Abstract
Vitellogenin (Vg) and vitellogenin receptor (VgR) play important roles in the vitellogenesis of insects. In this study, we cloned and characterized the two corresponding genes (TpVg and TpVgR) in an economically important insect, Thitarodes pui (Lepidoptera: Hepialidae), from the Tibetan plateau. The full length of TpVg is 5566 bp with a 5373 bp open reading frame (ORF) encoding 1,790 amino acids. Sequence alignment revealed that TpVg has three conserved domains: a Vitellogenin_N domain, a DUF1943 domain, and a von Willebrand factor type D domain (VWD). The full length of TpVgR is 5732 bp, with a 5397 bp ORF encoding 1798 amino acids. BLASTP showed that TpVgR belongs to the low-density lipoprotein receptor (LDLR) gene superfamily. Structural analysis revealed that TpVgR has a group of four structural domains: a ligand-binding domain (LBD), an epidermal growth factor (EGF)–precursor homology domain, a transmembrane (TM) domain, and a cytoplasmic domain. In addition, TpVgR has four cysteine-rich LDL repeats in the first ligand-binding site and seven in the second. Quantitative real-time polymerase chain reaction analysis revealed that the expression levels of TpVg and TpVgR are much higher in later pupa than in either the larval or adult stage, implying that the synthesis and uptake of Vg in T. pui occurs in the later pupal stage. These results will help us to understand the molecular mechanism of the reproductive capacity and will provide new insight into the mass rearing and utilization of T. pui.
Keywords: vitellogenin, vitellogenin receptor, Thitarodes pui
Like other animals, insects require large amounts of vitellogenin (Vg) to meet the nutritional needs of egg development. Vg, the precursor of vitellin and the major yolk protein, is usually synthesized in the fat body, released into the hemolymph, and subsequently taken up by the vitellogenin receptor (VgR) located on the external surface of competent oocytes (Tufail and Takeda 2008, 2009). Vg has been studied extensively in many insects of different orders. The identification of insect Vgs revealed that these Vgs belong to a large family of lipid transfer genes and that they share similar structural motifs, including an N-terminal lipid binding domain (LPD_N), an unknown functional region (DUF1943), and a von Willebrand factor type D similar domain (VWD) (Trewitt et al. 1992, Chen et al. 1994, Yano et al. 1994a, Veerana et al. 2014, Upadhyay et al. 2016, Zhao et al. 2016, Ibanez et al. 2017).
VgR, the key element in the process of Vg uptake, plays an important role during vitellogenesis. To date, the molecular characteristics of VgRs have been well documented in vertebrates and invertebrates including many orders of insects, such as Diptera (Schonbaum et al. 1995, Cho and Raikhel 2001), Lepidoptera (Shu et al. 2011, Qian et al. 2015, Zhang et al. 2016), Hemiptera (Upadhyay et al. 2016), Coleoptera (Roy-Zokan et al. 2015), Hymenoptera (Chen et al. 2004), and Blattaria (Tufail and Takeda 2007, 2008). The deduced amino acid (aa) sequences of these VgRs indicate that they are members of the low-density lipoprotein receptor (LDLR) family. In general, the LDLR family members have five distinct, conserved motifs, including a ligand-binding domain (LBD) containing LDLR Class A cysteine-rich repeats, an epidermal growth factor (EGF)-like domain containing LDLR Class B cysteine-rich repeats and YWXD repeats, an O-linked carbohydrate domain with a serine/threonine-rich track, a transmembrane (TM) domain, and a cytoplasmic tail (Tufail and Takeda 2007, 2009; Shu et al. 2011; Upadhyay et al. 2016; Zhang et al. 2016).
Thitarodes pui (Lepidoptera: Hepialidae) is naturally distributed in Segrila Mountain of the Tibetan Plateau, which is characterized by a harsh climate with low temperatures and hypobaric hypoxia. T. pui larvae are the hosts for the famous traditional Chinese medicinal caterpillar fungus, the ascomycete Ophiocordyceps sinensis (Berk.) Sung, Sung, Hywel-Jones, and Spatafora (syn. Cordyceps sinensis).
Once infection occurs, the fungus grows within the body cavity of the insect larvae and finally kills and mummifies them in their underground burrows. A fruiting fungal body emerges from the front end of the caterpillar and protrudes from the ground in spring and early summer (Buenz et al. 2005). The medicinal activity is derived from the parasite complex of the mummified caterpillar and the fungal stromata (Zhang et al. 2009).
The complete life cycle of T. pui lasts at least 3 yr under natural conditions (Li et al. 2011). A better understanding of the molecular mechanisms regulating reproduction can indicate potential approaches for the mass rearing of these economically important insects. Vg and VgR are important molecular markers for insect fecundity. In this study, we cloned the full lengths of vitellogenin (TpVg) and the vitellogenin receptor (TpVgR) of T. pui, characterized the molecular structure of the corresponding deduced proteins, and analyzed their gene expression profiles at different developmental stages. Our results are expected to provide the theoretical basis for the conservation and utilization of this economically important insect.
