Summary
Oral lichen planus (OLP) is considered a chronic inflammatory immune‐mediated disease of the oral mucosa. Immunopathogenesis of OLP is thought to be associated with cell‐mediated immune dysregulation. O‐GlcNAcylation is a form of reversible glycosylation. It has been demonstrated that O‐GlcNAcylation promoted nuclear factor kappa B (NF‐κB) signalling. Activation of NF‐кB can induce expression of nucleotide‐binding domain‐like receptor family pyrin domain containing 3 (NLRP3) inflammasome, which is a large intracellular multi‐protein complex involving an immune response. Dysregulated expression of the NLRP3 inflammasome was reported to be associated with autoinflammatory diseases. No integrative studies between O‐GlcNAcylation and NLRP3 inflammasome in OLP patients have been reported. The present study aimed to determine the immunohistochemical expression of O‐GlcNAcylation, NF‐κB signalling molecules and NLRP3 inflammasome in oral mucosae of OLP patients. Oral tissue samples were collected from 30 OLP patients and 30 healthy individuals. Immunohistochemical staining and analyses of immunostaining scores were performed to evaluate expression of O‐GlcNAcylation, NF‐κB signalling molecules and NLRP3 inflammasome. According to observations in this study, significantly higher levels of O‐GlcNAcylation, NF‐κB signalling molecules and NLRP3 inflammasome were demonstrated in OLP patients compared with control subjects (P < 0·001). Positive correlations among O‐GlcNAcylation, NF‐κB signalling molecules and NLRP3 inflammasome were also observed in OLP samples (P < 0·01). In conclusion, the present study provides supportive evidence that increased O‐GlcNAcylation is associated with increased expression of NLRP3 inflammasome via the NF‐κB signalling pathway. These findings provide a new perspective on immunopathogenesis of OLP in relation to autoinflammation.
Keywords: inflammasome, inflammation, O‐GlcNAcylation, oral lichen planus, pathogenesis
Introduction
Oral lichen planus (OLP) is a chronic immune‐mediated inflammatory disease of the oral mucosa with unknown aetiology 1, 2. OLP is usually found in middle‐aged or older women 3. OLP is clinically heterogeneous, presenting as reticular, papular, plaque, atrophic, erosive and bullous forms with typical white striations called Wickham striae on the oral mucosa 4. Molecular mechanisms that induce various forms of OLP have never been clarified. OLP is associated with an increased risk of malignant transformation 5. Histopathological characteristics of OLP consist of degeneration of basal epithelial cells, basement membrane disruption and intense subepithelial infiltration of T lymphocytes 6. Increased expressions of nuclear factor kappa B (NF‐κB) signalling molecules and various inflammatory cytokines such as tumour necrosis factor (TNF)‐α and interleukin (IL)‐1β have been demonstrated previously in OLP 7, 8, 9. The current concepts of immunopathogenesis of OLP are related to cell‐mediated immune dysregulation. Some mechanisms behind the dysregulated immune response have been elucidated 10, 11. Very little, however, is known about the role of post‐translational modifications in relation to oral immune responses in OLP patients.
O‐GlcNAcylation is a single attachment of N‐acetylglucosamine (GlcNAc) to the hydroxyl site of serine or threonine residue of cytoplasmic, nuclear and mitochondrial proteins. This modification is a reversible process regulated by O‐GlcNAc transferase (OGT) and O‐GlcNAcase (OGA) for addition and removal of GlcNAc residue from target proteins 12. It was reported that O‐GlcNAcylation promotes transcriptional activity, nuclear translocation and DNA binding affinity of NF‐κB signalling molecules 13, 14. Growing evidence suggests that O‐GlcNAcylation modulates activation of NF‐κB signalling in several chronic inflammatory diseases. DNA binding activity of p65, a functional component of NF‐κB, is regulated by O‐GlcNAcylation during inflammatory processes and tumorigenesis of the colon 15. In addition, O‐GlcNAcylation promotes transcriptional activity of NF‐κB and up‐regulated expression of TNF‐α in pancreatitis 16. Expressions of O‐GlcNAcylated proteins (OGP) and O‐GlcNAcylation associated enzymes (OGT and OGA) have never been investigated in the oral mucosae of OLP patients.
The nucleotide oligomerization domain (NOD)‐like receptor family pyrin domain containing 3 (NLRP3) inflammasomes is a large intracellular multi‐protein complex consisting of three components: NLRP3, apoptosis‐associated speck‐like protein containing C‐terminal caspase‐recruitment domain (CARD) adaptor protein (ASC) and caspase‐1 17. NLRP3 inflammasome was reported to play a role in activation of the innate immune response 18. In addition, NLRP3 inflammasome is associated with an inflammatory form of programmed cell death known as pyroptosis 17. Increased expression of NLRP3 inflammasomes was observed in a variety of chronic inflammatory diseases, such as Crohn's disease 19 and periodontal diseases 20, 21, upon activation of the NF‐кB signalling pathway 22, 23. In addition, dysregulated inflammasome‐mediated expression of cytokines such as IL‐1β was reported to be associated with autoinflammatory diseases 24, 25, 26. No prior studies have reported expression of NLRP3 inflammasome in OLP. Moreover, no integrative studies between O‐GlcNAcylation and NLRP3 inflammasome have been performed in OLP patients. Taking these research gaps into account, it was hypothesized that changes in expression of O‐GlcNAcylation and NLRP3 inflammasome could be demonstrated at the sites of inflamed oral mucosae of OLP patients. Thus, the present study was aimed to investigate the immunohistochemical expressions of O‐GlcNAcylation (OGP, OGT and OGA), NF‐κB signalling molecules (NF‐кB and TNF‐α) and NLRP3 inflammasome (NLRP3, ASC, caspase‐1 and IL‐1β) in the inflamed oral mucosae of OLP patients, compared with normal oral mucosae of control subjects.
