J, et al. Perineural invasion is an independent predictor of outcome in colorectal cancer. J Clin Oncol. 2009;27:5131–7. doi: 10.1200/JCO.2009.22.4949. [Liebig C, Ayala G, Wilks J, et al. Perineural invasion is an independent predictor of outcome in colorectal cancer. J Clin Oncol 2009;27:5131-7. <DOI: 10.1200/JCO.2009.22.4949> <PMID: 19738119>] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huh JW, Kim HR, Kim YJ. Prognostic value of perineural invasion in patients with stage II colorectal cancer. Ann Surg Oncol. 2010;17:2066–72. doi: 10.1245/s10434-010-0982-7. [Huh JW, Kim HR, Kim YJ. Prognostic value of perineural invasion in patients with stage II colorectal cancer. Ann Surg Oncol 2010;17:2066-72. <DOI: 10.1245/s10434-010-0982-7> <PMID: 20182809>] [DOI] [PubMed] [Google Scholar]
- 5.Suzuki T, Suwa K, Ogawa M, et al. Adjuvant chemotherapy for the perineural invasion of colorectal cancer. J Surg Res. 2015;199:84–9. doi: 10.1016/j.jss.2015.03.101. [Suzuki T, Suwa K, Ogawa M, et al. Adjuvant chemotherapy for the perineural invasion of colorectal cancer. J Surg Res 2015;199:84-9. <DOI: 10.1016/j.jss.2015.03.101> <PMID: 25935467>] [DOI] [PubMed] [Google Scholar]
- 6.Yang Y, Huang X, Sun J, et al. Prognostic value of perineural invasion in colorectal cancer: a meta-analysis. J Gastrointest Surg. 2015;19:1113–22. doi: 10.1007/s11605-015-2761-z. [Yang Y, Huang X, Sun J, et al. Prognostic value of perineural invasion in colorectal cancer: a meta-analysis. J Gastrointest Surg 2015;19:1113-22. <DOI: 10.1007/s11605-015-2761-z> <PMID: 25663635>] [DOI] [PubMed] [Google Scholar]
- 7.Barton JB, Langdale LA, Cummins JS, et al. The utility of routine preoperative computed tomography scanning in the management of veterans with colon cancer. Am J Surg. 2002;183:499–503. doi: 10.1016/S0002-9610(02)00841-3. [Barton JB, Langdale LA, Cummins JS, et al. The utility of routine preoperative computed tomography scanning in the management of veterans with colon cancer. Am J Surg 2002;183:499-503. <DOI: 10.1016/S0002-9610(02)00841-3> <PMID: 12034380>] [DOI] [PubMed] [Google Scholar]
- 8.Leufkens AM, van den Bosch MA, van Leeuwen MS, et al. Diagnostic accuracy of computed tomography for colon cancer staging: a systematic review. Scand J Gastroenterol. 2011;46:887–94. doi: 10.3109/00365521.2011.574732. [Leufkens AM, van den Bosch MA, van Leeuwen MS, et al. Diagnostic accuracy of computed tomography for colon cancer staging: a systematic review. Scand J Gastroenterol 2011;46:887-94. <DOI: 10.3109/00365521.2011.574732> <PMID: 21504379>] [DOI] [PubMed] [Google Scholar]
- 9.Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. <DOI: 10.1038/ncomms5006> <PMID: 24892406>] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images are more than pictures, they are data. Radiology. 2016;278:563–77. doi: 10.1148/radiol.2015151169. [Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images are more than pictures, they are data. Radiology 2016;278:563-77. <DOI: 10.1148/radiol.2015151169> <PMID: 26579733>] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuo MD, Jamshidi N. Behind the numbers: Decoding molecular phenotypes with radiogenomics -- guiding principles and technical considerations. Radiology. 2014;270:320–5. doi: 10.1148/radiol.13132195. [Kuo MD, Jamshidi N. Behind the numbers: Decoding molecular phenotypes with radiogenomics -- guiding principles and technical considerations. Radiology 2014;270:320-5. <DOI: 10.1148/radiol.13132195> <PMID: 24471381>] [DOI] [PubMed] [Google Scholar]
