Abstract
The Polycomb repressive complex 2 (PRC2) catalyzes H3K27 methylation and is required for maintaining transcriptional patterns and cellular identity, but the specification and maintenance of genomic PRC2 binding and H3K27 methylation patterns remain incompletely understood. Epigenetic mechanisms have been proposed, wherein pre-existing H3K27 methylation directs recruitment and regulates the catalytic activity of PRC2 to support its own maintenance. Here we investigate if such mechanisms are required for specifying H3K27 methylation patterns in mouse embryonic stem cells (mESCs). Through re-expression of PRC2 subunits in genetic knockouts that have lost all H3K27 methylation, we demonstrate that methylation patterns can be accurately established de novo. We find that regional methylation kinetics correlate with original methylation patterns even in their absence, and specification of the genomic PRC2 binding pattern is retained and specifically dependent on the PRC2 core-subunit SUZ12. Thus, the H3K27 methylation patterns in mESCs are not dependent on self-autonomous epigenetic inheritance.
The Polycomb repressive complex 2 (PRC2) methylates lysine 27 on histone H3 (H3K27) to produce cell type-specific H3K27 mono- (H3K27me1), di- (H3K27me2), and tri-methylation (H3K27me3) patterns 1,2. The three core PRC2 subunits, paralogs EZH1 or EZH2, SUZ12 and EED, co-purify from cells in stoichiometric amounts together with several substoichiometric partners 3–6, and are all essential for the methyltransferase activity of the complex and for proper organismal development 7–14. Several cancers have driver mutations in PRC2 or its H3K27 substrate, with a resulting perturbed H3K27 methylation pattern 15.
H3K27me2 is the most abundant state, present on 50% of H3 and widely distributed across the genome in mouse embryonic stem cells (mESCs) 2,16,17. H3K27me1 and H3K27me3 each account for 10-15% of H3 in mESCs, but, unlike H3K27me2, they associate with distinct genomic features: H3K27me1 is predominant within gene bodies of active genes 2,18, while H3K27me3 is strongly enriched in regions centered on CpG islands (CGIs) at silent genes 15. Regions with less distinct methylation patterns may also have functional importance. For example, H3K27M mutant pediatric brain cancers are characterized by a loss of H3K27me3 from non-CGI regions that have intermediate levels of H3K27me3 while H3K27me3 levels at CGIs are unchanged 19.
During cell divisions, H3K27 methylation is halved as the genome is copied and newly synthesized histones are added to the replicated DNA 20. It has been proposed that recycling of parental histones carrying H3K27 methylation at the original genomic location 20–23 may help copying the mark from the modified nucleosomes to the nascent un-modified, neighboring nucleosomes 24,25. This is strengthened by observations that the core PRC2 subunit EED can bind H3K27me3 25,26, and this binding allosterically stimulates the methyltransferase activity of the complex 26.
It is of great interest whether this combination of locally inherited H3K27 methylated histones and their feedback to PRC2 is sufficient to give rise to self-maintenance, such that specification for H3K27 methylation in a subset of genomic regions may be dependent on its own pre-existence 27,28. A few studies have directly probed for H3K27me3 self-maintenance capacity with reporter gene systems, where removal of a PRC2 recruiting element can be induced 22,23,25. These show that H3K27me3 alone is insufficient to ensure long-term self-maintenance, but this may be due to simultaneous transcriptional activation of the reporter gene. Detailed analysis shows that disappearance of H3K27me3 occurs slower than would be expected by replication associated dilution 23, providing some evidence of self-maintenance activity even in this system. Because different genomic contexts can be permissive or restrictive of potential self-maintenance activities, genome-wide approaches are required to assert if self-maintenance specifies any part of the H3K27 methylation patterns. Therefore, we have investigated the dependence on pre-existing methylation for the entire H3K27 methylation patterns in mammalian cells, which include H3K27me3 at regions with abundant PRC2 occupancy as well as remaining genome-wide patterns of H3K27me1, H3K27me2 and H3K27me3. We find that specification of methylation patterns is retained by cells independently of pre-existing H3K27 methylation.
Results
PRC2 is responsible for all H3K27 methylation states
Studies of PRC2 core subunit knockout cells have shown that PRC2 is responsible for H3K27me2 and H3K27me3 but have differed in their conclusions regarding the role of PRC2 in generating H3K27me1 2,8–10,29–32. To resolve this, we have generated new knockout mouse embryonic stem cells (mESCs) using CRISPR/Cas9 and find that knockout of either Suz12, Eed or combined knockout of the catalytic subunits Ezh1 and Ezh2 (Ezh1/Ezh2 dKO) leads to abolishment of all degrees of H3K27 methylation (Supplementary Fig. 1a). Thus, these PRC2 knockout mESCs provide a suitable model system for studying de novo establishment of all H3K27 methylation states.
PRC2 can accurately establish all H3K27 methylation de novo
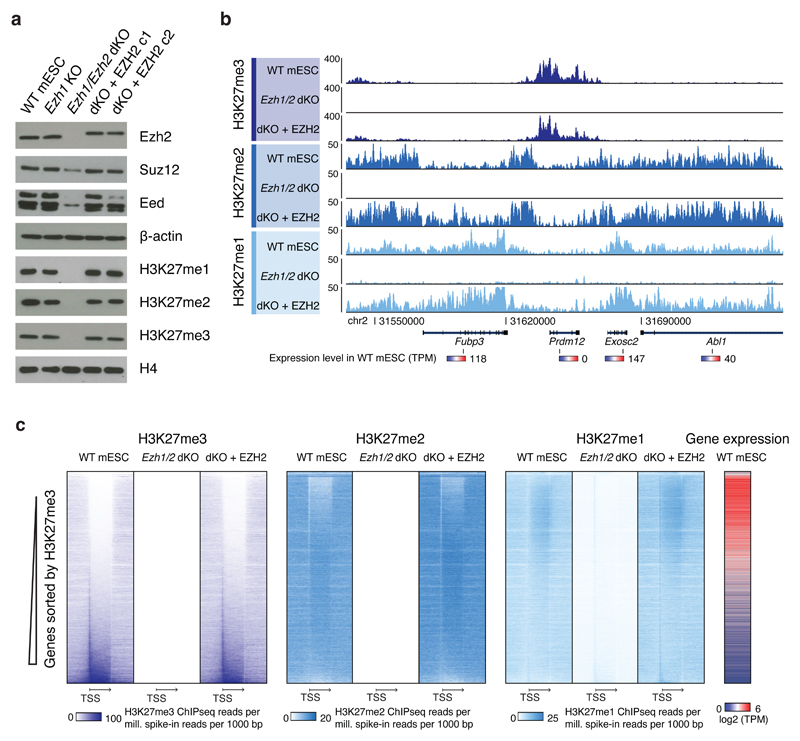
If self-propagating mechanisms are involved in maintenance of H3K27 methylation, parts of the methylation patterns in mESCs may be dependent on these and would not get re-established following erasure of the modifications. Alternatively, if PRC2 activity is specified in all regions independently of pre-existing methylation, correct methylation patterns should be established de novo following erasure (Fig. 1a). In order to determine if methylation can be established de novo in mESCs, we expressed Flag-tagged EZH2 in Ezh1/Ezh2 dKO mESCs and observed restoration of global methylation levels (Fig. 1b, Fig. 2a). As expected, the methylation patterns of parental WT mESCs are characterized by enrichment of H3K27me3 at promoters and in gene bodies of silent genes, H3K27me2 distributed broadly (but depleted in H3K27me3- and H3K27me1-dominant regions), and H3K27me1 enriched in gene bodies of active genes (Fig. 2b, c). These patterns are erased in Ezh1/Ezh2 dKO cells, where only background level ChIP-seq signals are detected with no trace of original patterns (Fig. 2b, c, Supplementary Fig. 1b-d). In cells with re-expression of EZH2, we find accurately restored methylation patterns across genes (Fig. 2b, c) as well as across the entire genome, where quantification of H3K27 methylation within all regions show strong correlation between parental WT mESCs and EZH2-rescued cells (Supplementary Fig. 1d). Thus, the H3K27 methylation patterns are not dependent on self-propagation, but can be established de novo.
