Abstract
Proteases are involved in almost every important cellular activity, from embryonic morphogenesis to apoptosis. To study protease activity in situ, hydrogels provide a synthetic mimic of the extracellular matrix (ECM) and have utility as a platform to study activity, such as those related to cell migration, in three-dimensions. While 3-dimensional visualization of protease activity could prove quite useful to elucidate the proteolytic interaction at the interface between cells and their surrounding environment, there has been no versatile tool to visualize local proteolytic activity in real time. Here, micron-sized gels were synthesized by inverse suspension polymerization using thiolene photo-click chemistry. The size distribution was selected to avoid cellular uptake and to lower cytotoxicity, while simultaneously allowing the integration of peptide-based FRET sensors of local cell activity. Proteolytic activity of collagenase was detected within an hour via changes in fluorescence of embedded microgels; incubation of microgel sensors with A375 melanoma cells showed upregulated MMP activity in the presence of soluble fibronectins in media. The microgel sensors were readily incorporated into both gelatin and poly(ethylene glycol) (PEG) hydrogels and used to successfully detect spatiotemporal proteolytic activity of A375 melanoma cells. Finally, a tumor model was constructed from a hydrogel microwell array that was used to aggregate A375 melanoma cells, and local variations in proteolytic activity were monitored as a function of distance from the cell aggregate center.
Keywords: protease sensors, hydrogels, 3-dimensional cell culture, cancer, tumor remodeling
Graphical Abstract
Introduction
Cells are known to respond plastically to microenvironmental cues (e.g., matrix elasticity,1–4 adhesive ligand density,3,5 dimensionality6,7 and interfacial geometry8), which collectively influence differentiation, morphology, proliferation, motility, and apoptosis. In these studies, synthetic hydrogel scaffolds have been widely used as physiologically relevant in vitro ECM mimics, and the ability to tune hydrogel properties has led to a versatile tool for studying cell behavior under defined microenvironmental conditions.9 In this regard, we have become particularly interested in measuring local cellular protease activity, and understanding the dynamic interplay between the influence of local matrix signals on protease secretion (e.g., outside-in signaling) and the feedback loop related to subsequent matrix remodeling (e.g., inside-out signaling). Proteases are involved in many essential cell processes, including growth,10 migration,10,11 proliferation,12 receptor activation,13,14 and apoptosis.12,15,16 High expression of proteases is often associated with various disease states, including inflammation,17 arthritis,18 tumor growth and invasion,10,19–21 affecting disease progression and drug resistance. Abnormal protease activity can be further correlated to the microenvironment of diseased tissues; for example, there is evidence showing that expression of proteases in disease cells can be responsive to stiffness,7 dimensionality of the culture platform,7,8 and cell-cell interactions.21–26 At the tumor-host interface, diffusion of secreted proteins and hormones affects protease activity of both tumor and host cells, altering cancer etiology, progression and metastasis of breast,23,27 melanoma,24 pulmonary25 and ovarian26 tumors.
With a growing interest in cell-matrix interactions and how this influences the dynamics of proteolytic activity and matrix remodeling, new methods to visualize and measure spatiotemporal activity of proteases in real time are valuable. To date, gel zymography has been a workhorse for characterizing hydrolytic activity of proteases.28 Despite its broad applicability, the technique requires electrophoresis and mass calibration of gel substrates, both requiring careful sample preparation and processing before evaluation, and rendering the method unsuitable for real time monitoring of protease activity.28,29 Enzyme-linked immunosorbent assays (ELISAs) are useful for obtaining quantitative information about the presence of proteases, but no information is gained about their function and activity. In a similar manner, gene expression data obtained by PCR is a useful quantitative tool, but it does not provide post-translational regulation information, such as activation of the zymogen to an active protease.28
To complement existing methods and address some of the shortcomings, recent studies have explored the development of protease sensing molecules, comprised of cleavable proteolytic substrates and covalently bonded FRET (Förster resonance energy transfer) fluorophore pairs29–36 to monitor proteolytic activity of caspase,30,31 MMPs,29,32,33 Alzheimer’s disease-associated proteases34 and Bacillus anthracis proteases35,36 in real time. However, in many tissue engineering applications (e.g., in vitro models of tumor microenvironments, 3D stem cell culture systems), significant challenges remain with respect to spatiotemporal monitoring of protease activity. Although several studies have attempted to monitor spatiotemporal protease activity, most have been limited to detecting membrane bound or intracellular proteases, rather than extracellular activity. Developing tools to enable spatiotemporal monitoring of secreted protease activity could prove complementary and quite useful, considering that many disease-related proteases are secreted rather than membrane bound (e.g., MMP-2, MMP-9, Cathepsin-b, etc.). Furthermore, observing extracellular signaling can be important when studying cell-matrix signaling or designing degradable scaffold for 3D cell culture and tissue regeneration. Some challenges for developing methods to detect local protease activity in 3D environments include (i) prevention of cellular uptake or diffusion of the sensor molecules, (ii) facile integration of the sensor molecules in various types of bioscaffolds, (iii) reducing cytotoxicity of the sensor molecules, and (iv) enhancing the sensitivity of the sensor.
To address some of these issues, we incorporated protease sensor molecules into micron-sized hydrogel spheres that could be easily incorporated into cell-laden biomaterial systems. The size of the microgel particles was selected based on previous reports37,38 to avoid cellular uptake. Since the diameter of the microgel sensors is larger than the mesh size of natural ECM components39,40 or synthetic hydrogels,41 they are readily embedded in 3-dimensional cell culture matrices by simply mixing them during gel preparation. In addition, proteolytic activity can be detected in real time and in situ without disturbing the surrounding cellular microenvironment. By conjugating the fluorogenic substrates to the microgel, background fluorescence is reduced and cost-effective in situ monitoring is achieved since the bulk hydrogel does not need to be modified with expensive dyes. By functionalizing the microgels, one can easily introduce varying concentrations of the sensor peptides by reaction of the thiol group in the cysteine residue to achieve desired levels of sensitivity or detection of targeted protease activity. To demonstrate an application for these microgel sensors in cancer biology, we prepared aggregates of a A375 melanoma cell line in a hydrogel well array. The array was laden with our micro-sensors, and protease activity was monitored as a function of time and space in a tumor-like microenvironment.
Materials and Methods
Synthesis of MMP Sensor Peptides
MMP degradable sensor peptides, which have a fluorophore and a quencher molecule at each end, were synthesized as previously reported.29 Briefly, an MMP degradable peptide, GGPQG ↓ IWGQK(Dde)-AhxC was synthesized on a Tribute Peptide Synthesizer (Protein Technologies) at 0.25 mmol scale on MBHA Rink amide resin using Fmoc solid phase chemistry with 2-(1H- benzotriazolyl)-1,1,3,3-tetramethyluronium hexafluoro- phosphate (HBTU) as a coupling reagent. To conjugate the dabcyl group to the amine group on the N-terminus of a glycine unit, dabcyl-NHS ester and the synthesized peptides were reacted with 1% (v/v) N,N-Diisopropylethylamine (DIPEA) in DMF overnight. The orthogonally protected Dde group on the lysine residue was de-protected using 10 ml of 2% hydrazine monohydrate in DMF for 10 min, twice. The exposed free amine group was confirmed by a positive ninhydrin test and covalently functionalized with fluorescein using the same methods for dabcyl functionalization. After washing with DMF, methanol and DCM, the peptides were cleaved from the beads using a cleavage cocktail (trifluoroacetic acid: triisopropyl silane: water = 90:2.5:2.5 (v/v) and 50 mg/ml phenol). These peptides were precipitated three times in chilled diethyl ether and dried under vacuum for 4 hours. The crude fluorogenic peptide G(dabcyl)GPQG ↓ IWGQK(Fluorescein)AhxC was purified by HPLC (Waters Delta Prep 4000) on a C18 column with linear 20% to 90% gradient of acetonitrile to water. Purified peptides were lyophilized and stored in a −80 °C freezer before usage. Synthesis and purification were verified by MALDI-TOF mass spectrometry.
