Abstract
Background
A percent brain volume change (PBVC) cut-off of −0.4% per year has been proposed to distinguish between pathological and physiological changes in multiple sclerosis (MS). Unfortunately, standardized PBVC measurement is not always feasible on scans acquired outside research studies or academic centers. Percent lateral ventricular volume change (PLVVC) is a strong surrogate measure of PBVC, and may be more feasible for atrophy assessment on real-world scans. However, the PLVVC rate corresponding to the established PBVC cut-off of −0.4% is unknown.
Objective
To establish a pathological PLVVC expansion rate cut-off analogous to −0.4% PBVC.
Methods
We used three complementary approaches. First, the original follow-up-length-weighted receiver operating characteristic (ROC) analysis method establishing whole brain atrophy rates was adapted to a longitudinal ventricular atrophy dataset of 177 relapsing-remitting MS (RRMS) patients and 48 healthy controls. Second, in the same dataset, SIENA PBVCs were used with non-linear regression to directly predict the PLVVC value corresponding to −0.4% PBVC. Third, in an unstandardized, real world dataset of 590 RRMS patients from 33 centers, the cut-off maximizing correspondence to PBVC was found. Finally, correspondences to clinical outcomes were evaluated in both datasets.
Results
ROC analysis suggested a cut-off of 3.09% (AUC = 0.83, p < 0.001). Non-linear regression R2 was 0.71 (p < 0.001) and a − 0.4% PBVC corresponded to a PLVVC of 3.51%. A peak in accuracy in the real-world dataset was found at a 3.51% PLVVC cut-off. Accuracy of a 3.5% cut-off in predicting clinical progression was 0.62 (compared to 0.68 for PBVC).
Conclusions
Ventricular expansion of between 3.09% and 3.51% on T2-FLAIR corresponds to the pathological whole brain atrophy rate of 0.4% for RRMS. A conservative cut-off of 3.5% performs comparably to PBVC for clinical outcomes.
Keywords: Brain atrophy, Ventricular volume, Pathological cutoff, Multiple sclerosis, NEDA
Highlights
-
•
Pathological atrophy in MS can be measured on clinical T2-FLAIR images alone.
-
•
Ventricular enlargement of 3.5% per year separates MS/HC as well as PBVC on T1 images.
-
•
Ventricular cut-offs also correspond to clinical outcome.
-
•
This cut-off can substitute in NEDA-4 when only clinical T2-FLAIR images are available.
1. Introduction
Multiple sclerosis (MS) is the most common debilitating neurological disease in young adults worldwide (Alonso and Hernán, 2008). Although there is currently no cure, there are a number of approved first- and second-line therapies that can potentially slow disability progression, lower relapse rates, and reduce accumulation of new or enlarging T2- and T1-enhancing lesions (Torkildsen et al., 2016). In this context, monitoring each of these disease activity markers is important both in clinical trials (van Munster and Uitdehaag, 2017) and on an individual basis (Giovannoni et al., 2015). In fact, these outcomes have recently been combined into a single, dichotomous measure called ‘no evidence of disease activity’ (NEDA) (Giovannoni et al., 2015; Havrdova et al., 2010).
At the same time, brain atrophy in MS has recently emerged as one of the most important predictors of future disability progression (Popescu et al., 2013; Lukas et al., 2010; Miller et al., 2002; Zivadinov et al., 2016a, Zivadinov et al., 2016b), and one of the strongest correlates of cognitive dysfunction (Summers et al., 2008; Sanfilipo et al., 2006; Tekok-Kilic et al., 2007; Benedict and Zivadinov, 2011). However, brain atrophy also occurs in healthy individuals as a normal function of aging – albeit at a slower rate. It is therefore necessary to differentiate pathological atrophy rates from normal physiological rates. De Stefano et al., 2016 established appropriate cutoffs by using a receiver operating characteristic (ROC) analysis on longitudinal, whole-brain data from 206 MS patients and 35 healthy controls (HC). In that work, −0.4% per year percent brain volume change (PBVC) was found to have 80% specificity and 65% specificity in discriminating between MS and HC, and atrophy rates higher than this were predictive of disability progression. Based on this, a −0.4% PBVC threshold was proposed as a mechanism for incorporation of brain volume loss into a NEDA-4 extension of the original NEDA metric (Kappos et al., 2016).
Unfortunately, standardized PBVC measurement is not always possible on clinical routine scans. T1-weighted anatomical images suitable for analysis with SIENA (Smith et al., 2002) or similar techniques may not be acquired in many cases. Scans may also be subject to more artifacts, or be more likely to lack coverage of the entire brain (which, although still calculable with SIENA, would limit the ability to standardize cut-offs). To address this, measurement of lateral ventricular volume (LVV) and percent LVV change (PLVVC) over time has been proposed as a broadly applicable proxy for whole brain atrophy measurement (Zivadinov et al., 2016a, Zivadinov et al., 2016b; Dwyer et al., 2017). PLVVC has been shown to correlate strongly with PBVC and to relate similarly to disability progression (Zivadinov et al., 2016a, Zivadinov et al., 2016b; Popescu et al., 2013; Dwyer et al., 2017). In the recent, real-world MS-MRIUS study, estimation of brain atrophy using PLVVC on T2-FLAIR was feasible in 94% of patients, compared to <50% for PBVC (and PLVVC) on 2D T1-WI and <20% on 3D T1-WI (Zivadinov et al., 2018).