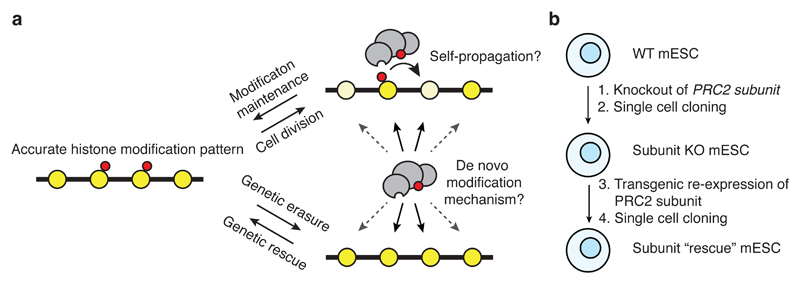
Figure 1. Models of histone methylation maintenance.
a, Histone modifications (red) are retained locally during replication but diluted by nascent histones (pale yellow). Maintenance can occur via self-propagation or via a de novo modification mechanism. When modifications are erased through genetic knockout of modifier followed by re-expression, only de novo methylation mechanisms are possible.
b, Knockout and rescue strategy to probe de novo methylation capacity.
Figure 2. Propagation independent establishment of H3K27 methylation patterns.
a, Western blot of extracts from indicated cell lines for PRC2 subunits and H3K27 methylation. Experiment has been repeated at least three times with the same result. Uncropped Western blot images are shown in Supplementary Data Set 1.
b, ChIP-seq signals generated with H3K27 methylation specific antibodies. The shown region includes genes indicated at the bottom together with their expression level in WT mESCs (Ezh2f/f). ChIP-seq signal is spike-in normalized (reads per million Drosophila-mapped reads).
c, Heatmaps of spike-in normalized ChIP-seq data of H3K27 methylation in genic regions. Vertical axis contains all RefSeq genes > 250bp and horizontal axis is centered on genes represented by relative gene length. Gene expression levels from RNAseq of WT mESCs (Ezh2f/f) cells are shown on right. TSS: Transcription start site. Arrow ends at end of genes.
Mouse ESCs with PRC2 KO can be derived and maintained stably in mESC culture conditions. Gene deregulation has been reported for cells with complete PRC2 loss (Eed KO or Ezh2 KO combined with knockdown of Ezh1) 10, but not in mESCs containing a slightly leaky Suz12 gene trap or Ezh2 KO alone 33. Consistent with this, the Ezh1/Ezh2 dKO cells show deregulation of gene expression, and this is reverted in the EZH2-rescued cells (Supplementary Fig. 2a, b). Thus, the genome-wide erasure of H3K27 methylation in mESCs is reversible despite a challenged gene expression pattern in the intermittent knockout cells.
De novo methylation is immediately correctly specified
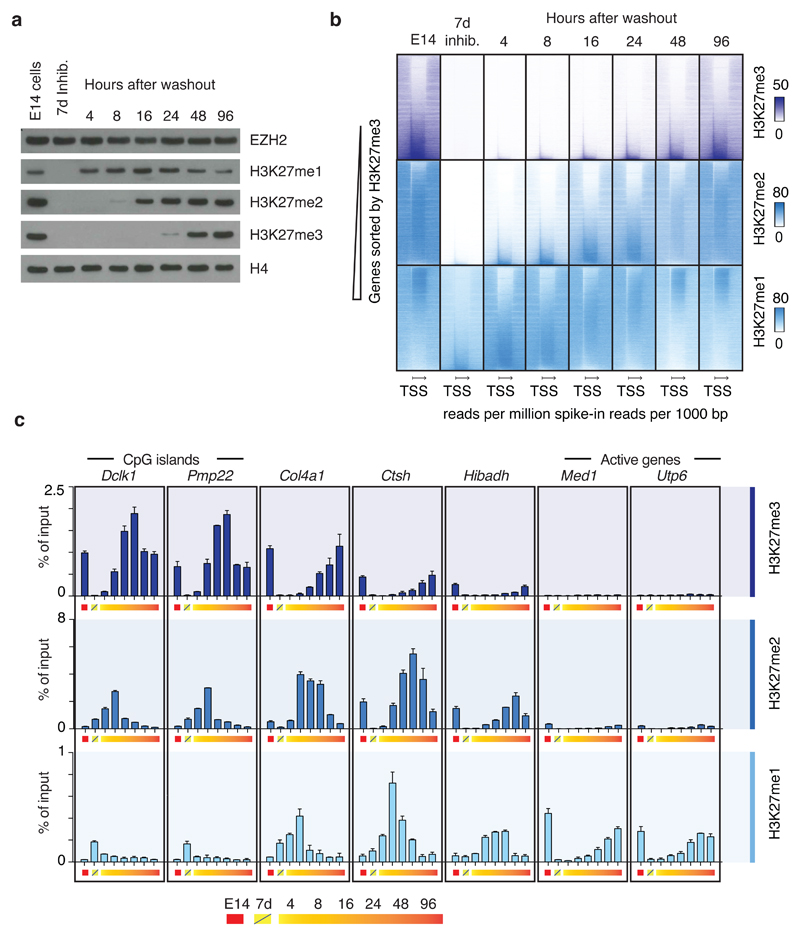
To determine if the erased methylation patterns are restored with immediate accuracy, or perhaps through selection of cells with suitable methylation, we studied the kinetics of re-methylation in cells severely depleted for H3K27 methylation through the use of high concentration of an EZH2 inhibitor 34. The mESC line E14 was treated with 10 µM of inhibitor for seven days, during which the cells undergo more than 10 cell divisions and the pool of parental histones thus is diluted more than 1000-fold. The inhibitor treatment, which does not affect differentiation capacity, proliferation, or expression of pluripotency markers (Supplementary Fig. 3a-g), is followed by inhibitor washout and observations of re-methylation over four days. Both global methylation levels (Fig. 3a) and patterns (Fig. 3b, c, Supplementary Fig. 4a, b) are restored in these four days. Importantly, H3K27me3 does not appear temporarily in regions, where it is not found in the parental cells, and H3K27me1 and H3K27me2 do so only as intermediate stages before conversion to a higher methylation state that matches the original pattern (Fig. 3b, c, Supplementary Fig. 4a, b). These results show that the specification of correct methylation patterns is independent of pre-existing H3K27 methylation.
