Abstract
We describe the preparation, evaluation, and application of an S100A12 protein-conjugated solid support, hereafter the “A12-resin,” that can remove 99% of Zn(II) from complex biological solutions without significantly perturbing the concentrations of other metal ions. The A12-resin can be applied to selectively deplete Zn(II) from diverse tissue culture media and from other biological fluids, including human serum. To further demonstrate the utility of this approach, we investigated metabolic, transcriptomic, and metallomic responses of HEK293 cells cultured in medium depleted of Zn(II) using S100A12. The resulting data provide insight into how cells respond to acute Zn(II) deficiency. We expect that the A12-resin will facilitate interrogation of disrupted Zn(II) homeostasis in biological settings, uncovering novel roles for Zn(II) in biology.
Elucidating roles of Zn(II) in biology benefits from methods to image,1–3 quantify,4–5 and perturb6 both labile and bound7 Zn(II) in living cells. Fluorescent sensors have revealed that labile Zn(II) concentrations can be tightly controlled at sub-nM levels.8–9 Stimulation of cells with high concentrations of Zn(II) was instrumental in defining the metazoan response to Zn(II) overload.10 However, a major obstacle impeding progress in Zn(II) biology is the inability to selectively and efficiently deplete Zn(II) from complex biological media. One strategy is to nonspecifically remove metal ions using resin-supported chelators, such as Chelex® (Figures 1a and b, Table S1), iminodiacetate on a solid support, and then add back all metal ions except Zn(II).11 This strategy requires quantitation of metal ions before and after Chelex® treatment. Moreover, even careful metal repletion may not restore the metal ion speciation of untreated media.
Figure 1.
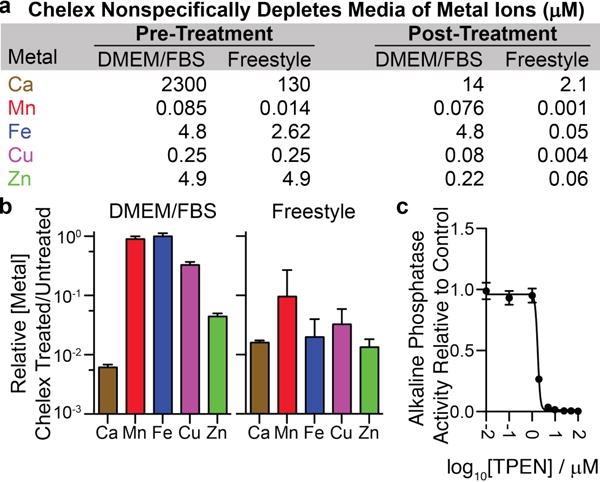
(a/b) Chelex® resin nonspecifically depletes cations from media (n=4, ±SEM). (c) TPEN treatment of alkaline phosphatase secreted from transfected HEK293T cells diminishes the activity of the enzyme (n=3, ±SEM)
Another approach used to study Zn(II) deficiency is to treat cells with a chelator such as N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN).12–13 However, this reagent has a high affinity for other d-block metal ions,14 and it can inhibit the activity of metalloproteins. For instance, TPEN strongly blocks the Zn(II)-dependent hydrolytic activity15 of tissue nonspecific alkaline phosphatase (Figure 1c). The risk of TPEN inhibiting metalloenzymes or affecting other metal-dependent processes precludes its application to cells as a means to effect Zn(II) deficiency. Moreover, incubation of cells with TPEN or any other small-molecule chelator is not equivalent to Zn(II) deficiency. Cells may be able to recover TPEN-complexed Zn(II) and/or TPEN itself may have unappreciated biological activities.16
A third strategy to study Zn(II) deficiency is to obtain a custom-made, chemically defined cell culture medium that lacks Zn(II).17 This approach is time-consuming, expensive, and only a subset of cells can be cultured in such media.18
None of the above approaches or related alternatives allow researchers to address the generic issue of Zn(II) deficiency in cells. A robust Zn(II) depletion method must (1) selectively deplete Zn(II) from diverse and complex biological media, (2) be easy to use, and (3) be cost-effective. Here we describe a protocol that meets these criteria, enabling precise modulation of Zn(II) content in biological media and facilitating the investigation of many aspects of biology.
