Abstract
Purpose
Children with Hirschsprung disease (HD) who have a history of enterocolitis (HAEC) have a shift in colonic microbiota, many of which are necessary for short chain fatty acid (SCFA) production. As SCFAs play a critical role in colonic mucosal preservation, we hypothesized that fecal SCFA composition is altered in children with HAEC.
Methods
A multicenter study enrolled 18 HD children, abstracting for history of feeding, antibiotic/probiotic use, and enterocolitis symptoms. HAEC status was determined per Pastor et al. criteria (12). Fresh feces were collected for microbial community analysis via 16S sequencing as well as SCFA analysis by gas chromatography–mass spectrometry.
Results
Nine patients had a history of HAEC, and nine had never had HAEC. Fecal samples from HAEC children showed a 4-fold decline in total SCFA concentration vs. non-HAEC HD patients. We then compared the relative composition of individual SCFAs and found reduced acetate and increased butyrate in HAEC children. Finally, we measured relative abundance of SCFA-producing fecal microbiota. Interestingly, 10 of 12 butyrate-producing genera as well as 3 of 4 acetate-producing genera demonstrated multi-fold expansion.
Conclusion
Children with HAEC history have reduced fecal SCFAs and altered SCFA profile. These findings suggest a complex interplay between the colonic metabolome and changes in microbiota, which may influence the pathogenesis of HAEC.
Keywords: Hirschsprung disease, Hirschsprung-associated enterocolitis, Aganglionosis, Pediatric, Short chain fatty acids, Microbiome
1. Background
Hirschsprung-associated enterocolitis (HAEC) is a life-threatening complication of congenital aganglionic megacolon, or Hirschsprung disease (HD). While surgical pull-through achieves satisfactory restoration of functional intestinal continuity in most patients, up to 40% of children experience at least one episode of HAEC after surgery [1]. Despite being the primary cause of morbidity for children with HD, the pathophysiology of HAEC is poorly understood. Several contributing factors have been identified, including dysfunctional host immunity, diminished epithelial barrier function, and altered gut microbiota, although no unifying causative agent has been identified [2].
A microbial etiology of HAEC has been postulated since initial reports of elevated Clostridium difficile toxin titers in children with HAEC [3], although carriage rates have since proven highly variable [4]. Analysis of changes in the gut microbiome using molecular microbiological techniques has shown decreased colonization of Bifidobacterium and Lactobacillus in children with HD who developed HAEC [5], while genomic approaches have found increased bacterial population diversity during HAEC episodes in children with HD [6], yet modest differences in children with a history of HAEC [7]. Similar findings have been noted in animal models of HD using neural crest cell specific endothelin receptor B (Ednrb) knockout mice [8]. At the genus level, Ednrb knockout mice demonstrated decreased Lactobacillus and increased Bacteroides and Clostridium prior to enterocolitis. These findings suggest that disequilibriumin the gutmicrobiome – or dysbiosis – may result in an altered microbial ecosystem that leads to HAEC development.
The mechanisms by which altered colonic microbiota relate to the development of HAEC is not known. One important physiologic role of gut bacteria is the production of short chain fatty acids (SCFAs). Complex oligosaccharides and other organic, indigestible fiber matter not absorbed in the upper intestinal tract are fermented by the anaerobic microbial community of the colon, producing SCFAs (primarily acetate, propionate, and butyrate) and gas [9]. These SCFAs, of which butyrate is the best-studied, play a key role in maintaining gut homeostasis and epithelial integrity [10]. Butyrate serves as a principal energy source for colonocytes, regulates host gene expression via histone deacetylase inhibition [11]. This latter role has led to both an increase in IL-10 production and an anti-inflammatory role by inhibiting NF-ĸB signaling [12]. We hypothesized that children with a history of HAEC would have altered fecal SCFA composition, as well as disequilibrium of SCFA-producing microbiota.
This study was designed to evaluate the fecal SCFA makeup of patients who had completed definitive surgical treatment for HD. We compared 9 children with a history of at least one episode of HAEC to 9 children with no episodes of HAEC. We first performed gas chromatography–mass spectrometry (GC-MS) based quantification of fecal SCFA contents, and then compared this to 16S-based microbiota analysis of the same samples to elucidate the relationship between an altered microbiome and SCFA production.