Figure 3. Regional de novo methylation kinetics.
a, Western blots of extracts from E14 cells treated with 10 µM EPZ6438 EZH2 inhibitor for 7 days and sampled at time points following inhibitor washout. Blots were probed with the indicated antibodies. Experiment has been repeated at least three times with same result. Uncropped Western blot images are shown in Supplementary Data Set 1
b, Heatmaps of spike-in normalized ChIP-seq data probing restoration kinetics of H3K27me3 (top row), H3K27me2 (middle row) and H3K27me1 (bottom row) in genic regions.
c, ChIP-qPCR data probing kinetics of H3K27me3 (top row), H3K27me2 (middle row) and H3K27me1 (bottom row) at seven representative loci labeled (above graph) according to nearest gene and in approximate order of decreasing methylation rates (% of input/hr washout). Data values come from a single biological (n=1) experiment, with error bars depicting s.d. of technical duplicates. Data from a biological replicate of the experiment is shown in Supplementary Fig. 4c, and the experiments have also been performed once in Ezh2f/f and Ezh1 KO mESC lines with comparable result.
Regional kinetics correspond to original methylation state
The inhibitor washout experiment also provides new insight into regional PRC2 methylation activity. We find that the global level of H3K27me1 reaches its original level sooner than H3K27me2 and H3K27me3 (Fig. 3a). This observation is in agreement with recent studies on histone methylation maintenance following DNA replication 20, but our ChIP analysis shows that this does not reflect that the H3K27me1 pattern is finalized prior to those of the higher methylation states. Rather, the majority of the early deposited H3K27me1 is found in regions that will become H3K27me2 and H3K27me3, while the regions that are normally predominantly carrying H3K27me1 are restored to their original level much later (Fig. 3b, c, Supplementary Fig. 4a, b). In fact, the methylation rate in different genomic regions directly correlate with the steady-state methylation level of the region, such that H3K27me3 positive regions have faster methylation than H3K27me2 regions, and H3K27me1-dominant regions displaying the slowest methylation (Fig. 3b, c, Supplementary Fig. 4a, b).
Despite the use of high concentrations of the EZH2 inhibitor, it is insufficient to remove all methylation. Residual H3K27me1 and H3K27me2 can be detected by ChIP analysis, but it is now found at regions that are bound by PRC2 and normally associated with H3K27me3 (Supplementary Fig. 4a, c), further demonstrating the higher methylation activities in these regions. The inhibitor is reported to be selective for Ezh2 over Ezh1 34, but at these high concentrations of inhibitor this selectivity is not the cause of residual methylation as we detect similar residual H3K27me1 at high PRC2-occupant regions in Ezh1 KO cells treated with the inhibitor (data not shown).
Suz12 binds CGIs independently of H3K27 methylation
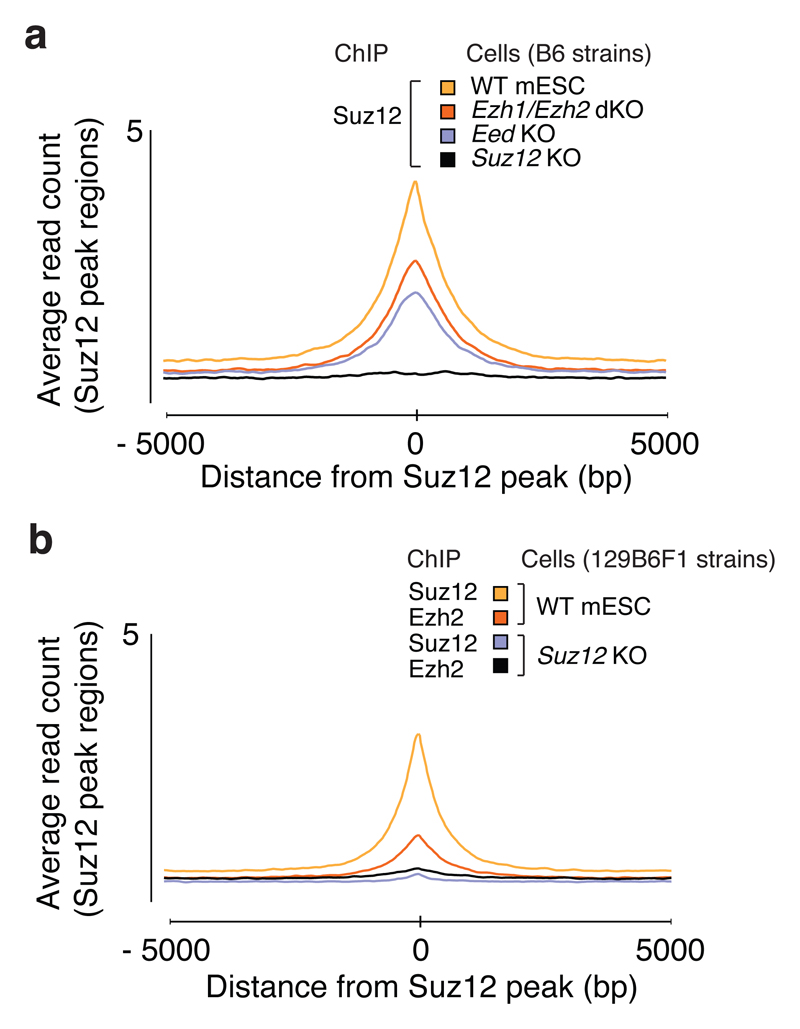
In the E14 mESCs treated with EZH2 inhibitor, where H3K27 methylation is severely depleted (Fig. 3a), Suz12 binding pattern is largely unaffected (Supplementary Fig. 4a, c), i.e. it is not dependent on H3K27 methylation. Likewise, we find that Suz12 binds to its normal target regions in Ezh1/Ezh2 dKO and Eed KO cells (Fig. 4a). Thus, the preferential binding to CGIs at silent genes by PRC2 is observed for Suz12 independently of H3K27 methylation and independently of Eed. The Ezh2 KO model used is a functional knockout with only the methyltransferase domain (SET) genetically removed 35, and thus a shorter truncated Ezh2∆SET protein remains. However, Ezh2∆SET is unstable and present at low levels in the cells. In Eed KO cells, full-length Ezh2 is likewise unstable and low in abundance (Supplementary Fig. 1a). Thus, Suz12 can bind DNA in the absence of the other core PRC2 proteins.