Despite this potential proxy value of LVV and PLVVC, it remains unknown what PLVVC cut-off best approximates the established PBVC pathological threshold. Therefore, we set out in this study to determine the LVV expansion rate best corresponding to the established PBVC cut-off of −0.4% per year. We did this first by replicating the approach of De Stefano et al., secondly by establishing a direct relationship between PBVC and PLVVC in a regression model, and finally by validating the findings in a multi-center, real world dataset.
2. Methods
2.1. Subjects and datasets
For this investigation, we used data from both the Avonex–Steroid–Azathioprine (ASA) (Havrdova et al., 2009) and Multiple Sclerosis and MRI in the US study (MS-MRIUS) (Zivadinov et al., 2017) studies. The ASA study included 181 early relapsing–remitting multiple sclerosis (RRMS) patients at a single site treated with intramuscular (IM) interferon beta-1a who were followed with serial yearly MRI using the same protocol on the same 1.5 T scanner that did not undergo major hardware upgrades over 10 years. During the same time period, a group of 48 normal controls was recruited prospectively from among healthy hospital volunteers and scanned with the same MRI protocol (Horakova et al., 2008). Therefore, this combined dataset was used to establish PLVVC cut-offs in a well-controlled setting. In contrast, the MS-MRIUS included 590 RRMS patients from 33 sites initiating fingolimod who were scanned according to local MRI protocols without any pre-standardization or prohibition on scanner changes (Zivadinov et al., 2017).This dataset was used to evaluate the newly determined PLVVC cut-off in a real-world setting.
2.2. MRI analysis
For both datasets, whole-brain SIENA PBVCs (Smith et al., 2002) were previously calculated from all available longitudinal scan pairs. In the case of ASA, high-resolution 1 mm cubic 3D T1-weighted images were used as inputs. In the case of MS-MRIUS, high-resolution 3D T1-weighted images were used when available, and 2D T1-weighted images were used otherwise. Images were oriented axially, and lesions were inpainted prior to segmentation to reduce the impact of T1 hypointensities (Gelineau-Morel et al., 2012). All SIENA analyses underwent quality control by a trained operator (N.B.) to ensure proper execution, including raw image review, review of inpainting, manual correction of deskulling if necessary, and evaluation of final segmentation and edge flow maps.
PLVVC measures between timepoints were calculated on T2-FLAIR weighted images using the recently described NeuroSTREAM technique, which operates on clinical-quality 2D- or 3D–FLAIR images and performs segmentation of the lateral ventricles (Dwyer et al., 2017). Briefly, each subject's T2-FLAIR image was pre-processed and then non-linearly aligned to three previously derived multi-subject, multi-scanner atlases. The inverse transforms were then used to bring standard space ventricular masks into the subject space, and to combine them via a locally optimized joint-label fusion scheme. Finally, the fused mask was used to initialize and guide a tuned level set algorithm incorporating supersampling-based partial volume estimation. Final quality control images were reviewed by a trained operator (N.B.).
2.3. Receiver operating curve (ROC) cut-off estimation
Given the resulting PBVC and PLVVC measures for the ASA dataset, we first sought to replicate the work of De Stefano et al., 2016 directly with PBVC, and then to perform a similar analysis using PLVVC rather than PBVC. As in that study, annualized PBVC was determined for each subject as the slope of the regression line fit to all PBVC measurements for that subject, based on the assumption of a linear change over time. Also similarly, case weights were used for subsequent analyses, where the weights were proportional to exact follow-up duration. Because HC had only 2 years of follow-up, analyses were performed on 2 year data to ensure similar timing and conditions. A t-test was employed to confirm differences between groups, and the annualized PBVC rate maximizing sensitivity and in discriminating MS from HC at 80% specificity was determined using a weighted ROC, including determination of area under the curve (AUC). Then, we independently repeated the entire process using PLVVC measures instead of PBVC measures – analogously determining annualized PLVVC based on individual subject regressions, testing group differences, and performing follow-up duration-weighted ROC analysis. Thus, we obtained a lateral ventricular expansion rate cut-off similarly maximizing sensitivity at 80% specificity in discriminating MS from HC, and derived independently from PBVC.
2.4. Regression-based cut-off estimation
As a complementary approach, and because a PBVC cut-off of −0.4% per year has already been proposed and adopted by others (De Stefano et al., 2016; Kappos et al., 2016), we sought to confirm the validity of the PLVVC ROC-based results by direct calculation from PBVC. Here, rather than investigating PLVVC independently of PBVC, we used non-linear regression with PLVVC as the dependent variable and PBVC as the only predictor. A quadratic model was used, and the intercept was a priori fixed at 0 to constrain lack of measured change to be equivalent in both cases. Overall model fit was evaluated and tested for significance via F test. The resulting model coefficients were then used to compute the PLVVC value corresponding to an input PBVC value of −0.4%. Cross-fold validation with 10 folds was used to determine robustness and 95% confidence interval of the calculated PLVVC cut-off.
2.5. Application to an unstandardized, multi-center dataset
Next, to explore the generalizability of analyses from the well-controlled ASA dataset, we also analyzed the MS-MRIUS multi-center, real world imaging study (Zivadinov et al., 2017). In order to again use a complementary approach, we computed the accuracy of all possible PLVVC thresholds against pathological/non-pathological status as determined by PBVC on T1-weighted images. This was done on a subset of the database consisting of those subjects who had both index and post-index MRI exams, and of those exams for which 2D or 3D T1-weighted images and T2-FLAIR images were available and analyzable. From this analysis, we then identified the threshold or thresholds maximizing agreement between PLVVC-derived pathological status and PBVC-derived pathological status.
