Abstract
Background
P53 mutation status is a pivotal biomarker for gliomas. Here, we developed a machine-learning model to predict p53 status in lower-grade gliomas based on radiomic features extracted from conventional magnetic resonance (MR) images.
Methods
Preoperative MR images were retrospectively obtained from 272 patients with primary grade II/III gliomas. The patients were randomly allocated in a 2:1 ratio to a training (n = 180) or validation (n = 92) set. A total of 431 radiomic features were extracted from each patient. The lest absolute shrinkage and selection operator (LASSO) method was used for feature selection and radiomic signature construction. Subsequently, a machine-learning model to predict p53 status was established using the selected features and a Support Vector Machine classifier. The predictive performance of all individual features and the model was calculated using receiver operating characteristic curves in both the training and validation sets.
Results
The p53-related radiomic signature was built using the LASSO algorithm; this procedure consisted of four first-order statistics or related wavelet features (including Maximum, Median, Minimum, and Uniformity), a shape and size-based feature (Spherical Disproportion), and ten textural features or related wavelet features (including Correlation, Run Percentage, and Sum Entropy). The prediction accuracies based on the area under the curve were 89.6% in the training set and 76.3% in the validation set, which were better than individual features.
Conclusions
These results demonstrate that MR image texture features are predictive of p53 mutation status in lower-grade gliomas. Thus, our procedure can be conveniently used to facilitate presurgical molecular pathological diagnosis.
Keywords: p53, Lower-grade gliomas, Radiogenomics, Prediction, Machine learning
Highlights
-
•
We established a p53-related radiomic signature in lower-grade gliomas based on LASSO algorithm.
-
•
We developed a machine-learning model using the radiomic signature and a support vector machine.
-
•
P53 mutation status of lower-grade gliomas was predicted effectively based on our machine-learning model.
1. Introduction
Lower-grade gliomas (World Health Organization grade II/III gliomas) are diffusely infiltrative tumors that originate most often in the cerebral hemispheres of adults (Zhang and Brat, 2016). For a century, glioma diagnosis has been based on histologic appearance, but recent discoveries of molecular biomarkers have led to the reassessment of diagnostic definitions and criteria (Appin and Brat, 2015).
TP53 is a pivotal gene that often mutates in diffuse gliomas and especially in astrocytomas (Sarkar et al., 2005). TP53 encodes p53, a transcription factor that regulates the cell cycle to suppress the proliferation of cells with oncogenic properties (Gillet et al., 2014). Mutant p53 promotes tumor cell proliferation, invasion, and survival (Muller and Vousden, 2013), and is a critical biomarker of diffuse gliomas (Appin and Brat, 2015, Suzuki et al., 2015). TP53 sequencing is the primary method used to detect the p53 mutation (Gillet et al., 2014). Immunohistochemical analyses to detect overexpression of p53 are generally regarded as a surrogate marker for p53 mutation (Levidou et al., 2010). However, both methods are invasive assays based on craniotomy.
Non-invasive detection of p53 status has been attempted. For example, mutant p53 was found to be specifically associated with certain tumor locations and enhancement patterns in lower-grade gliomas based on preoperative magnetic resonance (MR) images (Mut et al., 2007, Wang et al., 2015b). More recently, machine-learning algorithms have been used to predict genotype based on quantitative imaging features derived from conventional MR images (Ellingson et al., 2011, Macyszyn et al., 2016). A series of pioneer studies have revealed that quantitative imaging features have great potentials in predicting the diagnosis and prognosis of diseases(Chaddad et al., 2016a, Chaddad et al., 2016b, Chaddad and Tanougast, 2016). Moreover, several pivotal glioma molecular biomarkers such as MGMT and IDH1 (Korfiatis et al., 2016, Zhou et al., 2017) have already been predicted efficiently with quantitative imaging features, which greatly increases the impetus for preoperative determination of the p53 genotype.
Here, we extracted radiological features from the preoperative MR images of patients with gliomas and we hypothesized that a machine-learning approach to analyze radiomic signatures could predict p53 status.
2. Materials and methods
2.1. Patients
Two hundred and seventy-two patients who were treated at the Beijing Tiantan Hospital from August 2005 to August 2012 were enrolled in this retrospective study. MR images and clinical information were collected based on the Chinese Glioma Genome Atlas database (http://www.cgga.org.cn). The inclusion criteria were as follows: 1) histologic confirmation of primary grade II/III gliomas according to the World Health Organization criteria; 2) no history of preoperative therapy; 3) available preoperative T2-weighted MR images; 4) available p53 status based on immunohistochemical detection; and 5) available clinical characteristics. The patients were randomly allocated in a 2:1 ratio to a training (n = 180) or validation (n = 92) set. The machine-learning model for p53 prediction was built using the training set and evaluated using the validation set. Our study was approved and reviewed by the institutional review board.