- 12.Ahn SJ, Kim JH, Park SJ, et al. Prediction of the therapeutic response after FOLFOX and FOLFIRI treatment for patients with liver metastasis from colorectal cancer using computerized CT texture analysis. Eur J Radiol. 2016;85:1867–74. doi: 10.1016/j.ejrad.2016.08.014. [Ahn SJ, Kim JH, Park SJ, et al. Prediction of the therapeutic response after FOLFOX and FOLFIRI treatment for patients with liver metastasis from colorectal cancer using computerized CT texture analysis. Eur J Radiol 2016;85:1867-74. <DOI: 10.1016/j.ejrad.2016.08.014> <PMID: 27666629>] [DOI] [PubMed] [Google Scholar]
- 13.Huang YQ, Liang CH, He L, et al. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol. 2016;34:2157–64. doi: 10.1200/JCO.2015.65.9128. [Huang YQ, Liang CH, He L, et al. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol 2016;34:2157-64. <DOI: 10.1200/JCO.2015.65.9128> <PMID: 27138577>] [DOI] [PubMed] [Google Scholar]
- 14.Lubner MG, Stabo N, Lubner SJ, et al. CT textural analysis of hepatic metastatic colorectal cancer: pre-treatment tumor heterogeneity correlates with pathology and clinical outcomes. Abdom Imaging. 2015;40:2331–7. doi: 10.1007/s00261-015-0438-4. [Lubner MG, Stabo N, Lubner SJ, et al. CT textural analysis of hepatic metastatic colorectal cancer: pre-treatment tumor heterogeneity correlates with pathology and clinical outcomes. Abdom Imaging 2015;40:2331-7. <DOI: 10.1007/s00261-015-0438-4> <PMID: 25968046>] [DOI] [PubMed] [Google Scholar]
- 15.Miles KA, Ganeshan B, Griffiths MR, et al. Colorectal cancer: texture analysis of portal phase hepatic CT images as a potential marker of survival. Radiology. 2009;250:444–52. doi: 10.1148/radiol.2502071879. [Miles KA, Ganeshan B, Griffiths MR, et al. Colorectal cancer: texture analysis of portal phase hepatic CT images as a potential marker of survival. Radiology 2009;250:444-52. <DOI: 10.1148/radiol.2502071879> <PMID: 19164695>] [DOI] [PubMed] [Google Scholar]
- 16.Ng F, Ganeshan B, Kozarski R, et al. Assessment of primary colorectal cancer heterogeneity by using whole-tumor texture analysis: contrast-enhanced CT texture as a biomarker of 5-year survival. Radiology. 2013;266:177–84. doi: 10.1148/radiol.12120254. [Ng F, Ganeshan B, Kozarski R, et al. Assessment of primary colorectal cancer heterogeneity by using whole-tumor texture analysis: contrast-enhanced CT texture as a biomarker of 5-year survival. Radiology 2013;266:177-84. <DOI: 10.1148/radiol.12120254> <PMID: 23151829>] [DOI] [PubMed] [Google Scholar]
- 17.Rao SX, Lambregts DM, Schnerr RS, et al. CT texture analysis in colorectal liver metastases: A better way than size and volume measurements to assess response to chemotherapy? United European Gastroenterol J. 2016;4:257–63. doi: 10.1177/2050640615601603. [Rao SX, Lambregts DM, Schnerr RS, et al. CT texture analysis in colorectal liver metastases: A better way than size and volume measurements to assess response to chemotherapy? United European Gastroenterol J 2016;4:257-63. <DOI: 10.1177/2050640615601603> <PMID: 27087955>] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edge SB, Byrd DR, Compton CC, et al. AJCC cancer staging manual. 7th ed. New York: Springer, 2010.
- 19.Liu Z, Zhang XY, Shi YJ, et al. Radiomics analysis for evaluation of pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Clin Cancer Res. 2017;23:7253–62. doi: 10.1158/1078-0432.CCR-17-1038. [Liu Z, Zhang XY, Shi YJ, et al. Radiomics analysis for evaluation of pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Clin Cancer Res 2017;23:7253-62. <DOI: 10.1158/1078-0432.CCR-17-1038> <PMID: 28939744>] [DOI] [PubMed] [Google Scholar]
- 20.Harrell FE Jr. Regression Modeling Strategies With Applications to Linear Models, Logistic and Ordinal Regression, and Survival Analysis. New York: Springer, 2015.