Materials and Methods
Insect Rearing and Collection
T. pui samples were collected in the Tibetan Plateau Peculiar Bio-resources Research Station of Sun Yat-sen University at Nyingchi in the Tibet Autonomous Region (4,156 m altitude, 29°36′N, 94°36′E) from June to August in 2014. The larvae were reared as previously described (Sun et al. 2011, Wu et al. 2015). The larvae of different instars (third through eighth), female pupae of different ages (1, 9, 17, and 33 d old), and 1-d-old adult females were used for the analysis of gene expression. All of the samples at different developmental stages were freshly frozen in a Sample Protector (Takara, Tokyo, Japan) at −80°C until RNA isolation.
RNA Isolation and cDNA Synthesis
Total RNA was extracted from the whole body of T. pui using TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions, and RNA purity and degradation were checked on 1% agarose gels and Bioanalyzer 2100 system (Agilent Technologies). For tissue-specific expression profiles, total RNAs were extracted from the thorax and abdomen parts of newly emerged female adults which were stored separately in a refrigerator at −80°C. The RNA sample was dissolved in 20 µl diethylpyrocarbonate (DEPC)-treated H2O and assessed by spectrophotometry (Nanodrop 2000, Thermo Fisher Scientific, DE). A PrimeScript Reagent Kit with gDNA Eraser (TaKaRa, Tokyo, Japan) was used to synthesize first-strand complementary DNA (cDNA) with 1µg total RNA in a 20 µl reaction mixture following the manufacturer’s recommendations.
Molecular Cloning of TpVg and TpVgR
Two partial sequences of both Vg and VgR were identified from the pupal transcriptome of T. pui (unpublished data). The fragment sequences of TpVg and TpVgR were further verified by polymerase chain reaction (PCR) amplification using the primers listed in Supplementary Table 1. The PCR program was performed as follows: one cycle of predenaturing at 95°C for 5 min; 32 cycles of 95°C for 30 s, 53°C for 30 s, and 72°C for 2 min; and then 72°C for 5 min for elongation. The PCR products were then gel purified, cloned into the pGEM-T vector (Takara, Dalian, China), and transformed into Escherichia coli DH5α cells for amplification. Positive clones were assessed by PCR and sequenced. To obtain the full-length cDNA sequences of TpVg and TpVgR, a SMARTer RACE (rapid amplification of cDNA ends) cDNA Amplification Kit (Clontech) was used according to the manufacturer’s instructions. The first round of RACE-PCR was performed using outer primer (Supplementary Fig. 1 and Supplementary Table 1) and a universal primer mix (UPM) with the following protocol: one cycle of predenaturing at 94°C for 5 min,; 35 cycles of 30 s at 95°C, 30 s at 65°C, and 2 min at 72°C; and a final extension at 72°C for 5 min. The second round of RACE-PCR was performed using inner primers (Supplementary Fig. 1 and Supplementary Table 1) and a nested universal primer (NUP). The second RACE-PCR protocol was performed as follows: one cycle of predenaturing at 94°C for 5 min; 35 cycles of 30 s at 95°C, 30 s at 68°C, and 2 min at 72°C; and a final extension at 72°C for 5 min. The RACE products were gel purified and sequenced as described above.
Sequence Comparisons and Phylogenetic Analysis
The sequence similarities were analyzed using the online BLASP program on the NCBI website (https://blast.ncbi.nlm.nih.gov/). The open reading frames (ORFs) of TpVg and TpVgR were predicted using the NCBI ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/). The molecular weights and isoelectric points (pIs) of the deduced protein sequences were predicted using the ExPASy proteomics server (http://www.expasy.org). The domain architecture and the conserved domains were analyzed using the online servers of Scan-Prosite (http://prosite.expasy.org/scanprosite/), SMART (http://smart.embl-heidelberg.de/), and InterProScan (http://ebi.ac.uk/Tools/pfa/iprscan/). The signal peptide was predicted by using the SignalP 4.1 Server (http://www.cbs.dtu.dk/services/SignalP). The potential O-linked glycosylation sites were evaluated using the GPP Prediction Server (http://comp.chem.nottingham.ac.uk/glyco/), and the TM regions were predicted using the TMHMM server v2.0 (http://cbs.dtu.dk/services/TMHMM/). All the computational programs were run with the recommended default values set up by the programs. The phylogenetic tree was constructed with MEGA 6.0 by using the neighbor-joining method with a bootstrap of 1,000 replicates.
