Abstract
PAX9 is a transcription factor of the PAX family characterized by a DNA-binding paired domain. Previous studies have suggested a potential role of PAX9 in squamous cell differentiation and carcinogenesis of the oro-esophageal epithelium. However, its functional roles in differentiation and carcinogenesis remain unclear. In this study, Pax9 deficiency in the mouse esophagus promoted cell proliferation, delayed cell differentiation and altered the global gene expression profile. Ethanol exposure down-regulated PAX9 expression in human esophageal epithelial cells in vitro and mouse forestomach and tongue in vivo. We further showed that PAX9 was down-regulated in human oro-esophageal squamous cell carcinoma (OESCC), and its down-regulation was associated with alcohol drinking and promoter hypermethylation. Moreover, ad libitum feeding with a liquid diet containing ethanol for 40 weeks or Pax9 deficiency promoted N-nitrosomethylbenzylamine-induced squamous cell carcinogenesis in mouse tongue, esophagus, and forestomach. In conclusion, PAX9 regulates squamous cell differentiation in the oro-esophageal epithelium. Alcohol drinking and promoter hypermethylation are associated with PAX9 silencing in human OESCC. PAX9 down-regulation may contribute to alcohol-associated oro-esophageal squamous cell carcinogenesis.
Keywords: PAX9, Esophagus, Ethanol, Promoter hypermethylation, Squamous cell carcinoma
Introduction
PAX9 is a transcription factor of the PAX family characterized by a DNA-binding paired domain. Being expressed in somites, pharyngeal pouches, and mesenchyme, PAX9 is essential for the development of thymus, parathyroid, limb, palate, and teeth during mouse embryogenesis. Homozygous Pax9 knockout mice die shortly after birth due to cleft palate [1]. Subsequent studies have also clearly demonstrated an essential role of PAX9 in the development of filiform papilla and taste bud of the tongue, lip, and intervertebral disc [2–5]. However, in adults, PAX9 expression is restricted to the endocrine tissues (e.g., thymus, parathyroid) and the upper gastrointestinal tract (e.g., tongue, esophagus, salivary gland, cheek epithelium), with the strongest expression in the tongue [6].
Previous studies have suggested a potential role for the absence of PAX9 in human esophageal squamous cell carcinoma (ESCC) and oral squamous cell carcinoma (OSCC). In human ESCC tissue samples, PAX9 was found to be lost or down-regulated, and progressive loss of PAX9 expression was correlated with increasing malignancy [7]. PAX9 expression was associated with better survival and radiosensitivity [8]. In human OSCC cells, PAX9 expression was essential for cell growth and survival [9]. In Barrett’s esophagus, a human disease characterized by intestinal metaplasia of esophageal squamous epithelium, PAX9 was found to be down-regulated [10], suggesting its involvement in squamous differentiation. Pax9 and its downstream genes were involved in the terminal maturation of esophageal epithelium of mouse esophagus [11]. Moreover, morpholino-knockdown of pax9 in zebrafish resulted in loss or disorganization of the squamous epithelium in the tongue [12]. These evidence support the hypothesis that PAX9 may regulate squamous epithelial cell differentiation in the adult oro-esophageal epithelium.
In this study, we aimed to understand the functional role of PAX9 in the oro-esophageal epithelium using tissue-specific PAX9-deficient mice, human cells and tissues.
Materials and Methods
Human tissue samples
Human ESCC and OSCC tissue samples were obtained from Duke University Medical Center and Beijing Stomatological Hospital, respectively, with informed written consent and institutional IRB approval (Pro39682 and Pro56638). All human samples (n=26) were coded with patient identifiers removed. Clinical data including alcohol exposure were collected from the medical record. Patients with a self-reported history of alcohol drinking (n=15) were regarded as "drinker" (5 drinks/week on average), and those who reported never drinking alcohol (n=5) as "non-drinkers". Occasional drinkers (n=6) were excluded from further analysis on the association between alcohol drinking and PAX9 expression. A human ESCC tissue array with tumors and matched normal adjacent tissues (Cat #: HEso-Squ060PG-01) and a human OSCC array with tumors and non-matched normal tissues (Cat #: OR601b) were purchased from US Biomax (Derwood, MD, USA).
Cell culture and treatments
Human ESCC cells, KYSE510, KYSE450, and KYSE70, were obtained from ATCC (Manassas, VA, USA) and ECACC (Porton Down, Salisbury, UK) with proper authentication. KYSE510 and KYSE450 cells were exposed to ethanol. KYSE70 cells were treated with 5-aza-2’-deoxycytidine (DAC; Sigma-Aldrich, St. Louis, MO, USA) for 72 h.
Animal experiments
All animal experiments were approved by the IACUC at the North Carolina Central University. Krt5Cre mice [13] and Pax9loxP/loxP mice [14] were crossed to generate tissue-specific Pax9-deficient mice (Krt5Cre;Pax9loxP/loxP). Tissue samples (tongue, esophagus, forestomach) were harvested for analysis of morphology, gene expression and transmission electron microscopy (TEM).
In an ethanol exposure study, wild-type C57BL/6J mice (Jackson Laboratory, Bar Harbor, ME) were given ad libitum water, or 20% ethanol (plus 0.3% saccharin) in the drink for 4 weeks, or 15% alcohol (plus 0.3% saccharin) in the drink for 40 weeks.
In a carcinogenesis study, N-nitrosomethylbenzylamine (NMBA) was used to induce tumorigenesis in mouse esophagus and forestomach [15]. Mice were given six intragastric doses of NMBA (Ash Stevens, Riverview, MI, USA) at 2 mg/kg body weight twice each week for 3 weeks. Wild-type C57BL/6J mice were given ad libitum the Lieber-DeCarli Regular Control diet (710027; Dyets Inc., Bethlehem, PA, USA) (Group A), or an isocaloric Lieber-DeCarli diet plus 6.4% v/v ethanol (710260; Dyets) (Group B). Lieber-DeCarli diet is a high-fat, nutritionally complete liquid diet, which was designed to partially overcome the murine dislike of ethanol [16]. Pax9-deficient mice were given the control liquid diet (Group C) (Table 1). All mice were sacrificed at 40 weeks and analyzed for tumorigenesis in tongue, esophagus, and forestomach. The number of macroscopically visible tumors (≥0.5 mm in diameter) in mouse forestomach and tongue was scored. Tissues were processed routinely for paraffin sectioning (5 µm), H&E staining and histopathology. Microscopic lesions including dysplasia, papilloma, and SCC were diagnosed according to established criteria [15].