- 21.Collins GS, Reitsma JB, Altman DG, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ. 2015;350:g7594. doi: 10.1136/bmj.g7594. [Collins GS, Reitsma JB, Altman DG, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ 2015;350:g7594. <DOI: 10.1136/bmj.g7594> <PMID: 25569120>] [DOI] [PubMed] [Google Scholar]
- 22.Kramer AA, Zimmerman JE. Assessing the calibration of mortality benchmarks in critical care: The Hosmer-Lemeshow test revisited. Crit Care Med. 2007;35:2052–6. doi: 10.1097/01.CCM.0000275267.64078.B0. [Kramer AA, Zimmerman JE. Assessing the calibration of mortality benchmarks in critical care: The Hosmer-Lemeshow test revisited. Crit Care Med 2007;35:2052-6. <DOI: 10.1097/01.CCM.0000275267.64078.B0> <PMID: 17568333>] [DOI] [PubMed] [Google Scholar]
- 23.Santos C, López-Doriga A, Navarro M, et al. Clinicopathological risk factors of Stage II colon cancer: results of a prospective study. Colorectal Dis. 2013;15:414–22. doi: 10.1111/codi.12028. [Santos C, López-Doriga A, Navarro M, et al. Clinicopathological risk factors of Stage II colon cancer: results of a prospective study. Colorectal Dis 2013;15:414-22. <DOI: 10.1111/codi.12028> <PMID: 22974322>] [DOI] [PubMed] [Google Scholar]
- 24.Benson AB 3rd, Schrag D, Somerfield MR, et al. American Society of Clinical Oncology recommendations on adjuvant chemotherapy for stage II colon cancer. J Clin Oncol. 2004;22:3408–19. doi: 10.1200/JCO.2004.05.063. [Benson AB 3rd, Schrag D, Somerfield MR, et al. American Society of Clinical Oncology recommendations on adjuvant chemotherapy for stage II colon cancer. J Clin Oncol 2004;22:3408-19. <DOI: 10.1200/JCO.2004.05.063> <PMID: 15199089>] [DOI] [PubMed] [Google Scholar]
- 25.Birkhahn M, Mitra AP, Cote RJ. Molecular markers for bladder cancer: the road to a multimarker approach. Expert Rev Anticancer Ther. 2007;7:1717–27. doi: 10.1586/14737140.7.12.1717. [Birkhahn M, Mitra AP, Cote RJ. Molecular markers for bladder cancer: the road to a multimarker approach. Expert Rev Anticancer Ther 2007;7:1717-27. <DOI: 10.1586/14737140.7.12.1717> <PMID: 18062746>] [DOI] [PubMed] [Google Scholar]
- 26.Riley RD, Hayden JA, Steyerberg EW, et al. Prognosis Research Strategy (PROGRESS) 2: prognostic factor research. PLoS Med. 2013;10:e1001380. doi: 10.1371/journal.pmed.1001380. [Riley RD, Hayden JA, Steyerberg EW, et al. Prognosis Research Strategy (PROGRESS) 2: prognostic factor research. PLoS Med 2013;10:e1001380. <DOI: 10.1371/journal.pmed.1001380> <PMID: 23393429>] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Royston P, Moons KG, Altman DG, et al. Prognosis and prognostic research: Developing a prognostic model. BMJ. 2009;338:b604. doi: 10.1136/bmj.b604. [Royston P, Moons KG, Altman DG, et al. Prognosis and prognostic research: Developing a prognostic model. BMJ 2009;338:b604. <DOI: 10.1136/bmj.b604> <PMID: 19336487>] [DOI] [PubMed] [Google Scholar]
- 28.Steyerberg E. Clinical Prediction Models: A Practical Approach to Development, Validation, and Updating. New York: Springer, 2009.