2.2. MR imaging acquisition and preprocessing
T2-weighted MR images were used for the extraction of radiomics features, as these images are well accepted in the identification of abnormal hyperintense signals that represent the involved regions of low-grade gliomas (Kinoshita et al., 2016, Ricard et al., 2007, Wang et al., 2015a). T2-weighted MR imaging of the patients was mainly performed on a Magnetom Trio 3 T scanner (Siemens, Erlangen, Germany). The imaging protocol is generally in consistent with the international standardized brain tumor imaging protocol (Ellingson et al., 2015), and the detailed parameters were as follows: 24 slices; echo time = 110 ms; repetition time = 5800 ms; flip angle = 150 degrees; voxel size = 0.6 × 0.6 × 5.0 mm3; field of view = 240 × 188 mm2; matrix = 384 × 300. Two experienced neurosurgeons delineated tumor masks using MRIcron software (http://www.mccauslandcenter.sc.edu/mricro). Hyperintense signals on the T2-weighted images were regarded as tumor areas. The Dice coefficient was used to measure the discrepancy between tumor masks, and a senior neuroradiologist made a final decision about the tumor border when the discrepancy was > 5%.
2.3. Quantitative radiological feature extraction and univariate analyses
Using an in-house MATLAB process, all the imaging data were normalized (Z score transformation) before feature extraction to reduce the bias. For each glioma case, we extracted four groups of radiological features from volume masks as the previous study described and detailed descriptions of each feature was listed in the supplementary material of the previous study (Aerts et al., 2014). Fourteen indexes that quantitatively described the distribution of voxel intensities were categorized into group 1. Group 2 included eight radiological features based on the shape and size of the tumors. Group 3 features were textural features that quantified intra-tumor heterogeneity, calculated from gray level co-occurrence (22 indexes) and gray level run-length (11 indexes) texture matrices. Texture matrices were determined considering 26-connected voxels. Features in group 4 were derived from wavelet decompositions of group 1 and 3 features (376 indexes). The original feature was decomposed into 8 decompositions, by directional low-pass (i.e. a scaling) and high-pass (i.e. a wavelet) filtering. All processes of feature extraction were conducted using MATLAB version 2014a, (Natick, MA, USA). Univariate analyses were performed by using individual features to predict p53 status with receiver operating characteristic (ROC) curve analysis in the training set. The 10-fold cross validation was performed and then the average of the AUCs were calculated.
2.4. Machine-learning model for p53 prediction
The lest absolute shrinkage and selection operator (LASSO) algorithm, which is suitable for the regression of high-dimensional data (Sauerbrei et al., 2007), was used to select the most predictive features in the training set to prevent overfitting and improve generalization (Huang et al., 2016). The misclassification error is determined by a tuning parameter (Lambda). As the Lambda gets smaller, some coefficients may be shrunk towards zero (Kumamaru et al., 2016, Vasquez et al., 2016). We then selected the Lambda for which the cross-validation error is the smallest. In this way, most of the coefficients of the covariates are decreased to zero and the remaining non-zero coefficients are selected by LASSO. The machine-learning model predictive of p53 was constructed based on the selected radiological features and the support vector machine (SVM) in the training set, and was evaluated in the validation set. The SVM classifier was based on transforming the feature space to a high-dimensional space where a separating hyperplane maximized the margin between classes (Korfiatis et al., 2016); this approach is effective for many pattern recognition problems (Huang et al., 2017). For each patient, the SVM classifier generated an estimated value output. ROC curve analysis was performed in both the training and validation sets to evaluate the predictive efficiency of the machine-learning model.
2.5. p53 immunohistochemistry
Immunoperoxidase staining for p53 mutants was conducted following the standard procedure described in our previous study (Wang et al., 2015b). Briefly, formalin-fixed, paraffin-embedded tissue was cut to 5 μm and deparaffinized, rehydrated, and incubated in tris-buffered saline. The samples were heated in sodium citrate buffer for 10 min at 100 °C, and non-specific protein binding was blocked in 5% horse serum in phosphate-buffered saline (PBS). Rabbit polyclonal p53 IgG was then applied to the samples for 60 min at room temperature (25 °C). After being washed in PBS, the tumor samples were incubated in secondary antibody for 30 min at room temperature. The sections were washed again and treated with Elite ABC (Vectastain; Vector Laboratories, Burlingame, CA, USA), and washed and developed with 3,3′-diaminobenzidine tetrahydrochloride (50 mg 3,3′-diaminobenzidine and 150 μl 3% H2O2 in 100 ml PBS). After being rinsed in PBS, the samples were dehydrated in graded alcohols, cleared in xylene, and permanently covered. Two pathologists independently examined the immunostained sections. When > 10% of positive immunostained cells appeared in the section, the tumor was considered to be p53-mutated (Ando et al., 2015).
2.6. Statistical analysis
The LASSO algorithm, SVM classifier, and ROC curve analysis were implemented in R software version 3.3.2 (The R Foundation, Salt Lake City, UT, USA). T-statistical test and Chi-square test were used to compare clinical characteristics between training and validation sets, and p < 0.05 was considered statistically significant. ROC curve analysis was conducted to demonstrate the performance of the p53 predictive model, and an optimal cutoff value was identified when the sensitivity plus specificity was maximal. The prediction accuracy and area under the curves (AUC) were calculated for both the training and validation sets.