Developmental Expression Profile of TpVg and TpVgR
Equal quantities of cDNA (1 µg) from different developmental stages were used for a relative expression analysis of TpVg and TpVgR messenger RNA (mRNA) using a real-time PCR machine (LightCycler 480 real-time PCR machine, Roche Applied Science, Switzerland) with SYBR Green (Invitrogen). β-actin was used as an internal control based on a previous study (Sun et al. 2011). Primers used for quantitative PCR (qPCR) are listed in Supplementary Table 1. The qPCR amplifications were conducted in a total volume of 10 µl, including 1 µg of cDNA, 5 µl of SYBR Green I Master mix, 0.5 µl of each primer (10 µM), and a certain amount of ddH2O. The qPCR program included one cycle of 95°C for 10 s, followed by 40 cycles of 95°C for 5 s, 60°C for 20 s, and 72°C for 20 s. To ensure reliability, each sample included three biological replicates. The melt curve genotyping analysis was performed to confirm the homogeneity of the PCR products. Reactions were performed in triplicate for each sample, and gene expression levels were normalized against T. pui β-actin according to the 2−∆∆CT method (Livak and Schmittgen 2001).
Results
Sequence and Structural Analysis of TpVg and TpVgR
The full sequence of TpVg was 5566 bp, including an 83 bp 5′ untranslated region (UTR), a 5373 bp ORF encoding 1,790 aa residues, and a 110 bp 3′ UTR (GenBank accession no. MF622538). The theoretical molecular weight was 204.43 kDa, and the pI was 6.51. A BLASTP analysis showed that the aa sequence of TpVg had a 30 to 40% similarity with other insects from different orders. TpVg showed a 34 % and 33 % similarity with Operophtera brumata and Actias selene, respectively. As with other insects, TpVg in T. pui also contained the typical conserved domains of the Vg protein.
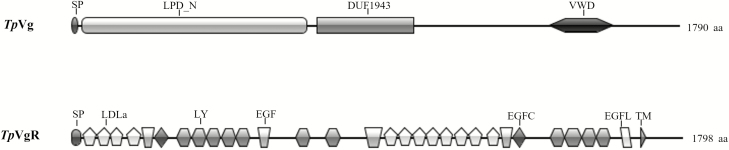
Domain architecture analysis by SMART showed that the TpVg aa sequence has three conserved domains (Fig. 1): the vitellogenin_N domain, the DUF1943 domain, and the VWD. A signal peptide with 18 amino acids (MRTFVLFALLATAFCANS) was found in the analysis of the predicted aa sequence of TpVg. In addition, the deduced aa sequences of TpVg included four N-glycosylation sites (NXS/T) and seven K/RXXR cleavage sites.
Fig. 1.
Conserved domain architecture of vitellogenin (TpVg) and vitellogenin receptor (TpVgR) in T. pui. SP, signal peptide; LPD-N, lipoprotein N-terminal domain; DUF1943, domains of unknown function protein families; VWD, von Willebrand factor type D domain; LDLa, low-density lipoprotein receptor domain class A; EGF, epidermal growth factor-like domain; EGFC, calcium-binding EGF-like domain; LY, low-density lipoprotein receptor YWTD protein; EGFL, epidermal growth factor-like domain.
The full sequence of TpVgR was 5732 bp, including a 74 bp 5′ UTR, a 5397 bp ORF encoding 1,798 aa residues, and a 261 bp 3′ UTR (GenBank accession no. MF622539). The theoretical molecular weight was 200.47 kDa, and the pI was 5.46. The analysis of the predicted aa sequence revealed that a signal peptide with 27 amino acids (MGHADLLHIALIIVVNVFLLKWKMAGA) was located at the N-terminal of TpVgR.
The protein sequence analysis indicated that TpVgR contained the characteristic features of the LDLR family (Fig. 1). TpVgR showed two LBDs with four class A (LDLa) cysteine-rich repeats in the first domain and eight repeats in the second domain. Each repeat had six cysteine residues, and each LDLa was followed by an EGF-like domain with/without a calcium-binding region. The EGF-like domain contained repeats of the LDLR YWTD motif. After the second EGF-like domain, a TM domain spanning amino acids 1674–1691 and a cytoplasmic domain spanning amino acids 1692–1798 were predicted by the TMHMM server v. 2.0.