2. Methods
2.1. Patient selection
This was a multi-institution study of children younger than 18 years of age who had completed definitive pull-through surgical treatment for a histopathological diagnosis of HD. Twenty children were enrolled by four member institutions of the HAEC Collaborative Research Group (HCRG): Cedars-Sinai Medical Center (CSMC), Los Angeles, CA; Astrid Lindgren Children's Hospital, Karolinska University Hospital, Stockholm, Sweden; Children's Hospital Los Angeles, Los Angeles, CA; Children's Hospital of Oakland, Oakland, CA. Of these, 10 children had a history of HD without documented enterocolitis, and 10 had at least one episode of HAEC as defined by Pastor et al. criteria [13]. Two of these 20 children were excluded from analysis owing to the presence of a diverting ileostomy and active HAEC at the time of stool collection, respectively. Thus, a total of 9 HD and 9 HAEC samples were analyzed. Medical records were reviewed and parent interview was performed using standardized questionnaires. Data collected included demographics, medical/surgical history, diet in the first year of life, medications including antibiotics, probiotic use, and enterocolitis symptoms. This study was approved by the CSMC institutional review board (IRB no. CR00008054) as a multicenter study, as well as individual approval by all participating sites, and the University of Michigan IRB (HUM00079878). Stool was collected within one week of enrollment and immediately snap-frozen at −80 °C in air-tight containers to prevent the loss of volatile SCFAs. Frozen sample aliquots were shipped to the University of Michigan for SCFA analysis and to CSMC for bacterial DNA isolation.
2.2. Fecal SCFA analysis
Sample extraction was performed using aqueous extraction solvent containing 3% 1 M HCl (v/v) and isotope-labeled internal standards (d7-buytric acid and d11-hexanoic acid). Samples were then homogenized and centrifuged. Supernatants were transferred to new Eppendorf tubes for extraction by diethyl ether. After layer separation, the upper layer was transferred to an autosampler vial for GC-MS analysis. GC (Agilent 6890, Wilmington, DE) separation was performed using a ZB-Wax plus column, 0.25 µm × 0.25 mm × 30 m (Phenomenex, Torrance, CA). A single quadrupole mass spectrometer (Agilent, 5973 inert MSD) was used to identify and quantify SCFAs using Agilent Masshunter software, version B.06 [14]. Absolute quantities of SCFAs were normalized to sample mass.
2.3. Bacterial DNA isolation and amplicon preparation
Fecal samples were suspended in 50 mM Tris buffer (pH 7.5) containing 1 mM EDTA, 0.2% β-mercaptoethanol (Sigma) and 1000 U/ml of lyticase (Sigma). The mix was incubated at 37 °C for 30 min, and DNA was isolated using QIAamp DNA Stool Mini Kit (Qiagen). Bacterial 16S rRNA gene amplicons spanning variable regions 1–4 were generated in 20 µL PCR reactions using 20 ng of fecal DNA with 25 cycles using high-fidelity Phusion Polymerase (New England Biolabs, Beverly, MA) at 52.7 °C annealing using with degenerate 8F (AGAGTTTGATCM TGGCTCAG) and R357 (CTGCTGCCTYCCGTA) primers. All PCR reactions were purified using Agencourt AmPure Magnetic Beads (Beckman), resuspended in 20 µL of nuclease-free water and quantified using a Qubit fluorometer (Invitrogen, Carlsbad, CA).
2.4. 16S sequencing
Paired-end adapters with unique indexes were ligated to 100 ng of 16S amplicons and used to generate Ion Torrent sequencing libraries using the Ion Xpress Library Kit (Life Technologies, Carlsbad, CA). Library enrichment was performed with 10 cycles of PCR and purified using Agencourt Ampure Magnetic Beads (Beckman). All libraries were subjected to quality control using qPCR, DNA 1000 Bioanalyzer (Agilent), and Qubit (Life Technologies, Carlsbad, CA). Pooled libraries were assayed on Agilent Bioanalyzer (Santa Clara, CA) to check final sizing and KAPA Biosciences qPCR for quantitation. 16S samples were multiplexed and sequenced on the Ion Torrent PGM on a 318 chip with 400 bp chemistry. 250 single-end sequencing-by-synthesis was performed using the MiSeq Illumina sequencer (Illumina, San Diego, CA). Torrent reads shorter than 200 bp, or not containing the designed 16S primers (>2 nt mismatches) were discarded. 300 bp sequences of remaining high-quality reads were aligned to the Greengenes reference database (February 2011 release) using BLAST v2.2.22 in QIIME v1.5 wrapper [15] with an identity percentage ≥97% to select the operational taxonomic units (OTUs). Taxonomy for each sequence was assigned using the Ribosomal Database Project (RDP) classifier v2.2.
