Summary
To date, the pathogenesis of Ménière's disease (MD) remains unclear. This study aims to investigate the possible relationship between potential immune system‐related genes and sporadic MD. The whole RNA‐sequencing (RNA‐seq) technology was used to analyse the transcriptome of peripheral blood mononuclear cells of three MD patients and three control individuals. Of 366 differentially expressed genes (DEGs), 154 genes were up‐regulated and 212 genes were down‐regulated (|log2 fold change| > 1 and P < 0·05). Gene ontology (GO) enrichment analysis illustrated that immune relevant factors played a key role in the pathogenesis of MD. Of 366 DEGs, we focused upon analysing the possible immune‐related genes, among which the significantly up‐regulated genes [glutathione S‐transferase mu 1 (GSTM1), transmembrane protein 176 (TMEM176)B, TMEM176A] and down‐regulated genes [solute carrier family 4 member (SLC4A)10 and SLC4A1] especially drew our attention. The mRNA expression levels of GSTM1, TMEM176B, TMEM176A, SLC4A1 and SLC4A10 were analysed by quantitative reverse transcription–polymerase chain reaction (qRT–PCR). The serum concentration of GSTM1, TMEM176B and SLC4A10 proteins were measured by enzyme‐linked immunosorbent assay (ELISA). Considering the results of qRT–PCR and ELISA, it was noteworthy that GSTM1 exhibited the highest fold change between two groups, which was consistent with the deep sequencing results by RNA‐seq. In conclusion, our study first offers a new perspective in MD development on the basis of RNA expression patterns, suggesting that immune factors might be involved in the MD pathogenesis. Remarkably, GSTM1 might be a possible candidate gene for the diagnostic biomarker of MD and provides the basis for further biological and functional investigations.
Keywords: immune system, Ménière's disease, RNA sequencing, transcriptome
Introduction
Ménière's disease (MD), a chronic and complex disease of the inner ear, is characterized by episodic vertigo, fluctuating hearing loss, ear fullness, tinnitus and progressive vestibular dysfunction. MD was found mainly in the 40–60‐year age group 1, 2. A sex difference has been reported in MD, with the ratio of women to men 1·3 : 1 in Japan and 1·89 : 1 in the United States 3, 4. Epidemiological studies showed that the estimated prevalence of MD varied from 17 to 513 cases per 100 000 individuals, which was higher than the prevalence of systemic lupus erythematosus and multiple sclerosis 5, 6, 7, 8. Notably, it was estimated that definite familial MD might be found in 6·3% of South Koreans and 8·4% in Spain. Most cases were shown with an autosomal‐dominant inheritance pattern 9, 10. Recent findings showed missense variants in protein kinase C beta (PRKCB), dermatopontin (DPT) and semaphorin 3D (SEMA3D) genes in familial MD patients influencing low‐to‐middle frequency sensorineural hearing loss (SNHL) 11, 12. In addition to the genetic factors, excess production of free radicals or oxidative stress were involved in the development of endolymphatic hydrops and chromogranin A (CgA) changes in the homeostatic mechanisms might have a role in hydrops and vertigo clustering attack 13, 14.
However, the pathogenesis of MD is still poorly understood. It was speculated that MD might be an immune‐mediated or even an autoimmune disease 15, 16, 17. The prevalence of systemic autoimmune diseases such as rheumatoid arthritis, ankylosing spondylitis and systemic lupus erythematosus in patients with MD were three‐ to eightfold higher than in the general population 18, 19. Approximately one‐third of MD patients seemed to be of an autoimmune origin, although the immunological mechanisms involved were not clear 20, 21. Autoimmunity and human leucocyte antigen (HLA) associations were focused on early investigations into the causes of MD 22, 23. Studies found that the frequency of human leucocyte antigen (HLA)Cw*04, HLA‐DRB1*1101 and the allelic group HLA‐DRB1*11 might suggest an increased susceptibility to develop MD 22, 24, 25. It has been shown that some major histocompatibility complexes (MHCs) and HLAs were related to MD, supporting that the autoimmune mechanism might be involved in the pathogenesis of MD 26, 27. Intratympanic injection of dexamethasone has been adapted as an anti‐immune or anti‐inflammatory therapy for patients with intractable MD, which preserved the inner ear function probably by its anti‐inflammatory and ion‐homeostatic effects 17, 28, 29.
RNA‐seq can quantify accurately the expression levels of genes and establish a global view of the whole genome 30, 31. RNA‐seq has become a powerful tool to resolve the global pattern of gene expression, including the discovery of an unprecedented capability of new genes, expression and sequence variation of allele‐specific expression 32, 33. The most common use of RNA‐seq has identified that genes are expressed differentially between two or more conditions. High‐throughput RNA‐seq offers the ability to discover new genes or transcription groups and detects transcript expression 34.
RNA‐seq technology has been reported on the potential molecular aetiology or therapeutic targets in a variety of malignancies, including liver cancer 35, prostate cancer 36 and lung cancer 37, etc. However, there are no reports on the application of RNA‐seq in the study of MD. Here, we use RNA‐seq technology to characterize and identify those differentially expressed genes in MD. The study of MD blood samples may help to detect the underlying pathogenesis of MD, understand the causes more clearly and even develop more effective and targeted treatment of MD in the future.
Materials and methods
Patients and controls
The protocols and informed consent forms used in this study were approved by the Ethics Committee of the Shandong Provincial Hospital affiliated to Shandong University, Jinan, China. All the participants signed a written informed consent. The diagnostic criteria of definite unilateral MD were formulated by the Classification Committee of the Bárány Society in 2015 38. Sporadic patients who we selected from the Department of Otolaryngology – Head and Neck Surgery (Shandong Provincial Hospital affiliated to Shandong University) met the 2015 diagnostic criteria. All patients we chose met unilateral MD type 1, which was defined as sporadic and classic MD without family factors, migraine and autoimmune disorder (AD) 39, 40. A complete neuro‐otology assessment was carried out to exclude patients with tympanic membrane perforation, infection or tumour of ear and acoustic nerve disease. Brain magnetic resonance imaging (MRI) was performed to rule out any concomitant neurological lesions. The controls were volunteers without cochlea‐vestibular disorders and systemic autoimmune diseases.
