SUMMARY
Interactions between sperm and egg proteins can occur physically between gamete surface-binding proteins, and genetically between gamete proteins that work in complementary pathways in which they may not physically interact. Physically interacting sperm–egg proteins have been functionally identified in only a few species, and none have been verified within mammals. Candidate genes on both the sperm and egg surfaces exist, but gene deletion studies do not support functional interactions between these sperm–egg proteins; interacting sperm–egg proteins thus remain elusive. Cooperative gamete proteins undergo rapid evolution, and it is predicted that these sperm–egg proteins will also have correlated evolutionary rates due to compensatory changes on both the sperm and egg. To explore potential physical and genetic interactions in sperm–egg proteins, we sequenced four candidate genes from diverse primate species, and used regression and likelihood methods to test for signatures of coevolution between sperm–egg gene pairs. With both methods, we found that the egg protein CD9 coevolves with the sperm protein IZUMO1, suggesting a physical or genetic interaction occurs between them. With regression analysis, we found that CD9 and CRISP2 have correlated rates of evolution, and with likelihood analysis, that CD9 and CRISP1 have correlated rates. This suggests that the different tests may reflect different levels of interaction, be it physical or genetic. Coevolution tests thus provide an exploratory method for detecting potentially interacting sperm–egg protein pairs.
INTRODUCTION
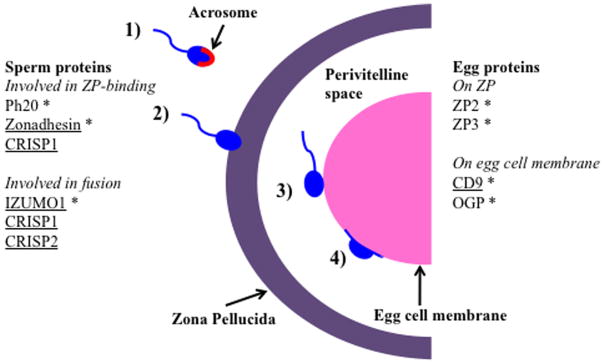
Mammalian fertilization is a complex process that involves direct and indirect interactions between proteins from the male and female reproductive tracts. Direct sperm–egg interactions occur initially as sperm bind to the zona pellucida, an outer glycoprotein coat surrounding the plasma membrane of eggs, and subsequently as acrosome-reacted sperm bind and fuse with the egg’s plasma membrane (Fig. 1). Gene-deletion studies in mice have identified many proteins that are important during this process (Nishimura et al., 2001; Rubinstein et al., 2006; Da Ros et al., 2008), however, it is still unclear which ones physically interact on the surface of sperm and egg cells. There are also many sperm–egg proteins that have un-characterized functions, and these proteins may interact indirectly with each other in complementary pathways on the sperm and egg. For example, ZP3 is a major component of the human zona pellucida, and is hypothesized to be a major player in sperm-zona pellucida binding (Harris et al., 1994). The three other proteins that make up the zona pellucida (ZP1, ZP2, and ZP4) may not physically interaction with a sperm protein, although they still may facilitate gamete binding and be subject to similar evolutionary pressures as ZP3 through indirect or genetic interactions.
Figure 1.
Stages of mammalian fertilization. (1) Sperm swims towards the egg. (2) Sperm undergoes the acrosome reaction, in which the proteins in the acrosome are released and sperm is able to then cross the zona pellucida. (3) Sperm enters the perivitelline space and binds to the egg’s plasma membrane. (4) Sperm and egg become cytoplasmically contiguous. Asterisks indicate proteins that have previously been shown to be under positive selection. Underlined proteins are used in this study.
Interacting sperm–egg proteins, along with reproduction proteins in general, often evolve rapidly by adaptive evolution between species (Swanson and Vacquier, 2002). It is hypothesized that these rapidly evolving proteins may be involved in maintaining species-specificity at the level of sperm–egg fusion, and that the amino-acid residues targeted by positive selection are located in their binding interfaces. To maintain this interaction, the evolution of these amino acids should be constrained such that adaptive mutations in the binding site of one protein select for compensatory mutations in the binding site of its interacting partner. Such a coevolutionary process predicts a long-term correlation of evolutionary rates along phylogenic lineages between two interacting proteins; these predictions are supported by several studies (Pazos and Valencia, 2001; Hakes et al., 2007). For example, lysin and VERL, a well-characterized pair of interacting sperm–egg proteins, have experienced correlated rates of evolution among abalone phylogenetic lineages (Clark et al., 2009). In subsequent work, this correlated evolutionary-rate signature was further found between proteins of similar biological function and between proteins with similar gene expression profiles across species (Clark et al., 2012).
