Abstract
We have previously reported that overexpression of Programmed Death -1 Homolog (PD-1H) in human monocytes leads to activation and spontaneous secretion of multiple pro inflammatory cytokines. Here we evaluate changes in monocytes gene expression after enforced PD-1H expression by gene array. The results show that there are significant alterations in 51 potential candidate genes that relate to immune response, cell adhesion and metabolism. Genes corresponding to pro-inflammatory cytokines showed the highest upregulation, 7, 3.2, 3.0, 5.8, 4.4 and 3.1 fold upregulation of TNF-α, IL-1 β, IFN-α, γ, λ and IL-27 relative to vector control. The data are in agreement with cytometric bead array analysis showing induction of proinflammatory cytokines, IL-6, IL-1β and TNF-α by PD-1H. Other genes related to inflammation, include transglutaminase 2 (TG2), NF-κB (p65 and p50) and toll like receptors (TLR) 3 and 4 were upregulated 5, 4.5 and 2.5 fold, respectively. Gene set enrichment analysis (GSEA) also revealed that signaling pathways related to inflammatory response, such as NFκB, AT1R, PYK2, MAPK, RELA, TNFR1, MTOR and proteasomal degradation, were significantly upregulated in response to PD-1H overexpression. We validated the results utilizing a standard inflammatory sepsis model in humanized BLT mice, finding that PD-1H expression was highly correlated with proinflammatory cytokine production. We therefore conclude that PD-1H functions to enhance monocyte activation and the induction of a pro-inflammatory gene expression profile.
Keyword: Immunology
1. Introduction
With FDA approval of anti-PD-1 treatment for a variety of cancer types, immune “checkpoint” molecules of the B7/CD28 superfamily of receptors have been shown to be attractive targets for cancer immunotherapy. The B7 family of co-receptors, either co-stimulatory or co-inhibitory, orchestrate the activation of immune cells via a complex signaling mechanism, the outcome of which is mainly determined by the balance of expression and signaling of these co-stimulatory and co-inhibitory co-receptors [1]. Instead of directly targeting tumor cells, the intervention of immune “checkpoint” molecules modulates the host's immune response within the tumor microenvirionment [2], thus improving the tumor-killing capacity of host immune cells. Given the apparent success of such therapeutic strategies, the identification of additional immune regulatory pathways has potentially important therapeutic implications.
Programmed death-1 homologue (PD-1H, also known as VISTA) is a newly-discovered molecule of the CD28/B7 family [3, 4, 5]. PD-1H is a 309 amino acid type I transmembrane protein and contains an extracellular IgV domain, which resembles the CD28 family member, PD-1 [3]. The cytoplasmic tail of PD-1H is highly conserved between mice and humans, however, unlike PD-1, PD-1H does not contain the immunoreceptor tyrosine-based inhibitory motif (ITIM) or the immunoreceptor tyrosine-based switch motif (ITSM) [6], suggesting that the function of PD-1H may have evolved independently and is distinct from other co-signaling molecules within the family. Recently, two independent research groups identified PD-1H as a novel co-inhibitory receptor/ligand that can suppress T cell function in mouse models [5, 7]. The mechanism of action of PD-1H, however, remains unresolved.
Monocytes play critical roles in the innate immune system to defend against pathogens. They can eliminate pathogens by phagocytosis, production of proinflammatory cytokines, and modulation of T-cell immune response [8]. Although human studies of PD-1H expression of monocytes are limited, one study showed PD-1H expression in both patrolling and inflammatory monocytes [9]. We have found that there is a significant elevation of PD-1H expression in HIV-infected patients, either naïve or ART-treated, which is significantly correlated with spontaneous secretion of multiple pro-inflammatory cytokines [10]. In this report, we transduced human monocytes with PD-1H overexpressing plasmid or vector control plasmid via nucleofection, then isolated RNA and performed gene array analysis to investigate how PD-1H overexpression in monocytes influences the global expression of genes and gene sets related to immune response.
2. Results
-
1.
KEGG gene sets differentially expressed in response to PD-1H over-expression in monocytes.
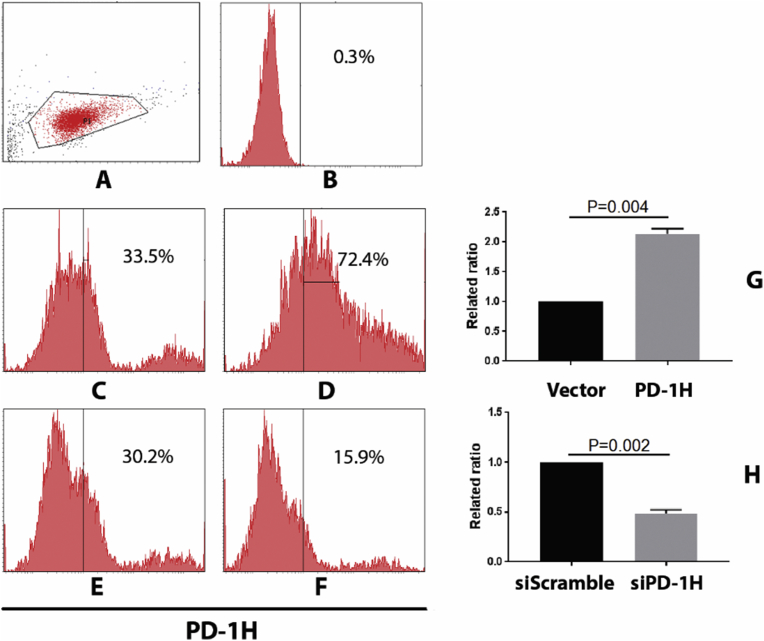
To understand genome-wide influences of PD-1H on the expression of genes and gene sets in transfected monocytes, we performed RNA-seq contrasting both PD-1H over-expression to vector control and PD-1H knockdown (siPD-1H) to non-targeting control (siScramble). After PD-1H overexpression vector transfection, PD-1H expression in monocytes increased 1.2 fold, compared to vector control (from an average expression of 32%–71%). While after siPD-1H transfection, PD-1H expression was reduced by more than 50% when compared to siScramble transfection as control (from an average expression of 29.7%–15%) (Fig. 1). We then analyzed differential expression of PD-1H by GSEA analysis using Biocarta database and Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Gene sets with significant enrichment scores showing either up- or down-regulation were further characterized by statistical analysis.
Fig. 1.
