Abstract
The application of biomarkers for ‘recent’ infection in cross-sectional HIV incidence surveillance requires the estimation of critical biomarker characteristics. Various approaches have been employed for using longitudinal data to estimate the Mean Duration of Recent Infection (MDRI) – the average time in the ‘recent’ state. In this systematic benchmarking of MDRI estimation approaches, a simulation platform was used to measure accuracy and precision of over twenty approaches, in thirty scenarios capturing various study designs, subject behaviors and test dynamics that may be encountered in practice. Results highlight that assuming a single continuous sojourn in the ‘recent’ state can produce substantial bias. Simple interpolation provides useful MDRI estimates provided subjects are tested at regular intervals. Regression performs the best – while ‘random effects’ describe the subject-clustering in the data, regression models without random effects proved easy to implement, stable, and of similar accuracy in scenarios considered; robustness to parametric assumptions was improved by regressing ‘recent’/‘non-recent’ classifications rather than continuous biomarker readings. All approaches were vulnerable to incorrect assumptions about subjects’ (unobserved) infection times. Results provided show the relationships between MDRI estimation performance and the number of subjects, inter-visit intervals, missed visits, loss to follow-up, and aspects of biomarker signal and noise.
Keywords: HIV, incidence estimation, duration of recent infection, cross-sectional incidence surveys, biomarkers for recent infection
1 Introduction
The reliable estimation of HIV incidence is essential for monitoring the epidemic and targeting and assessing interventions. The cross-sectional estimation of HIV incidence using biomarkers for ‘recent’ infection has attracted much interest since its introduction in 1995 (Brookmeyer and Quinn 1995). Over the last two decades, numerous biomarkers have been developed and applied in incidence surveys, and various working groups and funding opportunities have been established to support data generation and accelerate implementation of the surveillance approach (Busch, Pilcher & Mastro 2010; Centers for Disease Control and Prevention, United States Department of Health and Human Services 2015; Kassanjee, Pilcher & Keating 2014; Laeyendecker, Brookmeyer & Cousins 2013; Le Vu, Pillonel & Semaille 2008; Mastro, Kim & Hallett 2010; Murphy & Parry 2008; Parekh, Kennedy & Dobbs 2002; Sharma, Schito & Welte 2012; The Consortium for the Evaluation and Performance of HIV Incidence Assays (CEPHIA) 2015; WHO Technical Working Group on HIV Incidence Assays 2015). Despite considerable progress, questions remain around how best to analyze data at various stages along the pathway from biomarker discovery to surveillance application.
The principle behind cross-sectional incidence surveillance is that a particular weighted average of past incidence can be estimated from (i) a single survey’s counts of HIV-negative, ‘recently’ infected and ‘non-recently’ infected subjects, and (ii) a small number of well-defined parameters describing the properties of the test for recent infection (TRI) in the population of interest. Under a general framework for incidence estimation, two parameters are required (Kassanjee, McWalter & Barnighausen 2012): The Mean Duration of Recent Infection (MDRI) – the average time subjects spend ‘recently’ infected within some time T post infection; and the False-Recent Rate (FRR) – the probability that a subject who is infected for longer than T will return a ‘recent’ result. While the FRR should ideally be zero, it is non-negligible for many currently available TRIs and is understood to vary by time and place (Busch, Pilcher & Mastro 2010; Hallett, Ghys & Barnighausen 2009; Kassanjee, Pilcher & Keating 2014; Le Vu, Pillonel & Semaille 2008; Longosz, Mehta & Kirk 2014; Mastro, Kim & Hallett 2010; Murphy & Parry 2008). The MDRI, typically required to be at least half a year for a TRI to begin to show promise (Incidence Assay Critical Path Working Group 2011), should ideally remain constant so that a once-calibrated TRI would be useful when transferred to other contexts. This work focuses solely on estimation of the MDRI.
Conventionally, the MDRI is estimated using longitudinal data, which captures biomarker measurements at multiple times after infection for each of a number of subjects. Constructing such datasets requires the costly and difficult collection of specimens from (initially HIV-negative) subjects regularly over time until becoming infected and for some time after. A number of approaches have been utilized for analyzing the resulting data (Braunstein, Nash & Kim 2011; Brookmeyer, Konikoff & Laeyendecker 2013; Curtis & Hanson 2013; Duong, Kassanjee & Welte 2015; Duong, Qiu & De 2012; Hargrove, Eastwood & Mahiane 2012; Hargrove, Humphrey & Mutasa 2008; Janssen, Satten & Stramer 1998; Keating, Hanson & Lebedeva 2012; Mahiane, Fiamma & Auvert 2014; McDougal, Parekh & Peterson 2006; Parekh, Hanson & Hargrove 2011; Parekh, Kennedy & Dobbs 2002; Sommen, Commenges & Le Vu 2011; Sweeting, De Angelis & Parry 2010; Wang & Lagakos 2009).
Unbiased incidence estimation requires unbiased estimates of TRI characteristics, and therefore robust and widely accepted methods for estimating the MDRI are essential. It is also important to be able to distinguish between variation in a chosen TRI’s MDRI estimates caused by true study population differences (such as HIV subtype), or different testing or laboratory conditions, from that caused by differences in MDRI estimation approaches. Also, to design studies for reliably characterizing TRIs, the relationships between data features (such as sample sizes and frequencies of specimen draws) and the performance of MDRI estimation need to be understood.
Consequently, in 2012, the HIV Modelling Consortium, funded by the Bill and Melinda Gates Foundation, requested that an international collaboration be established to investigate and compare the performances of MDRI estimation approaches (HIV Modelling Consortium Work Package on Characterisation of Tests for Recent Infection 2015). The resulting benchmarking exercise is presented below. A defining feature of this project is the use of a simulation platform: not only is the true underlying MDRI computable (against which MDRI estimates can be compared), but experiments can be replicated thousands of times. Through this replication, the accuracy and precision of a large number of MDRI estimation methods were measured in a number of modelled scenarios that capture essential features of what could be encountered in practice, namely different study designs, subject behaviors and underlying biomarker dynamics.