Phylogenetic Analyses of TpVg and TpVgR
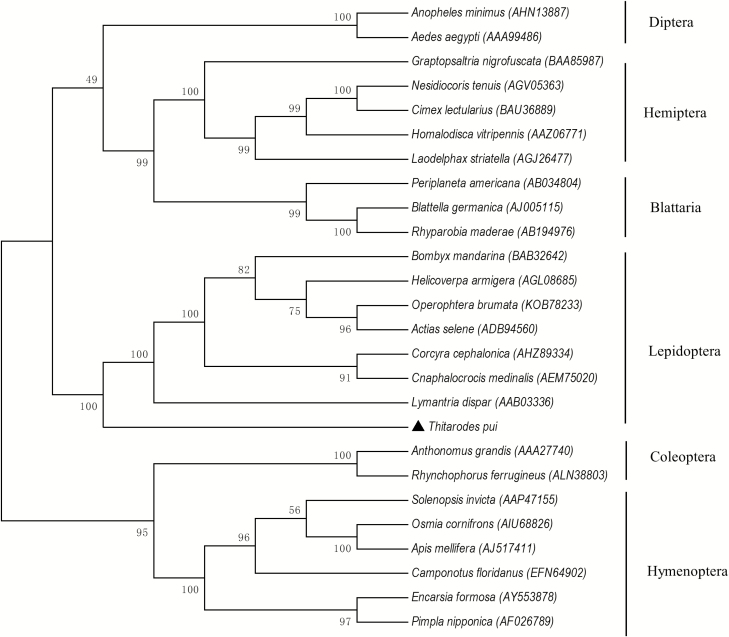
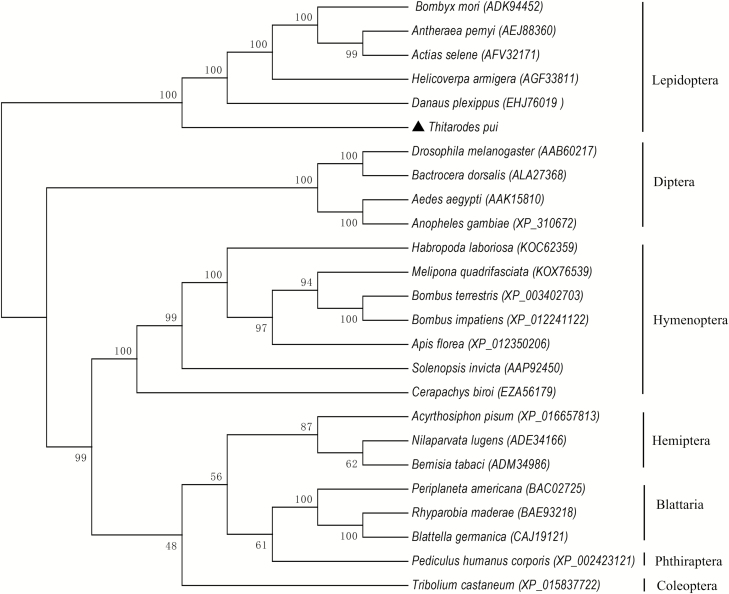
A phylogenetic tree was constructed based on the distances of aa sequences of TpVg and TpVgR. The phylogenetic tree showed that TpVg and Vgs of other Lepidoptera species clustered together (Fig. 2). Vgs from the same insect order also clustered in the same group, implying the conservation of Vgs and the closer relationships within the same taxonomic status. A similar phylogenetic tree was constructed by comparing VgR aa sequences from different groups of insects (Fig. 3). TpVgR and other VgRs of moth species formed a cluster. Other VgRs from the same insect orders were also clustered. In the phylogenetic tree of VgRs, Lepidoptera VgRs formed a single branch and was separated from other species (Fig. 3). However, the dendrogram of Vgs showed that the Vgs of Diptera and Lepidoptera species were close together (Fig. 2), suggesting that the Vgs of those two insect orders share a closer ancestry than other insects.
Fig. 2.
Phylogenetic analysis of vitellogenin in T. pui with that of insects from different orders. The phylogenetic tree was constructed by Mega 6 with a statistical method of neighbor joining. Numbers indicate bootstrap support values (%) based on 1,000 replicates. The numbers in the brackets indicate the GenBank accession for each insect.
Fig. 3.
Phylogenetic analysis of the vitellogenin receptor in T. pui with that of insects from different orders. The phylogenetic tree was constructed by Mega 6 with a statistical method of neighbor joining. Numbers indicate bootstrap support values (%) based on 1,000 replicates. The numbers in the brackets indicate the GenBank accession for each insect.