PD-1H expression after transfection of PD-1H overexpression vector or siPD-1H vector. (A) Gating strategy. (B) Isotype stained monocytes. (C) Empty vector Neon-transfected monocytes. (D) PD-1H overexpressing vector transfected monocytes. (E) siScramble transfected monocytes. (F) siPD-1H transfected monocytes. (G) Related ratio of PD-1H expression after vector vs. PD-1H transfection using average MFIs from three different experiments represented in C and D. (H) Related ratio of PD-1H expression after siScramble vs. siPD-1H transfection using average MFIs from three different experiments represented in E and F.
For KEGG gene sets, we first analyzed 186 gene sets comprising a total of 19,695 genes and compared RNAseq data from control vs. PD-1H over-expression with similar microarray data. We then filtered the gene set size at a range of 15–500 genes (the filtration criteria also include fold change >2, and p value <0.05), identifying 177 gene sets for the analysis. We found that 149 out of 177 gene sets were up-regulated after PD-1H over-expression, while 28 sets were down-regulated. Setting the normalized p value <0.05, 98 sets were significantly up-regulated, while 13 sets were significantly down-regulated. When we set a cut-off value of normalized enrichment score (NES) at ±2.0, only 4 gene sets in the list were down-regulated comprising olfactory transduction, retinal metabolism, taste transduction and neuroactive ligand receptor interaction, all of which are related to odorant, taste or visual sensing. Among the 98 upregulated gene sets, 30 gene sets met the threshold criteria (FDR ≤ 0.001, Table 1). Among 20 of the most significant up-regulated gene sets, most relate to immune and inflammatory response. Specifically, the top 3 pathways are important for immune response to infection. Ubiquitin mediated proteolysis (ranked 1) plays an important role in p53-mediated apoptosis during HPV infection, and is regulated by IFN-γ [11]. Spliceosome function (ranked 2) is required for alternative splicing of numerous cytokines and their receptors (e.g. IL-1, IL-5, IL-6, IL-7, IL-15 and CD33) which are important for chronic inflammation [12]. While proteasome degradation (ranked 3) participates in the proteolysis of pro-ligands to form mature ligands, such as IL-1β, IL-6, IFN-β and IL-12, during inflammatory responses [13].
Table 1.
Gene sets enriched in PD-1H overexpressed monocytes, analyzed by KEGG (Kyoto Encyclopedia of Genes and Genomes) gene sets.
| NAME | SIZE | ES | NES | NOM p-val | FDR q-val | FWER p-val |
|---|---|---|---|---|---|---|
| Upregulated gene sets | ||||||
| Ubiquitin mediated proteolysis | 132 | 0.628471 | 2.688491 | 0 | 0 | 0 |
| Spliceosome | 113 | 0.617208 | 2.597225 | 0 | 0 | 0 |
| Proteosome | 43 | 0.716289 | 2.578759 | 0 | 0 | 0 |
| Lysosome | 121 | 0.588662 | 2.497777 | 0 | 0 | 0 |
| RNA degradation | 57 | 0.658668 | 2.473238 | 0 | 0 | 0 |
| Leishmania infection | 66 | 0.601159 | 2.350985 | 0 | 0 | 0 |
| Aminoacyl tRNA biosynthesis | 41 | 0.650459 | 2.321397 | 0 | 0 | 0 |
| Citrate cycle TCA cycle | 30 | 0.684975 | 2.269259 | 0 | 0 | 0 |
| B cell receptor signaling pathway | 75 | 0.561486 | 2.244579 | 0 | 0 | 0 |
| Neurotrophin signaling pathway | 126 | 0.52788 | 2.243486 | 0 | 0 | 0 |
| NOD-like receptor signalining pathway | 62 | 0.579527 | 2.242351 | 0 | 0 | 0 |
| Fc gamma R mediated phagocytosis | 92 | 0.538345 | 2.21778 | 0 | 0 | 0 |
| Apoptosis | 87 | 0.538775 | 2.176365 | 0 | 0 | 0 |
| TOLL-like receptor signalining pathway | 100 | 0.525158 | 2.147492 | 0 | 0 | 0 |
| Oxidative phosphorylation | 115 | 0.50918 | 2.131876 | 0 | 0 | 0 |
| Renal cell carcinoma | 70 | 0.545899 | 2.131357 | 0 | 0 | 0 |
| Alzheimers diseases | 155 | 0.490106 | 2.125872 | 0 | 0 | 0 |
| Acute myeloid leukemia | 57 | 0.557474 | 2.105689 | 0 | 6.13E-05 | 0.001 |
| Long term potentiation | 70 | 0.531729 | 2.089761 | 0 | 1.14E-04 | 0.002 |
| NOTCH signaling pathway | 47 | 0.567842 | 2.089182 | 0 | 1.08E-04 | 0.002 |
| Huntingtons disease | 169 | 0.475458 | 2.088653 | 0 | 1.03E-04 | 0.002 |
| Epithelial cell signaling in helicobacter infection | 68 | 0.52987 | 2.082758 | 0 | 9.82E-05 | 0.002 |
| SNARE interaction in vesicular transport | 37 | 0.59472 | 2.062443 | 0 | 1.87E-04 | 0.004 |
| Endocytosis | 181 | 0.461407 | 2.037321 | 0 | 2.73E-04 | 0.006 |
| Chronic myeloid leukemia | 73 | 0.524357 | 2.029166 | 0 | 2.62E-04 | 0.006 |
| T cell receptor signaling pathway | 108 | 0.491484 | 2.024237 | 0 | 3.37E-04 | 0.008 |
| MTOR signaling pathway | 52 | 0.543775 | 2.022957 | 0 | 3.64E-04 | 0.009 |
| Pancreatic cancer | 70 | 0.524255 | 2.022309 | 0 | 3.51E-04 | 0.009 |
| Vibrio cholerae infection |
54 |
0.538295 |
2.010241 |
0 |
4.57E-04 |
0.012 |
| Downregulated gene sets | ||||||
| Olfactory transduction | 365 | -0.62038 | -3.78277 | 0 | 0 | 0 |
| Retinol metabolism | 59 | -0.50072 | -2.37898 | 0 | 0 | 0 |
| Taste transduction | 50 | -0.50267 | -2.24919 | 0 | 2.46E-04 | 0.001 |
| Neuroactive ligand receptor interaction | 268 | -0.36353 | -2.1765 | 0 | 3.59E-04 | 0.002 |
Similar results were obtained comparing RNAseq data from non-targeting control vs. PD-1H knockdown with similar microarray data. When we use a cut-off value of FDR ≤ 0.25, compared to siScramble groups, the expression of 87 gene sets were significantly down-regulated in siPD-1H groups (Table S1). Among significant gene sets that were down-regulated, apoptosis (Ranked 1), spliceosome function (Ranked 2) and Endocytosis (Ranked 3) are typical pathological processes involved inflammation. Other pathways, such as spliceosome assembly, proteolysis, proteasomal degradation and several other signaling pathways known to be directly involved in inflammation were also down-regulated, but to a lesser extent. It is worth noting that most of these pathways overlap with pathways identified in PD-1H over-expression data. Both data sets validate the hypothesis that PD-1H overexpression may elicit an inflammatory response.
