Abstract
Antibody affinity maturation occurs in germinal centres (GC) where B cells cycle between the light zone (LZ) and the dark zone. In the LZ GC B cells bearing immunoglobulins with the highest affinity for antigen receive positive selection signals from T helper cells that promotes their rapid proliferation. Here we show that the RNA binding protein PTBP1 is necessary for the progression of GC B cells through late S-phase of the cell cycle and for affinity maturation. PTBP1 is required for the proper expression of the c-MYC-dependent gene program induced in GC B cells receiving T cell help and directly regulates the alternative splicing and abundance of transcripts increased during positive selection to promote proliferation.
Keywords: Alternative splicing, germinal centre, light zone, dark zone, RNA Binding Proteins, cell cycle proliferation, antibody affinity maturation, Polypyrimidine Track Binding Proteins, iCLIP
Introduction
Germinal centres (GCs) are specialized areas of secondary lymphoid tissues where B cells undergo antibody affinity maturation. The GC can be divided into a dark zone (DZ) characterized by extensive proliferation and somatic hypermutation (SHM) and a light zone (LZ) where B cells are less proliferative and make contacts with follicular dendritic cells and T cells. There, B cells are positively selected to survive and undergo further rounds of proliferation in the DZ1,2. T cell help promotes faster cell cycle progression, a greater number of cell divisions and increased SHM frequency resulting in increased antibody affinity maturation3,4. Positive selection in the LZ results in the transient expression of the transcription factor c-MYC necessary for the progression of selected cells through the cell cycle5,6. c-MYC also induces the transcription of genes important for anabolic metabolism3,7. Moreover, mTORC1 activity is increased during positive selection and promotes the anabolic gene expression program8. In addition to c-MYC, the transcription factors AP4, FOXO1 and BATF are important during the LZ-DZ transition of GC B cells9–11. These findings highlight the existence in B cells of relays of transcription factors that are responsive to T cell help and promote B cell proliferation and mutation.
Alternative splicing (AS), alternative polyadenylation (APA), mRNA decay and translation have the potential to regulate cell fate. However, we know relatively little about the relevant changes and regulation of AS during immune responses12–15 and molecular regulation of APA is only beginning to be appreciated16,17. The impact of RNA binding proteins (RBP) that have the potential to integrate multiple aspects of gene expression in B cells undergoing selection in the GC is only emerging13,14.
One class of RBP with pleiotropic function are the Polypyrimidine Tract Binding Proteins (PTBP)18–20. Most cell types express PTBP1, whereas PTBP2 is abundant in differentiated neurons. During neuronal differentiation there is a switch between the expression of PTBP1 and PTBP2 which drives changes in AS patterns of genes important for neuronal function18,21. PTBP3 is highly expressed in hematopoietic cells and previously shown to be co-expressed with PTBP1 in B lymphocytes22. PTBP1 is both a repressor and an activator of AS18 and has been implicated in APA, mRNA decay and translational regulation19,23,24. In CD4 T cells PTBP1 has been shown to enhance CD40L-mRNA stability25 and in human B cells to be responsive to toll-like receptor 926. Although PTBP1 has the potential to regulate posttranscriptional gene expression programs in lymphocytes its physiological functions in lymphocyte development and activation are unknown.
Here we show that PTBP1 acts in concert with c-MYC to ensure the selection of B cell clones with the highest affinity for antigen by promoting the proliferation of GC B cells. PTBP1 guarantees proper posttranscriptional processing and expression of genes induced as part of the c-MYC-dependent gene expression program.
Results
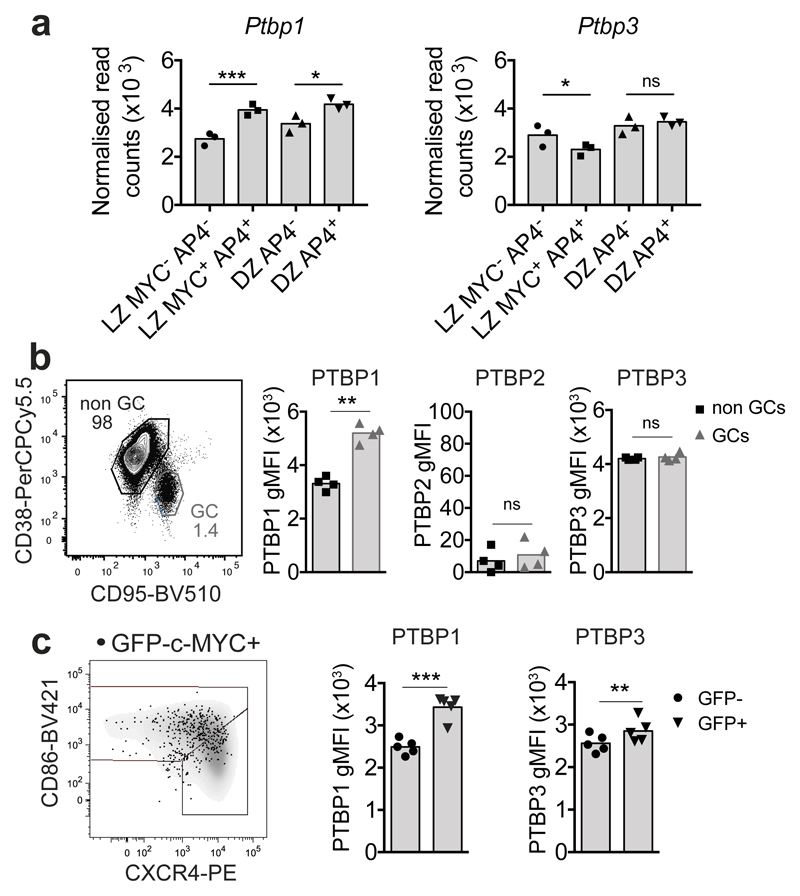
PTBP1 expression is increased in positively selected GC B cells
Uniquely amongst Ptb-family members the expression of Ptbp1-mRNA was increased ~1.5-fold in GC B cells compared to naive B cells (Supplementary Fig. 1a,b). Ptbp2 transcripts were rare and showed evidence of skipping exon 10 (Supplementary Fig. 1b) generating mRNAs degraded by nonsense-mediated RNA decay (NMD)27. Ptbp1- but not Ptbp3-mRNA was increased 1.4-fold upon c-MYC expression in LZ B cells (Fig. 1a) and was also increased in GC B cells that had received the greatest levels of T cell help (Supplementary Fig. 1c,d). A validated a panel of monoclonal antibodies recognizing PTBP1, PTBP2 and PTBP3 that were specific for each PTBP and able to detect the proteins by flow cytometry (Supplementary Fig. 2a,b) detected PTBP1 and PTBP3 but not PTBP2 in B cells (Fig. 1b, Supplementary Fig. 1f and 2a,b). PTBP1 protein was increased (~1.6-fold) in GC B cells compared to non-GC B cells (Fig. 1b) and increased ~1.4-fold in GFP-c-MYC reporter transgene (MycGFP)-positive GC B cells compared to GFP-c-MYC-negative GC B cells (Fig. 1c). Previous studies showed that PTBP1 expression correlates positively with c-MYC28 and that c-MYC binds to the promoter of PTBP1 in B cells29. Thus PTBP1 may act downstream of, or in parallel with c-MYC, in GC B cells responding to T cell help.
Figure 1. PTBP1 expression is increased in positively selected GC B cells.
(a) Ptbp1 and Ptbp3 mRNAseq normalised DESeq2 read counts in sorted GC B cell populations by c-MYC and AP4 expression7. P***<0.001, P*<0.05 and ns (non-significant) P >0.1. P-adjusted values were calculated with DESeq2 for the comparisons indicated by the horizontal lines
(b) Gating strategy for GC and non-GC B cells and expression of different PTBPs within the gated populations analysed by flow cytometry 7 days after NP-KLH immunisation. Full gating strategy is shown in Supplementary Fig. 1e. Data shown is representative from three independent experiments.
(c) Flow cytometry analysis of PTBP1 and PTBP3 in GFP-c-MYC+ and GFP-c-MYC- GC B cells from MycGFP/GFP mice immunised with SRBC for 6 days. Cytometry plot shows CXCR4 and CD86 expression of GFP-c-MYC+ (dots) and GFP-c-MYC- (density plot) GC B cells.
In b and c, graphs show geometric mean fluorescence intensity (gMFI) for each anti-PTBP antibody after subtraction of background staining determined with isotype control antibodies as shown in Supplementary Fig. 1f. Each symbol shows data from an individual mouse and bars represent the mean. Two-tailed paired Student’s t-test. ns (not significant) P>0.1, **P<0.01 and ***P<0.001. Numbers in cytometry plots show percentages of the gated populations.
PTBP1 is dispensable for B cell development
We validated conditional knockout (cKO) mice targeting Ptbp130 from the pro-B cell stage by using the Cd79acre allele (Supplementary Fig. 2b,c,d). B cell development was normal in the absence of PTBP1 (Supplementary Fig. 2c,e,f). Moreover, in lethally-irradiated CD45.1+ B6.SJL mice reconstituted with a 1:1 mixture of bone marrow cells from B6.SJL and Cd79acre/+Ptbp1fl/fl mice the numbers of follicular B cells arising from the Cd79acre/+Ptbp1fl/fl cKO bone marrow were not reduced compared to those arising from B6.SJL bone marrow (Data not shown). In cells that had deleted Ptbp1 the expression of PTBP2 was evident from the pro-B cell stage onwards (Supplementary Fig. 2d). The loss of PTBP1 and expression of PTBP2 was confirmed by immuno-blotting (Supplementary Fig. 2a). As expected31, Ptbp1-deficient B cells also contained increased amounts of the long-isoform of PTBP3 (Supplementary Fig. 2a). This indicates that although PTBP1 is not necessary for B cell development its deletion is consequential at the molecular level.