Table 1.
| Group | Genotype | Treatment | No. | Esophagus | Forestomach | Tonguec | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Dysplasia | Papilloma | SCC | Dysplasia | Papilloma | SCC | Dysplasia | Papilloma | SCC | ||||
| A | Wild-type | Liquid diet | 18 | 0 | 0 | 0 | 22.2% | 72.2% | 5.6% | 0 | 0 | 0 |
| (4/18) | (13/18) | (1/18) | ||||||||||
| B | Wild-type | Isocaloric diet with 6.4% v/v ethanol | 24 | 4.2% | 50% | 0 | 16.7% | 95.8% | 8.3% | 0 | 0 | 0 |
| (1/24) | (12/24) | (4/24) | (23/24) | (2/24) | ||||||||
| (P=0.0003) | ||||||||||||
| C | Krt5Cre;Pax9loxP/loxP | Liquid diet | 22 | 18.2% | 77.3% | 0 | 81.8% | 90.9% | 31.8% | 4.5% | 9.1% | 22.7% |
| (4/22) | (17/22) | (18/22) | (20/22) | (7/22) | (1/22) | (2/22) | (5/22) | |||||
| (P=0.0001) | (P=0.0003) | (P=0.046) | (P=0.053) | |||||||||
Microscopically detected lesions including dysplasia, papilloma, and SCC were diagnosed according to established criteria.
All P values were calculated by comparing with Group A and determined using Student’s t-test.
P=0.005 when tongue dysplasia, papilloma and SCC are combined for comparison.
Western blotting
Protein was isolated from the human ESCC cells and mouse tissues using a standard method. In brief, tissue lysates were prepared by homogenizing tissue with the TissueLyser bead mill (Qiagen, Valencia, CA, USA) in RIPA buffer (Sigma-Aldrich). Cell debris was removed by a short centrifugation and an aliquot of cleared lysate was kept for protein quantitation using the BCA Protein Assay Kit (Biorad, Hercules, CA, USA). Proteins were detected with a rabbit monoclonal anti-PAX9 (1:600, Cat #: 12847, Cell Signaling Technology, Danvers, MA, USA) and a mouse monoclonal anti-GAPDH antibody (1:50000, ab8245, Abcam, Cambridge, MA, USA).
Histochemical staining, immunohistochemical staining (IHC), TUNEL assay and immunofluorescent staining
Haematoxylin & eosin staining was conducted based on a routine protocol. For IHC staining, deparaffinized sections were pre-treated to retrieve antigens using a Tris-based Antigen Unmasking Solution (Vector Laboratories, Burlingame, CA, USA), prior to incubation with either a rabbit monoclonal anti-PAX9 (1:400, Cell Signaling Technology), a rat monoclonal anti-BrdU (1:1000, ab6326, Abcam), a rabbit polyclonal anti-loricrin (1:500, ab24722, Abcam), a mouse monoclonal anti-proliferating cell nuclear antigen (PCNA; 1:3000, P8825, Sigma-Aldrich), a rabbit monoclonal anti-SOX2 (1:100, ab92494, Abcam), a rabbit polyclonal anti-FOXA3 (1:100, PA1-17038, Affinity Bioreagents, Golden, CO, USA), a rabbit polyclonal anti-WNT3 (1:100, ab32249, Abcam), a rabbit polyclonal anti-SFRP5 (1:50, LS-C169017, LifeSpan Biosciences, Seattle, WA), a guinea pig polyclonal anti-KRT35 (1:200, LS-C20268, LifeSpan Biosciences), a rabbit monoclonal anti-KRT5 (1:200, RM-2106-S0, Thermo Scientific, Rockford, IL), a rabbit polyclonal anti-P63 (1:100, GTX102425, GeneTex, Irvine, CA, USA), a rabbit polyclonal anti-involucrin (1:500, 924401, BioLegend, San Diego, CA, USA) or a rabbit polyclonal anti-filaggrin (1:100, GTX37695, GeneTex) overnight at 4 °C. The percentage of positively stained epithelial cells was calculated. Three sections were counted for each sample.
A terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay was conducted according to the protocol provided by the manufacturer of the TACSTM TDT kit (R&D Systems, Minneapolis, MN, USA). The apoptotic index was calculated by dividing the number of TUNEL positive cells by the total number of epithelial cells.
We performed immunofluorescent double staining for BrdU and Ki67 to test whether the BrdU-positive cells that were retained in the epithelial layer were still proliferating 5 days after pulse-labeling (50 mg/kg, i.p.) [17]. Immunofluorescent double staining was conducted based on standard procedures. Ki67 and BrdU were detected by a rabbit anti-Ki67 (1:200, RM-9106, Thermo Scientific) and a rat monoclonal anti-BrdU (1:1000, ab6326, Abcam) overnight at 4 °C, and then by a goat-anti-rabbit Alexa 568 and goat-anti-rat Alexa 488 (1:1000, A-11008 and A-11077, Thermo Scientific).
Transmission electron microscopy (TEM)
Mouse tissue was fixed with 4% glutaraldehyde in cacodylate buffer for at least one hour. Thick and thin sections were prepared on a Reichert-Jung ultramicrotome (Leica, Bannockburn, IL). Thick sections were stained with 1% toluidine blue-borax for selection using light microscopy of the appropriate areas for thin sections. Thin sections were prepared, mounted on copper grids, and double stained for examination using a Philips CM 12 electron microscope (FEI, Hillsboro, OR).
Gene microarray analysis
For gene microarray analysis, total RNA was extracted from mouse forestomach, or mouse esophageal epithelium with an RNeasy Fibrous Tissue Mini Kit (Qiagen), according to the manufacturer’s instructions. Microarray experiments were performed with Agilent mouse 4x44k microarrays. The raw data has been submitted to NCBI’s GEO database (series GSE75373, GSE96735). As previously described [11], differentially expressed genes were obtained from two-class significance analysis of microarrays (SAM) in Excel with the median number of false positives less than 1. Gene set analysis (GSA) was carried out as an add-in in Excel with curated gene sets in three major categories: Canonical Pathway (CP), Gene Ontology (GO) and Transcription Factor (TF). Knowledge-based gene sets (KB) were generated based on the literature and have been successfully used in our previous microarray study [11]. The Pax9 gene set was based on a list of genes differentially expressed in Pax9 knockout mice [2]. Hierarchical clustering analysis and principal component analysis (PCA) were performed using the R package. Microarray data of four GEO datasets (GSE23400, GSE20347, GSE13601, GSE6631) were downloaded and analyzed to compare PAX9 mRNA expression in cancer versus matched normal tissue.
Pyrosequencing of the human PAX9 gene promoter and mouse Pax9 gene promoter
Genomic DNA was purified using a DNeasy Tissue kit (Qiagen) according to the manufacturer’s instructions. DNA methylation level in the PAX9 gene promoter region was determined by EpigenDx Inc (Worcester, MA, USA) using the PSQ™96 HS system (Qiagen) according to standard procedures with a unique set of primers. The methylation status of each CpG site in the promoter regions (supplementary material, Figure S7) was determined individually as an artificial C/T SNP using QCpG software (Qiagen). A series of unmethylated and methylated DNA were included as controls in each PCR.
RT-PCR, qRT-PCR and chromatin immunoprecipitation (ChIP)
For regular RT-PCR, total RNA was reverse transcribed using the Advantage RT-for-PCR kit (Clontech), according to the manufacturer’s instructions. Pax9 exon 2 was detected with the following primer pairs (predicted size: 278 bp): Pax9F (5’-ACCACATTTACTCATATCCCAGTCCCA-3’) and Pax9R (5’–GGCTCCCTTCTCCAATCCATTCA-3’).