-
2.
Biocarta gene sets differentially expressed in response to PD-1H over-expression in monocytes
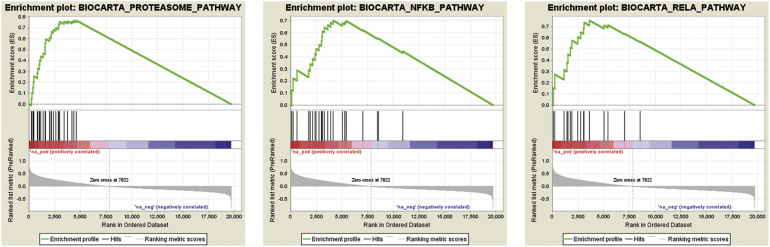
Analysis of differential Biocarta gene set expression, we identified 148 gene sets that responded to PD-1H overexpression, 144 were up-regulated and 4 were down-regulated. When we set the nominal p value at 5%, 107 gene sets were enriched as up-regulated, while one gene set was down-regulated. However, when the stringency was to set up NES value of ±2.0 (FDR ≤ 0.001), 31 gene sets were shown to be significantly up-regulated by PD-1H overexpression, while none were significantly down-regulated (Table 2). After analyzing these 31 individual up-regulated pathways, we found that most pathways are involved in inflammation, similar to the analysis using the KEGG database. Among the top-20-ranked gene sets, proteasome pathway (28 genes), AT1R pathway (32 genes), PYK2 pathway (28 genes), MAPK pathway (86 genes), NF-κB pathway (23 genes), RELA pathway (16 genes), TNFR1 pathway (29 genes), mTOR pathway (21 genes) and EGF pathway (31 genes) have all been extensively reported to be directly involved in inflammatory response by signaling inflammatory cytokines or other molecules. Other less significant pathways identified are also revealed to be associated with inflammatory response and more specifically to infection-induced inflammation, such as HIV/NEF pathway. To illustrate the GSEA output, three signaling pathways are shown as representative (Fig. 2).
Table 2.
Gene sets enriched in PD-1H overexpressed monocytes, analyzed by Biocarta gene sets.
| NAME | SIZE | ES | NES | NOM p-val | FDR q-val | FWER p-val |
|---|---|---|---|---|---|---|
| Upregulated gene sets | ||||||
| Proteasome pathway | 28 | 0.7669194 | 2.5176857 | 0 | 0 | 0 |
| HIVNEF pathway | 57 | 0.6349662 | 2.4080927 | 0 | 0 | 0 |
| AT1R pathway | 32 | 0.6698542 | 2.2998908 | 0 | 0 | 0 |
| PYK2 pathway | 28 | 0.6876869 | 2.2334847 | 0 | 0 | 0 |
| PDGF pathway | 32 | 0.6528511 | 2.232296 | 0 | 0 | 0 |
| RACCYCD pathway | 26 | 0.6897389 | 2.2234564 | 0 | 0 | 0 |
| MAPK pathway | 86 | 0.5364569 | 2.2062736 | 0 | 0 | 0 |
| NFKB pathway | 23 | 0.7023099 | 2.1759067 | 0 | 0 | 0 |
| RELA pathway | 16 | 0.7558813 | 2.1715233 | 0 | 1.57E-04 | 0.001 |
| NTHI pathway | 24 | 0.6685817 | 2.157457 | 0 | 1.41E-04 | 0.001 |
| TNFR1 pathway | 29 | 0.6512108 | 2.1534717 | 0 | 1.28E-04 | 0.001 |
| FCER1 pathway | 38 | 0.6169635 | 2.1517484 | 0 | 1.18E-04 | 0.001 |
| MTOR pathway | 21 | 0.7118477 | 2.1505196 | 0 | 1.09E-04 | 0.001 |
| MET pathway | 37 | 0.6128663 | 2.1445467 | 0 | 1.01E-04 | 0.001 |
| NDKDYNAMIN pathway | 18 | 0.7299176 | 2.1436856 | 0 | 9.41E-05 | 0.001 |
| B cell survival pathway | 16 | 0.7273845 | 2.0980804 | 0 | 2.79E-04 | 0.003 |
| EGF pathway | 31 | 0.6259823 | 2.0772097 | 0 | 4.42E-04 | 0.005 |
| PPARA pathway | 56 | 0.5467193 | 2.0593934 | 0 | 5.76E-04 | 0.007 |
| ARAP pathway | 17 | 0.6829583 | 2.04499 | 0 | 6.26E-04 | 0.008 |
| EIF4 pathway | 22 | 0.658423 | 2.0433595 | 0 | 5.95E-04 | 0.008 |
| CCR5 pathway | 17 | 0.7157682 | 2.0381536 | 0 | 6.33E-04 | 0.009 |
| VIP pathway | 26 | 0.6309993 | 2.0313003 | 0 | 9.47E-04 | 0.014 |
| TCR pathway | 44 | 0.56018 | 2.0273864 | 0.001381216 | 9.70E-04 | 0.015 |
| RARRXR pathway | 15 | 0.7322379 | 2.0261118 | 0 | 9.30E-04 | 0.015 |
| IGF1-MTOR pathway | 20 | 0.6649683 | 2.0245264 | 0 | 9.50E-04 | 0.016 |
| P38MAPK pathway | 39 | 0.5725741 | 2.0215604 | 0 | 9.13E-04 | 0.016 |
| TNFR2 pathway | 18 | 0.691175 | 2.0208588 | 0 | 8.80E-04 | 0.016 |
| MEF2D pathway | 18 | 0.6841997 | 2.018628 | 0 | 9.04E-04 | 0.017 |
| HCMV pathway | 17 | 0.7023321 | 2.0160334 | 0.001587302 | 0.0010264 | 0.02 |
| CREB pathway | 27 | 0.614528 | 2.0083513 | 0 | 0.0010831 | 0.022 |
| Biopeptides pathway | 42 | 0.5644003 | 2.0060863 | 0 | 0.0010924 | 0.023 |
Fig. 2.