Materials and methods
Tissue specimens
This study was approved for the use of tissues from human subjects by the Institutional Human Ethics Committee of Khon Kaen University (HE602071). The diagnostic criteria of OLP in this study were based on the modified 2003 World Health Organization (WHO) diagnostic criteria 27. The inclusion criteria for selection of biopsy specimens with histopathological diagnosis of OLP made by an oral pathologist were as follows: OLP patients with no history of systemic diseases, cigarette smoking, receiving medication or being pregnant. The exclusion criteria were as follows: OLP patients with a history of systemic diseases, cigarette smoking, receiving medications or being pregnant and OLP patients who received topical or systemic steroids for treatment of OLP in the past 3 months.
Formalin‐fixed and paraffin‐embedded biopsy specimens of OLP and normal oral mucosa (NOM) were collected from the archives of the Oral Pathology Division, Faculty of Dentistry, Khon Kaen University. All original haematoxylin and eosin (H&E)‐stained slides of OLP and NOM from 2007 to 2016 were examined under a light microscope for the quality and suitability of oral mucosal tissue. OLP specimens demonstrating epithelial dysplasia were excluded. Then, OLP (n = 30) and NOM (n = 30) with intact epithelia and connective tissue were selected for immunohistochemical staining.
In the control group, tissue specimens were obtained from oral mucosae located at the retromolar area of the surgical procedure of tooth extraction due to the ethical limitations in collecting normal tissue samples at the same sites of surgical biopsy and with ages matching those in the OLP group. Control subjects had no history of systemic diseases, cigarette smoking, receiving medication or being pregnant. Selected biopsy specimens in the control group were diagnosed clinically and histopathologically as having normal oral mucosa.
Immunohistochemical method
The procedure of immunostaining was modified from previous studies by the present authors 28, 29. Briefly, all biopsy specimens were fixed in 10% neutral buffered formalin upon collection, embedded in paraffin wax, sectioned at 5‐µm thicknesses and incubated in a hot‐air oven at 60°C for 24 h. Deparaffinization of tissue specimens was performed. Antigen retrieval was performed by heating the tissue sections, which were immersed in sodium citrate buffer pH 6.0 in a microwave for 10 min. Endogenous peroxidase activity was eliminated by 3% hydrogen peroxide. Non‐specific bindings were blocked with protein block free serum (Dako, Carpinteria, CA, USA). Sections were incubated with primary antibodies in a humidifying chamber overnight at 4°C. Details of primary antibodies (OGP, OGT, OGA, NF‐кB, TNF‐α, NLRP3, ASC, caspase‐1 and IL‐1β) used in this study are shown in Supporting information, Table S1. Sections were incubated with the second antibodies (Dako EnVision + system‐horseradish peroxidase (HRP)‐labelled polymer anti‐rabbit or anti‐mouse) and then with 3,3'‐diaminobenzidine (DAB; Dako) and counterstained with Mayer's haematoxylin. The slides were dehydrated in ethanol, cleared in xylene and mounted. Biopsy specimens derived from patients with periodontitis, colon cancer and oral squamous cell carcinoma were used as positive controls. Negative controls were established by incubating with phosphate‐buffered saline (PBS) solution instead of primary antibodies.
Morphometric analysis
Immunostained slides were examined and immunostaining scoring was performed under a light microscope at ×200 magnification by one examiner (T. T. D.) after calibrating with an expert (P. C.). The whole area of oral epithelial layers and the lamina propria beneath the basement membrane were selected for immunohistochemical analyses. Immunostaining scores were established for semiquantitative analyses as follows: 0 = no immunostained cells; 1 = fewer than 25% positively stained cells, 2 = 25–50% positively stained cells, 3 = 50–75% positively stained cells and 4 = more than 75% positively stained cells. All specimens were assessed, and immunostaining scores were conducted twice with an interval of 2 weeks.
Statistical analyses
Statistical analyses were performed using spss Statistics version 17.0. The Mann–Whitney U‐test was performed to compare differences in age and differences in immunostaining scores of OGP, OGT, OGA, NF‐кB, TNF‐α, NLRP3, ASC, caspase‐1 and IL‐1β between OLP patients and control subjects. Ordinal logistic regression was performed to eliminate confounding effects of age. Spearman's correlation coefficient was used to evaluate correlations among these investigated molecules in each group. A two‐tailed P‐value less than 0·05 was considered statistically significant.
Search Tool for Interacting Chemicals (STITCH) analyses
STITCH version 5.0 30, 31 was used to predict relationships among investigated molecules, including OGT, NF‐кB, TNF‐α, NLRP3, ASC, caspase‐1 and IL‐1β.