We studied the coevolution of several mammalian proteins involved in the sperm–egg fusion step of fertilization (CD9, CRISP1, CRISP2, and IZUMO1) (Fig. 1) to predict pairs of proteins that interact physically or genetically. Based on mouse models, CD9 has been proposed as a sperm receptor on the egg cell membrane (Chen et al., 1999) while epididymal protein DE (CRISP1), TXP2 (CRISP2), and IZUMO1 have been identified as putative egg receptors on the surface of sperm (Okabe et al., 1987; Rochwerger et al., 1992; Busso et al., 2007a). CD9 is a tetraspanin containing four transmembrane domains and two extracellular loops of unequal size (Wright and Tomlinson, 1994). It is highly expressed on the egg cell surface, as well as several other types of hematopoietic and epithelial cells (Rubinstein et al., 1993). CRISP1 and CRISP2 are members of the cysteine-rich secretory proteins, antigen 5 and pathogenesis-related 1 proteins (CAP) superfamily, and each contains 16 conserved cysteine residues. CRISP1 is expressed exclusively in the epididymis, attaches loosely and tightly to the spermatozoa during epididymal maturation, and is proposed to be involved in both sperm-zona pellucida binding and sperm–egg fusion through a Signature 2 egg-binding site (Ellerman et al., 2006; Cohen et al., 2008). CRISP2 is expressed in developing spermatids in the testes, and is able to bind to the egg-cell surface (Busso et al., 2005; Cohen et al., 2008). IZUMO1 is a member of the immunoglobulin superfamily, and contains an extracellular immunoglobulin domain. It is expressed specifically in sperm and testis tissue (Inoue et al., 2005). Of these proteins, CD9 and IZUMO1 are the only protein pairs that show a complete absence of sperm–egg fusion when deleted, thus they are essential to fertilizaton (Kaji et al., 2000; Le Naour et al., 2000; Miyado et al., 2000; Inoue et al., 2005) and are likely to interact (although no functional experiments have unequivocally confirmed this). On the other hand, the role of CRISP1 and CRISP2 in sperm–egg fusion has yet to be fully characterized. As each of these membrane-associated proteins has been strongly implicated in sperm–egg fusion, they were chosen for our tests of selection and coevolution in primates.
RESULTS
Evolutionary Analysis of Sperm–Egg Fusion Genes
To detect selection in the sperm–egg proteins, we sequenced the exons (or obtained their sequences from publicly available reference genomes) of 4 candidate genes on the sperm (IZUMO1, CRISP1, and CRISP2) and egg (CD9) cell surfaces from 12 diverse primate species (Fig. 2). We tested each of these genes for evidence of positive selection using likelihood models (Nielsen and Yang, 1998; Yang et al., 2000). Calculating the ratio of the number of nonsynonymous substitutions per non-synonymous site (dN) to the number of synonymous substitutions per synonymous site (dS), or dN/dS, is a robust method to test for positive selection without a priori knowledge for each candidate protein (Yang, 1997). A ratio of dN/dS = 1 indicates that neutral evolution occurs; a ratio in which dN/dS < 1 indicates that purifying selection (conserved evolution) occurs; and a ratio in which dN/dS > 1 indicates that positive selection (rapid evolution) occurs. Using maximum-likelihood methods in the codeml program of the PAML 4.4 package, the overall gene and lineage-specific dN/dS values, which were allowed to vary across codons, were calculated for all candidate proteins and controls (Yang, 2007). We found evidence of adaptive evolution acting across all branches of the phylogeny for CRISP1 and CRISP2, but not for CD9 and IZUMO1. For CRISP1, we find evidence of positive selection acting on 7% of the sites (dN/dS = 3.77, χ2 =9.00; P <0.01) in Exon 3, which did not correspond to the Signature 2 binding domain. CRISP2 is evolving under positive selection with 4% of sites under selection (dN/dS = 5.48, χ2 =6.95; P <0.01), although we could not identify specific residues with a Bayes empirical Bayes posterior probability >95%. In addition, we found that both CD9 (fixed versus free model, χ2 =33.69; P <0.05) and CRISP1 (fixed versus free model, χ2 =41.28; P <0.01) show significantly different evolutionary rates between individual lineages.