Gene set enrichment analysis of Biocarta pathways. Genes were ranked according to the difference between average transcript expression of PD-1H overexpressed cases and controls. Each pathway in Biocarta database was then assigned the (normalized) enrichment score using GSEA (version 2.2.4). Three enrichment plots with highest enrichment scores are shown for representatively.
We also analyzed the siPD-1H or siScramble microarray data using Biocarta data sets. Similar to the vector vs PD-1H data, 59 out of 148 gene sets are significantly down-regulated in siPD-1H groups (nominal p value <5%) when compared with siScramble groups (Table S2). No gene sets were enriched as significantly up-regulated. When looking at the top 20 down-regulated gene sets, most are related to cytokine signaling during inflammation, such as IL-1R, Fas, NF-κB, IL-17, Il-10, TNFR, NI-HI, NO2IL-12. Some are associated with pathological processes of inflammation, such as proteasomal degradation, caspase activation and subsequent apoptosis. Others are related to pathogen-induced inflammation, such as HIV/NEF and HCMV. Overall, this transcriptome profile is similar to our data analyzing expression changes between vector control and PD-1H over-expression, displaying a typical inflammatory response profile.
-
3.
Individual genes up- or down-regulated by PD-1H overexpression
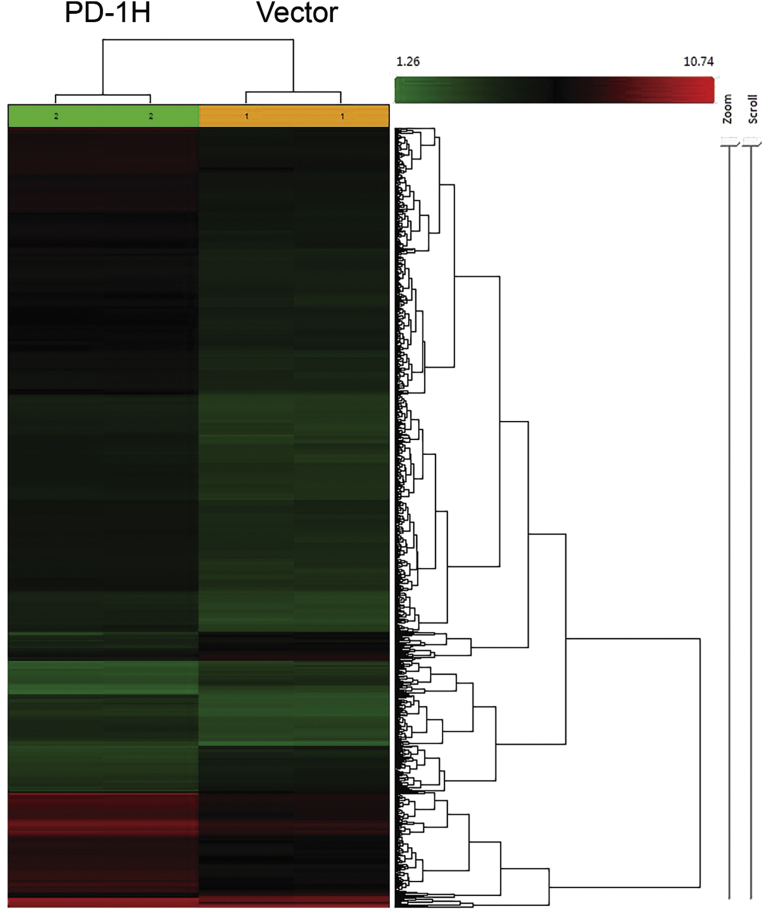
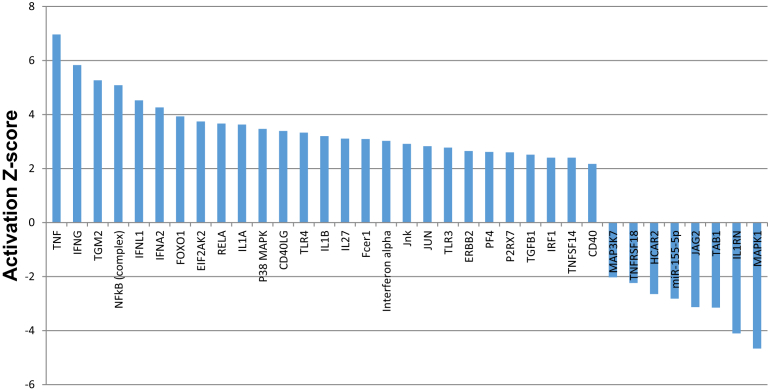
To identify the most significantly affected individual gene expression change in response to PD-1H overexpression, we performed Qiagen IPA analysis (Ingenuity Pathway Analysis) with the microarray data. There are more than 4,000 individual genes up- or down-regulated in response to PD-1H overexpression, as shown in the heatmap (Fig. 3). When comparing with vector control, the pro-inflammatory cytokines-related genes, such as TNF-α, IL-1 β, IFN-α, γ, λ and IL-27 showed 7, 3.2, 3.0, 5.8, 4.4 and 3.1 fold upregulation, as shown in Table 3. Other genes related to inflammation, include transglutaminase 2 (TG2), NF-κB (p65 and p50) and toll like receptors (TLR) 3 and 4 were upregulated 5, 4.5 and 2.5 fold, respectively. When using a cut-off of activation z-score ≥2 (for up-regulated genes) or ≤-2 (for down-regulated genes), there are 27 genes significantly up-regulated and 8 genes down-regulated. Table 3 and Fig. 4 shows that the majority of genes up-regulated are inflammatory pathway related, specifically TNF-α, IFN-γ, NF-κB, IFN-L1, IFN-A2, REL-A, P38MAPK, CD-40LG, CD-40, TLR3, TLR4, IL-1β and IFN-α. Notably, tgm2 (ranked 3 among up-regulated genes) is a marker of M2 macrophage and has also been reported to be activated by inflammation and hypoxia, and specifically includes regulatory elements that respond to IL-6 [14] and HIF-1 [15].
