Abstract
Robust innate immune detection of rapidly evolving pathogens is critical for host defense. Nucleotide-binding domain leucine-rich repeat (NLR) proteins function as cytosolic innate immune sensors in plants and animals. However, the structural basis for ligand-induced NLR activation has so far remained unknown. NAIP5 (NLR family, apoptosis inhibitory protein 5) binds the bacterial protein flagellin and assembles with NLRC4 to form a multiprotein complex called an inflammasome. Here we report the cryo–electron microscopy structure of the assembled ~1.4-megadalton flagellin-NAIP5-NLRC4 inflammasome, revealing how a ligand activates an NLR. Six distinct NAIP5 domains contact multiple conserved regions of flagellin, prying NAIP5 into an open and active conformation. We show that innate immune recognition of multiple ligand surfaces is a generalizable strategy that limits pathogen evolution and immune escape.
The innate immune system deploys germline-encoded receptors to detect conserved pathogen-encoded molecules (1, 2). However, large population sizes and short generation times provide pathogens with the capacity for rapid evolution to evade immune detection. It is unclear how, over evolutionary time, innate immune receptors maintain the recognition of pathogen ligands, especially relatively mutable ligands such as proteins. Members of the nucleotide-binding domain leucine-rich repeat (NLR) superfamily function as cytosolic pathogen sensors in plants and animals (3). One of the best-characterized NLRs is mouse NAIP5 (NLR family, apoptosis inhibitory protein 5), which binds to the D0 domain of flagellin, a protein that forms the bacterial flagellum (4–6). Family members NAIP1 and NAIP2 detect the needle and inner rod proteins, respectively, of bacterial type III secretion systems (T3SSs) (4, 6–8). Upon binding their cognate bacterial ligands, NAIPs co-oligomerize with NLRC4 [NLR family, CARD (caspase activation and recruitment domain)–containing 4] to form a high-molecular-weight complex (4, 9, 10) called an inflammasome, which recruits and activates the caspase-1 protease. Active caspase-1 initiates immune responses by cleaving and activating interleukins IL-1β and IL-18 and by triggering a lytic form of cell death called pyroptosis (11).
The mechanism by which NAIPs bind microbial ligands to nucleate inflammasome formation has so far been unknown. No NAIP structure has been reported, and prior cryo–electron microscopy (cryo-EM) studies of the NAIP2-NLRC4 inflammasome applied symmetry that assumed the NAIP and NLRC4 protomers to be identical and thus failed to reveal the structure of the NAIP or its bound ligand (9, 10, 12). Here we report the structure of the flagellin-NAIP5-NLRC4 inflammasome complex, determined without applying symmetry, which reveals the mechanism by which NAIP5 detects flagellin and oligomerizes with NLRC4.
Overall structure of the NAIP5 inflammasome
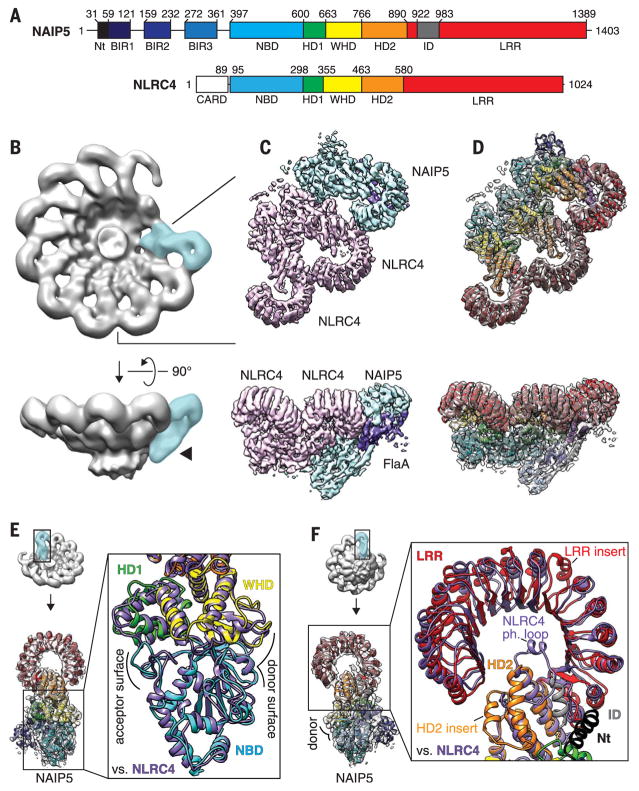
As with all mammalian NLRs, NAIP5 and NLRC4 both contain a conserved nucleotide-binding domain (NBD), helical domain 1 (HD1), winged helix domain (WHD), helical domain 2 (HD2), and leucine-rich repeat (LRR) domain (Fig. 1A). In addition, NLRC4 contains a CARD, which recruits caspase-1, whereas NAIP5 contains three baculovirus inhibitor-of-apoptosis repeat (BIR) domains of unknown function. We used cryo-EM to determine the structure of the assembled NAIP5-NLRC4 inflammasome bound to Legionella pneumophila flagellin (FlaA) (Fig. 1B and figs. S1 to S3). Our three-dimensional (3D) reconstructions reveal assemblies that contain a single NAIP5 subunit (Fig. 1B, blue), easily identified by its bulky BIR domains (Fig. 1B, arrowhead), bound to a single FlaA monomer (Fig. 1C, purple). These results are consistent with prior evidence that the oligomer is nucleated by a single NAIP (9, 10, 13), which then associates with a variable number of NLRC4 subunits (fig. S2D).