Results
Demographic and clinical data
Details of OLP patients and control subjects are shown in Table 1.
Table 1.
Demographic and clinical data of oral lichen planus (OLP) patients and control subjects
| OLP patients (n = 30) | Control subjects (n = 30) | |
|---|---|---|
| Gender | ||
| Male | 7 | 13 |
| Female | 23 | 17 |
| Age * (years) | ||
| Mean ± s.d. | 48·73 ± 11·82 | 22·27 ± 7·21 |
| Range | 32–75 | 17–57 |
| Biopsy sites | ||
| Buccal mucosa | 30 | – |
| Retromolar area of buccal mucosa | – | 30 |
| Clinical diagnoses | ||
| Reticular OLP | 4 | – |
| Erosive OLP | 13 | – |
| Atrophic OLP | 13 | – |
| Normal oral mucosa | – | 30 |
| Duration of OLP lesions (months) | ||
| Mean ± s.d. | 15·93 ± 21·23 | – |
| Range | 0·25–84 | – |
*Significant differences in age between OLP patients and control subjects were observed (P = 0.001, Mann‐Whitney U‐test); s.d. = standard deviation.
Agreement levels in the judgement of immunostaining scores
The level of agreement on immunostaining scores for intra‐observer reliability by the kappa statistics was 0·819.
Expression of OGP, OGT and OGA in the oral mucosae of OLP patients and control subjects
According to immunohistochemical analyses, higher expressions of OGP, OGT and OGA were observed in oral epithelia and connective tissues of OLP patients compared with control subjects (all P < 0·001) (Fig. 1) (Table 2). Due to a marked difference in age between OLP patients and control subjects, multivariable analyses after adjusting for age were performed and the results confirmed that expressions of OGP, OGT and OGP in the OLP group were significantly higher than those in the control group [OGP: odds ratio (OR) = 16·94, 95% confidence interval (CI) = 2·67–90·74 and P = 0·002; OGT: OR = 6·47, 95% CI = 1·24–33·85 and P = 0·027; OGA: OR = 19·12, 95% CI = 3·13–116·62 and P = 0·001].
Figure 1.
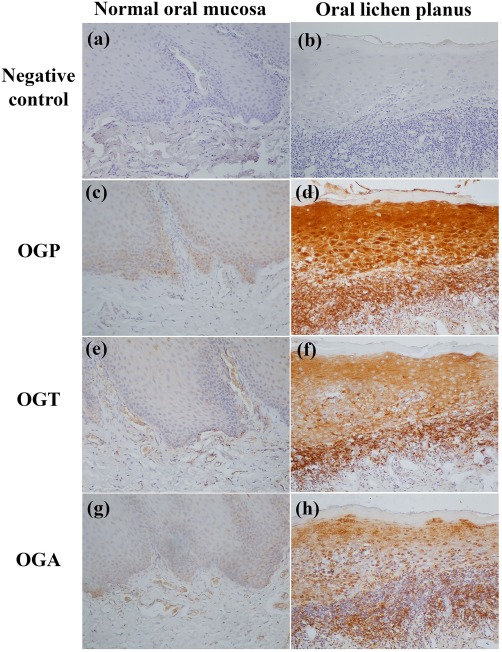
Expression of O‐GlcNAcylated protein (OGP), O‐linked N‐acetylglucosamine (GlcNAc) transferase (OGT) and O‐GlcNAcase (OGA) in normal oral mucosae (c,e,g) and oral lichen planus (OLP) lesions (d,f,h). (a,b) Negative controls. Stronger immunoreactivities of OGP, OGT and OGA are observed in OLP lesional mucosae. Original magnification ×200. [Colour figure can be viewed at wileyonlinelibrary.com]
Table 2.
Immunostaining scores of O‐GlcNAcylated proteins (OGP), O‐GlcNAc transferase (OGT), O‐GlcNAcase (OGA), nuclear factor kappa B (NF‐κb), tumour necrosis factor (TNF)‐α, nucleotide‐binding domain‐like receptor family pyrin domain containing 3 (NLRP3), apoptosis‐associated speck‐like protein containing a C‐terminal caspase recruitment domain (ASC), caspase‐1 p10, interleukin (IL)‐1β in normal oral mucosae (NOM) from control subjects (n = 30) and inflamed oral mucosae from oral lichen planus (OLP) patients (n = 30)
| Investigated molecules | Immunostaining scores | P‐values * | |||||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | |||
| OGP | OLP | 0 | 4 | 7 | 6 | 13 | 0·002 |
| NOM | 0 | 23 | 4 | 3 | 0 | ||
| OGT | OLP | 0 | 6 | 6 | 6 | 12 | 0·027 |
| NOM | 1 | 18 | 10 | 1 | 0 | ||
| OGA | OLP | 0 | 7 | 8 | 7 | 8 | 0·001 |
| NOM | 2 | 22 | 6 | 0 | 0 | ||
| NF‐кB | OLP | 2 | 7 | 9 | 7 | 5 | 0·001 |
| NOM | 20 | 10 | 0 | 0 | 0 | ||
| TNF‐α | OLP | 1 | 5 | 8 | 8 | 8 | 0·004 |
| NOM | 5 | 23 | 1 | 1 | 0 | ||
| NLRP3 | OLP | 1 | 7 | 8 | 5 | 9 | 0·012 |
| NOM | 1 | 25 | 3 | 1 | 0 | ||
| ASC | OLP | 0 | 4 | 6 | 5 | 15 | < 0·001 |
| NOM | 1 | 27 | 2 | 0 | 0 | ||
| Caspase‐1 | OLP | 0 | 10 | 9 | 4 | 7 | 0·011 |
| NOM | 0 | 29 | 1 | 0 | 0 | ||
| IL‐1β | OLP | 0 | 5 | 7 | 5 | 13 | 0·002 |
| NOM | 0 | 17 | 11 | 2 | 0 | ||
*P‐values were established using logistic regression to eliminate confounding effects of age.