Quantitative PCR was performed on cDNA pools to assess the expression levels of genes of interest with primers and probes obtained from Applied Biosystems Inc. (Foster City, CA) using TaqMan and 96-well optical plate using A ABI 7900HT Fast Real-Time PCR system in triplicate.
PAX9 binding sites within the 5’-upstream DNA sequence of Krt35 (ENSMUSG00000048013) were predicted using Anchored Combination TFBS Cluster Analysis (oPOSSUM version 3.0). ChIP analysis was performed in triplicate using an EZ-ChIP kit (Millipore, Billerica, MA, USA) according to the manufacturer's instructions with the above-mentioned anti-PAX9. Immunoprecipitated DNA or input was PCR amplified with the following primer pairs: Krt35 (5’-CCACATCCTGAGTTCAATCC-3’ and 5’-CCAAGACAGGATCTCTCATTAC-3’; predicted size=174 bp); negative control P63 (5’-CAAATGTTGCTTGTCTGGTG-3’ and 5’-GTCAGTCGAGTGCACAGTTT-3’; predicted size=210 bp).
Statistical analyses
GraphPad Prism 6 (GraphPad Software, La Jolla, CA, USA) was used for Student’s t-test, Fisher’s exact test and McNemar’s test, with the statistical significance level set at 0.05. All comparisons were two-sided.
Results
Esophageal phenotypes of Krt5Cre;Pax9loxP/loxP mice
To elucidate the function of PAX9 in the oro-esophageal epithelium in vivo, Pax9 was inactivated in stratified squamous epithelial cells by crossing Krt5Cre mice and Pax9loxP/loxP mice resulting in Krt5Cre;Pax9loxP/loxP mice. Whereas Pax9 knockout mice die after birth [1], Krt5Cre;Pax9loxP/loxP mice appeared healthy and bred normally. Pax9 deficiency in the esophagus was validated by RT-qPCR (supplementary material, Figure S1A) and IHC (supplementary material, Figure S1B–E). IHC for PCNA showed epithelial hyperproliferation in Pax9-deficient esophagus (supplementary material, Figure S1F–H).
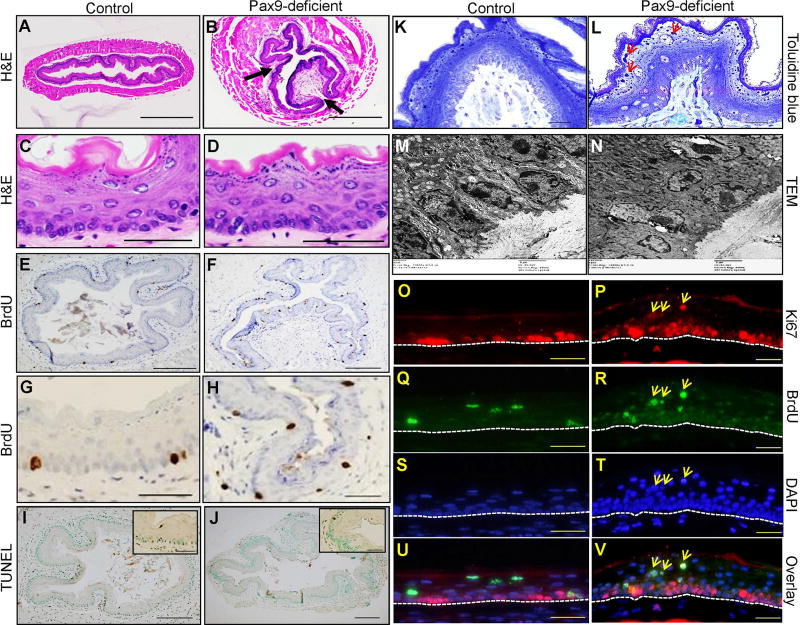
Papilloma-like structures were formed in the squamous epithelium of Pax9-deficient esophagus (Figure 1B, D) as compared with control esophagus (Figure 1A, C). Hyperproliferation was evident with an increased number of BrdU-positive cells (Figure 1E–H). Apoptosis as detected by a TUNEL assay was slightly different between Pax9-deficient esophagus (0.7±0.4%, Figure 1J) and control esophagus (0.2±0.1%, Figure 1I). Toluidine blue staining showed increased keratohyalin granules in the keratinized cells and palisading of basal cells in the mutant esophagus (Figure 1K, L). Under TEM, in the basal layer of mutant esophagus, the intercellular space, desmosomes and adhesion fingers between cells were disorganized (Figure 1M, N). At the fifth day following a BrdU pulse injection, some suprabasal cells of mutant esophagus remained strongly positive for BrdU and Ki67 (Figure 1P, R, T, V; yellow arrows) as compared with the control esophagus (Figure 1O, Q, S, U), suggesting that migration of these cells toward the lumen was impaired.
Figure 1. Esophageal phenotypes of Krt5Cre;Pax9loxP/loxP mice.
The papilloma-like structures were formed in the squamous epithelium (indicated by arrows) of Pax9-deficient esophagus (B, D) as compared with control esophagus on H&E-stained sections (A, C). Proliferation was increased in Pax9-deficient mice (F, H) as compared with controls (E, G). Apoptosis was similar in Pax9-deficient esophagus (J) and control esophagus (I), as detected by TUNEL assay. Toluidine blue staining (K, L) and transmission electron microscopy (M, N) showed an increase in keratohyalin granules (red arrows) and disorganization of the basal cell layer in Pax9-deficient esophagus as compared with control esophagus. Double IF showed that Ki67 was exclusively expressed in the basal cells of esophageal epithelium in control esophagus (O, Q, S, U) at the fifth day after BrdU pulse injection. However, in the mutant esophagus, some Ki67 positive cells appeared in the parabasal and superficial layers, and they were also BrdU-positive with the same staining intensity as the BrdU-positive cells in the basal cell layer (P, R, T, V) (yellow arrows). The base membrane of the squamous epithelium was marked with white dotted lines. (A, B, E, F, I, J: Scale bar=200 µm. C, D, G, H, O, P, Q, R, S, T, U, V: Scale bar=50 µm. K, L: Scale bar=20 µm. M, N: Scale bar=2 µm).