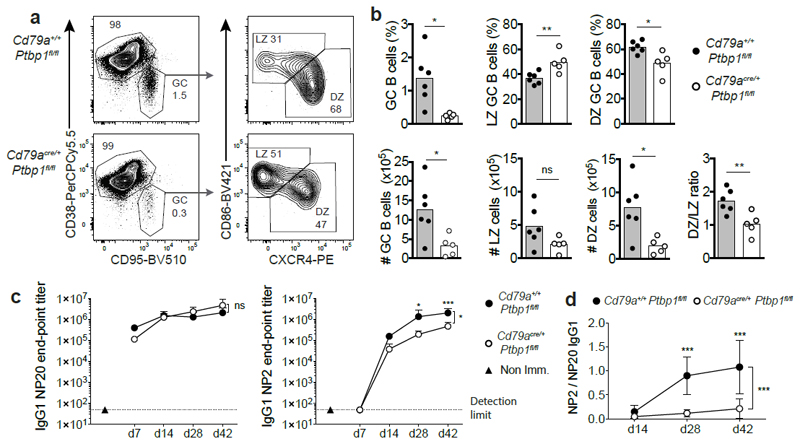
PTBP1 in B cells is necessary for the GC B cell response
We immunised Cd79acre/+Ptbp1fl/fl and Cd79a+/+Ptbp1fl/fl mice with 4-hydroxy-3-nitrophenyl-acetyl conjugated to keyhole limpet hemocyanin (NP-KLH). Seven days later the proportions and absolute numbers of GC B cells per spleen were reduced (5.9- and 3.9-fold, respectively) in Cd79acre/+Ptbp1fl/fl cKO compared to Cd79a+/+Ptbp1fl/fl control mice (Fig. 2a,b). The proportions of GC B cells with a DZ phenotype were reduced in Ptbp1-deficient GC B cells compared to control GC B cells (Fig. 2a,b) despite efficient depletion of the protein (Supplementary Fig. 3a). By contrast, Cd79acre/+Ptbp1fl/+ mice immunised with NP-KLH showed similar GC B cell responses to those of Cd79a+/+Ptbp1fl/fl mice (Supplementary Fig. 3b,c). The same GC B cell defects were found in Cd79acre/+Ptbp1fl/fl cKO GC B cells from bone marrow chimeras where B6.SJL mice were reconstituted with a 1:1 mixture of bone marrow cells from CD45.1+ B6.SJL and CD45.2+ Cd79acre/+Ptbp1fl/fl cKO mice (Supplementary Fig. 3d,e). Therefore, the defect in Ptbp1-deficient GC B cells was cell-autonomous. These data indicate an indispensable role for PTBP1 function in GC B cells.
Figure 2. PTBP1 is necessary for GC B cell responses.
(a) Representative flow cytometry plots showing gating strategy for GC B cells, DZ and LZ GC B cells from the spleen of mice immunised with NP-KLH for 7 days. Events shown on the left have been pre-gated on B220+CD19+ cells. Numbers indicate the percentages of the gated populations.
(b) Percentages and numbers of GC B cells, DZ and LZ GC B cells identified as shown in a. Data shown is representative of more than four independent experiments. Bar graphs show data from individual mice from one experiment and the mean is depicted with the bar. ns P>0.05, *P<0.05, **P<0.01 from two-tailed unpaired Student’s t-test are shown for the indicated comparisons.
(c) Anti-NP20 (high + low affinity) or anti-NP2 (high affinity only) IgG1 end-point titers measured by ELISA in the sera of mice at different days following immunization with NP-KLH.
(d) High affinity (NP2) vs. high and low affinity (NP20) anti-NP IgG1 ratio calculated from the data shown in c. Data in c and d are from one out of two independent experiments with similar results. The experiment shown had 3 Cd79a+/+Ptbp1fl/fl control mice and 6 Cd79acre/+Ptbp1fl/fl cKO mice. Shown is the mean + SD in c and ± SD in d. Differences between control and cKO mice were analysed with two-way ANOVA plus Sidak’s multiple comparison test. ns P>0.05, *P≤0.05 and ***P≤0.0002.
PTBP1 is necessary for antibody affinity maturation
Cd79acre/+Ptbp1fl/fl cKO mice produced reduced amounts of high affinity antibodies compared to Cd79a+/+Ptbp1fl/fl control mice (Fig. 2c). In Cd79a+/+Ptbp1fl/fl control mice the ratio of high affinity versus total affinity antibodies increased over time, but this ratio remained low in Cd79acre/+Ptbp1fl/fl cKO mice (Fig. 2d). Antibodies from mice lacking Ptbp2 in B cells (Cd79acre/+Ptbp2fl/fl) showed no defect in affinity maturation compared to Cd79a+/+Ptbp2fl/fl mice (Supplementary Fig. 3f,g). Cd79acre/+Ptbp1fl/fl cKO GC B cells had switched to IgG1 in vivo at greater frequencies compared to Cd79a+/+Ptbp1fl/fl control GC B cells (Supplementary Fig. 3h), indicating the presence of functional AID in Ptbp1-deficient GC B cells. Additionally, mutations were found at similar frequencies in the Jh4-intronic region of GC B cells sorted from Cd79acre/+Ptbp1fl/fl and Cd79a+/+Ptbp1fl/fl mice (Supplementary Fig. 3i,j). Thus, PTBP1 is necessary in B cells for optimal antibody affinity maturation, but this is unlikely to stem from reduced function of AID.
PTBP2 partially compensates for the loss of PTBP1 in GC B cells
The expression of PTBP2 in Ptbp1-deficient GC B cells could compensate for the absence of PTBP1 in GC cells31,32. To address this in the absence of confounding effects due to the deletion of genes during the bone marrow stages of B cell development, we introduced transgenic-Aicda-cre (AicdaTg-cre) to generate Ptbp1 single and Ptbp1:Ptbp2 double conditional knockout (dcKO) mice. After immunisation with sheep red blood cells (SRBCs) the numbers and proportions of GC B cells of AicdaTg-cre Ptbp1fl/fl cKO mice were reduced compared to Ptbp1fl/flPtbp2fl/fl control mice and the remaining GC B cells had the altered LZ/DZ phenotype seen in Cd79acre/+Ptbp1fl/fl mice (Fig. 3a,b). AicdaTg-cre Ptbp1+/+Ptbp2+/+ mice showed similar LZ and DZ B cell numbers compared to Ptbp1fl/flPtbp2fl/fl control mice (Supplementary Fig. 3k). Thus, PTBP1 function in B cells is required subsequent to B cell activation and expression of AID.
Figure 3. PTBP2 is partially redundant with PTBP1 in GC B cells.
(a) Representative flow cytometry plots showing gating strategy of for GC B cells, DZ and LZ GC B cells following SRBC immunisation (day 8). Events shown on the left have been pre-gated on CD19+ cells. Numbers indicate the percentages of the gated populations.
(b) Percentages and numbers of GC B cells, DZ GC B cells and LZ GC B cells identified as shown in a from spleens of mice after immunisation with SRBC (day 8). Bar graphs show data from individual mice and the mean is depicted with the bar. **P<0.01, ***P<0.001 from two-tailed unpaired Student’s t-test are shown for the indicated comparisons. Data in a and b is representative from two independent experiments.
(c) PTBP1, PTBP2 and PTBP3 expression in GC B cells (CD95+CD38lowCD19+) from spleens of mice 8 days post SRBC immunisation. Numbers shown in plots are the percentages of cells within the gates.
(d) Geometric mean fluorescence intensity of PTBP1 analysed by flow cytometry in GC B cells shown in c. Each symbol represents data from a different mouse. **P<0.01, ***P<0.001 from unpaired two-tailed Student’s t-test are shown for the comparisons indicated.
Compared to Ptbp1fl/flPtbp2fl/fl control and AicdaTg-cre Ptbp1fl/fl single cKO mice very few GC B cells were detected in AicdaTg-cre Ptbp1fl/flPtbp2fl/fl dcKO mice (Fig. 3a,b and Supplementary Fig. 3k). The residual AicdaTg-cre Ptbp1fl/flPtbp2fl/fl dcKO GC B cells had the phenotype typical of LZ B cells (Fig. 3a,b) and they expressed PTBP3, reduced amounts of PTBP1 but no PTBP2 (Fig. 3c,d). No NP-specific IgG1 antibody secreting cells were detected in AicdaTg-cre Ptbp1fl/flPtbp2fl/fl dcKO mice 21 days after immunisation with NP-KLH (Data not shown). Thus, PTBP2, but not PTBP3, partially compensates for the loss of PTBP1.
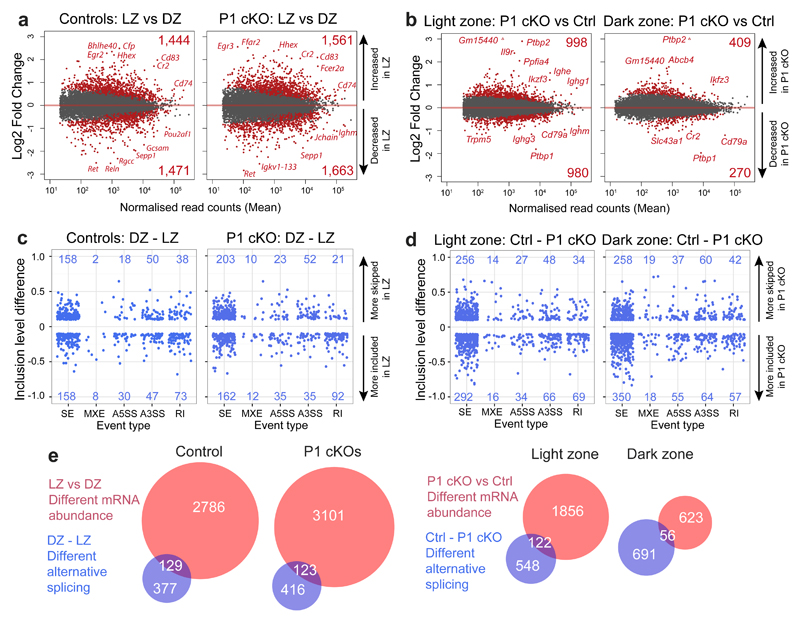
PTBP1 regulates mRNA abundance and alternative splicing in GC B cells
We performed mRNAseq on LZ and DZ B cells of Cd79a+/+Ptbp1fl/fl control and Cd79acre/+Ptbp1fl/fl cKO (PTBP1 cKO) mice. In addition, we generated a transcriptome-wide inventory of PTBP1 binding sites in mitogen-activated B cells using individual-nucleotide resolution Cross-Linking and ImmunoPrecipitation (iCLIP) (Supplementary Table 1). By combining mRNAseq with PTBP1 iCLIP we can discern direct from indirect effects of PTBP1 on the transcriptome and, through the consideration of positional information, deduce the likely mechanism of action of PTBP1. Changes in mRNA abundance at the whole gene level in control LZ B cells compared to control DZ B cells were consistent with those seen previously33 and were conserved in Ptbp1-deficient GC B cells (Fig. 4a, Supplementary Fig. 4a,b and Supplementary Table 2).