Expression Profile of TpVg and TpVgR
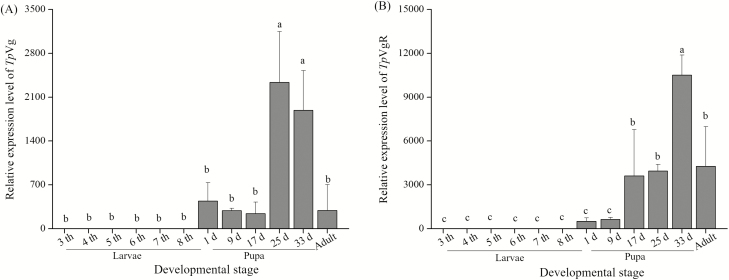
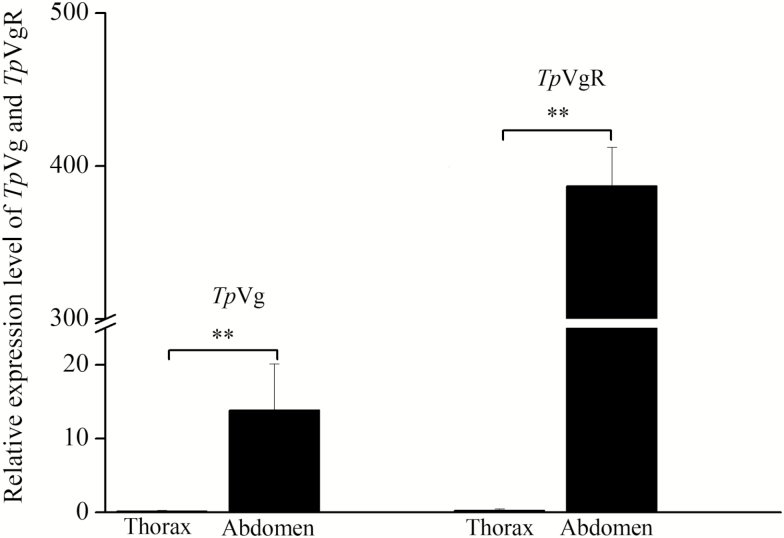
The expression analysis of TpVg and TpVgR was conducted during the larval, pupal, and adult stages by real-time PCR (Fig. 4). The expression level of TpVg was normalized with a reference gene, β-actin of T. pui. The results showed that TpVg had no detectable expression in the larval stage, but it had a high expression during the pupae stage, decreasing significantly in adult females (Fig. 4). The expression of TpVg began to increase in the pupal stage and reached its peak in the 25 and 33-d-old pupae, i.e., the final stage of pupae. Then, the expression level of TpVg decreased significantly in the adult stage. TpVgR had an expression profile similar to that of TpVg. TpVgR was found to have lower expression in the early pupal stage and then a remarkably high expression in the 17-d-old pupae, subsequently reaching a maximum expression level in the later pupae stage. Then, a lower expression level was maintained in the adult period. In addition, the tissue-specific expression profile revealed that TpVg and TpVgR were highly expressed in the abdomen which contained the reproductive organ-ovary (Fig. 5).
Fig. 4.
Relative expression of TpVg and TpVgR at different developmental stages. Samples include cDNA from third, fourth, fifth, sixth, seventh, and eighth instar larvae, 1-d-old pupae, 9-d-old pupae, 17-d-old pupae, 25-d-old pupae, 33-d-old pupae, and 1-d-old female adults. The mRNA levels were normalized using β-actin as a reference gene. Different letters indicate statistically significant differences at P < 0.05 (One-way ANOVA followed by LSD multiple comparison tests). The bars represent SD (n = 3).
Fig. 5.
Relative expression of TpVg and TpVgR in the thorax and abdomen of T. pui. The mRNA levels were normalized using β-actin as a reference gene. Double asterisks indicate statistically significant differences at P < 0.01 (Student’s t-test). The bars represent SD (n = 3).
Discussion
T. pui is a holometabolous insect with a long life history. The complete life cycle includes 41–47 d egg stage, 990–1,350 d larval stage, 35–41 d pupal stage, and 3–8 d adult stage (Li et al. 2011). The reproductive success of all species, including insects, relies on Vg biosynthesis and its uptake by VgR in the developing oocytes. As an important host of traditional Chinese medicine caterpillar fungus, it is of great significance to study the Vg and VgR in T. pui.
In this study, the full-length cDNA corresponding to the TpVg and TpVgR genes was obtained, and the molecular characteristics of these two genes were also analyzed. The GL/ICG, DGXR, and K/RXXR motifs, considered to be the conserved domains, have been reported in many insect Vg proteins (Sappington and Raikhel 1998, Tufail and Takeda 2008, Upadhyay et al. 2016). In earlier studies, the GL/ICG motif was assumed to be essential for the proper function of Vg during embryogenesis (Tufail et al. 2001). As is the case with other insects, a GLCG motif was found in the aa sequence of TpVg (Supplementary Fig. 2). Apart from the class Insecta, the GLCG motif was also present in Arachnoidea species, such as Tetranychus urticae (Kawakami et al. 2009), Panonychus citri (Zhong et al. 2015), and Ornithodoros moubata (Horigane et al. 2010). A DGXR motif is usually located 17–19 residues upstream of the GL/ICG motif in most insect Vg sequences (Tufail and Takeda 2008), but that motif was not found in Rhyparobia maderae or in T. pui in this study (Supplementary Fig. 2). In addition, some aa residues, such as E (acidic), G, T and P (hydrophobic), and Y (hydrophilic) were also highly conserved at the C-terminus of all the tested Vgs (Supplementary Fig. 3). However, the specific roles of these conservative sites remain unclear.