2.6. Clinical relevance in both datasets
Finally, we also looked at the relationship between the obtained PLVVC cut-off and clinical outcomes in both datasets, and compared it to the −0.4% PBVC cut-off. In the ASA dataset, we evaluated the accuracy, sensitivity, and specificity of the obtained PLVVC cut-off in determining confirmed disability progression (CDP) at 120 months (Zivadinov et al., 2016a, Zivadinov et al., 2016b). CDP was defined as a sustained (confirmed after 48 weeks) increase of 1.5 point or more in EDSS if the starting EDSS was 0, or a sustained (confirmed after 48 weeks) increase of 1 point of more if the starting EDSS was not 0. Significant association between PBVC and PLVVC categorical thresholds and CDP data were assessed via Chi-square tests. In the MS-MRIUS dataset, we evaluated accuracy, sensitivity, and specificity for PBVC and PLVVC with regard to both and disability progression (DP) and to timed 25-ft walking score progression (Hobart et al., 2013). Again, associations were tested for significance via Chi-square tests. DP was defined as an increase of 1.5 or higher in EDSS if the starting EDSS was 0, 1 or higher if the starting score was between 1 and 5, and 0.5 or higher if the starting score was greater than 5. Walking score progression was defined as a 20% increase in mobility score.
2.7. Alternate cut-offs
Although −0.4% PBVC has been incorporated into the NEDA-4 criteria, other thresholds have also been proposed to meet different trade-offs in sensitivity vs. specificity. In particular, (De Stefano et al., 2016) also proposed −0.52% as the cut-off with 95% specificity and −0.46% as the cut-off with 90% specificity. Since the choice of trade-off may be different in different settings (e.g. clinical routine vs. clinical trials), we also estimated the appropriate PLVVC cut-offs and associated sensitivities for 90% and 95% specificity levels.
3. Results
3.1. Receiver operating curve (ROC) cut-off estimation
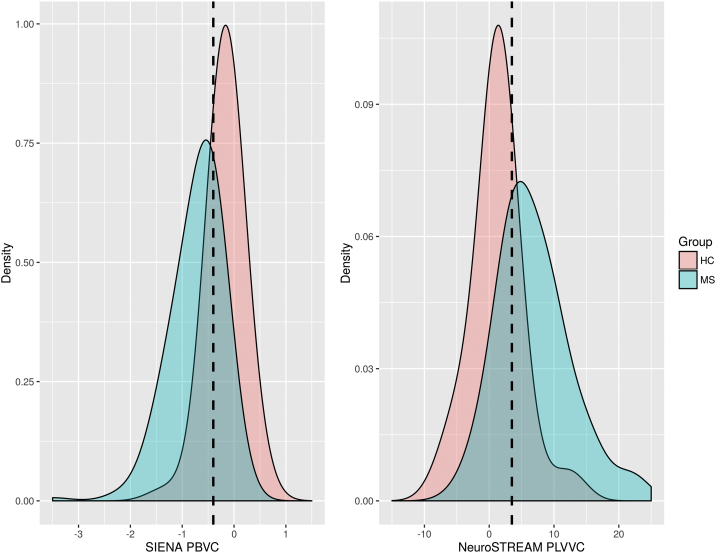
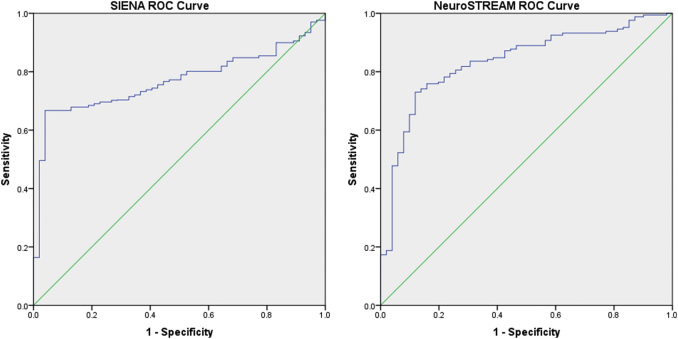
Results from the ROC fitting technique are presented in Table 1. Visualizations of the histogram overlaps between groups for each method are shown in Fig. 1, and the resulting ROC curves are shown in Fig. 2. For annualized PBVC, MS and HC groups differed significantly (p < 0.001), and 80% specificity was achieved at a brain volume loss cut-off of −0.33% per year. For annualized PLVVC, MS and HC again differed significantly (p < 0.001), and 80% specificity was achieved at a ventricular expansion rate cut-off of 3.09% per year. AUC for PLVVC was 0.835 (p < 0.001), compared with 0.769 (p < 0.001) for PBVC.
Table 1.
Replication (first row) and extension to ventricular volume (second row) of the approach described in De Stefano et al. (2016) for establishing pathological cutoffs for brain volume loss. PBVC – percent brain volume change, PLVVC – percent lateral ventricular volume change, MS – multiple sclerosis, HC – healthy control, ROC – receiver operating characteristic, AUC – area under the curve.
| MS mean (SD) | HC mean (SD) | Group diff. p | ROC AUC | ROC p | 80% specificity cut-off | Sensitivity | |
|---|---|---|---|---|---|---|---|
| SIENA PBVC | −0.72 (0.43) | −0.16 (0.27) | <0.001 | 0.769 | <0.001 | −0.35 | 0.69 |
| NeuroSTREAM PLVVC | 8.83 (11.81) | 1.19 (3.66) | <0.001 | 0.835 | <0.001 | 3.09 | 0.76 |
Fig. 1.