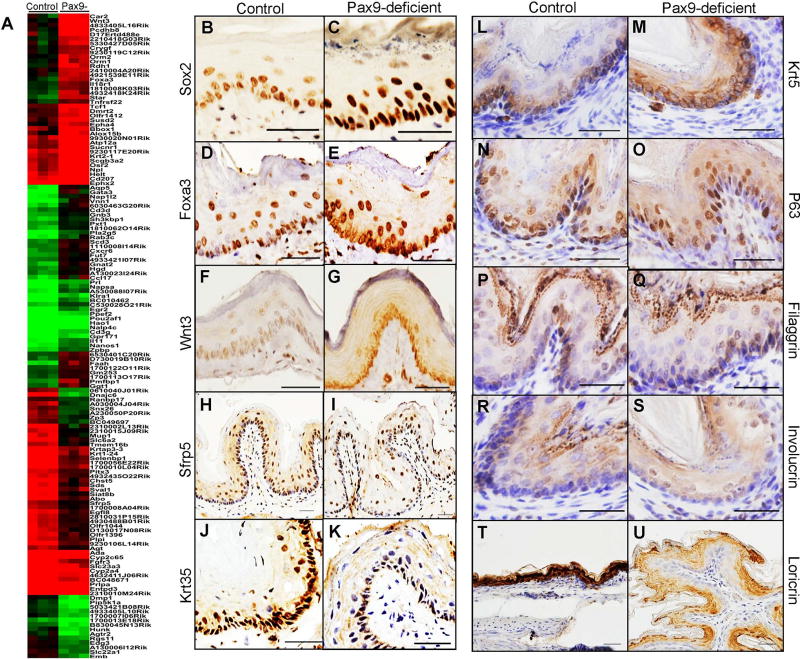
Gene microarray analysis showed that 60 genes were down-regulated of which some were associated with squamous cell differentiation (e.g., Krtap3-3, Krt1-24/Krt35, Sfrp5), and 83 genes were up-regulated in Pax9-deficient mice (e.g., Wnt3, Foxa3, Gata3) (Figure 2A). Multiple gene sets were enriched in control esophagus including “Pax9 target genes”. Several gene sets were enriched in the Pax9-deficient esophagus, e.g., “basal layer”, “Sox2 target genes”, “cell-cell junction” (supplementary material, Table S1). RT-qPCR showed up-regulation of hedgehog pathway genes (Gli1, Gli2), Wnt pathway genes (Wnt3, Gata3), and genes highly expressed in basal cells (Sox2, P63), and down-regulation of Pax9 and its downstream genes (Krt35) (supplementary material, Figure S2A). ChIP-PCR confirmed PAX9 binding to the 5’ upstream DNA sequence of Krt35 gene (supplementary material, Figure S2B). IHC for the selected genes involved in epithelial differentiation showed up-regulation of SOX2 (Figure 2B, C), FOXA3 (Figure 2D, E), WNT3 (Figure 2F, G) and KRT5 (Figure 2L, M), slightly increase of P63 expression (Figure 2N, O), and down-regulation of SFRP5 (Figure 2H, I), KRT35 (Figure 2J, K) and loricrin (Figure 2T, U) in the esophageal epithelium of Pax9-deficient mice. An obvious change in expression of filaggrin (Figure 2P, Q) or involucrin (Figure 2R, S) was not observed.
Figure 2. Differentially expressed genes in Krt5Cre;Pax9loxP/loxP esophagus.
Control and mutant esophagus were clustered based on differential expression of genes identified by SAM (A). IHC showed expression of SOX2 (B, C: Scale bar=50 µm), FOXA3 (D, E: Scale bar=50 µm), WNT3 (F, G: Scale bar=50 µm), SFRP5 (H, I: Scale bar=100 µm), KRT35 (J, K: Scale bar=50 µm), KRT5 (L, M: Scale bar=50 µm), P63 (N, O: Scale bar=50 µm), filaggrin (P, Q: Scale bar=50 µm), involucrin (R, S: Scale bar=50 µm) and loricrin (T, U: Scale bar=50 µm) in the mutant esophagus as compared with the wild-type esophagus. Bar represents mean ± SD.
PAX9 is down-regulated in human ESCC and OSCC
To examine the clinical relevance of PAX9 down-regulation, we analyzed microarray data of four GEO datasets (GSE23400, GSE20347, GSE13601, and GSE6631) and these showed significant down-regulation of PAX9 mRNA in human ESCC (supplementary material, Figure S3A, B) and human OSCC (supplementary material, Figure S3C, D), as compared with matched normal tissues.
Using two commercially available tissue microarrays, we obtained positive PAX9 staining in 2/29 ESCC and 25/29 matched normal esophagus (p<0.0001), and in 6/50 OSCC and 6/10 normal oral mucosa (p=0.0026).
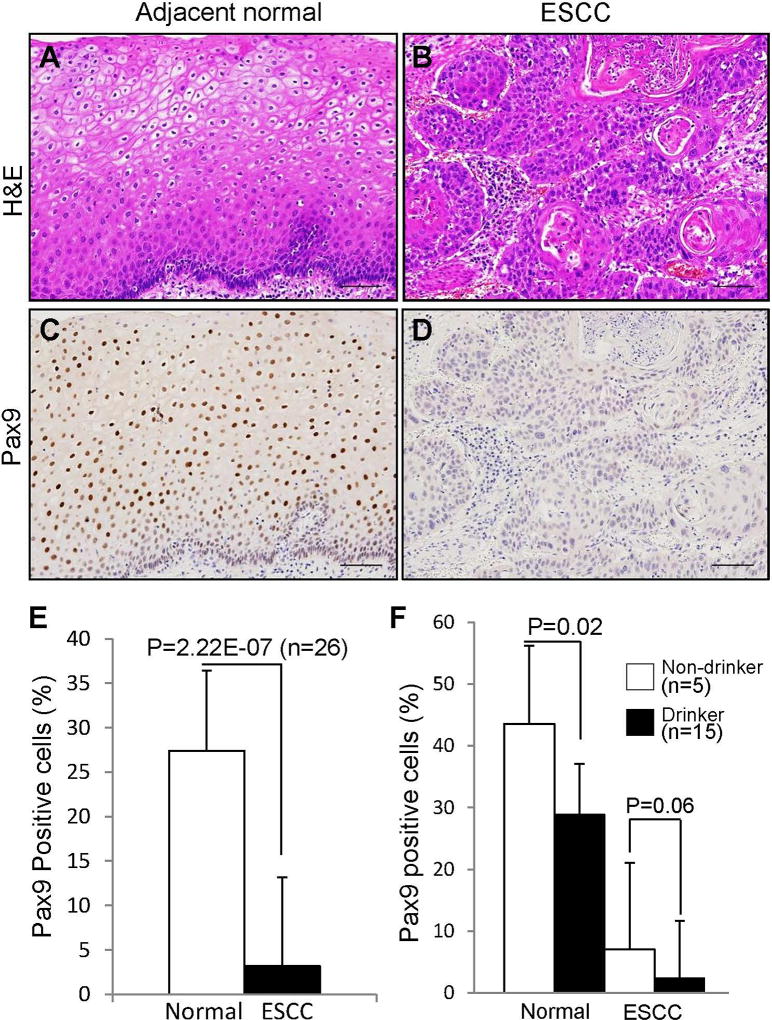
Using our human samples, IHC showed a dramatic decrease of PAX9 expression in ESCC versus adjacent normal esophageal tissues (Figure 3C, D, E). ESCC and adjacent normal esophageal tissue were confirmed by H&E staining (Figure 3A, B). Interestingly, based on the alcohol-drinking status of these patients (drinker or non-drinker), a significant decrease of PAX9-positive cells was observed in drinker’s normal tissue as compared to normal tissue from the non-drinkers (Figure 3F). Even in cancer tissues, a slight decrease of PAX9 expression was also observed in drinker’s when compared to non-drinker’s tissue.