TpVgR also showed sequence similarities with other insect VgRs. The phylogenetic analysis indicated that TpVgR shares a homology with other Lepidopteran VgRs, such as Bombyx mori, A. selene, Helicoverpa armigera, Danaus plexippus, and Antheraea pernyi (Fig. 3). An analysis of the domain conservation of TpVgR suggests that TpVgR in a member of the LDLR family and shares several modular elements with other insect Vgs. TpVgR was shown to contain the classical LBDs, EGF-like domains, and a TM domain. TpVgR was found to have two ligand-binding sites that were characterized by cysteine-rich LDLa repeats. Most insect Vgs have two to four LDLa repeats in the first ligand-binding site and seven to eight in the second (Tufail and Takeda 2009, Lu et al. 2015). There were four LDLa repeats found in the first ligand-binding site of T. pui, which is a typical feature in Lepidoptera species (Shu et al. 2011, Lin et al. 2013, Zhang et al. 2016). However, TpVgR bore eight LDLa repeats in the second ligand-binding site, which differs from other Vgs of Lepidoptera species; they generally have seven LDLa repeats. There are also eight LDLa repeats in the second ligand-binding site of the VgRs in other insects, including species of Hymenoptera, Coleoptera, Diptera, Hemiptera, Blattaria, which are also found in Acarina species (Mitchell et al. 2007, Boldbaatar et al. 2010, Zhong et al. 2015).
In some insect VgRs, the O-linked sugar domain (OLSD), which is rich in S and/or T residue, is always between the last EGF precursor domain and the TM domain (Tufail and Takeda 2007). No OLSD was found in TpVgR, which is consistent with VgRs of Solenopsis invicta, H. armigera, and Drosophila melanogaster but not with B. mori, A. pernyi, and Nilaparvata lugens (Chen et al. 2004, 2010; Lu et al. 2015; Zhang et al. 2016), implying that the motif is not a typical feature in insect VgRs. OLSDs have also been shown to be absent in vertebrate VgRs (Prat et al. 1998, Hiramatsu et al. 2004).
In the current study, TpVg and TpVgR had similar expression patterns (Fig. 4). The transcription profiles of TpVg and TpVgR showed that the mRNA levels of these two genes began to increase in the pupae stage and reached their peak during that stage; then expression decreased after emergence. The difference in transcription lay in the fact that TpVg reached its peak in the 25-d-old pupae, whereas TpVgR reached its peak in the 33-d-old pupae, demonstrating that the maximum expression of TpVgR occurs later than that of TpVg. Despite the different expression profiles in different species, the expression peaks of Vgs and VgRs of insects primarily occur in the pupae or adult stage. For example, B. mori Vg first appeared in the sixth instar larvae and ended in the late pupal stage (Yano et al. 1994a,b). In Spodoptera litura, the Vg mRNA was first detected in late pupal stage and reached its maximum expression in adult females 2 d after emergence (Shu et al. 2009).
The expression of VgR is closely related to ovarian development, which has been investigated in many species. In Periplaneta americana, Leucophaea maderae, N. lugens, and S. litura, the VgR transcript level was low until it increased during the early ovarian development, reaching its peak during the vitellogenic period, after which expression was subsequently low again (Tufail and Takeda, 2005; Tufail and Takeda 2007, Shu et al. 2011, Lu et al. 2015). The expression of TpVgR mRNA appeared to peak on the 33rd d of the pupae, and it distinctly declined in the adults (Fig. 4), indicating that the vitellogenic period of T. pui occurs in the late pupae stage before emergence.
Conclusion
In conclusion, we identified the cDNAs and analyzed the molecular characteristics of TpVg and TpVgR. Those two genes were structurally conserved when compared with the analogous genes of other species. The development expression profiles indicated that the synthesis and uptake of Vg in T. pui occur in the late pupal stage. Our study will provide insights for the molecular regulation of high fecundity in T. pui. In the future, we will focus on how TpVg interacts with TpVgR to regulate oocyte maturation.
Conflict of interest
These authors have no conflict of interest to declare.
Supplementary Data
Supplementary data are available at Journal of Insect Science online.
Supplementary Material
Acknowledgments
We would like to thank Cirentajie for the insect collection and rearing. This work was supported by the Special Fund for Agro-scientifc Research in the Public Interest (201403030) and the National Key Technology Research and Development Program of China (2011BAI13B06). The funders had no role in study design, data collection and analysis, decision to publish, or the preparation of the manuscript.