Our approach was inspired by the existence of proteins that sequester nutrient metal ions from invading pathogens. Such proteins are important components of the mammalian innate immune system. Human S100A12 is one such protein that harbors two His3Asp sites that coordinate Zn(II) with sub-nM affinity.19–20 Moreover, S100A12 can deplete Zn(II) from microbial growth medium.20
We therefore wondered whether the selectivity of S100A12 for Zn(II) could facilitate the development of a Zn(II) depletion method meeting the above requirements. To test this possibility, we first evaluated whether recombinant S100A12 depletes Zn(II) from chemically-defined, protein-free Free-style™ mammalian cell culture medium. We incubated S100A12 (25 μM) with Freestyle™ medium for 4 h prior to filtering it through a 10-kDa molecular weight cutoff filter to remove the protein and protein-bound metal. ICP-MS measurements of the metal ion concentrations in untreated versus S100A12-treated media revealed selective depletion of >99% of total Zn from Freestyle™ medium (Figure 2a, Table S2). Chelex treatment, in contrast, removed multiple metal ions (Figure 1a).
Figure 2.
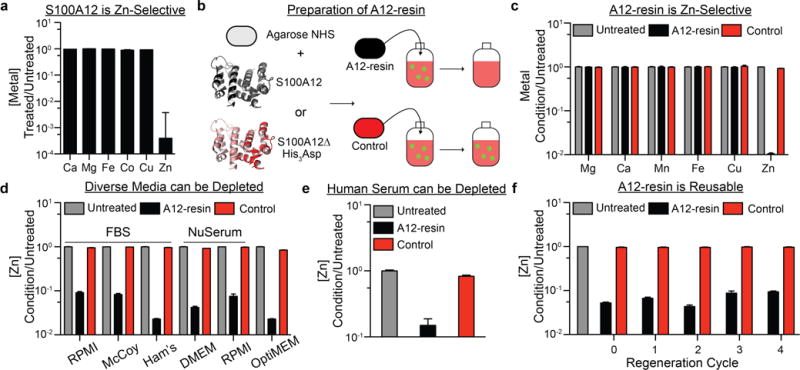
(a) ICP-MS analysis indicating that Freestyle™ medium is depleted of Zn(II) by direct addition of S100A12 followed by removal of Zn(II)-bound S100A12 with a 10-kDa molecular weight cutoff filter (n=3, ±SEM). (b) S100A12 or S100A12 ΔHis3Asp can be conjugated to agarose to afford two distinct resins. (c) A12-resin specifically depletes Zn(II) from DMEM/FBS, whereas the S100A12Δ His3Asp (control) resin does not affect the metal ion concentrations of the medium (n=3, ±SEM). (d) A12-resin depletes Zn(II) from an array of diverse cell culture medium formulations (n=4, ±SEM). (e) A12-resin selectively depletes human serum of zinc. (f) Regeneration of the A12-resin in 1.0 M acetic acid (pH=3.8) enables multiple cycles of resin reuse (n=4, ±SEM).
We next sought to deplete Zn(II) from more widely used media formulations that are not chemically defined and contain high concentrations of protein or fetal bovine serum (FBS). The method described above could not be used to selectively deplete Zn(II) from such complex media formulations because separation of Zn(II)-bound S100A12 from the treated media using a molecular weight cutoff filter would also deplete other high molecular weight biomolecules, thereby perturbing the composition of the media.
To solve this problem, we immobilized S100A12 on a solid support by incubating recombinant protein (5 mg/mL) with NHS-agarose for 2 h. As anticipated, the resulting A12-resin was capable of selectively removing Zn(II) from complex cell culture medium. Specifically, ICP-MS analysis indicated that treatment of the commonly employed Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% FBS with A12-resin, followed by removal of the resin by passage of treated medium through a 0.2 μm filter, selectively depleted 99% of Zn(II) from the medium (Figure 2c, Table S3). Considering that Zn(II) binding by S100A12 requires the association of two polypeptides,21 it is remarkable that covalent, nonspecific immobilization of S100A12 did not prevent Zn(II) binding. We confirmed by Western blot analysis of A12-resin-treated DMEM/FBS using an anti-S100A12 antibody that no detectable quantity of protein leached from the resin (Figure S1).