Expression of NF‐кB signalling molecules and NLRP3 inflammasome in oral mucosae of OLP patients and control subjects
NF‐кB and TNF‐α were localized in the nuclei and cytoplasm of oral epithelial cells and infiltrated immune cells (Fig. 2). Expressions of NF‐кB and TNF‐α were significantly higher in OLP patients compared to control subjects (P < 0·001) (Table 2). Multivariate analyses after adjusting for age demonstrated that expressions of NF‐кB and TNF‐α in the OLP group were significantly higher than those in the control group (NF‐кB: OR = 27·68, 95% CI = 3·70–206·43 and P = 0·001; TNF‐α: OR = 16·05, 95% CI= 2·46–95·87 and P = 0·004).
Figure 2.
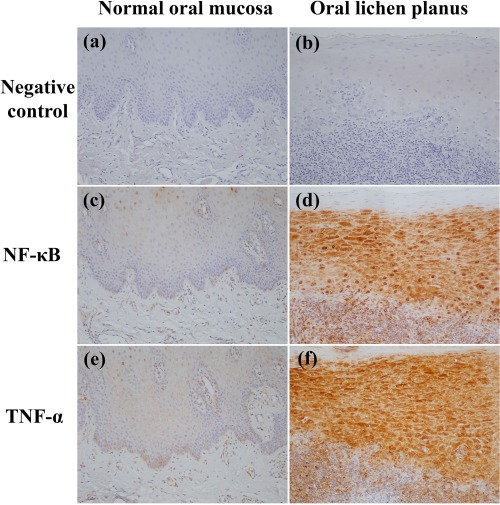
Expression of nuclear factor kappa B (NF‐кB) and tumour necrosis factor (TNF)‐α in normal oral mucosae (c,e) and oral lichen planus (OLP) lesions (d,f). (a,b) negative controls. Stronger immunoreactivities of NF‐кB and TNF‐α are observed in OLP lesional mucosae. Original magnification ×200. [Colour figure can be viewed at wileyonlinelibrary.com]
Expressions of NLRP3, ASC, caspase‐1 and IL‐1β were increased significantly in oral epithelia and connective tissues of OLP patients compared to those in control subjects (P < 0·001) (Table 2) (Fig. 3). After adjusting for age, multivariate analyses demonstrated that expression of NLRP3, ASC, caspase‐1 and IL‐1β in the OLP group were significantly higher than those in the control group (NLRP3: OR = 9·66, 95% CI = 1·66–56·32 and P = 0·012; ASC: OR = 74·67, 95% CI = 7·94–702·05 and P < 0·001; caspase‐1: OR = 54·38, 95% CI = 4·16–710 and P = 0·011; IL‐1β: OR = 8·80, 95% CI = 1·64 – 47·18 and P = 0·002).
Figure 3.
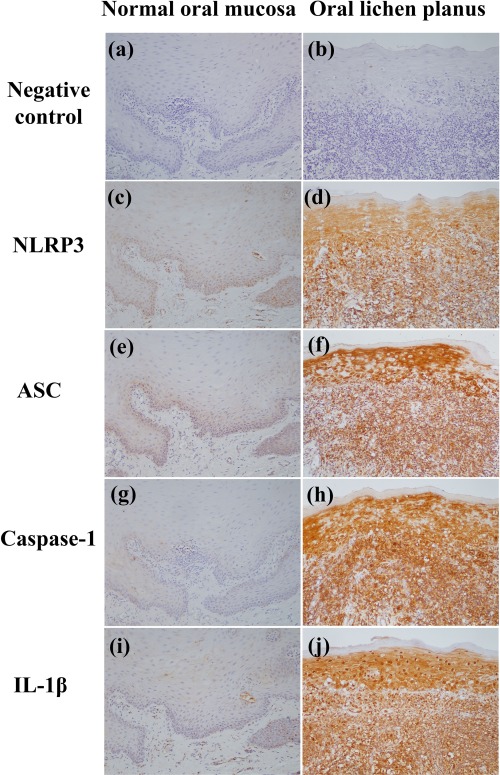
Expression of nucleotide‐binding domain‐like receptor family pyrin domain containing 3 (NLRP3), apoptosis‐associated speck‐like protein containing a C‐terminal caspase recruitment domain (ASC), caspase‐1 and interleukin (IL)‐1β in normal oral mucosae (c,e,g,i) and oral lichen planus (OLP) lesions (d,f,h,j). (a,b) Negative controls. Stronger immunoreactivities of NLRP3, ASC, caspase‐1 and IL‐1β are observed in OLP lesional mucosae. Original magnification ×200. [Colour figure can be viewed at wileyonlinelibrary.com]
Correlations among expression of O‐GlcNAcylation, NF‐кB signalling molecules and NLRP3 inflammasome in OLP patients
Positive correlations among O‐GlcNAcylation (OGP, OGT, OGA), NF‐кB signalling molecules (NF‐кB and TNF‐α) and NLRP3 inflammasome (NLRP3, ASC, caspase‐1 and IL‐1β) were observed in OLP patients (P < 0·01). No correlations of these investigated molecules were demonstrated in control subjects. Details of Spearman's rank correlation analyses among these investigated molecules in OLP patients are demonstrated in the Supporting information, Table S2. STITCH analyses revealed predictive relationships among OGT, NF‐кB, TNF‐α, NLRP3, ASC, caspase‐1 and IL‐1β (Fig. 4). In addition, these investigated molecules are associated with danger associated molecular pattern (DAMP) molecules such as heat shock protein (HSP)60 and HSP90. Details of these analysed molecules are demonstrated in the Supporting information, Table S3.