Figure 3. Down-regulation of PAX9 expression in human ESCC and OSCC.
Paraffin sections of human ESCC were confirmed for histopathology by H&E staining (A, B) and stained for PAX9 (C, D). The percentage of PAX9-positive cells was determined as the number of PAX9-positive cells divided by the number of epithelial cells (E). Based on the drinking status of these patients (drinker or non-drinker), PAX9 expression was compared between drinker’s normal tissue and non-drinker’s normal tissue, and between drinker’s cancer tissue and non-drinker’s cancer tissue (F). Bar represents mean ± SD. Scale bar=100 µm. P values were determined using Student’s t-test.
Ethanol down-regulates PAX9 expression in oro-esophageal squamous epithelial cells in vitro and in vivo
Time- and dose-dependent exposure of KYSE510 and KYSE450 cells to ethanol confirmed down-regulation of PAX9 expression as shown by Western blotting (supplementary material, Figure S4). To further understand the effects of ethanol on PAX9 and global gene expression in vivo, mice were exposed to 20% sweetened ethanol for 4 weeks, or 15% sweetened ethanol for 40 weeks. H&E staining showed the thickness of the epithelium increased (Figure 4A, B, C) in those exposed to 15% ethanol for 40 weeks. Proliferation increased in response to ethanol at both 4 weeks (20% alcohol) and 40 weeks (15% alcohol): IHC for BrdU showed a significant increase of BrdU-positive epithelial cells in mouse forestomach (Figure 4D, E, F, G), indicating increased cell proliferation as a result of ethanol exposure in vivo.
Figure 4. Down-regulation of PAX9 expression in squamous epithelial cells of mouse forestomach due to alcohol drinking in vivo.
Ethanol caused thickening and basal cell hyperproliferation in mouse forestomach (A, B, C; H&E staining). Epithelial hyperproliferation in mouse forestomach was validated by BrdU IHC (D, E, F) and quantitation (G). Hierarchical clustering analysis of gene microarray data showed that Pax9 gene set was enriched in the non-exposed mouse forestomach as compared with those exposed to ethanol (H). Loricrin IHC showed that ethanol inhibited differentiation of the squamous epithelial cells in mouse forestomach (I, J, K). PAX9 IHC (L, M, N), PAX9 Western blotting (O, P), and RT-qPCR (Q) confirmed down-regulation of Pax9 mRNA and protein in the squamous epithelial cells in mouse forestomach. Broken lines are placed to indicate alignment of the group lanes. * P <.05, ** P <.01. Bar represents mean ± SD. Scale bar=50 µm. P values were determined using Student’s t-test.
Thirty-two genes were down-regulated by 20% ethanol for 4 weeks, and 581 genes up-regulated and 1,506 genes down-regulated by 15% ethanol for 40 weeks, as compared with control (supplementary material, Table S2). Using GSA analysis, we found enrichment of multiple gene sets including “Pax9 target genes” and “epidermal differentiation complex” in control forestomach, suggesting downregulation of Pax9 target genes by ethanol exposure (Figure 4H). PCA analysis and clustering analysis also supported distinct gene expression profiles of three groups (supplementary material, Figure S5).
IHC staining showed reduced expression of loricrin (a marker of squamous cell differentiation) in ethanol-exposed forestomach as compared with control (Figure 4I, J, K), indicating that squamous differentiation was suppressed by ethanol. Down-regulation of PAX9 expression in mouse forestomach was also observed (Figure 4L, M, N), especially in those exposed to 15% ethanol for 40 weeks. Pax9 down-regulation was further confirmed by Western blotting and RT-qPCR (Figure 4O, P, Q). In addition to the forestomach, ethanol-exposed mouse tongue also expressed PAX9 at a reduced level as shown by Western blotting and IHC (Supplementary material, Figure S6). No obvious change of PAX9 expression was observed in mouse esophagus (data not shown).
Promoter hypermethylation is associated with PAX9 silencing in human ESCC cells and OSCC tissues
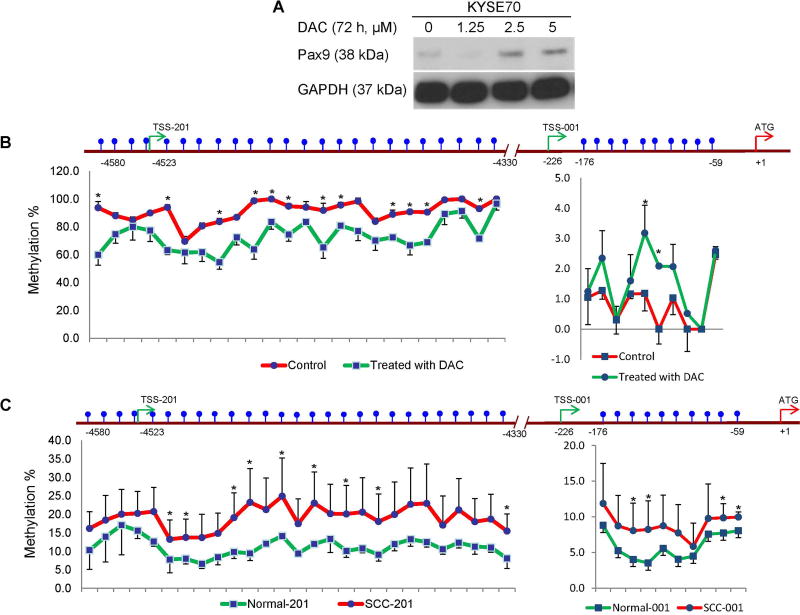
To understand the molecular mechanism through which alcohol drinking down-regulates PAX9 expression, CpG sites in the promoter regions of two different PAX9 transcriptional start sites (supplementary material, Figure S7) were pyrosequenced in control and DAC-treated KYSE70 cells. Consistent with an increase in PAX9 expression (Figure 5A), the methylation level of DAC-treated KYSE70 cells was lower than that of control (Figure 5B). More importantly, the methylation levels of both transcripts were higher in OSCC tissue samples than in matched normal samples (Figure 5C).
Figure 5. PAX9 promoter hypermethylation in KYSE70 cells and human OSCC samples.
KYSE70 cells treated with different concentrations of DAC were analyzed by Western blotting for PAX9 expression (A). The percentages of methylation at CpG sites in the promoter regions of two different PAX9 transcriptional start sites were analyzed by pyrosequencing for comparison between control and DAC-treated KYSE70 cells (B, n=3; * P <.05). The percentage of methylation at CpG sites in the promoter regions of two different PAX9 transcriptional start sites were analyzed by pyrosequencing for comparison between human OSCC and matched normal samples (C, n=6; * P <.05). P values were determined using Student’s t-test.