Figure 4. Suz12 binds CGIs independently of Eed, Ezh1 and Ezh2.
a, Mean Suz12 ChIP-seq signal for indicated cell lines in regions centered on 7480 Suz12 peak regions identified in WT mESCs (B6).
b, Mean Suz12 and Ezh2 ChIP-seq signal for WT mESC (129B6F1) and Suz12 KO (c2) cell line in regions centered on 7480 Suz12 peak regions identified in WT mESCs (B6).
De novo PRC2 binding restores methylation and pluripotency
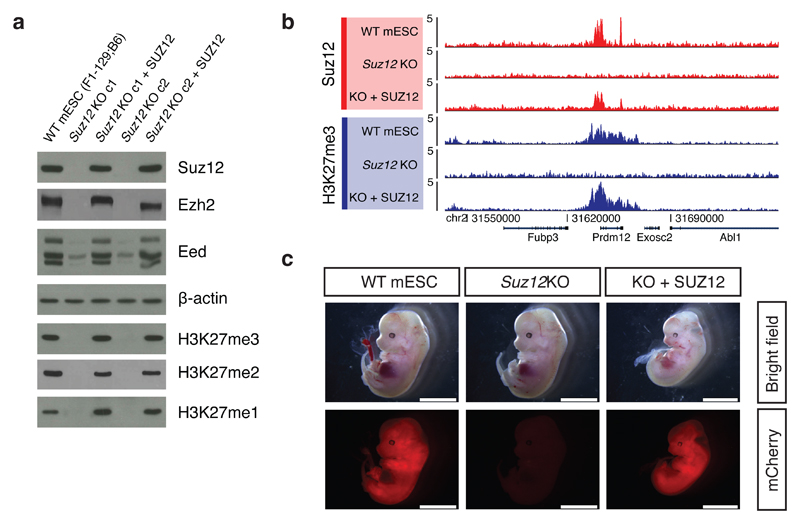
In order to address if pre-bound Suz12 is required for the de novo re-establishment of H3K27 methylation patterns, we expressed SUZ12 in Suz12 KO cells, in which H3K27 methylation is erased and neither Suz12 nor Ezh2 binding can be detected at regions normally bound by PRC2 (Fig. 4b). Re-expression of SUZ12 in these cells resulted in stabilization of Ezh2 and Eed (Fig. 5a) and accurate restoration of H3K27 methylation and PRC2 binding patterns (Fig. 5a, b, Supplementary Fig. 5a).
Figure 5. Accurate H3K27 methylation and pluripotency established from Suz12 KO cells with both methylation and PRC2 binding erased.
a, Western blot of extracts from indicated cell lines probing levels of PRC2 subunits and H3K27 methylation levels. Experiment has been repeated at least three times in individual knockout clones with the same result. Uncropped Western blot images are shown in Supplementary Data Set 1.
b, ChIP-seq signals within a representative genomic region from indicated cell lines generated with H3K27me3 and Suz12 antibodies. ChIP-seq signal is sequence-depth normalized (reads per million mapped reads).
c, Images of embryos dissected at E13.5. Embryos are derived from injection of morulae aggregated with mCherry labeled WT mESCs (129B6F1), Suz12 KO or SUZ12 rescue (KO + SUZ12) cells. Scale bar: 5 mm.
It remains a possibility that we have not uncovered rare regions in mESCs that are dependent on self-propagation of methylation or PRC2 binding. Although we have not identified such regions, we decided to test whether re-expression of SUZ12 in Suz12 KO mESCs restored pluripotency. To do this, we performed morula injection experiments with Suz12 KO and rescued cells. Knockout of PRC2 core subunits in mice leads to embryonic lethality around gastrulation (E6.5) 8,12,13. Accordingly, we observed no chimeric contribution of Suz12 KO cells in E13.5 embryos (Fig. 5c, Supplementary Fig. 5b-d). The SUZ12-rescued cells, in contrast, gave high chimeric contribution in tissues throughout the embryos (Fig. 5c, Supplementary Fig. 5b-d), thereby showing that they have regained pluripotency.
Suz12∆VEFS binds CGIs and directs PRC2 and H3K27me3 patterns
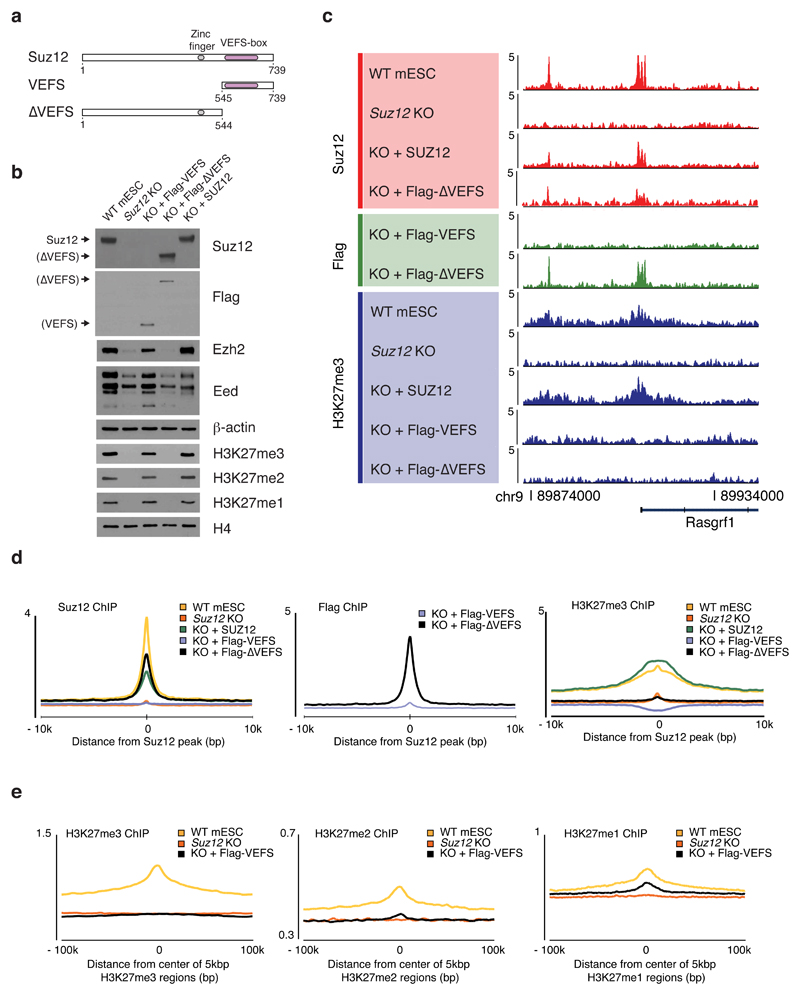
Suz12 interacts with Ezh2 via its C-terminal VEFS domain, which has been shown capable of forming a truncated PRC2 complex together with Eed and Ezh2 that is both catalytically active and has demonstrated rigidity to facilitate the first crystal structures of a partial PRC2 complex with full-length Eed and Ezh2 36–39. To determine if a PRC2 complex with just the Suz12 VEFS domain can localize to CGIs, or if the binding is mediated by a separate region of Suz12, we expressed a SUZ12 fragment containing the VEFS domain (VEFS) or a Suz12 fragment lacking the VEFS domain (SUZ12∆VEFS) in Suz12 KO cells (Fig. 6a, b).