Figure 4. PTBP1 regulates mRNA abundance and AS in GC B cells.
(a) MA plot comparing changes in mRNA abundance at the gene level when comparing LZ to DZ GC B cells in Cd79a+/+Ptbp1fl/fl (Controls) or Cd79acre/+Ptbp1fl/fl (P1 cKO) mice.
(b) MA plot comparing changes in mRNA abundance at the gene level in LZ and DZ GC B cells from Cd79a+/+Ptbp1fl/fl (Ctrl) and Cd79acre/+Ptbp1fl/fl (P1 cKO) mice.
In a and b genes with significant (padj value < 0.1) different mRNA abundance and expressed with at least 1 fragments per kilobase of exon per million reads mapped (FPKM) in one of the conditions analysed are shown in red, genes with a padj value ≥0.1 are shown in dark grey. The number of significantly changed genes are given in the right-hand corners of each plot.
(c) Changes in AS when comparing LZ to DZ GC B cells in controls (Cd79a+/+Ptbp1fl/fl) and P1 cKO (Cd79acre/+Ptbp1fl/fl) cells for skipped exons (SE), mutually exclusive exons (MXE), alternative 5’ and 3’ splice sites (A5SS and A3SS, respectively) and retained introns (RI) (Supplementary Fig. 4e).
(d) Changes in AS due to Ptbp1 deletion in LZ and DZ GC B cells. Ctrl: Cd79a+/+Ptbp1fl/fl and P1 cKO: Cd79acre/+Ptbp1fl/fl.
c and d show inclusion level differences of significantly (FDR <0.05) differentially AS events with an inclusion level difference bigger than 10% (≥0.1 or ≤-0.1) for the comparisons made. Supplementary Fig. 4f shows an example of inclusion level difference calculation. Numbers in the graphs show the number of events that are differentially alternatively spliced.
(e) Overlaps of the number of genes with different mRNA abundance (a,b) and different AS (at any of the event types shown in c,d).
Comparison of PTBP1 cKO to control GC B cells revealed 998 and 980 genes with increased and decreased mRNA abundance, respectively, in the LZ, and 409 and 270 genes with increased and decreased mRNA abundance, respectively, in the DZ (Fig. 4b, Supplementary Table 2). The changes in genes that were differentially expressed in LZ and DZ B cells due to Ptbp1 deletion showed a strong positive correlation (Supplementary Fig. 4c) indicating that the absence of PTBP1 affects the same genes in LZ and DZ B cells. PTBP1 can increase mRNA stability when bound to 3’UTRs19,23. Amongst genes with either increased or decreased mRNA abundance the proportions of genes bound by PTBP1 in their 3’UTRs were similar (~20%, Supplementary Fig. 4d). Thus, PTBP1-binding to 3’UTRs does not appear to have a marked preference towards increasing or decreasing mRNA stability in GC B cells.
We used rMATS to determine inclusion level differences between two conditions for five different types of AS events (Supplementary Fig. 4e,f). We identified 266 AS events more skipped and 316 events more included (from 506 genes) with an inclusion level difference greater than 10% in control LZ compared to control DZ B cells (Fig. 4c, Supplementary Table 3) indicating extensive AS changes in the transition between LZ and DZ. The changes in AS due to Ptbp1 deficiency were greater in both the magnitude and the number of events (Fig. 4d, 856 AS events in LZ and 960 AS events in DZ GC B cells) than the changes seen between LZ and DZ control B cells (Fig. 4c, 582 events). The 52% overlap of AS events that change between LZ and DZ in control B cells and those affected by Ptbp1 deletion (Supplementary Fig. 4g) indicates that PTBP1 affects a substantial proportion of the AS events that differ between LZ and DZ B cells.
In LZ B cells 379 AS events were more skipped and 477 AS events were more included (in 670 genes) due to Ptbp1 deletion (Fig. 4d, Supplementary Table 3). In DZ B cells, 416 AS events were more skipped and 544 AS events were more included (in 747 genes) due to Ptbp1 deletion (Fig. 4d, Supplementary Table 3). The proportions of AS events directly regulated by PTBP1, inferred from the binding of PTBP1 in the vicinity of the event, varied depending on the type of AS event and ranged from ~50% in mutually exclusive exons (MXEs) to ~7% in alternative 3’ splice sites (A3SS) (Supplementary Fig. 4h). The proportions of skipped and included AS events due to Ptbp1 deletion bound by PTBP1 were similar (Supplementary Fig. 4h) indicating that PTBP1 both represses and activates AS in GC B cells. The overlap between genes that have different mRNA abundance and those that have different AS due to Ptbp1 deletion is 6% in LZ and in 8% DZ (Fig. 4e). Similarly, there was a small overlap between genes with changes in abundance and AS when control LZ B cells are compared to control DZ B cells (Fig. 4e). Taken together, our data shows that PTBP1 is a posttranscriptional regulator in GC B cells where it controls mRNA abundance and five different types of AS events.
PTBP1 is necessary for the c-MYC-dependent program induced upon positive selection
To determine if PTBP1 regulated changes in annotated gene ontology (GO) pathways we examined genes with changes at the mRNA abundance or AS level separately. Several GO terms relevant to the biology of GC B cells were enriched amongst genes with different mRNA abundance (Supplementary Fig. 5 and Supplementary Table 4). No pathways were enriched (FDR<0.1) when AS changes were analysed (Supplementary Table 4). Instead, individual genes of different pathways are regulated by PTBP1 at the AS level.
Pathways involved in nucleotide biosynthesis and cell proliferation were enriched amongst genes with differential mRNA abundance in LZ and DZ B cells (Supplementary Fig. 5a-c). In LZ B cells most of the differentially expressed genes within these pathways had decreased mRNA abundance (Supplementary Fig. 5b). Genes involved in the regulation of Ras protein signaling were also enriched in LZ B cells, but most of these genes had increased mRNA abundance (Supplementary Fig. 5b). Pathways implicated in B cell differentiation, lymphocyte migration, cholesterol metabolism and the regulation of apoptosis in leukocytes were enriched in DZ B cells (Supplementary Fig. 5a,c). Among the 243 genes with different mRNA abundance that belong to the enriched pathways we found 64 bound by PTBP1 in their 3’UTR and only 10 to be AS in the absence of PTBP1 (Supplementary Fig. 5b,c). Thus PTBP1 directly regulates some of these genes, but also has an indirect effect on the expression of genes in these pathways. We investigated further how the lack of PTBP1 affects the expression of genes that are part of the c-MYC hallmark34 and also of genes induced in response to T cell help3,7. In LZ B cells lacking PTBP1 there was a global decrease in mRNA abundance of the genes induced upon positive selection compared to control LZ B cells (Fig. 5a). 21% of the genes with reduced mRNA abundance in LZ B cells due to Ptbp1 deletion are part of the gene expression program induced upon positive selection (Supplementary Fig. 6a and Supplementary Table 5).
Figure 5. Positive selection gene expression program is reduced due to Ptbp1 deletion.
(a) Genes which have different mRNA abundance (DA) in LZ GC B cells due to Ptbp1 deletion which are c-MYC targets (GSEA HALLMARK_MYC_TARGETS_V1), increased in GC B cells receiving high levels of T cell help compared to CG B cells which do not receive high levels of T cell help and increased in c-MYC+AP4+ LZ GC B cells compared to c-MYC-AP4- LZ GC B cells. DS means differentially AS (inclusion level difference >10%). Numbers in the MA plots indicate the number of genes that are part of the studied gene sets and have DA in LZ GC B cells due to the lack of PTBP1.
(b) Myc and AP4 (Tfap4) mRNAseq data from LZ and DZ GC B cells.
(c) GFP-c-MYC+ GC B cells gating strategy. Events shown correspond to GC B cells (CD19+CD38lowCD95high) from spleens of Rag2-/- mice reconstituted with Cd79a+/+ Ptbp1fl/flMycGFP/GFP or Cd79acre/+Ptbp1fl/flMycGFP/GFP bone marrow cells 8 days post SRBC immunisation.
(d) Percentages of GFP-c-MYC+ cells within GC B cells and c-MYC gMFI of GFP-c-MYC+ GC B cells as shown in c. Dots show data from individual mice. Bars show the means. ns P>0.05, *P<0.05 from unpaired two-tailed Student’s t-tests are shown.
(e) Differentially AS events (FDR <0.05) with an inclusion level difference greater than 10% due to Ptbp1 deletion in LZ GC B cells from genes that are part of the gene sets shown in a. Labels indicate the gene name followed by the type of alternatively spliced event and the rMATS id number, which can be found in Supplementary Table 3. Genes bound by PTBP1 nearby the AS event can be found in Supplementary Table 5.
Myc mRNA abundance and AS pattern were the same in Cd79acre/+Ptbp1fl/fl cKO and Cd79a+/+Ptbp1fl/fl control LZ B cells (Fig. 5b and Supplementary Table 3). The proportions of GFP-cMYC+ GC B cells and c-MYC protein levels following SRBC immunisation in Rag2-/- mice reconstituted with Cd79acre/+Ptbp1fl/flMycGFP/GFP bone marrow cells were similar to those of Rag2–/– mice reconstituted with Cd79a+/+Ptbp1fl/flMycGFP/GFP bone marrow cells (Fig. 5c,d). Furthermore, Tfap4 mRNA encoding AP4 (Fig. 5b) and the levels of Ser240/244 phosphorylation of ribosomal protein S6 in Cd79acre/+Ptbp1fl/fl cKO LZ GC B cells were similar to those of Cd79a+/+Ptbp1fl/fl control LZ GC B cells (Supplementary Fig. 6b). Additionally, Cxcr4, Cxcr5, Bcl-6, Bach2, Aicda, IL21r, Foxo1 and Batf and had equivalent expression and AS patterns in Ptbp1-deficient and control GC B cells (Supplementary Fig. 6c-e and Supplementary Table 3). Thus GC B cells do not require PTBP1 for the sensing of T cell help or the induced expression of c-MYC.