2.5. Statistical analysis
Statistical analysis was performed using GraphPad Prism 5 (GraphPad Software, Inc., San Diego, CA). Comparison between two groups used the two-tailed, unpaired Student's T-test. All results are expressed as mean ± standard deviation unless otherwise specified. A p value of <0.05 was considered significant.
3. Results
3.1. Patient characteristics
The two groups (HD and HAEC) each included 9 patients, with an equal distribution of 8 males and 1 female (Table 1). Median age was 2.7 years (3 months to 8 years) for all children, 2.3 years for the HAEC group, and 3.5 years for the HD group (p = 0.40). There were no significant differences in length of aganglionosis, non-HAEC complications, early feeding type, or probiotics received. While no children in the HD group received antibiotics within 2 months of stool collection, three patients in the HAEC group had received antibiotics (two for prior HAEC treatment, and one for sickle cell prophylaxis). Trisomy 21 was present in one patient in the HD group and two patients in the HAEC group.
Table 1.
Patient demographic characteristics.
| Demographics | HD | HAEC |
|---|---|---|
| N | 9 | 9 |
| Male | 8 | 8 |
| Median age (years) | 2.3 | 3.5 |
| Trisomy 21 | 1 | 2 |
| HD characteristics | ||
| Rectosigmoid transition zone | 8 | 8 |
| Postop complicationsa | 0 | 0 |
| Dietb | ||
| Breast milk | 5 | 5 |
| Formula | 7 | 7 |
| Medications | ||
| Probioticsc | 4 | 3 |
| Antibioticsd | 0 | 3 |
Complications occurring within 30 days of pull-through operation, not including HAEC.
Diet in the first year of life.
Probiotics included Lactobacillus spp. and Bifidobacterium spp.
Received within 2 months prior to stool collection.
3.2. Fecal SCFA composition
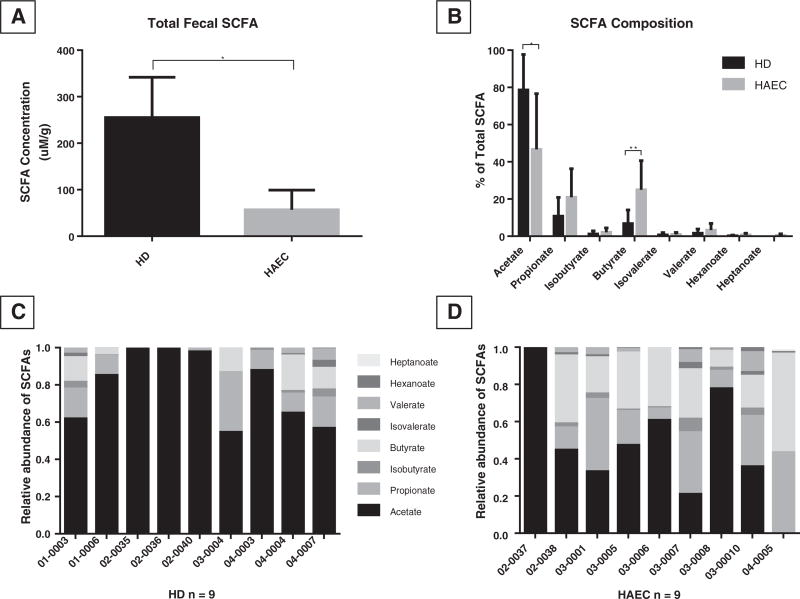
Total fecal SCFA composition of children with a history of HAEC was 4-fold lower than that of HD patients without a history of HAEC (56.98 ± 42.30 mM/g stool vs. 255.20 ± 259.70 mM/g; p = 0.038; Fig. 1A). The composition of individual SCFAs was then secondarily examined and was markedly altered in HAEC children. The concentration of each SCFA detected by GC/MS in each group is shown in Table 2. Fecal samples from the HAEC group had a significantly lower concentration of acetate (34.04 ± 40.40 µM/g vs. 224.59 ± 266.12 µM/g; p = 0.049) compared to the HD group, without a significant change in the absolute concentration of other detected SCFAs. When comparing pooled SCFA compositions (expressed as % of total SCFAs) between the two groups, a significant reduction in the proportion of acetate (46.70 ± 29.89% vs. 78.66 ± 19.08%; p=0.016) and an increase in the proportion of butyrate (24.99 ± 15.64% vs. 6.75 ± 7.40%; p=0.006; Fig. 1B) was found in samples from the HAEC group versus the HD group. While individual SCFA composition varied within the HD group, a preponderance of acetate was present, followed by propionate and butyrate (Fig. 1C). In children with a history of HAEC, however, the fecal SCFA profile became markedly more varied between individuals (Fig. 1D), reflecting a loss of acetate and increased proportion of other SCFAs.