We then pyrosequenced the promoter regions of two different PAX9 transcriptional start sites in ethanol-exposed KYSE510 cells (100 mM for 72 h) and mouse forestomach (15% for 40 weeks). Despite the decrease in PAX9 expression, no significant difference of methylation levels was found between control samples and ethanol-exposed samples (supplementary material, Figure S8).
Increased susceptibility to NMBA-induced oro-esophageal squamous cell carcinogenesis in Krt5Cre;Pax9loxP/loxP mice
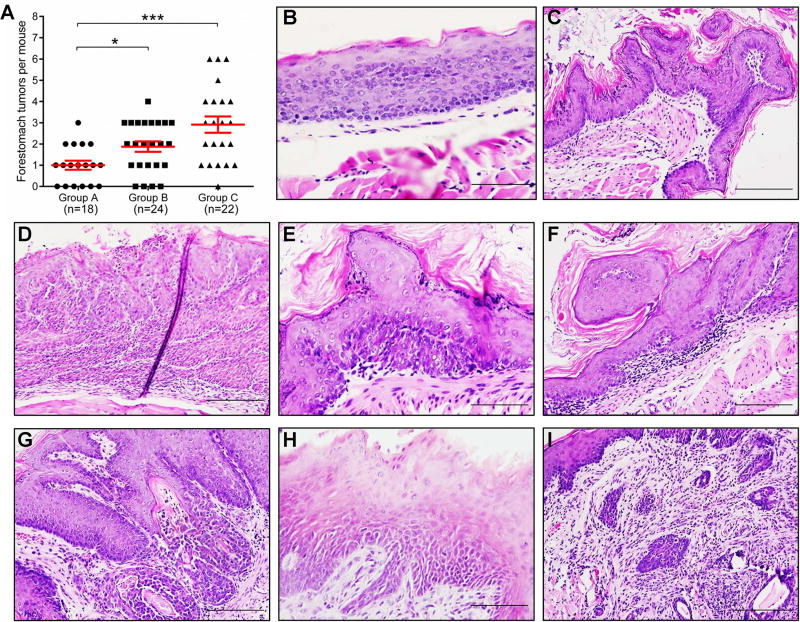
To determine whether Pax9 deficiency promotes OESCC, wild-type or Pax9-deficient mice were given six intragastric doses of NMBA and sacrificed at 40 weeks. The number of macroscopically visible tumors in mouse forestomach was significantly increased in Pax9-deficient mice, as compared with control (Figure 6A). Seven Pax9-deficient mice developed tongue tumors. Similar to Pax9 deficiency, an isocaloric Lieber-DeCarli diet containing ethanol for 40 weeks also significantly enhanced forestomach tumorigenesis in wild-type mice as compared with a control liquid diet.
Figure 6. Promotion of NMBA-induced tumorigenesis in mouse forestomach due to Pax9 deficiency or alcohol drinking.
The number of macroscopically visible tumors in mouse forestomach increased significantly in mice fed with an isocaloric Lieber-DeCarli diet containing ethanol (Group B) and Pax9-deficient mice (Group C), as compared with control (Group A) (A, * P <.05, *** P <.001). The middle red lines show the averages. Esophageal dysplasia (B), papilloma (C), SCC (D), forestomach dysplasia (E), papilloma (F), SCC (G), and tongue dysplasia (H), SCC (I) are shown here (B, E, H, Scale bar=50 µm; C, D, F, G, I, Scale bar=100 µm). P values were determined using Student’s t-test.
Under the microscope, the incidence of esophageal papilloma was significantly increased in ethanol-exposed mice, as compared with control. Incidences of forestomach dysplasia and SCC significantly increased from 22.2% and 5.6% in control mice to 81.8% and 31.8% in Pax9-deficient mice. The incidence of tongue lesions (dysplasia, papilloma, and SCC) was also significantly increased from 0% in control mice to 36.4% in Pax9-deficient mice (Table 1) (Figure 6B–I). IHC showed that PAX9 expression was not dramatically altered in histologically normal epithelium of NMBA-treated mice (supplementary material, Figure S9A, B). However, PAX9 expression was down-regulated during forestomach carcinogenesis as shown by decreasing expression from histologically normal to dysplasia and cancer (supplementary material, Figure S9B–D)
Discussion
This study is the first to demonstrate functional roles of PAX9 in the adult oro-esophageal epithelium. Tissue-specific Pax9 deficiency was associated with squamous epithelial cell hyperproliferation, delayed cell differentiation and altered global gene expression profile in mouse esophagus. PAX9 down-regulation was confirmed in human OESCC and associated with alcohol drinking. Ethanol exposure down-regulated PAX9 expression in oro-esophageal squamous epithelial cells in vitro and in vivo. Promoter hypermethylation was found to be associated with PAX9 silencing in human OESCC. Furthermore, both Pax9 deficiency and ethanol exposure promoted OESCC in vivo.
As a transcription factor, PAX9 is expressed in adult squamous epithelial cells in the upper gastrointestinal tract with the strongest expression in the tongue [6]. Clinically, PAX9 mutations have been reported in patients with oligodontia and cleft palate [18,19]. Previous studies by us and others have suggested a potential role of PAX9 in squamous cell differentiation and carcinogenesis [7,9–12]. In this study, Pax9 deficiency enhanced epithelial cell proliferation (papilloma-like structure, an increase of BrdU-labeling index and PCNA expression) and inhibited cell differentiation (loricrin downregulation, delayed maturation of BrdU-labelled cells) in mouse esophagus (Figure 1, 2).
Using GEO datasets and our own tissue samples, we further confirmed PAX9 down-regulation in human ESCC and OSCC (supplementary material, Figure S3 and Figure 3). Although human ESCC with a low level of PAX9 expression does share similarities in gene expression with Pax9-deficient esophagus (data not shown), it should be noted that mouse array and human array did not test the same genes, particularly in the early days. Therefore, such a relevance should be treated with caution. An interesting association between PAX9 down-regulation and alcohol drinking in human ESCC led us to investigate whether alcohol drinking, a major risk factor for OESCC [20–22], may cause PAX9 down-regulation. In fact, OSCC has a stronger association with alcohol exposure than cancers of any other organ sites [23–25]. A case-control study on the risk of ESCC showed a higher odds ratio (OR) for heavy drinkers (OR=10) compared to tobacco smokers (OR=5.8). Heavy drinking led to an increased numbers of SCC and dysplastic lesions in the human esophagus [26,27]. Acetaldehyde, an ethanol metabolite, is believed to be a carcinogen that contributes to the development of ESCC. Genetic polymorphisms of ethanol-metabolizing genes, such as acetaldehyde dehydrogenase and alcohol dehydrogenase, are also strongly and consistently associated with OESCC [21,22]. Our data showed that ethanol exposure caused time- and dose-dependent decrease of PAX9 expression in esophageal epithelial cells in vitro (supplementary material, Figure S4). More importantly, ethanol exposure reduced PAX9 expression in mouse forestomach (Figure 4) and tongue (supplementary material, Figure S6). It was unexpected that PAX9 expression in mouse esophagus was not significantly affected by ethanol exposure. Presumably, short duration of contact may account for relatively weak effect of ethanol on the esophageal epithelium.