3. Results
3.1. Clinical characteristics
MR images were obtained from 272 patients with primary WHO grade II/III gliomas, including 179 with grade II (male, 106; female, 73; age range, 4–68 years) and 93 with grade III (male, 64; female, 29; age range, 17–71 years). The proportion of patients with a p53 mutation was 42.2% and 48.9% in the training (n = 180) and validation (n = 92) sets, respectively. No significant difference was found in age, sex, grade, p53 status, or tumor location between the two sets of patients. Detailed demographic characteristics of the patients are presented in Table 1.
Table 1.
Patient characteristics.
| Total (n = 272) | Training (n = 180) | Validation (n = 92) | p value | |
|---|---|---|---|---|
| Age (years; mean) | 40.1 | 39.2 | 41.9 | 0.054a |
| Sex (male/female) | 169/103 | 111/69 | 58/34 | 0.825b |
| Grade II/Grade III | 179/93 | 123/57 | 56/36 | 0.220b |
| P53 wild type/P53 mutation | 151/121 | 104/76 | 47/45 | 0.293b |
| Tumor location (left/right/bilateral) | 142/113/17 | 95/73/12 | 47/40/5 | 0.306b |
Legends.
T-statistical test.
Chi-square test.
3.2. MRI features and univariate analyses
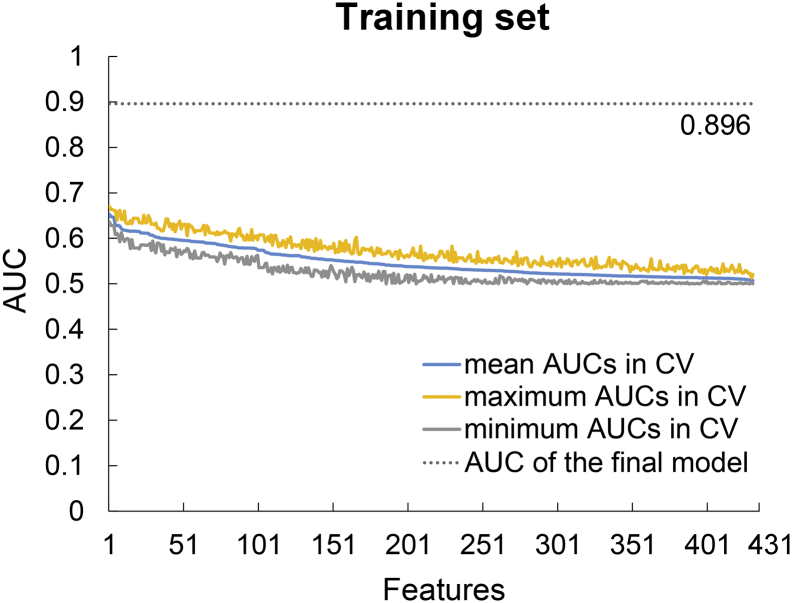
A total of 431 quantitative radiological features were extracted from the images of each patient, including 14 first-order statistics, 8 shape- and size-based features, 33 textural features, and 376 wavelet features derived from first-order statistics and textural features. We also evaluated the predictive efficiency of individual features in the training set using ROC curve analysis and 10-fold cross validation. The results showed that AUCs of individual features were lower than the AUC of the final machine-learning model established later (Fig. 1). All quantitative radiological features and their corresponding mean AUCs after 10-fold cross validation in the training set are listed in Supplementary Table 1.
Fig. 1.
Area under the curves (AUCs) of individual features after 10-fold cross validation (CV) in the training set. The features were listed based on the mean AUCs from the largest to the smallest. The results indicated that AUCs of individual features were lower than the AUC of the final model.
3.3. Machine-learning model for p53 prediction
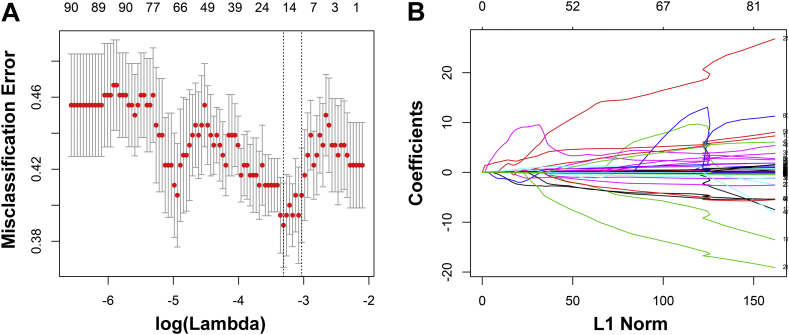
Based on the p53 mutation status, 431 radiological features extracted from T2-weighted MR images were reduced to 15 potential predictors in the training set (Fig. 2A and B). The 15 selected predictors were features with nonzero coefficients in the LASSO regression model, including first-order statistics-derived wavelet features such as Maximum, Minimum, and Uniformity; shape- and size-based features such as Spherical Disproportion; and textural features derived wavelet features such as Autocorrelation, Correlation, and Run Percentage (Table 2).
Fig. 2.