Preparation of Hydrogel Precursors
The monomer, PEG-norbornene, was synthesized from amine functionalized PEG using the HBTU/DIPEA pair as an activator. 8-arm PEG amine (hexaglycerol) (MW 40 kDa or 10 kDa) was purchased from JenKem Technology and dried in a vacuum oven overnight before modification. The carboxyl group of 5-norbornene-2-carboxylic acid (2 equivalents per amine group) was activated with HBTU (2 equivalents per amine group) and DIPEA (4 equivalents per amine group) for 10 min in amine-free DMF. PEG amine was separately dissolved in amine-free DMF and added to the above solution; the mixture was left to react overnight. This solution was precipitated twice in pre-chilled diethyl ether and dialyzed against deionized water in dialysis tubing (MWCO 5 kDa) for 2 days to remove unreacted species. The product was then lyophilized and dissolved in deionized water to make stock solutions at a final concentration of 20 wt% and stored in a −80 °C freezer. The photoinitiator, lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP), was synthesized as previously reported.42
Synthesis of Microgels by Inverse Suspension Polymerization
Microgels were prepared using an inverse-suspension polymerization where the hydrophilic aqueous phase contained all monomer, initiator and crosslinker. The aqueous phase was suspended in a hydrophobic continuous phase, which was composed of hexane with 3 wt% of 3:1 weight ratio of SPAN™ 80 (sorbitan monooleate) and Tween™ 80 (polyethyleneglycol-sorbitanmonooleate).43 The aqueous phase, which contained 100 μl of 10 mM 10 kDa 8-arm PEG-norbrnene, 36 mM Dithiothreitol (DTT), 10 mM LAP and 4 mM sensor peptide dissolved in PBS buffer solution, was added to 900 μl of continuous phase. A micrometer sized suspension was prepared by quickly vortexing this solution, and the suspension was stabilized during polymerization by a stabilizer (SPAN™ 80 and Tween™ 80) and continuous mechanical stirring at 150 rpm. The aqueous phase of the suspension was polymerized into insoluble microgels by irradiating with 4 mW/cm2 of 365 nm light for 3 min using an Omnicure series 1000 (EXFO) light. Excess SPAN™ and TWEEN™ was removed by washing with hexanes, 3 times, isopropanol once and PBS twice. Finally, 1 wt% microgels solutions were refrigerated, stored, and used within a month.
Culture of Melanoma Cells
The A375 cell line was cultured in RPMI 1640 media without phenol red, supplemented with 10% fetal bovine serum (FBS), 100 U ml−1 penicillin and 100 μg ml−1 streptomycin. Cells were cultured on tissue culture treated flasks at 37 °C in 5% CO2 atmosphere an incubator.
Characterization of Microgel Size and Analysis of the Fluorescence Reporter
Fluorescent images of microgel sensors were taken from a spinning disk confocal setup (Operetta PerkinElmer® High Content Screening System), and both the size and fluorescence intensity were analyzed using the ‘Find Nuclei’ function in the Harmony® software, and only the particles that had a roundness of higher than 0.7 were utilized for analysis. A Cermax Xenon Fiber (Excelitas) was used as the optic light source, and the fluorescence of microgels was scanned by excitation at 460~490 nm and measurement of emission at 500~550 nm for 200 ms with an excitation power of 50%. To analyze the size distribution of the microgel sensors, 2 μl of a 1 wt% solution of particles and 100 μl of PBS solution were added to a well plate (384 PerkinElmer® cell carrier), and this solution remained static for 30 min before taking images to allow the microparticles to settle to the bottom of the wells. The fluorescent images of microgel sensors were acquired by taking a z-stack at 5 μm intervals. The diameter of over 900 microgel sensors was analyzed from their maximum z-stack projection images assuming a circular shape.
Measurement of cellular uptake, cell viability, number, and morphology
100 μl of concentrated A375 melanoma cell solutions (≥ 105 cells/ml) was transferred to a glass bottom well plate (384 PerkinElmer® cell carrier). After 4 hours, unattached cells and media were aspirated and the wells were refilled with 100 μl of fresh RPMI media supplemented with 10% FBS. In each well, the microgel sensors (synthesized with 0.1 mM, 0.4 mM, or 4 mM sensor peptides) or soluble sensor peptides (0.1 mM, 1 mM) were added. To test cellular uptake of sensor molecules, bright-field and z-stack fluorescent images were collected immediately after the addition of sensor molecules and again at one-day after.
To test cell viability, dead cells were identified by ethidium homodimer uptake (Molecular Probes). After 24 hours of incubation with the sensor molecules, cell culture media was aspirated, and the A375 cells were exposed to the ethidium homodimer solution (2 μl/ml) for 30 min, and then rinsed in RPMI media. The total number of cells were identified by the bright-field image, and the number of dead cells were identified by the red fluorescence of DNA-bound ethidium homodimer. After 3 days of incubation, cells were labelled with CellTracker™ Red (Molecular Probes), and cellular morphology and number were quantified from the fluorescence images by Harmony® software.
Monitoring Proteolytic Activity in Collagenase Solutions
As a positive control, changes in the fluorescence intensity of the microgel sensors was followed during exposure to various collagenase solutions. 10 ng/ml, 100 ng/ml, 200 ng/ml, 1 μg/ml, 5 μg/ml of Collagenase Type-1 (Worthington, 300 U/mg) solutions were prepared in PBS and 100 μl of this was added to each well (384 PerkinElmer® cell carrier) containing 2 μl of 1 wt% microgel sensors. Fluorescence images (70 μm z-stack at 5 μm intervals) of microgel sensors were taken automatically every 5 min for an hour. The initial fluorescence intensity of the microgel sensors before adding collagenase solutions was denoted as F0 to calculate the fluorescence change (F/F0) of microgels over time.
Monitoring Proteolytic Activity of Cancer Cells as a Function of Culture Conditions
A375 cells were cultured using various media conditions (RPMI media supplemented with or without either 10% FBS or 33 μg/ml of human fibronectin (Alfa Aesar)), and the fluorescence change (F/F0) in the microgel sensors was monitored as a real time measure of MMP activity. To study some of the potential pathways involved, 10 μM FAK inhibitor-14 (Y15, Sigma)44 or 500 μM ERK inhibitor (3-(2-Aminoethyl)-5-((4-ethoxyphenyl)methylene)-2,4-thiazolidinedione, Calbiochem)45 was added to the fibronectin-supplemented culture solution. 10 μl of 106 A375 cells/ml of solution and 10 μl of 1 wt% microgel sensors were added to 80 μl of each culture media in the well plate (384 PerkinElmer cell carrier). As a negative control, the fluorescence changes in the microgels incubated in each culture media without A375 melanoma cells were monitored. Fluorescence images (70 μm z-stack at 5 μm intervals) of microgel sensors in each media condition with or without cells were collected by an Operetta PerkinElmer® High Content Screening System. The initial fluorescence of the microgel sensors, right after adding the culture media, was utilized as F0 to calculate fluorescence change (F/F0) of the microgels. To normalize fluorescence changes of microgels, F/F0 of the microgel sensors with A375 cells were divided by the fluorescence change of microgels without A375 cells in each media conditions.
Monitoring Spatiotemporal Collagenase Activity in 3D
For 4-dimensional (time and 3-dimensional space) monitoring of MMP activity, three types of matrix gel solutions (precursor solutions prior to gelation) were prepared. Non-degradable hydrogels were prepared with 5 mM 40 kDa 8-arm PEG-norbornene, 36 mM DTT, 10 mM LAP and 0.5 wt% microgels. MMP-degradable hydrogels were polymerized with 5 mM of 40 kDa 8-arm PEG-norbornene, 36 mM DTT, 10 mM LAP and 0.5 wt% microgels. 15 μl of each solution was added to the wellplate (384 PerkinElmer cell carrier, well dimension 3.26 mm (length) × 3.26 mm (width) × 13.1 mm (height)), and microgels were immediately encapsulated in the matrix gel solution by illuminating the solution under 20 mW/cm2 of 365 nm UV light for 3 min. Lastly, gelatin hydrogels were prepared from 15 wt% gelatin (Gelatin from porcine skin, gel strength 300, Type A, Sigma) solubilized in PBS with 0.5 wt% microgel sensors suspended in the solution. To solidify the gelatin, the mixture was transferred from a 97 °C dry block heater to a 4 °C refrigerator for 5 min. After swelling in 80 μl of PBS for more than 2 hours, 20 μl of concentrated collagenase solution (5 mg/ml) was added on top of the prepared hexahedral shaped hydrogels (3.26 mm × 3.26 mm × ~ 1 mm). Spatial information related to collagenase activity was acquired by imaging the sensor particles from the bottom of the gel to top every 20 min for 200 min, using an Operetta PerkinElmer® High Content Screening System.