Due to the large scope and complexity of this benchmarking exercise, only summaries of the methodology and results are provided below. More thorough documentation is available in the Web Appendix.
2 Methods
The MDRI estimation approaches were assessed in a base case scenario and each of a number of comparison scenarios. For each scenario, 1,000 datasets were generated and each MDRI estimation method was applied to each dataset – other than for the computationally expensive ‘mixed effects’ models, which were each applied to a common subset of 250 datasets per scenario. Data generation and MDRI estimation are each discussed below. All technical details required to reproduce the work are provided in Web Appendix A and B.
For each scenario and each estimation method, a distribution of MDRI point estimates was obtained. Two statistics are presented to summarize this distribution (and its relationship to the true MDRI): (i) accuracy is reported as the relative bias – the difference between the average MDRI estimate and true MDRI, divided by the true MDRI, and (ii) precision as the relative standard error – the standard deviation of estimates, divided by the mean estimate.
The simulation platform was developed (in Python, R and MySQL) to automate the data generation, application of the estimation methods (in the form of Matlab or R scripts) and storage of results. Due to the large run times involved, computing resources were procured from Amazon Web Services.
A central concept is that of ‘infection’: throughout this work, infection refers to detectable infection, which depends on the specific diagnostic algorithm used in any real-world setting. For example, if utilizing a Western Blot assay, a subject’s infection time is when she begins to test positive (or seroconverts) on Western Blot.
The scope of this benchmarking exercise is potentially very wide, and was therefore carefully limited to provide a feasible investigation focused on the most immediate needs of analysts in the field.
Firstly, only TRIs based on single biomarkers were considered, where the ‘biomarker’ measurement may itself be a complex summary metric of multiple measurements. A measurement below a chosen classification threshold is interpreted as indicating ‘recent’ infection, as in currently used ‘incidence assays’.
Secondly, a single HIV diagnostic test is used in the simulated longitudinal study, and provides no information beyond distinguishing HIV-positive from HIV-negative subjects. Therefore, each subject’s infection time is simply known to lie in the interval from his last HIV-negative visit to first HIV-positive visit, called the infection interval.
Thirdly, to limit the number of comparison scenarios, only a single aspect of the data generation process was varied at a time.
Lastly, confidence intervals were not explicitly investigated. For each estimation method, a number of approaches could be used to obtain confidence intervals. Within this simulation study, the accuracy and precision of point estimates could be directly measured, which inform the coverage and widths of confidence intervals that could be produced (and would be reported in real-world studies).
2.1 Data generation
The underlying processes that produce a real-world dataset for MDRI estimation can be considered in two parts. Firstly, the study design and subject behavior produce the observed visit and unobserved infection times of subjects. Secondly, particulars of the biological signal over time since infection, and noise around the signal arising from fluctuations within the host or imperfect measurement, govern the observed biomarker readings for the visits. In all scenarios considered, a subject’s adherence to the study design is independent of his biomarker values. Stochastic models were constructed for generating visit times, infection times and biomarker readings of subjects. The models draw on the experience of the team, and rely on few input parameters while providing sufficient flexibility for this investigation. The structures underlying the data generation models are represented in Figure 1.
Figure 1.
Representation of the data generation models. The visit and infection times are generated according to the parameters and distributions contained in (1) to (10) (grey box), while the subjects’ observed biomarker readings at visits are generated according to the specifications in (A) to (D). The values or distributional forms specified for (1) to (10) relate to the base case scenario.
The base case scenario was defined by specifying values for the data generation parameters (Figure 1), based on an ideal adherence to an optimistic study design, and insights into existing biomarkers. Within each investigation – comprising a set of comparison scenarios – one aspect of the data generation process was systematically varied, such as the number of subjects (a list of investigations and scenarios is contained in the tables in Results). To quantify the bias, the true MDRI was computed using the parameters fed into the data generation models – more specifically, the parameters defining the behavior of the biomarker with time after infection, as summarized by (A) to (D) in Figure 1.
The timing of infection within the infection interval will depend on both the study design and subject behavior. For example, as in the base case scenario, if visit times are strictly controlled by the study, it is reasonable to assume that a subject is equally likely to have been infected at any time in the interval. However, in other settings, subjects may exhibit test-seeking or test-deferring behaviors (Burchell, Calzavara & Ramuscak 2003; Centers for Disease Control and Prevention 2004; Schreiber, Glynn & Satten 2002), and therefore a Beta distribution (scaled to span the interval) was used to generate infection times from a skew distribution in the relevant modelled scenarios.
To explore the impact of the functional form of the biomarker signal, an alternative model for generating biomarker readings was also implemented. For the base case scenario (Figure 1), the biological signal follows a sigmoidal curve – namely, a three-parameter log-logistic curve starting at zero and defined by shape, scale and asymptote parameters. In the alternative model (the ‘power function’), the biomarker signal remains zero for some period after infection, and then experiences rapid growth that slows down over time. In this case, the signal equals some power, between zero and one, of time since signal growth.
Further technical details on the data generation and the parameter values for the scenarios are provided in Web Appendix A.
2.2 MDRI estimation approaches
The MDRI can be expressed mathematically as dt, where PR (t) is the probability of being (alive and) ‘recently’ infected at time t after infection (Kassanjee, McWalter & Barnighausen 2012). Estimation of the MDRI therefore entails inferring PR (t), either (i) directly by fitting a chosen model for PR (t) to the dichotomous ‘recent’ and ‘non-recent’ classifications, or (ii) indirectly by modelling the continuous biomarker measurements and then computing the probability of obtaining a measurement below the classification threshold. In the base case scenario, a subject’s biomarker signal increases from 0 at infection to an average asymptote of 85, and the classification threshold was set to 40. Throughout this work, T = 1 year and negligible mortality within T after infection was assumed. In practice, T would be carefully chosen based on biomarker dynamics, surveillance objectives and practical considerations (Kassanjee, McWalter & Barnighausen 2012).