Figure 6. An N-terminal Suz12 fragment is essential for CGI binding and correct H3K27 methylation patterns.
a, Domain architecture of SUZ12 and design of studied SUZ12 fragments.
b, Western blot probing levels of core PRC2 subunits, Ezh2 and Eed, and global H3K27 methylation levels in extracts from Flag-VEFS-expressing cells or Flag-∆VEFS-expressing cells. The ∆VEFS fragment contains the epitope for the SUZ12 antibody. Experiment has been repeated at least three times with same result. Uncropped Western blot images are shown in Supplementary Data Set 1.
c, ChIP-seq signals, within a representative region that include two Suz12 peaks (in WT mESCs), probing binding of Flag-VEFS (Flag ChIP) or Flag-∆VEFS (Flag and Suz12 ChIP) and H3K27me3. ChIP-seq signal is sequence-depth normalized (reads per million mapped reads).
d, Mean ChIP-seq signal generated with indicated antibodies (top left on graphs) for cell lines (top right of graphs) in 7480 Suz12 peak regions identified in WT mESCs (B6).
e, Mean ChIP-seq signal generated with indicated antibodies (top left on graphs) for cell lines (top right on graphs) in regions identified by cluster analysis (Supplementary Fig. 4b) as being enriched for H3K27me3 (left graph), H3K27me2 (middle graph) or H3K27me1 (right graph).
Expression of the SUZ12 VEFS domain stabilizes Ezh2 and Eed through complex formation (Fig. 6b) and is sufficient to restore global levels of all three H3K27 methylation states, while no such stabilization or detectable methylation is observed with SUZ12∆VEFS (Fig. 6b). However, the VEFS domain-stabilized PRC2 is unable to localize to Suz12 peak regions (Fig. 6c, d), and, despite establishing normal global H3K27 methylation levels, the VEFS-expressing cells show an aberrant methylation pattern (Fig. 6c, Supplementary Fig. 6a, b). Most notably, in VEFS-expressing cells H3K27me3 is not enriched in the areas found in wild type mESCs (Fig. 6c-e), but is found diffusely in all regions except the H3K27me1 regions and is most abundant in a large cluster (cluster 7) of normally H3K27me2 positive regions (Supplementary Fig. 6b), suggesting that this region may be the most accessible to PRC2 activity in absence of specific recruitment to CGIs. Some H3K27me1 regions have the same pattern in VEFS-expressing cells (Fig. 6e, Supplementary Fig. 6b, c), but the characteristic H3K27me1 pattern in gene bodies of active genes is weak (Supplementary Fig. 6a). Thus, part of the H3K27me1 pattern is also dependent on the N-terminal portion of Suz12.
In contrast to the VEFS domain, the SUZ12∆VEFS fragment localizes to the normal Suz12 binding regions, but with no detectable H3K27 methylation globally or in the bound regions (Fig. 6b-e). Thus, this N-terminal portion of Suz12 binds PRC2 target regions independently of the other core subunits, and is essential for guiding PRC2 to its correct genomic loci, and thereby directing the deposition of H3K27 methylation.
Discussion
There has been much speculation of whether histone modifications and histone modifying enzymes confer epigenetic inheritance. Direct evidence for epigenetic memory through self-maintenance of H3K27 methylation has not been convincingly demonstrated, but molecular mechanisms that could enable such activity have been identified: the local inheritance of parental histones during DNA replication provides a template for copying modifications to neighboring nascent histones 20–23 and the ability of H3K27me3 to bind and stimulate PRC2 may promote methylation of regions with pre-existing H3K27me3 25,26. In Drosophila, it was recently shown that H3K27me3 is lost when PRC2-recruiting cis-regulatory elements (Polycomb response elements (PREs)) were removed 22,23, suggesting that H3K27me3 alone is not sufficient for long-term self-maintenance, although evidence of maintenance activity was also observed 23. However, the simultaneous transcriptional activation of these reporter genes may have prevented self-maintenance, and such challenge from transcriptional activators will vary in both strength and type across genomes. PRC2 recruitment mechanisms are fundamentally different in mammalian cells as compared to Drosophila and could have a different involvement of its H3K27 methylation products in orchestrating its maintenance. Furthermore, regions other than those with high PRC2 occupancy (PREs in Drosophila and CGIs in mammalian cells) are H3K27 methylated and may depend on self-maintenance. In this study, we have investigated all regions, levels and states of H3K27 methylation in mammalian cells to address whether they rely on an autonomous self-propagative mechanism in any part.
One of the main conclusions of our work is that PRC2 binding and all H3K27 methylation patterns can be established independently of pre-existing H3K27-methylated nucleosomes, demonstrating that cells contain the instructive information for directing PRC2 activities to the correct genomic regions. By utilizing high concentration of an EZH2 inhibitor to deplete H3K27 methylation followed by short-term recovery, we show that the de novo methylation occurs rapidly rather than through selective pressure. Analysis of the early methylation activities demonstrates that regional methylation rates correlate with the original H3K27 methylation state. It remains possible that when the first H3K27me3 is generated, it may contribute to finalize and maintain the steady-state patterns in that region, e.g. through increased methylation activity or facilitation of broad spreading. Because the regions that are enriched for H3K27me3 in parental cells display immediate faster methylation at a time when this modification is depleted, this high activity is not dependent on allosteric stimulation of PRC2 by local pre-existing H3K27me3. Thus, our studies do not confirm a crucial role of this compelling model, and other strategies are needed to demonstrate a potential role of H3K27me3 in propagation.
Another main conclusion from our studies is that the core-subunit SUZ12 binds to CGIs through its N-terminal (∆VEFS) region, independently of the other core-subunits EZH2 and EED and H3K27 methylation, and that this stable recruitment to genomic loci is essential for correct H3K27 methylation patterns. We find that this important function of SUZ12 can be uncoupled from the function of its C-terminal VEFS domain, which, in agreement with previous studies, is critical for interacting with EZH2 (and indirectly EED) and for supporting a catalytically competent complex. Despite the loss of proper localization, the catalytically competent VEFS-PRC2 complex produces normal global levels of all three H3K27 methylation states, but lacks H3K27me3 enrichment at normal PRC2 target regions and lacks H3K27me1 at active gene bodies. Other parts of the H3K27 methylation patterns are, however, not dependent on the N-terminal SUZ12 function, since much of the H3K27me2 remains low in regions that are normally enriched for H3K27me1, and is similarly abundant in regions where it is enriched in WT cells. Likewise, a subset of H3K27me1 regions that overlap H3K9me3 is also retained in VEFS-PRC2 cells. Thus, specification of H3K27 methylation patterns occurs through both SUZ12-mediated recruitment and through mechanisms that directly affect the activity of a minimal catalytic complex.