-
4.
Elevated PD-1H expression is associated with inflammatory response during sepsis in humanized BLT mice
Fig. 3.
Global gene expression difference between PD-1H overexpressed cases and vectors. Two-way unsupervised clustering was performed on gene expression profiles of PD-1H overexpressed cases and vectors. Green and red color indicate relatively lower/higher expression of genes, respectively, while black indicates intermediated gene expression. Distance in the dendrogram tree was calculated using Ward's method.
Table 3.
Summary of the most significant differentially regulated genes/gene clusters in PD-1H overexpressed monocytes.
| Upstream regulator | Fold change | Molecule type | Predicted activation state | Activation z-score | Notes | p-value of overlap | Mechanistic network |
|---|---|---|---|---|---|---|---|
| TNF | 2.223 | cytokine | Activated | 6.963 | bias | 2.70E-19 | 160 (14) |
| MAPK1 | kinase | Inhibited | -4.666 | 5.43E-16 | |||
| IFNG | cytokine | Activated | 5.831 | bias | 2.27E-15 | 130 (10) | |
| TGM2 | enzyme | Activated | 5.266 | bias | 6.75E-14 | 94 (6) | |
| IFNL1 | cytokine | Activated | 4.523 | bias | 3.64E-12 | 32 (3) | |
| IFNA2 | cytokine | Activated | 4.264 | bias | 1.53E-11 | 133 (12) | |
| Interferon alpha | group | Activated | 3.023 | bias | 2.20E-11 | 138 (13) | |
| IL27 | cytokine | Activated | 3.109 | bias | 3.49E-11 | 135 (13) | |
| TGFB1 | growth factor | Activated | 2.512 | bias | 1.87E-10 | 96 (6) | |
| CD40LG | cytokine | Activated | 3.390 | bias | 6.80E-10 | 173 (16) | |
| Immunoglobulin | complex | Activated | 2.704 | bias | 7.47E-10 | ||
| CD40 | transmembrane receptor (TR) | Activated | 2.172 | 3.51E-09 | 129 (13) | ||
| NFκB (complex) | complex | Activated | 5.082 | bias | 3.53E-09 | 117 (12) | |
| ERBB2 | kinase | Activated | 2.646 | bias | 9.97E-09 | 48 (4) | |
| RELA | transcription regulator | Activated | 3.665 | bias | 1.22E-08 | 114 (10) | |
| FOXO1 | transcription regulator | Activated | 3.931 | bias | 4.68E-08 | ||
| TLR4 | TR | Activated | 3.329 | bias | 5.90E-08 | 118 (13) | |
| Jnk | group | Activated | 2.914 | bias | 6.02E-08 | 109 (10) | |
| MAP3K7 | kinase | Inhibited | -2.030 | bias | 6.54E-08 | 126 (12) | |
| IRF1 | 2.204 | transcription regulator | Activated | 2.400 | bias | 9.41E-08 | |
| TAB1 | enzyme | Inhibited | -3.148 | bias | 9.44E-08 | 61 (9) | |
| IL1RN | 2.806 | cytokine | Inhibited | -4.101 | bias | 1.24E-07 | 96 (11) |
| IL1A | 2.117 | cytokine | Activated | 3.626 | bias | 1.29E-07 | 128 (13) |
| miR-155-5p (miRNAs w/seed UAAUGCU) | mature microrna | Inhibited | -2.817 | bias | 1.33E-07 | ||
| Fcer1 | complex | Activated | 3.092 | bias | 1.59E-07 | 167 (16) | |
| P38 MAPK | group | Activated | 3.467 | bias | 4.99E-07 | 124 (12) | |
| JUN | transcription regulator | Activated | 2.827 | bias | 9.74E-07 | 59 (7) | |
| PF4 | cytokine | Activated | 2.615 | bias | 9.93E-07 | 142 (16) | |
| HCAR2 | GPCR | Inhibited | -2.646 | bias | 9.93E-07 | ||
| EIF2AK2 | kinase | Activated | 3.742 | bias | 1.59E-06 | ||
| P2RX7 | ion channel | Activated | 2.599 | bias | 2.05E-06 | 62 (7) | |
| TNFSF14 | cytokine | Activated | 2.400 | bias | 2.11E-06 | 53 (7) | |
| TLR3 | TR | Activated | 2.775 | bias | 2.13E-06 | 174 (16) | |
| IL1β | 6.916 | cytokine | Activated | 3.199 | bias | 2.16E-06 | 75 (10) |
| JAG2 | growth factor | Inhibited | -3.130 | bias | 2.91E-06 | ||
| TNFRSF18 | TR | Inhibited | -2.236 | bias | 3.01E-06 | 51 (5) |
Fig. 4.
IPA analysis of upstream transcription regulators in PD-1H overexpressed monocytes. IPA was used to identify the transcription regulators with significant target gene overlap with changes in PD-1H overexpressed monocytes transcriptome. The histogram showed the most significant activators and inhibitors using a Z-score cut-off of ≥2 (for activators) or ≤-2 (for inhibitors).
We have previously reported that PD-1H over-expression in monocytes leads to activation and spontaneous secretion of multiple inflammatory cytokines. We also found that in chronically infected HIV patients, chronic inflammation is highly correlated with elevated PD-1H level [10]. Because we observed significant up-regulation of pro-inflammatory signaling pathways following PD-1H over-expression in monocytes in vitro, we wished to determine whether PD-1H is up-regulated in humanized BLT mice during sepsis, where inflammatory monocytes are known to play major pathogenic roles.