28% of the mRNAs that are part of the c-MYC-dependent gene expression program induced upon positive selection and reduced in LZ B cells due to Ptbp1 deletion were bound by PTBP1 in their 3’UTR (Supplementary Fig. 6a and Supplementary Table 5). This indicates a direct role of PTBP1 in stabilising a fraction, but not all, of c-MYC-dependent transcripts. Moreover, 22 genes induced upon positive selection showed differential AS (inclusion level difference >10%) due to Ptbp1-deletion (Fig. 5e and Supplementary Fig. 6a) and six of these were bound by PTBP1 near the AS event (Supplementary Table 5). Pkm, Abcb1b, Tspan33, Phb2 and Dkc1 were the only genes with both reduced mRNA abundance and also differential AS (inclusion level difference >10%) due to the lack of PTBP1. Thus PTBP1 regulates directly and indirectly the expression of the c-MYC-dependent gene expression program in positively selected GC B cells through regulating mRNA abundance and AS.
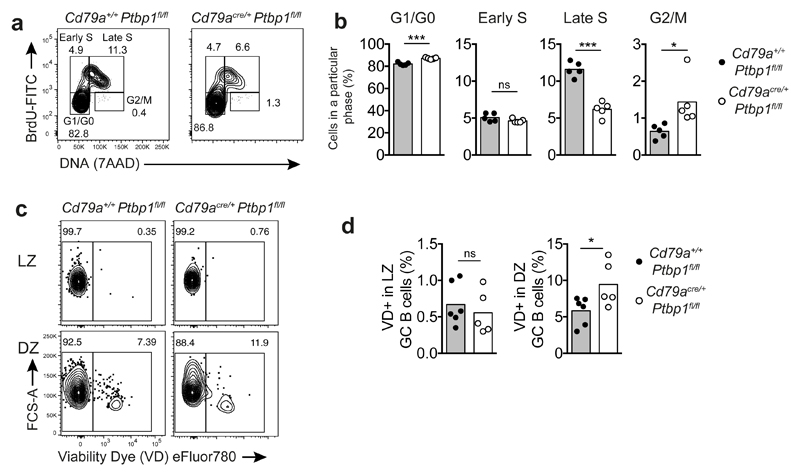
PTBP1 regulates proliferation of GC B cells
We analysed if PTBP1 was required for GC B cell proliferation by measuring DNA content and the incorporation of bromodeoxyuridine (BrdU) into GC B cells in vivo. The proportions of Cd79acre/+Ptbp1fl/fl cKO GC B cells in late S-phase (BrdU+ with high DNA content) were reduced compared to control Cd79a+/+Ptbp1fl/fl GC B cells in mice immunised with NP-KLH (Fig. 6a,b). By contrast, the proportions of cells that were in early S-phase (BrdU+ with low DNA content) were similar between Cd79acre/+Ptbp1fl/fl cKO and control Cd79+/+Ptbp1fl/fl GC B cells. Additionally, Cd79acre/+Ptbp1fl/fl cKO GC B cells had increased proportions of cells in G2/M phases compared to control Cd79a+/+Ptbp1fl/fl GC B cells (Fig. 6a,b). The proportions of cells in late S-phase were reduced in both LZ (5/5 experiments) and DZ (4/5 experiments) B cells from Cd79acre/+Ptbp1fl/fl cKO mice compared to Cd79a+/+Ptbp1fl/fl controls (Supplementary Fig. 7a). We also observed a reduction in the proportion of cells in late S-phase of GC B cells from AicdaTg-cre Ptbp1fl/fl cKOs compared to AicdaTg-cre Ptbp1+/+Ptbp2+/+ or Ptbp1fl/flPtbp2fl/fl controls immunised with SRBCs (Supplementary Fig. 7b). The proportions of GC B cells in late S-phase were also reduced amongst Ptbp1-deficient cells in bone marrow chimeras immunised with NP-KLH (Supplementary Fig. 7c) showing that this is a cell autonomous defect. In contrast to GC B cells, the proportions of cells at different stages of the cell cycle were normal in the highly proliferative early-pre B cells of Cd79acre/+Ptbp1fl/fl cKO mice (Supplementary Fig. 7d). Thus PTBP1 is not universally required in cells that are undergoing cell division but is necessary for the progression of GC B cells through late S-phase.
Figure 6. Late S-phase progression is impaired in the absence of PTBP1.
(a) Gating strategy for GC B cells (B220+CD19+CD38lowCD95high) at different stages of the cell cycle based on BrdU and DNA (7AAD) staining by flow cytometry in cells from spleens 7 days after immunisation with NP-KLH. Numbers shown are percentages.
(b) Percentages of cells in each phase of the cell cycle as shown in a. Individual data points represent individual mice. Bars depict the mean. ns P>0.05, *P<0.05 and ***P<0.001 from a two-tailed Student’s t-test are shown above the comparisons made. Data shown is representative from three independent experiments.
(c) Gating strategy for dead (viability dye eFluor780+) LZ (CD86highCXCR4low) or DZ (CD86lowCXCR4high) GC B cells (CD95+CD38lowCD19+B220+) by flow cytometry. Numbers are proportions of cells.
(d) Percentages of dead (viability dye eFluor780+) cells shown in c. Data points show data from individual mice of one out of three similar independent experiments. Bars depict means. ns P>0.05 and *P<0.05 shown are from two-tailed unpaired t-tests. Data shown is representative from three independent experiments.
Closer examination of the expression of genes important for cell cycle progression revealed that Ccnd2, Ccnd3 and Ccne2 were unaffected in Cd79acre/+Ptbp1fl/fl cKO GC B cells compared to Cd79a+/+Ptbp1fl/fl control GC B cells (Supplementary Fig. 7e) suggesting that the GO enrichment in proliferation and nucleotide biosynthetic pathways does not arise from a failure to express early cell cycle progression factors. Impaired nucleotide synthesis could cause replication stress as cells progress through S-phase and cause cell death35. Flow cytometric analyses using a fixable viability dye to detect non-viable cells showed a higher proportion of dead cells amongst Cd79acre/+Ptbp1fl/fl compared to Cd79a+/+Ptbp1fl/fl DZ GC B cells (Fig. 6c,d). By contrast, the proportions of dead cells in LZ B cells were similar between Ptbp1-deficient and sufficient cells (Fig. 6c,d). PTBP1 is thus necessary for progression through the S and G2/M phases of the cell cycle and promotes the survival of DZ cells.
PTBP1 controls alternative splicing of c-Myc target genes that regulate B cell proliferation
Given the impaired proliferation of Ptbp1-deficient GC B cells we looked for evidence of PTBP1-dependent AS isoforms amongst genes that are part of the c-MYC-dependent gene expression program that are important for proliferation. The mRNA encoding thymidylate synthase (Tyms), a c-MYC target gene (Fig. 5a) necessary for de novo nucleotide synthesis, is 30-fold increased in GC B cells compared to naive B cells and is increased upon positive selection in GC B cells (Fig. 7a). Tyms mRNA was differentially spliced in the absence of PTBP1 (Fig. 7b). Ptbp1 deletion resulted in a complex AS pattern with increased inclusion of exons and retained introns (yellow bins, Fig. 7b) that generate NMD-predicted transcript isoforms. We quantified the ratio of mRNAseq reads that map to segments generating NMD-predicted transcripts (NMD yellow bins, Fig. 7b) relative to mRNAseq reads that map to the first three exons of Tyms which encode the full-length protein (FL blue bins, Fig. 7b). The ratio of NMD/FL reads was reduced upon positive selection in GC B cells indicating an increase of protein coding Tyms mRNA in positively selected GC B cells (Fig. 7c). The NMD/FL read ratio was increased in LZ and DZ B cells (Fig. 7d) and Tyms mRNA abundance (at the whole gene level) was reduced due to Ptbp1 deletion in LZ B cells (Fig. 7e). TYMS protein was reduced due to Ptbp1 deletion in LZ and DZ B cells (Fig. 7f). Consistent with a direct role for PTBP1 in regulating these Tyms AS events, iCLIP showed PTBP1 bound to this region (Fig. 7a). These data indicate that PTBP1 ensures increased TYMS expression during positive selection by regulating Tyms AS.
Figure 7. PTBP1 regulates Tyms mRNA abundance through control of alternative splicing.
(a) Tyms FPKMs in GC B cells compared to naïve B cells49, in GC B cells receiving high levels of T cell help (Anti-DEC-Ctrl) compared to CG B cells which do not (Anti-Dec-OVA)3 or in LZ and DZ B cells which express c-MYC+AP4+ or not7.
(b) Tyms mRNAseq from LZ GC B cells and PTBP1 iCLIP data. Sashimi plots show read coverage and the reads that map to exon-exon junctions with arches. The numbers on the arches are the number of reads that map to that exon-exon junction. Scale on the PTBP1 iCLIP track shows the number of independent X-link sites identified.
(c) NMD/FL read ratio of reads mapping to exons and retained introns which will generate a NMD Tyms transcript (NMD, yellow bins in b) vs. reads mapping to the first three exons needed for the full-length Tyms (FL, blue bins in b) of mRNAseq libraries from GC B cells which receive high levels of T cell help (Anti-Dec-OVA) or not (Anti-DEC-Ctrl)3 and in LZ and DZ GC B cells which express c-MYC+AP4+ or not7.
(d) NMD/FL read ratio of mRNAseq libraries from LZ and DZ GC B cells.
(e) Tyms DESeq2 normalised read counts. ***Padj value<0.001 is shown. Not significant (ns) Padj>0.1.
(f) TYMS protein levels analysed by flow cytometry in non-GC B cells (CD19+CD95-CD38high) and LZ (CD86hiCXCR4low) or DZ (CD86lowCXCR4high) GC B (CD19+CD95+CD38low) cells from the spleens of mice 8 days after immunisation with SRBCs. gMFI of TYMS is shown after subtraction of background staining determined with isotype control staining. Student’s two-tailed t-tests. *P<0.05 and ***P<0.001.