Our data suggest that the PRC2 recruitment to CGIs occurs through SUZ12-∆VEFS, and it may be direct or mediated by other proteins that have been proposed to orchestrate CGI localization of PRC2. The Polycomb repressive complex 1 (PRC1), was initially proposed as a downstream factor recruited to CGIs via PRC2-deposited H3K27me3. Recently, it has been demonstrated that a variant PCGF1-PRC1 complex targeted to CGIs by KDM2B could play the opposite role and initiate recruitment of PRC2 40, and PRC2 sub-stoichiometric subunit JARID2 was identified as a candidate to mediate such PRC1-PRC2 crosstalk 41. However, the overlap between PRC1 and PRC2 is only partial 42,43, and a strong depletion of PRC1 does not entirely disrupt PRC2 binding 40. Alternatively, Polycomb-like (PCL) proteins, which are also sub-stoichiometric subunits of PRC2, were recently shown to bind CpG-DNA preferentially, and are thus also candidates for mediators of PRC2 recruitment to CGIs 44. Interestingly, there is some evidence that PCL proteins may interact with the SUZ12∆VEFS region 45. In mESCs, knockout of the predominant PCL homolog, Mtf2, only partially affected PRC2 binding and H3K27me3 deposition 44. The incomplete dependence of PRC2-CGI localization on PRC1, JARID2 or PCL2 is in contrast to the absolute requirement for SUZ12∆VEFS. Taken together, this indicates that while these proteins may not be sole recruiters of PRC2, they may well contribute to PRC2 recruitment by acting in concert with each other, with additional unidentified factors or in region-specifc contexts. It will be critical to determine which potential recruiters can interact with SUZ12∆VEFS or if this domain can bind CpG-DNA directly.
Despite the prior suggestion of various mechanisms that could facilitate self-maintenance of H3K27 methylation, we find no part of the methylation pattern that is dependent on inheriting the methylation. While we cannot exclude such inheritance through self-maintenance in some cell types, we find that genome-wide recruitment of PRC2 occurs independently of H3K27-methylated nucleosomes and high methylation rates are measured in these regions in situations where allosteric stimulation of PRC2 by H3K27me3 is not possible, arguing against a critical involvement of these proposed mechanisms in specification of H3K27 methylation patterns.
Online Methods
Cell lines
Ezh2f/f mice were obtained from the Tarakhovsky lab 35, bred in-house to Rosa26CreERT2 (B6 strain) and used for the derivation of OHT-inducible Ezh2 KO mESCs. Additional knockout mESCs were generated by CRISPR-Cas9n targeting of Eed or Suz12 in wild type WT mESCs (B6 and 129B6F1 backgrounds) or Ezh1 in Ezh2f/f;Rosa26CreERT2 mESCs. Resulting clones were tested for expression and genomic DNA was PCR-amplified and sequenced to verify out-of-frame deletions/insertions. Ezh1/Ezh2 dKO was achieved by OHT-treatment of Ezh1-/-;Ezh2f/f;Rosa26CreERT2, followed by single cell cloning. EZH2 inhibitor washout experiments were carried out using the mESC line E14. Cell lines were regularly tested for mycoplasma. See Supplementary Table 1 and Supplementary Table 2 for further details.
Cell culture
mESCs were derived from timed matings: 4-6 week-old females were super-ovulated by IP injection of PMS and hCG (app. 46 hours post-PMS administration) prior to mating. Pregnant females were sacrificed at 3.5 dpc by cervical dislocation and the uteruses dissected out and flushed by injecting medium through the top part of the uterus using a syringe in order to retrieve blastocysts. Single blastocysts were isolated and plated in 2i/LIF on gelatin-coated 24-well plates and left for 5-7 days to allow outgrowth of the ICM. Outgrown clones were picked and dissociated to establish cultures. All mESCs were cultured on gelatin-coated dishes in 2i/LIF-containing medium (1:1 mix of DMEM/F12 and Neurobasal media (Gibco), supplemented with 1x Pen-Strep (Gibco), 2 mM Glutamax (Gibco), 50 µM β-mercaptoethanol (Gibco), 0.1 mM Non-essential amino acids (Gibco), 1 mM Sodium Pyruvate (Gibco), N2+B27 (ThermoFisher), GSK3i (CHIR99021), MEKi (PD0325901) and Leukemia Inhibitory Factor (LIF, produced in the lab). Cells were passaged every 2(-3) days by removal of medium, wash in PBS, dissociation by 0.25% trypsin-EDTA (Gibco) or Accutase (Sigma) with gentle disruption of colonies by pipetting, resuspension in medium and pelleting by centrifugation, followed by resuspension and plating at a density of app. 1 mio cells/10 cm dish.
Ezh2 inhibition and washout
E14 mESCs were treated with high concentrations (10 µM) of the EPZ6438 EZH2 inhibitor 34 (MedChem Express) for 7 days. For washout, cells were first washed three times with PBS, followed by dissociation of cells and resuspension in growth medium. Medium was changed every 5 minutes in three further washes, to allow inhibitor to diffuse out of cells. Cells were harvested at full inhibition (with inhibitor present during dissociation) and at the indicated time points following release from inhibitor treatment.
Transgenic expression in mESCs
For rescue experiments, pCAG or piggyBAC vectors encoding Flag-HA-tagged human EZH2 or SUZ12 were transfected into mESCs using Lipofectamine 2000 (Thermo Fisher) according to the manufacturer’s specifications. Stable clones of pCAG-transfected cells were derived through puromycin-selection and expansion of single clones with stable integration of the construct. For piggyBAC-transfected cells, pools of Blasticidin-selected cells were used for experiments. PiggyBAC-mCherry was transfected into cells for in vivo experiments and sorted on a FACS-Aria III (BD Biosciences).
Morula injection to test for pluropotency
All animal work was carried out in compliance with ethical regulation. All work was authorized by and carried out under license by the Danish Regulatory Authority. E2.5 morulae from wild type superovulated female mice were microinjected with 7-8 mESCs and transferred to the oviduct of 0.5 dpc pseudo-pregnant females. Pregnant females were sacrificed and embryos collected at E13.5, photographed (bright field and mCherry to visualize chimerism). Morula were derived from C57BL/6N (4 weeks old). Aggregated morula were transferred to CD1 females (8-13 weeks old)
Isolation of MEFs and NSCs from E13.5 embryos
NSCs were isolated from the dorsal forebrain of embryonic-day 13.5 (E13.5) mouse embryos. After removal of the skin, dorsal forebrains were dissected out and incubated with 0.25% trypsin-EDTA (GIBCO) at 37 °C for 20 min. The tissue was dissociated thoroughly with pipette, spun down, washed and cultured on poly-D-lysine (PDL, Sigma-Aldrich)- and laminin (Sigma-Aldrich)-coated plates in neural stem cell medium (50% DMEM-F12, 50% neurobasal medium, N2 and B27 supplements, sodium pyruvate, glutamax, HEPES, β-mercaptoethanol, nonessential amino acids, bovine serum albumin, heparin, 100 U/ml penicillin, 100 μg/ml streptomycin, human recombinant epidermal and basic fibroblast growth factors). After 3 days, the expanded cells were trypsinized and cultured for one more passage before they were analyzed by FACS for mCherry expression.