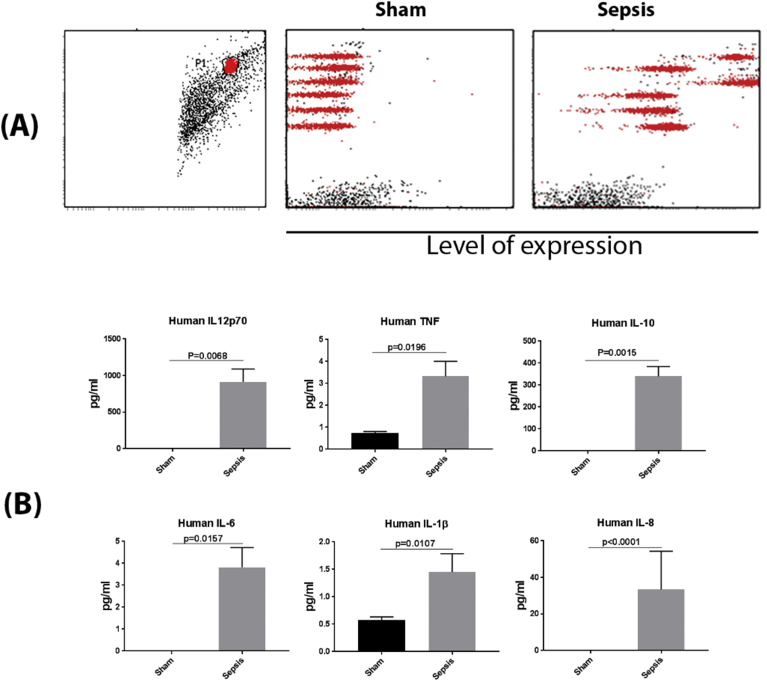
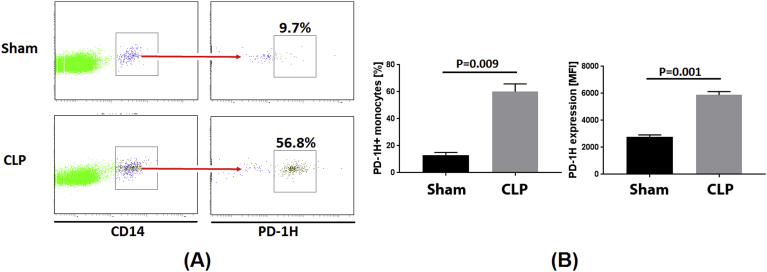
We generated BLT mice using severely immune-deficient non-obese diabetic (NOD)/SCID/IL2Rγ-/- mice. Reconstitution of human hematopoietic cells was done by fetal liver CD34 + HSC transplantation after surgical implantation of fetal liver and thymic tissue under the kidney capsule and was confirmed 12 weeks post-transplantation. Monocytes were well differentiated in these mice. After we sacrificed one mouse and obtained its spleen, we analyzed monocyte frequency by staining the spleen cells with CD45 and CD14 antibody, and found that 9.2% of the whole human cell population (CD45 staining) were monocytes (CD14 staining) (Fig. S1), similar to frequencies previously reported [16]. Sepsis was induced by cecal ligation and puncture (CLP), animals were sacrificed after 24 h and spleen cells evaluated for PD-1H expression on CD14 + gated cells within the human CD45 + cell population. As shown in Fig. 5A and B, all human inflammatory cytokines, including IL-12p70, IL-1β, IL-6, IL-8, IL-10 and TNF, were significantly increased in the sepsis model of BLT mice, as compared to sham controls. We then investigated how PD-1H expression in monocytes associates with cytokine levels. Mean basal levels of PD-1H positivity in CLP animals (56.8%) was approximately 6-fold greater than in sham treated controls (9.7%). The mean fluorescent intensity of PD-1H expression on monocytes was also higher in the CLP group (Fig. 6A and B). Consistent with our gene array analysis, our model of sepsis in vivo further demonstrates that PD-1H expression may function in activating monocytes and triggering inflammatory responses in monocytes to infection.
Fig. 5.
Inflammatory cytokine secretion in sepsis humanized BLT mice. The humanized BLT mouse Sepsis model was established using Cecal Ligation Puncture (CLP). After 24 h of establishment of sepsis model, blood was collected from sham-treated (n = 3) or sepsis mice (n = 7), and the sera were used to detect the inflammatory cytokines using CBA Human inflammatory cytokine beads array. Briefly, sera from mice was incubated with tagged beads as per manufacturer's instructions. Standards were used to determine concentrations of the cytokines. Six cytokines were detected. FCAP array v3 was used to analyze the data gated on beads (Red) with each line representing a cytokine scatter plot (from top to bottom: IL-8, IL-1β, IL-6, IL-10, TNF-α, and IL12-p70). Binding to cytokines led to displacement of these to the right side at varying intensities representing amounts of cytokine present in the sample. These amounts were calculated back of the standards used in the assay as per manufacturer's instructions. Black dots represent debris or non-bead events. (A) Representative result from one of the sham-treated or sepsis mouse. Left figure showed gating strategy. In the Sham and Sepsis samples, the red population respectively represent human IL-8, IL-1β, IL-6, IL-10, TNF-α and IL-12p70 from the top to the bottom. (B) Different cytokine levels in the sham-treated or sepsis mice.
Fig. 6.
PD-1H expression in sham-treated or sepsis mice. (A) Representative PD-1H expression level from one of the sham-treated or sepsis mouse. (B) Average percentages (left histogram) and MFI (right histogram) of PD-1H expression. CLP: Cecal Ligation and Puncture.
3. Discussion
Our study provides the first snapshot of the monocytic transcriptome by investigating genome-wide regulation of biological pathways in response to PD-1H overexpression in monocytes. Gene array analysis showed an overall pro-inflammatory response profile upon PD-1H expression. Additionally, we also observed significant up-regulation of PD-1H in an in vivo model of sepsis in a humanized mouse model, which corroborated the observation that PD-1H expression correlates with pro-inflammatory response. We conclude that PD-1H potentially activates monocytes and triggers systematic inflammatory response.
The most significantly up-regulated signaling pathway was TNF-α/NF-κB. TNF-α signaling results in a wide spectrum of outcomes from pro-survival/pro-inflammatory via downstream activation of NF-κB to pro-apoptotic/pro-necroptotic, via activation of the ripoptosome or activation of the necrosome, respectively. However, it appears likely that the primary outcome in response to PD-1H over-expression is pro-inflammatory.
Consistent with observed up-regulation of pro-inflammatory genes, there were many anti-inflammatory molecules found to be down-regulated. For instances, Hcar2 (hydroxycarboxylic acid receptor 2, also known as G-protein coupled receptor 109A or Gpr109a) has been shown to be an anti-inflammatory receptor and plays an anti-inflammatory role in retinal pigment epithelium and is upregulated in diabetes [17]. Interestingly, the down-regulated genes in the KEGG data set are all related to neuron sensing, most likely can be explained by down-regulating the G-protein coupled receptor pathways [11, 18, 19]. Another molecule, miR-155, acts as an anti-inflammatory factor in atherosclerosis-associated foam cell formation by repressing calcium-regulated heat stable protein 1 [20]. TAB1 is critical for activated macrophage survival [21]. IL1RN (interleukin-1 receptor antagonist) is reported to be associated with suppression of early carcinogenic events in human oral malignancies [22]. The down-regulation of anti-inflammatory molecules further demonstrates that PD-1H overexpression does trigger an inflammatory response in monocytes.