Sample collection and RNA extraction
Blood was collected from three volunteers and three patients with unilateral MD type 1 between 7:00 a.m. and 10:00 a.m. to limit the effect of circadian variation of cytokine production. For each individual, 4 ml blood was put into an ethylenediamine tetraacetic acid (EDTA) BD Vacutainer tube (BD, New York, NY, USA) and diluted with an equal volume of sterile phosphate‐buffered saline (PBS). Peripheral blood mononuclear cells (PBMCs) from the venous blood of healthy volunteers and MD patients were isolated by Ficoll‐Hypaque density‐gradient separation (Lympholyte‐H; Cedarlane Laboratories, Burlington, Ontario, Canada). Briefly, blood samples were first centrifuged at 800 g for 20 min at room temperature. The well‐defined lympholyte layer at the second interface (from the top) was then transferred carefully to another new centrifuge tube. Lymphocytes were washed three times and centrifuged at 250 g for 10 min at −80°C before RNA extraction. Total RNA of PMBCs from MD patients and healthy volunteers were isolated using Trizol reagent (Life Technologies, Carlsbad, CA, USA). RNA quality was assessed using the Agilent 2100 Bioanalyzer and the RNA 6000 Nano Kit (Agilent Technologies Inc., Santa Clara, CA, USA).
RNA preparation, library construction and Illumina sequencing
A total amount of 2 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, Ipswich, MA, USA), following the manufacturer's recommendations, and index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly‐T oligo‐attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEB Next first‐strand synthesis reaction buffer (×5). First‐strand cDNA was synthesized using random hexamer primer and RNase H. Second‐strand cDNA synthesis was performed subsequently using buffer, dNTPs, DNA polymerase I and RNase H. The cDNAs were assessed using Agilent Bioanalyzer 2100 system (Agilent Technologies, USA) and ABI StepOnePlus™ real‐time PCR system (ABI, Life Technologies, Carlsbad, CA, USA). The libraries were sequenced on an Illumina Hiseq 4000 platform, and 150 base pairs (bp) paired‐end reads were generated.
Reverse transcription–polymerase chain reaction (RT–PCR)
Total RNA was extracted from PMBCs of 30 paired patients with unilateral MD type 1 and control individuals using the Trizol Reagent (Life Technologies), according to the manufacturer's instructions. In the presence of random primers, 1 μg of total mRNA was reverse‐transcribed into complementary DNA using the Revert Aid First‐Strand cDNA Synthesis Kit (Thermo Scientific, Fremont, CA, USA), following the manufacturer's protocols. Gene expression was examined by quantitative RT–PCR (qRT–PCR) with the SYBR Premix Ex Taq (TaKaRa, Shiga, Japan) and an Eppendorf PCR machine (Hamburg, Germany). The PCR conditions were as follows: initial denaturation at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 40s; annealing at 60°C for 40s; and extension at 72°C for 45s. The primer sets are described in Table 1. The specificity of each PCR reaction was confirmed by melting curve analysis. The expression levels of genes were normalized by 18S rRNA. Each group contained three samples and each PCR was repeated in triplicate. The expression of gene was analysed using the 2−ΔΔCt method.
Table 1.
Polymerase chain reaction (PCR) primer sequences used in the experiment.
| Gene | Forward sequence | Reverse sequence |
|---|---|---|
| GSTM1 | GGGACGCTCCTGATTATGAC | TTGCTCTGGGTGATCTTGTG |
| TMEM176B | GGCGAAGTCAAGAGAACCAA | CAAGACACAGACAGCCAGGA |
| TMEM176A | TACACCCTCCTCGTCACCTC | CAGGGCCCAGTATGTACCAC |
| NSF‐P1 | CGACAAGATGGCAGCAGAAT | GGCTTCAATGTCCTTCACCA |
| MYL4 | GCCAGAACCCTACCAATGC | CCCTCCACGAAGTCCTCATA |
| SLC4A1 | ATGGAGGAGAATCTGGAGCA | GGGTGTGATGTGGTGTGGTA |
| SLC4A10 | AGATTCCTCCAGGTGCTGAA | CTTCAGCCAGTCCTTGAAGC |
| UTY | ACTGGAATGGTGGCCAGAGT | TGCTCGCAGTTGTTCCAAGT |
| 18S rRNA | CGCGGTTCTATTTTGTTGGT | AGTCGGCATCGTTTATGGTC |
GSTMI = glutathione S‐transferase mu 1; TMEM176 = transmembrane protein 176; NFS‐P1 = N‐ethylmaleimide sensitive factor, vesicle fusing ATPase; MYL4 = myosin light‐chain 4; SLC4A = solute carrier family 4 member; UTY = ubiquitously transcribed tetratricopeptide repeat containing, Y‐linked (UTY).
Enzyme linked immunosorbent assay (ELISA)
Blood was collected from another 30 paired patients with unilateral MD type 1 and control individuals at 7:00 am to 10:00 am in order to eliminate the effect of circadian variation of cytokine secretion. After samples were centrifuged at 1000 g for 20 min at room temperature, blood sera were collected from the top and stored at −80°C before ELISA. Serum concentrations of glutathione S‐transferase mu 1 (GSTM1), transmembrane protein 176 (TMEM176)B and solute carrier family 4 member (SLC4A)10 protein products were measured using ELISA kits (MyBioSource, San Diego, CA, USA), according to the manufacturer's instructions. All analyses and calibrations were performed in triplicate. The concentrations were determined by comparing the optical density (OD) values of the samples to the standard curve using a spectrophotometer at 450 nm.
Statistical analysis
Data were analysed statistically using spss version 19.0 software (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism Software (San Diego, CA, USA). All measurements and calculations are presented as mean ± standard error of the mean (s.e.m.). Student's t‐test was used to compare data between MD and control groups; P < 0·05 was considered statistically significant.
Results
General information of the RNA sequencing data
The expression of every gene was measured by fragments per kilobase of transcript per million fragments mapped (FPKM). An average of 42 007 genes were detected in sequenced samples, by requiring that the FPKM value was greater than 0·1. The correlation analysis showed that the average global profiles of gene expressions between MD and control samples were correlated highly (correlation coefficient r = 0·98, Fig. 1a). After excluding the following FPKM baseline gene, 939 and 806 genes were expressed in the volunteer and MD groups (Fig. 1b). A total of 366 genes were expressed differentially between control and MD samples, with 154 genes up‐regulated and 212 down‐regulated in the MD group (|log2 fold change| > 1 and P < 0·05) (Fig. 1c). The expression patterns of these 366 DEGs were shown as a heat‐map using hierarchical cluster analysis (Fig. 1d). These results indicated that the RNA‐seq system was of good quality and accurate.