Fig. 1.
(A) Fecal SCFA concentration was significantly reduced in HAEC children versus those with HD without HAEC. (B) Individual SCFAs expressed as % of total SCFAs from HD children with a history of HAEC and those with no history of HAEC. (C) Histograms demonstrating the fecal SCFA composition of individual subjects with HD and HAEC (D). Individual subject numbers are labeled on the X axis and expressed as relative SCFA abundance per each subject. *p < 0.05, *p < 0.01.
Table 2.
SCFA concentration in feces (µM/g).
| HD (mean ± SD; n = 9) | HAEC (mean ± SD; n = 9) | p value | |
|---|---|---|---|
| SCFAs (all) | 255.20 ± 259.70 | 56.98 ± 42.30 | 0.038* |
| Acetate | 224.59 ± 266.12 | 34.04 ± 40.40 | 0.049* |
| Propionate | 16.93 ± 16.07 | 7.54 ± 5.81 | 0.119 |
| Butyrate | 9.35 ± 8.33 | 11.98 ± 12.82 | 0.613 |
| Isobutyrate | 1.35 ± 1.76 | 0.99 ± 0.98 | 0.605 |
| Isovalerate | 0.77 ± 1.20 | 0.45 ± 0.53 | 0.481 |
| Valerate | 1.90 ± 2.08 | 1.65 ± 2.03 | 0.800 |
| Hexanoate | 0.25 ± 0.32 | 0.30 ± 0.51 | 0.827 |
| Heptanoate | 0.01 ± 0.04 | 0.03 ± 0.05 | 0.359 |
SCFA = short chain fatty acid.
p < 0.05.
3.3. SCFA-producing bacterial analysis
A detailed analysis of the broad changes in the fecal microbiome using 16S analyses has been reported [7]; however, a subanalysis of this dataset was used in the present paper to examine the anaerobic microbiota which produce fecal SCFAs. Therefore, we compared the populations of SCFA-producing bacterial strains between HAEC and HD group fecal samples. As the full fecal microbiome was dominated by non-SCFA-producing bacteria, we identified genera that are known to have butyrogenic [16] and acetogenic [17] strains. While no genus-level changes reached statistical significance (Table 3), 10 out of 12 butyrogenic genera, identified by 16S sequencing, demonstrated elevated OTUs (operational taxonomic units) with a mean increase of 640% in the HAEC group compared with the HD group (Fig. 2A). This mirrored the relative increase in butyrate found on SCFA analysis of HAEC samples. Interestingly, despite a decrease in the proportion of acetate in HAEC samples, 3 of 4 acetogenic genera had increased OTUs in the HAEC group, with a mean increase of 1710% (Fig. 2B).
Table 3.
SCFA-producing bacterial genera in feces.