MEFs were isolated from E13.5 embryos. After evisceration and removal of the heads, the embryos were finely minced using a sterile scalpel and collected in 0.5-1 ml of 0.25% trypsin-EDTA (Gibco) and incubated at 37 °C for 30 minutes with occasional mixing. The digested embryonic tissue was further dissociated by pipetting, followed by washing in growth medium (DMEM (Gibco) supplemented with 10% FBS (Hyclone), 1x Pen-Strep (100 µg/ml penicillin, 100 µg/ml streptomycin, Gibco) and 50 µM β-mercaptoethanol (Gibco)) and plating at a density of 1 embryo per 10 cm dish. Cells were trypsinized and cultured for one additional passage, before they were analysed by FACS for mCherry expression.
Western blotting
Western blotting was carried out according to standard protocols with the primary antibodies listed in Supplementary Table 3 and HRP-conjugated secondary antibodies (Vector Laboratories). Bands were visualized using Super Signal West Pico chemiluminiscent ECL substrate (Thermo Scientific) for exposing Amersham hyperfilm ECL films (GE Healthcare), which were developed (Ferrania Imaging Technologies).
Processing and analysis of RNA-seq data
Total cellular RNA was isolated from cells and sequencing adapters added using the TrueSeq RNA library prep kit v2 (Illumina) according to the manufacturer’s instructions. Three biological replicates of each sample were used. Library qualities were checked on a Bioanalyzer 2100 and sequenced on Illumina HiSeq 2000 (50bp single-end). Reads were processed within the Galaxy environment 46. Reads were trimmed with Trimmomatic (version 0.32.2) 47 using illuminaclip and sliding window trimming with average quality of 2 required. Trimmed reads were aligned to mouse (mm10) genome with STAR 48 using default parameters. Aligned reads were counted with HTSeq 49 and differential expression analyzed using DESeq2 50. Three biological replicates were used for DESeq2 analysis. Gene expression levels were calculated based on RNA-seq data from mESCs (Ezh2f/f) as TPM (Transcripts Per kilobase Million) values: HTSeq counts were first divided by gene length (kb) to give reads per kilobase (RPK). Gene lengths were extracted using GenomicFeatures package from Bioconducter using the following line of commands 51:
>library(GenomicFeatures)
>txdb <- makeTranscriptDbFromGFF("gtffile.gtf",format="gtf")
>exons.list.per.gene <- exonsBy(txdb,by="gene")
>exonic.gene.sizes <- lapply(exons.list.per.gene,function(x){sum(width(reduce(x)))}).
RPK values for all genes were summed and divided by one million as a normalization factor. RPK values were divided by this normalization factor to give TPM value for each gene. TPM values from three replicate RNA-seq samples were averaged.
Chromatin immunoprecipitation
ChIP experiments were carried out according to standard protocols. Chromatin was cross-linked by addition of 1 % formaldehyde (Sigma-Aldrich), either in single cell suspension or directly in the dish and DNA sheared to 200-500 bp fragments by sonication (Bioruptor, Diagenode). Each ChIP was carried out using 50-200 µg of chromatin from mESCs with spike-in of 5% Drosophila chromatin prepared in same manner (from S2 cells) and 2-3 µg of the indicated antibodies (Supplementary Table 3). For the inhibitor washout experiment, qPCR analysis was carried out on ChIP’ed and input DNA (primer sequences in Supplementary Table 4). Data is presented as percent of input DNA and error bars represent standard deviations for technical duplicates of the qPCR analysis.
Processing and analysis of ChIP-seq data
For ChIP-seq analysis, libraries were built using NEBNext ultra kits according to the manufacturer’s specifications with Ampure XP beads (Beckman) for the size selection step. Libraries were sequenced on Illumina HiSeq 2000 (50bp single-end) or Illumina NextSeq 550 (75bp single-end). Reads were processed within the Galaxy environment 46. Reads were trimmed with Trimmomatic 47 using illuminaclip and sliding window trimming with standard settings. Trimmed reads were mapped to mouse (mm10) or Drosophila (dm3) genomes using Bowtie2 52 using (--sensitive) preset settings. Duplicate mouse mapped reads were removed. Normalization factors used in analysis were obtained for each sample by dividing with total number of non-duplicate, mapped mouse read (sequence depth normalized) or total number of mapped Drosophila reads (spike-in normalized) for the sample.
Mapped reads were analyzed and visualized with Easeq 53. ChIP-seq tracks show signal segmented in 400 bins without further smoothening. Heatmaps show signal segmented in 200 binds. Mean signal plots show signal segmented in 400 binds with 4 bins smoothings. Heatmaps of genic regions contain all RefSeq genes larger than 250 bp on vertical axis displayed centered and in units of relative gene length along horizontal axis, which is 3 relative gene lengths wide. The genes were sorted according to intragenic H3K27me3 signals quantified within Easeq. Heatmaps of clustered regions show 50 000 random genomic 5000 bp regions on vertical axis. These regions were generated with bedtools (2.26.0) random command 54, and were clustered using k-means clustering within Easeq (k=10) on H3K27me1, H3K27me2 and H3K27me3 signals in E14 cells quantified for each region within Easeq. The resulting clusters were manually ordered according to methylation state in E14 cells. Heatmaps in Supplementary Fig. 6c show sub-clustering (k-means clustering (k=6)) of 6670 H3K27me1 enriched 5000 bp regions (clusters 8 and 9) with H3K9me3 and H3K36me3 ChIP-seq data published for 2i-grown mESCs 55. For scatterplots of H3K27 methyation (Supplementary Fig. 1d), the entire mouse genome was divided into 5000 bp regions (bedtools 2.26.0 makewindows 54) and spike-in normalized H3K27 methylation signals quantified for each region. Suz12 peak calling was done with Easeq’s Adaptive Local Thresholding (Window size 200bp, merge peaks within 800 bp, FDR < 0.0001, log2-fold difference > 2) based on Suz12 ChIP-seq reads from WT mESCs (129B6F1) with reads from Suz12 KO cells (129B6F1) as negative control. This yielded 7480 peaks.
Assays of mESC properties
Proliferation was assayed by seeding 30,000 cells in wells of 6-well plates and counting cells in each of two wells at 6 time points. Capacity of cells to differentiate into beating clusters was determined by culturing cells as embroid bodies in GMEM + 10% FBS for 7 days, followed by plating out cells on gelatin in GMEM + 10% FBS. After 6 days, clusters were observed under microscope for beating movement. Expression analysis of pluripotency marker genes as well as Hoxa10 was determined by standard RT-qPCR using primers in Supplementary Table 5.