M-type pyruvate kinase (Pkm) catalyses the conversion of phosphoenolpyruvate to pyruvate in glycolysis and is also a c-Myc target28 that is differentially spliced due to Ptbp1 deletion (Fig. 5e). Pkm encodes two protein isoforms generated from mutually exclusive inclusion of exon 9 (PKM1) or exon 10 (PKM2). Whereas PKM1 exists only as a highly active tetrameric form, PKM2 interchanges between less active monomeric and dimeric forms as well as a fully active tetramer in response to nutrient availability and energy demands36. B cells express Pkm2 almost exclusively and Pkm2, but not Pkm1, is induced upon positive selection in GC B cells (Fig. 8a). In the absence of PTBP1 the inclusion level of exon 9 is increased from 0.05 to 0.2 (Fig. 8b,c). Immunoblot of proteins from naive and mitogen-activated B cells in vitro revealed that PKM1 was hardly detected in Cd79a+/+Ptbp1fl/fl B cells but readily detected in Cd79acre/+Ptbp1fl/fl cKO B cells (Fig. 8d). This AS event has been previously shown to be regulated by PTBP1 in human cell lines to favour the production of PKM228,31. Our iCLIP data revealed PTBP1 binding close to the intronic 3’ splice site of exon 9 in B cells (Fig. 8c). Thus, PTBP1 promotes skipping of this exon favouring inclusion of exon 10 and suppression of Pkm1 mRNA in GC B cells (Fig. 8e).
Figure 8. PTBP1 regulates alternative splicing of Pkm.
(a) Pkm1 and Pkm2 transcript levels (FPKMs) calculated with Cuffnorm in GC B cells and naïve B cells49, in GC B cells receiving high levels of T cell help (Anti-DEC-Ctrl) and CG B cells which do not receive high levels of T cell help (Anti-Dec-OVA)3 or in LZ and DZ GC B cells which express c-MYC+AP4+ or not7.
(b) rMATS inclusion values for exon 9 (Pkm1) of Pkm.
(c) Visualisation of mRNAseq and PTBP1 iCLIP data as in Fig. 7b but for Pkm exons 8 to10.
(d) Immunoblots of PKM1 and PKM2 proteins in B cells freshly isolated from the spleen (ex vivo) or stimulated in vitro for 48h with LPS. Protein lysates from B cells isolated from different mice (numbers above the plots) are shown. Immunoblot images were cropped. Full images can be found in Supplementary Fig. 8.
(e) Pkm1 and Pkm2 transcript levels (FPKMs) calculated with Cuffnorm in our mRNAseq libraries from LZ and DZ GC B cells.
(f) CellTrace Violet profile from in vitro stimulated splenic B cells stimulated for 62 hours as indicated above the histogram overlays in the presence of DASA-58 or vehicle control (as the highest concentration of DASA-58).
(g) Quantification of the proliferation profiles shown in f and absolute cell numbers recovered from the cultures. Each dot shows data from B cells isolated from an individual mouse. Bar graphs depict the mean. ns P>0.05, *P<0.05, **P<0.01 and ***P<0.001 from unpaired Student’s two-tailed t-tests are shown.
Expression of PKM1 has been shown to impair the proliferation of malignant cells and non-transformed fibroblasts and the small molecule DASA-58 activates PKM2 tetramerization and inhibits the proliferation of transformed cells36. DASA-58 reduced the proliferation of mouse B cells in response to in vitro stimulation with anti-CD40+IL-4+IL-5 or anti-IgM+IL-4 (Fig. 8f,g). These findings are consistent with the hypothesis that increased activity of pyruvate kinase, as would be expected upon PKM1 expression, is detrimental for B cell proliferation.
Discussion
PTBP1 is either induced by and acts downstream of c-MYC, or forms part of a previously unrecognised pathway that acts in parallel with c-MYC to drive GC B cell proliferation. We favour the former hypothesis as there is evidence that PTBP1 is a c-MYC-responsive gene in other cell systems28,29. A proliferation defect of Ptbp1-deficient ES cells and human CD4 T cells with reduced PTBP1 levels has been observed30,37,38 indicating that PTBP1 is necessary for cell proliferation in other systems but the basis for the reported effects was not clear. However, the requirement for PTBP1 in proliferation is not universal as the proliferation of Ptbp1-deficient early-Pre B cells was normal. This difference could reflect compensatory mechanisms or the distinct environments of the GC compared to the bone marrow39,40.
PTBP1 controls gene expression by regulating multiple processes in the biogenesis and fate of mRNA. The mRNA abundance or AS of 213 genes that are part of the c-MYC-dependent gene expression program induced upon positive selection was PTBP1-dependent, but there are additional direct and indirect targets of PTBP1 that are not part of the MYC-dependent program. Amongst these, we observed changes in AS of Sema4d, Pdlim7, Pbrm1, Acly and Ikzf3. Changes in Semaphorin 4d (CD100, Sema4d) could affect the migration of GC B cells through the LZ and DZ and interactions with T cells41. Changes in enigma (Pdlim7) could alter p53 expression42. Altered splicing of ATP citrate lyase (Acly) could have an impact on lipid biosynthesis43. Polybromo-1 (Pbrm1 or BAF180) is part of the SWI/SNF-B (PBAF) chromatin-remodelling complex and a c-MYC cofactor which could help propagate the c-MYC-induced gene expression program44 and alterations in Aiolos (Ikzf3) might influence plasma cell formation45. Despite the potential for these genes to have roles in GC B cell biology most of the alternative isoforms found have not been studied before. Elucidation of the functions of these AS transcripts in GC B cell biology will require careful examination.
Pkm and Tyms are but two examples of c-MYC regulated genes that are subject to PTBP1-dependent AS. TYMS inhibition by 5-flurouracil blocks primary T cell46 and B cell proliferation in vitro (data not shown). Splicing of the retained intron upstream of exon 2 from the full-length Tyms transcript is necessary for increased Tyms mRNA expression in cultured cells47. Reduced amounts of TYMS could be one limiting factor for the proliferation of positively selected GC B cells. Other PTBP1 regulated events must also promote GC B cell proliferation and our results implicate the regulation of PKM activity as important for B cell proliferation. PTBP1, by regulating Pkm AS, may limit glycolytic flux and thereby contribute to biosynthetic pathways through the accumulation of glycolytic intermediates36. Consistent with this, PKM1 expression is growth inhibitory when expressed in cancer xenograft tumour models36 and B cell proliferation in vitro was inhibited by PKM2 activators.
Previous studies reported a function of PTBP2 in antibody class switch recombination (CSR)48. We did not detect PTBP2 expression in GC B cells unless Ptbp1 is deleted, and antigen-specific IgG1 secretion was unaffected in mice with Ptbp2-deficient B cells. Therefore it is unlikely that PTBP2 promotes CSR in GC B cells. Nonetheless, the increase in the frequency of IgG1+ GC B cells in Ptbp1-deficient GC B cells could indicate that, if expressed, PTBP2 might indeed promote antibody CSR.
There may be additional PTBP1-mediated post-transcriptional processes of importance to the GC reaction. Changes in alternative polyadenylation (APA) analysed with DaPars found that Ptbp1 deletion affected the APA of 4 genes in LZ and 7 genes in DZ B cells (Data not shown), suggesting a limited role of PTBP1 in regulating APA in GC B cells compared to its roles controlling AS and mRNA abundance. However, to fully address the roles of PTBP1 in APA analysis of RNAseq libraries specifically targeted at capturing 3’-ends are required. In the present study we were unable to measure the impact of PTBP1 on the tempo of translation, e.g. through IRES-mediated regulation, within GC B cells and this must await improved techniques for measuring translational regulation in rare cell populations.
In summary, we have observed that the regulation of gene expression in B cells by PTBP1 is necessary for GC B cell proliferation. At the cellular level PTBP1 functions in GC B cells to promote the rapid progression through the late S-phase of the cell cycle. At the molecular level we have identified the role of PTBP1 in regulating the quantitative and qualitative changes in the transcriptome that are part of the c-MYC-dependent gene expression program. Post-transcriptional regulation by PTBP1 acts in concert with transcription factors such as c-MYC to integrate anabolic metabolism and cell cycle progression and drive the production of high affinity antibodies.
Online Methods
Mice
All mice were on a C57BL/6 background. For bone marrow chimera experiments, B6.SJL were used as recipients. Conditional knockout mice used in this study derive from crossing the following transgenic strains: Ptbp1fl/fl (Ptbp1tm1Msol)30, Ptbp2fl/fl (Ptbp2tm1.1Dblk)50, Cd79acre(Cd79atm1(cre)Reth)51 and AicdaTg-cre (Tg(Aicda-cre)9Mbu)52 as specified in the results section. GFP-c-MYC reporter mice Myctm1Slek53 and Rag2-/- knockout mice (Rag2tm1Fwa)54 were also used in this study.
Rats
RT7b rats were used for the generation of anti-PTBP3 monoclonal antibodies.
Immunisation Protocols and in vivo BrdU administration
All procedures performed were approved by the Babraham Institute's Animal Welfare and Experimentation Committee and the UK Home Office and are in compliance with all relevant ethical regulations. Mice immunised with alum-NP-KLH received 100 μg NP-KLH (Biosearch Technologies) intraperitoneally (ip). Mice immunised with SRBCs received 2*108 SRBCs ip. For in vivo BrdU incorporation experiments mice received 2 mg BrdU ip 1.5 hours before they were culled. Bone marrow competitive chimeras were generated by reconstituting lethally irradiated (2 x 500Rad) B6.SJL mice with 3*106 bone marrow cells derived from B6.SJL in a 1:1 ratio with CD79acre/+Ptbp1fl/fl cKO or CD79a+/+Ptbp1fl/fl control bone marrow cells administered intravenously. Rag2-/- knockout bone marrow chimeras were generated by reconstituting sub-lethally irradiated (1x 500Rad) Rag2-/- knockout mice with 3*106 bone marrow cells from Cd79a+/+ Ptbp1fl/flMycGFP/GFP or Cd79acre/+Ptbp1fl/flMycGFP/GFP mice administered intravenously. Mice used in immunisation experiments with different genotypes were sex and age matched. Whenever possible littermates of the same sex but different genotypes were kept in the same cages to avoid confounding effects. Male and female mice were used in this study. Except in bone marrow reconstitution experiments, immunisations were carried out on 8 to 14 week-old mice. Bone marrow competitive chimeras were immunised 15 weeks post reconstitution. Rag2-/- knockout bone marrow chimeras were immunised 13 weeks post reconstitution.