In addition to the experiment described above, we prepared a control resin composed of agarose-conjugated S100A12 ΔHis3Asp, a variant of S100A12 that lacks the His3Asp Zn(II)-binding sites. Treatment of DMEM/FBS with this A12Δ-modified resin did not significantly affect the concentration of Zn(II) or other metal ions (Figures 2b and c, Table S3).
To define the scope of Zn(II) depletion using our method, we tested the A12-resin against six additional complex media formulations, including RPMI/FBS, McCoy’s 5A/FBS, Ham’s F-12K/FBS, DMEM/NuSerum™, RPMI/Nu-Serum™, and OptiMEM™. A12-resin selectively depleted Zn(II) from all of these diverse cell culture media (Figure 2d, Table S4). We also found that the A12-resin could be applied to deplete Zn(II) from other complex, biologically relevant fluids. For example, the A12-resin depleted up to 85% of Zn(II) from human serum (Figure 2e, Table S5).
Not only does A12-resin remove Zn(II) from a range of biological media, it can also be regenerated. After depleting DMEM/FBS of Zn(II) with the A12-resin, we treated the recovered material with 1.0 M acetic acid (pH = 3.8) for 15 min at room temperature, and then washed the resin with PBS and DMEM to neutralize the pH of the resin suspension. Strikingly, the protein retained its Zn(II)-binding capacity after this harsh treatment, yielding a refreshed A12-resin that could remove Zn(II) from subsequent batches of medium. This recycling protocol was repeated four times with negligible loss of Zn(II) removal efficacy (Figure 2f, Table S6).
Having demonstrated the selectivity of the A12-resin, we next studied the response of cells to growth in Zn(II)-depleted and Zn(II)-repleted media, providing insight into the consequences of prolonged Zn(II) depletion. In particular, we characterized the metabolic, transcriptomic, and metallomic consequences of Zn(II) deficiency for HEK293 cells.
The metabolic activity of two HEK293-derived cell lines cultured in Zn(II)-depleted medium decreases significantly upon 3 d of culture compared to control. This defect was revealed by either a CellTiter-Glo® assay (Figure 3a; luminescent readout proportional to total ATP) or a resazurin assay (Figure 3b; fluorescent readout proportional to metabolic activity). In both cases, Zn(II) repletion rescues the observed metabolic defects.
Figure 3.
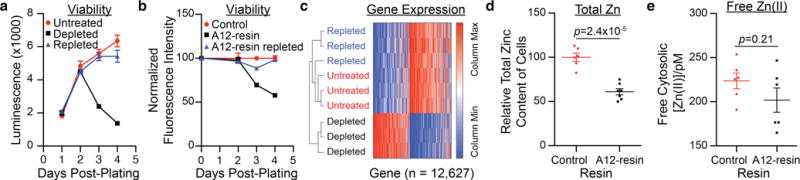
(a) HEK293 Freestyle cells cultured in a Zn(II)-depleted medium exhibit a loss of metabolic activity, as assessed by CellTiter-Glo® assay over 4 d of culture (n=3, ±SEM). (b) HEK293T cells cultured in a Zn(II)-depleted DMEM/FBS exhibit a loss of metabolic activity, as assessed by a resazurin assay over 4 d of culture (n=3, ± SEM). (c) Heat map from RNA Seq of HEK-293 Freestyle cells cultured in untreated, depleted, and repleted medium reveals the specificity of S100A12 medium depletion for Zn(II). (d) Total cellular Zn(II) content of HEK293T cultured in Zn(II)-depleted medium is diminished relative to cells cultured in Zn(II)-adequate medium (n≥6, ±SEM). (e) Culture of HEK293T cells in Zn(II) depleted medium does not affect intracellular free Zn(II) concentration (n=6, ±SEM).
