Abstract
Fibronectin (Fn) is an extracellular matrix protein that orchestrates complex cell adhesion and signaling through cell surface integrin receptors during tissue development, remodeling, and disease, such as fibrosis. Fn is sensitive to mechanical forces in its tandem type III repeats, resulting in extensive molecular enlongation. As such, it has long been hypothesized that cell- and tissue-derived forces may activate an “integrin switch” within the critical integrin-binding ninth and 10th type III repeats—conferring differential integrin-binding specificity, leading to differential cell responses. Yet, no direct evidence exists to prove the hypothesis nor demonstrate the physiological existence of the switch. We report direct experimental evidence for the Fn integrin switch both in vitro and ex vivo using a scFv engineered to detect the transient, force-induced conformational change, representing an opportunity for detection and targeting of early molecular signatures of cell contractile forces in tissue repair and disease.
Keywords: mechano-biology, antibody phage display, integrins, fibronectin, fibrosis, angiogenesis
Graphical abstract
The extracellular matrix (ECM) forms the complex niche of structural elements surrounding cells in vivo. Cells interact with and are instructed by the ECM via cellular structures known as focal adhesions, large protein complexes composed of transmembrane receptors (integrins) and intracellular adaptor proteins that mechanically couple the cell’s cytoskeleton to fibrillar ECM proteins such as fibronectin (Fn). Protein–protein interactions within focal adhesions are dynamic; mechanical forces play important roles for focal adhesion maturation and development, as well as for force-sensitive cell signaling via mechano-sensory proteins. Recent work has shown that conformations of both intracellular focal adhesion constituents (e.g., vinculin, integrins)1–3 as well as extracellular components (e.g., Fn)4–6 are altered by forces transmitted to and from the ECM. In the latter case, previous work from our group and others demonstrates that Fn within the ECM exhibits distinct but undefined altered structural states in response to cellular forces both in vitro and in vivo.5,7
Fn is composed of three types of tandem repeating units, each containing two antiparallel β-sheets. Type I and II repeats are structurally stabilized by disulfide bonds, whereas type III repeats are stabilized only by hydrogen bonding and van der Waals forces, making them sensitive to unfolding due to physiologically relevant forces.8–12 These findings, when coupled with the active role of Fn’s ninth and 10th type III repeats (FnIII9–10) in mediating integrin-specific interactions, inspired the theory that mechanical forces could trigger a “switch” in the integrin-binding profile of Fn.8 Fn–integrin interactions are known to drive critical cell behaviors and are mediated primarily through the canonical and promiscuous integrin-binding sequence Arg-Gly-Asp (RGD) within the 10th type III repeat.13 A subset of integrins, including integrin α5β1, is additionally dependent on the sequence motif PHSRN within the neighboring ninth type III repeat.14–17 Integrin specificity to Fn can be modulated in vitro by altering the structural stability of the integrin-binding domain (i.e., the ninth and 10th type III repeats) via directed mutation,18 resulting in the regulation of developmentally and pathologically relevant cell differentiation pathways19,20 and, importantly, cellular responses to microenvironmental mechanics (e.g., stiffness).21 Despite these findings, the integrin switch theory and its potential relevance to biological processes in vivo remain undefined.
To prove the existence of Fn’s integrin switch and to demonstrate its relevance in vivo, we sought to develop single-chain antibodies (scFv) capable of detecting a ∼1 nm extension in the conformation of Fn’s integrin-binding domain through directed evolution and screening against recombinant Fn fragments that mimic the force-induced structural states predicted in silico. Engineered Fn fragments spanning the ninth and 10th type III repeats of Fn were produced as reported,20 in which a Leu1408Pro mutation18 was created to stabilize the spatial and angular orientation of PHSRN with respect to RGD (denoted FnIII9*10). A second extended conformation variant was created by inserting a 4-glycine (4G) linker between the ninth and 10th type III repeats, increasing the separation between the PHSRN and RGD sites from 3.4 nm to approximately 4.3 nm (denoted FnIII9-4G-10; Figure 1A). Surface plasmon resonance (SPR) experiments demonstrate that the insertion of the 4G linker is sufficient to decrease in vitro α5β1 integrin-binding affinity from 12 nM to ∼2.5 μM, whereas integrin αvβ3 affinity predictably remains relatively constant at ∼40 nM (Figure 1B). Immunofluorescence staining of integrins on human foreskin fibroblasts cultured on these Fn fragments confirms that this molecular modification results in a switch in the integrin-binding profile from α5β1 and αvβ3 to predominantly αvβ3 at the cellular level (Figure 1C and Supporting Information Figure S1), supporting earlier reports of FnIII9*10 fragment’s ability to engage α5β1 integrins preferentially.20 Cell adhesion assays were additionally performed on decellularized Fn-rich ECMs formed on elastic PDMS membranes, which enable artificial straining or relaxing. Cells exclusively expressing α5β1 integrin22 adhere to a lesser extent on strained Fn ECMs compared to relaxed, whereas cells exclusively expressing αvβ3 do not exhibit differential adhesion (Figure 1D,E). The overall enhanced adhesion of cells expressing α5β1 is likely due to binding the PHSRN sequence, which enhances cell adhesion over binding of RGD alone.16 Furthermore, the decreased adhesion of cells exclusively expressing αvβ3 could be due to the transfection efficiency of the ITGAV vector into CHO B2 cells, which do not naturally express α5 or αv. By contrast, the CHO K1 cells endogenously express hamster α5β1 integrin.22 Taken together, these results support the hypothesis of a force-sensitive “integrin switch” in Fn fibers, yet fall short of demonstrating a specific conformational change in the integrin-binding domain.