- 29.Park IJ, Choi GS, Lim KH, et al. Serum carcinoembryonic antigen monitoring after curative resection for colorectal cancer: clinical significance of the preoperative level. Ann Surg Oncol. 2009;16:3087–93. doi: 10.1245/s10434-009-0625-z. [Park IJ, Choi GS, Lim KH, et al. Serum carcinoembryonic antigen monitoring after curative resection for colorectal cancer: clinical significance of the preoperative level. Ann Surg Oncol 2009;16:3087-93. <DOI: 10.1245/s10434-009-0625-z> <PMID: 19629600>] [DOI] [PubMed] [Google Scholar]
- 30.Steyerberg EW, Moons KG, van der Windt DA, et al. Prognosis Research Strategy (PROGRESS) 3: prognostic model research. PLoS Med. 2013;10:e1001381. doi: 10.1371/journal.pmed.1001381. [Steyerberg EW, Moons KG, van der Windt DA, et al. Prognosis Research Strategy (PROGRESS) 3: prognostic model research. PLoS Med 2013;10:e1001381. <DOI: 10.1371/journal.pmed.1001381> <PMID: 23393430>] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yun HR, Lee WY, Lee WS, et al. The prognostic factors of stage IV colorectal cancer and assessment of proper treatment according to the patient’s status. Int J Colorectal Dis. 2007;22:1301–10. doi: 10.1007/s00384-007-0315-x. [Yun HR, Lee WY, Lee WS, et al. The prognostic factors of stage IV colorectal cancer and assessment of proper treatment according to the patient’s status. Int J Colorectal Dis 2007;22:1301-10. <DOI: 10.1007/s00384-007-0315-x> <PMID: 17486358>] [DOI] [PubMed] [Google Scholar]
- 32.Zhou Y, Wang H, Gong H, et al. Clinical significance of perineural invasion in stages II and III colorectal cancer. Pathol Res Pract. 2015;211:839–44. doi: 10.1016/j.prp.2015.09.001. [Zhou Y, Wang H, Gong H, et al. Clinical significance of perineural invasion in stages II and III colorectal cancer. Pathol Res Pract 2015;211:839-44. <DOI: 10.1016/j.prp.2015.09.001> <PMID: 26429129>] [DOI] [PubMed] [Google Scholar]
- 33.Mimae T, Miyata Y, Mimura T, et al. Radiologic findings to predict low-grade malignant tumour among clinical T1bN0 lung adenocarcinomas: lessons from histological subtypes. Jpn J Clin Oncol. 2015;45:767–73. doi: 10.1093/jjco/hyv078. [Mimae T, Miyata Y, Mimura T, et al. Radiologic findings to predict low-grade malignant tumour among clinical T1bN0 lung adenocarcinomas: lessons from histological subtypes. Jpn J Clin Oncol 2015;45:767-73. <DOI: 10.1093/jjco/hyv078> <PMID: 26056324>] [DOI] [PubMed] [Google Scholar]
- 34.Kim DW, Kim HJ, Kim KW, et al. Neuroendocrine neoplasms of the pancreas at dynamic enhanced CT: comparison between grade 3 neuroendocrine carcinoma and grade 1/2 neuroendocrine tumour. Eur Radiol. 2015;25:1375–83. doi: 10.1007/s00330-014-3532-z. [Kim DW, Kim HJ, Kim KW, et al. Neuroendocrine neoplasms of the pancreas at dynamic enhanced CT: comparison between grade 3 neuroendocrine carcinoma and grade 1/2 neuroendocrine tumour. Eur Radiol 2015;25:1375-83. <DOI: 10.1007/s00330-014-3532-z> <PMID: 25465713>] [DOI] [PubMed] [Google Scholar]
- 35.Han K, Song K, Choi BW. How to develop, validate, and compare clinical prediction models involving radiological parameters: study design and statistical methods. Korean J Radiol. 2016;17:339–50. doi: 10.3348/kjr.2016.17.3.339. [Han K, Song K, Choi BW. How to develop, validate, and compare clinical prediction models involving radiological parameters: study design and statistical methods. Korean J Radiol 2016;17:339-50. <DOI: 10.3348/kjr.2016.17.3.339> <PMID: 27134523>] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmoll HJ, Van Cutsem E, Stein A, et al. ESMO Consensus Guidelines for management of patients with colon and rectal cancer. a personalized approach to clinical decision making. Ann Oncol. 2012;23:2479–516. doi: 10.1093/annonc/mds236. [Schmoll HJ, Van Cutsem E, Stein A, et al. ESMO Consensus Guidelines for management of patients with colon and rectal cancer. a personalized approach to clinical decision making. Ann Oncol 2012;23:2479-516. <DOI: 10.1093/annonc/mds236> <PMID: 23012255>] [DOI] [PubMed] [Google Scholar]