Next, we interrogated the transcriptional response of HEK293 cells to Zn(II) deficiency. We first verified that HEK293 cells cultured for 1.5 d in Zn(II)-depleted medium have a diminished level of metallothionein mRNA, which is an established marker of Zn(II) starvation (Figures S2 and S3).22 Subsequently, we surveyed the transcriptome of HEK293 Freestyle cells cultured in untreated, S100A12-treated (Figure 2a), or Zn(II)-repleted medium using RNA-Seq after the same duration of culture. Hierarchical clustering of all expressed genes exposed two clades in the RNA-Seq data (Figure 3c). The first clade clusters transcriptomic information from HEK293 Freestyle cells cultured in Zn(II)-depleted Freestyle™ medium. The second clade clusters transcriptomic information from cells cultured in repleted and untreated medium. The co-clustering of the latter two conditions is consistent with highly Zn(II)-selective metal depletion by S100A12. Moreover, the extensive remodeling of the transcriptome after 1.5 d of culture in Zn(II)-depleted medium suggests a massive cellular response to zinc deprivation, despite the lack of a metabolic phenotype after 1.5 d (Figure 3a). Indeed, the expression of >75% of transcripts is significantly altered (padj < 0.01) in Zn(II)-depleted HEK293 Freestyle cells (Figure S4, see Supporting Information).
Finally, we assessed the metallomic consequences for HEK293T cells cultured in A12-resin-treated DMEM/FBS. We first examined how total cellular Zn(II) changed as a consequence of 1 d of growth in Zn(II)-depleted medium. We chose this time point because HEK293T cells double approximately once per day. Consistent with this growth rate and the fact that little extracellular Zn(II) was available for uptake, cells cultured in Zn(II)-depleted medium exhibit a decrease in total cellular levels of the ion compared to control cells by ~40% (Figure 3d). The total concentrations of other metals were unperturbed by culture in Zn(II)-depleted medium (Figure S5).
A 40% reduction in total cellular Zn(II) content might be expected to alter labile Zn(II) levels inside cells. Labile Zn(II) comprises a pool of zinc in cells for which roles in intra-23–24 and intercellular25 signaling and regulation have been hypothesized and demonstrated. Labile Zn(II) levels can be interrogated using a variety of protein-based sensors, including the BLZinCh-1 sensor,4 which has 160 pM affinity for Zn(II). Remarkably, the difference between labile Zn(II)-levels in cells grown for 24 h in A12-resin-treated medium,4 and control cells is not statistically significant, as measured using BLZinCh-1 (Figure 3e). Although further work is required to rigorously evaluate this observation, it suggests, intriguingly, that cells may have mechanisms to maintain labile Zn(II) concentrations even when Zn(II) is a limiting nutrient.
In conclusion, we report a method for perturbing Zn(II) levels in complex biological media. A12-resin is selective for Zn(II), is straightforward to use – requiring only incubation with biological medium followed by filtration – can be applied in diverse and complex biological media, and is recyclable. Phenotypic characterization of cells cultured in A12-resintreated medium supports the argument that the method is highly specific for Zn(II). Beyond studies of severe Zn(II) deficiency, we note that this method will facilitate precise titration of different concentrations of Zn(II) into medium, as well as enable enrichment of medium with a stable or radioisotope of Zn(II). In sum, this method provides researchers with an improved capacity to tune Zn(II) levels and interrogate mechanistic consequences of dysregulated Zn(II) homeostasis in complex biological settings.
Supplementary Material
Acknowledgments
This work was supported by the 56th Edward Mallinckrodt Jr. Foundation Faculty Scholar Award, a Pilot Grant from the MIT Center for Environmental Health Sciences supported by NIEHS P30-ES002109, and NIH grant R01-AR071443 (all to M.D.S.), NIH grant R01-GM065519 (S.J.L.), the NSF (CHE-1352132; E.M.N.), and NIH T32-EB019940 (to C.E.R.R). This work was also supported in part by the Koch Institute Support (core) Grant P30-CA14051 from the NCI.
Footnotes
Supporting Information is available free of charge on the ACS Publications website and includes complete experimental methods, data for all ICP-MS experiments (Tables S1–S6), supplementary figures, and differential expression analysis of RNA-Seq data. Raw RNA-Seq data is available on the Gene Expression Omnibus (GSE108923).
Notes
The authors have submitted a patent application on the A12-resin.