| HD (meana ± SD; n = 9) |
HAEC (mean ± SD; n = 9) |
p value | |
|---|---|---|---|
| Butyrogens (all) | 4.9% ± 6.4% | 4.2% ± 3.6% | 0.783 |
| Roseburia | 4.0% ± 6.4% | 1.5% ± 1.9% | 0.282 |
| Odoribacter | 0.4% ± 0.9% | 1.0% ± 1.9% | 0.450 |
| Faecalibacterium | 0.2% ± 0.2% | 0.3% ± 0.5% | 0.499 |
| Eubacterium | 0.08% ± 0.2% | 0.4% ± 0.7% | 0.261 |
| Subdoligranulum | 0.05% ± 0.06% | 0.3% ± 0.6% | 0.221 |
| Peptoniphilus | 0.04% ± 0.1% | 0.001% ± 0.002% | 0.337 |
| Coprococcus | 0.03% ± 0.05% | 0.2% ± 0.3% | 0.135 |
| Fusobacterium | 0.01% ± 0.02% | 0.2% ± 0.7% | 0.354 |
| Porphyromonas | 0.01% ± 0.03% | 0.02% ± 0.05% | 0.750 |
| Clostridium | 0.008% ± 0.02% | 0.2% ± 0.5% | 0.303 |
| Anaerotruncus | 0.007% ± 0.01% | 0.01% ± 0.03% | 0.619 |
| Megasphaera | 0.002% ± 0.01% | 0.04% ± 0.1% | 0.337 |
| Acetogens (all) | 0.1% ± 0.2% | 0.7% ± 0.9% | 0.065 |
| Eubacterium | 0.08% ± 0.2% | 0.3% ± 0.7% | 0.261 |
| Clostridium | 0.008% ± 0.02% | 0.2% ± 0.5% | 0.303 |
| Ruminococcus | 0.003% ± 0.006% | 0.1% ± 0.2% | 0.053 |
| Syntrophococcus | 0.001% ± 0.003% | <0.0001% | 0.151 |
Mean % of total operational taxonomic units (OTUs).
Fig. 2.
Genus-level identification of fecal microbiota via 16s rRNA sequencing. Percent change in mean OTUs of (A) butyrogenic bacteria and (B) acetogenic bacteria in fecal samples from children with a history of HAEC versus HD children without HAEC.
4. Discussion
HAEC is a highly morbid, though poorly understood complication of HD. While a role for altered colonic microbiota has been suggested by prior studies [5–8,18], the mechanism by which changes in bacterial community characteristics might impact susceptibility to or influence the development of HAEC is not clear. In this study, we found an over 4-fold reduction in total fecal SCFA content in children with a history of HAEC compared to those who never had HAEC. This may suggest that the colonic environment associated with HAEC is associated with a microbial community that has reduced capacity to produce SCFAs. Given that SCFAs are critical nutrients in the maintenance of healthy colonic mucosa, the loss of SCFAs may adversely affect enterocyte homeostasis thereby contributing to HAEC development.
Specifically, acetate was found in this study to be the most markedly reduced among children with HAEC, although it has been less vigorously studied in the context of colitis pathophysiology. The SCFA butyrate is also a major source of energy for colonic epithelial cells and has both immunomodulatory and anti-inflammatory properties [19–21]. Interestingly, a reduction in butyrogenic bacteria has been shown in infectious and active inflammatory colitis versus healthy subjects, suggesting a role of intestinal dysbiosis and butyrate reduction in colitis development [22,23]. Butyrate has also been shown to ameliorate the impairment of mucosal immunity seen with total parenteral nutrition [24]. The trophic effect of mixed SCFAs (including acetate) has been shown to exceed that of butyrate alone [25], it is likely that these other SCFAs play an important role in colonic mucosal integrity.
This study found a relative increase in the proportion of fecal butyrate in children with a history of HAEC, and that the net loss of fecal SCFAs found among these patients was primarily owing to a significant reduction in acetate. However, the absolute butyrate concentration was maintained, thereby making up a larger proportion of the remaining SCFA pool. The significant net loss of acetate may represent decreased acetate production via fermentation, or it may be the result of increased acetate absorption by the colonic mucosa versus increased acetate metabolism by colonic microbiota. Acetate is the most prevalent SCFA in the healthy colon [26]. Bacteria isolated from the human colon have been shown to utilize acetate for the production of butyrate [27]. This might provide an explanation for the reduction in acetate found in children with a history of HAEC. More metabolically active butyrogenic strains, as observed in our study, may have led to lower acetate levels in these patients, leading to a higher proportion of butyrate while decreasing the total concentration of SCFAs primarily via a reduction in remaining acetate.
While the etiology of the change in SCFA concentrations observed in this study is unknown, the relative increase in butyrate production may represent a compensatory mechanism to decrease the associated inflammatory state driven by other unknown factors. For example, epithelial inflammation may lead to elevated butyrate requirements by the colonic mucosa of children with a history of HAEC. This may drive the relative expansion of butyrogenic strains in these patients, which maintain butyrate concentrations, perhaps metabolizing acetate in this process. Future work to specifically examine the metabolic activity of the colonic microbiota and epithelium in children with HD might provide key insights into the pathogenesis of HAEC.