References Cited
- Boldbaatar D., Umemiya-Shirafuji R., Liao M., Tanaka T., Xuan X., and Fujisaki K.. 2010. Multiple vitellogenins from the Haemaphysalis longicornis tick are crucial for ovarian development. J. Insect Physiol. 56: 1587–1598. [DOI] [PubMed] [Google Scholar]
- Buenz E. J., Bauer B. A., Osmundson T. W., and Motley T. J.. 2005. The traditional Chinese medicine Cordyceps sinensis and its effects on apoptotic homeostasis. J. Ethnopharmacol. 96: 19–29. [DOI] [PubMed] [Google Scholar]
- Chen J. S., Cho W. L., and Raikhel A. S.. 1994. Analysis of mosquito vitellogenin cDNA. Similarity with vertebrate phosvitins and arthropod serum proteins. J. Mol. Biol. 237: 641–647. [DOI] [PubMed] [Google Scholar]
- Chen S. A., Lee T. Y., and Ou Y. Y.. 2010. Incorporating significant amino acid pairs to identify O-linked glycosylation sites on transmembrane proteins and non-transmembrane proteins. BMC Bioinformatics. 11: 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. E., Lewis D. K., Keeley L. L., and Pietrantonio P. V.. 2004. cDNA cloning and transcriptional regulation of the vitellogenin receptor from the imported fire ant, Solenopsis invicta Buren (Hymenoptera: Formicidae). Insect Mol. Biol. 13: 195–204. [DOI] [PubMed] [Google Scholar]
- Cho K. H., and Raikhel A. S.. 2001. Organization and developmental expression of the mosquito vitellogenin receptor gene. Insect Mol. Biol. 10: 465–474. [DOI] [PubMed] [Google Scholar]
- Hiramatsu N., Chapman R. W., Lindzey J. K., Haynes M. R., and Sullivan C. V.. 2004. Molecular characterization and expression of vitellogenin receptor from white perch (Morone americana). Biol. Reprod. 70: 1720–1730. [DOI] [PubMed] [Google Scholar]
- Horigane M., Shinoda T., Honda H., and Taylor D.. 2010. Characterization of a vitellogenin gene reveals two phase regulation of vitellogenesis by engorgement and mating in the soft tick Ornithodoros moubata (Acari: Argasidae). Insect Mol. Biol. 19: 501–515. [DOI] [PubMed] [Google Scholar]
- Ibanez F., Levy J., and Tamborindeguy C.. 2017. Identification and expression analyses of vitellogenin in Bactericera cockerelli (Šulc). J. Insect Physiol. 98: 205–213. [DOI] [PubMed] [Google Scholar]
- Kawakami Y., Goto S. G., Ito K., and Numata H.. 2009. Suppression of ovarian development and vitellogenin gene expression in the adult diapause of the two-spotted spider mite Tetranychus urticae. J. Insect Physiol. 55: 70–77. [DOI] [PubMed] [Google Scholar]
- Li J. F., Zou Z. W., Liu X., and Zhang G. R.. 2011. Biology of Thitarodes pui (Lepidoptera, Hepialidae), a host species of Ophiocordyceps sinensis. J. Environ. Entomol. 33: 195–202. [Google Scholar]
- Lin Y., Meng Y., Wang Y. X., Luo J., Katsuma S., Yang C. W., Banno Y., Kusakabe T., Shimada T., and Xia Q. Y.. 2013. Vitellogenin receptor mutation leads to the oogenesis mutant phenotype “scanty vitellin” of the silkworm, Bombyx mori. J. Biol. Chem. 288: 13345–13355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., and Schmittgen T. D.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lu K., Shu Y., Zhou J., Zhang X., Zhang X., Chen M., Yao Q., Zhou Q., and Zhang W.. 2015. Molecular characterization and RNA interference analysis of vitellogenin receptor from Nilaparvata lugens (Stål). J. Insect Physiol. 73: 20–29. [DOI] [PubMed] [Google Scholar]
- Mitchell R. D., Ross E., Osgood C., Sonenshine D. E., Donohue K. V., Khalil S. M., Thompson D. M., and Michael Roe R.. 2007. Molecular characterization, tissue-specific expression and RNAi knockdown of the first vitellogenin receptor from a tick. Insect Biochem. Mol. Biol. 37: 375–388. [DOI] [PubMed] [Google Scholar]
- Prat F., Coward K., Sumpter J. P., and Tyler C. R.. 1998. Molecular characterization and expression of two ovarian lipoprotein receptors in the rainbow trout, Oncorhynchus mykiss. Biol. Reprod. 58: 1146–1153. [DOI] [PubMed] [Google Scholar]
- Qian C., Fu W. W., Wei G. Q., Wang L., Liu Q. N., Dai L. S., Sun Y., Zhu B. J., and Liu C. L.. 2015. Identification and expression analysis of vitellogenin receptor from the wild silkworm, Bombyx mandarina. Arch. Insect Biochem. Physiol. 89: 181–192. [DOI] [PubMed] [Google Scholar]
- Roy-Zokan E. M., Cunningham C. B., Hebb L. E., McKinney E. C., and Moore A. J.. 2015. Vitellogenin and vitellogenin receptor gene expression is associated with male and female parenting in a subsocial insect. P. R. Soc. B. 282: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sappington T. W., and Raikhel A. S.. 1998. Molecular characteristics of insect vitellogenins and vitellogenin receptors. Insect Biochem. Mol. Biol. 28: 277–300. [DOI] [PubMed] [Google Scholar]