Fig. 1. Structure of the NAIP5-NLRC4 inflammasome.
(A) Schematic of domain architecture for NAIP5 and NLRC4. NAIP5 domains were defined in this work (methods); NLRC4 domains were previously defined (19). Residue numbers are shown. Nt, N terminus. (B) 3D reconstruction of inflammasomes containing a single NAIP5-FlaA (blue) and nine NLRC4 protomers (gray). The arrowhead highlights extra density that identified the NAIP5 protomer. (C) Refined 3D reconstruction for NAIP5 and the first two NLRC4 protomers at higher resolution (NAIP5 segmented in blue, the two NLRC4 protomers in pink, and FlaA in purple). (D) Modeled structures of NAIP5 and two NLRC4 protomers, all colored by domains as in (A), fitted within the EM map. In (B) to (D), two orthogonal views are shown. (E and F) The structure of NAIP5 [colored as in (A)] is aligned with that of an NLRC4 protomer (purple). (E) The NBD, HD1, and WHD oligomerization domains of NAIP5 and NLRC4 are highly similar. Oligomerization donor and acceptor surfaces [(9, 10) and fig. S5] are indicated. For clarity, the NAIP5 BIR domains were omitted from the inset view (right). (F) The HD2 and LRR of NAIP5 diverge from those of NLRC4. NAIP5-specific insertions, including an extra leucine-rich repeat (LRR insert; residues 1102 to 1138) and the modeled helix of the inserted domain (ID; gray), are indicated. The NLRC4 S533 phosphorylation loop is replaced by two alpha helices in NAIP5 (HD2 insert; residues 818 to 851).
We carried out focused refinement on a region of the inflammasome containing the single NAIP5 and two of the adjacent NLRC4 subunits (Fig. 1, C and D; fig. S2; and methods), yielding a reconstruction with an overall resolution of 5.2 Å (fig. S4). Modeling and advanced fitting techniques (14–17) (methods) enabled the characterization of NAIP5 and its binding interfaces with NLRC4 and flagellin (Figs. 1 and 2 and fig. S5A). NAIP5 contacts NLRC4 with an oligomerization “donor” surface, contributed by the WHD and NBD, which is composed primarily of basic and hydrophobic residues (Fig. 1E and fig. S5A), as previously predicted (9, 10). The cognate “acceptor” surface of NLRC4 consists of complementary acidic and hydrophobic residues. We found that multiple residues in the NLRC4 acceptor surface are required for NLRC4 to bind to NAIP5 (fig. S5B). Several of these residues, particularly I124 and D125, are also required for NLRC4 to bind a neighboring NLRC4 (9). The conformations of the two adjacent NLRC4 molecules are identical, even though symmetry was not imposed. Thus, the acceptor surface of NLRC4 is the same regardless of whether it contacts a NAIP5 or NLRC4 donor surface. In contrast to prior averaged reconstructions of the NAIP-NLRC4 inflammasome and the structures of related complexes such as apoptosomes (18), we did not observe a substantial number of rings that appear to be closed (Fig. 1B and fig. S2). The lack of ring closure is consistent with the prior proposal that although the donor surface on NAIP5 can recruit and activate the cognate acceptor surface on NLRC4, the donor surface on NLRC4 can only interact with additional NLRC4 protomers and cannot close the ring by interacting with an acceptor surface of NAIP5 (9, 10).