Figure 1.
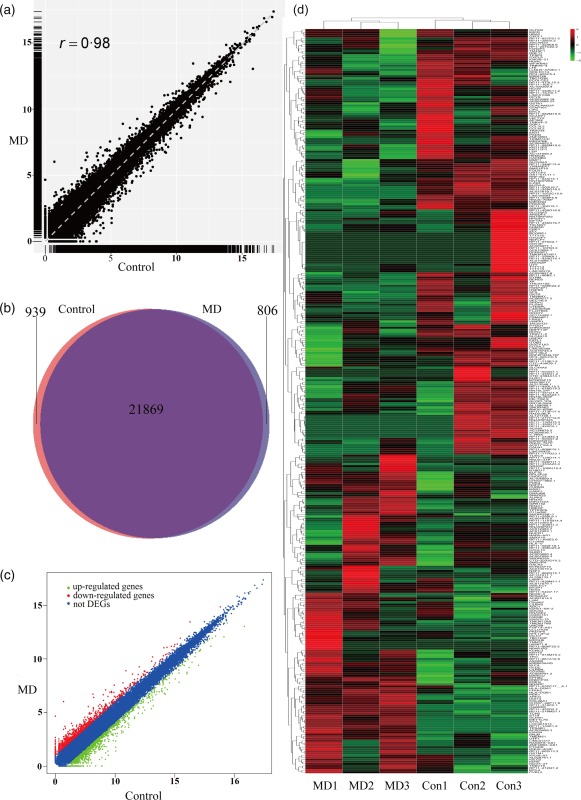
Analysis of homogeneity between peripheral blood monocular cells of volunteers and Ménière's disease (MD) patients. (a) Expression analysis of volunteer and MD samples. The Pearson correlation coefficient is shown. (b) Venn diagram showing 939 and 806 genes expressed in volunteer and MD group. (c) Scatterplot of differentially expressed genes and sample clustering analysis for all replicates of volunteer and MD samples. Results show the expression levels of 154 genes are observed up‐regulated and 212 genes are observed down‐regulated (P < 0·05 and |log2 fold change| >1). (d) Heat map is generated from normalized fragments per kilobase (FPKM) of 366 consistent differentially expressed genes (DEGs) among three paired samples. The expression level of each transcript is represented by a colour range from green (low) to red (high). [Colour figure can be viewed at wileyonlinelibrary.com]
Expression levels of the top 200 most abundant genes in MD or control groups
In order to describe the characterization of gene expression profiles in the control and MD groups, genes that were expressed most abundantly in two populations were analysed, respectively. The expression levels of the top 200 most abundant transcripts in control group are shown in Fig. 2a. For comparison, expression levels and abundance rankings for the same transcripts in the MD group are presented at the same time. Figure 2b shows the 200 most abundant transcripts in MD group in a similar manner. As shown in both figures, the majority of the transcripts expressed richly in one group were also expressed abundantly in the other. Notably, among those most abundantly expressed genes, HBB, FCGR3B and MAP3K7CL were only expressed richly in the control group (Fig. 2a), and LRP1 and SLC11A1 were only expressed richly in the MD group (Fig. 2b).
Figure 2.
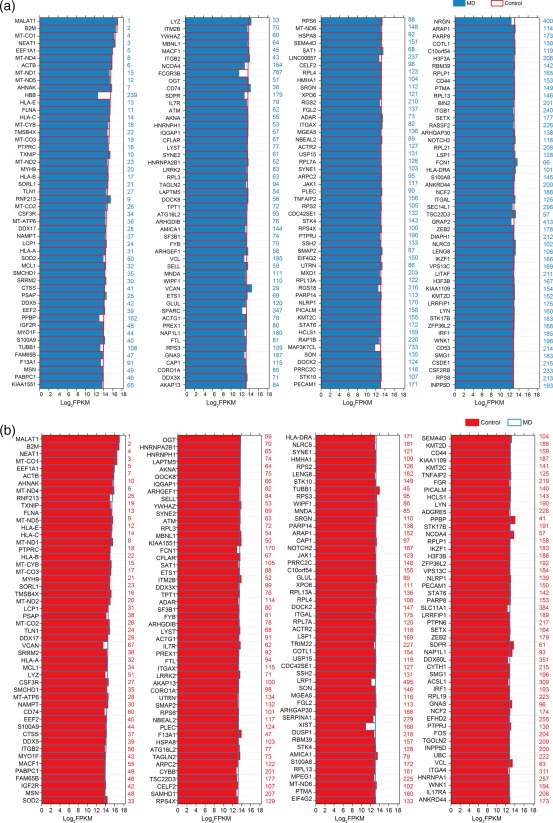
Expression levels of the top 200 genes in volunteer and Ménière's disease (MD) samples. (a) Expression levels of the top 200 genes in the volunteer group in descending order. Numbers in blue on the right side of each panel represents the ranking of the same genes in MD group. (b) Expression levels of the top 200 genes in the MD group in descending order. Numbers in red on the right side of each panel represents the ranking of the same genes in volunteer group. [Colour figure can be viewed at wileyonlinelibrary.com]
Differentially expressed genes in MD and control groups
To determine the genes that were expressed differentially in the control and MD groups, the expression levels of all transcripts detected in the control group were compared with those in the MD group, and the top differentially expressed genes (DEGs) in both populations were selected. Figure 3a shows the general view of the detected transcripts in both populations. DEGs were classified as those whose expression levels were above background and at least twofold different in two groups (P < 0·05). Figure 3b,c illustrates the top 150 DEGs in the control and MD groups, respectively. Notably, the DEGs highly expressed in the MD group, such as NSFP1, TMEM176B, TMEM176A and MYL4, have not been characterized previously and need to be studied further in future.
Figure 3.