Figure 1.
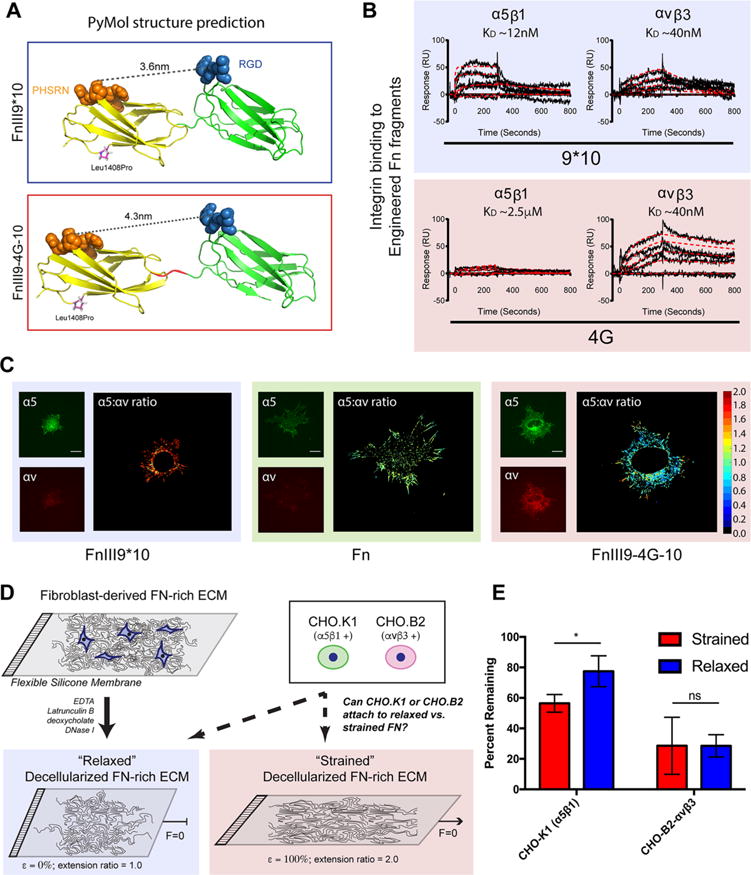
Fibronectin strain drives differential integrin affinity. (A) PyMol structure predictions of engineered recombinant fragments of Fn’s integrin-binding domain. FnIII9*10, represents a stabilized native structure through a Leu-to-Pro point mutation at position 1408. In this conformation, the PHSRN-to-RGD distance is approximately 36 Å. FnIII9–4G-10 is a mutation of the FnII9*10 variant that contains a 4xGly insertion between the 9th and 10th type III repeats. This mutation increases domain separation and the PHSRN-to-RGD distance to approximately 43 Å. These fragments have been employed in the past to predict potential biological consequences of the theorized integrin switch. B) SPR binding characterization of integrins α5β1 and αVβ3 to the FnIII9*10 and FnIII9–4G-10 fragments demonstrate a nearly complete loss of binding of α5β1 upon domain separation, whereas αvβ3 binding is predictably unaffected. Black curves show experimental sensogram traces, and red curves show computationally fitted data. Equilibrium dissociation constants (KD) are shown above the curves. (C) Fibroblasts cultured on FnIII9*10 and FnIII9-4G-10 fragments and immunostained for integrin α5 and αv demonstrate the fragments’ capability of skewing cellular binding toward specific integrins corroborating SPR analysis. Scale bar is 20 μm. (D) Schematic of cell attachment experiments designed to demonstrate the impact of strain of a decellularized Fn-rich matrix on integrin binding. (E) Quantitation of cell adhesion on strained and relaxed Fn-rich ECM demonstrates that strain of the ECM significantly impacts cell binding via α5β1 but not αvβ3. Error bars reflect SD, *p < 0.05, N = 6, ANOVA with Tukey’s post-test.
By performing phage display panning and selection using scFv antibody libraries23,24 on the recombinant Fn fragments FnIII9*10 and FnIII9-4G-10, we were able to isolate specific scFv with both high binding affinities and discrimination between the two model Fn fragments. Forty antibody clones were isolated, and eight were carried forward for subsequent validation. Clone H5 displayed high selectivity toward the extended (FnIII9-4G-10), but not the native, stabilized (FnIII9*10) conformation (Supporting Information Figure S2).