Fifteen texture features were selected using the least absolute shrinkage and selection operator algorithm (LASSO). (A) The misclassification error is shown versus log(lambda), with the lowest misclassification point indicating the optimal number of features remaining to fit the model. Dotted vertical lines are drawn at the best lambda values based on the minimum criteria and the 1 standard error criteria by 10-fold cross-validation. (B) LASSO coefficient profiles are shown for the 431 texture features. A vertical line is drawn at the value where the optimal lambda results in 15 nonzero coefficients.
Table 2.
Fifteen radiological features selected by the LASSO algorithm.
| Features | Descriptions |
|---|---|
| Autocorrelation_3 | A wavelet feature derived from autocorrelation. Autocorrelation evaluates the linear spatial relationship between texture primitives and measures the coarseness of an image. |
| Correlation_3 | A wavelet feature derived from correlation. Correlation is a measure of gray level linear dependence between the pixels at the specified positions relative to each other. |
| Informational measure of correlation 2_2 | Wavelet features derived from informational measure of correlation2. Informational measure of correlation2 measures nonlinear gray-level dependence. |
| Informational measure of correlation 2_7 | |
| Long run low gray level emphasis_3 | A wavelet feature derived from long run low gray level emphasis. It measures the joint distribution of long runs and low gray level values. |
| Maximum_6 | A wavelet feature derived from maximum. It describes the maximum value. |
| Maximum probability_2 | A wavelet feature derived from maximum probability. It describes the maximum value probability. |
| Median_6 | A wavelet feature derived from median. The median is the value that separates the lower and upper half of the sorted array of pixel values. |
| Minimum_1 | A wavelet feature derived from minimum. It describes the maximum gray value. |
| Run length nonuniformity_8 | A wavelet feature derived from run length nonuniformity. Run length nonuniformity examines the distribution of run lengths, higher when the texture is dominated by a few run lengths outliers. |
| Run percentage_4 | A wavelet feature derived from run percentage. Run percentage indicates the homogeneity and the distribution of runs of an image in a given direction. |
| Spherical disproportion | Spherical disproportion indicates how close the shape is to a sphere. |
| Sum average | Sum average measures overall image brightness. |
| Sum entropy_3 | A wavelet feature derived from sum entropy. Sum entropy provides the texture pattern of inhomogeneity inside the tumors. |
| Uniformity_4 | A wavelet feature derived from Uniformity. It describes the uniformity of the Image. |
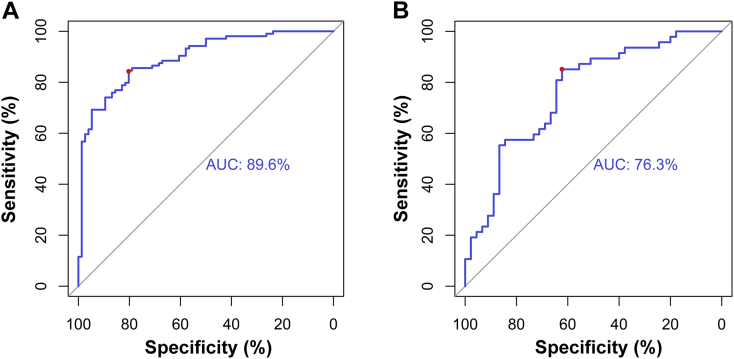
Training set. A p53 predictive machine-learning model was constructed using the training set based on the selected features and the SVM classifier. The AUC was 89.6% in the ROC curve analysis (Fig. 3A), and the sensitivity, specificity, and accuracy were 80.3%, 84.6%, and 80.0% respectively at the optimal cutoff point (0.1379). The estimated values calculated by the SVM classifier from patients in the training set are shown in Supplementary Fig. S1A.
Fig. 3.
Receiver operating characteristic curve for p53 genotype prediction in the training and validation sets. (A) In the training set, the area under the curve (AUC) was 89.6%. The optimal cutoff value (0.138), determined as the point when the sensitivity plus specificity was maximal, exhibited a sensitivity, specificity, and accuracy of 80.3%, 84.6%, and 80.0%, respectively (red dot). (B) The AUC was 76.3% in the validation set. At the optimal cutoff value (0.181), the sensitivity, specificity, and accuracy were 62.2%, 85.1%, and 70.7%, respectively (red dot).
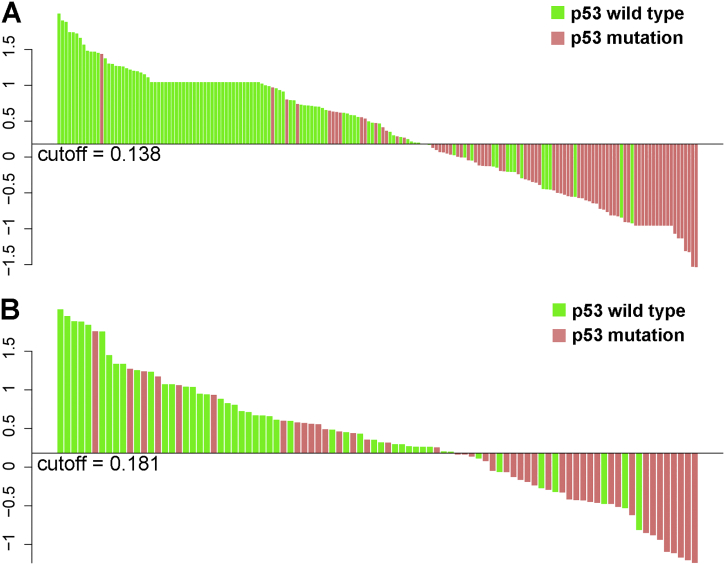
Supplementary Fig. S1.
