Abstract
Gadd45a (growth arrest and DNA-damage-inducible protein 45 alpha) plays a pivotal role in cellular stress responses and is implicated in DNA repair, cell cycle arrest and apoptosis.1 Recently, it was proposed that GADD45A is a key regulator of active DNA demethylation by way of its role in DNA repair.2 Barreto et al. reported that Gadd45a overexpression activated transcription from methylation-silenced reporter plasmids and promoted global DNA demethylation. siRNA-mediated knockdown of Gadd45a levels resulted in increased levels of DNA methylation at specific endogenous loci. Based on these exciting results, Gadd45a-/- mice might be predicted to have a hypermethylation phenotype. We report here that neither global nor locus-specific methylation is increased in Gadd45a-/- mice.
Keywords: DNA methylation, DNA demethylation, Gadd45a, DNA repair, mouse
DNA methylation is an essential epigenetic modification in mammals that plays an important role in gene silencing and occurs predominantly on cytosines in the context of CG dinucleotides.3 In somatic cells, patterns of DNA methylation are relatively stable and are heritably maintained after DNA replication by DNA methyltransferase 1 (DNMT1). During early development, however, methylation profiles undergo sweeping changes.4 Programming of DNA methylation has been suggested to be crucial for proper developmental regulation, and in theory, this would require establishment of the patterns by sequence-specific factors that recruit DNA methyltransferases to target genes. On the other hand, erasure of methylation patterns can occur passively in a DNA-replication-dependent fashion. Some instances have been documented in which demethylation occurs by an active, replication-independent mechanism,5,6 though the factors involved in this pathway have remained elusive. Surprisingly, two recent reports claim that the methyltransferases DNMT3A and DNMT3B are capable of acting as demethylases,7-9 although the genetic data do not support the biochemical evidence.10
Gadd45a (growth arrest and DNA-damage-inducible protein 45 alpha) plays a key role in DNA repair, cell cycle arrest and apoptosis in response to environmental and physiological stress.1 Interestingly, Baretto et. al. reported that transient transfection of Gadd45a into NIH/3T3 cells led to demethylation of the endogenous Pou5f1(Oct4) promoter. More recently, however, a report was published in which these results were not reproduced.12 Regardless of these in vitro observations, we wished to determine the effect of Gadd45a loss of function at the endogenous locus on DNA methylation levels in vivo. We first studied the methylation levels of the Pou5f1 gene by methylation-sensitive PCR of DNA from tails of wild-type and Gadd45a-/- mice (Fig. 1A). Four CG sites upstream of Pou5f1 were assayed.11 No gain of methylation was detected in the Gadd45a-/- DNA relative to the wild-type levels. On the contrary, Gadd45a-/- DNA exhibited less methylation than the wild-type DNA at sites 3 and 4. Additionally, we assayed the H19 imprinting control region, a CG-rich domain that is methylated exclusively on the paternal allele. Methylation-sensitive PCRs showed equivalent presence of both methylated and unmethylated alleles in both wild-type and Gadd45a-/- DNA.
Figure 1.
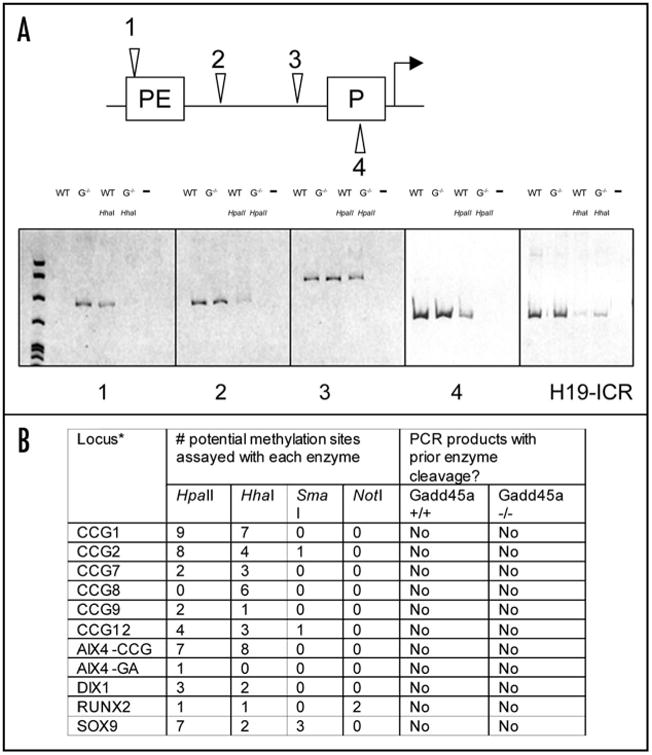
(A) Top, map of the 5′regulatory region of the Pou5f1(Oct4) gene. The arrow indicates transcription initiation. HhaI (site 1) and HpaII (2–4) restriction sites are represented by arrowheads. The promoter (P) and proximal enhancer (PE) are boxed. Primers flanking the HhaI and HpaII sites were used for the methylation-sensitive PCR assay. Bottom, DNA samples were digested with either HhaI or HpaII, which will not cut methylated DNA. PCR was carried out and products were run out on 7% polyacrylamide gels. The presence of PCR products indicates resistance to digestion due to DNA methylation. WT, wild-type; G-/-, Gadd45a-/-. (B) CpG-rich loci tested by cleavage with methylation-sensitive restriction endonucleases followed by PCR.