Flow cytometry
Single cell suspensions were prepared from tissues by passing the tissues though cell strainers with 70 and 40 μm pore sizes. After Fc receptors (CD16/32) were blocked with the monoclonal rat antibody 2.4G2 cells were stained with different antibodies listed in Supplementary Table 6 table in 1% FCS PBS at 4 °C. Cell viability was assessed by staining cells with the Fixable Viability Dye eFluor® 780 dye from eBioscience. Unless otherwise stated, dead cells were always excluded from the analysis. For intracellular staining cells were fixed and permeabilized with the BD Cytofix/Cytoperm™ Fixation and Permeabilization Solution from BD Biosciences. Nuclear permeabilization was carried out with the Permeabilization Buffer Plus from BD Biosciences when cells were prepared from the spleen or by freezing fixed cells in FCS containing 10% DMSO at -80 °C when cells were isolated from the bone marrow. Intracellular staining was carried out by incubating permeabilized cells with combinations of antibodies (listed in Supplementary Table 6) diluted in BD Perm/Wash Buffer. For some intracellular stains such as for those detecting PTBP1, the incubation with antibodies was carried out for 4 hours at room temperature. Foxp3/Transcription Factor Staining Buffer Set from eBioscience was also occasionally used. BrdU and DNA stainings were carried out using the FITC BrdU Flow Kit from BD Biosciences. Anti PTBP1, PTBP2 and PTBP3 monoclonal antibodies were directly conjugated using Alexa Fluor® 488, Alexa Fluor® 647 or Pacific Blue Antibody Labeling Kits from ThermoFisher Scientific.
Detection of NP specific immunoglobulins
NP-specific antibodies of different affinities were detected as previously described14 using two different conjugation ratios of NP to BSA. Detection of NP-specific antibodies with low and high affinity was done by coating ELISA plates with a ratio of at least 20 NP per 1 BSA molecule (NP20, Biosearch Technologies). Detection of NP-specific antibodies with only high affinity was performed by coating ELISA plates with a ratio of 2 NP per 1 BSA molecules. End-point titres were calculated from serial dilutions of serum samples.
Isolation of B cells and ex vivo stimulation
B cells were isolated from spleens of mice after preparing a single cell suspension by MACS negative depletion with a B cell isolation kit (Cat. # 130-090-862 from Miltenyi). When stimulated ex vivo B cells were cultured in IMDM media (Cat # 21980, ThermoFisher Scientific) with L-Glutamine and 25mM HEPES, supplemented with 10% heat inactivated FCS, 50 μM β-Mercaptoethanol, penicillin and streptomycin (GIBCO). Cells were stimulated with LPS (10 μg/ml 127:B0, Sigma); anti-CD40 antibody (FGK4.2, 10 μg/ml), IL-4 (10 ng/ml) and IL-5 (10 ng/ml) or anti-IgM antibody (B7.6, 9 μg/ml) and IL-4 (10 ng/ml). In proliferation assays B cells were labeled with CellTrace™ Violet (ThermoFisher) before culturing them. 1.5*105 B cells were added per well in 96 well plates. Cells were counted using counting beads analysed by flow cytometry. In PKM stimulation experiments DASA-5855 (MedChem Express, Cat. # HY-19330-1ml) was added to the cultures. The same amount of DMSO as the highest concentration of DASA-58 was added to the cultures as vehicle control.
PTBP1 iCLIP
iCLIP reveals the site of direct binding of a RBP to RNA at single nucleotide resolution. This is achieved first by the covalent binding of the RBP to its cognate RNA in intact cells by UV irradiation and second, through the truncation of cDNA synthesis when reverse polymerase encounters residual peptides derived from the RBP at the site of cross-linking to the RNA56. PTBP1 immunoprecipitations were carried out with the CLONE 1 monoclonal antibody (ThermoFisher Scientific, Cat. # 32-4800) coupled to protein A/G magnetic beads (Pierce, Cat. #88802) and UV-cross-linked (150mJ/cm2) cell extracts of primary B cells stimulated ex vivo for 48h with LPS as described above. B cells were isolated from several C57BL/6 females and pooled together before stimulation. 30*106 cells were used in each immunoprecipitation and were lysed in 50 mM Tris-HCL pH7.4, 100mM NaCl, 1% NP-40 and 0.1% SDS lysis buffer. Before immunoprecipitating PTBP1, cell extracts were treated with Turbo DNAse (Ambion, #AM2239) and low amounts of RNAse I (1.5 to 3 units) from Ambion (Cat. # AM2294). Immunoprecipitates were run on a SDS-PAGE and RNA-protein complexes were transferred to a nitrocellulose membrane. Protein-RNA complexes were isolated from the nitrocellulose membrane after cutting only the areas (from ~75 to ~120 kDa) where PTBP1 was expected to be cross-linked to long RNAs. After protein digestion, RNA was isolated and cDNA synthetized by reverse transcription. At this time barcoded primers were used which allow first, the identification of cDNAs generated from the same RNA molecule (with a random unknown 4 nucleotide barcode) which allows to discriminate PCR duplicates and second, multiplexing of several samples together. Amplification of iCLIP cDNA libraries was done with 20 to 27 PCR cycles. We carried out five replicates. Multiplexed iCLIP cDNA libraries were sequenced on an Illumina HiSeq 2000 platform on a 50 bp single-ended mode. Negative controls (PTBP1 immunoprecipitations from non-UV cross-linked lysates and immunoprecipitations with a mouse IgG1 negative isotype control antibody) resulted in very little isolated RNA, from which no cDNA library could be generated.
Computational processing of iCLIP data
Identification of transcriptome-wide PTBP1 binding sites was done as previously described56. Briefly, mapping of cDNAs to the mouse genome (mm10) was carried out with Bowtie and those reads that mapped to the same location and had the same random 4 nt barcode were considered PCR duplicates and collapsed as a single cDNA molecule. A PTBP1 binding site (or X-link site) is the nucleotide before the first nucleotide of a mapped cDNA molecule. A FDR value, which determines the probability of a X-link site to appear by chance, was calculated (as described in56) for each X-link site. iCLIP is highly dependent on RNA abundance and therefore, is not an absolute measurement of RNA-protein interactions. For this reason we pooled the five replicates together before calculating FDRs for each X-link site.
Generation of mRNAseq libraries
LZ and DZ GC B cells from CD79acre/+Ptbp1fl/fl cKO or CD79a+/+Ptbp1fl/fl control mice were FACS-sorted from animals immunised with alum-NP-KLH seven days before. Single cell suspensions were prepared from spleens of immunised mice. Erythrocytes were lysed and GC B cells were enriched before FACS sorting by depleting cells stained with biotinylated anti IgD, CD3e, Gr1 and Ter119 antibodies by MACS. Four biological replicates were used per condition. In each biological replicate, GC B cell-enriched splenocytes from 3 to 5 animals of the same genotype and sex were pooled together before FACS sorting. LZ and DZ GC B cells were sorted from the same GC B cell enriched samples. LZ and DZ GC B cells from control CD79a+/+Ptbp1fl/fl and CD79acre/+Ptbp1fl/fl cKO mice were sorted on the same day. Two biological replicates per condition were from females and two from males.
RNA was prepared using the RNeasy Micro Kit kit from Qiagen (Cat. #74004) from ~25.000 up to ~200.000 cells. RNA quality was analysed on a 2100 Bioanalyzer (Agilent). RNA integrity numbers ranged from 9.1 to 10. 2 ng of total RNA was used to generate cDNA from polyadenylated transcripts using the SMART-Seq v4 Ultra low input RNA kit from Clontech (Cat. #634888). cDNA quality was analysed on a 2100 Bioanalyzer (Agilent). 0.5 ng cDNA were used to prepare the mRNAseq libraries with 8 cycles of PCR using the Ultra Low library preparation kit v2 from Clontech (Cat. #634899). Compatible barcoded libraries were multiplexed and sequenced across three lanes on an Illumina HiSeq 2500 platform on a 100bp paired-end mode.
Analysis of mRNAseq libraries generated in this study
Trimming of libraries was carried out with Trimgalore (v0.4.2) using default parameters. After that, reads were mapped to the mouse Mus_musculus.GRCm38 genome with Hisat257 using -p 7 -t --phred33-quals --no-mixed --no-discordant parameters and providing a known splice sites file generated from the Mus_musculus.GRCm38.70.gtf annotation. Counting of reads mapping to all exons of a particular gene was done with HTSeq58 using the Mus_musculus.GRCm38.84.gtf annotation and default parameters. DESeq2 (v1.12.1)59 was used to calculate differential RNA abundance between two conditions at the whole gene level by quantifying differences in RNA complementary to all annotated exons of a particular gene in the RNAseq data. Information on the genotype and sex of the animals was included in the design formula in order to control for variation in the data due to the sex differences in the samples. DESeq2 results were only considered for genes that are expressed with at least 1 FPKM in any of the conditions. Significant differentially abundant genes are those that have an adjusted p-value <0.1.
Differential alternative splicing (AS) was analysed with rMATS (v3.2.2)60. rMATS uses an exon-centric approach to discover both annotated and unannotated AS events in a reference transcriptome. To compare changes in AS between two conditions rMATS calculates first inclusion levels (defined as the proportion of transcripts containing that particular AS segment) for five different types of AS events: skipped exons (SE), mutually exclusive exons (MXE), alternative 5’ splice sites (A5SS) alternative 3’ splice sites (A3SS) and retained introns in each of the two conditions (Supplementary Fig. 4e). Subsequently, rMATS calculates inclusion level differences by subtracting the inclusion levels of condition one from the inclusion levels of condition two (see Supplementary Fig. 4f). The version used of rMATS only accepts mapped reads of a particular length. For this reason, libraries trimmed with Trimgalore as described above were further trimmed with Trimmomatic v0.3561 so that all reads had a length of 98 bp. Reads shorter than 98 bp were discarded. These reads of only 98bp were then mapped to the mouse genome using Hisat2 as described above. rMATS was run on a paired mode (-analysis P) to analyse differential AS between two conditions using the Mus_musculus.GRCm38.84.gtf annotation. Only results obtained with reads that map to exon-exon junctions were used for further analysis. Genes that have less than 1 FPKM in the analysed conditions were discarded. Significantly differentially spliced events are those that have an FDR <0.05. A cut-off of an inclusion level difference greater than 10% (0.1) was introduced for significant differentially alternatively spliced events.