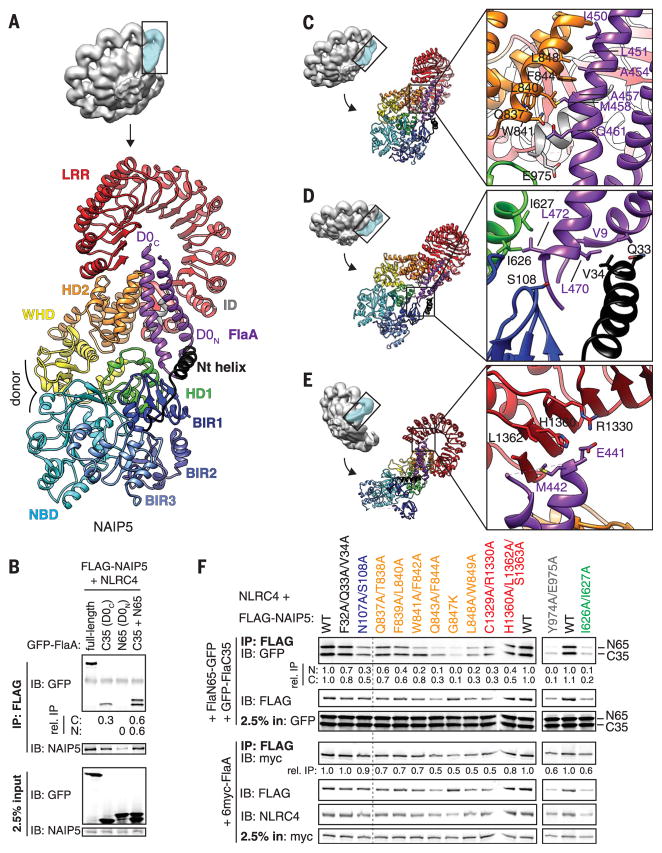
Fig. 2. Multiple NAIP5 domains contact extended surfaces on both helices of the flagellin D0 domain.
(A) The D0 domain (purple) is locked into place by multiple NAIP5 domains. (B) Both D0 helices bind to NAIP5. (C to E) Detailed interactions between the flagellin D0 helices and the NAIP5 domains HD2 and ID (C); Nt, BIR1, and HD1 (D); and LRR (E). Side chains shown correspond to mutated residues in (F). (F) Mutagenesis confirms the importance of NAIP5 residues in binding D0N, D0C, or full-length FlaA. Point mutations in NAIP5 residues that contact D0C disrupt both D0N and D0C binding because association of D0N requires D0C binding [(B) and fig. S9A]. In (B) and (F), relative IP strength was quantified by densitometry [IP signal normalized to input and then GFP-FlaA (B) or WT NAIP5 (F)]. Results are representative of at least three independent experiments. IP, immunoprecipitation; IB, immunoblot; WT, wild type. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
Although the structural homology between NAIP5 and NLRC4 is high in the regions that mediate oligomerization (Fig. 1E), some regions of NAIP5 and NLRC4 are relatively divergent, particularly the HD2 and LRR (Fig. 1F). Consequently, our model of NLRC4 differs slightly from previous structures that averaged the NAIP with NLRC4 (9, 10), particularly in the phosphorylation loop (19, 20), which is replaced with two helices in NAIP5 (Fig. 1F, HD2 insert), in addition to other smaller deviations (fig. S6).
Flagellin recognition by NAIP5
The single FlaA protomer in the complex is recognized solely by NAIP5, with no apparent contribution to binding by NLRC4 (Fig. 1, C and D). NAIP5 makes numerous contacts with both helices of the FlaA D0 domain (Fig. 2A). The N-terminal D0 helix (D0N; residues 1 to 33) makes fewer contacts with NAIP5 than the C-terminal D0 helix (D0C; residues 441 to 475), which is contacted along its length, as well as at three C-terminal leucines (L470, L472, and L473) previously shown to be important for flagellin recognition (5). Consistent with these contacts, both the D0N and D0C helices are capable of binding to NAIP5 when expressed as separate polypeptides fused to green fluorescent protein (GFP), although association of D0N with NAIP5 requires the binding of D0C (Fig. 2B). Control experiments confirmed that the D0N and D0C helices do not bind to each other in the absence of NAIP5 (fig. S7B), suggesting an important role for D0N-NAIP5 contacts in flagellin binding (13).
Many of the NAIP5 regions contacting FlaA are divergent from the corresponding areas of NLRC4. For example, one of the inserted helices in HD2 (residues H835 to L850) makes numerous, primarily hydrophobic, contacts with the D0C helix (Fig. 2C). Similarly, NAIP5 contains a large insertion in the N-terminal region of the LRRs, which we term the inserted domain (ID; residues 922 to 983) (Fig. 1, A and F). Although the ID is not fully resolved in our cryo-EM density, a helix in this domain (residues 964 to 976) appears to contact both helices of the FlaA D0 domain (Fig. 2C). One helix of HD1 also appears to bind D0C (Fig. 2D). These three flagellin-binding regions are poorly conserved between NAIP2 and NAIP5 (fig. S8), consistent with prior data suggesting that these regions confer ligand specificity to NAIPs (21).