H5 binding kinetics were evaluated using SPR (Figure 2A,B). Equilibrium dissociation constants (KD) were determined to be 107 nM for H5 binding to FnIII9*10 and 16 nM for binding to FnIII9-4G-10. Selectivity of H5 binding to the FnIII9-4G-10 fragment was thus approximately an order of magnitude greater compared to that of FnIII9-10. Competitive ELISAs of H5 binding to FnIII9-4G-10 in the presence of increasing concentrations of soluble Fn fragments displaying only the ninth (FnIII6-9) or 10th (FnIII10-14) type III repeats demonstrated that increasing concentrations of FnIII6-9 blocked binding of H5 to Fn9-4G-10, which was not observed with increasing concentrations of FnIII10-14 (Figure 2C). Additionally, SPR analysis of H5 binding to these fragments revealed that the equilibrium dissociation constant (KD) for H5 binding to FnIII6-9 was 34.8 nM, a higher affinity interaction than H5 binding to FnIII10-14 (5.1 μM; Figure 2D,E). Together these data indicate that H5’s epitope is located within Fn’s ninth type III repeat. Binding characteristics of H5 to Fn and the Fn fragments were then evaluated by Western blot analysis in parallel with a commercially available control antibody clone (HFN7.1) against Fn’s integrin-binding domain (Figure 2F–K). The increased affinity of H5 to FnIII9-4G-10 versus FnIII9*10 is observed in ELISA but not in dot blots or traditional denaturing Western blots, suggesting that the epitope bound by H5 is likely a linear peptide sequence and not a more complex 3D structure. We hypothesized that if H5 bound to the open (or “off”) state of the switch that it should selectively inhibit αvβ3–Fn interactions as the antibody only weakly recognizes the closed/“on” α5β1-binding conformation. Standard cell attachment assays to Fn-adsorbed substrates, which present multiple conformations of Fn due to surface-mediated unfolding, confirm the H5 clone selectively inhibits Fn–integrin αvβ3, but not α5β1, interactions with Fn (Supporting Information Figure S3).
Figure 2.

H5 antibody recognizes conformational change of the integrin-binding domain by binding an epitope specifically on the ninth type III repeat that can be exposed by denaturation. (A,B) SPR analysis of binding of H5 antibody to recombinant FnIII9*10 (A) and FnIII9-4G-10 (B) fragments demonstrates the conformation selectivity of its binding. H5 binds preferentially to the molecularly extended conformation. Black lines show experimental sensogram traces, and red lines show fitted data. Equilibrium dissociation constants (KD) are shown above the curves. (C) Competitive binding of the H5 antibody to FnIII9-4G-10 in the presence of soluble FnIII9 (FnIII6-9) and FnIII10 (FnIII10-14) fragments. FnIII9-4G-10 was immobilized on ELISA plates, and the H5 antibody was co-incubated with the indicated soluble Fn fragments prior to incubation with FnIII9-4G-10. N = 8 for each group; error bars reflect SEM. (D,E) Domain mapping studies were performed by SPR using Fn fragments including either the ninth type III repeat (FnIII6-9) (B) or the 10th type III repeat (FnIII10-14) (C). Binding of H5 was only observed by SPR when fragments including the ninth Fn type III repeat were immobilized, suggesting that the epitope for H5 is located within FnIII9. (F) ELISA of H5 binding to full-length Fn and Fn fragments. H5 bound increasingly to increasing amounts of surface-adsorbed FnIII9-4G-10 to a significantly greater degree than FnIII9*10 or full-length Fn at all concentrations (p < 0.0001) (two-way ANOVA with Tukey’s post-test, N = 3). (G,H) Nitrocellulose dot blot (G) and Western blot (H) of H5 binding to full-length Fn and Fn fragments. Under these denaturing conditions, H5 binds FnIII9-4G-10 and FnIII9*10 to a similar degree. (I–K) ELISA (I), dot blot (J), and Western blot (K) of HFN7.1, a commercially available antibody targeting FnIII9-10. In the ELISA, HFN7.1 only bound FnIII9-4G-10 to a significantly greater degree than FnIII9*10 at 1 μM (p < 0.05) and was not significant at higher concentrations (two-way ANOVA with Tukey’s post-test, N = 3). HFN7.1 did not show preferential affinity for FnIII9*10 or FnIII9-4G-10 in the dot blot or Western blot.
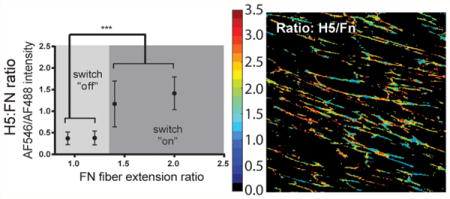
Validation of H5’s recognition to FnIII9 and its selective inhibition of αvβ3 binding to Fn-adsorbed surfaces led us to use the H5 clone to test the hypothesis that Fn’s integrin-binding domain displays a force-induced conformational change (i.e., extension) when Fn fibers are mechanically strained. Manually deposited Fn fibers on micropatterned and stretched PDMS membranes display unfolding of Fn’s type III repeats, demonstrated by the exposure of cryptic cysteines.5,6,25 Using this system, the binding of H5 to Fn fibers under increasing strain was found to display switch-like behavior, with H5 signal increasing significantly at an extension ratio of ∼1.4–1.5, and plateauing at additional strain (Figure 3A). This discrete signature suggests an abrupt transition from a native folded to extended conformation upon the application of tensile force.