We next aimed to understand the molecular mechanism leading to PAX9 silencing in OESCC. PAX9 has been identified as a gene subject to promoter methylation in human ESCC and in lung cancer [28,29]. A genome-wide DNA methylome analysis revealed widespread ethanol-induced alterations with significant hypermethylation of many regions of chromosomes in human embryonic stem cells [30]. We, therefore, pyrosequenced the promoter regions of two different PAX9 transcriptional start sites (supplementary material, Figure S7). Consistent with our hypothesis, the methylation percentages of multiple CpG sites in OSCC samples were significantly higher than those of matched normal tissues (Figure 5C). A demethylating agent increased PAX9 expression in KYSE70 cells and meanwhile reduced CpG methylation percentages (Figure 5A, B). However, ethanol exposure of KYSE510 cells or mouse forestomach did not result in PAX9 promoter hypermethylation even though PAX9 expression was down-regulated (supplementary material, Figure S8). Our data suggested alternative mechanisms are responsible for PAX9 down-regulation by ethanol in our experimental settings. However, we cannot exclude the possibility that our experimental settings, i.e., ethanol exposure of cells and feeding mice with ethanol, may not exactly reproduce sequential changes leading to PAX9 silencing in human OESCC. It remains unknown whether promoter hypermethylation may be associated with PAX9 down-regulation due to alcohol drinking.
Our last question was whether Pax9 deficiency contributes to OESCC in vivo. In Pax9-deficient mice, we observed a significant increase in the number of forestomach tumors and the incidences of esophageal and forestomach lesions (papilloma, dysplasia, SCC) (Table 1, Figure 6). Because NMBA is not expected to induce tumors in the tongue of wild-type mice [15], development of tongue lesions in Pax9-deficient mice further supported the importance of PAX9 down-regulation in the development of OESCC. Consistent with the association between alcohol drinking and PAX9 down-regulation, feeding mice with an isocaloric liquid diet containing ethanol significantly increased the number of forestomach papilloma and the incidence of esophageal papilloma (Figure 6A, Table 1). Based on the gene expression data, we speculate that PAX9 down-regulation may allow activation of multiple oncogenic pathways, and thus facilitates OESCC. For example, Shh signaling activation results in the expansion of epithelial precursor cell compartment [17]. Of note, hedgehog ligand is required for the growth of digestive tract tumors including esophageal cancer [31]. Wnt signaling is involved in early differentiation of esophageal epithelium [11]. In OESCC, poor histological differentiation and clinical outcome are associated with increased expression of nuclear β-catenin [32]. Exposure to carcinogen and ethanol increases overall β-catenin level in the tongue [33]. Sox2 and P63 colocalized at genetic loci and co-regulated gene expression to facilitate the development of ESCC [34]. Transgenic overexpression of SOX2 itself was found to induced SCC in mouse forestomach [35] and lung [36]. It should be noted that humans do not have forestomach. Therefore enhanced forestomach carcinogenesis by Pax9 deficiency may or may not be relevant to human ESCC.
Taken together, PAX9 regulates squamous cell differentiation and carcinogenesis in the oro-esophageal epithelium. Our data support a novel mechanism that PAX9 down-regulation may also contribute to alcohol-associated OESCC. Further studies are warranted to understand upstream events leading to PAX9 down-regulation by ethanol, and downstream mechanisms which promote OESCC.
Supplementary Material
Table S1. Differentially expressed genes and enriched gene sets in Pax9-deficient esophagus as compared with control esophagus
Table S2. Differentially expressed genes and enriched gene sets in mouse forestomach due to alcohol drinking (20% in the drink for 4 weeks or 15% in the drink for 40 weeks)
Acknowledgments
The authors acknowledge excellent microarray service by Dr. Yan Shi and her staff at the Genomics Core Facility, Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill, NC. The authors also thank Dr. Sara E. Miller and Mr. Phillip Christopher at the Department of Pathology, Duke University, Durham, NC, for their excellent electron microscopy service. The pyroseqencing service provided by Dr. Liying Yan at EpigenDx Inc. is greatly appreciated. This work was supported by research grants from the National Natural Science Foundation of China (grant numbers 30973325, 81372897) and the National Institutes of Health (grant numbers U54 AA019765, U54 CA156735).
Abbreviations
- DAC
5-aza-2′-deoxycytidine
- ESCC
esophageal squamous cell carcinoma
- OESCC
Oro-esophageal squamous cell carcinoma
- OR
odds ratio
- SAM
significance analysis of microarrays
Footnotes
Conflict of interest: The authors declare no potential conflicts of interest.
Author Contributions: ZX, SR, HC, YL, CH, LZ, and JO conducted the experiments and analyzed the data. ZX, TC, RK, HK, KG, ZS and XC wrote and revised the manuscript. TC helped with the mouse experiments. RK and HK provided the Pax9loxP/loxP mouse line. KG provided human ESCC tissue sections and clinical data. ZS and XC designed the experiments and supervised the whole process.