Estimated values derived from the machine-learning model for every patient in each cohort. The cutoff value was identified when the sensitivity plus specificity was optimal in the receiver operating characteristic curve analysis. (A) Estimated values are shown for each patient in the training set. (B) Estimated values are shown for each patient in the validation set. The p53 mutation status is indicated using different colors.
Validation set. The model was then applied to the validation set, and the p53 mutation status was effectively predicted. Estimated values derived from patients in the validation set are shown in Supplementary Fig. S1B. In the ROC curve analysis (Fig. 3B), the AUC was 76.3%. In addition, the optimal cutoff value (0.1812) exhibited a sensitivity, specificity, and accuracy of 62.2%, 85.1%, and 70.7%, respectively. Hence, the 15 radiological features that constituted our model were regarded as a p53-associated radiomic signature.
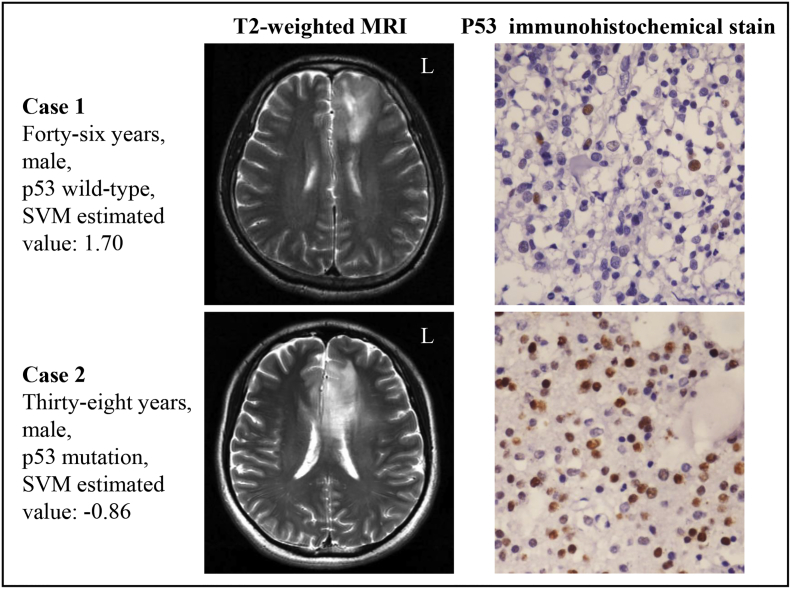
Supplementary Fig. S2 presents two representative cases of patients with lower grade gliomas. The first case was a 46-year-old male patient with p53 wild-type, who was correctly classified into the p53 wild-type group based on the SVM estimated value being high (1.70). Case 2 was a 38-year-old male patient with p53 mutation, who was correctly classified into the p53 mutation group according to the SVM estimated value being low (− 0.86).
Supplementary Fig. S2.
Magnetic resonance images of two lower-grade glioma patients with different p53 status. Case 1 was a 46-year-old man with p53 wild-type, who was correctly assigned to the p53 wild-type group based on the SVM classifier. Case 2 was a 38-year-old male patient with p53 mutation, who was correctly assigned to the p53 mutation group.
4. Discussion
We extracted several radiological features from the preoperative MR images, and assessed the p53 status predictive efficiency of each feature. Subsequently, p53-associated features were screened using the LASSO algorithm, and an SVM classifier was created to integrate the selected features to predict p53 genotype in lower-grade gliomas. Our model achieved an AUC of 89.6% in the training set data and an AUC of 76.3% in the validation set data. Notably, this result was better than the predictive efficiency of any individual feature. Overall, our results indicate that the p53 mutation status can be predicted using non-invasive radiological data, and that a machine-learning approach that integrates multivariate features is more effective and robust than individual features.
Our analysis expands the work of several recent studies that revealed novel associations between radiological features and the glioma expression profile. In a previous study of glioblastomas, tumors that were highly positive for the p53 mutation possessed typical lesions with well-defined borders and a ring enhancement pattern in T1-weighted images with contrast (Mut et al., 2007). In addition, the p53 mutation status has been associated with tumor location based on a voxel-based lesion-symptom mapping analysis; p53-mutated glioblastomas tended to be located in the frontal lobe surrounding the rostral extension of the lateral ventricles (Zhang et al., 2014), and p53-mutated low-grade gliomas were specifically associated with the left medial temporal lobe and right anterior temporal lobe (Wang et al., 2015b). With the development of radiogenomics, p53 status can be quantitatively predicted. One previous study enrolled 31 glioma patients and extracted 86 radiomic features from preoperative MRI to predict p53 status, with an accuracy of 65.2% (AUC = 71.9%) (Liu et al., 2012). Our study enrolled 272 lower-grade glioma patients and extracted 431 radiomic features, with accuracies of 80.0% (AUC = 89.6%) and 70.7% (AUC = 76.3%) in the training and validation sets respectively. As for the comparison between our study and another radiogenomics study focused on MGMT (Korfiatis et al., 2016), 155 glioblastoma patients were collected and 18 texture features were extracted in the later study, with an AUC of 85%.