This is the first study to investigate SCFA changes in children with HD compared with those with a history of HAEC, Ward et al. performed a nuclear magnetic resonance (NMR)-based fecal metabolome analysis using a mouse model of HD [18]. This untargeted investigation found a significant reduction in formate, a minor SCFA product of bacterial fermentation, in endothelin receptor B knockout mice prior to onset of HAEC. However, no targeted SCFA metabolomic analysis was performed in this mouse study, and changes in other SCFAs were not characterized. We did not detect formate in the SCFA analysis which was not surprising, given that formate is not a major human fecal SCFA; although the parallel to the Ednrb-null mouse is noteworthy.
The types of SCFAs produced by bacterial fermentation are determined by the relative abundance of undigested carbohydrates, proteins, and amino acids in the colonic lumen [26]. We therefore used matched cohorts of patients with equal distributions of breast milk and formula feeding histories within the first year of life to minimize differences in nutrients on microbial community development. A more detailed nutrition assessment, quantifying caloric intake with protein:carbohydrate ratio at the time of sample collection was not performed, although this may have detected differences in oral intake between the two groups. Another variable which may impact SCFA production is gut motility – as shorter colonic transit has been shown to increase SCFA production [28]. While no patients had active HAEC at the time of sample collection, colonic transit time was not assessed in this study.
While probiotic prophylaxis has been shown to decrease the risk of other inflammatory diseases of the intestine such as necrotizing enterocolitis [29], such intervention has not been shown to reduce the risk of HAEC in children [30]. This study suggests that the altered microbiota seen with HAEC may mediate their influence on the colonic mucosa via dysfunctional SCFA metabolism. Future work will be required to better understand how the complex interplay between diet, colonic bacteria, and mucosal metabolism in the setting of HD leads to a unique SCFA profile in children with a history of HAEC. As butyrate enemas have shown promise in the treatment of colitis [26], a therapeutic role may exist for such therapy directed at normalization of the colonic SCFA profile in the prevention of HAEC, or perhaps by delivering substrate (nondigestible fibers) for increased butyrate production by this altered microbiome.
This study demonstrates a strong association between prior episodes of HAEC and an altered SCFA profile, however, it does not establish the initiating cause. It is possible that prior HAEC episodes lead to the altered microbiome and resultant SCFA changes, perhaps contributing to further episodes of HAEC. A prospective study of patients after surgical correction of HD without prior HAEC would address this question by comparing changes in the microbiome and SCFA profile between patients who do and do not later develop HAEC.
A limitation of this study is a lack of long-term antibiotic history, prior to the 2-month window before sample collection. Patients with a history of HAEC may have received several courses of antibiotics to treat prior HAEC episodes, which may have influenced the development of an altered colonic microbial community. Post-hoc analysis of SCFA profiles after exclusion of the three patients who had received antibiotics in the HAEC group demonstrated continued reduction in total SCFA content, as well as consistent changes in acetate and butyrate content. Another limitation of this study is the small sample size of each group. While this study is the first to date to investigate SCFA content and related microbiota in humans with HD, a more robustly powered study may have detected more significant changes in SCFA concentrations as well as genus-level microbial shifts. Additionally, this study evaluated fecal microbial changes as a proxy for direct measurement of colonic microbiota. Substantial variation has been reported between fecal and colonic mucosa-associated microbiomes [31]. Though more invasive specimen collection would be required for colonic microbiome and metabolome assessment, such an investigation might reveal more physiologically relevant data. Finally, bacterial 16S sequencing in this study did not allow identification beyond the genus level. The identification of specific species with known capacity to produce SCFAs was therefore not performed, but rather the genera including these strains were identified. Future work might be directed at complete genome sequencing to confirm the ability of specific strains to produce butyrate, acetate, and other SCFAs.
In summary, children with HAEC history were found to have markedly reduced fecal SCFAs, and an altered SCFA profile. These findings suggest a complex interplay between altered local environment and changes in intestinal microbiota, which may influence the pathogenesis of HAEC.
Acknowledgments
This work was supported by NIH grant DK090281 (P.K.F.) and by the National Center for Advancing Translational Sciences, grant UL1TR000124. This work used Core Services supported by NIH grant DK097153 to the University of Michigan.
Appendix A
Discussions
Presented by Farokh R. Demehri, Ann Arbor, MI
UNIDENTIFIED SPEAKER This is really interesting. The thing that we worry about in the kids who have recurrent enterocolitis, we have always thought of it as a stasis problem. Which comes first? Do you have the altered microbiome because you have stasis, or do you have – which is the chicken and which is the egg here?