- Schonbaum C. P., Lee S., and Mahowald A. P.. 1995. The Drosophila yolkless gene encodes a vitellogenin receptor belonging to the low density lipoprotein receptor superfamily. Proc. Natl. Acad. Sci. U. S. A. 92: 1485–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y. H., Zhou J. L., Tang W. C., Lu K., Zhou Q., and Zhang G. R.. 2009. Molecular characterization and expression pattern of Spodoptera litura (Lepidoptera: Noctuidae) vitellogenin, and its response to lead stress. J. Insect. Physiol. 55: 608–616. [DOI] [PubMed] [Google Scholar]
- Shu Y. H., Wang J. W., Lu K., Zhou J. L., Zhou Q., and Zhang G. R.. 2011. The first vitellogenin receptor from a Lepidopteran insect: molecular characterization, expression patterns and RNA interference analysis. Insect Mol. Biol. 20: 61–73. [DOI] [PubMed] [Google Scholar]
- Sun Z. X., Wu W. J., and Zhang G. R.. 2011. Structure and expression of beta-1,3-glucan recognition proteins from the ghost moth, Thitarodes pui (Hepialidae), and their response to Beauveria bassiana infection. J. Insect. Physiol. 57: 1660–1669. [DOI] [PubMed] [Google Scholar]
- Trewitt P. M., Heilmann L. J., Degrugillier S. S., and Kumaran A. K.. 1992. The boll weevil vitellogenin gene: nucleotide sequence, structure, and evolutionary relationship to nematode and vertebrate vitellogenin genes. J. Mol. Evol. 34: 478–492. [DOI] [PubMed] [Google Scholar]
- Tufail M., and Takeda M.. 2005. Molecular cloning, characterization and regulation of the cockroach vitellogenin receptor during oogenesis. Insect Mol. Biol. 14: 389–401. [DOI] [PubMed] [Google Scholar]
- Tufail M., and Takeda M.. 2007. Molecular cloning and developmental expression pattern of the vitellogenin receptor from the cockroach, Leucophaea maderae. Insect Biochem. Mol. Biol. 37: 235–245. [DOI] [PubMed] [Google Scholar]
- Tufail M., and Takeda M.. 2008. Molecular characteristics of insect vitellogenins. J. Insect Physiol. 54: 1447–1458. [DOI] [PubMed] [Google Scholar]
- Tufail M., and Takeda M.. 2009. Insect vitellogenin/lipophorin receptors: molecular structures, role in oogenesis, and regulatory mechanisms. J. Insect Physiol. 55: 87–103. [DOI] [PubMed] [Google Scholar]
- Tufail M., Hatakeyama M., and Takeda M.. 2001. Molecular evidence for two vitellogenin genes and processing of vitellogenins in the American cockroach, Periplaneta americana. Arch. Insect Biochem. Physiol. 48: 72–80. [DOI] [PubMed] [Google Scholar]
- Upadhyay S. K., Singh H., Dixit S., Mendu V., and Verma P. C.. 2016. Molecular characterization of vitellogenin and vitellogenin receptor of Bemisia tabaci. Plos One. 11: e0155306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerana M., Kubera A., and Ngernsiri L.. 2014. Analysis of the vitellogenin gene of rice moth, Corcyra cephalonica STAINTON. Arch. Insect Biochem. Physiol. 87: 126–147. [DOI] [PubMed] [Google Scholar]
- Wu W., Sun H., Guo J., Jiang F., Liu X., and Zhang G.. 2015. De novo transcriptome characterization of the ghost moth, Thitarodes pui, and elevation-based differences in the gene expression of its larvae. Gene. 574: 95–105. [DOI] [PubMed] [Google Scholar]
- Yano K., Sakurai M. T., Izumi S., and Tomino S.. 1994a. Vitellogenin gene of the silkworm, Bombyx mori: structure and sex-dependent expression. FEBS Lett. 356: 207–211. [DOI] [PubMed] [Google Scholar]
- Yano K., Sakurai M. T., Watabe S., Izumi S., and Tomino S.. 1994b. Structure and expression of mRNA for vitellogenin in Bombyx mori. Biochim. Biophys. Acta. 1218: 1–10. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Xu L., Zhang S., Liu X., An Z., Wang M., and Guo Y.. 2009. Genetic diversity of Ophiocordyceps sinensis, a medicinal fungus endemic to the Tibetan Plateau: implications for its evolution and conservation. BMC Evol. Biol. 9: 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Ma L., Xiao H., Xie B., Smagghe G., Guo Y., and Liang G.. 2016. Molecular characterization and function analysis of the vitellogenin receptor from the cotton bollworm, Helicoverpa armigera (Hübner) (Lepidoptera, Noctuidae). Plos One. 11: e0155785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Sun Y., Xiao L., Tan Y., and Bai L.. 2016. Molecular characterization and expression of vitellogenin gene from Spodoptera exigua exposed to cadmium stress. Gene. 593: 179–184. [DOI] [PubMed] [Google Scholar]
- Zhong R., Ding T. B., Niu J. Z., Xia W. K., Liao C. Y., Dou W., and Wang J. J.. 2015. Molecular characterization of vitellogenin and its receptor genes from citrus red mite, Panonychus citri (McGregor). Int. J. Mol. Sci. 16: 4759–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.