References
- 1.Peters H, Neubuser A, Kratochwil K, et al. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998;12:2735–2747. doi: 10.1101/gad.12.17.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jonker L, Kist R, Aw A, et al. Pax9 is required for filiform papilla development and suppresses skin-specific differentiation of the mammalian tongue epithelium. Mech Dev. 2004;121:1313–1322. doi: 10.1016/j.mod.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Kist R, Watson M, Crosier M, et al. The formation of endoderm-derived taste sensory organs requires a Pax9-dependent expansion of embryonic taste bud progenitor cells. PLoS Genet. 2014;10:e1004709. doi: 10.1371/journal.pgen.1004709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakatomi M, Wang XP, Key D, et al. Genetic interactions between Pax9 and Msx1 regulate lip development and several stages of tooth morphogenesis. Dev Biol. 2010;340:438–449. doi: 10.1016/j.ydbio.2010.01.031. [DOI] [PubMed] [Google Scholar]
- 5.Sivakamasundari V, Kraus P, Sun W, et al. A developmental transcriptomic analysis of Pax1 and Pax9 in embryonic intervertebral disc development. Biol Open. 2017;6:187–199. doi: 10.1242/bio.023218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters H, Schuster G, Neubuser A, et al. Isolation of the Pax9 cDNA from adult human esophagus. Mamm Genome. 1997;8:62–64. doi: 10.1007/s003359900351. [DOI] [PubMed] [Google Scholar]
- 7.Gerber JK, Richter T, Kremmer E, et al. Progressive loss of PAX9 expression correlates with increasing malignancy of dysplastic and cancerous epithelium of the human oesophagus. J Pathol. 2002;197:293–297. doi: 10.1002/path.1115. [DOI] [PubMed] [Google Scholar]
- 8.Tan B, Wang J, Song Q, et al. Prognostic value of PAX9 in patients with esophageal squamous cell carcinoma and its prediction value to radiation sensitivity. Mol Med Rep. 2017;16:806–816. doi: 10.3892/mmr.2017.6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JC, Sharma M, Lee YH, et al. Pax9 mediated cell survival in oral squamous carcinoma cell enhanced by c-myb. Cell Biochem Funct. 2008;26:892–899. doi: 10.1002/cbf.1522. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, Qin R, Ma Y, et al. Differential gene expression in normal esophagus and Barrett's esophagus. J Gastroenterol. 2009;44:897–911. doi: 10.1007/s00535-009-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen H, Li J, Li H, et al. Transcript profiling identifies dynamic gene expression patterns and an important role for Nrf2/Keap1 pathway in the developing mouse esophagus. PloS One. 2012;7:e36504. doi: 10.1371/journal.pone.0036504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H, Beasley A, Hu Y, et al. A zebrafish model for studies on esophageal epithelial biology. PloS One. 2015;10:e0143878. doi: 10.1371/journal.pone.0143878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sano S, Itami S, Takeda K, et al. Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. EMBO J. 1999;18:4657–4668. doi: 10.1093/emboj/18.17.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kist R, Greally E, Peters H. Derivation of a mouse model for conditional inactivation of Pax9. Genesis. 2007;45:460–464. doi: 10.1002/dvg.20295. [DOI] [PubMed] [Google Scholar]
- 15.Aqeilan RI, Hagan JP, Aqeilan HA, et al. Inactivation of the Wwox gene accelerates forestomach tumor progression in vivo. Cancer Res. 2007;67:5606–5610. doi: 10.1158/0008-5472.CAN-07-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkin RJ, Lalor PF, Parker R, et al. Murine models of acute alcoholic hepatitis and their relevance to human disease. Am J Path. 2016;186:748–760. doi: 10.1016/j.ajpath.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 17.van Dop WA, Rosekrans SL, Uhmann A, et al. Hedgehog signalling stimulates precursor cell accumulation and impairs epithelial maturation in the murine oesophagus. Gut. 2013;62:348–357. doi: 10.1136/gutjnl-2011-301141. [DOI] [PubMed] [Google Scholar]
- 18.Klein ML, Nieminen P, Lammi L, et al. Novel mutation of the initiation codon of PAX9 causes oligodontia. J Dent Res. 2005;84:43–47. doi: 10.1177/154405910508400107. [DOI] [PubMed] [Google Scholar]
- 19.Zhou J, Gao Y, Lan Y, et al. Pax9 regulates a molecular network involving Bmp4, Fgf10, Shh signaling and the Osr2 transcription factor to control palate morphogenesis. Development. 2013;140:4709–4718. doi: 10.1242/dev.099028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamangar F, Chow WH, Abnet CC, et al. Environmental causes of esophageal cancer. Gastroenterol Clin N Am. 2009;38:27–57. vii. doi: 10.1016/j.gtc.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Chen H, Sun Z, et al. Molecular mechanisms of ethanol-associated oro-esophageal squamous cell carcinoma. Cancer Lett. 2015;361:164–173. doi: 10.1016/j.canlet.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohashi S, Miyamoto S, Kikuchi O, et al. Recent advances from basic and clinical studies of esophageal squamous cell carcinoma. Gastroenterology. 2015;149:1700–1715. doi: 10.1053/j.gastro.2015.08.054. [DOI] [PubMed] [Google Scholar]
- 23.Bagnardi V, Blangiardo M, La Vecchia C, et al. A meta-analysis of alcohol drinking and cancer risk. Br J Cancer. 2001;85:1700–1705. doi: 10.1054/bjoc.2001.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Mao Y, Zhang Y, et al. Alcohol drinking and upper aerodigestive tract cancer mortality: a systematic review and meta-analysis. Oral Oncol. 2014;50:269–275. doi: 10.1016/j.oraloncology.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 25.Marron M, Boffetta P, Moller H, et al. Risk of upper aerodigestive tract cancer and type of alcoholic beverage: a European multicenter case-control study. Europ J Epidemiol. 2012;27:499–517. doi: 10.1007/s10654-012-9699-1. [DOI] [PubMed] [Google Scholar]
- 26.Morita M, Kumashiro R, Kubo N, et al. Alcohol drinking, cigarette smoking, and the development of squamous cell carcinoma of the esophagus: epidemiology, clinical findings, and prevention. Intl J Clin Oncol. 2010;15:126–134. doi: 10.1007/s10147-010-0056-7. [DOI] [PubMed] [Google Scholar]
- 27.Mwachiro MM, Burgert SL, Lando J, et al. Esophageal squamous dysplasia is common in asymptomatic Kenyans: a prospective, community-based, cross-sectional study. Am J Gastroenterol. 2016;111:500–507. doi: 10.1038/ajg.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rauch T, Li H, Wu X, et al. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res. 2006;66:7939–7947. doi: 10.1158/0008-5472.CAN-06-1888. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka K, Imoto I, Inoue J, et al. Frequent methylation-associated silencing of a candidate tumor-suppressor, CRABP1, in esophageal squamous-cell carcinoma. Oncogene. 2007;26:6456–6468. doi: 10.1038/sj.onc.1210459. [DOI] [PubMed] [Google Scholar]
- 30.Khalid O, Kim JJ, Kim HS, et al. Gene expression signatures affected by alcohol-induced DNA methylomic deregulation in human embryonic stem cells. Stem Cell Res. 2014;12:791–806. doi: 10.1016/j.scr.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berman DM, Karhadkar SS, Maitra A, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 32.Santoro A, Pannone G, Papagerakis S, et al. Beta-catenin and epithelial tumors: a study based on 374 oropharyngeal cancers. Biomed Res Int. 2014;2014:948264. doi: 10.1155/2014/948264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osei-Sarfo K, Tang XH, Urvalek AM, et al. The molecular features of tongue epithelium treated with the carcinogen 4-nitroquinoline-1-oxide and alcohol as a model for HNSCC. Carcinogenesis. 2013;34:2673–2681. doi: 10.1093/carcin/bgt223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe H, Ma Q, Peng S, et al. SOX2 and p63 colocalize at genetic loci in squamous cell carcinomas. J Clin Invest. 2014;124:1636–1645. doi: 10.1172/JCI71545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu K, Jiang M, Lu Y, et al. Sox2 cooperates with inflammation-mediated Stat3 activation in the malignant transformation of foregut basal progenitor cells. Cell Stem Cell. 2013;12:304–315. doi: 10.1016/j.stem.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu Y, Futtner C, Rock JR, et al. Evidence that SOX2 overexpression is oncogenic in the lung. PloS One. 2010;5:e11022. doi: 10.1371/journal.pone.0011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Differentially expressed genes and enriched gene sets in Pax9-deficient esophagus as compared with control esophagus
Table S2. Differentially expressed genes and enriched gene sets in mouse forestomach due to alcohol drinking (20% in the drink for 4 weeks or 15% in the drink for 40 weeks)