Our structure unexpectedly reveals at least three additional regions of NAIP5 that contact FlaA (Fig. 2, D and E): (i) BIR1 (residues N107 and S108) appears to directly contact the D0C helix of FlaA, which would be the first known specific function attributable to the NAIP BIR domains; (ii) a helix, just N-terminal to BIR1, contains residues (Q33 and V34) that appear to contact one or both D0 helices of FlaA; and (iii) a small portion of the LRR domain contains residues (R1330, H1360, and S1363) that appear to contact the D0C helix. To validate all the observed FlaA-NAIP5 interactions, we assessed the effect of NAIP5 mutations on binding to either the D0N or D0C FlaA helices (expressed as separate polypeptides fused to GFP) or to full-length FlaA (Fig. 2F). We found that point mutations in the N-terminal helix, BIR1, HD1, HD2, ID, and LRR each affected, to varying degrees, the ability of NAIP5 to bind flagellin (Fig. 2F). Taken together, our results suggest that the binding pocket for flagellin is formed from these six NAIP5 domains (Fig. 2).
Mechanism of inflammasome assembly
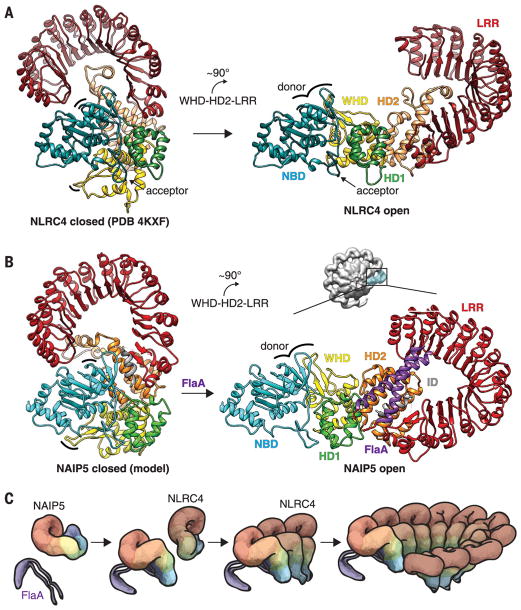
Activation of NLRC4 results from a rigid-body rotation hinged between the HD1 and WHD (9, 10, 19). This conformational change unfurls NLRC4 and positions its donor surface to recruit and activate the next incoming NLRC4 (Fig. 3A). On the basis of the high degree of homology between NAIP5 and NLRC4 in the NBD, HD1, and WHD domains (Fig. 1E), we propose that NAIP5 also undergoes a rigid-body rotation hinged around the WHD upon binding flagellin (Fig. 3B). If we make the parsimonious assumption that the inactive form of NAIP5 is similar to that of NLRC4 (Fig. 3, A and B), then crucial parts of the flagellin-binding surface, particularly those in HD2, would be sterically occluded by the NBD and LRR (Fig. 3B). Flagellin binding would necessarily displace the occluding LRR and HD2 from the NBD, freeing the latter to serve as a donor surface for NLRC4 to initiate inflammasome assembly (Fig. 3C).
Fig. 3. Model of NAIP5-NLRC4 inflammasome assembly.
(A) Structures of inactive (left) and active (right, determined in this work) NLRC4, showing a ~90° rigid-body rotation of the WHD-HD2-LRR module (9, 10) triggered by interaction with an activated NAIP5 or another already activated NLRC4. (B) To generate the inactive NAIP5 conformation [left; modeled based on NLRC4 (methods)], the HD2 insert was moved to avoid collision with the NBD. We propose that flagellin binding induces a ~90° rigid-body rotation of the WHD-HD2-LRR module, analogous to the rotation of NLRC4, which displaces the occluding LRR and HD2 from the NBD to complete and expose the donor oligomerization surface (indicated by curved lines in left and right panels) for interaction with NLRC4. (C) Proposed events of inflammasome assembly. The flagellin D0 domain (purple) binds to NAIP5 and unfurls the protein for subsequent NLRC4 recruitment and activation. Active NLRC4 recruits further NLRC4 protomers for self-propagating oligomerization and completion of a caspase-1 recruitment platform. Colors are as in Fig. 1A.