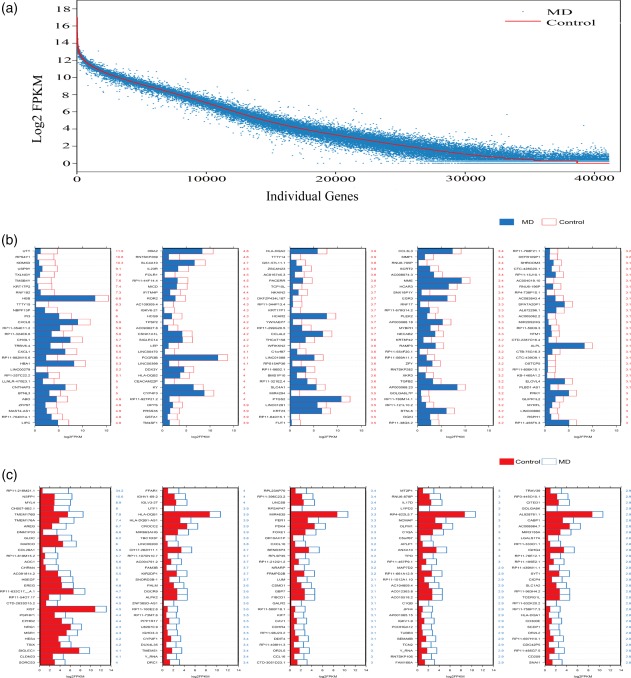
Differentially expressed genes (DEGs) in control and Ménière's disease (MD) groups. (a) All DEGs in control and MD samples. The red line indicates the expression level of 36 333 transcripts in the control group, and each blue dot represents the expression level of the same transcripts in the MD group. (b) The 150 most DEGs in the control group. The numerical values in red on the right side of each panel represent the fold difference in expression for control versus MD. (c) The 150 most DEGs in the MD group. The numerical values in blue on the right side of each panel represent the fold difference in expression for MD versus control. [Colour figure can be viewed at wileyonlinelibrary.com]
Figure 4a,b shows the top 50 uniquely expressed genes (UEGs) in the control and MD groups, the majority of which were expressed at relatively low levels in the other group, with approximately half being uncharacterized genes and half the known genes being non‐coding RNAs. It was noteworthy that GSTM1 was expressed specifically in all MD RNA‐seq data, which was in the first place in the top50‐UEGs list and was selected to be validated in the following validation.
Figure 4.
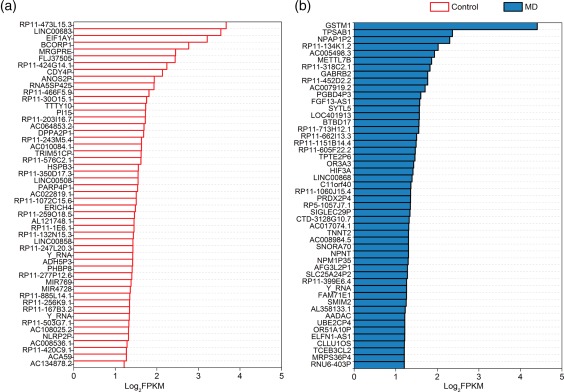
The top 50 uniquely expressed genes in control and Ménière's disease (MD) groups. (a) The top 50 uniquely expressed genes in control group in descending order. (b) The top 50 uniquely expressed genes in MD group in descent order. Glutathione S‐transferase mu 1 (GSTM1) exhibits highest fold change between MD patients and controls. [Colour figure can be viewed at wileyonlinelibrary.com]
Genes related to ion homeostasis
In order to alleviate vertigo in MD patients, it was important to restore and maintain ion homeostasis. Figure 5 shows the possible genes related to ion homeostasis in the control and MD groups. We found that the solute carrier (SLC) family members were expressed differentially in both populations, with SLC4A10, SLC4A1 being down‐regulated significantly as well as SLC9A2 and SLC8A2 up‐regulated in MD versus control (Fig. 5).
Figure 5.
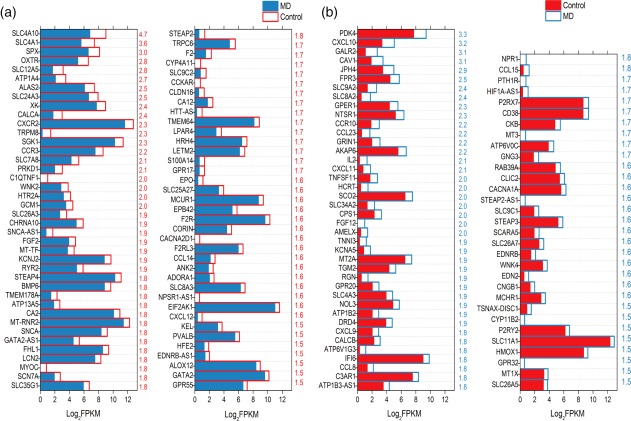
Genes related to ion homeostasis. (a) Comparison of expression levels for genes that encode proteins associated with ion homeostasis by dividing the value of control with that of Ménière's disease (MD). Numerical values in red in the right side of each panel represent the ratio of control versus MD. (b) Comparison of expression levels for genes that encode proteins associated with ion homeostasis by dividing the value of MD with that of control. Numerical values in blue in the right side of each panel represent the ratio of MD versus control. [Colour figure can be viewed at wileyonlinelibrary.com]
Gene ontology (GO) enrichment of the genes expressed differentially in MD and control groups
In order to gain overall insight into the function of annotation genes, the GO functional classification was performed. A total of 298 clusters (P < 0·05) was annotated significantly with the GO function of biological process (BP). GO analysis demonstrated that the up‐regulated enrichment was enriched in specific biological processes, including cellular response to corticotrophin‐releasing hormone stimulus, negative regulation of dendritic cell differentiation, cellular response to histamine and down‐regulated with the chemokine‐mediated signalling pathway and fatty acid elongation (P < 0·01) (Fig. 6). The top four of the up‐regulated enrichments were related closely to the immune system. These results suggest that further studies may be required to elucidate whether the immune system is involved in the pathogenesis of MD.
Figure 6.