Monitoring of MMP Activity of Cancer Cell Aggregates in 4D
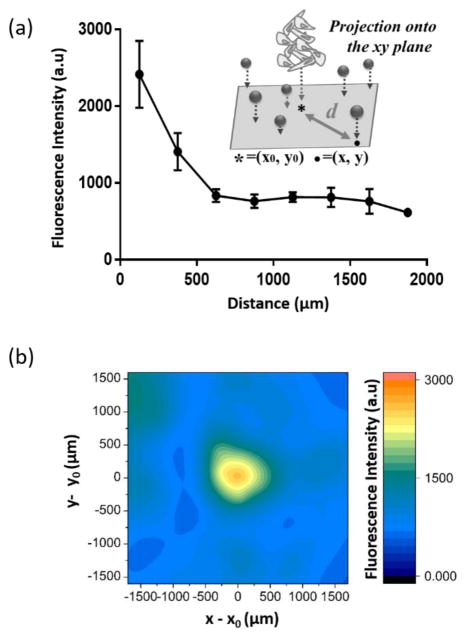
15 wt% gelatin was solubilized in PBS using a 97 °C dry block heater, and 0.5 wt% microgels was suspended in the solution. 20 μl of this suspension was then added to well plates (384PerkinElmer cell carrier) and placed in a 4 °C refrigerator for 5 min to solidify the gelatin. To make a cylindrical empty well inside the gelatin hydrogel, hydrogels were punctured with a syringe needle (BD PrecisionGlide Needle) which was cut to a length of 1cm. 100 μl of a A375 cell suspension (106 cells/ml) was then added to the top of each hydrogel, and the cells were centrifuged down to the empty well inside the hydrogel at 1200 rpm for 2 min to from an aggregate. Next, the remaining supernatant was aspirated, and 10 μl of the 15 wt% gelatin solution was added to the top of each cell-laden hydrogel and quickly placed in 4 °C refrigerator for 5 min. 100 μl of cell culture media was added to each well, and the prepared hydrogel constructs, embedding the cancer cell aggregate, were incubated in the incubator for 3 days before taking images. Fluorescence of microgel sensors surrounding the cell aggregates were monitored by collecting z-stack images using a spinning disk confocal microscope (Perkin Elmer, Operetta). From the fluorescence images, the x, y position of each microgel center and cell aggregate center (x0, y0) was calculated, and the distance from the cell aggregate center to each microgel was calculated as .
Statistical Analyses
All of the statistical analyses used in the experiments were conducted using a one-way ANOVA coupled with Tukey’s multiple comparison tests (Prism 6; GraphPad Software Inc). The mean microgels fluorescence intensity of each maximum z-stack projection image (0.014 cm2 or 0.0035 cm2 area), containing at least 5 microgel sensors per image, was taken as an individual data-point. Data were collected from at least three replicate measurements of more than two individual experiments, and presented as mean ± SEM. Asterisk indicates a statistically significant difference between each group (* P<0.05, ** P<0.01, *** P<0.001, ****P<0.0001).
Results and Discussion
Design and Synthesis of the Microgel Sensors
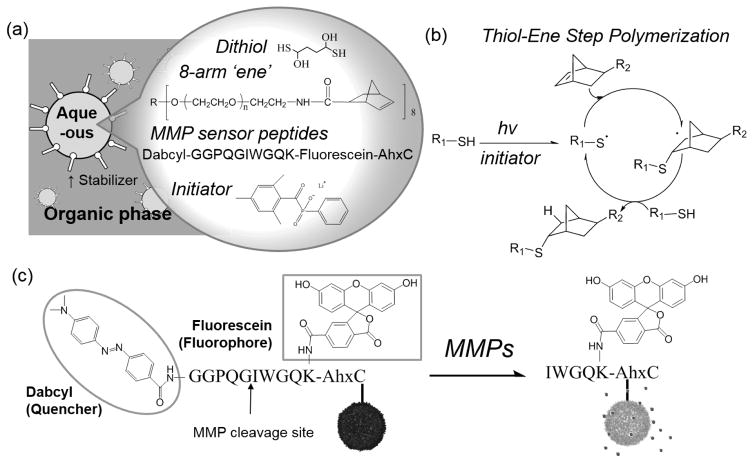
Figure 1 illustrates the general formulation and methods for synthesizing the microgel sensors. To fabricate micron-sized particles, an inverse suspension polymerization method was utilized, where all reagents (i.e., monomers, crosslinkers, protease sensor peptides and initiators) were solubilized in an aqueous solution, and the resulting solution was suspended in a hydrophobic continuous phase (Figure 1-a). To combine the advantages of a step-growth polymerization with the facile introduction of cysteine-containing sensor peptides, a thiol-ene photoclick reaction was used, specifically reacting PEG-norbornene with DTT (Figure 1-b). The step growth thiol-ene photopolymerization was conducted under ambient temperature and atmosphere by illumination with UV light (4 mW/cm2, centered around 365 nm) for 3 min. Here, initiators form radicals under UV light illumination and react with thiols to form thiyl radicals. Thiyl radicals attack on the site of electron rich norbornenes to begin the step growth polymerization. The reaction propagates as thiyl radicals are regenerated by norbornene radicals. Since the hydrogel microspheres synthesized by this method incorporate the functional peptides uniformly and throughout the bulk of the particle, the microgels have a high loading capacity for sensor peptides. As a model fluorogenic substrate that can detect proteolytic activity, we used an MMP- cleavable FRET-based sensor peptide that has been previously reported.29 This sensor peptide is composed of a proteolytically cleavable GPQG ↓ IWGQ motif and a fluorophore (fluorescein) - quencher (dabcyl) FRET pair (Figure 1-c). When the fluorescein and dabcyl FRET pairs are in close proximity (e.g., usually under 5~10 nm), the transfer of excited energy from the donor to acceptor molecule is efficient46 (i.e., EFRET = 1/(1 + (R/R0)6)) and fluorescence of the fluorescein is effectively quenched. Proteolytic activity was detected by monitoring any increases in the fluorescence of the microgel sensors as cleaved quencher molecules (dabcyl) diffuse away from the microgels and FRET from fluorescein to dabcyl disappears.
Figure 1.
(a) Schematic illustration of the inverse suspension polymerization method along with the chemical composition of the microgel sensors. Dithiothreitol (DTT) is the dithiol, 8-arm 10 kDa PEG-norbornene is the multi-ene, G(dabcyl)GPQG↓IWGQK(Fluorescein)AhxC acts as the functional sensor peptide and lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) serves as the photoinitiator. (b) Mechanism of the thiol-ene reaction. Reproduced with permission from ref 5. Copyright 2009 Wiley. (c) Protease sensing mechanism of the microgel sensors.