We assigned PTBP1 binding to the vicinity of a differentially alternatively spliced event if we found at least a significant PTBP1 binding site in our iCLIP data in the following cases. Skipped exons (SE) were considered to be bound by PTBP1 if a binding site was identified on the SE, on the intronic 500 nucleotides upstream and downstream of the SE 3’ splice site (SS) and 5’SS, respectively; on the upstream flanking constitutive exon and the intronic 500 nucleotides downstream of its 5’SS or on the downstream flanking constitutive exon and the intronic 500 nucleotides upstream of its 3’SS. Mutually exclusive exons (MXE) were bound by PTBP1 if a binding site was found on any of the MXE and the intronic 500 nucleotides upstream and downstream of their 3’SS and 5’SS, respectively; on the upstream flanking constitutive and the intronic 500 nucleotides downstream of its 5’SS or on the downstream flanking constitutive exon and the intronic 500 nucleotides upstream of its 3’SS. Alternative 5’ splice sites (A5SS) were bound by PTBP1 if a binding site was found on the longer exon generated by the A5SS, the intronic 500 nucleotides downstream of the 5SS of the longest A5SS exon or on the flanking constitutive exon downstream of the A5SS and the intronic 500 nucleotides upstream of its 3SS. Alternative 3’ splice sites (A3SS) were bound by PTBP1 if a binding site was found on the longer exon generated by the A3SS, the intronic 500 nucleotides upstream of the 3SS of the longest A3SS exon or on the flanking constitutive exon upstream of the A3SS and the intronic 500 nucleotides downstream of its 5SS. A retained intron was bound by PTBP1 if a binding site was found on the exon resulting from intron retention.
Differential alternative polyadenylation site usage
Alternative polyadenylation (APA) usage was assessed with the 'Dynamic analysis of Alternative PolyAdenylation from RNA-Seq' algorithm (DaPars v0.9.162). DaPars computes first the Percentage of Distal polyadenylation (polyA) site Usage Index (PDUI) and, subsequently, computes changes in PDUI between two conditions (ΔPDUI). Annotated gene models were generated from mouse genome build GRCm38/mm10, facilitating the prediction of proximal de novo APA sites as well as long and short 3′ UTR expression levels. Bedtools v2.25.0 was used to convert the RNA-seq BAM files to BedGraph format, and these files were then used as input for DaPars to identify dynamic APA usage between the control and knockout mice. Differences in APA were considered for those genes with an absolute change in percentage of distal polyA site usage of >20% (ΔPDUI >|0.2|), an adjusted P-value of <0.05 and a fitted value of the regression model used to identify the proximal polyA site > 500.
Analysis of mRNAseq libraries previously published
RNAseq data from Hogenbirk et al.49 was trimmed with Trimgalore (v0.3.8) using default parameters. Trimmed reads were mapped to the mouse genome using Tophat2(v2.0.12)63 with -p 6 -g 2 parameters and the Mus_musculus.GRCm38.70.gtf annotation. Mapped reads to exons of a particular gene were counted with HTSeq58 using the Mus_musculus.GRCm38.73.gtf annotation and default parameters. Differential RNA abundance was calculated with DESeq2 (v1.4.5).
RNAseq data generated by Gitlin et al.3 was trimmed with Trimgalore (v0.4.1). Trimmed reads were mapped to the mouse genome with Tophat2 (v2.0.12)63 with -p 6 -g 2 parameters and the Mus_musculus.GRCm38.70.gtf annotation. Mapped reads were counted with HTSeq using the Mus_musculus.GRCm38.75.gtf annotation and default parameters. DESeq2 (v1.8.1) was used to analyse differential RNA abundance between GC B cells that received high levels of T cell help (Anti-DEC-OVA) and GC B cells that did not receive high levels of T cell help (Anti-DEC-Ctrl). Genes with a p-adjusted value <0.05 were considered to have significant differential RNA abundance. Genes with a positive log2 fold change (Anti-DEC-OVA vs Anti-DEC-Ctrl) were chosen as genes that are increased in GC B cells receiving high levels of T cell help compared to GC B cells that are not receiving high levels of T cell help (used in Fig. 5).
RNAseq data published by Chou and colleagues7 was trimmed with Trimgalore (v0.4.2_dev). Trimmed reads were mapped to the mouse genome using Hisat2(v2.0.5)57 and the Mus_musculus.GRCm38 genome. Reads mapping to all exons of a particular gene were counted with HTSeq using the Mus_musculus.GRCm38.84.gtf annotation and default parameters. Differential RNA abundance between two conditions was calculated with DESeq2 v1.12.4. Genes with an increased RNA abundance in AP4+Myc+ LZ GC B cells compared to AP4-Myc- LZ GC B cells used in Fig. 5 were defined as genes with a p-adjusted value <0.05 whose log2 fold change (AP4+ Myc+ LZ GC B cells vs AP4- Myc- LZ GC B cells) is equal or greater than 1.5.
Calculation of FPKMs
FPKMs (Fragments per kilobase of exon per million reads mapped) were used to compare expression values of different genes or transcripts and to filter genes based on their expression level. FPKMs were calculated with the Cuffnorm option of Cufflinks (v2.2.1)64 using the geometric normalisation method and the mapped reads of the different RNAseq libraries described above. The following annotations were used to calculate FPKMS from the different studies: Mus_musculus.GRCm38.84.gtf was used with our RNAseq data and data from Chou et al.7, Mus_musculus.GRCm38.83.gtf was used with the data from Hogenbirk et al.49 and Mus_musculus.GRCm38.75.gtf was used with data published by Gitlin et al.3.
GO analysis
Gene ontology terms enrichment analysis was carried out with GOrilla65. A background list of genes (genes which were expressed with at least 1 FPKM in the conditions analysed) was included in the analysis. For visualisation purposes when several related terms were significantly enriched the term which had a higher percentage of significant genes with different mRNA abundance or AS was chosen to be shown in Supplementary Fig. 5.
RNAseq data visualisation
Sashimi plots, which show RNAseq coverage and reads mapping across exon-exon junctions were generated using the IGV genome browser.
Analysis of Jh4 mutations
Mutations in the Jh4 intronic region were analysed as previously described4. Briefly, genomic DNA was isolated from FACS-sorted GC B cells from spleens of mice that were immunised with alum-NP-KLH 7 days before. GC B cells from 2 to 4 mice of the same genotype and sex were pooled together. Jh4 intronic regions were amplified with the primers Jh4-intron Forward and Jh4-intron Reverse in using the PfuUltra II Fusion HS DNA Polymerase (Agilent Technologies) in a PCR with 35 cycles, 57 °C annealing temperature and 15 seconds extension time at 72 °C. PCR amplified Jh4 intronic regions were cloned using the Zero Blunt® TOPO® PCR Cloning Kit (ThermoFischer scientific). Mutation frequencies were calculated by dividing the total number of mutations identified in each replicate by the total length of amplified DNA (565 bp per clone analysed, which is the genomic region amplified by PCR without taking into account the regions complementary to the primers).
Generation of anti PTBP3 mAb
RT7b rats were immunised with a GST-PTBP3 fusion protein containing amino acids 279 to 359 of the full-length mouse PTBP3 generated in E. coli. After several immunisations spleens were isolated and fused to the IR983F rat myeloma cell line. Hybridomas were screened by immunoblot using HEKT cell lysates expressing mouse PTBP3, PTBP1 or PTBP2-GFP fusion proteins. Two hybridomas secreted IgG2a antibodies specific for PTBP3: MAC454 and MAC455. Monoclonal antibodies secreted by these two hybridomas were purified by affinity chromatography using protein G.
Immunoblot
In order to analyse PKM1 and PKM2 expression, 12 μg of proteins extracted from B cells isolated as described above from individual mice and stimulated or not with LPS for 48h were run on a 10% SDS-PAGE and transferred to a nitrocellulose membrane. PKM1 detection was carried out with a rabbit monoclonal antibody (clone D30G6, Cell Signalling) and PKM2 was detected with a rabbit monoclonal antibody (clone D78A4, Cell signalling). Visualisation of anti PKM1 and PKM2 antibodies was carried out with an HRP-conjugated goat anti-rabbit antibody (Cat # 2020-10, Dako). Immunoblot analysis of the different PTBPs was carried out with 15 μg of proteins extracted from B cells isolated as described above from individual mice run on a 10 % SDS-PAGE and transferred to a nitrocellulose membrane. Mouse monoclonal primary antibodies were detected with an anti-mouse TrueBlot HRP-conjugated antibody (eBioscience, Cat. #18-8817-31). Rat monoclonal primary antibodies were detected with anti-rat goat HRP-conjugated antibodies (Cat. #P0450, Dako). Rabbit primary antibodies were detected with HRP-conjugated goat anti-rabbit antibody (Cat. #2020-10, Dako).
Quantification and statistical analysis
Flow cytometry data was analysed using FlowJo (versions 10.0.8r1 or 9.8.3). Analysis and quantification of RNAseq and iCLIP experiments is detailed in the methods detail section. Statistical significance of flow cytometry data was assessed using Prism (versions 7 or 6). The details of the tests used in different experiments can be found in the figure legends).
Data availability
mRNAseq and iCLIP data that support the findings of this study have been deposited in GEO with the GSE100969 accession code.
Supplementary Material
Acknowledgments
We thank D. L. Black for the Ptbp2fl/fl mice, M. Reth for the Cd79acre mice, M. Busslinger for the AicdaTg-cre mice, B. P. Sleckman for the MycGFP/GFP mice, and F. W. Alt for the Rag2–/– mice; J. Ule, N. Haberman and T. Curk for help with iCLIP data; G. Butcher for assistance in generating anti-PTBP3 monoclonal antibodies; K. Bates, D. Sanger, A. Davis, the Biological Support Unit, Flow Cytometry and Bioinformatics Facilities for technical assistance; M. Spivakov and members of the Turner and Smith laboratories for helpful discussions. Supported by The Biotechnology and Biological Sciences Research Council (BB/J004472/1 and BB/J00152X/1 to M.T), the Wellcome Trust (200823/Z/16/Z to M.T.) and Bloodwise (14022 to M.T.).
Footnotes
Accession codes
Data availability
mRNAseq and iCLIP data that support the findings of this study have been deposited in GEO with the GSE100969 accession code.