Figure 3.
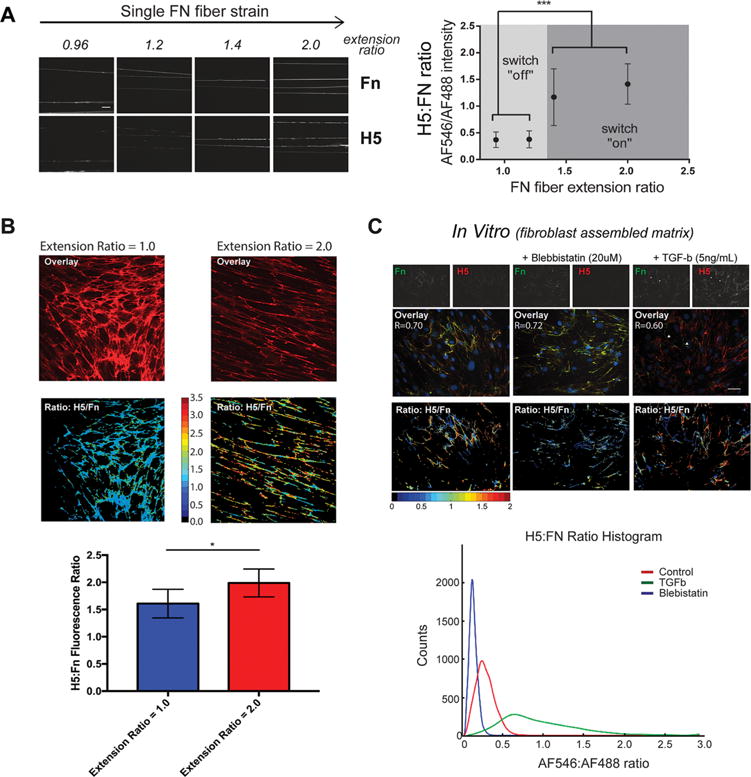
H5 antibody is capable of discriminating conformational changes of Fn’s integrin-binding domain (i.e., the integrin switch) in multiple model systems. (A) Staining of H5 on in vitro Fn fibers deposited on PDMS membranes demonstrates the strain-dependent conformation of Fn’s integrin-binding domain. A discrete transition from modest to high binding occurs between an extension ratio of 1.25 and 1.5. N = 10, error bars reflect SD, ***p < 0.001, one-way ANOVA with Tukey’s post-test. (B) Staining of strained or relaxed decelluarized ECM assembled by human foreskin fibroblasts by H5 indicates that the Fn integrin-binding domain within complex (anisotropic) Fn-rich ECM undergoes a conformational change in response to strain. N = 6, error bars reflect SD, *p < 0.01, Wilcoxon sum-rank test. (C) Furthermore, the conformation of Fn’s integrin-binding domain within primary lung fibroblast-laden Fn-rich ECM is sensitive to modulation of fibroblast contractility through agonists (TGF-β) and inhibitors (blebbistatin). Scale bar, 50 μm.
Similarly, on decellularized Fn-rich ECMs assembled on elastic PDMS membranes (Figure 3B), H5 was able to both globally discriminate strained from relaxed Fn ECMs and locally discriminate between fibers in both a relaxed and strained ECM. H5 staining intensity of Fn-rich ECM also correlated strongly with the contractile state of resident fibroblasts, as induced by the addition of contractility agonists (TGF-β) or inhibitors (blebbistatin), suggesting that cell-generated forces are sufficient to activate the integrin switch (Figure 3C).
We then used H5 to probe Fn integrin-binding domain conformations within native tissues. Stained tissues from two model systems were used: a mouse model of pulmonary fibrosis26,27 and a mouse model of postnatal retinal angiogenesis. Using frozen tissue sections from saline control and bleomycin-treated fibrotic mouse lungs, staining of H5, but not a negative control scFv, localized to fibrillar Fn (assessed using a polyclonal anti-Fn antibody) within the ECM (Figure 4 and Supporting Information Figure S4). Ratiometric image analysis revealed that the H5 staining patterns within these lung samples were spatially distinct, suggesting distinct conformational states of Fn within the matrix. Notably, the fibrotic time points (2–8 weeks) show areas of high H5/Fn ratio, correlating with the severity of fibrosis observed in the corresponding H&E staining. By week 10, at which point the fibrosis is resolved, the H5/Fn ratio resembles that of the saline control. H5 was also used to probe the Fn-rich ECM during postnatal retinal vascularization. The retinas of newborn mice are avascular but become vascularized in a reproducible manner over the first 10 days after birth.28 During angiogenesis, migration of endothelial tip cells is guided by fibronectin fibers deposited by the astrocytic network.29 H5/Fn ratio was observed to be increased at the filopodia of tip cells, suggesting that cell-generated forces during vascular expansion is sufficient to alter the conformation of the integrin-binding domain of surrounding Fn fibers. In contrast, H5 showed minimal staining in regions of mature vessels (Figure 5).