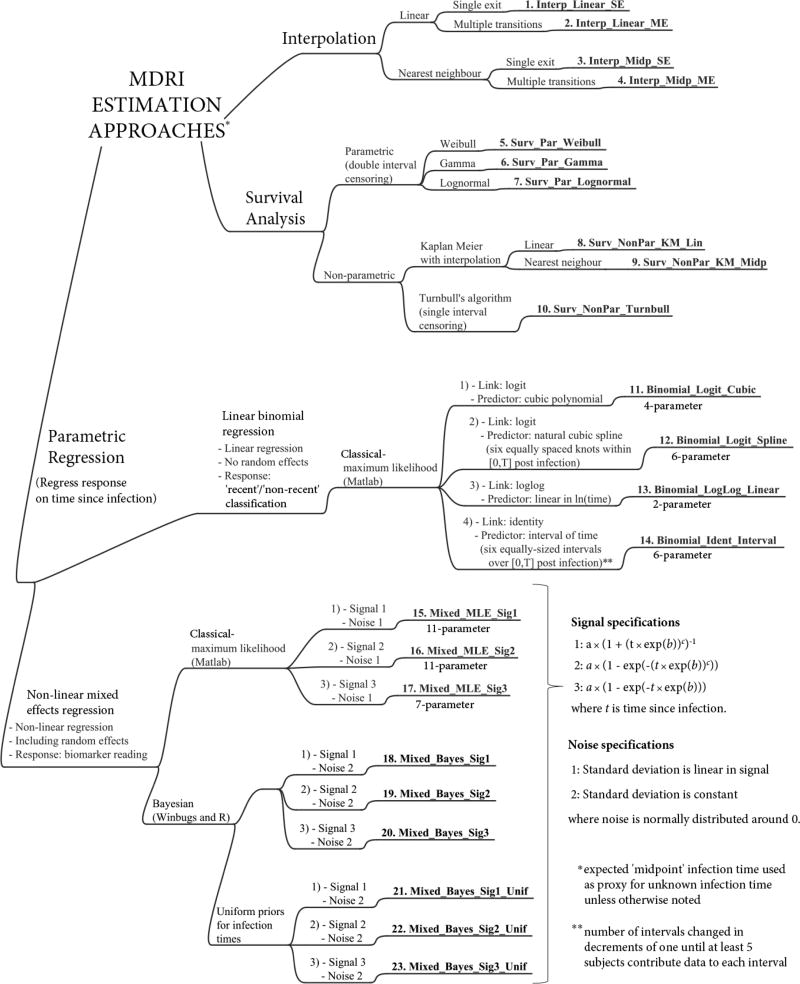
Numerous MDRI estimation methods were implemented, to represent those published and to explore extensions (Braunstein, Nash & Kim 2011; Brookmeyer, Konikoff & Laeyendecker 2013; Curtis & Hanson 2013; Duong, Kassanjee & Welte 2015; Duong, Qiu & De 2012; Hargrove, Eastwood & Mahiane 2012; Hargrove, Humphrey & Mutasa 2008; Janssen, Satten & Stramer 1998; Keating, Hanson & Lebedeva 2012; Mahiane, Fiamma & Auvert 2014; McDougal, Parekh & Peterson 2006; Parekh, Hanson & Hargrove 2011; Parekh, Kennedy & Dobbs 2002; Sommen, Commenges & Le Vu 2011; Sweeting, De Angelis & Parry 2010; Wang & Lagakos 2009). The methods, captured in Figure 2, fall into three categories: (i) interpolation, (ii) survival analysis, and (iii) parametric regression. A number of statistical groups contributed the MDRI estimation tools used in this benchmarking exercise, at times apparent by the subtle differences in analysis design and implementation – all thoroughly documented in Web Appendix B. A brief summary of the approaches follows.
Figure 2.
Mind map of analysis approaches for estimation of the MDRI. Each of the twenty three approaches benchmarked is numbered and labelled in bold (these identifiers are used in Results).
A challenge faced when estimating the MDRI is the unknown infection times. In studies where visit times are stipulated, it is reasonable to assume that the infection time is uniformly distributed in the infection interval. Therefore, the expected infection time lies at the midpoint of the interval. Unless otherwise stated, the approaches below treat this ‘midpoint infection time’ as a proxy for the (unobserved) infection time.
Interpolation (Methods 1–4)
These methods provide a basic, informal analysis approach. For each subject, biomarker readings between visits are obtained using linear or nearest neighbor interpolation (assuming a zero reading at infection). The approach either (i) uses data as is, allowing multiple transitions between the ‘recent’ and ‘non-recent’ states, or (ii) assumes that once a subject’s reading moves above the classification threshold, it will remain above it, thus enforcing a single exit from the ‘recent’ state, as in some of the literature (Braunstein, Nash & Kim 2011; Duong, Qiu & De 2012; Hargrove, Eastwood & Mahiane 2012; Janssen, Satten & Stramer 1998; Keating, Hanson & Lebedeva 2012; Parekh, Hanson & Hargrove 2011; Sweeting, De Angelis & Parry 2010; Wang & Lagakos 2009). The function PR (t) is then estimated as the proportion of measurements that are below the threshold at time t.