Figure 4.
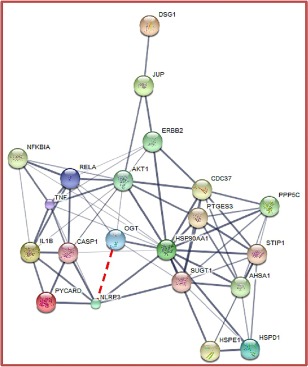
Data analysis by Search Tool for Interacting Chemicals (STITCH) version 5.0 for prediction of relationships among: O‐GlcNAcylation (OGT: O‐GlcNAc transferase); nuclear factor kappa B (NF‐Κb) signaLling (RELA: transcription factor p65; TNF: tumour necrotic factor); danger‐associated molecular patterns [hear shock protein (HSP)D1: HSP60; HSP90AA1: HSP90]; nucleotide‐binding domain‐like receptor family pyrin domain containing 3 (NLRP3) inflammasome [NLRP3, cASP1: caspase‐1, interleukin (IL)‐1β)]; epithelial cell differentiation and cell adhesion (DSG1: desmoglein1; JUP: junction plakoglobin) (http://stitch.embl.de/cgi/network.pl?taskId=1t51EemYpTt0). The red dashed line was established to demonstrate the correlation between OGT and NLRP3 according to the immunohistochemical analyses. [Colour figure can be viewed at wileyonlinelibrary.com]
Discussion
In the present study, increased immunohistochemical expression of O‐GlcNAcylated proteins (OGP) and O‐GlcNAcylation associated enzymes (OGT and OGA) was detected in OLP lesional mucosae. One previous study demonstrated that an increase in O‐GlcNAcylation in tissue specimens from patients with cholangiocarcinoma was due to increased expression of OGT but decreased expression of OGA 32. These findings suggest that regulation of O‐GlcNAcylation is dynamic and may depend not only on the expression levels of its cycling enzymes (OGT and OGA), but also on its enzymatic activities 12. O‐GlcNAcylation was reported to play a role in epithelial cell differentiation and cell adhesion 33, 34, 35. Regarding the clinicopathogenesis of OLP, it would be beneficial to identify specific O‐GlcNAcylated proteins at the sites of the OLP lesional mucosa and to investigate whether changes in O‐GlcNAcylated proteins during chronic inflammation might interfere with homeostasis of oral epithelial cell differentiation and cell adhesion, contributing to clinical heterogeneity of OLP. In addition, further studies are needed to clarify the role of O‐GlcNAcylation in activation of immune responses at the site of inflamed oral mucosa.
Regarding the expression of NF‐κB signalling molecules, these results are similar to previous studies 7, 8, 9, demonstrating an increase in expression of NF‐кB and TNF‐α in OLP. Moreover, increased levels of NLRP3 inflammasome (NLRP3, ASC, caspase‐1 and IL‐1β) in OLP are in agreement with previous studies in other chronic inflammatory diseases 19, 20, 21, 36. Correlations between NF‐кB signalling and NLRP3 inflammasome activation have been described in some studies 22, 37. It was suggested that NF‐κB signalling was important for correct activation of the NLRP3 inflammasome 23. In other studies, ASC and caspase‐1 were reported to modulate NF‐кB activation 38, 39, 40. Dysregulated NLRP3 inflammasome expression was reported to be associated with autoinflammatory diseases 24, 25, 41. Considering these observations, an increased expression of NLRP3 inflammasome in oral epithelial cells and infiltrated immune cells at the sites of OLP lesional mucosae possibly reflects cellular responses to pathological stimuli. DAMP molecules such as HSPs have been demonstrated to play a role in NLRP3 inflammasome activation 42, 43. Previous studies have demonstrated aberrant expression of HSP60 and HSP90 in OLP lesions 44, 45. Taking these findings into account, it is temping to postulate that altered expression of DAMP molecules such as HSP60 and HSP90 due to pathological stimuli may induce activation of the NLRP3 inflammasome in OLP. Future studies should elucidate the role of HSP60, HSP90, O‐GlcNAcylation and NLRP3 inflammasome in the pathogenesis of OLP.