In the current study, the most predictive features of p53 status were selected based on the LASSO algorithm, and the biological correlations between these features and p53 status could also be revealed preliminarily. For example, it was reported that the microvessel count is higher in p53-mutated tumors than in p53-wildtype tumors (Guo et al., 2008), as the vascular endothelial growth factor is overexpressed in p53-mutated tumors (Riedel et al., 2000, Uchida et al., 1998). Differences in microvascularity could result in differences in signal on T2 weighted images due to differences in water content (Dang et al., 2015), which could explain why Maximum_6 and Median_6 are higher in p53-mutant tumors than those in p53-wildtype tumors. Further, our results indicate that Uniformity_4, a radiological indicator for the consistency of the image (Aerts et al., 2014), could serve as a predictive factor of the p53 status. As previously noted, the p53 mutation promotes tumor malignancy and heterogeneity (Gillet et al., 2014), thereby leading to the expressive discrepancy of Uniformity.
At present, the p53 status of lower-grade gliomas is mainly used for diagnosis and prognosis. IDH-mutant gliomas can be subdivided into diffuse astrocytic and oligodendroglial tumors based on the ATRX and p53 mutation status (Malzkorn and Reifenberger, 2016). Strong nuclear p53 positivity is frequently observed in IDH-mutant astrocytic gliomas, but almost never detected in IDH-mutant and 1p/19q-codeleted oligodendrogliomas (Ceccarelli et al., 2016). Regarding the prognostic significance of the p53 status, a previous study reported that patients with wild-type p53 survive longer than those with mutant p53 in triple-negative (i.e., 1p/19q codeletion, IDH mutation, and TERT promoter mutation) lower-grade gliomas (Chan et al., 2016).
To develop the p53-related radiomic signature, a LASSO logistic regression model was used to reduce the 431 candidate radiological features to a set of 15 features that are predictive of the p53 mutation status. LASSO avoids overfitting and can be used to analyze multiple radiological features from a relatively small sample size (Gui and Li, 2005, Hepp et al., 2016). In addition, the SVM is an effective and robust machine-learning approach (Han and Jiang, 2014) that has been broadly applied within the field of medicine. The SVM has been used to differentiate benign and malignant pulmonary nodules (Zhu et al., 2010), predict grades (Zacharaki et al., 2009, Zollner et al., 2012) and survival (Emblem et al., 2014) of gliomas, and diagnose thyroid cancer (Gopinath and Shanthi, 2013). The LASSO and SVM matched well in our study, achieving a favorable prediction of the p53 mutation status in lower-grade gliomas.
A limitation of our study is that our machine-learning approach relied on respectively collected single-institution data. Therefore, our results should be evaluated further in a prospective study. Moreover, limited by the retrospectively collected data, how different imaging protocols and field intensities (1.5 T and 3 T) affect radiomic features have not been revealed in the present study. However, many studies have already demonstrated the robustness of radiomics feature extraction in terms of repeatability and reproducibility in test/re-test settings (Aerts et al., 2014, Fried et al., 2014, Parmar et al., 2015). In addition, we did not thoroughly reveal the biological process behind each selected radiological feature. Further radiogenomics analysis and experiments are necessary to solve this problem and further our understanding of the disease. Lastly, multi-model radiological data should be integrated into our model to improve prediction of the p53 status.
5. Conclusion
Here, we found a correlation between p53 phenotype and radiomic features in lower-grade gliomas. Using LASSO regression and SVM, we established a radiomic signature to achieve non-invasive and efficient prediction of the p53 status in gliomas.
The following are the supplementary data related to this article.
All quantitative radiological features and their corresponding mean AUCs after 10-fold cross validation in the training set.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Acknowledgments
Acknowledgment
This study was supported by the National Natural Science Foundation of China (No. 81601452), Beijing Natural Science Foundation (No. 7174295), the National Key Research and Development Plan (No. 2016YFC0902500), Capital Medical Development Research Fund (2016-1-1072), Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (ZYLX201708).
Contributor Information
Tao Jiang, Email: taojiang1964@163.com.
Xing Liu, Email: 15846591696@126.com.
Yinyan Wang, Email: tiantanyinyan@126.com.