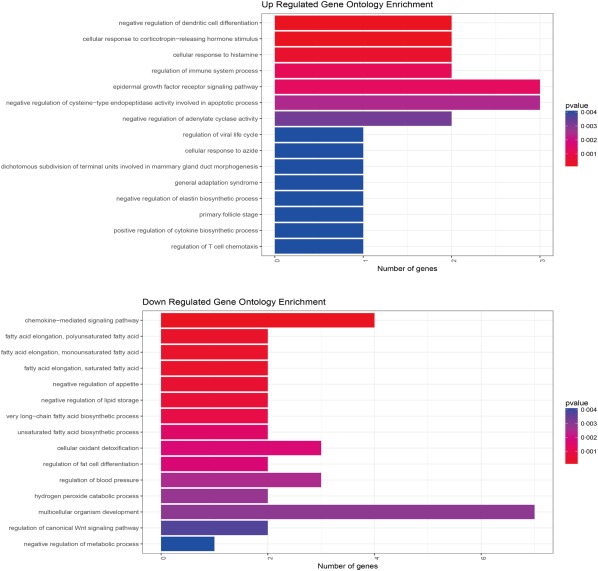
Top gene ontology (GO) analysis of differentially expressed genes (DEGs) in Ménière's disease (MD) patients. The top enriched GO terms of the up‐regulated and down‐regulated DEGs in biological progress are shown. The adjusted enrichment P‐ values are labelled in different colours in the graph. [Colour figure can be viewed at wileyonlinelibrary.com]
Protein–protein interaction (PPI) network for the protein products of the differentially expressed genes in MD and control groups
The PPI network was constructed based on the Search Tool for the Retrieval of Interacting Genes (STRING) database to depict their complex relationship between control and MD patients. As shown in Fig. 7a, proteins encoded by DEGs in both populations presented a complicated interaction network, which might facilitate the further exploration of molecular mechanisms underlying MD development. The specific PPI networks for protein products of targeted DEGs, GSTM1, SLC4A10, TMEM176A and TMEM176B are shown in Fig. 7b, supplying us with feasible candidates for further research concerning PPI.
Figure 7.
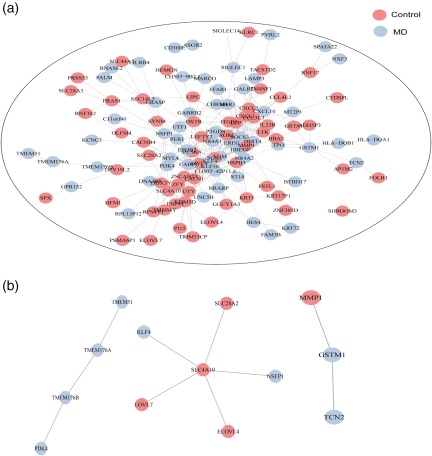
Protein–protein interaction network analysis of the differentially expressed genes (DEGs) in control and Ménière's disease (MD) groups. (a) PPI network for DEGs in both groups supplying a complicated interaction network. (b) Specific protein–protein interaction (PPI) network for protein products of glutathione S‐transferase mu 1 (GSTM1), solute carrier family 4 member (SLC4)A10, transmembrane protein 176 (TMEM176)A and TMEM176B. [Colour figure can be viewed at wileyonlinelibrary.com]
Verification of RNA sequencing data by qRT–PCR and ELISA
qRT‐PCR was first applied to verify the expression levels of candidate genes. As shown in Fig. 8, the relative mRNA expression levels of GSTM1, TMEM176B, TMEM176A, NSFP1 and MYL4 were increased differentially and SLC4A1, SLC4A10, UTY were decreased in MD patients, which were consistent with the RNA‐seq data.
Figure 8.
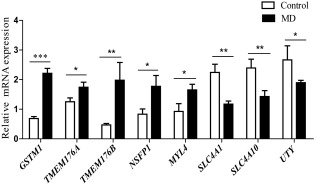
Expression levels of differentially expressed genes (DEGs) in validation cohort by quantitative reverse transcription–polymerase chain reaction (qRT–PCR) in 30 paired samples. The mRNA expression levels of glutathione S‐transferase mu 1 (GSTM1), T transmembrane protein 176 (TMEM176)A, TMEM176B, N‐ethylmaleimide‐sensitive factor pseudogene 1 (NSF‐P1), myosin light‐chain 4 (MYL4), solute carrier family 4 member (SLC4A)1, SLC4A10 and ubiquitously transcribed tetratricopeptide repeat containing, Y‐linked (UTY) are presented, respectively. Data are presented as the relative fold change in expression. Asterisk indicates a significant difference when compares with controls. *P < 0·05; **P < 0·01; ***P < 0·001. DEGs: differentially expressed genes.
Human serum samples were analysed for the levels of GSTM1, TMEM176B and SLC4A10 protein products by ELISA. As shown in Fig. 9, the total GSTM1, TMEM176B and SLC4A10 protein levels are shown in MD serum compared to those in the control group, respectively (P < 0·05), consistent with the results of qRT–PCR. The serum concentration of GSTM1 protein was increased substantially in MD patients. These indicate that the gene expression observed in blood transcriptome between the two groups was highly credible.
Figure 9.
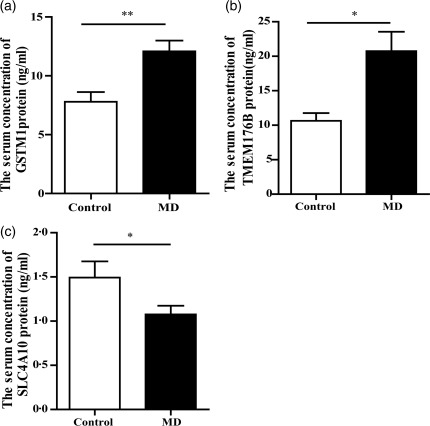
Validation of protein products by enzyme‐linked immunosorbent assay (ELISA) in another 30 paired samples. (a‐c) The serum levels of glutathione S‐transferase mu 1 (GSTM1), transmembrane protein 176 (TMEM176)B and solute carrier family 4 member (SLC4A)10 protein products are shown for each group studied. Asterisk indicates a significant difference when compared with controls. *P < 0·05; ** P < 0·01·
Discussion
To our knowledge, this is the first description of the global transcriptome of differential genes between controls and sporadic MD patients using RNA‐seq technology. In the present study, we found that a total of 366 genes were expressed differentially between control and MD samples, in which 154 genes were up‐regulated and 212 were down‐regulated in the MD group. These findings indicate that the initial data are obtained successfully with RNA‐seq technology, which lays a solid foundation for the subsequent experiments.
Of these top 150 DEGs, certain genes, i.e. TMEM176B and TMEM176 highly expressed in MD, especially drew our attention. Previous reports have suggested that TMEM176B was an immature dendritic cell marker and its expression was up‐regulated in blood of patients with acute rejection, indicating that it was associated preferentially with alloantigen presentation and inflammation 41, 42. Moreover, studies have demonstrated that TMEM176A and TMEM176B were expressed highly in immature monocytes and dendritic cells (DCs) of rat and human cancer tissues 43, 44. It is conceivable that TMEM176B and TMEM176A may contribute to the pathogenesis of MD via regulation of the activities in the immune system.