Regarding correlations among O‐GlcNAcylation, NF‐κB signalling molecules and NLRP3 inflammasome, previous studies have demonstrated that increased O‐GlcNAcylation enhanced the activation of NF‐кB, and promoted an inflammatory response in several chronic inflammatory diseases 14, 15. In addition, it was reported that O‐GlcNAcylation played a role in activation of T lymphocytes 46, 47, and OGT activity was necessary for functions of effector T cells 48. Previous studies have also demonstrated NLRP3 inflammasome‐regulated activation of T helper cells 49, 50. The current study demonstrated increased levels of OGT and NLRP3 inflammasome in infiltrated lymphocytes at the sites of OLP lesions. Thus, it is possible that O‐GlcNAcylation and NLRP3 inflammasome may be associated with activation of T lymphocytes in OLP. In addition, NLRP3 inflammasome was reported to be involved in pyroptosis, which was shown to be a form of programmed cell death under an inflammatory condition 51. This molecular mechanism could be one possible explanation for the degeneration of basal epithelial cells in OLP.
In conclusion, there are two important findings from these current observations. First, increased levels of O‐GlcNAcylation, NF‐κB signalling molecules and NLRP3 inflammasome in OLP lesional mucosae were demonstrated. Secondly, positive correlations among O‐GlcNAcylation, NF‐κB signalling molecules and NLRP3 inflammasome were also observed in OLP. The present findings provide supportive evidence of a connection between O‐GlcNAcylation and NLRP3 inflammasome in relation to autoinflammation. In addition, this study provides incitement for innovative future research to unravel the immune‐mediated inflammatory signalling processes in the oral mucosal environment that would help to clarify the immunopathogenesis of OLP.
Author contributions
T. T. D. contributed to collection of tissue samples and clinical data, design of experiments and analysis of the data. C. P., V. C. and A. S. contributed to design of the experiments and analysis of the data. P. C. contributed to design of the experiments, analysis of the data and interpretation of the results. T. T. D. and P. C. wrote the manuscript. All the authors critically reviewed the manuscript and approved the final version for submission.
Ethical clearance
This study was approved for the use of tissues from human subjects by the Institutional Human Ethics Committee of Khon Kaen University (HE602071).
Disclosure
The authors declare that they have no disclosures related to this manuscript.
Supporting information
Additional Supporting information may be found in the online version of this article at the publisher's web‐site:
Table S1. List of antibodies used for immunohistochemical staining.
Table S2. Analyses of correlations among the expression of O‐GlcNAcylation, nuclear factor kappa B (NF‐кB) signalling molecules and nucleotide‐binding domain‐like receptor family pyrin domain containing 3 (NLRP3) inflammasome in the oral lichen planus (OLP) group using spss Statistics version 17.0.
Table S3. Full names and functions of investigated molecules analysed by Search Tool for Interacting Chemicals (STITCH) version 5.0.
Acknowledgements
We would like to express our sincere appreciation to Professor Sopit Wongkham from Department of Biochemistry, Faculty of Medicine, Khon Kaen University for her kind guidance on Glycobiology. We wish to thank Associate Professor Tanapat Palaga, Department of Microbiology, Faculty of Science, Chulalongkorn University for his advice on autoinflammation and Dr Patcharee Ritprajak, Department of Microbiology and Immunology and DRU of Oral Microbiology, Faculty of Dentistry, Chulalongkorn University for her suggestions on innate immunity. We would also like to thank Dr Rajda Noisombut from the Department of Community Dentistry, Faculty of Dentistry, Khon Kaen University for her advice on statistical analyses and Professor James A. Will, University of Wisconsin, Madison, Wisconsin, for editing the manuscript and suggestions via Publication Clinic KKU, Thailand. This research work was supported by Graduate School, Khon Kaen University (no. 59212102) and by Khon Kaen University (no. 2560A10301042).
References
- 1. Carrozzo M. Understanding the pathobiology of oral lichen planus. Curr Oral Health Rep 2014; 1:173–9. [Google Scholar]
- 2. Kurago ZB. Etiology and pathogenesis of oral lichen planus: an overview. Oral Surg Oral Med Oral Pathol Oral Radiol 2016; 122:72–80. [DOI] [PubMed] [Google Scholar]
- 3. Alrashdan MS, Cirillo N, McCullough M. Oral lichen planus: a literature review and update. Arch Dermatol Res 2016; 308:539–51. [DOI] [PubMed] [Google Scholar]
- 4. Cheng YSL, Gould A, Kurago Z, Fantasia J, Muller S. Diagnosis of oral lichen planus: a position paper of the American Academy of Oral and Maxillofacial Pathology. Oral Surg Oral Med Oral Pathol Oral Radiol 2016; 122:332–54. [DOI] [PubMed] [Google Scholar]
- 5. Shirasuna K. Oral lichen planus: malignant potential and diagnosis. Oral Sci Int 2014; 11:1–7. [Google Scholar]