References
- Aerts H.J., Velazquez E.R., Leijenaar R.T. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando K., Oki E., Saeki H. Discrimination of p53 immunohistochemistry-positive tumors by its staining pattern in gastric cancer. Cancer Med. 2015;4:75–83. doi: 10.1002/cam4.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appin C.L., Brat D.J. Biomarker-driven diagnosis of diffuse gliomas. Mol. Asp. Med. 2015;45:87–96. doi: 10.1016/j.mam.2015.05.002. [DOI] [PubMed] [Google Scholar]
- Ceccarelli M., Barthel F.P., Malta T.M. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaddad A., Tanougast C. Extracted magnetic resonance texture features discriminate between phenotypes and are associated with overall survival in glioblastoma multiforme patients. Med. Biol. Eng. Comput. 2016;54:1707–1718. doi: 10.1007/s11517-016-1461-5. [DOI] [PubMed] [Google Scholar]
- Chaddad A., Desrosiers C., Bouridane A. Multi texture analysis of colorectal cancer continuum using multispectral imagery. PLoS One. 2016;11 doi: 10.1371/journal.pone.0149893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaddad A., Desrosiers C., Toews M. Radiomic analysis of multi-contrast brain MRI for the prediction of survival in patients with glioblastoma multiforme. Conf Proc IEEE Eng Med Biol Soc. 2016;2016:4035–4038. doi: 10.1109/EMBC.2016.7591612. [DOI] [PubMed] [Google Scholar]
- Chan A.K., Mao Y., Ng H.K. TP53 and histone H3.3 mutations in triple-negative lower-grade gliomas. N. Engl. J. Med. 2016;375:2206–2208. doi: 10.1056/NEJMc1610144. [DOI] [PubMed] [Google Scholar]
- Dang M., Lysack J.T., Wu T. MRI texture analysis predicts p53 status in head and neck squamous cell carcinoma. AJNR Am. J. Neuroradiol. 2015;36:166–170. doi: 10.3174/ajnr.A4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellingson B.M., Malkin M.G., Rand S.D. Volumetric analysis of functional diffusion maps is a predictive imaging biomarker for cytotoxic and anti-angiogenic treatments in malignant gliomas. J. Neuro-Oncol. 2011;102:95–103. doi: 10.1007/s11060-010-0293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellingson B.M., Bendszus M., Boxerman J. Consensus recommendations for a standardized brain tumor imaging protocol in clinical trials. Neuro-Oncology. 2015;17:1188–1198. doi: 10.1093/neuonc/nov095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emblem K.E., Due-Tonnessen P., Hald J.K. Machine learning in preoperative glioma MRI: survival associations by perfusion-based support vector machine outperforms traditional MRI. J. Magn. Reson. Imaging. 2014;40:47–54. doi: 10.1002/jmri.24390. [DOI] [PubMed] [Google Scholar]
- Fried D.V., Tucker S.L., Zhou S. Prognostic value and reproducibility of pretreatment CT texture features in stage III non-small cell lung cancer. Int. J. Radiat. Oncol. Biol. Phys. 2014;90:834–842. doi: 10.1016/j.ijrobp.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet E., Alentorn A., Doukoure B. TP53 and p53 statuses and their clinical impact in diffuse low grade gliomas. J. Neuro-Oncol. 2014;118:131–139. doi: 10.1007/s11060-014-1407-4. [DOI] [PubMed] [Google Scholar]
- Gopinath B., Shanthi N. Support vector machine based diagnostic system for thyroid cancer using statistical texture features. Asian Pac. J. Cancer Prev. 2013;14:97–102. doi: 10.7314/apjcp.2013.14.1.97. [DOI] [PubMed] [Google Scholar]
- Gui J., Li H. Penalized Cox regression analysis in the high-dimensional and low-sample size settings, with applications to microarray gene expression data. Bioinformatics. 2005;21:3001–3008. doi: 10.1093/bioinformatics/bti422. [DOI] [PubMed] [Google Scholar]
- Guo R., Li Q., Meng L. P53 and vascular endothelial growth factor expressions are two important indices for prognosis in gastric carcinoma. West Indian Med J. 2008;57:2–6. [PubMed] [Google Scholar]
- Han H., Jiang X. Overcome support vector machine diagnosis overfitting. Cancer Informat. 2014;13:145–158. doi: 10.4137/CIN.S13875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepp T., Schmid M., Gefeller O. Approaches to regularized regression — a comparison between gradient boosting and the LASSO. Methods Inf. Med. 2016;55:422–430. doi: 10.3414/ME16-01-0033. [DOI] [PubMed] [Google Scholar]
- Huang Y.q., Liang C.h., He L. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J. Clin. Oncol. 2016;34:2157–2164. doi: 10.1200/JCO.2015.65.9128. [DOI] [PubMed] [Google Scholar]
- Huang M.W., Chen C.W., Lin W.C. SVM and SVM ensembles in breast cancer prediction. PLoS One. 2017;12 doi: 10.1371/journal.pone.0161501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M., Sakai M., Arita H. Introduction of high throughput magnetic resonance T2-weighted image texture analysis for WHO grade 2 and 3 gliomas. PLoS One. 2016;11 doi: 10.1371/journal.pone.0164268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korfiatis P., Kline T.L., Coufalova L. MRI texture features as biomarkers to predict MGMT methylation status in glioblastomas. Med. Phys. 2016;43:2835. doi: 10.1118/1.4948668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamaru K.K., Saboo S.S., Aghayev A. CT pulmonary angiography-based scoring system to predict the prognosis of acute pulmonary embolism. J Cardiovasc Comput Tomogr. 2016;10:473–479. doi: 10.1016/j.jcct.2016.08.007. [DOI] [PubMed] [Google Scholar]