Figure 4.

Fn’s integrin mechano-switch displays spatially distinct patterns of activation in a model of resolving lung fibrosis. (A–D) Mouse lung tissue sections (scale bar = 50 μm) were immunostained for H5 (red), Fn (green), and DNA (blue) at the indicated time points after bleomycin-induced fibrotic injury. In this model, bleomycin induces fibrosis by 14–21 days, and is typically resolved by 56 days. H5/Fn ratio images displaying the heterogeneity of Fn fiber conformation within the ECM at the tissue scale are shown, along with H&E staining of a corresponding serial section at the same time points. The early repair (B) and fibrotic (C) time points show areas of higher H5/Fn ratio, indicating the unfolding of Fn’s integrin-binding domain, perhaps indicative of active fibrosis. By resolution (D), the H5/Fn ratiometric image resembles that of the saline control.
Figure 5.
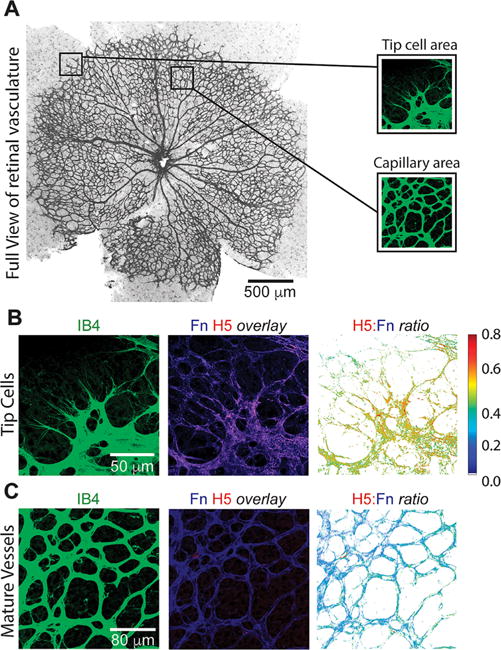
Fn’s integrin mechano-switch is activated at tip cells during retinal angiogenesis. (A) Whole mount immunostaining for retinal endothelial cells during postnatal retinal angiogenesis with tip cell area and capillary area (i.e., mature vessels) identified (scale bar = 500 μm). (B) Tip cell region (scale bar = 50 μm) and (C) mature vessels (scale bar = 80 μm) were immunostained with isolectin B4 (IB4, green) and H5 (red) and anti-Fn (blue) at postnatal day 6. Tip cells and blood vessels were visualized using isolectin B4. H5/Fn ratiometric images were generated for each region. The tip cell area (B) shows regions of high H5/Fn ratio, suggestive of endothelial tip cell force generation during angiogenesis. The mature vessel area, where forces are predicted to be low, displays a low H5/Fn ratio.
These combined results both demonstrate the in vivo existence and activation of the long-theorized Fn conformational switch within the integrin-binding domain and suggest its influence in skewing integrin specificity in both developmental processes, as well as in pathological tissue fibrosis. H5 thus represents an attractive approach to detecting and targeting key developmental and disease processes.
Reports have suggested that the relative separation distance between the “synergy” PHSRN sequence in the ninth Fn type III repeat and the RGD site in the 10th Fn type III repeat is critical for engagement and activation of integrin α5β120 and α3β1,30 with an optimal PHSRN-RGD distance of 3.7 nm for high affinity integrin α5β1 engagement.31 Furthermore, recent findings from the Smith group demonstrate that Fn fiber extension decreases cell spreading and adhesion,32 suggesting a mechanism by which Fn strain regulates the affinity of cell surface receptors, such as integrins, involved in cell adhesion. These observations led us to strive for the development of a molecular probe that would help prove the long-standing hypothesis8 and establish the physiological/pathophysiological relevance of this direct mechano-transductive event.