Survival analysis (Methods 5–10)
These techniques model the time from entering to exiting a state of interest (here ‘recent’ infection). The techniques easily accommodate data censoring (unknown event times) and have been applied in this area (Curtis & Hanson 2013; Duong, Qiu & De 2012; Hargrove, Eastwood & Mahiane 2012; Keating, Hanson & Lebedeva 2012; Parekh, Hanson & Hargrove 2011; Sweeting, De Angelis & Parry 2010; Wang & Lagakos 2009). However, many of the existing survival analysis approaches ignore fluctuations between states that can result from either non-monotonic evolution of the biomarker signal or simply measurement noise. To utilize a single continuous sojourn framework, all data points beyond a subject’s first ‘non-recent’ result were discarded. When there is no ‘non-recent’ visit, exit occurs at some subsequent time.
Three parametric distributions (Weibull, Gamma and Lognormal) for the time spent ‘recent’ were fitted to the data by maximum likelihood (ML) and under double interval censoring (the entry time lies uniformly in the infection interval, and then the exit time lies uniformly in an appropriately defined ‘exit interval’).
For non-parametric ML estimation, a Kaplan-Meier estimator was used (Kaplan & Meier 1958), using a midpoint infection time and an exit time obtained by linear or nearest neighbor interpolation. The MDRI estimate will equal that produced by the corresponding single-exit interpolation method when there are no right censored exit times within T after infection. To eliminate the use of proxy entry and exit times, Turnbull’s extension of the Kaplan-Meier estimator (Turnbull 1976) was also applied. Here, a subject’s time spent ‘recent’ is uniformly distributed between the minimum and maximum possible times implied by the data. This is theoretically inconsistent with the double interval censoring occurring – which would imply a non-uniform distribution – but reproduces a previous application (Duong, Qiu & De 2012).
Parametric regression (Methods 11–23)
An appropriately parameterized functional form for the expected response (either the dichotomous classification or continuous biomarker reading), as a function of predictors (time since infection), is fitted to the data. Three classes of models were employed (each discussed below), namely (i) linear binomial regression, (ii) non-linear (normal-response) mixed effects models using ML estimation and (iii) non-linear (normal-response) mixed effects models using a Bayesian framework and allowing for uncertainty in infection times through prior distributions
The linear binomial regression models (Methods 11–14) are fitted using the ‘recent’/‘non-recent’ classifications and were of the form g (PR (t)) = βT x (t), where g (.) is the link function, and η = βT x (t) is the linear predictor. The linear predictor contains both the model parameters in β (estimated by ML) and the predictors in x (t), which are functions of time since infection t. These models neglect the subject-level clustering of data points. Four forms of the model were implemented, using the link functions and linear predictors indicated in Figure 2. In practice, based on the density of data, not all forms may be appropriate.
In the non-linear mixed effects models, a parametric form is chosen for the biomarker signal as a function of time since infection, as well as the measurement noise structure. Subject-level clustering of data manifests as subject-specific deviations (random effects) of signal parameters from some average parameter values (fixed effects). The parameters to be estimated are the noise specification parameters, fixed effects and covariance matrix for the random effects, which are normally distributed around 0. Three forms for the signal and two forms for the noise were used (Figure 2).
In the classical model implementation (Methods 15–17), a Markov Chain Monte Carlo (MCMC) approach was used to search for the ML parameters using Matlab’s ‘nlmefitsa’ function. For a real dataset, convergence criteria would be carefully assessed; however, the present estimation was performed for several thousands of datasets. Based on initial investigations into run times and stability of results, three MCMC chains were used, of different starting points and 1,000 steps each.
The Bayesian implementation of the mixed models (Methods 18–23) makes use of the MCMC approach provided by WinBUGS to derive the posterior distribution of the unknown parameters, as described elsewhere (Sweeting, De Angelis & Parry 2010). The models use either a midpoint infection time or a uniform prior distribution for infection times. The estimated MDRI corresponds to the mean of the posterior distribution for the MDRI – appropriately modifying the methodology in earlier work (Sweeting, De Angelis & Parry 2010) to utilize the MDRI definition above.
Each method of estimation above provides an estimate of PR (t). The MDRI estimate equals the area under this curve, from t = 0 to t = T, either analytically or numerically.
3 Results
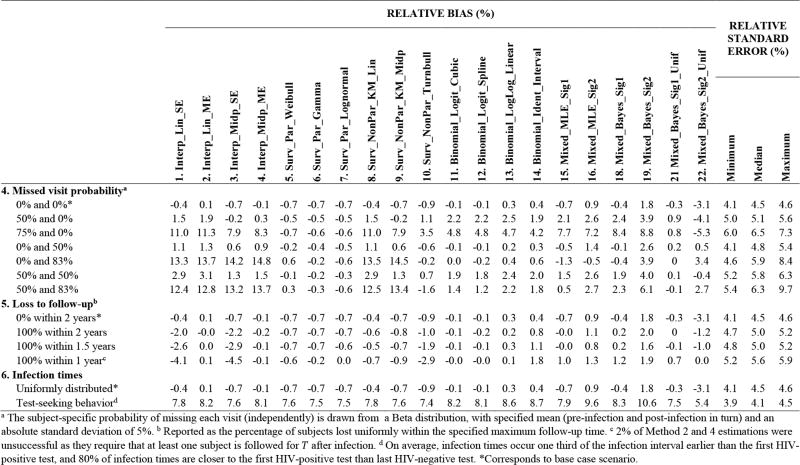
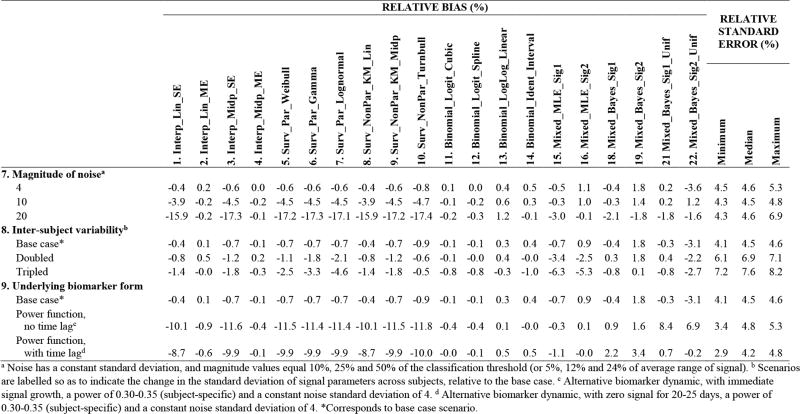
The MDRI estimates for the base case scenario are plotted in Figure 3, and performance statistics for the various investigations are provided in the tables. The impact of the study protocol and subject behavior (which determine visit and infection times) is explored in Figure 4 and Figure 5, and that of the biomarker dynamic (which governs biomarker readings) is considered in Figure 6. In Figure 4, Figure 5 and Figure 6: Methods 17, 20 and 23 are excluded (explained below). Investigations about the number of subjects and the mean inter-visit intervals, while HIV-negative or HIV-positive, are summarized in Figure 4 (Investigations 1–3); and about the extent of missed visits, loss to follow-up and non-uniformity of infection times in Figure 5 (Investigations 4–6). In Figure 6, the magnitude of noise, extent of inter-subject variability and form of the biomarker signal are varied (Investigations 7–9). More detailed results are provided in Web Appendix C.