It was reported that a genetically induced dysfunction of ionic transport, which affected the ion homeostasis in the inner ear, might act as a predisposing factor to develop MD 45, 46. In this study, we found that SLC4A10 and SLC4A1 were down‐regulated significantly in MD. A previous report suggested that SLC4A10 participated in the regulation of pH homeostasis in leucocytes, which indicated that inflammation was associated with a local pH reduction and might be a ‘danger signal’ to activate immune responses 47. Recently, another SLC family member, SLC4A1, had been hypothesized to be a candidate gene in the pathogenesis of lupus and lupus nephritis, which stimulated the functional activity of innate immune cells and cell‐specific functions 48. Interestingly, SLC4A1 was reported to be over‐expressed significantly in the human endolymphatic sac, compared to adjacent dura mater 49, which was contrary to the result seen in our study. Possible explanations might be that the methods and the sources of the organizations are different and thus require further investigation to understand the potential differences.
Next, we analysed the PPI network for genomewide RNA‐seq of two groups. The analysis showed that DEGs were enriched in different ways, supporting the hypothesis that these genes might have different networks of mechanisms mediated by a diverse array of signalling molecules. The specific PPI network for protein products of targeted DEGs, GSTM1, SLC4A10, TMEM176A and TMEM176B were also presented, which indicated that the regulation of those genes could be governed by the interaction with other members of the components. Later, the results were confirmed by qRT–PCR and ELISA, which demonstrated the accuracy and reliability of the RNA‐seq data.
Interestingly, it is noteworthy that MD patients demonstrated higher levels of GSTM1 transcripts relative to controls. Several studies have suggested that GSTM1 was associated with higher oxidative stress and increased risk for inflammatory diseases, including rheumatoid arthritis 50, systemic lupus erythematosus 51, ulcerative colitis 52, ankylosing spondylitis 5 and inflammatory bowel disease 6. In addition, the increased frequency of HLA and GSTM1 plays an important role in the alloimmune responses of renal transplant recipients 53. Moreover, null genotypes of GSTM1 and GSTT1 involved in oxidative stress and mitochondrial dysfunction were susceptible to noise‐induced temporary threshold shifts for high frequencies and were found to have an 8·88‐fold risk for the onset of presbycusis 54, 55.
Recent studies have demonstrated that several allelic variations in immune response genes such as MHC class I polypeptide‐related sequence A (MICA), Toll‐like receptor (TLR)‐10 or nuclear factor kappa B1 (NF‐κB1) were associated with hearing loss progression in MD patients. Some data suggested that allelic variants of MICA and TLR‐10 genes, involved in the innate immune response, might influence the susceptibility and time–course of hearing loss of MD in European populations 56, 57. A study showed that allelic variants in NF‐κB1 influenced the hearing outcome in patients with unilateral MD. Moreover, steroids were potent blockers of the NF‐κB1 pathway, which might explain the observed response to systemic steroid treatment in patients with sudden SNHL or MD 58. Notably, a meta‐analysis demonstrated that the functional GSTM1 and NF‐κB1 polymorphisms were associated with SLE risk in Asians 59.
However, there are no reports concerning the expression of GSTM1 in MD study. Our research shows that GSTM1 exhibits a high fold change between two groups and has been proposed to have a functional association with MD. The findings from the current study provide convincing evidence that the immune system is involved in the pathogenesis of MD, which is consistent with a previous study 60. Our results demonstrate that a high level of GSTM1 expression detected in MD blood indicates a higher probability of immune system involvement in MD development. Biological information analyses suggest that immune factors might be involved in the development of MD and GSTM1 might be a candidate potential biomarker for MD diagnosis.
In conclusion, the present study indicates that the RNA‐seq system is of good quality, accurate and reproducible. The RNA expression patterns suggest that immunological factors may be involved in MD pathogenesis. Additionally, the present work points out possible target genes related with MD pathogenesis through layers of screening. To our knowledge, the top UEG in MD, GSTM1, is probably a candidate for the diagnostic biomarkers of MD and provides as basis for further biological and functional investigations. However, our results from RNA‐seq were based on a small sample size, which might not provide sufficient information to support our hypothesis. Further studies with large samples are needed to identify the accuracy of GSTM1 in MD diagnosis and the relationship between GSTM1 level and future treatment modalities, and to determine the true prevalence of immune pathological mechanisms underlying MD.
Disclosure
The authors declare that they have no competing interests.
Author contributions
H. W., Z. F. and J. L. helped conceive the project. Y. S. and D. Z. designed and performed experiments, analysed data and wrote the manuscript. G. S. and Y. S. evaluated all results, helped the project and wrote the manuscript. Y. L., Y. L. and X. L. selected patients and characterized the study cohort. Finally, all authors revised the manuscript. All authors obtained permission to acknowledge from all those mentioned in the Acknowledgements.
Acknowledgements
H. Wang, Z. Fan and J. Li helped conceive the project. Y. Sun and D. Zhang designed and performed experiments, analyzed data and wrote the manuscript. G. Sun and Y. Song evaluated all results, helped the project and wrote the manuscript. Y. Lv, Y. Li and X. Li selected patients and characterized the study cohort.