The development of conformation-specific antibodies by phage display is well-established, as work by Lefkowitz and co-workers have used phage display to isolate a conformation specific Fab to activated β-arrestin-1.33 Yet, the specific challenges of our goal were that (1) the conformational change of the integrin-binding domain is due to the application of force, (2) the application of force to Fn fibers leads to multiple conformational changes along the length of the 440 kDa protein that are impossible to distinguish, and (3) the conformational change is highly labile due to the ability of Fn type III repeats to refold in the absence of force. Here, we utilized predicted structures from steered molecular dynamics simulations coupled with molecular engineering to produce a mimetic of the strained integrin-binding domain in order to perform phage display to discover the H5 clone. It is likely that the two model Fn fragments differ not only in separation between RGD and PHSRN but also in relative conformational stability. FnIII9*10 is stabilized by a Leu1408Pro mutation between FnIII9 and FnIII10,18 whereas FnIII-4G-10 is separated by a 4-glycine linker between the two domains. Polyglycine sequences are commonly used in the design of flexible linker regions.34 In this context, the flexibility conferred by the 4-glycine linker may recapitulate the inherent sensitivity of FnIII repeats to force-mediated unfolding.8–12
One application of the H5 scFv could be to probe pathologic ECMs, specifically tumor stroma and fibrotic ECMs which contain highly contractile myofibroblasts. Recent reports suggest that αv integrins on myofibroblasts are implicated in fibrogenesis in a broad range of fibrotic diseases, and that pharmacological blockade of αv integrins ameliorates liver and lung fibrosis.35 The data regarding H5 staining of bleomycin-treated lungs are particularly interesting in the context of idiopathic pulmonary fibrosis (IPF), a fatal form of progressive lung fibrosis in humans. The lungs of IPF patients are mechanically and biochemically heterogeneous, with areas of soft, normal lung tissue and stiffer regions of mature fibrosis. The H5 scFv may be used to delineate regions of high ECM strain that also present an enhanced αv integrin-binding character due to the conformation of the integrin-binding domain, perhaps indicative of ongoing fibrosis.
The ability of the H5 antibody to extract structural information from the ECM was also demonstrated in a model of retinal angiogenesis, the process by which new blood vessels form by from endothelial sprouting.36 In mouse tissue sections, regions of high H5/Fn ratio were found at the extensions of endothelial tip cells, suggesting that Fn is unfolded in these regions. Fn is known to be a mediator of retinal angiogenesis, wherein astrocytes deposit fibronectin prior to differentiation of angioblasts to endothelial cells.29 Our results build on this foundation by suggesting that forces from endothelial tip cells unfold Fn, presenting an αvβ3 binding character within the provisional matrix that may influence the formation of new blood vessels, a result further supported by the work of Segura et al.37 using engineered Fn fragments of the integrin-binding domain in reparative angiogenesis.
We have developed a conformation-sensitive single-chain antibody (clone H5) to the integrin-binding FnIII9-10 domain of Fn and demonstrated its mechano-sensitive binding to Fn in multiple model systems in vitro and ex vivo. We believe these force-sensitive conformational changes observed in the integrin-binding domain of Fn are evidence of the long theorized Fn “integrin-switch” which likely regulate integrin-specific cell responses based on controlling the presentation and accessibility of Fn epitopes in vivo and in engineered contexts. We also believe that H5 specifically detects a force-induced conformational change within a protein. As mechanics of tissues are becoming increasingly implicated in pathogenesis of fibrotic diseases, there is a nascent opportunity to explore targeting the mechano-chemical character of ECM as a paradigm for tissue imaging and disease diagnosis.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health, National Heart, Lung and Blood Institute (R01HL127283) to T.H.B., the National Science Foundation Graduate Research Fellowship Program (DGE-1650044) to J.N., and the Swedish Cancer Society, the Swedish Childhood Cancer Foundation, Cancer Society of Stockholm, Swedish Heart and Lung foundation and the Swedish Research Council to L.H.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.7b02755.
Experimental details and Figures S1–S4 (PDF)
ORCID
Ashley C. Brown: 0000-0001-6995-1785
Thomas H. Barker: 0000-0002-3218-8111
Notes
The authors declare no competing financial interest.
References