Density plots showing the distributions of whole brain change via PBVC (left panel) and lateral ventricular change via PLVVC (right panel) for healthy controls (HC) and multiple sclerosis (MS) patients in the ASA dataset. Dotted lines mark the previously proposed −0.4% PBVC threshold (left) and the newly derived 3.5% PLVVC threshold (right). PBVC – percent brain volume change, PLVVC – percent lateral ventricular volume change, MS – multiple sclerosis, HC – healthy control.
Fig. 2.
Comparison of ROC curves between annualized SIENA PBVC and annualized NeuroSTREAM PLVVC. PBVC – percent brain volume change, PLVVC – percent lateral ventricular volume change, ROC – receiver operating characteristic.
3.2. Regression-based cut-off estimation
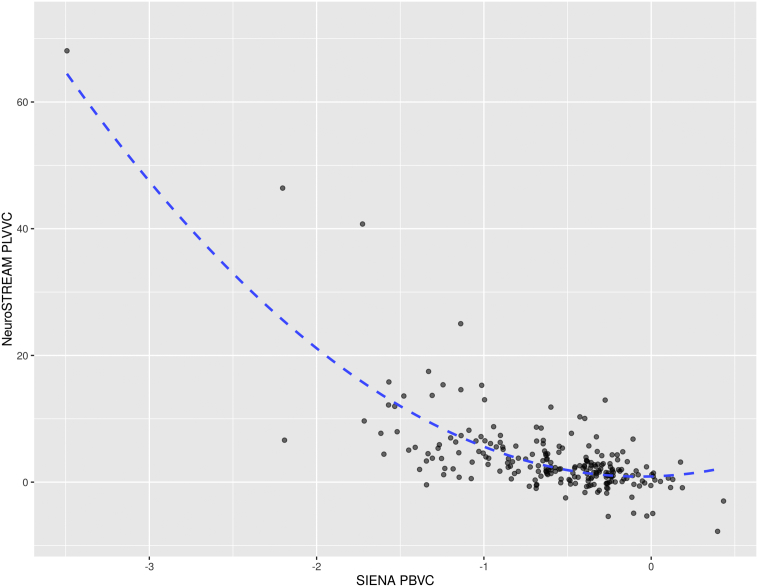
Pearson correlation between PLVVC and PBVC was r = −0.65 (p < 0.001). For the regression-based approach, the final quadratic model was:
PLVVC = − 8.51 × PBVC + 0.53 × PBVC2 + ε
Adjusted R2 was 0.58 (p < 0.001). A scatterplot with the final model fit is shown in Fig. 3. Based on these estimated coefficients, an annualized PBVC of −0.4% was found to correspond to a PLVVC of 3.49% per year. Mean cross-validation value was 3.48%, with a 95% confidence interval between 3.30 and 3.64.
Fig. 3.
Scatterplot showing the relationship between whole brain change via PBVC and lateral ventricular change via PLVVC in the ASA dataset. The quadratic fit line, with R2 = 0.58 (p < 0.001) is overlaid. Applying the fitted coefficients to the −0.4% established PBVC cutoff yields and equivalent PLVVC cut-off of 3.49%. PBVC – percent brain volume change, PLVVC – percent lateral ventricular volume change.
3.3. Application to an unstandardized, multi-center dataset
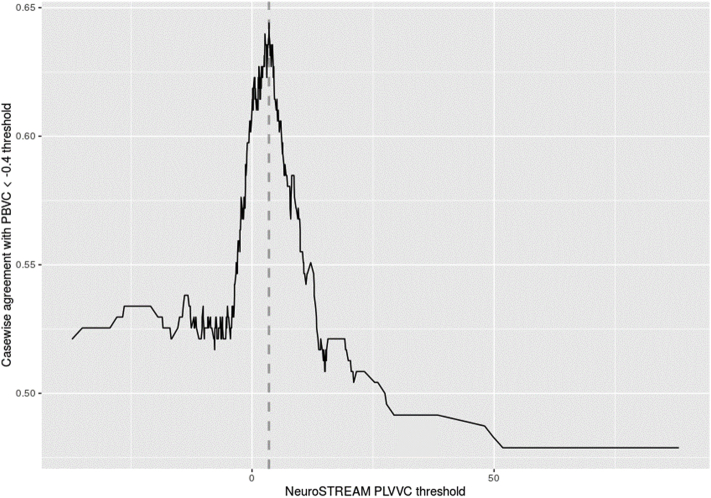
In the real-world MS-MRIUS dataset PBVC was available on 259 2D T1 exam pairs (44% of subjects) and only 110 3D T1 exam pairs (19% of subjects), whereas PLVVC was available on 554 T2-FLAIR pairs (94% of subjects) (Zivadinov et al., 2018). In total, 247 subjects (42%) had both valid PBVC and T2-FLAIR PLVVC and were included in this analysis. The resulting relationship between choice of PLVVC threshold and concordance with PBVC-derived pathological status is shown in Fig. 4, and resulted in accuracy of 0.64 with cut-off of 3.51% per year.
Fig. 4.
Graph of accuracy vs. pathological PBVC (<−0.4%) status as a function of PLVVC threshold in the MS-MRIUS study. The independently determined threshold of 3.5% PLVVC (dashed line) corresponds closely to a peak in accuracy at 3.51% in this dataset. PBVC – percent brain volume change, PLVVC – percent lateral ventricular volume change.