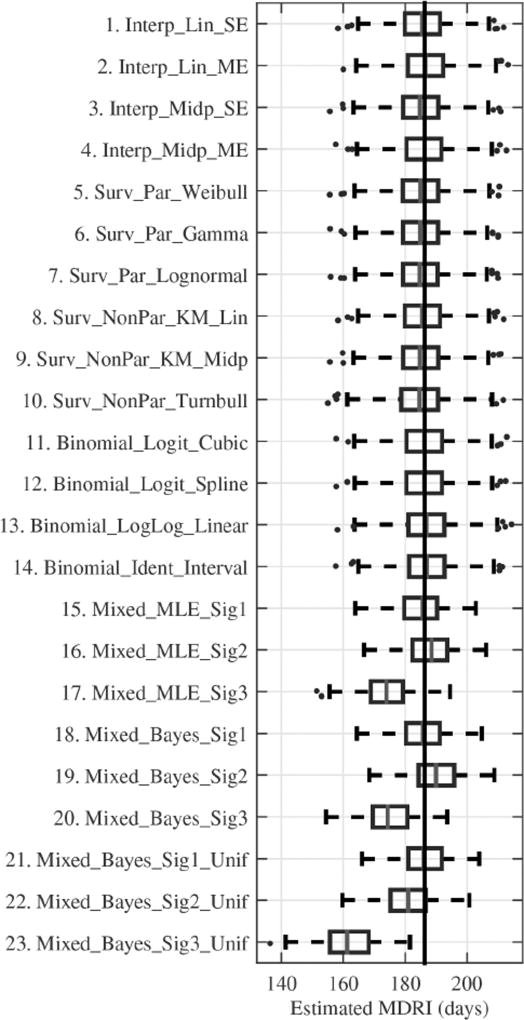
Figure 3.
Box-and-whisker plots of the Mean Duration of Recent Infection point estimates (days) for each estimation method, for the base case scenario. The box and dividing line indicate the central 50 % and median of estimates respectively, and whiskers and dots capture remaining estimates and outliers respectively (outliers are more than 1.5 times the box length away from the central box). The vertical line indicates the true MDRI. For Methods 15–23, fewer experiments were replicated (250 instead of 1 000).
Figure 4.
Relative bias (%) and relative standard error (%) of MDRI estimation in scenarios capturing various study designs (number of subjects and visit gaps).
Figure 5.
Relative bias (%) and relative standard error (%) of MDRI estimation in scenarios capturing varying subject behavior (missed visit probabilities, loss to follow-up and infection time distributions).
Figure 6.
Relative bias (%) and relative standard error (%) of MDRI estimation in scenarios capturing varying biomarker dynamics (magnitude of noise, inter-subject variability, form of signal).
In the tables, the observed relative bias and relative standard error are reported, based on 1,000 experiment replications (or 250 for the mixed effects models). In the base case scenario, a 95 % confidence interval for the relative bias would extend out by about 0.5 % (or 1 %) to each side of the observed measure, in absolute terms (treating estimates as normally distributed, based on the asymptotic behavior of the estimates).
Base case scenario findings
For the base case scenario, in which 50 subjects are visited monthly for two years post infection and the infection interval is three months, all classes of estimation approaches can provide reasonable accuracy (Figure 3). However, results are sensitive to the parametric assumptions underlying biomarker regression (Methods 15–23).
Parametric assumptions for biomarker regression (Methods 15–23)
It is important to understand the relationship between the true biomarker dynamic which generates the data and the assumed biomarker dynamic when analyzing the data using biomarker regression (Methods 15–23, see Figure 2). The models assume three different biomarker signals: Signals 1 and 2 are both flexible sigmoidal curves, where Signal 1 matches the true form (although the regression model uses a subtly different distribution to capture inter-subject variability) and Signal 2 does not; Signal 3 is much more restrictive form of Signal 1. None of the assumed noise structures exactly match the true noise structure, as would be expected to occur in practice.
Biomarker Signal 3, which is concave downwards and does not allow for an initial period of little growth, produces substantial bias (Figure 3). The corresponding computationally expensive Methods 17, 20 and 23 were therefore eliminated from subsequent investigations. A more general comparison of the results for Signal 1, which matches data generation, and Signal 2, which does not, suggests that similar inferences can be made by choosing a reasonable and sufficiently flexible model form. Ironically, allowing for a movable infection time through a uniform prior has the potential to substantially increase the bias caused by the wrong parametric assumptions (for example, Investigation 2, 3 and 9). The way the model is fitted to the data, and therefore the biases from incorrect parametric assumptions, also depends on factors such as visit gaps, frequency of missed visits and magnitude of noise.