Size Distribution and Cellular Uptake Analysis
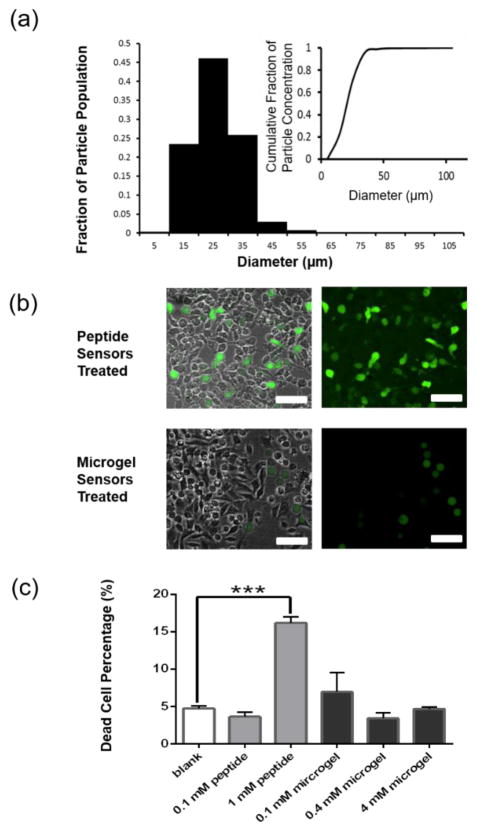
The diameter of over 900 microgel sensors was analyzed based on their maximum cross-sectional area taken from z-stack images of microgel sensors. The number average diameter of the synthesized microgels (28 ± 5 μm, n>900) was similar to the size of A375 melanoma cells (24 ± 11 μm, n>2000). Over 98.6% of the particles had a number averaged diameter between 10 μm and 50 μm (Figure 2-a). We targeted microgel sensors in this size range to minimize internalization by cells, and experiments were conducted to verify this.
Figure 2.
Size distribution and cellular uptake of the microgel sensors. (a) The size distribution of the microgel sensors was analyzed from fluorescent images (n=990). (b) Fluorescent images of A375 cells which were incubated with 1 mM Peptide sensors or microgel sensors (synthesized with 4 mM sensor peptides). Scale bars, 100 μm. (c) Percentage of dead cells detected by staining with ethidium homodimer.
To compare cellular uptake of sensor peptides, when solubilized in media or incorporated in microgel sensors, A375 melanoma cells were incubated with 0.1 mM sensor peptides, 1 mM sensor peptides or sensor peptides incorporated into microgel sensors (synthesized with 0.1 mM, 0.4 mM or 4 mM sensor peptides) with a negative control without any sensing molecules. Figure 2-b shows fluorescent images of the sensing molecules overlayed with a bright-field image of the plated cells. The image was taken immediately (< 5 min) after either 1 mM of soluble sensor peptides or 754 ± 97 sensor microgels (containing 4 mM sensor peptides) were introduced to the A375 cell culture. This representative image shows a high degree of cellular uptake of the sensor peptides by A375 cells, and the uptake is observed as the high intracellular fluorescence viewed in the combined brightfield and fluorescent images. After 1 day of incubation with 1 mM of the soluble sensor peptides, localized intracellular fluorescence, as well as alteration in the A375 cell shape were observed (Figure S1-a, left, Supporting information). In contrast, when the sensor peptides were immobilized in the microgels, there was negligible uptake by the cells. The microgels remained in the extracellular space, and negligible fluorescence was observed intracellularly in the A375 cells (Figure 2-b, right, Figure S1-a, right, Supporting information). The microgels can be observed as the spherical objects in the fluorescent images (when a high excitation power is used). This results demonstrates that the microgels prevent cellular uptake and preserves the ability of the microgels to potentially detect spatial proteolytic information of cell-secreted enzymes.
Further analysis of cell viability, using a live/dead cell viability assay (dead only for this assay), showed possible cytotoxicity of the sensor peptides when they were introduced at higher concentrations (i.e., >1 mM). Here, dead cells were fluorescently labeled red by ethidium homodimer-1, and the total cell population was observed by bright field images. Figure 2-c and Figure S1-b (Supporting Information) quantify the cellular viability when A375 cells were incubated with microgel sensors and sensor peptides for 3 days. When A375 cells were incubated with 1 mM of sensor peptides in solution, >16% of the cells fluoresced red. In comparison to this, sensor peptides immobilized in microgels could be incorporated at high levels without measurable increases in cytotoxicity. When A375 cells were incubated with microgel sensors, which were synthesized with 0.1 mM, 0.4 mM, or 4 mM sensor peptide concentrations, the percentage of dead cells was not significantly different from the percentage of dead cells under normal culture conditions.
In addition, we observed that the morphology and number of cells can be affected by the sensor peptides when the sensor peptides are introduced solubly. After 3 days of incubation with a 1 mM sensor peptide solution, there was a significantly lower average cell number (1400 ± 200 cells/cm2) and cell area (180 ± 10 μm2) compared to A375s cultured without the soluble peptide (average cell number (11000 ± 900 cells/cm2) and cell area (230 ± 10 μm2)) (Figure S1-c, Supporting Information). This result further motivated our approach to immobilize the peptide sensors into the microgels, as highly concentrated, soluble peptide sensors can have unintended effects on cell morphology and survival.
Monitoring Proteolytic Activity in Solution in Real Time
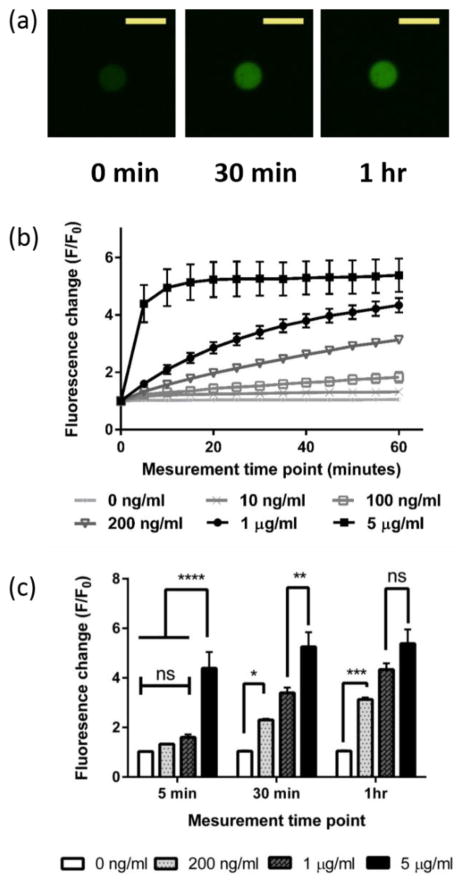
The fluorescence change (F/F0) in the microgel sensors was measured in real time when exposed to various concentrations of collagenase solutions. Figure 3-a shows a typical image of the microgel sensors when exposed to a 1 μg/ml collagenase solution. As collagenase cleaves the sensor peptides incorporated in the microgels, the fluorescence intensity increases with time. A fluorescence intensity profile, within the microgels and along the diameter, showed evenly distributed fluorescence before and after cleavage of the quencher group by collagenase. This supports the assumption that the sensor peptides were incorporated evenly throughout the microgels and that the MMP activity was not sterically excluded from any regions within the particles (Figure S2, Supporting Information).
Figure 3.
Real time monitoring of fluorescence change (F/F0) in the microgel sensors in a collagenase solution. (a) Typical image of microgel sensors in a 1 μg/ml collagenase solution right after exposure (left), after 30 minutes (middle) and after an hour (right). Scale bars, 50 μm. (b) Fluorescence change (F/F0) monitored for 1 hour in various concentrations of collagenase solution. (c) Fluorescence changes (F/F0) in the microgel sensors in 0 ng/ml, 200 ng/ml, 1 μg/ml and 5 μg/ml collagenase concentrations after 5 min, 30 min and after an hour.
Figure 3-b shows the fold change in fluorescence intensity of the microgel sensors as a function of time, normalized to the initial fluorescence intensity of the microgels. When the microgel sensors were exposed to collagenase solutions of varying concentrations, from 10 ng/ml to 5 μg/ml, a maximum ~5 fold increase in F/F0 was observed. Although the maximum F/F0 was smaller than that of the soluble form sensor peptide (F/F0 = 9.2 ± 0.6, n=5), possibly due to radical mediated damage of the fluorophore or a quencher, the lowest limit of detection (LOD) was under 10 ng/ml throughout all measurements taken (Table S1, Supporting Information). In comparison, no significant change in fluorescence was observed from the 0 ng/ml control over 1 hour. In the low collagenase concentration range (10 ~ 200 ng/ml), a linear increase of F/F0 was observed over the course of an hour. In the higher collagenase concentration range (1 ~ 5 μg/ml), the slope of F/F0 was steeper at the initial time point, but eventually reached a plateau as most of the fluorogenic sensor peptides were cleaved. The dabcyl (quencher) containing peptide fragments diffuse away and only the fluorophore (fluorescein) containing peptide fragments remained in the microgel sensors.