Figure 2.
Consensus tree of 12 primate species included in our analyses.
Coevolution between Sperm–Egg Fusion Genes
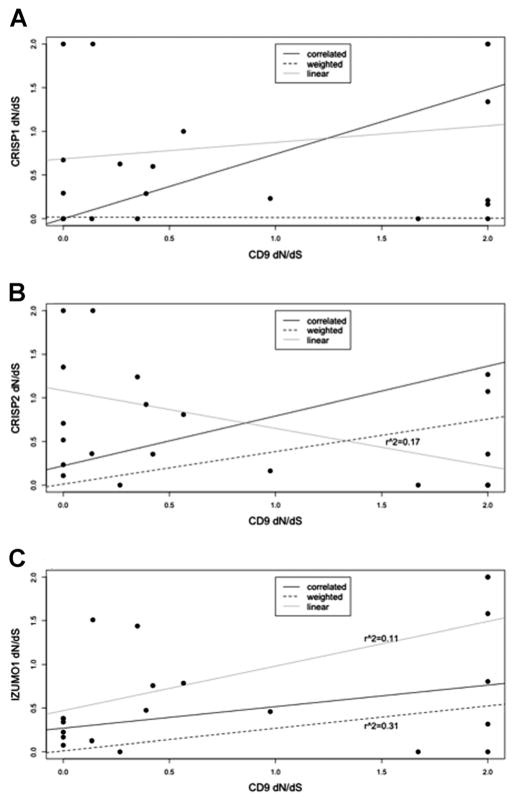
To explore the coevolution of potential sperm–egg interacters, we first estimated lineage-specific dN/dS values and tested their correlation across the branches of each sperm–egg gene pair (CD9-CRISP1, CD9-CRISP2, and CD9-IZUMO1) with standard and weighted linear regressions (Fig. 3). Both the standard and weighted linear regression methods are conservative in detecting correlations between gene-pairs (Clark et al., 2009). Using standard linear regression as an initial first test, we did not detect significant correlations between any sperm–egg gene pairs. Undefined branches were rounded up to the highest integer in the datasets to reduce their effect on the regression, but multiple modified branches in the data may have created spurious correlations. This simple method would be a powerful approach if the undefined points were limited, but otherwise could provide false correlations. In our weighted analysis, however, both CD9-CRISP2 (adjusted r2 =0.17; P =0.04; two-sided t-test) and CD9-IZUMO1 (adjusted r2 =0.31; P =0.005; two-sided t-test) show a significant correlation of lineage-specific dN/dS values. Weighted values for the remaining sperm–egg gene pairs were not significantly correlated in our weighted analysis. The amount of divergence on each lineage was used to weigh each branch, and thus eliminated the occurrence of undefined branches that result from the standard linear regression method used above. Because we use divergence instead of the ratio of synonymous sites to calculate the data points, the weighted method would be a more rigorous way to detect correlations.
Figure 3.
Linear regressions of sperm–egg correlations. A: CD9 and CRISP1 dN/dS values were estimated for each branch of a 12-species phylogeny and plotted on the graph. The line of regression for each correlation model was plotted using the points, with adjustments as stated in methods section: Correlation likelihood model (black line), standard linear regression (gray line), weighted linear regression (hashed line). B: CD9 and CRISP2 dN/dS values were estimated on each branch of a 12-species phylogeny. C: CD9 and IZUMO1 dN/dS values were estimated on each branch of a 12-species phylogeny.
To account for uncertainty in our dN/dS estimates, we used likelihood models that jointly estimate lineage-specific dN/dS values and their correlation between gene pairs (Clark et al., 2009). We compared a correlated model, which constrains the dN/dS values to fall on a best-fit line of regression, with a null model, which sets the slope of this line to zero to represent an uncorrelated relationship. Statistical methods may find correlations that are significant rather than relying solely on data points on a graph as in regression analyses, which may reflect genetic interactions rather than physical interactions. The correlated model was found to fit the data significantly better than the null for CD9-CRISP1 (P <0.001, slope =0.666428) and CD9-IZUMO1 (P <0.05, slope =0.254498) (Table 1). This is consistent with the idea that CD9 and IZUMO1 are coevolving and likely interacting. CD9 and CRISP1 also show a strong correlation of evolutionary rates, suggesting that they may also interact during the sperm–egg fusion process. These interactions may occur physically (as between sperm–egg binding proteins) or genetically (as between proteins that work in complementary male and female pathways, i.e., assisting in sperm–egg binding but not physically interacting). As the identification of new reproductive proteins becomes easier with proteomics, we can now use the exploratory methods described above to discover novel physically or genetically interacting proteins.