- 1.Grashoff C, Hoffman BD, Brenner MD, Zhou RB, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, Schwartz MA. Measuring Mechanical Tension Across Vinculin Reveals Regulation of Focal Adhesion Dynamics. Nature. 2010;466:263–U143. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carisey A, Tsang R, Greiner AM, Nijenhuis N, Heath N, Nazgiewicz A, Kemkemer R, Derby B, Spatz J, Ballestrem C. Vinculin Regulates the Recruitment Release of Core Focal Adhesion Proteins in a Force-Dependent Manner. Curr Biol. 2013;23:271–81. doi: 10.1016/j.cub.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu J, Luo BH, Xiao T, Zhang C, Nishida N, Springer TA. Structure of a Complete Integrin Ectodomain in a Physiologic Resting State Activation and Deactivation by Applied Forces. Mol Cell. 2008;32:849–61. doi: 10.1016/j.molcel.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemmon CA, Ohashi T, Erickson HP. Probing the Folded State of Fibronectin Type III Domains in Stretched Fibrils by Measuring Buried Cysteine Accessibility. J Biol Chem. 2011;286:26375–82. doi: 10.1074/jbc.M111.240028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao LZ, Zeller MK, Fiore VF, Strane P, Bermudez H, Barker TH. Phage-Based Molecular Probes that Discriminate Force-Induced Structural States of Fibronectin in vivo. Proc Natl Acad SciUS A. 2012;109:7251–7256. doi: 10.1073/pnas.1118088109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith ML, Gourdon D, Little WC, Kubow KE, Eguiluz RA, Luna-Morris S, Vogel V. Force-Induced Unfolding of Fibronectin in the Extracellular Matrix of Living Cells. PLoS Biol. 2007;5:e268. doi: 10.1371/journal.pbio.0050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandler EM, Saunders MP, Yoon CJ, Gourdon D, Fischbach C. Adipose Progenitor Cells Increase Fibronectin Matrix Strain Unfolding in Breast Tumors. Phys Biol. 2011;8:015008. doi: 10.1088/1478-3975/8/1/015008. [DOI] [PubMed] [Google Scholar]
- 8.Krammer A, Craig D, Thomas WE, Schulten K, Vogel V. Structural Model for Force Regulated Integrin Binding to Fibronectin’s RGD-synergy site. Matrix Biol. 2002;21:139–147. doi: 10.1016/s0945-053x(01)00197-4. [DOI] [PubMed] [Google Scholar]
- 9.Gee EP, Ingber DE, Stultz CM. Fibronectin Unfolding Revisited: Modeling Cell Traction-Mediated Unfolding of the Tenth Type-III Repeat. PLoS One. 2008;3:e2373. doi: 10.1371/journal.pone.0002373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L, Huang HH, Badilla CL, Fernandez JM. Mechanical Unfolding Intermediates Observed by Single-Molecule Force Spectroscopy in a Fibronectin Type III Module. J Mol Biol. 2005;345:817–26. doi: 10.1016/j.jmb.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 11.Craig D, Gao M, Schulten K, Vogel V. Tuning the Mechanical Stability of Fibronectin Type III Modules Through Sequence Variations. Structure. 2004;12:21–30. doi: 10.1016/j.str.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 12.Craig D, Krammer A, Schulten K, Vogel V. Comparison of the Early Stages of Forced Unfolding for Fibronectin Type III Modules. Proc Natl Acad SciUS A. 2001;98:5590–5595. doi: 10.1073/pnas.101582198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruoslahti E, Pierschbacher MD. New Perspectives in Cell Adhesion: RGD Integrins. Science. 1987;238:491–7. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 14.Mardon HJ, Grant KE. The Role of the Ninth Tenth Type III Domains of Human Fibronectin in Cell Adhesion. FEBS Lett. 1994;340:197–201. doi: 10.1016/0014-5793(94)80137-1. [DOI] [PubMed] [Google Scholar]
- 15.Mould AP, Askari JA, Aota S, Yamada KM, Irie A, Takada Y, Mardon HJ, Humphries MJ. Defining the Topology of Integrin alpha5beta1-Fibronectin Interactions Using Inhibitory Anti-alpha5 Anti-beta1Monoclonal Antibodies Evidence that the Synergy Sequence of Fibronectin is Recognized by the Amino-Terminal Repeats of the alpha5 Subunit. J Biol Chem. 1997;272:17283–92. doi: 10.1074/jbc.272.28.17283. [DOI] [PubMed] [Google Scholar]
- 16.Aota S, Nomizu M, Yamada KM. The Short Amino Acid Sequence Pro-His-Ser-Arg-Asn in Human Fibronectin Enhances Cell-Adhesive function. J Biol Chem. 1994;269:24756. [PubMed] [Google Scholar]
- 17.Garcia AJ, Schwarzbauer JE, Boettiger D. Distinct Activation States of alpha5beta1 Integrin Show Differential Binding to RGD Synergy Domains of Fibronectin. Biochemistry. 2002;41:9063–9. doi: 10.1021/bi025752f. [DOI] [PubMed] [Google Scholar]