Figure 3-c demonstrates that the sensitivity of the microgel sensors was dependent on the time point and concentration of the collagenase solution (Figure 3-c). In highly concentrated collagenase solutions, the microgel sensors showed the highest sensitivity at earlier measurement time points, before most of the sensor peptides were cleaved. In dilute collagenase solutions, higher sensitivity was obtained by extending the measurement time. For example, microgel sensors in 5 μg/ml collagenase solution showed significantly higher fluorescence changes (p ≤ 0.0001) than the microgel sensors in 1 μg/ml collagenase solution after 5 min, but showed negligible fluorescence changes after 1 hour. In contrast, microgel sensors in a 200 ng/ml collagenase solution did not show a significant fluorescence change after 5 min of exposure. As the exposure time to collagenase solution increased, the level of significance also increased. The p-value for this was ≤0.05 after 30 min and ≤0.001 after 1 hour. These results show that the optimal measurement time point can be chosen depending on the target collagenase concentration. These results also show that higher sensitivity at the lower protease concentration can be obtained by choosing longer exposure time to the protease solution.
Measuring Proteolytic Activity of Cancer Cells in Serum-Free versus Serum-Supplemented Media
To better assess whether the microgel sensors can detect changes in cellular MMP activities in 2-dimensional culture environments, A375 cells were cultured in media supplemented with either serum, fibronectin, or without any serum components. Culture conditions were chosen based on published reports showing upregulated gelatinase (MMP-2 and MMP-9) activities in the presence of fibronectin47–49. In these reports, MMP activity was detected using conventional zymography, thus MMP activity could not be monitored in real time.
Here, A375 melanoma cells were cultured with microgel sensors in RPMI media supplemented with either 10% serum, 33 μg/ml of fibronectin, or without any serum components. A375 melanoma cells, which show upregulated MMP-2 expression, were chosen as model cancer cells to compare our results with published results47. Additionally, microgel sensors were incubated without cells in each culture condition to ensure that there was no significant cleavage or inhibition of cleavage of the microgel sensors due to serum components (Figure S3, Supporting Information). After 2 days of incubation, the microgel fluorescence intensity increased by 1.8 ± 0.5 fold or 2.5 ± 0.2 fold in serum supplemented or fibronectin supplemented media, respectively. In comparison, no significant fluorescence change was detected for the microgel sensors when they were incubated with A375 cells without serum components. Without serum components, the microgels showed a delayed and lower fluorescence change (F/F0 = 2.0 ± 0.1) after 4 days of incubation in comparison to the serum supplemented (F/F0 = 2.7 ± 0.8) or fibronectin supplemented (F/F0=3.0 ± 0.5) conditions. This result shows the ability to detect the difference in MMP activity in cells as a function of culture environment and over time. Here, delayed MMP activation and lower proteolytic activities were observed in the absence of serum, as serum-components likely interact with tumor cells to promote MMP expression and activation.
Among the serum components, we found that fibronectin is one of the key factors that can activate MMPs. The microgel sensors in fibronectin supplemented media with A375 cells showed significantly higher (p<0.01) fluorescence changes after only two days compared to the microgel sensors in serum supplemented media. As most of the sensor peptides are cleaved in the microgels by day 4, the difference decreases. To examine potential signaling pathways, A375 cancer cells were incubated with either FAK or ERK inhibitors. In both cases, a significant decrease in MMP activity was observed (Figure 4-b) after 12 hours of culture, supporting the notion that MMP activation is related to FAK/ERK pathway and is consistent with the previous reports based on zymography techniques.50,51
Figure 4.
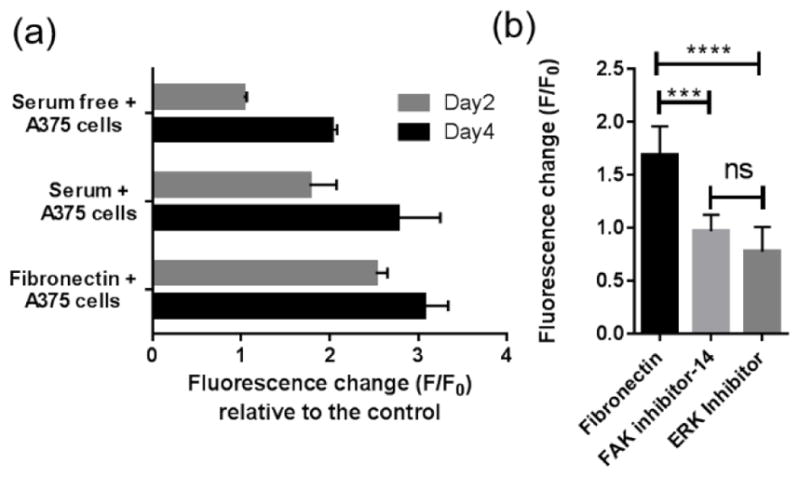
(a) Fluorescence changes (F/F0 ) of microgel sensors in various culture media after two days and after four days of incubation with A375 cells. 0.1 wt% microgel sensors and 105 cells/ml of A375 cells were incubated in 100 μl culture media supplemented with either 10% FBS, 33 μg/ml of fibronectins, or without any serum components. The highest fluorescence changes was observed in the fibronectin supplemented media indicating MMP activation in presence of this adhesion protein. (b) Treatment with either FAK inhibitor (FAK inhibitor 14) or ERK inhibitor (3-(2-Aminoethyl)-5-((4-ethoxyphenyl)methylene)-2,4-thiazolidinedione) led to decreased MMP activation for A375s in the fibronectin supplemented media.
Monitoring Spatiotemporal Collagenase Activity in 3D
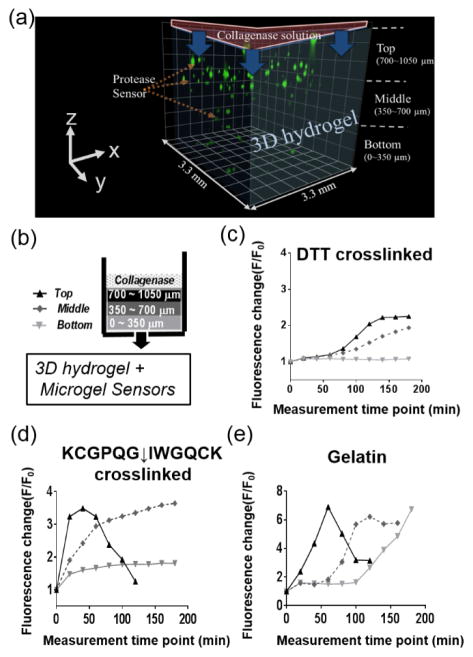
Microgel sensors were used to detect spatiotemporal MMP activity by embedding them in either synthetic hydrogels (PEG) or naturally derived hydrogels (gelatin). To explore spatiotemporal MMP activity, first the gels were exposed to collagenase solutions, introduced at the top gel surface, and the fluorescence change of the microgel sensors was monitored every 10 min for 2 hours as the collagenase solution diffused through the gel. The microgels were categorized by their location in the z-direction within the 3-dimensional hydrogels. Microgels located at the top (700~1050 um), middle (350~700 um), and bottom (0~350 um) planes were analyzed in real time (Figure 5-a,b). The synthetic hydrogels used to encapsulate the microgel sensors were prepared by photo-initiated thiol-ene polymerization and crosslinked with either non-degradable DTT or proteolytically degradable KCGPQG ↓ IWGQCK. By simply suspending the microgel sensors within the pre-polymer solution, the microgels were easily immobilized throughout the bulk of the hydrogel and at a spacing that was controlled by the initial microgel loading concentration.
Figure 5.