NAIPs recognize multiple ligand surfaces
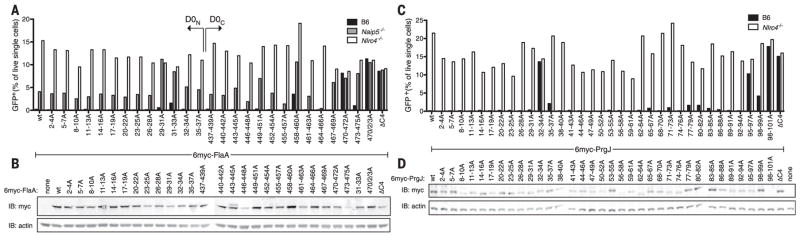
To identify regions of flagellin recognized by NAIP5, we performed an unbiased alanine scanning mutagenesis of the FlaA D0 domain in three amino acid blocks. We assayed NAIP5 recognition of FlaA mutants using a retroviral lethality assay (5) in which FlaA is expressed in bone marrow–derived macrophages (BMMs) with a GFP-marked retrovirus. Wild-type FlaA robustly induces pyroptosis, preventing the recovery of GFP+ B6 BMMs, whereas GFP+ Nlrc4−/− BMMs fail to undergo pyroptosis and are readily detected (Fig. 4A). Naip5−/−BMMs produce an intermediate number of GFP+ cells, owing to partially redundant flagellin recognition by NAIP6 (22). Mutation of the C terminus of FlaA, including an L470A/L472A/L473A mutant previously reported to evade NAIP5 recognition (5), abrogated the activation of both NAIP5 and NAIP6 and permitted recovery of GFP+ transductants. In addition, motifs at residues 458 to 460 and 31 to 33 were partially required for NAIP5 and NAIP6 activation. The inability of FlaA mutants to activate NAIP5 and NAIP6 was not due to their lack of expression (Fig. 4B). The motifs at residues 458 to 460 and 470 to 473 are supported by our EM structure and make contacts with the NAIP5 N terminus, BIR1, and HD1 domains and the HD2 and ID, respectively, whereas the motif at residues 31 to 33 appears to be proximal to unmodeled regions of NAIP5.
Fig. 4. Multisurface ligand recognition is common to NAIPs.
(A) Full-length 6myc-tagged L. pneumophila FlaA, or variants with the indicated residues mutated to alanine, were transduced into BMMs by using a retrovirus marked with IRES (internal ribosomal entry site)–GFP.Transduction efficiency was assessed by flow cytometry for GFP expression at day 4. Failure to transduce B6 BMMs, as compared with transduction of Nlrc4−/− BMMs, is indicative of NAIP activation (5). Naip5−/− BMM responses to FlaA are NAIP6-dependent (22). (B) Transduced Nlrc4−/− BMM lysates were probed for FlaA expression by anti-myc IB. (C) Full-length 6myc-tagged S. typhimurium PrgJ, or variants with the indicated residues mutated to alanine, were transduced into BMMs as in (A) to assess NAIP2 recognition. (D) Constructs were transfected into human embryonic kidney–293 Tcells, and lysates were probed for PrgJ expression by anti-myc IB. Results are representative of at least two independent experiments (n = 1 per trial).
We also performed alanine scanning mutagenesis of PrgJ, the Salmonella typhimurium SPI1 T3SS inner rod protein that is recognized by NAIP2. Analogous to flagellin, mutations at the C terminus of PrgJ abolished NAIP2 recognition (Fig. 4C). In addition, an RLS motif at residues 32 to 34 was also required for NAIP2 activation but not for PrgJ expression (Fig. 4, C and D). This motif is particularly striking because of its positional correspondence to the RLL motif at residues 31 to 33 of FlaA. Lastly, the PrgJ region spanning residues 65 to 88 (with residues 82 to 85 positionally corresponding to the motif at residues 458 to 460 in FlaA) consistently contributed weakly to NAIP2 recognition. The motifs identified in PrgJ and FlaA are important for binding to their cognate NAIPs (fig. S9) and are well conserved (fig. S10). Mutation of these motifs in S. typhimurium flagellin abrogated its recognition by NAIP5 (fig. S11). Despite recognizing similar motifs in their respective ligands, NAIP5 and NAIP6 do not bind to either half of PrgJ, and NAIP2 does not bind to FlaA D0N or D0C (fig. S12). Thus, it is likely that the specificity of NAIPs for their cognate ligands is contributed by variable regions outside the motifs, consistent with the additional FlaA contacts that we observe with NAIP5 (Fig. 2). Collectively, these results indicate that NAIPs recognize multiple discrete surfaces on their cognate ligands.