FAROKH DEMEHRI Right. That's a great question. I think this study is really a hypothesis-generating study because of these two groups there was only one patient in the enterocolitis group who demonstrated stricture and stasis and required a dilation. None of the others did, but you are right that is often thought to be a contributing factor. I think going forward we need to understand actually what is the motility of these patients and then what is the function of the bacteria that we are finding in these patients?
BRADWARNER (St. Louis, MO) My question sort of gets to the metabolome and you focus on basically on short-chain fatty acids, but there are obviously a lot of other things that the bacteria are metabolizing to include amino acids and other things in carbohydrate metabolism that may play a role as well. If you focus specifically on short-chain fatty acids, which is an obvious first start – that is the fuel for the colonocytes. Have you looked at these other factors?
FAROKH DEMEHRI Thank you. That is an excellent question. Actually when we had these samples we had to decide are we going to do a targeted metabolomic analysis or untargeted. We decided to start with targeted because as you mentioned this is a good first step, but what we hope to do is an untargeted analysis and that might generate all kinds of other questions about not just what these bacteria are able to produce but also what the interplay is between the epithelium and the bacteria. Thank you.
Footnotes
Conflicts of Interest: The authors declare no conflicts of interest.
Level of Evidence: III.
Author contributions
FR Demehri, Z Cheng: Conceptualization, sample processing, data interpretation, and manuscript preparation.
C Ruan, V Funari: Sample processing, data interpretation, and manuscript preparation.
T Wester, A Nordenskjöld, A Kawaguchi, TT Hui, AL Granström: Conceptualization, specimen collection.
PK Frykman, DH Teitelbaum: Conceptualization, data interpretation, and manuscript preparation.
References
- 1.Demehri FR, Halaweish IF, Coran AG, et al. Hirschsprung-associated enterocolitis: pathogenesis, treatment and prevention. Pediatr Surg Int. 2013;29(9):873–81. doi: 10.1007/s00383-013-3353-1. [DOI] [PubMed] [Google Scholar]
- 2.Frykman PK, Short SS. Hirschsprung-associated enterocolitis: prevention and therapy. Semin Pediatr Surg. 2012;21(4):328–35. doi: 10.1053/j.sempedsurg.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas DF, Fernie DS, Malone M, et al. Association between Clostridium difficile and enterocolitis in Hirschsprung's disease. Lancet. 1982;1(8263):78–9. doi: 10.1016/s0140-6736(82)90216-1. [DOI] [PubMed] [Google Scholar]
- 4.Wilson-Storey D, Scobie WG, McGenity KG. Microbiological studies of the enterocolitis of Hirschsprung's disease. Arch Dis Child. 1990;65(12):1338–9. doi: 10.1136/adc.65.12.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen DH, Shi CR, Chen JJ, et al. Detection of intestinal bifidobacteria and lactobacilli in patients with Hirschsprung's disease associated enterocolitis. World J Pediatr. 2009;5(3):201–5. doi: 10.1007/s12519-009-0038-x. [DOI] [PubMed] [Google Scholar]
- 6.Yan Z, Poroyko V, Gu S, et al. Characterization of the intestinal microbiome of Hirschsprung's disease with and without enterocolitis. Biochem Biophys Res Commun. 2014;445(2):269–74. doi: 10.1016/j.bbrc.2014.01.104. [DOI] [PubMed] [Google Scholar]
- 7.Frykman PK, Nordenskjöld A, Kawaguchi A, et al. Characterization of bacterial and fungal microbiome in children with Hirschsprung disease with and without a history of enterocolitis: a multicenter study. PLoS One. 2015;10(4):e0124172. doi: 10.1371/journal.pone.0124172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pierre JF, Barlow-Anacker AJ, Erickson CS, et al. Intestinal dysbiosis and bacterial enteroinvasion in a murine model of Hirschsprung's disease. J Pediatr Surg. 2014;49(8):1242–51. doi: 10.1016/j.jpedsurg.2014.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christl SU, Murgatroyd PR, Gibson GR, et al. Production, metabolism, and excretion of hydrogen in the large intestine. Gastroenterology. 1992;102(4 Pt 1):1269–77. [PubMed] [Google Scholar]