We next selected 11 CG-rich loci that are normally hypomethylated (Fig. 1B) and tested their methylation levels in DNA from wild-type and Gadd45a-/- MEFs by cleavage with methylation-sensitive restriction endonucleases, followed by PCR. No qualitative differences were found at any of the loci. To examine the methylation of individual CG sites, bisulfite sequencing assays were performed on three of the loci. No increase of methylation was observed at 61 CG sites in the Gadd45a-/- samples relative to wild-type controls.
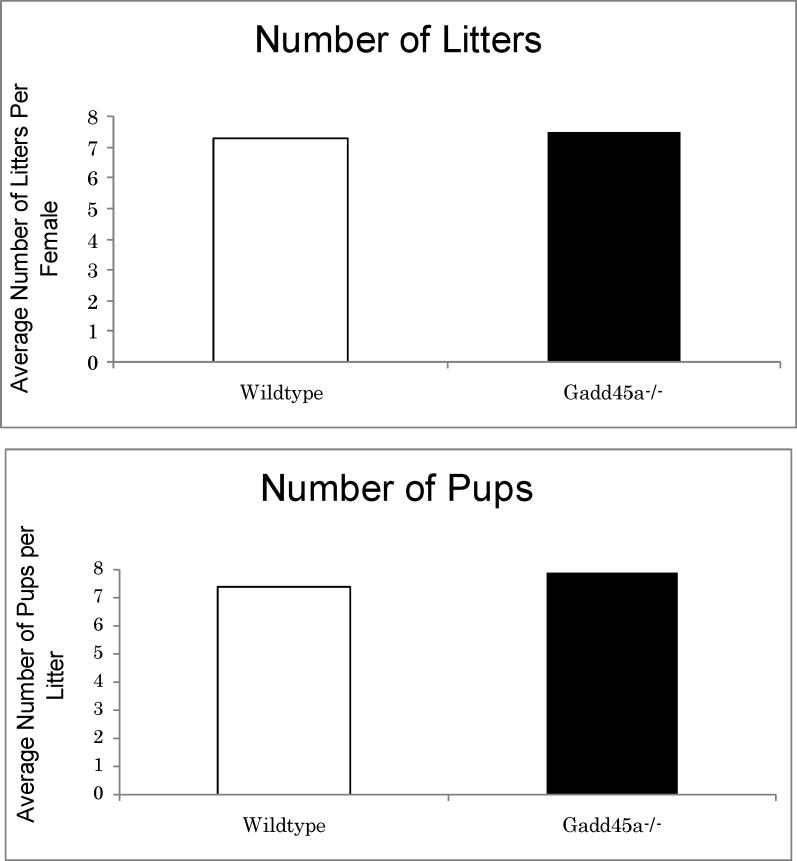
Baretto et al. suggest a model whereby GADD45A promotes active, targeted demethylation of DNA by promoting DNA repair. However, it is unlikely that the rapid, stress-induced responses mediated by GADD45A in vivo would involve reversal of silencing by DNA demethylation. In fact, active demethylation has been documented only during early embryogenesis, a stage in which Gadd45a is not expressed.12 While acute siRNA-mediated Gadd45a knockdown in tissue culture may induce hypermethylation of specific genes, the prediction that Gadd45a mutant mice would have DNA hypermethylation is not validated by any of the assays we performed. Our results do not preclude a role for Gadd45a-mediated DNA demethylation at specific loci. However, we were unable to detect hypermethylation in the Pou5f1 promoter region tested by Baretto et al. Even minor variations in levels of DNA methylation can cause severe defects or even lethality in mutant mice. We have found that Gadd45a-/- mice are fertile and have litters of normal size through multiple generations of breeding, which is inconsistent with alterations in DNA methylation or with the predicted deficiency of Pou5f1 expression (Fig. 2). We conclude that in vitro modeling, while useful for elucidating some mechanistic aspects of Gadd45a function, does not necessarily reflect the in vivo function. Although Gadd45a is not involved in global demethylation, as had been suggested, it will be interesting to see if the methylation status of a subset of genes is affected either directly or indirectly by Gadd45a and whether other cooperating factors are required.
Figure 2.
Fertility of Gadd45a-/- mice compared to wild-type mice. Breeding females were monitored for 8 months.
References
- 1.Hoffman B, Liebermann DA. Role of gadd45 in myeloid cells in response to hematopoietic stress. Blood Cells Mol Dis. 2007;39:344–7. doi: 10.1016/j.bcmd.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–5. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 3.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 4.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–32. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 5.Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, et al. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–8. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 6.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–2. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 7.Metivier R, Huet G, Gallais R, Finot L, Petit F, Tiffoche C, et al. Dynamics of estrogen receptor-mediated transcriptional activation of responsive genes in vivo: apprehending transcription in four dimensions. Adv Exp Med Biol. 2008;617:129–38. doi: 10.1007/978-0-387-69080-3_12. [DOI] [PubMed] [Google Scholar]
- 8.Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, et al. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–5. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 9.Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 10.Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–8. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Gidekel S, Bergman Y. A unique developmental pattern of Oct-3/4 DNA methylation is controlled by a cis-demodification element. J Biol Chem. 2002:34521–30. doi: 10.1074/jbc.M203338200. [DOI] [PubMed] [Google Scholar]
- 12.Jin S, Guo C, Pfeifer GP. Gadd45a does not promote DNA demethylation. PLoS Genet. 2008;4:1–9.w. doi: 10.1371/journal.pgen.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]