References
- 1.Que EL, Bleher R, Duncan FE, Kong BY, Gleber SC, Vogt S, Chen S, Garwin SA, Bayer AR, Dravid VP, Woodruff TK, O’Halloran TV. Nat Chem. 2015;7:130–9. doi: 10.1038/nchem.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walkup GK, Burdette SC, Lippard SJ, Tsien RY. J Am Chem Soc. 2000;122:5644–5645. doi: 10.1021/ja010059l. [DOI] [PubMed] [Google Scholar]
- 3.McRae R, Bagchi P, Sumalekshmy S, Fahrni CJ. Chem Rev. 2009;109:4780–827. doi: 10.1021/cr900223a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aper SJ, Dierickx P, Merkx M. ACS Chem Biol. 2016;11:2854–2864. doi: 10.1021/acschembio.6b00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miranda JG, Weaver AL, Qin Y, Park JG, Stoddard CI, Lin MZ, Palmer AE. PLoS One. 2012;7:e49371. doi: 10.1371/journal.pone.0049371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhammika Bandara HM, Kennedy DP, Akin E, Incarvito CD, Burdette SC. Inorg Chem. 2009;48:8445–55. doi: 10.1021/ic901062n. [DOI] [PubMed] [Google Scholar]
- 7.Labile zinc refers to a loosely bound pool of zinc in cells that is exchangeable and may be involved in signaling. Bound zinc refers to zinc that is tightly bound to biomacromolecules.
- 8.Vinkenborg JL, Koay MS, Merkx M. Curr Opin Chem Biol. 2010;14:231–7. doi: 10.1016/j.cbpa.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 9.Carter KP, Young AM, Palmer AE. Chem Rev. 2014;114:4564–601. doi: 10.1021/cr400546e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Westin G, Schaffner W. EMBO J. 1988;7:3763–70. doi: 10.1002/j.1460-2075.1988.tb03260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rayner MH, Suzuki KT. Biometals. 1995;8:188–92. doi: 10.1007/BF00143374. [DOI] [PubMed] [Google Scholar]
- 12.Homma K, Fujisawa T, Tsuburaya N, Yamaguchi N, Kadowaki H, Takeda K, Nishitoh H, Matsuzawa A, Naguro I, Ichijo H. Mol Cell. 2013;52:75–86. doi: 10.1016/j.molcel.2013.08.038. [DOI] [PubMed] [Google Scholar]
- 13.Wilson D, Varigos G, Ackland ML. Immunol Cell Biol. 2006;84:28–37. doi: 10.1111/j.1440-1711.2005.01391.x. [DOI] [PubMed] [Google Scholar]
- 14.v Anderegg G, Hubmann E, Podder NG, Wenk F. Helv Chim Acta. 1977;60:123–140. [Google Scholar]
- 15.Kim EE, Wyckoff HW. J Mol Biol. 1991;218:449–64. doi: 10.1016/0022-2836(91)90724-k. [DOI] [PubMed] [Google Scholar]
- 16.Bozym RA, Thompson RB, Stoddard AK, Fierke CA. ACS Chem Biol. 2006;1:103–11. doi: 10.1021/cb500043a. [DOI] [PubMed] [Google Scholar]
- 17.Yan M, Song Y, Wong CP, Hardin K, Ho E. J Nutr. 2008;138:667–73. doi: 10.1093/jn/138.4.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao T, Asayama Y. Reprod Med Biol. 2017;16:99–117. doi: 10.1002/rmb2.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dell’Angelica EC, Schleicher CH, Santomé JA. J Biol Chem. 1994;269:28929–36. [PubMed] [Google Scholar]
- 20.Cunden LS, Gaillard A, Nolan EM. Chem Sci. 2016;7:1338–1348. doi: 10.1039/c5sc03655k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moroz OV, Blagova EV, Wilkinson AJ, Wilson KS, Bronstein IB. J Mol Biol. 2009;391:536–51. doi: 10.1016/j.jmb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Ryu MS, Langkamp-Henken B, Chang SM, Shankar MN, Cousins RJ. Proc Natl Acad Sci U S A. 2011;108:20970–5. doi: 10.1073/pnas.1117207108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colvin RA, Holmes WR, Fontaine CP, Maret W. Metallomics. 2010;2:306–17. doi: 10.1039/b926662c. [DOI] [PubMed] [Google Scholar]
- 24.Maret W. Biometals. 2013;26:197–204. doi: 10.1007/s10534-013-9613-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan E, Zhang XA, Huang Z, Krezel A, Zhao M, Tinberg CE, Lippard SJ, McNamara JO. Neuron. 2011;71:1116–26. doi: 10.1016/j.neuron.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