- 6. Alves MG, Balducci I, Carvalho YR, Nunes FD, Almeida JD. Oral lichen planus: a histopathological study. Histopathology 2015; 66:463–4. [DOI] [PubMed] [Google Scholar]
- 7. Santoro A, Majorana A, Bardellini E, Festa S, Sapelli P, Facchetti F. NF‐kappaB expression in oral and cutaneous lichen planus. J Pathol 2003; 201:466–72. [DOI] [PubMed] [Google Scholar]
- 8. Thongprasom K, Dhanuthai K, Sarideechaigul W, Chaiyarit P, Chaimusig M. Expression of TNF‐alpha in oral lichen planus treated with fluocinolone acetonide 0.1%. J Oral Pathol Med 2006; 35:161–6. [DOI] [PubMed] [Google Scholar]
- 9. Zhou G, Xia K, Du GF et al Activation of nuclear factor‐kappa B correlates with tumor necrosis factor‐alpha in oral lichen planus: a clinicopathologic study in atrophic‐erosive and reticular form. J Oral Pathol Med 2009; 38:559–64. [DOI] [PubMed] [Google Scholar]
- 10. Roopashree MR, Gondhalekar RV, Shashikanth MC, George J, Thippeswamy SH, Shukla A. Pathogenesis of oral lichen planus–a review. J Oral Pathol Med 2010; 39:729–34. [DOI] [PubMed] [Google Scholar]
- 11. Wang H, Zhang D, Han Q et al Role of distinct CD4(+) T helper subset in pathogenesis of oral lichen planus. J Oral Pathol Med 2016; 45:385–93. [DOI] [PubMed] [Google Scholar]
- 12. Yang X, Qian K. Protein O‐GlcNAcylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol 2017; 18:452–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Allison DF, Wamsley JJ, Kumar M et al Modification of RelA by O‐linked N‐acetylglucosamine links glucose metabolism to NF‐κB acetylation and transcription. Proc Natl Acad Sci USA 2012; 109:16888–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim HB, Lee SW, Mun CH et al O‐linked N‐acetylglucosamine glycosylation of p65 aggravated the inflammation in both fibroblast‐like synoviocytes stimulated by tumor necrosis factor‐alpha and mice with collagen induced arthritis. Arthritis Res Ther 2015; 17:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang YR, Kim DH, Seo YK et al Elevated O‐GlcNAcylation promotes colonic inflammation and tumorigenesis by modulating NF‐kappaB signaling. Oncotarget 2015; 6:12529–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang D, Cai Y, Chen M, Gao L, Shen Y, Huang Z. OGT‐mediated O‐GlcNAcylation promotes NF‐kappaB activation and inflammation in acute pancreatitis. Inflamm Res 2015; 64:943–52. [DOI] [PubMed] [Google Scholar]
- 17. Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol 2016; 16:407–20. [DOI] [PubMed] [Google Scholar]
- 18. Guo H, Callaway JB, Ting JP. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med 2015; 21:677–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lazaridis LD, Pistiki A, Giamarellos‐Bourboulis EJ et al Activation of NLRP3 inflammasome in inflammatory bowel disease: differences Between Crohn's disease and ulcerative colitis. Dig Dis Sci 2017; 62:2348–56. [DOI] [PubMed] [Google Scholar]
- 20. Bostanci N, Emingil G, Saygan B et al Expression and regulation of the NALP3 inflammasome complex in periodontal diseases. Clin Exp Immunol 2009; 157:415–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Park E, Na HS, Song Y‐R, Shin SY, Kim Y‐M, Chung J. Activation of NLRP3 and AIM2 Inflammasomes by Porphyromonas gingivalis infection. Infect Immun 2014; 82:112–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bauernfeind FG, Horvath G, Stutz A et al Cutting edge: NF‐kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol 2009; 183:787–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Boaru SG, Borkham‐Kamphorst E, Van de Leur E, Lehnen E, Liedtke C, Weiskirchen R. NLRP3 inflammasome expression is driven by NF‐kappaB in cultured hepatocytes. Biochem Biophys Res Commun 2015; 458:700–6. [DOI] [PubMed] [Google Scholar]
- 24. Ozkurede VU, Franchi L. Immunology in clinic review series; focus on autoinflammatory diseases: role of inflammasomes in autoinflammatory syndromes. Clin Exp Immunol 2012; 167:382–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Menu P, Vince JE. The NLRP3 inflammasome in health and disease: the good, the bad and the ugly. Clin Exp Immunol 2011; 166:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goldbach‐Mansky R. Immunology in clinic review series; focus on autoinflammatory diseases: update on monogenic autoinflammatory diseases: the role of interleukin (IL)‐1 and an emerging role for cytokines beyond IL‐1. Clin Exp Immunol 2012; 167:391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van der Meij EH, van der Waal I. Lack of clinicopathologic correlation in the diagnosis of oral lichen planus based on the presently available diagnostic criteria and suggestions for modifications. J Oral Pathol Med 2003; 32:507–12. [DOI] [PubMed] [Google Scholar]
- 28. Chaiyarit P, Thongprasom K, Klanrit P, Giraud AS. Trefoil factor expression by immunohistochemistry in patients with oral lichen planus. Asian Biomed 2014; 8:743 –9. [Google Scholar]
- 29. Luengtrakoon K, Wannakasemsuk W, Vichitrananda V et al Increased melatonin in oral mucosal tissue of oral lichen planus (OLP) patients: a possible link between melatonin and its role in oral mucosal inflammation. Arch Oral Biol 2017; 78:13–9. [DOI] [PubMed] [Google Scholar]