Data availability
ChIP-Seq and RNA-Seq data have been submitted to the Gene Expression Omnibus under accession GSE103685 and to the NCBI Sequence Read Archive (SRA) with BioProject accession number PRJNA404057 and SRA accession number SRP117246. Source data for Fig. 3c and Supplementary Fig. 4c are available with the paper online. Additional data is available upon request.
Supplementary Material
Acknowledgments
We thank members of the Helin laboratory and A. Groth for advice and discussion and J. Martin and the Transgenic Core Facility staff for assistance with morula injection experiments. H.D. was supported by a postdoctoral fellowship from the Danish Cancer Society. The work in the Helin laboratory was supported by grants to K.H. from The European Research Council (294666_DNAMET), the 7th framework program of the European Union (4DCellFate), the Danish Cancer Society, the Danish National Research Foundation (DNRF82), the Danish Medical Research Council (DFF- 4183-00237), the Novo Nordisk Foundation (NNF16OC0023234), The Lundbeck Foundation, and through a center grant from the Novo Nordisk Foundation (NNF17CC0027852)).
Footnotes
Author contributions
J.W.H. and A.L. designed the study, performed the majority of experiments, performed data analysis and wrote the manuscript. K.H. designed the study, performed data analysis and wrote the manuscript. B.M.W. performed experiments; in particular regarding the generation of KO cell lines. H.D. performed experiments; in particular regarding the inhibitor study. L.H. and F.M performed experiments; in particular regarding the blastocyst injection experiment. A.T. and O.N.J. performed and analyzed experiments critical for development of the project.
Competing Financial Interests Statement:
The authors declare no competing financial interests.
References
- 1.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferrari KJ, et al. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol Cell. 2014;53:49–62. doi: 10.1016/j.molcel.2013.10.030. [DOI] [PubMed] [Google Scholar]
- 3.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 4.Muller J, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 5.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smits AH, Jansen PW, Poser I, Hyman AA, Vermeulen M. Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res. 2013;41:e28. doi: 10.1093/nar/gks941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 8.Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montgomery ND, et al. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol. 2005;15:942–947. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 10.Shen X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 12.Faust C, Schumacher A, Holdener B, Magnuson T. The eed mutation disrupts anterior mesoderm production in mice. Development. 1995;121:273–285. doi: 10.1242/dev.121.2.273. [DOI] [PubMed] [Google Scholar]
- 13.O'Carroll D, et al. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pengelly AR, Copur O, Jackle H, Herzig A, Muller J. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science. 2013;339:698–699. doi: 10.1126/science.1231382. [DOI] [PubMed] [Google Scholar]
- 15.Laugesen A, Hojfeldt JW, Helin K. Role of the Polycomb Repressive Complex 2 (PRC2) in Transcriptional Regulation and Cancer. Cold Spring Harb Perspect Med. 2016;6 doi: 10.1101/cshperspect.a026575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peters AH, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 17.Jung HR, Pasini D, Helin K, Jensen ON. Quantitative mass spectrometry of histones H3.2 and H3.3 in Suz12-deficient mouse embryonic stem cells reveals distinct, dynamic post-translational modifications at Lys-27 and Lys-36. Mol Cell Proteomics. 2010;9:838–850. doi: 10.1074/mcp.M900489-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steiner LA, Schulz VP, Maksimova Y, Wong C, Gallagher PG. Patterns of histone H3 lysine 27 monomethylation and erythroid cell type-specific gene expression. J Biol Chem. 2011;286:39457–39465. doi: 10.1074/jbc.M111.243006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohammad F, et al. EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat Med. 2017;23:483–492. doi: 10.1038/nm.4293. [DOI] [PubMed] [Google Scholar]
- 20.Alabert C, et al. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 2015;29:585–590. doi: 10.1101/gad.256354.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaydos LJ, Wang W, Strome S. Gene repression. H3K27me and PRC2 transmit a memory of repression across generations and during development. Science. 2014;345:1515–1518. doi: 10.1126/science.1255023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laprell F, Finkl K, Muller J. Propagation of Polycomb-repressed chromatin requires sequence-specific recruitment to DNA. Science. 2017 doi: 10.1126/science.aai8266. [DOI] [PubMed] [Google Scholar]
- 23.Coleman RT, Struhl G. Causal role for inheritance of H3K27me3 in maintaining the OFF state of a Drosophila HOX gene. Science. 2017;356 doi: 10.1126/science.aai8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramachandran S, Henikoff S. Replicating Nucleosomes. Sci Adv. 2015;1 doi: 10.1126/sciadv.1500587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen KH, et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 26.Margueron R, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ptashne M. Epigenetics: core misconcept. Proc Natl Acad Sci U S A. 2013;110:7101–7103. doi: 10.1073/pnas.1305399110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henikoff S, Greally JM. Epigenetics, cellular memory and gene regulation. Curr Biol. 2016;26:R644–648. doi: 10.1016/j.cub.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 29.Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schoeftner S, et al. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Margueron R, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riising EM, et al. Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol Cell. 2014;55:347–360. doi: 10.1016/j.molcel.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 34.Knutson SK, et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci U S A. 2013;110:7922–7927. doi: 10.1073/pnas.1303800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su IH, et al. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol. 2003;4:124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- 36.Rai AN, et al. Elements of the polycomb repressor SU(Z)12 needed for histone H3-K27 methylation, the interface with E(Z), and in vivo function. Mol Cell Biol. 2013;33:4844–4856. doi: 10.1128/MCB.00307-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiao L, Liu X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science. 2015;350:aac4383. doi: 10.1126/science.aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brooun A, et al. Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance. Nat Commun. 2016;7:11384. doi: 10.1038/ncomms11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Justin N, et al. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat Commun. 2016;7:11316. doi: 10.1038/ncomms11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blackledge NP, et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014;157:1445–1459. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooper S, et al. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat Commun. 2016;7:13661. doi: 10.1038/ncomms13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tavares L, et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 2012;148:664–678. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao Z, et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, et al. Polycomb-like proteins link the PRC2 complex to CpG islands. Nature. 2017;549:287–291. doi: 10.1038/nature23881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kloet SL, et al. The dynamic interactome and genomic targets of Polycomb complexes during stem-cell differentiation. Nat Struct Mol Biol. 2016;23:682–690. doi: 10.1038/nsmb.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Afgan E, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44:W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lawrence M, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lerdrup M, Johansen JV, Agrawal-Singh S, Hansen K. An interactive environment for agile analysis and visualization of ChIP-sequencing data. Nat Struct Mol Biol. 2016;23:349–357. doi: 10.1038/nsmb.3180. [DOI] [PubMed] [Google Scholar]
- 54.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marks H, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-Seq and RNA-Seq data have been submitted to the Gene Expression Omnibus under accession GSE103685 and to the NCBI Sequence Read Archive (SRA) with BioProject accession number PRJNA404057 and SRA accession number SRP117246. Source data for Fig. 3c and Supplementary Fig. 4c are available with the paper online. Additional data is available upon request.