Contributor Information
Z. Fan, Email: fanent@126.com
H. Wang, Email: whboto11@163.com.
References
- 1. Van Esch BF, Van Benthem PP, Van Der Zaag‐Loonen HJ, Bruintjes TD. Age of onset of Meniere's disease in the Netherlands: data from a specialised dizziness clinic. J Laryngol Otol 2016; 130:624–7. [DOI] [PubMed] [Google Scholar]
- 2. Iwasaki S, Yamasoba T. Dizziness and imbalance in the elderly: age‐related decline in the vestibular system. Aging Dis 2015; 6:38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harris JP, Alexander TH. Current‐day prevalence of Meniere's syndrome. Audiol Neurootol 2010; 15:318–22. [DOI] [PubMed] [Google Scholar]
- 4. Shojaku H, Watanabe Y, Fujisaka M et al Epidemiologic characteristics of definite Meniere's disease in Japan. A long‐term survey of Toyama and Niigata prefectures. ORL J Otorhinolaryngol Relat Spec 2005; 67:305–9. [DOI] [PubMed] [Google Scholar]
- 5. İnal EE, Görükmez O, Eroğlu S et al Association of GSTM1, GSTT1, GSTP1‐ILE105VAL and ACE I/D polymorphisms with ankylosing spondylitis. Rheumatol Int 2016; 36:17–23. [DOI] [PubMed] [Google Scholar]
- 6. Al‐Judaibi B, Schwarz UI, Huda N. Genetic predictors of azathioprine toxicity and clinical response in patients with inflammatory bowel disease. J Popul Ther Clin Pharmacol 2016; 23:e26–36. [PubMed] [Google Scholar]
- 7. Havia M, Kentala E, Pyykko I. Prevalence of Meniere's disease in general population of Southern Finland. Otolaryngol Head Neck Surg 2005; 133:762–8. [DOI] [PubMed] [Google Scholar]
- 8. Watanabe Y, Mizukoshi K, Shojaku H, Watanabe I, Hinoki M, Kitahara M. Epidemiological and clinical characteristics of Meniere's disease in Japan. Acta Otolaryngol Suppl 1995; 519:206–10. [DOI] [PubMed] [Google Scholar]
- 9. Lee JM, Kim MJ, Jung J, Kim HJ, Seo YJ, Kim SH. Genetic aspects and clinical characteristics of familial Meniere's disease in a South Korean population. Laryngoscope 2015; 125:2175–80. [DOI] [PubMed] [Google Scholar]
- 10. Requena T, Espinosa‐Sanchez JM, Cabrera S. Familial clustering and genetic heterogeneity in Meniere's disease. Clin Genet 2014; 85:245–52. [DOI] [PubMed] [Google Scholar]
- 11. Martin‐Sierra C, Requena T, Frejo L et al A novel missense variant in PRKCB segregates low‐frequency hearing loss in an autosomal dominant family with Meniere's disease. Hum Mol Genet 2016; 25:3407–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martín‐Sierra C, Gallego‐Martinez A, Requena T, Frejo L, Batuecas‐Caletrío A, Lopez‐Escamez JA. Variable expressivity and genetic heterogeneity involving DPT and SEMA3D genes in autosomal dominant familial Meniere's disease. Eur J Hum Genet 2017; 25:200–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Teranishi M, Uchida Y, Nishio N et al Polymorphisms in genes involved in the free‐radical process in patients with sudden sensorineural hearing loss and Meniere's disease. Free Radic Res 2013; 47:498–506. [DOI] [PubMed] [Google Scholar]
- 14. Teggi R, Colombo B, Trimarchi M et al Altered chromogranin A circulating levels in Meniere's disease. Dis Markers 2015; 2015:643420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fattori B, Nacci A, Dardano A et al Possible association between thyroid autoimmunity and Meniere's disease. Clin Exp Immunol 2008; 152:28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moller MN, Kirkeby S, Caye‐Thomasen P. Innate immune defense in the inner ear – mucines are expressed by the human endolymphatic sac. J Anat 2017; 230:297–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Suchan M, Kaliarik L, Krempaska S, Koval J. Immune‐mediated cochleovestibular disease. Bratisl Lek Listy 2016; 117:87–90. [DOI] [PubMed] [Google Scholar]
- 18. Piacentini S, Polimanti R, Moscatelli B, Re MA, Manfellotto D, Fuciarelli M. Lack of association between GSTM1, GSTP1, and GSTT1 gene polymorphisms and asthma in adult patients from Rome, central Italy. J Investig Allergol Clin Immunol 2012; 22:252–6. [PubMed] [Google Scholar]
- 19. Gazquez I, Soto‐Varela A, Aran I et al High prevalence of systemic autoimmune diseases in patients with Meniere's disease. PLoS One 2011; 6:e26759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Krzeska‐Malinowska I, Held‐Ziolkowska M, Kowalska M, Niemczyk K. The role of immunological factors in Meniere's disease. Otolaryngol Pol 2002; 56:583–7. [PubMed] [Google Scholar]
- 21. Mazor Y, Koifman E, Elkin H et al Risk factors for serious adverse effects of thiopurines in patients with Crohn's disease. Curr Drug Saf 2013; 8:181–5. [DOI] [PubMed] [Google Scholar]
- 22. Rawal SG, Thakkar KH, Ziai K, Santi PA, Djalilian HR. HLA‐B27‐associated bilateral Meniere disease. Ear Nose Throat J 2010; 89:122–7. [PubMed] [Google Scholar]
- 23. Klockars T, Kentala E. Inheritance of Meniere's disease in the Finnish population. Arch Otolaryngol Head Neck Surg 2007; 133:73–7. [DOI] [PubMed] [Google Scholar]
- 24. Khorsandi MT, Amoli MM, Borghei H et al Associations between HLA‐C alleles and definite Meniere's disease. Iran J Allergy Asthma Immunol 2011; 10:119–22. [PubMed] [Google Scholar]
- 25. Lopez‐Escamez JA, Vilchez JR, Soto‐Varela A et al HLA‐DRB1*1101 allele may be associated with bilateral Meniere's disease in southern European population. Otol Neurotol 2007; 28:891–5. [DOI] [PubMed] [Google Scholar]
- 26. Gazquez I, Moreno A, Aran I et al MICA‐STR A.4 is associated with slower hearing loss progression in patients with Meniere's disease. Otol Neurotol 2012; 33:223–9. [DOI] [PubMed] [Google Scholar]
- 27. Lopez‐Vazquez A, Mozo L, Alonso‐Arias R et al Autoantibodies against MHC class I polypeptide‐related sequence A are associated with increased risk of concomitant autoimmune diseases in celiac patients. BMC Med 2014; 12:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sennaroglu L, Sennaroglu G, Gursel B, Dini FM. Intratympanic dexamethasone, intratympanic gentamicin, and endolymphatic sac surgery for intractable vertigo in Meniere's disease. Otolaryngol Head Neck Surg 2001; 125:537–43. [DOI] [PubMed] [Google Scholar]
- 29. Beyea JA, Instrum RS, Agrawal SK, Parnes LS. Intratympanic dexamethasone in the treatment of Meniere's disease: a comparison of two techniques. Otol Neurotol 2017; 38:e173–8. [DOI] [PubMed] [Google Scholar]