Single sojourn assumptions
Methods that assume a single continuous sojourn in the ‘recent’ state (Methods 1 and 3, 5–10) underestimate the MDRI. The bias increases with more frequent visits and greater measurement noise as there is a greater chance of ‘early’ upward fluctuations of readings above the classification threshold (Investigations 3 and 7). Given these limitations, interpretations below focus on the remaining approaches.
Variability of estimates
In a given scenario, the methods exhibit similar precision; therefore, in the tables, the precision of MDRI estimation for each scenario is summarized by the minimum, median and maximum relative standard errors of the different methods. The standard deviation is approximately inversely proportional to the square root of the number of subjects (Investigation 1), and proportional to the standard deviation of individual durations in the ‘recent’ state (Investigation 8 and Web Appendix B). Larger visit gaps, more missed visits, increased loss to follow-up, and greater measurement noise increase variability in more nuanced ways (Investigations 2–5 and 7).
Loss to follow-up (Investigation 5)
When no subjects are followed until T after infection, only approaches that extrapolate beyond the latest data can be used (note the failure of Methods 2 and 4). When there is drop-out, those subjects observed to transition out of ‘recent’ will over-represent shorter sojourns, and therefore biases arise when naïvely averaging the data (Methods 1 and 3).
Binomial regression models versus biomarker mixed effect models (Methods 11–14 versus Methods 15–23)
While binomial regression does not account for the subject-level clustering of data, it was easy to implement, stable, relatively insensitive to parametric assumptions, and generally performed on par (or better) than the mixed models, which account for the clustering, in the scenarios considered.
HIV-negative visit gaps /missed visits
All methods performed poorly when there were some very large infection intervals relative to the duration of ‘recent’ infection (Investigation 4), with the exception of the biomarker mixed model that used the correct signal parametric form and a uniform prior for infection times (Method 21).
HIV-positive visit gaps /missed visits
The number of HIV-positive visits can be decreased by increasing the intended visit gap or increasing missed visits. The interpolation methods were vulnerable to high missed visit probabilities because interpolation between widely separated data points is unreliable. For parametric regression, it is desirable to have sufficient data at different times after infection to reliably fit the model to the data.
Unknown infection times
All methods performed poorly when the model assumptions about infection times were at odds with the data generation process (Investigation 6).
Data exclusion
For some of the parametric approaches above (Methods 5–7, 11–17), data points that were beyond T plus some margin after infection were discarded before model fitting (see Web Appendix B for details). In practice, biomarker dynamics may become less predictable years after infection, and therefore care should be taken in choosing data to train models intended to describe only early biomarker dynamics, also based on a strong understanding of the specific modelling approach used.
4 Discussion
This work presents a systematic benchmarking of currently used methods to estimate the critical Mean Duration of Recent Infection, as required for the application of a test for ‘recent’ infection in cross-sectional HIV incidence studies. The benchmarking uses a simulation approach, which could now be used to perform further MDRI estimation investigations, or even, with deeper adaptations, explore other aspects of incidence surveillance such as the performance of estimation of context-specific False-Recent Rates or the power to detect trends in incidence.
The results highlight the danger of using estimation procedures that assume single continuous sojourns in the ‘recent’ state. Simplistic approaches, such as the interpolation of biomarker readings, allowing for multiple transitions between states, are useful for obtaining ‘quick and dirty’ estimates provided the times between visits are sufficiently small.
Regression approaches performed well. While non-linear mixed models for the biomarker readings captured the subject-specific evolutions of the biomarker, they are complex and computationally demanding. Importantly, when analyzing any dataset, parametric assumptions should be carefully chosen and formally assessed to mitigate bias. While not fully capturing the data structure, the linear binomial regression models proved useful – algorithms were stable, and results were accurate in the scenarios considered and less sensitive to parametric assumptions. However, the extension of the binomial regression methods to include random effects would generally improve precision. Also, analysts should be warned against using the model-derived standard errors, which necessarily underestimate variability by incorrectly treating observations as independent, to construct confidence intervals, and subject-level bootstrapping could instead be adopted (Brookmeyer, Konikoff & Laeyendecker 2013). The performance of these models when including random effects could be investigated as part of further work, such as in the scenario when loss to follow-up is related to biomarker progression.
Uncertain infection times pose a particular challenge to MDRI estimation. Regardless of whether the unknown infection times are handled simplistically (using a proxy expected infection time) or formally accommodated (Brookmeyer, Konikoff & Laeyendecker 2013; Mahiane, Fiamma & Auvert 2014; Sommen, Commenges & Le Vu 2011; Sweeting, De Angelis & Parry 2010), incorrect assumptions about the underlying process will lead to bias. The assumptions about, and flexibilities allowed in, infection times also interact with other aspects of the estimation, such as the assumed form of the biomarker signal, to determine the overall MDRI bias. Some groups have attempted to use the biomarker readings themselves to estimate infection times (Curtis & Hanson 2013; Hargrove, Humphrey & Mutasa 2008; Keating, Hanson & Lebedeva 2012; Parekh, Hanson & Hargrove 2011; Parekh, Kennedy & Dobbs 2002) (not explored in this work). Also, in this exercise, a single dichotomous HIV diagnostic test was used at all visits. In some studies, staging information may be available – for example, detectable p24 antigens and undetectable antibodies at a visit would suggest infection within the preceding few weeks (Brookmeyer, Konikoff & Laeyendecker 2013; Fiebig, Wright & Rawal 2003; Lee, Giorgi & Keele 2009). The large biases caused by incorrect assumptions about infection times highlights the need for such staging data to be collected, and the methodology for their use to be appropriately developed.