3.4. Clinical relevance in both datasets
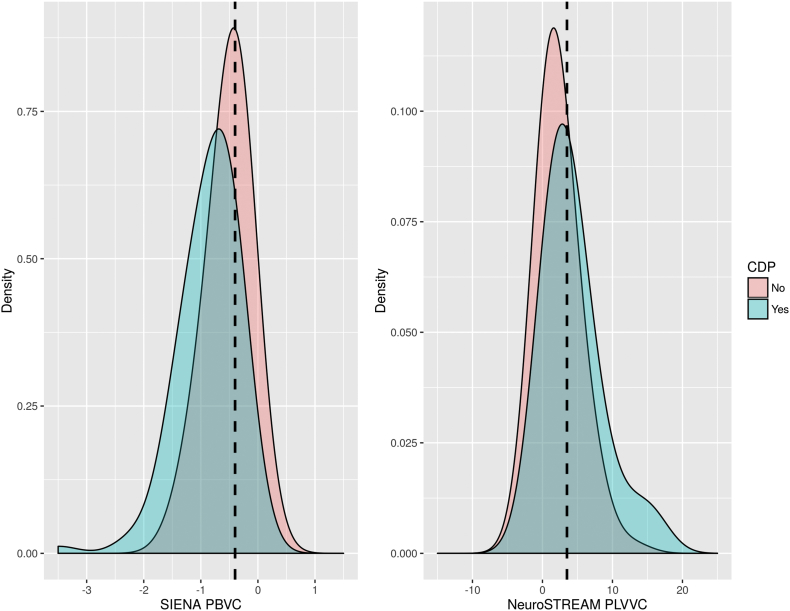
Based on the outcomes of the preceding analyses, we used a threshold of 3.5% per year to distinguish pathological from non-pathological ventricular enlargement. For the ASA dataset, this PLVVC threshold was significantly related to clinical CDP status (p < 0.001). Overall accuracy was 0.62, with a sensitivity of 0.76 and specificity of 0.52. A cut-off of −0.4% PBVC was also significantly related to CDP (p < 0.001). Accuracy for the PBVC threshold was 0.68, with a sensitivity of 0.50 and a specificity of 0.82. The distributions and overlaps are shown in Fig. 5. For the MS-MRIUS dataset, results are presented in Table 2. Accuracy in determining DP was 55.5% for PBVC (sensitivity/specificity = 88.5%/10.2%) and 63.0% for PLVVC (sensitivity/specificity = 88.9%/10.6%). However, neither association was statistically significant. Accuracy in determining walking score progression was 65.1% for PBVC (sensitivity/specificity = 90.3%/4.6%) and 73.8% for PLVVC (sensitivity/specificity = 95.8%/22.6%). Only the association with PLVVC was significant (p = 0.004).
Fig. 5.
Density plots showing the distributions of whole brain change via PBVC (left panel) and lateral ventricular change via PLVVC (right panel) for patients with and without confirmed disability progression (CDP) in the ASA dataset. Dotted lines mark the previously proposed −0.4% PBVC threshold (left) and the newly derived 3.5% PLVVC threshold (right). PBVC – percent brain volume change, PLVVC – percent lateral ventricular volume change, CPD – confirmed disability progression.
Table 2.
Cross-tabulations of EDSS and walking score progression vs. brain volume and ventricular volume cut-off status in the MS-MRIUS dataset. PBVC – percent brain volume change, PLVVC – percent lateral ventricular volume change, EDSS – expanded disability status scale. Results with p < 0.05 shown in bold.
| PBVC |
Accuracy (p⁎) |
PLVVC |
Accuracy (p⁎) |
||||
|---|---|---|---|---|---|---|---|
| <0.4% Annual loss | ≥0.4% Annual loss | <3.5% Annual gain | ≥3.5% Annual gain | ||||
| EDSS | Stable | 131 | 97 | 55.5%/0.742 | 152 | 76 | 63.0%/0.900 |
| Worsened | 17 | 11 | 19 | 9 | |||
| Walking score | Stable | 93 | 41 | 65.1%/0.310 | 69 | 24 | 73.8%/0.004 |
| Worsened | 10 | 2 | 3 | 7 | |||
Calculation of p-values performed via χ 2 test.
3.5. Alternate cut-offs
At a cut-off of −0.35%, SIENA PBVC had a specificity of 90% and a sensitivity of 69%. The analogous 90% specificity cut-off for NeuroSTREAM PLVVC was 4.19%, with a similar sensitivity of 65%. At a cut-off of −0.39%, SIENA PBVC had a specificity of 95% and a sensitivity of 67%. The analogous 95% specificity cut-off for NeuroSTREAM PLVVC was 6.87%, with a sensitivity of 48%.
4. Discussion
This study aimed to address the lack of pathological PLVVC cut-offs for ventricular enlargement, which is a proxy for whole brain volume loss. Using the same methodology that De Stefano et al., 2016 previously applied to PBVC, we found that a PLVVC cut-off of 3.09% separates MS from HC with 80% specificity. Next, by “plugging in” the −0.4% PBVC threshold into a model predicting PLVVC from PBVC, we arrived at a similar cut-off of 3.49%. Finally, in evaluating the threshold(s) providing maximum agreement in an unstandardized, multi-center, real-world dataset, we found a peak at 3.51% PLVVC. The fact that these rates agree so well despite different derivations and different datasets provides a strong measure of confidence that the observed range contains the correct point estimate of the true pathological cut-off.