Author Contributions
E.M.-C., M.T. and C.W.J.S. designed experiments. E.M.-C., M.Sc., M.D.D.-M., S.E.B. and G.L., performed experiments and analysed data. E.M.-C. carried out computational analysis. R.M.R.C. ran DaPars. M.So. provided Ptbp1tm1Msol mice and anti-PTBP2 antibody. E.M.-C. and M.T. wrote the manuscript with input from the co-authors.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Mesin L, Ersching J, Victora GD. Germinal Center B Cell Dynamics. Immunity. 2016;45:471–482. doi: 10.1016/j.immuni.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannard O, Cyster JG. Germinal centers: programmed for affinity maturation and antibody diversification. Current Opinion in Immunology. 2017;45:21–30. doi: 10.1016/j.coi.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Gitlin AD, et al. T cell help controls the speed of the cell cycle in germinal center B cells. Science. 2015;349:643–646. doi: 10.1126/science.aac4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gitlin AD, Shulman Z, Nussenzweig MC. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature. 2014;509:637–640. doi: 10.1038/nature13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calado DP, et al. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nat Immunol. 2012;13:1092–1100. doi: 10.1038/ni.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dominguez-Sola D, et al. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat Immunol. 2012;13:1083–1091. doi: 10.1038/ni.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou C, et al. The Transcription Factor AP4 Mediates Resolution of Chronic Viral Infection through Amplification of Germinal Center B Cell Responses. Immunity. 2016;45:570–582. doi: 10.1016/j.immuni.2016.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ersching J, et al. Germinal Center Selection and Affinity Maturation Require Dynamic Regulation of mTORC1 Kinase. Immunity. 2017;46:1045–1058.e6. doi: 10.1016/j.immuni.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sander S, et al. PI3 Kinase and FOXO1 Transcription Factor Activity Differentially Control B Cells in the Germinal Center Light and Dark Zones. Immunity. 2015;43:1075–1086. doi: 10.1016/j.immuni.2015.10.021. [DOI] [PubMed] [Google Scholar]
- 10.Dominguez-Sola D, et al. The FOXO1 Transcription Factor Instructs the Germinal Center Dark Zone Program. Immunity. 2015;43:1064–1074. doi: 10.1016/j.immuni.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 11.Inoue T, et al. The transcription factor Foxo1 controls germinal center B cell proliferation in response to T cell help. Journal of Experimental Medicine. 2017;214:1181–1198. doi: 10.1084/jem.20161263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez NM, Lynch KW. Control of alternative splicing in immune responses: many regulators, many predictions, much still to learn. Immunol Rev. 2013;253:216–236. doi: 10.1111/imr.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaub A, Glasmacher E. Splicing in immune cells - mechanistic insights and emerging topics. Int Immunol. 2017;29:173–181. doi: 10.1093/intimm/dxx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diaz-Muñoz MD, et al. The RNA-binding protein HuR is essential for the B cell antibody response. Nat Immunol. 2015;16:415–425. doi: 10.1038/ni.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang X, Li B, Rao A. RNA-binding protein hnRNPLL regulates mRNA splicing and stability during B-cell to plasma-cell differentiation. Proc Natl Acad Sci USA. 2015;112:E1888–97. doi: 10.1073/pnas.1422490112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berkovits BD, Mayr C. Alternative 3' UTRs act as scaffolds to regulate membrane protein localization. Nature. 2015;522:363–367. doi: 10.1038/nature14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pioli PD, Debnath I, Weis JJ, Weis JH. Zfp318 regulates IgD expression by abrogating transcription termination within the Ighm/Ighd locus. J Immunol. 2014;193:2546–2553. doi: 10.4049/jimmunol.1401275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keppetipola N, Sharma S, Li Q, Black DL. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Critical Reviews in Biochemistry and Molecular Biology. 2012;47:360–378. doi: 10.3109/10409238.2012.691456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawicka K, Bushell M, Spriggs KA, Willis AE. Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochemical Society Transactions. 2008;36:641–647. doi: 10.1042/BST0360641. [DOI] [PubMed] [Google Scholar]
- 20.Gooding C, Kemp P, Smith CWJ. A novel polypyrimidine tract-binding protein paralog expressed in smooth muscle cells. J Biol Chem. 2003;278:15201–15207. doi: 10.1074/jbc.M210131200. [DOI] [PubMed] [Google Scholar]
- 21.Baralle FE, Giudice J. Alternative splicing as a regulator of development and tissue identity. Nature Reviews Molecular Cell Biology. 2017;825:267. doi: 10.1038/nrm.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan L-Y, et al. Generation of functionally distinct isoforms of PTBP3 by alternative splicing and translation initiation. Nucleic Acids Res. 2015;43:5586–5600. doi: 10.1093/nar/gkv429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knoch K-P, et al. Polypyrimidine tract-binding protein promotes insulin secretory granule biogenesis. Nature Cell Biology. 2004;6:207–214. doi: 10.1038/ncb1099. [DOI] [PubMed] [Google Scholar]
- 24.Knoch K-P, et al. PTBP1 is required for glucose-stimulated cap-independent translation of insulin granule proteins and Coxsackieviruses in beta cells. Molecular Metabolism. 2014;3:518–530. doi: 10.1016/j.molmet.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vavassori S, Shi Y, Chen CC, Ron Y, Covey LR. In vivo post-transcriptional regulation of CD154 in mouse CD4+ T cells. European Journal of Immunology. 2009;39:2224–2232. doi: 10.1002/eji.200839163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porter JF, Vavassori S, Covey LR. A Polypyrimidine Tract-Binding Protein-Dependent Pathway of mRNA Stability Initiates with CpG Activation of Primary B Cells. The Journal of Immunology. 2008;181:3336–3345. doi: 10.4049/jimmunol.181.5.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boutz PL, et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sabò A, et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014;511:488–492. doi: 10.1038/nature13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suckale J, et al. PTBP1 is required for embryonic development before gastrulation. PLoS ONE. 2011;6:e16992. doi: 10.1371/journal.pone.0016992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spellman R, Llorian M, Smith CWJ. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Molecular Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vuong JK, et al. PTBP1 and PTBP2 Serve Both Specific and Redundant Functions in Neuronal Pre-mRNA Splicing. Cell Reports. 2016;17:2766–2775. doi: 10.1016/j.celrep.2016.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Victora GD, et al. Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood. 2012;120:2240–2248. doi: 10.1182/blood-2012-03-415380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liberzon A, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nature Cell Biology. 2013;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dayton TL, Jacks T, Vander Heiden MG. PKM2, cancer metabolism, and the road ahead. EMBO reports. 2016;17:e201643300–1730. doi: 10.15252/embr.201643300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.La Porta J, Matus-Nicodemos R, Valentín-Acevedo A, Covey LR. The RNA-Binding Protein, Polypyrimidine Tract-Binding Protein 1 (PTBP1) Is a Key Regulator of CD4 T Cell Activation. PLoS ONE. 2016;11:e0158708. doi: 10.1371/journal.pone.0158708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shibayama M, et al. Polypyrimidine tract-binding protein is essential for early mouse development and embryonic stem cell proliferation. The FEBS Journal. 2009;276:6658–6668. doi: 10.1111/j.1742-4658.2009.07380.x. [DOI] [PubMed] [Google Scholar]
- 39.Boothby M, Rickert RC. Metabolic Regulation of the Immune Humoral Response. Immunity. 2017;46:743–755. doi: 10.1016/j.immuni.2017.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan LN, et al. Metabolic gatekeeper function of B-lymphoid transcription factors. Nature. 2017;542:479–483. doi: 10.1038/nature21076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki K, Kumanogoh A, Kikutani H. Semaphorins and their receptors in immune cell interactions. Nat Immunol. 2008;9:17–23. doi: 10.1038/ni1553. [DOI] [PubMed] [Google Scholar]
- 42.Jung C-R, et al. Enigma negatively regulates p53 through MDM2 and promotes tumor cell survival in mice. J Clin Invest. 2010;120:4493–4506. doi: 10.1172/JCI42674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dufort FJ, et al. Glucose-dependent de novo lipogenesis in B lymphocytes a requirement for ATP-citrate lyase in lipopolysaccharide-induced differentiation. J Biol Chem. 2014;289:7011–7024. doi: 10.1074/jbc.M114.551051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hann SR. MYC cofactors: molecular switches controlling diverse biological outcomes. Cold Spring Harb Perspect Med. 2014;4:a014399–a014399. doi: 10.1101/cshperspect.a014399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cortés M, Georgopoulos K. Aiolos Is Required for the Generation of High Affinity Bone Marrow Plasma Cells Responsible for Long-Term Immunity. Journal of Experimental Medicine. 2004;199:209–219. doi: 10.1084/jem.20031571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quéméneur L, et al. Differential control of cell cycle, proliferation, and survival of primary T lymphocytes by purine and pyrimidine nucleotides. The Journal of Immunology. 2003;170:4986–4995. doi: 10.4049/jimmunol.170.10.4986. [DOI] [PubMed] [Google Scholar]
- 47.Ke Y, Ash J, Johnson LF. Splicing signals are required for S-phase regulation of the mouse thymidylate synthase gene. Mol Cell Biol. 1996;16:376–383. doi: 10.1128/mcb.16.1.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nowak U, Matthews AJ, Zheng S, Chaudhuri J. The splicing regulator PTBP2 interacts with the cytidine deaminase AID and promotes binding of AID to switch-region DNA. Nat Immunol. 2011;12:160–166. doi: 10.1038/ni.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hogenbirk MA, et al. Differential programming of B cells in AID deficient mice. PLoS ONE. 2013;8:e69815. doi: 10.1371/journal.pone.0069815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Q, et al. The splicing regulator PTBP2 controls a program of embryonic splicing required for neuronal maturation. eLife Sciences. 2014;3:e01201. doi: 10.7554/eLife.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hobeika E, et al. Testing gene function early in the B cell lineage in mb1-cre mice. PNAS. 2006;103:13789–13794. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kwon K, et al. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity. 2008;28:751–762. doi: 10.1016/j.immuni.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 53.Huang C-Y, Bredemeyer AL, Walker LM, Bassing CH, Sleckman BP. Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. European Journal of Immunology. 2008;38:342–349. doi: 10.1002/eji.200737972. [DOI] [PubMed] [Google Scholar]
- 54.Shinkai Y, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 55.Anastasiou D, et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nature Chemical Biology. 2012;8:839–847. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.König J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shen S, et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci USA. 2014;111:E5593–601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xia Z, et al. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nat Comms. 2014;5:5274. doi: 10.1038/ncomms6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
mRNAseq and iCLIP data that support the findings of this study have been deposited in GEO with the GSE100969 accession code.