(a) Three-dimensionally reconstituted image of microgel sensors encapsulated in various hydrogels. A collagenase solution was applied to the top of hydrogel and changes in F/F0 of the microgel sensors were analyzed as a function of space and time. Microgel sensors were categorized according to their location in the z-direction as top (700~1050 μm), middle (350~700 μm) and bottom (0~350 μm). (b,c,d) Spatiotemporal fluorescence change (F/F0) of microgel sensors after an exposure to a 5 mg/ml of collagenase solution on the top of (c) DTT crosslinked PEG hydrogels and (d) KCGPQG↓IWGQCK crosslinked PEG hydrogels and (e) gelatin hydrogels.
In the DTT crosslinked (MMP non-degradable) PEG hydrogels (Figure 5-c), rapid and large fluorescence changes were observed at the top compared to the middle and the bottom of the hydrogel, which provides a measure of the diffusion of collagenase from the top to the bottom of the ~1 mm thick gel. Fluorescence intensity of the microgel sensors increased at the middle of the hydrogel after 140 min. This result agrees well with the theoretically estimated diffusion time scales. Assuming the hydrodynamic radius (rH) of MMPs as 5 nm, the mesh size (ζ) of the thiol-ene polymer network as 15 nm,5,52,53 the diffusion coefficient of MMPs in free solution (Dwater) as 4.5×10−7 cm2 s−1,54 and the cross-sectional radius of the polymer chain (rf) as 0.51 nm,55 the diffusion coefficient of the MMPs (D) inside the hydrogel was approximated by the following equation.56
| (1) |
From this equation, the value for the diffusivity of collagenase (D) in the PEG hydrogel was calculated to be 1.6×10−7 cm2 s−1. Then, the approximate diffusion time scale for MMPs to travel 350 μm can be estimated using the following equation, time=distance2/2D, which is derived from Fick’s law.57 Solving this equation yield an estimated time scale for diffusion of 65 min, which agrees well with our results.
When the microgel sensors were encapsulated in a KCGPQG↓IWGQCK crosslinked (MMP-degradable) PEG hydrogel (Figure 5-d), the time scale for diffusion to the bottom of the hydrogel was much faster, as the diffusivity of the collagenase increases with gel degradation and the gel dimensions decrease with erosion. The microgels near the top of the hydrogel showed an abrupt fluorescence increase at the beginning, but the fluorescence intensity decreased as the hydrogel eroded and the microgel sensors settled down to the middle of the hydrogel. The fluorescence change of the microgel increased over 1.5 fold in the middle and bottom in 20 min and 40 min, respectively. The faster and higher change in fluorescence in the microgels shows a faster diffusion of MMP in proteolytically degradable hydrogels than in the non-degradable hydrogels. Similar spatiotemporal protease activity profiles (i.e., a spatiotemporal fluorescence changes in the microgels) were observed when the microgel sensors were encapsulated in gelatin (Figure 5-e), a denatured form of collagen that is a natural ECM component and contains MMP degradable motifs. The immediate fluorescence change was observed at the top of hydrogel followed by a fluorescence change at the middle after 40 min and at the bottom after 100 min. The fluorescence intensity at the top of the hydrogel decreased as the gelatin gel eroded. Collectively, these results show the ability of the microgel sensors to monitor local changes in protease activity, and that proteolytically degradable PEG hydrogels were degraded by collagenases in a manner that is very similar to a gelatin hydrogel, which is susceptible to collagenases. Finally, the technique allows one to study the diffusion of various enzymes throughout hydrogels, when non-degradable hydrogels are employed.
Spatiotemporal Proteolytic Activities Surrounding Tumor Cell Aggregates
It is often very difficult to predict spatial activity of proteases because protease activities are regulated by their natural activators and inhibitors. In addition, the diffusivity of proteases is dynamically changing as protease secreted from cells remodel local microenvironments. To address these problems, protease sensor microgels were utilized to visualize in situ protease activity in 3D from A375 melanoma cells. To mimic aspects of a tumor microenvironment, A375 cell aggregates were embedded in a gelatin hydrogel (Figure S4, Supporting Information). The gelatin gel was prepared to have a gel-sol transition temperature higher than 37 °C by using higher bloom and weight percent gelatin.58,59 The change in fluorescence over time (F/F0) of microgels located near the bottom of the hydrogels was monitored, and the correlation of F/F0 with the distance from the cell aggregate center in the xy plane was analyzed. After 3 days of incubation, we observed higher local fluorescence of microgel sensors near the cell aggregate center. Particles were grouped based on their distance from the cell aggregate center, and F/F0 of each particle was analyzed. Results in Figure 6-a and Figure S5 indicate that more proteolytic activity was observed near the gel-aggregate interface (distance<500 μm). Figure 6-b shows a contour map of the fluorescence of each microgel sensors (n=413) when co-encapsulated with a cell aggregate (radius = 230 ± 3 μm). In this graph, (0,0) indicates the cell aggregate center (x0, y0). This graph visualizes the secreted protease activity of a tumor aggregates and shows that significant proteolytic activities were observed in close proximity to cell aggregates (distance<500 μm). In addition, this result showed that MMP activity is a function of distance from the A375 cancer cell aggregate in the scale of a few hundred micrometers.
Figure 6.
(a) Fluorescence intensity in the microgel sensors with distance (d) from the cell aggregate center after 3 days’ incubation with a A375 melanoma cancer cell aggregate. Z-stack images were projected in the xy plane such that (x0,y0) is the location of the cell aggregate center and (x,y) represents the center of each microgel. The distance from the cell aggregate center to each microgel was calculated as . (b) Contour map depicting the fluorescence of each microgel surrounding a cell aggregate, where the origin of the graph (x0,y0), represents the center of the cell aggregate. The contour map was compiled from superimposed fluorescence images for all microgel sensors collected from three different A375 cell aggregates.
Conclusions
Microgel sensors were synthesized with proteolytically susceptible fluorogenic peptides to detect spatiotemporal proteolytic activity of cells. The number averaged diameter of the microgel sensors was 28 μm ± 5 μm and this size minimized cellular uptake by A375 melanoma cells. The microgel sensors were used to detect collagenase activities as low as 10 ng/ml and were further used to visualize differences in MMP activity of A375s in the presence and absence of fibronectins. The microgel sensors were easily incorporated in hydrogels used for 3D cell culture, and both synthetic (PEG) and natural (gelatin) hydrogels were prepared by simply suspending the microgel sensors in the precursor solutions prior to gelation. Studies with collagenase solutions, allowed spatiotemporal visualization of local collagenase activity on the order of hundreds of microns. Finally, a model cancer environment was constructed by aggregating A375 cells and embedding them in a gelatin hydrogel loaded with the microgel sensors. Results of spatial MMP activity allowed visualization of a high local activity near the cell aggregate, and spatially varying MMP activity several hundreds of microns away that depends on diffusion of the MMP from the cancer cell aggregate in 3D microenvironments.
Supplementary Material
Acknowledgments
We thank S. Tang for his help in revising the manuscript and discussions. We thank Howard Hughes Medical Institute and the National Institutes of Health research grant (R21 EB018505) for financial support of this work, and the U.S. Department of Education’s GAANN Program for fellowships to DSS.
Funding Sources
This research was supported by the National Institutes of Health Grant #R21EB018505.
Footnotes
Supporting Information. Additional characterization of microgel sensors, including cellular uptake, uniformity, fluorescence changes of microgel sensors in various media, illustration of cellular aggregate preparation inside 3-dimensional hydrogels, and typical images of microgel sensors surrounding A375 cell aggregates.