Based on these findings, it seems reasonable that a threshold of 3.5% (conservatively selecting the higher end of the range) may serve as an appropriate pathological cut-off for use in place of the established −0.4% PBVC in situations where PBVC is unavailable. This 3.5% threshold might also be incorporated into an alternative to NEDA-4 when PBVC is not available. In these cases, a ventricular expansion based NEDA criterion – NEDA-4 V – could be used to capture atrophy status information that would otherwise be lost entirely.
We also showed that use of a 3.5% cut-off for PLVVC behaves similarly to PBVC with regard to clinical outcomes. In the ASA dataset, accuracy of determination of CDP status from pathological atrophy status was comparable via either PBVC or PLVVC, statistically significant in both cases, and in line with previous studies of the predictive value of brain atrophy. Specificity was somewhat higher for the PBVC cut-off, but this was at least partially offset by a somewhat higher sensitivity for the PLVVC cut-off. In the MS-MRIUS dataset, neither PBVC nor PLVVC cut-offs were significantly associated with DP status – a discrepancy likely due to the much shorter follow-up (9–24 months, with an average of only 16 months) of the MS-MRIUS study compared to the ASA study (10 years). Even in this short timeframe, though, PLVVC status was significantly associated with walking score progression. Ultimately, though, MS is a complex disease and it is unlikely that any single metric can fully determine prognosis. Therefore, while it is clear that brain atrophy (via PBVC or PLVVC) can play an important role, it will likely be most valuable when used in concert with other predictors.
The use of ventricular enlargement as a potential proxy for brain volume loss is not a new idea – in fact, ventricular dilation was one of the earliest qualitative neuroimaging indicators of atrophy reported in the literature (Rao et al., 1985; Young et al., 1981; Gyldensted, 1976). More recently, it has been largely superseded by quantitative and highly precise measures such as PBVC derived from modern computational neuroimaging algorithms like SIENA (Smith et al., 2002) or whole-brain Jacobian integration (Nakamura et al., 2014). In this context, it is important to note that PLVVC is not a more accurate, sensitive, or specific disease marker than PBVC per se. In general, when protocols are kept constant and scanner changes are minimized, PBVC better separates groups, better correlates with disease outcomes, and has lower absolute error (Zivadinov et al., 2016a, Zivadinov et al., 2016b). Measures of lateral ventricular atrophy are also not directly sensitive to cortical changes, since the ventricles are physically far removed from cortical gray matter (although in some cases, they observationally correlate better with cortical gray matter atrophy than PBVC (Zivadinov et al., 2016a, Zivadinov et al., 2016b). Another potential issue with relying on ventricular rather than PBVC measures arises due to the fact that there may be more natural variability in ventricular size among healthy individuals than there is in whole brain volume, although these cross-sectional differences should be somewhat ameliorated by longitudinal analysis (Blatter et al., 1995). Despite these caveats, this corresponding ventricular expansion cut-off assessed via PLVVC may still be very useful in cases where more commonly employed PBVC measures are not reliably obtainable.
It is important to note that in absolute terms, this corresponding PLVVC cut-off of 3.5% is nearly an order of magnitude (8.75 times) larger than the −0.4% PBVC cut-off. This is due to the fact that ventricular expansion (by percentage) occurs more rapidly than overall brain volume shrinkage (again by percentage) (Zivadinov et al., 2016a, Zivadinov et al., 2016b), which must be considered when comparing variances and/or absolute error in methods, and should also be taken into account when considering distance from the threshold.
Our results also independently confirmed the validity of the previously proposed −0.4% threshold: follow-up weighted PBVC ROC analysis in this study led to an 80% specificity cut-off of −0.33% per year. This is very close to −0.4%, and given the overall variability of MS progression, scanner differences between studies, and inevitable measurement error, this level of concordance is both striking and reassuring. As with the previous analyses, though, we also saw inter-individual variability in PBVC, and overlap between MS and HC groups. Previously reported AUC was 0.77, and sensitivity was 67% at 80% specificity, and was similar or better in this dataset at an AUC of 0.88 with 80% specificity having 75% sensitivity. For PLVVC, the AUC was comparable at 0.83 with 80% specificity having 76% sensitivity. Results between PBVC and PLVVC were also comparable for clinical outcomes (CDP concordance AUC of 0.68 for PBVC vs 0.62 for PLVVC). These results are also in line with another recent study that looked at other brain regional cut-offs, including corpus callosum and thalamus (Uher et al., 2017).
In addition to the general limitations of PLVVC discussed above, there are a number of points that should be considered in interpreting the present study. Tools like VIENA (Vrenken et al., 2014) may also measure ventricular expansion more accurately than the NeuroSTREAM technique since NeuroSTREAM is intended to operate on low resolution, clinical-quality scans. However, one of the main aims of this study was to establish pathological ventricular expansion cut-offs for use as an alternative measure for clinical routine scans where SIENA (and by extension VIENA) may not be available. Therefore, we chose to focus on NeuroSTREAM. Although we expect that other measures of the lateral ventricles like ALVIN (Kempton et al., 2011) will yield similar cutoffs, it must be noted that VIENA incorporates all central CSF, not specifically restricting the analysis to the lateral ventricles per se. Therefore, the resulting cut-offs for VIENA may differ slightly.