As the field moves towards recent infection tests that rely on multiple biomarkers (Brookmeyer, Konikoff & Laeyendecker 2013; Laeyendecker, Brookmeyer & Cousins 2013), some methods for estimating the MDRI, such as those that utilize the dichotomous test classifications, may be more amenable to this extension than those that parametrically model biomarker evolution, in which parameters would proliferate. Also, novel biomarkers may represent complex processes that are not well-understood, and thus the selection of parametric assumptions may become more challenging. Another nuance is the assumption of guaranteed survival until T (typically a year or two) after infection implicit in most MDRI estimation approaches. In settings where early mortality is high, analyses to estimate the MDRI could incorporate data on survival.
The insights presented here contribute towards a deeper understanding of results already in the literature, future analysis decisions, nuances of MDRI estimation, and key choices needing to be made in the design of test characterization studies. The undertaking of this project has already led to a number of prominent research groups critically reviewing and improving their estimation tools. While limited resources will always create restrictions in fieldwork, useful MDRI estimates can clearly be obtained in a range of realistic scenarios.
Supplementary Material
Acknowledgments
Funding
South African National Research Foundation, (Grant /Award Number: ‘UID 44895’); Bill and Melinda Gates Foundation, (Grant /Award Number: ‘OPP1022972’); Medical Research Council, (Grant /Award Number: ‘Unit Programme number MC_UP_1302/3’, ‘Unit Programme number U105260566’); National Institutes of Health, (Grant /Award Number: ‘R01 AI095068’, ‘R37 AI51164’); Division of Intramural Research, National Institute of Allergy and Infectious Diseases.
Footnotes
Supplemental Material: The online version of this article (DOI:scid-2016-0002) offers supplementary material, available to authorized users.
References
- Braunstein SL, Nash D, Kim AA, et al. Dual Testing Algorithm of BED-CEIA and Axsym Avidity Index Assays Performs Best in Identifying Recent HIV Infection in a Sample of Rwandan Sex Workers. PLoS One. 2011;6(4):e18402. doi: 10.1371/journal.pone.0018402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookmeyer R, Konikoff J, Laeyendecker O, et al. Estimation of HIV Incidence Using Multiple Biomarkers. American Journal of Epidemiology. 2013;177(3):264–272. doi: 10.1093/aje/kws436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookmeyer R, Quinn TC. Estimation of Current Human Immunodeficiency Virus Incidence Rates from a Cross-Sectional Survey Using Early Diagnostic Tests. American Journal of Epidemiology. 1995;141(2):166–172. doi: 10.1093/oxfordjournals.aje.a117404. [DOI] [PubMed] [Google Scholar]
- Burchell AN, Calzavara L, Ramuscak N, et al. Symptomatic Primary HIV Infection or Risk Experiences? Circumstances Surrounding HIV Testing and Diagnosis among Recent Seroconverters. International Journal of STD & AIDS. 2003;14(9):601–608. doi: 10.1258/095646203322301059. [DOI] [PubMed] [Google Scholar]
- Busch MP, Pilcher CD, Mastro TD, et al. Beyond Detuning: 10 Years of Progress and New Challenges in the Development and Application of Assays for HIV Incidence Estimation. AIDS. 2010;24(18):2763–2771. doi: 10.1097/QAD.0b013e32833f1142. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. HIV Testing Survey, 2002. Atlanta, GA: U.S. Department of Health and Human Services; 2004. [Google Scholar]
- Centers for Disease Control and Prevention, United States Department of Health and Human Services. [Accessed November 27, 2015];Grant opportunity: Population-based HIV Impact Assessments in Resource-Constrained Settings under the President’s Emergency Plan for AIDS Relief (PEPFAR) 2015 http://www.grants.gov/web/grants/view-opportunity.html?oppId=252788.
- Curtis KA, Hanson DL, et al. Evaluation of a Multiplex Assay for Estimation of HIV-1 Incidence. PLoS One. 2013;8(5):e64201. doi: 10.1371/journal.pone.0064201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong YT, Kassanjee R, Welte A. Recalibration of the Limiting Antigen Avidity EIA to Determine Mean Duration of Recent Infection in Divergent HIV-1 Subtypes. PLoS One. 2015;10(2):e0114947. doi: 10.1371/journal.pone.0114947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong YT, Qiu M, De AK, et al. Detection of Recent HIV-1 Infection Using a New Limiting-Antigen Avidity Assay: Potential for HIV-1 Incidence Estimates and Avidity Maturation Studies. PLoS One. 2012;7(3):e33328. doi: 10.1371/journal.pone.0033328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig EW, Wright DJ, Rawal BD, et al. Dynamics of HIV Viremia and Antibody Seroconversion in Plasma Donors: Implications for Diagnosis and Staging of Primary HIV Infection. AIDS. 2003;17(13):1871–1879. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- Hallett TB, Ghys P, Barnighausen T, et al. Errors in ‘BED’-Derived Estimates of HIV Incidence Will Vary by Place, Time and Age. PLoS One. 2009;4(5):e5720. doi: 10.1371/journal.pone.0005720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrove J, Eastwood H, Mahiane G, et al. How Should We Best Estimate the Mean Recency Duration for the BED Method? PLoS One. 2012;7(11):e49661. doi: 10.1371/journal.pone.0049661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrove JW, Humphrey JH, Mutasa K, et al. Improved HIV-1 Incidence Estimates Using the BED Capture Enzyme Immunoassay. AIDS. 2008;22(4):511–518. doi: 10.1097/QAD.0b013e3282f2a960. [DOI] [PubMed] [Google Scholar]
- HIV Modelling Consortium Work Package on Characterisation of Tests for Recent Infection. [Accessed November 27, 2015];2015 http://www.hivmodelling.org/projects/incidence-estimation.