- 10.Pryde SE, Duncan SH, Hold GL, et al. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett. 2002;217(2):133–9. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- 11.Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr. 2003;133(7 Suppl):2485S–93S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- 12.Hamer HM, Jonkers D, Venema K, et al. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27(2):104–19. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 13.Pastor AC, Osman F, Teitelbaum DH, et al. Development of a standardized definition for Hirschsprung's-associated enterocolitis: a Delphi analysis. J Pediatr Surg. 2009;44(1):251–6. doi: 10.1016/j.jpedsurg.2008.10.052. [DOI] [PubMed] [Google Scholar]
- 14.Pogribna M, Freeman JP, Paine D, et al. Effect of Aloe vera whole leaf extract on short chain fatty acids production by Bacteroides fragilis, Bifidobacterium infantis and Eubacterium limosum. Lett Appl Microbiol. 2008;46(5):575–80. doi: 10.1111/j.1472-765X.2008.02346.x. [DOI] [PubMed] [Google Scholar]
- 15.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio. 2014;5(2):e00889. doi: 10.1128/mBio.00889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drake HL, Gossner AS, Daniel SL. Old acetogens, new light. Ann N Y Acad Sci. 2008;1125:100–28. doi: 10.1196/annals.1419.016. [DOI] [PubMed] [Google Scholar]
- 18.Ward NL, Pieretti A, Dowd SE, et al. Intestinal aganglionosis is associated with early and sustained disruption of the colonic microbiome. Neurogastroenterol Motil. 2012;24(9):874–84. doi: 10.1111/j.1365-2982.2012.01937.x. [DOI] [PubMed] [Google Scholar]
- 19.Segain JP, Raingeard de la Bletiere D, Bourreille A, et al. Butyrate inhibits inflammatory responses through NFkappaB inhibition: implications for Crohn's disease. Gut. 2000;47(3):397–403. doi: 10.1136/gut.47.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klampfer L, Huang J, Sasazuki T, et al. Inhibition of interferon gamma signaling by the short chain fatty acid butyrate. Mol Cancer Res. 2003;1(11):855–62. [PubMed] [Google Scholar]
- 21.Chang PV, Hao L, Offermanns S, et al. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A. 2014;111(6):2247–52. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sokol H, Seksik P, Furet JP, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15(8):1183–9. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 23.Antharam VC, Li EC, Ishmael A, et al. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J Clin Microbiol. 2013;51(9):2884–92. doi: 10.1128/JCM.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murakoshi S, Fukatsu K, Omata J, et al. Effects of adding butyric acid to PN on gut-associated lymphoid tissue and mucosal immunoglobulin A levels. JPEN J Parenter Enteral Nutr. 2011;35(4):465–72. doi: 10.1177/0148607110387610. [DOI] [PubMed] [Google Scholar]
- 25.Drozdowski LA, Dixon WT, McBurney MI, et al. Short-chain fatty acids and total parenteral nutrition affect intestinal gene expression. JPEN J Parenter Enteral Nutr. 2002;26(3):145–50. doi: 10.1177/0148607102026003145. [DOI] [PubMed] [Google Scholar]
- 26.Wong JM, de Souza R, Kendall CW, et al. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40(3):235–43. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 27.Duncan SH, Barcenilla A, Stewart CS, et al. Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl Environ Microbiol. 2002;68(10):5186–90. doi: 10.1128/AEM.68.10.5186-5190.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macfarlane GT, Macfarlane S. Fermentation in the human large intestine: its physiologic consequences and the potential contribution of prebiotics. J Clin Gastroenterol. 2011;45(Suppl):S120–7. doi: 10.1097/MCG.0b013e31822fecfe. [DOI] [PubMed] [Google Scholar]
- 29.Wang Q, Dong J, Zhu Y. Probiotic supplement reduces risk of necrotizing enterocolitis and mortality in preterm very low-birth-weight infants: an updated meta-analysis of 20 randomized, controlled trials. J Pediatr Surg. 2012;47(1):241–8. doi: 10.1016/j.jpedsurg.2011.09.064. [DOI] [PubMed] [Google Scholar]
- 30.El-Sawaf M, Siddiqui S, Mahmoud M, et al. Probiotic prophylaxis after pullthrough for Hirschsprung disease to reduce incidence of enterocolitis: a prospective, randomized, double-blind, placebo-controlled, multicenter trial. J Pediatr Surg. 2013;48(1):111–7. doi: 10.1016/j.jpedsurg.2012.10.028. [DOI] [PubMed] [Google Scholar]
- 31.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]