- 18.van der Walle CF, Altroff H, Mardon HJ. Novel Mutant Human Fibronectin FIII9–10 Domain Pair with Increased Conformational Stability Biological Activity. Protein Eng Des Sel. 2002;15:1021–4. doi: 10.1093/protein/15.12.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown AC, Rowe JA, Barker TH. Guiding Epithelial Cell Phenotypes with Engineered Integrin-Specific Recombinant Fibronectin Fragments. Tissue Eng Part A. 2011;17:139–50. doi: 10.1089/ten.tea.2010.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martino MM, Mochizuki M, Rothenfluh DA, Rempel SA, Hubbell JA, Barker TH. Controlling Integrin Specificity Stem Cell Differentiation in 2D 3D Environments Through Regulation of Fibronectin Domain Stability. Biomaterials. 2009;30:1089–97. doi: 10.1016/j.biomaterials.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Markowski MC, Brown AC, Barker TH. Directing Epithelial to Mesenchymal Transition Through Engineered Microenvironments Displaying Orthogonal Adhesive Mechanical Cues. J Biomed Mater Res Part A. 2012;100:2119–27. doi: 10.1002/jbm.a.34068. [DOI] [PubMed] [Google Scholar]
- 22.Schornberg KL, Shoemaker CJ, Dube D, Abshire MY, Delos SE, Bouton AH, White JM. Alpha5beta1-Integrin Controls Ebolavirus Entry by Regulating Endosomal Cathepsins. Proc Natl Acad SciUS A. 2009;106:8003–8. doi: 10.1073/pnas.0807578106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee CMY, Iorno N, Sierro F, Christ D. Selection of Human Antibody Fragments by Phage Display. Nat Protoc. 2007;2:3001–3008. doi: 10.1038/nprot.2007.448. [DOI] [PubMed] [Google Scholar]
- 24.de Wildt RMT, Mundy CR, Gorick BD, Tomlinson IM. Antibody Arrays for High-Throughput Screening of Antibody-Antigen Interactions. Nat Biotechnol. 2000;18:989–994. doi: 10.1038/79494. [DOI] [PubMed] [Google Scholar]
- 25.Little WC, Smith ML, Ebneter U, Vogel V. Assay to Mechanically Tune Optically Probe Fibrillar Fibronectin Conformations from Fully Relaxed to Breakage. Matrix Biol. 2008;27:451–461. doi: 10.1016/j.matbio.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Izbicki G, Segel MJ, Christensen TG, Conner MW, Breuer R. Time Course of Bleomycin-Induced Lung Fibrosis. Int J Exp Pathol. 2002;83:111–9. doi: 10.1046/j.1365-2613.2002.00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawson WE, Polosukhin VV, Stathopoulos GT, Zoia O, Han W, Lane KB, Li B, Donnelly EF, Holburn GE, Lewis KG, Collins RD, Hull WM, Glasser SW, Whitsett JA, Blackwell TS. Increased Prolonged Pulmonary Fibrosis in Surfactant Protein C-Deficient Mice Following Intratracheal Bleomycin. Am J Pathol. 2005;167:1267–77. doi: 10.1016/S0002-9440(10)61214-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pitulescu ME, Schmidt I, Benedito R, Adams RH. Inducible Gene Targeting in the Neonatal Vasculature Analysis of Retinal Angiogenesis in Mice. Nat Protoc. 2010;5:1518–34. doi: 10.1038/nprot.2010.113. [DOI] [PubMed] [Google Scholar]
- 29.Jiang B, Liou GI, Behzadian MA, Caldwell RB. Astrocytes Modulate Retinal Vasculogenesis: Effects on Fibronectin Expression. J Cell Sci. 1994;107:2499–2508. doi: 10.1242/jcs.107.9.2499. [DOI] [PubMed] [Google Scholar]
- 30.Brown AC, Dysart MM, Clarke KC, Stabenfeldt SE, Barker TH. Integrin alpha3beta1 Binding to Fibronectin Is Dependent on the Ninth Type III Repeat. J Biol Chem. 2015;290:25534–47. doi: 10.1074/jbc.M115.656702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Craig JA, Rexeisen EL, Mardilovich A, Shroff K, Kokkoli E. Effect of Linker Spacer on the Design of a Fibronectin-Mimetic Peptide Evaluated via Cell Studies AFM Adhesion Forces. Langmuir. 2008;24:10282–10292. doi: 10.1021/la702434p. [DOI] [PubMed] [Google Scholar]
- 32.Hubbard B, Buczek-Thomas JA, Nugent MA, Smith ML. Fibronectin Fiber Extension Decreases Cell Spreading Migration. J Cell Physiol. 2016;231:1728–36. doi: 10.1002/jcp.25271. [DOI] [PubMed] [Google Scholar]
- 33.Shukla AK, Manglik A, Kruse AC, Xiao K, Reis RI, Tseng WC, Staus DP, Hilger D, Uysal S, Huang LY, Paduch M, Tripathi-Shukla P, Koide A, Koide S, Weis WI, Kossiakoff AA, Kobilka BK, Lefkowitz RJ. Structure of Active Beta-arrestin-1 Bound to a G-Protein-Coupled Receptor Phosphopeptide. Nature. 2013;497:137–41. doi: 10.1038/nature12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen X, Zaro JL, Shen WC. Fusion Protein Linkers: Property Design and Functionality. Adv Drug Delivery Rev. 2013;65:1357–69. doi: 10.1016/j.addr.2012.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henderson NC, Arnold TD, Katamura Y, Giacomini MM, Rodriguez JD, McCarty JH, Pellicoro A, Raschperger E, Betsholtz C, Ruminski PG, Griggs DW, Prinsen MJ, Maher JJ, Iredale JP, Lacy-Hulbert A, Adams RH, Sheppard D. Targeting of AlphaV Integrin Identifies a Core Molecular Pathway that Regulates Fibrosis in Several Organs. Nat Med. 2013;19:1617–24. doi: 10.1038/nm.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patan S. Vasculogenesis and Angiogenesis. Cancer Treat Res. 2004;117:3–32. doi: 10.1007/978-1-4419-8871-3_1. [DOI] [PubMed] [Google Scholar]
- 37.Li S, Nih L, Bachman H, Fei P, Li Y, Nam E, Dimatteo R, Carmichael S, Barker TH, Segura T. Hydrogels with Precisely Controlled Integrin Activation Dictate Vascular Patterning and Permeability. Nat Mater. 2017 doi: 10.1038/nmat4954. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