- Incidence Assay Critical Path Working Group. More and Better Information to Tackle HIV Epidemics: Towards Improved HIV Incidence Assays. PLoS Medicine. 2011;8(6):e1001045. doi: 10.1371/journal.pmed.1001045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen RS, Satten GA, Stramer SL, et al. New Testing Strategy to Detect Early HIV-1 Infection for Use in Incidence Estimates and for Clinical and Prevention Purposes. Jama. 1998;280(1):42–48. doi: 10.1001/jama.280.1.42. [DOI] [PubMed] [Google Scholar]
- Kaplan EL, Meier P. Nonparametric Estimation from Incomplete Observations. Journal of the American Statistical Association. 1958;53(282):457–481. [Google Scholar]
- Kassanjee R, McWalter TA, Barnighausen T, et al. A New General Biomarker-Based Incidence Estimator. Epidemiology. 2012;23(5):721–728. doi: 10.1097/EDE.0b013e3182576c07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassanjee R, Pilcher CD, Keating SM, et al. Independent Assessment of Candidate HIV Incidence Assays on Specimens in the CEPHIA Repository. AIDS. 2014;28(16):2439–2449. doi: 10.1097/QAD.0000000000000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating SM, Hanson D, Lebedeva M, et al. Lower-Sensitivity and Avidity Modifications of the Vitros Anti-HIV 1+2 Assay for Detection of Recent HIV Infections and Incidence Estimation. Journal of Clinical Microbiology. 2012;50(12):3968–3976. doi: 10.1128/JCM.01454-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laeyendecker O, Brookmeyer R, Cousins MM, et al. HIV Incidence Determination in the United States: A Multiassay Approach. The Journal of Infectious Diseases. 2013;207(2):232–239. doi: 10.1093/infdis/jis659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Vu S, Pillonel J, Semaille C, et al. Principles and Uses of HIV Incidence Estimation from Recent Infection Testing - a Review. Euro Surveillance. 2008;13(36):11–16. [PubMed] [Google Scholar]
- Lee HY, Giorgi EE, Keele BF, et al. Modeling Sequence Evolution in Acute HIV-1 Infection. Journal of Theoretical Biology. 2009;261(2):341–360. doi: 10.1016/j.jtbi.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longosz AF, Mehta SH, Kirk GD, et al. Incorrect Identification of Recent HIV Infection in Adults in the United States Using a Limiting-Antigen Avidity Assay. AIDS. 2014;28(8):1227–1232. doi: 10.1097/QAD.0000000000000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahiane SG, Fiamma A, Auvert B. Mixture Models for Calibrating the BED for HIV Incidence Testing. Statistics in Medicine. 2014;33(10):1767–1783. doi: 10.1002/sim.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastro TD, Kim AA, Hallett T, et al. Estimating HIV Incidence in Populations Using Tests for Recent Infection: Issues, Challenges and the Way Forward. Journal of HIV AIDS Surveillance & Epidemiology. 2010;2(1):1–14. [PMC free article] [PubMed] [Google Scholar]
- McDougal JS, Parekh BS, Peterson ML, et al. Comparison of HIV Type 1 Incidence Observed during Longitudinal Follow-Up with Incidence Estimated by Cross-Sectional Analysis Using the BED Capture Enzyme Immunoassay. AIDS Research and Human Retroviruses. 2006;22(10):945–952. doi: 10.1089/aid.2006.22.945. [DOI] [PubMed] [Google Scholar]
- Murphy G, Parry JV. Assays for the Detection of Recent Infections with Human Immunodeficiency Virus Type 1. Euro Surveillance. 2008;13(36):4–10. [PubMed] [Google Scholar]
- Parekh BS, Hanson DL, Hargrove J. Determination of Mean Recency Period for Estimation of HIV Type 1 Incidence with the BED-Capture EIA in Persons Infected with Diverse Subtypes. AIDS Research and Human Retroviruses. 2011;27(3):265–273. doi: 10.1089/aid.2010.0159. [DOI] [PubMed] [Google Scholar]
- Parekh BS, Kennedy MS, Dobbs T, et al. Quantitative Detection of Increasing HIV Type 1 Antibodies after Seroconversion: A Simple Assay for Detecting Recent HIV Infection and Estimating Incidence. AIDS Research and Human Retroviruses. 2002;18(4):295–307. doi: 10.1089/088922202753472874. [DOI] [PubMed] [Google Scholar]
- Schreiber GB, Glynn SA, Satten GA, et al. HIV Seroconverting Donors Delay Their Return: Screening Test Implications. Transfusion. 2002;42(4):414–421. doi: 10.1046/j.1525-1438.2002.00084.x. [DOI] [PubMed] [Google Scholar]
- Sharma UK, Schito M, Welte A, et al. Workshop Summary: Novel Biomarkers for HIV Incidence Assay Development. AIDS Research and Human Retroviruses. 2012;28(6):532–539. doi: 10.1089/aid.2011.0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommen C, Commenges D, Le Vu S, et al. Estimation of the Distribution of Infection Times Using Longitudinal Serological Markers of HIV: Implications for the Estimation of HIV Incidence. Biometrics. 2011;67(2):467–475. doi: 10.1111/j.1541-0420.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- Sweeting MJ, De Angelis D, Parry J, et al. Estimating the Distribution of the Window Period for Recent HIV Infections: A Comparison of Statistical Methods. Statistics in Medicine. 2010;29(30):3194–3202. doi: 10.1002/sim.3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Consortium for the Evaluation and Performance of HIV Incidence Assays (CEPHIA) [Accessed November 27, 2015];2015 http://www.incidence-estimation.com/page/cephia.
- Turnbull BW. The Empirical Distribution Function with Arbitrarily Grouped, Censored and Truncated Data. Journal of the Royal Statistical Society, Series B (Statistical Methodology) 1976;38(3):290–295. [Google Scholar]
- Wang R, Lagakos SW. Augmented Cross-Sectional Prevalence Testing for Estimating HIV Incidence. Biometrics. 2009;66(3):864–874. doi: 10.1111/j.1541-0420.2009.01356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Technical Working Group on HIV Incidence Assays. [Accessed November 27, 2015];2015 http://www.who.int/diagnostics_laboratory/links/hiv_incidence_assay/en/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.