Multisurface recognition limits pathogen immune evasion
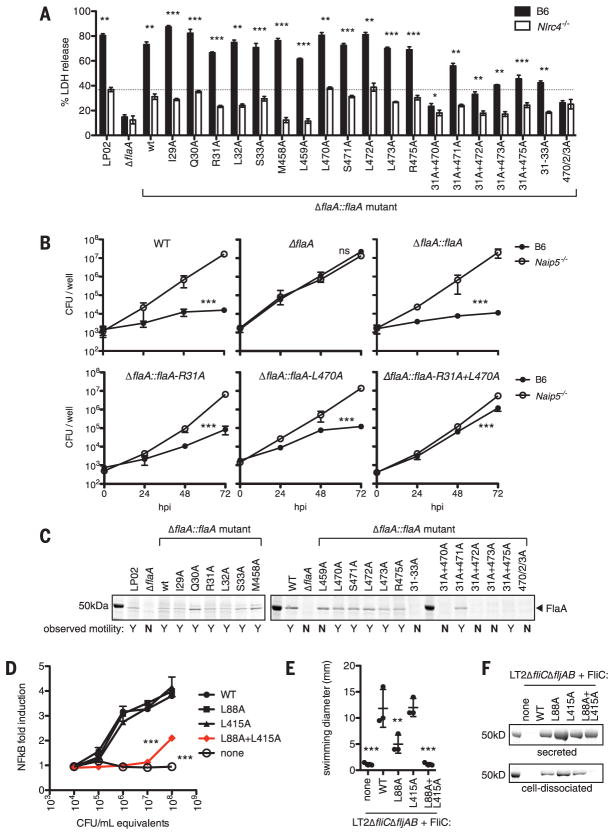
The D0 domain of the flagellin monomer is largely disordered (23, 24) but becomes ordered to form the core of the polymerized flagellum (25, 26). Notably, when bound to NAIP5, the flagellin D0 domain adopts a conformation that is nearly identical to its structure in the flagellar filament (fig. S13A). Furthermore, NAIP5 contacts similar residues to those buried in the flagellar filament (fig. S13, B and C). Thus, we hypothesized that mutations in FlaA that disrupt its interaction with NAIP5 might also disrupt flagellar filament formation. Because multiple flagellin surfaces are recognized by NAIP5, we further hypothesized that escape from NAIP5 recognition may require multiple mutations that would be more likely to disrupt flagellar function. Indeed, mutation of any single FlaA residue had little to no effect on NAIP5-mediated induction of pyroptosis, as measured by the retroviral lethality assay (fig. S14). However, simultaneous mutation of the N- and C-terminal recognition motifs reduced or abolished NAIP5 recognition. To confirm these results, we mutated the endogenously expressed L. pneumophila flaA gene and assayed NAIP5 and NLRC4 activation during bacterial infection of macrophages. We observed that even under these stringent conditions, single alanine substitutions in FlaA did not affect the ability of L. pneumophila to activate Nlrc4-dependent pyroptosis (Fig. 5A). In contrast, simultaneous mutation of the N- and C-terminal motifs affected NLRC4 activation, particularly the combination of R31A and L470A. Likewise, the R31A+L470A double mutant was only weakly restricted for intracellular replication, a sensitive assay for NAIP5 function, whereas the single mutants were robustly restricted (Fig. 5B).
Fig. 5. Simultaneous mutation of multiple recognition motifs is required to evade NAIP5 or TLR5 recognition but disrupts flagellar motility.
(A to C) The indicated mutations were introduced at the endogenous FlaA locus of L. pneumophila strain LP02. (A) BMMs were infected with L. pneumophila strains at multiplicity of infection (MOI) = 3, and cell death was measured by lactate dehydrogenase (LDH) release at 4 hours. The dashed line indicates Nlrc4-independent LDH release in wild-type LP02 infection. (B) NAIP5- and FlaA-dependent restriction of L. pneumophila replication in BMMs. BMMs were infected at MOI = 0.01, and colony-forming units (CFU) were measured at the indicated time points. hpi, hours post-infection. (C) L. pneumophila were classified as motile (Y) or nonmotile (N) on the basis of observation of swimming runs. Bacteria were vortexed to dissociate cell-surface flagella, and supernatants were analyzed by Coomassie stain. (D to F) S. typhimurium strain LT2ΔfliCΔfljAB was transformed with an expression vector encoding wild-type FliC or the indicated variants. (D) Overnight culture supernatants were incubated 6 hours with CHO cells expressing HsTLR5 and a nuclear factor κB (NFκB) luciferase reporter. Reporter cells were analyzed for luciferase expression. (E) Diameter of colonies incubated on 0.4% agarose plates for 8 hours. (F) Culture supernatants and the supernatants of vortexed bacteria were analyzed for the presence of secreted or cell-dissociated flagellin, respectively. Results are representative of at least three independent experiments (error bars, SD; n = 3 biological replicates). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001; ANOVA (analysis of variance) comparing across BMM genotype [(A) and (B)] or against wild-type FliC [(D) and (E)]).