References
- 1.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert PM, Havenstrite KL, Magnusson KEG, Sacco A, Leonardi NA, Kraft P, Nguyen NK, Thrun S, Lutolf MP, Blau HM. Substrate Elasticity Regulates Skeletal Muscle Stem Cell Self-Renewal in Culture. Science (80-) 2010;329(5995):1078–LP-1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang C, Tibbitt MW, Basta L, Anseth KS. Mechanical memory and dosing influence stem cell fate. Nat Mater. 2014;13(6):645. doi: 10.1038/nmat3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tokuda EY, Leight JL, Anseth KS. Modulation of matrix elasticity with PEG hydrogels to study melanoma drug responsiveness. Biomaterials. 2014;35(14):4310–4318. doi: 10.1016/j.biomaterials.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fairbanks BD, Schwartz MP, Halevi AE, Nuttelman CR, Bowman CN, Anseth KS. A Versatile Synthetic Extracellular Matrix Mimic via Thiol-Norbornene Photopolymerization. Adv Mater. 2009;21(48):5005–5010. doi: 10.1002/adma.200901808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker BM, Chen CS. Deconstructing the third dimension–how 3D culture microenvironments alter cellular cues. J Cell Sci. 2012;125(13):3015–3024. doi: 10.1242/jcs.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loessner D, Stok KS, Lutolf MP, Hutmacher DW, Clements JA, Rizzi SC. Bioengineered 3D platform to explore cell–ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials. 2010;31(32):8494–8506. doi: 10.1016/j.biomaterials.2010.07.064. DOI: http://dx.doi.org/10.1016/j.biomaterials.2010.07.064. [DOI] [PubMed] [Google Scholar]
- 8.Lee J, Abdeen AA, Wycislo KL, Fan TM, Kilian KA. Interfacial geometry dictates cancer cell tumorigenicity. Nat Mater. 2016;15(8):856–862. doi: 10.1038/nmat4610. [DOI] [PubMed] [Google Scholar]
- 9.Rosales AM, Anseth KS. The design of reversible hydrogels to capture extracellular matrix dynamics. Nat Rev Mater. 2016;1:15012. doi: 10.1038/natrevmats.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang C, Werb Z. The many faces of metalloproteases: cell growth, invasion, angiogenesis and metastasis. Trends Cell Biol. 2001;11(11):S37–S43. doi: 10.1016/s0962-8924(01)02122-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franco SJ, Huttenlocher A. Regulating cell migration: calpains make the cut. J Cell Sci. 2005;118(17):3829–3838. doi: 10.1242/jcs.02562. [DOI] [PubMed] [Google Scholar]
- 12.Lamkanfi M, Festjens N, Declercq W, Berghe T Vanden, Vandenabeele P. Caspases in cell survival, proliferation and differentiation. Cell Death Differ. 2007;14(1):44–55. doi: 10.1038/sj.cdd.4402047. [DOI] [PubMed] [Google Scholar]
- 13.Macfarlane SR, Seatter MJ, Kanke T, Hunter GD, Plevin R. Proteinase-activated receptors. Pharmacol Rev. 2001;53(2):245–282. [PubMed] [Google Scholar]
- 14.Ossovskaya VS, Bunnett NW. Protease-activated receptors: contribution to physiology and disease. Physiol Rev. 2004;84(2):579–621. doi: 10.1152/physrev.00028.2003. [DOI] [PubMed] [Google Scholar]
- 15.Kidd VJ. Proteolytic activities that mediate apoptosis. Annu Rev Physiol. 1998;60(1):533–573. doi: 10.1146/annurev.physiol.60.1.533. [DOI] [PubMed] [Google Scholar]
- 16.Cardone MH, Roy N, Stennicke HR, Salvesen GS, Franke TF, Stanbridge E, Frisch S, Reed JC. Regulation of cell death protease caspase-9 by phosphorylation. Science (80-) 1998;282(5392):1318–1321. doi: 10.1126/science.282.5392.1318. [DOI] [PubMed] [Google Scholar]
- 17.Rutschow S, Li J, Schultheiss H-P, Pauschinger M. Myocardial proteases and matrix remodeling in inflammatory heart disease. Cardiovasc Res. 2006;69(3):646–656. doi: 10.1016/j.cardiores.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 18.Burrage PS, Mix KS, Brinckerhoff CE. Matrix metalloproteinases: role in arthritis. Front Biosci. 2006;11(1):529–543. doi: 10.2741/1817. [DOI] [PubMed] [Google Scholar]
- 19.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141(1):52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Overall CM, Kleifeld O. Validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat Rev Cancer. 2006;6(3):227–239. doi: 10.1038/nrc1821. [DOI] [PubMed] [Google Scholar]
- 21.Noël A, Jost M, Maquoi E. Matrix metalloproteinases at cancer tumor–host interface. Semin Cell Dev Biol. 2008;19(1):52–60. doi: 10.1016/j.semcdb.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 22.Liotta LA, Kohn EC. The microenvironment of the tumour–host interface. Nature. 2001;411(6835):375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 23.Iyengar P, Espina V, Williams TW, Lin Y, Berry D, Jelicks LA, Lee H, Temple K, Graves R, Pollard J, Chopra N, Russell RG, Sasisekharan R, Trock BJ, Lippman M, Calvert VS, Petricoin EF, III, Liotta L, Dadachova E, Pestell RG, Lisanti MP, Bonaldo P, Scherer PE. Adipocyte-derived collagen VI affects early mammary tumor progression in vivo, demonstrating a critical interaction in the tumor/stroma microenvironment. J Clin Invest. 115(5):1163–1176. doi: 10.1172/JCI23424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moro N, Mauch C, Zigrino P. Metalloproteinases in melanoma. Eur J Cell Biol. 2014;93(1):23–29. doi: 10.1016/j.ejcb.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 25.Houghton AM, Grisolano JL, Baumann ML, Kobayashi DK, Hautamaki RD, Nehring LC, Cornelius LA, Shapiro SD. Macrophage Elastase (Matrix Metalloproteinase-12) Suppresses Growth of Lung Metastases. Cancer Res. 2006;66(12):6149–6155. doi: 10.1158/0008-5472.CAN-04-0297. [DOI] [PubMed] [Google Scholar]
- 26.Noskova V, Ahmadi S, Åsander E, Casslén B. Ovarian cancer cells stimulate uPA gene expression in fibroblastic stromal cells via multiple paracrine and autocrine mechanisms. Gynecol Oncol. 2009;115(1):121–126. doi: 10.1016/j.ygyno.2009.06.026. DOI: http://dx.doi.org/10.1016/j.ygyno.2009.06.026. [DOI] [PubMed] [Google Scholar]
- 27.Mauro L, Catalano S, Bossi G, Pellegrino M, Barone I, Morales S, Giordano C, Bartella V, Casaburi I, Andò S. Evidences that Leptin Up-regulates E-Cadherin Expression in Breast Cancer: Effects on Tumor Growth and Progression. Cancer Res. 2007;67(7):3412–3421. doi: 10.1158/0008-5472.CAN-06-2890. [DOI] [PubMed] [Google Scholar]
- 28.Vandooren J, Geurts N, Martens E, Van den Steen PE, Opdenakker G. Zymography methods for visualizing hydrolytic enzymes. Nat Methods. 2013;10(3):211–220. doi: 10.1038/nmeth.2371. [DOI] [PubMed] [Google Scholar]
- 29.Leight JL, Alge DL, Maier AJ, Anseth KS. Direct measurement of matrix metalloproteinase activity in 3D cellular microenvironments using a fluorogenic peptide substrate. Biomaterials. 2013;34(30):7344–7352. doi: 10.1016/j.biomaterials.2013.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.To T-L, Piggott BJ, Makhijani K, Yu D, Jan YN, Shu X. Rationally designed fluorogenic protease reporter visualizes spatiotemporal dynamics of apoptosis in vivo. Proc Natl Acad Sci U S A. 2015;112(11):3338–3343. doi: 10.1073/pnas.1502857112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rehm M, Dussmann H, Janicke RU, Tavare JM, Kogel D, Prehn JHM. Single-cell fluorescence resonance energy transfer analysis demonstrates that caspase activation during apoptosis is a rapid process. Role of caspase-3. J Biol Chem. 2002;277(27):24506–24514. doi: 10.1074/jbc.M110789200. [DOI] [PubMed] [Google Scholar]