- 30. Kuhn M, von Mering C, Campillos M, Jensen LJ, Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res 2008; 36:D684–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Szklarczyk D, Santos A, von Mering C, Jensen LJ, Bork P, Kuhn M. STITCH 5: augmenting protein‐chemical interaction networks with tissue and affinity data. Nucleic Acids Res 2016; 44:D380–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Phoomak C, Silsirivanit A, Wongkham C, Sripa B, Puapairoj A, Wongkham S. Overexpression of O‐GlcNAc‐transferase associates with aggressiveness of mass‐forming cholangiocarcinoma. Asian Pac J Cancer Prev 2012; 13:101–5. [PubMed] [Google Scholar]
- 33. Bektas M, Rubenstein DS. The role of intracellular protein ‐glycosylation in cell adhesion and disease. J Biomed Res 2011; 25:227–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hu P, Berkowitz P, Madden VJ, Rubenstein DS. Stabilization of plakoglobin and enhanced keratinocyte cell‐cell adhesion by intracellular O‐glycosylation. J Biol Chem 2006; 281:12786–91. [DOI] [PubMed] [Google Scholar]
- 35. Sohn KC, Lee EJ, Shin JM et al Regulation of keratinocyte differentiation by O‐GlcNAcylation. J Dermatol Sci 2014; 75:10–5. [DOI] [PubMed] [Google Scholar]
- 36. Kim EH, Park MJ, Park S, Lee ES. Increased expression of the NLRP3 inflammasome components in patients with Behcet's disease. J Inflamm (Lond) 2015; 12:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang A, Wang P, Ma X et al Mechanisms that lead to the regulation of NLRP3 inflammasome expression and activation in human dental pulp fibroblasts. Mol Immunol 2015; 66:253–62. [DOI] [PubMed] [Google Scholar]
- 38. Sarkar A, Duncan M, Hart J, Hertlein E, Guttridge DC, Wewers MD. ASC directs NF‐kappaB activation by regulating receptor interacting protein‐2 (RIP2) Caspase‐1 interactions. J Immunol 2006; 176:4979–86. [DOI] [PubMed] [Google Scholar]
- 39. Lamkanfi M, Kalai M, Saelens X, Declercq W, Vandenabeele P. Caspase‐1 activates nuclear factor of the kappa‐enhancer in B cells independently of its enzymatic activity. J Biol Chem 2004; 279:24785–93. [DOI] [PubMed] [Google Scholar]
- 40. Staal J, Bekaert T, Beyaert R. Regulation of NF‐kappaB signaling by caspases and MALT1 paracaspase. Cell Res 2011; 21:40–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Park H, Bourla AB, Kastner DL, Colbert RA, Siegel RM. Lighting the fires within: the cell biology of autoinflammatory diseases. Nat Rev Immunol 2012; 12:570–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell 2014; 157:1013–22. [DOI] [PubMed] [Google Scholar]
- 43. Mayor A, Martinon F, De Smedt T, Petrilli V, Tschopp J. A crucial function of SGT1 and HSP90 in inflammasome activity links mammalian and plant innate immune responses. Nat Immunol 2007; 8:497–503. [DOI] [PubMed] [Google Scholar]
- 44. Chaiyarit P, Kafrawy AH, Miles DA, Zunt SL, Dis MLV, Gregory RL. Oral lichen planus: an immunohistochemical study of heat shock proteins (HSPs) and cytokeratins (CKs) and a unifying hypothesis of pathogenesis. J Oral Pathol Med 2007; 28:210–5. [DOI] [PubMed] [Google Scholar]
- 45. Chaiyarit P, Jintakanon D, Klanrit P, Siritapetawee M, Thongprasom K. Immunohistochemical analyses of survivin and heat shock protein 90 expression in patients with oral lichen planus. J Oral Pathol Med 2009; 38:55–62. [DOI] [PubMed] [Google Scholar]
- 46. Golks A, Tran TT, Goetschy JF, Guerini D. Requirement for O‐linked N‐acetylglucosaminyltransferase in lymphocytes activation. Embo J 2007; 26:4368–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kearse KP, Hart GW. Lymphocyte activation induces rapid changes in nuclear and cytoplasmic glycoproteins. Proc Natl Acad Sci USA 1991; 88:1701–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lund PJ, Elias JE, Davis MM. Global analysis of O‐GlcNAc glycoproteins in activated human T cells. J Immunol 2016; 197:3086–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen M, Wang H, Chen W, Meng G. Regulation of adaptive immunity by the NLRP3 inflammasome. Int Immunopharmacol 2011; 11:549–54. [DOI] [PubMed] [Google Scholar]
- 50. Arbore G, West EE, Spolski R et al T helper 1 immunity requires complement‐driven NLRP3 inflammasome activity in CD4(+) T cells. Science 2016; 352:aad1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. He Y, Hara H, Núñez G. Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem Sci 2016; 41:1012–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting information may be found in the online version of this article at the publisher's web‐site:
Table S1. List of antibodies used for immunohistochemical staining.
Table S2. Analyses of correlations among the expression of O‐GlcNAcylation, nuclear factor kappa B (NF‐кB) signalling molecules and nucleotide‐binding domain‐like receptor family pyrin domain containing 3 (NLRP3) inflammasome in the oral lichen planus (OLP) group using spss Statistics version 17.0.
Table S3. Full names and functions of investigated molecules analysed by Search Tool for Interacting Chemicals (STITCH) version 5.0.