It is worth noting that our global transcriptome analysis of PD-1H overexpressing monocytes suggests that PD-1H may play a stimulatory role on monocyte activation. This result appears to contradict with studies in mouse models, in which PD-1H was identified as a co-inhibitory receptor suppressing T cell function. This is not necessarily surprising given that studies performed by the same group appear to have reported opposing results [3, 5]. Although the reason for this seeming discrepancy is not clear, it is possible that the interpretation of the results could vary depending on whether PD-1H serves as a receptor or as a ligand for the particular function studied. In our experimental studies, PD-1H appears to activate monocytes and enhance T cell responses. This is in agreement with the postulation by Flies et al., which states that PD-1H may potentially function as both a receptor and a ligand. It is also possible that the effect of PD-1h engagement on different cell types may differ. We have observed that PD-1H expression beyond a certain threshold is sufficient to activate monocytes, leading to spontaneous cytokine production [10]. In this setting, signaling via PD-1H appears to upregulate genes encoding pro inflammatory cytokines, as validated by a sepsis model in humanized BLT mice.
The exact mechanism of PD-1H mediated signaling is unclear. As both mouse and human myeloid populations express high densities of PD-1H, it is conceivable that in addition to PD-1H potentially serving as a ligand for T cells, it may also be involved in reverse signaling within the myeloid compartment. We have shown that transient overexpression of human PD-1H in CD14 + monocytes resulted in increased production of IL-6, IL-8, IL-1β, and TNFα. This activity requires the presence of the cytoplasmic domain of PD-1H, suggesting that PD-1H engagement on monocytes results in signal transduction. Thus, our results are in agreement with other reports that suggest PD-1H may work both as a receptor and as a ligand [3, 23].
Based on GSEA analysis, we hypothesize that the mechanism of how PD-1H regulates inflammatory response as follows: Although it is unclear if TNF-α is the primary driver of the inflammatory response or if other cytokines are more critical, our results suggest that PD-1H can directly or indirectly activate TNF- α expression, and sequentially activate NF-κB, as well as other pro-inflammatory signaling pathways, leading to production of a number of pro-inflammatory cytokines, which ultimately leads to inflammation. Therefore, it would be of interest to determine what effects blockade of TNF-α or other critical cytokines have on the response of cells to PD-1H overexpression.
4. Methods
4.1. Ethics statement
All work including animal studies were conducted following the guidelines for the Care and Use of Laboratory Animals of National Research Council, USA. The authors received approval from the IACUC of the Texas Tech University Health Sciences Center (protocol number 8007).
4.2. Cell culture and nucleofection
Monocytes were isolated from PBMCs (purchased from Astarte Biologics, Inc.) using EasySep human monocytes enrichment kit without CD16 depletion (StemCell Technologies, Inc.) as per the manufacturer's instructions. The viability of cells after isolation was 90% and the purity was ≥95% according to flow cytometry analysis staining with human CD14 antibody. One million of monocytes were nucleofected with 10 μg of pLVX-PD-1H plasmid (or pLVX vector plasmid as negative control), or siPD-1H (and siScramble as negative control). The above experiments were duplicated. Lonza 4D nucleofection system was used for the nucleofection according to parameters recommended by the manufacturer. The nucleofected monocytes was cultured in PBMC supplemented with 5% human serum (heat-inactivated) for 24 h. Then, the cells were harvested and sorted by FACS using the endogenous GFP expression by the pLVX vector. The GFP-positive cells were then used for RNA extraction. Monocytes were isolated from three different donors, and the experiments were repeated twice each time with the same three donors.
4.3. Total RNA extraction and IPA analysis
The total RNA was isolated using RNeasy kit (Qiagen, Germantown, MD) and the concentration was measured by Nanodrop. The RNA samples were sent to DNA microarray core facility at UCLA for microarray analysis using human Illumina HT-12 v4 Expression BeadChip. The microarray data were then run an IPA analysis to characterize the global expression profile and the fold changes, activation prediction were calculated using the IPA software.
4.4. Gene set enrichment analysis (GSEA)
Differentially expressed genes between two conditions, PD-1H over-expression vs. vector control or PD-1H knockdown (siPD-1H) vs. non-targeting control (siScramble), were subjected to GSEA analysis (http://software.broadinstitute.org/gsea/index.jsp). The enriched signaling pathways and individual genes were defined in the biological database KEGG and BioCarta.
4.5. Sepsis model in humanized mouse model
NOD.cg PrkdcscidIL2rgtm/Wjl/Sz (NOD/SCIDIL2rγ-/-, NSG mice) mice were purchased from the Jackson Laboratory (Bar Harbor, ME). BLT mice were generated as described [24]. Briefly, irradiated (180 rads) NSG mice were implanted with human liver and thymus tissue (Advanced Bioscience Resources, Alameda, CA). Fetal liver tissue was also minced and used for isolation of CD34 + HSCs. 2 × 105 fetal liver HSCs were injected intravenously into the animals to generate humanized BLT mice. After 12 weeks, the engraftment of human cells was determined by flow cytometric analysis of peripheral blood mononuclear cells (PBMCs) stained with human CD45, CD3, CD4, CD8, and CD14 antibodies. The humanized mouse Sepsis model was established using Cecal Ligation Puncture (CLP) procedure as described elsewhere [25].
4.6. PD-1h expression on human monocytes in sepsis mice and inflammatory cytokine assay
We used 3 BLT mice for Sham control group and 7 BLT mice for CLP group (justified with power analysis). After generation of sepsis mice for 24 h, the blood was collected to analyze the PD-1H expression in monocytes by staining the ficoll-isolated human PBMCs with anti-human PD-1H antibody (R&D systems, Inc.), as well as human CD45, CD3, CD4, CD8, and CD14 antibodies. The PD-1H expression was gated on CD14 positive cells. The sera were separated from the blood and were used to analyze inflammatory cytokines using The CBA Human Inflammatory Cytokines Kit (BD Biosciences Inc.) as per the manufacturer's instructions. The cytokine concentration was calculated using the FCAP software according to the instructions.