Finally, in the current study, we only determined a single, population-wide PLVVC cut-off. However, a recent study demonstrated the value of calculating individual brain volume cut-offs adjusted for age, disease duration, sex, baseline disability, and T2-lesion volume (Sormani et al., 2016). By considering those additional factors, the investigators were better able to determine whether volumes were truly pathological. It is likely that a similar approach would also benefit longitudinal atrophy analysis. However, that analysis was based on a very large database containing images from 2342 patients, and a database of that size was not available here to support this type of exploratory analysis. Future work should use a larger database with broad coverage of demographic and clinical characteristics and treatments to expand on these findings by differentiating pathological cut-offs for specific combinations of covariates, and should potentially also include additional MRI metrics to help quantify the balance between gray matter and white matter atrophy. Similarly, repeated acquisition across different scanners and/or acquisition protocols would also allow for determination of and possible correction for the potential impact of factors like field strength and scan parameters such as echo time or inversion time.
5. Conclusion
The ventricular expansion rate best corresponding to the established whole brain atrophy rate of −0.4% per year is 3.5% per year in RRMS. Thus, a 3.5% cut-off measured with NeuroSTREAM on clinical quality T2-FLAIR scans can potentially be used in place of traditional SIENA measures when suitable T1 scans are not available.
Disclosure of conflict of interest
Analysis of the second dataset in this study was supported by Novartis.
This project was also supported by the Czech Ministry of Education project Progres Q27/LF1 and by RVO-VFN 64165.
Author disclosure
Dr. Michael G. Dwyer has received consultant fees from Claret Medical and EMD Serono and research grant support from Novartis.
Jonathan R. Korn and NasreenKhan are employees of QuintilesIMS.
Drs. Diego Silva and Jennie Medin are employees of Novartis.
Drs. Jesper Hagemeier and Niels Bergsland have nothing to disclose.
Dr. Manuela Vaneckova was supported by Czech Ministry of Health, grant RVO-VFN 64165. She received compensation for speaker honoraria, travel and consultant fees from Biogen Idec, Sanofi Genzyme, Novartis, Merck and Teva, as well as support for research activities from Biogen Idec.
Dr. Horakova received compensation for travel, speaker honoraria and consultant fees from Biogen Idec, Novartis, Merck, Bayer, Sanofi Genzyme, Roche, and Teva, as well as support for research activities from Biogen Idec.
Dr. T. Uher received financial support for conference travel and honoraria from Biogen Idec, Novartis, Sanofi Genzyme, Roche, and Merck, as well as support for research activities from Biogen Idec.
Dr. Kubala Havrdova received speaker honoraria and consultant fees from Actelion, Biogen, Celgene, Merck, Novartis, Roche, Sanofi Genzyme and Teva.
Dr. R Zivadinov received personal compensation from EMD Serono, Genzyme-Sanofi, Novartis, Claret-Medical, Celgene for speaking and consultant fees. He received financial support for research activities from Claret Medical, Genzyme-Sanofi, QuintilesIMS Health, Intekrin-Coherus, Novartis and Intekrin-Coherus.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2018.02.009.
Appendix A. Supplementary data
The following is the supplementary data related to this article.
Additional data tables
References
- Alonso A., Hernán M.A. Temporal trends in the incidence of multiple sclerosis: a systematic review. Neurology. 2008;71(2):129–135. doi: 10.1212/01.wnl.0000316802.35974.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict R.H.B., Zivadinov R. Risk factors for and management of cognitive dysfunction in multiple sclerosis. Nat. Rev. Neurol. 2011;7(6):332–342. doi: 10.1038/nrneurol.2011.61. [DOI] [PubMed] [Google Scholar]
- Blatter D.D. Quantitative volumetric analysis of brain MR: normative database spanning 5 decades of life. Am. J. Neuroradiol. 1995;16(2) [PMC free article] [PubMed] [Google Scholar]
- De Stefano N. Establishing pathological cut-offs of brain atrophy rates in multiple sclerosis. J. Neurol. Neurosurg. Psychiatry. 2016;87:93–99. doi: 10.1136/jnnp-2014-309903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer M.G. Neurological software tool for reliable atrophy measurement (NeuroSTREAM) of the lateral ventricles on clinical-quality T2-FLAIR MRI scans in multiple sclerosis. NeuroImage: Clin. 2017;15:769–779. doi: 10.1016/j.nicl.2017.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelineau-Morel R. The effect of hypointense white matter lesions on automated gray matter segmentation in multiple sclerosis. Hum. Brain Mapp. 2012;33(12):2802–2814. doi: 10.1002/hbm.21402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni G. Multiple Sclerosis and Related Disorders. 4(4) 2015. Is it time to target no evident disease activity (NEDA) in multiple sclerosis? pp. 329–333. [DOI] [PubMed] [Google Scholar]
- Gyldensted C. Computer tomography of the cerebrum in multiple sclerosis. Neuroradiology. 1976;12(1):33–42. doi: 10.1007/BF00344224. [DOI] [PubMed] [Google Scholar]
- Havrdova E. Randomized study of interferon beta-1a, low-dose azathioprine, and low-dose corticosteroids in multiple sclerosis. Mult. Scler. 2009;15(8):965–976. doi: 10.1177/1352458509105229. [DOI] [PubMed] [Google Scholar]