- 30. Wang Z, Gerstein M, Snyder M. RNA‐Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009; 10:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA‐seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 2008; 18:1509–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Djebali S, Davis CA, Merkel A et al Landscape of transcription in human cells. Nature 2012; 489:101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 2011; 12:87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bruno VM, Wang Z, Marjani SL et al Comprehensive annotation of the transcriptome of the human fungal pathogen Candida albicans using RNA‐seq. Genome Res 2010; 20:1451–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guo Y, Su ZY, Zhang C et al Mechanisms of colitis‐accelerated colon carcinogenesis and its prevention with the combination of aspirin and curcumin: transcriptomic analysis using RNA‐seq. Biochem Pharmacol 2017; 135:22–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nikitina AS, Sharova EI, Danilenko SA et al Novel RNA biomarkers of prostate cancer revealed by RNA‐seq analysis of formalin‐fixed samples obtained from Russian patients. Oncotarget 2017; 8:32990–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang L, Li S, Choi YL et al Systematic identification of cancer‐related long noncoding RNAs and aberrant alternative splicing of quintuple‐negative lung adenocarcinoma through RNA‐Seq. Lung Cancer 2017; 109:21–7. [DOI] [PubMed] [Google Scholar]
- 38. Lopez‐Escamez JA, Carey J, Chung WH et al Diagnostic criteria for Meniere's disease. J Vestib Res 2015; 25:1–7. [DOI] [PubMed] [Google Scholar]
- 39. Frejo L, Soto‐Varela A, Santos‐Perez S et al Clinical subgroups in bilateral Meniere disease. Front Neurol 2016; 7:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Frejo L, Martin‐Sanz E, Teggi R et al Extended phenotype and clinical subgroups in unilateral Meniere disease: a cross‐sectional study with cluster analysis. Clin Otolaryngol 2017; 42:1172–80. [DOI] [PubMed] [Google Scholar]
- 41. Viklicky O, Krystufkova E, Brabcova I et al B‐cell‐related biomarkers of tolerance are up‐regulated in rejection‐free kidney transplant recipients. Transplantation 2013; 95:148–54. [DOI] [PubMed] [Google Scholar]
- 42. Segovia M, Louvet C, Charnet P et al Autologous dendritic cells prolong allograft survival through Tmem176b‐dependent antigen cross‐presentation. Am J Transplant 2014; 14:1021–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Condamine T, Le Texier L, Howie D et al Tmem176B and Tmem176A are associated with the immature state of dendritic cells. J Leukoc Biol 2010; 88:507–15. [DOI] [PubMed] [Google Scholar]
- 44. Cuajungco MP, Podevin W, Valluri VK, Bui Q, Nguyen VH, Taylor K. Abnormal accumulation of human transmembrane (TMEM)‐176A and 176B proteins is associated with cancer pathology. Acta Histochem 2012; 114:705–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Teggi R, Zagato L, Delli Carpini S et al Genetics of ion homeostasis in Meniere's Disease. Eur Arch Otorhinolaryngol 2017; 274:757–63. [DOI] [PubMed] [Google Scholar]
- 46. Nishio N, Teranishi M, Uchida Y et al Polymorphisms in genes encoding aquaporins 4 and 5 and estrogen receptor alpha in patients with Meniere's disease and sudden sensorineural hearing loss. Life Sci 2013; 92:541–6. [DOI] [PubMed] [Google Scholar]
- 47. Rajamaki K, Nordstrom T, Nurmi K et al Extracellular acidosis is a novel danger signal alerting innate immunity via the NLRP3 inflammasome. J Biol Chem 2013; 288:13410–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. AlFadhli S, Ghanem AA, Nizam R. Genome‐wide differential expression reveals candidate genes involved in the pathogenesis of lupus and lupus nephritis. Int J Rheum Dis 2016; 19:55–64. [DOI] [PubMed] [Google Scholar]
- 49. Moller MN, Kirkeby S, Vikesa J, Nielsen FC, Caye‐Thomasen P. Gene expression in the human endolymphatic sac: the solute carrier molecules in endolymphatic fluid homeostasis. Otol Neurotol 2015; 36:915–22. [DOI] [PubMed] [Google Scholar]
- 50. Ji JD, Lee WJ. Association between the polymorphisms of glutathione S‐transferase genes and rheumatoid arthritis: a meta‐analysis. Gene 2013; 521:155–9. [DOI] [PubMed] [Google Scholar]
- 51. Glesse N, Rohr P, Monticielo OA et al Genetic polymorphisms of glutathione S‐transferases and cytochrome P450 enzymes as susceptibility factors to systemic lupus erythematosus in southern Brazilian patients. Mol Biol Rep 2014; 41:6167–79. [DOI] [PubMed] [Google Scholar]
- 52. Varzari A, Deyneko IV, Tudor E, Turcan S. Polymorphisms of glutathione S‐transferase and methylenetetrahydrofolate reductase genes in Moldavian patients with ulcerative colitis: genotype‐phenotype correlation. Meta Gene 2016; 7:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Akgul SU, Oguz FS, Çalişkan Y et al The effect of glutathione S‐transferase polymorphisms and anti‐GSTT1 antibodies on allograft functions in recipients of renal transplant. Transplant Proc 2012; 44:1679–84. [DOI] [PubMed] [Google Scholar]
- 54. Lin CY, Shih TS, Guo YL, Wu JL, Sun YM, Tsai PJ. Effects of gene–environmental interaction on noise‐induced hearing threshold levels for high frequencies (HTLHF). Environ Sci Technol 2011; 45:7128–34. [DOI] [PubMed] [Google Scholar]
- 55. Manche SK, Jangala M, Putta P, Koralla RM, Akka J. Association of oxidative stress gene polymorphisms with presbycusis. Gene 2016; 593:277–83. [DOI] [PubMed] [Google Scholar]
- 56. Requena T, Gazquez I, Moreno A et al Allelic variants in TLR10 gene may influence bilateral affectation and clinical course of Meniere's disease. Immunogenetics 2013; 65:345–55. [DOI] [PubMed] [Google Scholar]
- 57. Hinds DA, McMahon G, Kiefer AK et al A genome‐wide association meta‐analysis of self‐reported allergy identifies shared and allergy‐specific susceptibility loci. Nat Genet 2013; 45:907–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cabrera S, Sanchez E, Requena T et al Intronic variants in the NFKB1 gene may influence hearing forecast in patients with unilateral sensorineural hearing loss in Meniere's disease. PLoS One 2014; 9:e112171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lee YH, Song GG. Association between glutathione S‐transferase M1, P1, and NFKB1 polymorphisms and systemic lupus erythematosus susceptibility: a meta‐analysis. Cell Mol Biol (Noisy‐le‐grand) 2016; 62:21–6. [PubMed] [Google Scholar]
- 60. Kim SH, Kim JY, Lee HJ, Gi M, Kim BG, Choi JY. Autoimmunity as a candidate for the etiopathogenesis of Meniere's disease: detection of autoimmune reactions and diagnostic biomarker candidate. PLOS ONE 2014; 9:e111039. [DOI] [PMC free article] [PubMed] [Google Scholar]