4.7. Statistical analysis
Data in Figs. 4 and 5 were analyzed using prism 6.0 (GraphPad). For comparison of data with two variables, non-parametric Mann- Whitney test was used. For all statistical tests, p value <0.05 were considered significant. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Declarations
Author contribution statement
Guohua Yi, Premlata Shankar, N. Manjunath: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Preeti Bharaj, Chunting Ye, Sean Petersen: Performed the experiments.
Qianghu Wang: Performed the experiments; Analyzed and interpreted the data.
Baoli Hu: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work is supported by a grant from NIH (R01 HL125016) to G.Y.
Competing interest statement
The authors declare no conflict of interest.
Additional information
Acknowledgements
Research reported in this publication was supported by the National Heart, Lung, And Blood Institute of the National Institutes of Health under Award Number R01HL125016. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Contributor Information
Premlata Shankar, Email: shankarprema66@gmail.com.
Guohua Yi, Email: g.yi@ttuhsc.edu.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Collins M., Ling V., Carreno B.M. The B7 family of immune-regulatory ligands. Genome Biol. 2005;6(6):223. doi: 10.1186/gb-2005-6-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spranger S. Tumor and host factors controlling antitumor immunity and efficacy of cancer immunotherapy. Adv. Immunol. 2016;130:75–93. doi: 10.1016/bs.ai.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flies D.B. Cutting edge: a monoclonal antibody specific for the programmed death-1 homolog prevents graft-versus-host disease in mouse models. J. Immunol. 2011;187(4):1537–1541. doi: 10.4049/jimmunol.1100660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L. VISTA, a novel mouse Ig superfamily ligand that negatively regulates T cell responses. J. Exp. Med. 2011;208(3):577–592. doi: 10.1084/jem.20100619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flies D.B. Coinhibitory receptor PD-1H preferentially suppresses CD4(+) T cell-mediated immunity. J. Clin. Invest. 2014;124(5):1966–1975. doi: 10.1172/JCI74589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flies A.S. Comparative analysis of immune checkpoint molecules and their potential role in the transmissible tasmanian devil facial tumor disease. Front. Immunol. 2017;8:513. doi: 10.3389/fimmu.2017.00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L. Disruption of the immune-checkpoint VISTA gene imparts a proinflammatory phenotype with predisposition to the development of autoimmunity. Proc. Natl. Acad. Sci. U. S. A. 2014;111(41):14846–14851. doi: 10.1073/pnas.1407447111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muralidharan S., Mandrekar P. Cellular stress response and innate immune signaling: integrating pathways in host defense and inflammation. J. Leukoc. Biol. 2013;94(6):1167–1184. doi: 10.1189/jlb.0313153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lines J.L. VISTA is an immune checkpoint molecule for human T cells. Canc. Res. 2014;74(7):1924–1932. doi: 10.1158/0008-5472.CAN-13-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bharaj P. Characterization of programmed death-1 homologue-1 (PD-1H) expression and function in normal and HIV infected individuals. PLoS One. 2014;9(10):e109103. doi: 10.1371/journal.pone.0109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciechanover A., Orian A., Schwartz A.L. The ubiquitin-mediated proteolytic pathway: mode of action and clinical implications. J. Cell. Biochem. 2000;34(Suppl):40–51. doi: 10.1002/(sici)1097-4644(2000)77:34+<40::aid-jcb9>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 12.Shakola F., Suri P., Ruggiu M. Splicing regulation of pro-inflammatory cytokines and chemokines: at the interface of the neuroendocrine and immune systems. Biomolecules. 2015;5(3):2073–2100. doi: 10.3390/biom5032073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qureshi N., Morrison D.C., Reis J. Proteasome protease mediated regulation of cytokine induction and inflammation. Biochim. Biophys. Acta. 2012;1823(11):2087–2093. doi: 10.1016/j.bbamcr.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suto N., Ikura K., Sasaki R. Expression induced by interleukin-6 of tissue-type transglutaminase in human hepatoblastoma HepG2 cells. J. Biol. Chem. 1993;268(10):7469–7473. [PubMed] [Google Scholar]
- 15.Jang G.Y. Transglutaminase 2 suppresses apoptosis by modulating caspase 3 and NF-kappaB activity in hypoxic tumor cells. Oncogene. 2010;29(3):356–367. doi: 10.1038/onc.2009.342. [DOI] [PubMed] [Google Scholar]
- 16.Billerbeck E. Humanized mice efficiently engrafted with fetal hepatoblasts and syngeneic immune cells develop human monocytes and NK cells. J. Hepatol. 2016;65(2):334–343. doi: 10.1016/j.jhep.2016.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gambhir D. GPR109A as an anti-inflammatory receptor in retinal pigment epithelial cells and its relevance to diabetic retinopathy. Invest. Ophthalmol. Vis. Sci. 2012;53(4):2208–2217. doi: 10.1167/iovs.11-8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao C.A., Carlson J.R. Role of G-proteins in odor-sensing and CO2-sensing neurons in Drosophila. J. Neurosci. 2010;30(13):4562–4572. doi: 10.1523/JNEUROSCI.6357-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Y., Thathiah A. Regulation of neuronal communication by G protein-coupled receptors. FEBS Lett. 2015;589(14):1607–1619. doi: 10.1016/j.febslet.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Li X. miR-155 acts as an anti-inflammatory factor in atherosclerosis-associated foam cell formation by repressing calcium-regulated heat stable protein 1. Sci. Rep. 2016;6:21789. doi: 10.1038/srep21789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mihaly S.R. Activated macrophage survival is coordinated by TAK1 binding proteins. PLoS One. 2014;9(4):e94982. doi: 10.1371/journal.pone.0094982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shiiba M. Interleukin-1 receptor antagonist (IL1RN) is associated with suppression of early carcinogenic events in human oral malignancies. Int. J. Oncol. 2015;46(5):1978–1984. doi: 10.3892/ijo.2015.2917. [DOI] [PubMed] [Google Scholar]
- 23.Nowak E.C. Immunoregulatory functions of VISTA. Immunol. Rev. 2017;276(1):66–79. doi: 10.1111/imr.12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishikawa F. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor {gamma} chain(null) mice. Blood. 2005;106(5):1565–1573. doi: 10.1182/blood-2005-02-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toscano M.G., Ganea D., Gamero A.M. Cecal ligation puncture procedure. J. Vis. Exp. 2011;(51) doi: 10.3791/2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.