In addition to failing to evade NAIP5 recognition, most single alanine mutations in FlaA did not affect flagellar assembly or motility. However, the combined mutations that allowed evasion of NAIP5 recognition also abrogated flagellar assembly and motility (Fig. 5C). Nonconservative single mutations (L470P, L470N, L470D, and L470R) did reduce NAIP5-mediated pyroptosis, but these mutations also disrupted bacterial motility (fig. S15). Taken together, these data suggest that single mutations in flagellin that do not affect its function are insufficient to escape NAIP5 detection, whereas more severe mutations that do escape recognition result in a fitness cost to the bacterium. Thus, we propose that multisurface recognition imposes a constraint on the mutational paths that pathogens can use to escape immune recognition.
Generalizability of multisurface innate immune recognition
Analogous to our results with FlaA, single point mutations in the N- and C-terminal motifs of PrgJ did not affect its recognition by NAIP2 (fig. S16, A and B); nor, with one exception (E96A), did single mutations affect the ability of PrgJ to assemble a functional T3SS (fig. S16, C and D). Combined point mutations were sufficient for PrgJ to evade NAIP2 recognition, but many of these mutations also disrupted PrgJ function. Unlike with FlaA, we were able to identify two double mutants of PrgJ (L33A+V95A and L33A+R100A) that escape NAIP2 recognition but appear to retain at least some native function, suggesting either that the evolutionary constraint of multi-surface recognition is not always complete or that our functional assays were not sufficiently sensitive to detect a modest loss of function.
The strategy of multisurface recognition may extend to other innate immune receptors that detect protein ligands (27–29). In particular, the binding of multiple surfaces has been proposed to contribute to robust detection of flagellin by TLR5 (30). We found that simultaneous point mutation of two motifs in S. typhimurium flagellin (FliC) substantially reduced TLR5 activation and disrupted bacterial motility, whereas each single point mutation was largely tolerated (Fig. 5, D to F). Thus, our data indicate that multisurface recognition is a general strategy to limit the evolutionary paths available for pathogens to evade innate immune detection.
Conclusion
The combinatorial use of antibiotics and antivirals is based on the principle that the simultaneous presence of dual selective pressures provides a greater constraint on the evolution of escape mutants than does each selective pressure individually (31). Our results provide structural and functional evidence that the innate immune system exploits this general principle and uses a multisurface recognition strategy to constrain bacterial immune escape (fig. S17). Although many pathogens do at least partially evade innate immunity, our results suggest that evasion of innate immunity likely involves more complex mechanisms than simple ligand modifications. These mechanisms may include the evolution of compensatory mutations to regain function of immune-evading mutants (32) or the acquisition of alternative virulence factors (33), such as intracellular actin–mediated rather than flagellin-mediated motility (34). The multisurface recognition strategy that we describe likely complements additional strategies that constrain pathogen evolution. For example, targeting the most conserved features on microbial ligands helps to constrain mutagenic escape, although even constrained sites can tolerate some mutations (Fig. 5C). Pathogen immune evasion can also be counteracted by diversifying selection, observed in both TLR5 (35) and NAIPs (21), at the interface with microbial ligands (36). Thus, we propose that multisurface recognition is one strategy in the arsenal deployed by hosts to counteract the intrinsic advantage held by large populations of rapidly evolving pathogens in their “arms race” with eukaryotic immune systems.
Supplementary Material
Acknowledgments
We thank T. Houweling and A. Chintangal for computer support. The EM data was collected in the EM facility of the Howard Hughes Medical Institute Janelia Research Campus. We are thankful to C. Hong and Z. Yu for expert EM assistance; B. Greber, R. Louder, A. Patel, and A. Sandstrom for discussion; P. Dietzen for technical support; K. Namba for the coordinates of the R-type flagellar filament (26); and A. Truxal for generating the illustration in Fig. 3C. We acknowledge the use of the LAWRENCIUM computing cluster at Lawrence Berkeley National Laboratory and the resources of the National Energy Research Scientific Computing Center, a U.S. Department of Energy (DOE) Office of Science user facility supported by the Office of Science of the DOE under contract no. DE-AC02-05CH11231. We thank K. Smith for the contribution of a CHO cell line stably expressing HsTLR5 and NFκB luciferase reporters, G. Barton for S. typhimurium strains, S. Brubaker and D. Monack for P22 transducing phage, and H. Darwin for advice on analysis of flagellin expression. This work was funded by the Spanish Ministry of Economy, Industry and Competitiveness grant BFU2016-76220-P (P.C.) and NIH grants AI075039 and AI063302 (R.E.V.). N.H. and J.L.T. were supported by the NSF Graduate Research Fellowship Program. R.E.V. and E.N. are Howard Hughes Medical Institute Investigators. The cryo-EM map has been deposited in the Electron Microscopy Databank with accession code EMD-7055. The atomic coordinate model has been deposited in the Protein Data Bank (PDB) with accession code 6B5B.