- Havrdova E. Freedom from disease activity in multiple sclerosis. Neurology. 2010;74(Suppl. 3):S3–7. doi: 10.1212/WNL.0b013e3181dbb51c. (17 Suppl 3) [DOI] [PubMed] [Google Scholar]
- Hobart J. Timed 25-foot walk direct evidence that improving 20% or greater is clinically meaningful in MS. Neurology. 2013;80(16):1509–1517. doi: 10.1212/WNL.0b013e31828cf7f3. [DOI] [PubMed] [Google Scholar]
- Horakova D. Evolution of different MRI measures in patients with active relapsing-remitting multiple sclerosis over 2 and 5 years: a case-control study. J. Neurol. Neurosurg. Psychiatry. 2008;79(4):407–414. doi: 10.1136/jnnp.2007.120378. [DOI] [PubMed] [Google Scholar]
- Kappos L. Inclusion of brain volume loss in a revised measure of “no evidence of disease activity” (NEDA-4) in relapsing–remitting multiple sclerosis. Mult. Scler. J. 2016;22(10):1297–1305. doi: 10.1177/1352458515616701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempton M.J. A comprehensive testing protocol for MRI neuroanatomical segmentation techniques: evaluation of a novel lateral ventricle segmentation method. NeuroImage. 2011;58(4):1051–1059. doi: 10.1016/j.neuroimage.2011.06.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C., Minneboo A., De Groot V. Early central atrophy rate predicts 5 year clinical outcome in multiple sclerosis. J. Neurol. Neurosurg. Psychiatry. 2010;81(12):1351–1356. doi: 10.1136/jnnp.2009.199968. [DOI] [PubMed] [Google Scholar]
- Miller D.H. Measurement of atrophy in multiple sclerosis: pathological basis, methodological aspects and clinical relevance. Brain. 2002;125(8):1676–1695. doi: 10.1093/brain/awf177. [DOI] [PubMed] [Google Scholar]
- van Munster C.E.P., Uitdehaag B.M.J. Outcome measures in clinical trials for multiple sclerosis. CNS Drugs. 2017;31(3):217–236. doi: 10.1007/s40263-017-0412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K. Jacobian integration method increases the statistical power to measure gray matter atrophy in multiple sclerosis. NeuroImage: Clin. 2014;4:10–17. doi: 10.1016/j.nicl.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu V. Brain atrophy and lesion load predict long term disability in multiple sclerosis. J. Neurol. Neurosurg. Psychiatry. 2013;84(10):1082–1091. doi: 10.1136/jnnp-2012-304094. [DOI] [PubMed] [Google Scholar]
- Rao S.M. Chronic progressive multiple sclerosis: relationship between cerebral ventricular size and neuropsychological impairment. Arch. Neurol. 1985;42(7):678. doi: 10.1001/archneur.1985.04060070068018. [DOI] [PubMed] [Google Scholar]
- Sanfilipo M.P. Gray and white matter brain atrophy and neuropsychological impairment in multiple sclerosis. Neurology. 2006;66(5):685–692. doi: 10.1212/01.wnl.0000201238.93586.d9. [DOI] [PubMed] [Google Scholar]
- Smith S.M. Accurate, robust, and automated longitudinal and cross-sectional brain change analysis. NeuroImage. 2002;17(1):479–489. doi: 10.1006/nimg.2002.1040. [DOI] [PubMed] [Google Scholar]
- Sormani M.P. Defining brain volume cutoffs to identify clinically relevant atrophy in RRMS. Mult. Scler. J. 2016;23(5):656–664. doi: 10.1177/1352458516659550. [DOI] [PubMed] [Google Scholar]
- Summers M.M. Cognitive impairment in relapsing - remitting multiple sclerosis can be predicted by imaging performed several years earlier. Mult. Scler. 2008;14(2):197–204. doi: 10.1177/1352458507082353. [DOI] [PubMed] [Google Scholar]
- Tekok-Kilic A. Independent contributions of cortical gray matter atrophy and ventricle enlargement for predicting neuropsychological impairment in multiple sclerosis. NeuroImage. 2007;36(4):1294–1300. doi: 10.1016/j.neuroimage.2007.04.017. [DOI] [PubMed] [Google Scholar]
- Torkildsen Ø., Myhr K.-M., Bø L. Disease-modifying treatments for multiple sclerosis - a review of approved medications. Eur. J. Neurol. 2016;23(S1):18–27. doi: 10.1111/ene.12883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uher T. Pathological cut-offs of global and regional brain volume loss in multiple sclerosis. Mult. Scler. J. 2017 doi: 10.1177/1352458517742739. (ePub ahead of print) [DOI] [PubMed] [Google Scholar]
- Vrenken H. Validation of the automated method VIENA: an accurate, precise, and robust measure of ventricular enlargement. Hum. Brain Mapp. 2014;35(4):1101–1110. doi: 10.1002/hbm.22237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young I.R. Nuclear magnetic resonance imaging of the brain in multiple sclerosis. Lancet. 1981;318(8255):1063–1066. doi: 10.1016/s0140-6736(81)91273-3. [DOI] [PubMed] [Google Scholar]
- Zivadinov R., Uher T., Hagemeier J. A serial 10-year follow-up study of brain atrophy and disability progression in RRMS patients. Mult. Scler. 2016;22(13):1709–1718. doi: 10.1177/1352458516629769. [DOI] [PubMed] [Google Scholar]
- Zivadinov R. Clinical relevance of brain atrophy assessment in multiple sclerosis. Implications for its use in a clinical routine. Expert. Rev. Neurother. 2016;16(7):777–793. doi: 10.1080/14737175.2016.1181543. [DOI] [PubMed] [Google Scholar]
- Zivadinov R. An observational study to assess brain MRI change and disease progression in multiple sclerosis clinical practice-the MS-MRIUS study. J. Neuroimaging. 2017;27(3):339–347. doi: 10.1111/jon.12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivadinov R. Feasibility of brain atrophy measurement in clinical routine without prior standardization of the MRI protocol: results from MS-MRIUS, a longitudinal. Am. Soc. Neuroradiol. 2018 doi: 10.3174/ajnr.A5442. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional data tables