- 32.Packard BZ, Artym VV, Komoriya A, Yamada KM. Direct visualization of protease activity on cells migrating in three-dimensions. Matrix Biol. 2009;28(1):3–10. doi: 10.1016/j.matbio.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu S, Wang Y, Huang H, Pan Y, Chaney EJ, Boppart SA, Ozer H, Strongin AY, Wang Y. Quantitative FRET Imaging to Visualize the Invasiveness of Live Breast Cancer Cells. PLoS One. 2013;8(3):e58569. doi: 10.1371/journal.pone.0058569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang GT, Krafft GA. Automated synthesis of fluorogenic protease substrates: design of probes for alzheimer’s disease-associated proteases. Bioorg Med Chem Lett. 1992;2(12):1665–1668. doi: 10.1016/S0960-894X(00)80452-7. [DOI] [Google Scholar]
- 35.Kimura RH, Steenblock ER, Camarero JA. Development of a cell-based fluorescence resonance energy transfer reporter for Bacillus anthracis lethal factor protease. Anal Biochem. 2007;369(1):60–70. doi: 10.1016/j.ab.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 36.Kaman WE, Hulst AG, Van Alphen PTW, Roffel S, Van Der Schans MJ, Merkel T, Van Belkum A, Bikker FJ. Peptide-based fluorescence resonance energy transfer protease substrates for the detection and diagnosis of Bacillus species. Anal Chem. 2011;83(7):2511–2517. doi: 10.1021/ac102764v. [DOI] [PubMed] [Google Scholar]
- 37.Champion JA, Walker A, Mitragotri S. Role of Particle Size in Phagocytosis of Polymeric Microspheres. Pharm Res. 2008;25(8):1815–1821. doi: 10.1007/s11095-008-9562-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zauner W, Farrow NA, Haines AMR. In vitro uptake of polystyrene microspheres: effect of particle size, cell line and cell density. J Control Release. 2001;71(1):39–51. doi: 10.1016/s0168-3659(00)00358-8. DOI: http://dx.doi.org/10.1016/S0168-3659(00)00358-8. [DOI] [PubMed] [Google Scholar]
- 39.Kaufman LJ, Brangwynne CP, Kasza KE, Filippidi E, Gordon VD, Deisboeck TS, Weitz DA. Glioma Expansion in Collagen I Matrices: Analyzing Collagen Concentration-Dependent Growth and Motility Patterns. Biophys J. 2005;89(1):635–650. doi: 10.1529/biophysj.105.061994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Polackwich RJ, Koch D, Arevalo R, Miermont AM, Jee KJ, Lazar J, Urbach J, Mueller SC, McAllister RG. A novel 3D fibril force assay implicates src in tumor cell force generation in collagen networks. PLoS One. 2013;8(3):e58138. doi: 10.1371/journal.pone.0058138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hahn MS, Liao H, Munoz-Pinto D, Qu X, Hou Y, Grunlan MA. Influence of hydrogel mechanical properties and mesh size on vocal fold fibroblast extracellular matrix production and phenotype. Acta Biomater. 2008;4(5):1161–1171. doi: 10.1016/j.actbio.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fairbanks BD, Schwartz MP, Bowman CN, Anseth KS. Photoinitiated polymerization of PEG-diacrylate with lithium phenyl-2,4,6-trimethylbenzoylphosphinate: polymerization rate and cytocompatibility. Biomaterials. 2009;30(35):6702–6707. doi: 10.1016/j.biomaterials.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewis KJR, Tibbitt MW, Zhao Y, Branchfield K, Sun X, Balasubramaniam V, Anseth KS. In vitro model alveoli from photodegradable microsphere templates. Biomater Sci. 2015;3(6):821–832. doi: 10.1039/c5bm00034c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zang M, Zhang Y, Zhang B, Hu L, Li J, Fan Z, Wang H, Su L, Zhu Z, Li C. CEACAM6 promotes tumor angiogenesis and vasculogenic mimicry in gastric cancer via FAK signaling. Biochim Biophys Acta (BBA)-Molecular Basis Dis. 2015;1852(5):1020–1028. doi: 10.1016/j.bbadis.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 45.Chong ZZ, Shang YC, Wang S, Maiese K. PRAS40 is an integral regulatory component of erythropoietin mTOR signaling and cytoprotection. PLoS One. 2012;7(9):e45456. doi: 10.1371/journal.pone.0045456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim Y, Shin SA, Lee J, Yang KD, Nam KT. Hybrid system of semiconductor and photosynthetic protein. Nanotechnology. 2014;25(34):342001. doi: 10.1088/0957-4484/25/34/342001. [DOI] [PubMed] [Google Scholar]
- 47.Banerji A, Das S, Chatterjee A. Culture of human A375 melanoma cells in the presence of fibronectin causes expression of MMP-9 and activation of MMP-2 in culture supernatants. J Environ Pathol Toxicol Oncol. 2008;27(2) doi: 10.1615/jenvironpatholtoxicoloncol.v27.i2.60. [DOI] [PubMed] [Google Scholar]
- 48.Esparza J, Vilardell C, Calvo J, Juan M, Vives J, Urbano-Márquez A, Yagüe J, Cid MC. Fibronectin Upregulates Gelatinase B (MMP-9) and Induces Coordinated Expression of Gelatinase A (MMP-2) and Its Activator MT1-MMP (MMP-14) by Human T Lymphocyte Cell Lines. A Process Repressed Through RAS/MAP Kinase Signaling Pathways. Blood. 1999;94(8):2754–LP-2766. [PubMed] [Google Scholar]
- 49.Das S, Banerji A, Frei E, Chatterjee A. Rapid expression and activation of MMP-2 and MMP-9 upon exposure of human breast cancer cells (MCF-7) to fibronectin in serum free medium. Life Sci. 2008;82(9):467–476. doi: 10.1016/j.lfs.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 50.Meng XN, Jin Y, Yu Y, Bai J, Liu GY, Zhu J, Zhao YZ, Wang Z, Chen F, Lee K-Y, Fu SB. Characterisation of fibronectin-mediated FAK signalling pathways in lung cancer cell migration and invasion. Br J Cancer. 2009;101(2):327–334. doi: 10.1038/sj.bjc.6605154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boukerche H, Aissaoui H, Prevost C, Hirbec H, Das SK, Su Z-Z, Sarkar D, Fisher PB. Src kinase activation is mandatory for MDA-9/syntenin-mediated activation of nuclear factor-[kappa]B. Oncogene. 2010;29(21):3054–3066. doi: 10.1038/onc.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh SP, Schwartz MP, Lee JY, Fairbanks BD, Anseth KS. A peptide functionalized poly(ethylene glycol) (PEG) hydrogel for investigating the influence of biochemical and biophysical matrix properties on tumor cell migration. Biomater Sci. 2014;2(7):1024–1034. doi: 10.1039/C4BM00022F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee S, Tong X, Yang F. The effects of varying poly(ethylene glycol) hydrogel crosslinking density and the crosslinking mechanism on protein accumulation in three-dimensional hydrogels. Acta Biomater. 2014;10(10):4167–4174. doi: 10.1016/j.actbio.2014.05.023. [DOI] [PubMed] [Google Scholar]
- 54.Woods J, Docker PT, Dyer CE, Haswell SJ, Greenman J. On-chip integrated labelling, transport and detection of tumour cells. Electrophoresis. 2011;32(22):3188–3195. doi: 10.1002/elps.201100172. [DOI] [PubMed] [Google Scholar]
- 55.Hagel V, Haraszti T, Boehm H. Diffusion and interaction in PEG-DA hydrogels. Biointerphases. 2013;8(1):36. doi: 10.1186/1559-4106-8-36. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y, Amsden BG. Application of an Obstruction-Scaling Model To Diffusion of Vitamin B12 and Proteins in Semidilute Alginate Solutions. Macromolecules. 2006;39(3):1073–1078. doi: 10.1021/ma0522357. [DOI] [Google Scholar]
- 57.Thurber GM, Schmidt MM, Wittrup KD. Antibody tumor penetration: Transport opposed by systemic and antigen-mediated clearance. Adv Drug Deliv Rev. 2008;60(12):1421–1434. doi: 10.1016/j.addr.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Osorio FA, Bilbao E, Bustos R, Alvarez F. Effects of concentration, bloom degree, and pH on gelatin melting and gelling temperatures using small amplitude oscillatory rheology. Int J Food Prop. 2007;10(4):841–851. [Google Scholar]
- 59.Paguirigan A, Beebe DJ. Gelatin based microfluidic devices for cell culture. Lab Chip. 2006;6(3):407–413. doi: 10.1039/b517524k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.