- Levidou G., El-Habr E., Saetta A.A. P53 immunoexpression as a prognostic marker for human astrocytomas: a meta-analysis and review of the literature. J. Neuro-Oncol. 2010;100:363–371. doi: 10.1007/s11060-010-0204-y. [DOI] [PubMed] [Google Scholar]
- Liu C., Zhang H., Pan Y. Towards MIB-1 and p53 detection in glioma magnetic resonance image: a novel computational image analysis method. Phys. Med. Biol. 2012;57:8393–8404. doi: 10.1088/0031-9155/57/24/8393. [DOI] [PubMed] [Google Scholar]
- Macyszyn L., Akbari H., Pisapia J.M. Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro-Oncology. 2016;18:417–425. doi: 10.1093/neuonc/nov127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malzkorn B., Reifenberger G. Practical implications of integrated glioma classification according to the World Health Organization classification of tumors of the central nervous system 2016. Curr. Opin. Oncol. 2016;28:494–501. doi: 10.1097/CCO.0000000000000327. [DOI] [PubMed] [Google Scholar]
- Muller P.A., Vousden K.H. p53 mutations in cancer. Nat. Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- Mut M., Turba U.C., Botella A.C. Neuroimaging characteristics in subgroup of GBMs with p53 overexpression. J. Neuroimaging. 2007;17:168–174. doi: 10.1111/j.1552-6569.2007.00112.x. [DOI] [PubMed] [Google Scholar]
- Parmar C., Leijenaar R.T., Grossmann P. Radiomic feature clusters and prognostic signatures specific for lung and head & neck cancer. Sci Rep. 2015;5 doi: 10.1038/srep11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard D., Kaloshi G., Amiel-Benouaich A. Dynamic history of low-grade gliomas before and after temozolomide treatment. Ann. Neurol. 2007;61:484–490. doi: 10.1002/ana.21125. [DOI] [PubMed] [Google Scholar]
- Riedel F., Gotte K., Schwalb J. Vascular endothelial growth factor expression correlates with p53 mutation and angiogenesis in squamous cell carcinoma of the head and neck. Acta Otolaryngol. 2000;120:105–111. doi: 10.1080/00016480060203334. [DOI] [PubMed] [Google Scholar]
- Sarkar C., Karak A.K., Nath N. Apoptosis and proliferation: correlation with p53 in astrocytic tumours. J. Neuro-Oncol. 2005;73:93–100. doi: 10.1007/s11060-004-3015-1. [DOI] [PubMed] [Google Scholar]
- Sauerbrei W., Royston P., Binder H. Selection of important variables and determination of functional form for continuous predictors in multivariable model building. Stat. Med. 2007;26:5512–5528. doi: 10.1002/sim.3148. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Aoki K., Chiba K. Mutational landscape and clonal architecture in grade II and III gliomas. Nat. Genet. 2015;47:458–468. doi: 10.1038/ng.3273. [DOI] [PubMed] [Google Scholar]
- Uchida S., Shimada Y., Watanabe G. In oesophageal squamous cell carcinoma vascular endothelial growth factor is associated with p53 mutation, advanced stage and poor prognosis. Br. J. Cancer. 1998;77:1704–1709. doi: 10.1038/bjc.1998.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez M.M., Hu C., Roe D.J. Least absolute shrinkage and selection operator type methods for the identification of serum biomarkers of overweight and obesity: simulation and application. BMC Med. Res. Methodol. 2016;16:154. doi: 10.1186/s12874-016-0254-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Fan X., Li H. Tumor border sharpness correlates with HLA-G expression in low-grade gliomas. J. Neuroimmunol. 2015;282:1–6. doi: 10.1016/j.jneuroim.2015.02.013. [DOI] [PubMed] [Google Scholar]
- Wang Y.Y., Zhang T., Li S.W. Mapping p53 mutations in low-grade glioma: a voxel-based neuroimaging analysis. AJNR Am. J. Neuroradiol. 2015;36:70–76. doi: 10.3174/ajnr.A4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharaki E.I., Wang S., Chawla S. Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn. Reson. Med. 2009;62:1609–1618. doi: 10.1002/mrm.22147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.M., Brat D.J. Genomic profiling of lower-grade gliomas uncovers cohesive disease groups: implications for diagnosis and treatment. Chin J Cancer. 2016;35:12. doi: 10.1186/s40880-015-0071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Wang Y., Fan X. Anatomical localization of p53 mutated tumors: a radiographic study of human glioblastomas. J. Neurol. Sci. 2014;346:94–98. doi: 10.1016/j.jns.2014.07.066. [DOI] [PubMed] [Google Scholar]
- Zhou H., Vallieres M., Bai H.X. MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro-Oncology. 2017;19:862–870. doi: 10.1093/neuonc/now256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Tan Y., Hua Y. Feature selection and performance evaluation of support vector machine (SVM)-based classifier for differentiating benign and malignant pulmonary nodules by computed tomography. J. Digit. Imaging. 2010;23:51–65. doi: 10.1007/s10278-009-9185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollner F.G., Emblem K.E., Schad L.R. SVM-based glioma grading: optimization by feature reduction analysis. Z. Med. Phys. 2012;22:205–214. doi: 10.1016/j.zemedi.2012.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All quantitative radiological features and their corresponding mean AUCs after 10-fold cross validation in the training set.