Footnotes
REFERENCES AND NOTES
- 1.Janeway CA., Jr Cold Spring Harb Symp Quant Biol. 1989;54:1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Takeuchi O, Akira S. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 3.Jones JD, Vance RE, Dangl JL. Science. 2016;354:aaf6395. doi: 10.1126/science.aaf6395. [DOI] [PubMed] [Google Scholar]
- 4.Kofoed EM, Vance RE. Nature. 2011;477:592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lightfield KL, et al. Nat Immunol. 2008;9:1171–1178. doi: 10.1038/ni.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Y, et al. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 7.Rayamajhi M, Zak DE, Chavarria-Smith J, Vance RE, Miao EA. J Immunol. 2013;191:3986–3989. doi: 10.4049/jimmunol.1301549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J, Zhao Y, Shi J, Shao F. Proc Natl Acad Sci USA. 2013;110:14408–14413. doi: 10.1073/pnas.1306376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Z, et al. Science. 2015;350:399–404. doi: 10.1126/science.aac5489. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L, et al. Science. 2015;350:404–409. doi: 10.1126/science.aac5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broz P, Dixit VM. Nat Rev Immunol. 2016;16:407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- 12.Diebolder CA, Halff EF, Koster AJ, Huizinga EG, Koning RI. Structure. 2015;23:2349–2357. doi: 10.1016/j.str.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Halff EF, et al. J Biol Chem. 2012;287:38460–38472. doi: 10.1074/jbc.M112.393512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J, et al. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopéz-Blanco JR, Chacón P. J Struct Biol. 2013;184:261–270. doi: 10.1016/j.jsb.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 16.Adams PD, et al. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.López-Blanco JR, Canosa-Valls AJ, Li Y, Chacón P. Nucleic Acids Res. 2016;44:W395–W400. doi: 10.1093/nar/gkw395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan S, Akey CW. Structure. 2013;21:501–515. doi: 10.1016/j.str.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Z, et al. Science. 2013;341:172–175. doi: 10.1126/science.1236381. [DOI] [PubMed] [Google Scholar]
- 20.Qu Y, et al. Nature. 2012;490:539–542. doi: 10.1038/nature11429. [DOI] [PubMed] [Google Scholar]
- 21.Tenthorey JL, Kofoed EM, Daugherty MD, Malik HS, Vance RE. Mol Cell. 2014;54:17–29. doi: 10.1016/j.molcel.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rauch I, et al. J Exp Med. 2016;213:657–665. doi: 10.1084/jem.20151809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aizawa SI, Vonderviszt F, Ishima R, Akasaka K. J Mol Biol. 1990;211:673–677. doi: 10.1016/0022-2836(90)90064-S. [DOI] [PubMed] [Google Scholar]
- 24.Vonderviszt F, Kanto S, Aizawa S, Namba K. J Mol Biol. 1989;209:127–133. doi: 10.1016/0022-2836(89)90176-9. [DOI] [PubMed] [Google Scholar]
- 25.Gugolya Z, Muskotál A, Sebestyén A, Diószeghy Z, Vonderviszt F. FEBS Lett. 2003;535:66–70. doi: 10.1016/s0014-5793(02)03859-0. [DOI] [PubMed] [Google Scholar]
- 26.Yonekura K, Maki-Yonekura S, Namba K. Nature. 2003;424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- 27.Chou S, et al. Proc Natl Acad Sci USA. 2011;108:13323–13328. doi: 10.1073/pnas.1109791108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Y, et al. Science. 2013;342:624–628. doi: 10.1126/science.1243825. [DOI] [PubMed] [Google Scholar]
- 29.Yoon SI, et al. Science. 2012;335:859–864. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith KD, et al. Nat Immunol. 2003;4:1247–1253. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]
- 31.Hughes D, Andersson DI. Nat Rev Genet. 2015;16:459–471. doi: 10.1038/nrg3922. [DOI] [PubMed] [Google Scholar]
- 32.Andersen-Nissen E, et al. Proc Natl Acad Sci USA. 2005;102:9247–9252. doi: 10.1073/pnas.0502040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miao EA, et al. Proc Natl Acad Sci USA. 2010;107:3076–3080. doi: 10.1073/pnas.0913087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choe JE, Welch MD. Pathog Dis. 2016;74:ftw099. doi: 10.1093/femspd/ftw099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith SA, et al. BMC Evol Biol. 2012;12:122. doi: 10.1186/1471-2148-12-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daugherty MD, Malik HS. Annu Rev Genet. 2012;46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.