TABLE 1.
Likelihood Model Results to Detect Correlated Evolutionary Rates Between the Egg Protein CD9 and 5 Sperm Proteins
| n | Chi-squared (2 ×(lnL))
|
y-intercept | Slope | ||
|---|---|---|---|---|---|
| Free vs. correlated | Correlated vs. null | ||||
| CD9-CRISP1 | 12 | 22.4 | 11.92*** | 0.00 | 0.67 |
| CD9-CRISP2 | 12 | 16.88 | 1.18 | 0.22 | 0.57 |
| CD9-IZUMO1 | 12 | 21.4 | 5.06* | 0.27 | 0.25 |
| CD9-PKDREJ | 12 | 23.2 | ns | 0.42 | 0.00 |
| CD9-ZAN | 11 | 32.2* | 0.00 | 0.18 | 0.00 |
The y-intercept and slope are shown for the correlated model. Control genes are highlighted in gray.
P value <0.05.
P value <0.01.
P value <0.001.
To determine if each protein has different evolutionary rates, we also compared a free model, which allows dN/dS values to vary between protein pairs, to the correlated model. Among all sperm–egg gene pairs, none of our candidate genes showed significant evidence that the free model fit the data better than the correlated model. We found no significant correlation of evolutionary rates for CD9 with either PKDREJ or ZAN (Table 1 and Fig. S1), two sperm surface proteins that are not known to be involved in sperm–egg fusion. These negative data control for the possibility that correlated evolution between CD9 and the sperm proteins is simply a general correlation seen between all reproductive proteins.
DISCUSSION
We identified positive selection acting on two of our candidate proteins (CRISP1 and CRISP2), and have added them to the list of rapidly evolving, mammalian reproductive proteins. CD9 was previously identified as being under positive selection (Swanson et al., 2003), though we do not find statistical evidence of this in our analysis. This may be due to the low number of species (n =5) and partial coding sequence used in the previous study or due to lineage-specific bursts of selection, which is consistent with our CD9 codeml free-ratio-model results. In our current analysis, we included the majority of coding sequence for each protein in 12 primate species, and expected to have better resolution than in previous studies. It is interesting that CD9 exhibited significantly different dN/dS values across primate lineages (free-ratio model), and may indicate that CD9 has experienced different selective pressures among primates. Our IZUMO1 results correspond to a previous study that found no evidence of selection in the primate lineages, although evidence for positive selection was found in the Laurasiatheria group (Grayson and Civetta, 2012). This previous study evaluated the entire IZUMO family of genes (IZUMO1-4), and found no consistent pattern of selection across its four members, although IZUMO3 did show a bout of positive selection within primates. It is unclear what drives the patterns of evolution of reproductive proteins, but species-specific selection patterns for reproductive proteins have clearly evolved. With the addition of more primate species and population data, we will be able to better study the long-term evolution of reproductive proteins.
CD9, CRISP1, CRISP2, and IZUMO1 may interact in a complex network of proteins that facilitates sperm–egg fusion. Our analyses suggest that the egg protein CD9 interacts with sperm proteins CRISP1 and IZUMO1, based on evolutionary-rate-correlation tests. How the proteins interact still remains uncertain, as previous studies have shown that correlated evolutionary rates can occur between proteins with the same function and/or tissue-specific expression or that physically interact (Clark et al., 2012). As egg (CD9) and sperm (CRISP1 and IZUMO1) proteins are all expressed in different tissues, we hypothesize that the correlated evolutionary rates are a result of these proteins having the same function or are interacting. Recent experimental results show that IZUMO1 alone does not facilitate sperm–egg fusion with CD9, and may interact with other egg-cell-surface proteins instead (Inoue et al., 2013). Coevolution driven by sexual conflict between CD9 and CRISP1-IZUMO1 predicts a strong correlation in evolutionary-rate covariation. It may be that CRISP1 assists IZUMO1 (or vice versa) in binding to the egg membrane, and thus CRISP1 must adapt whenever IZUMO1 changes in response to the amino acid changes in CD9. Alternatively, CD9 may be interacting separately with both CRISP1 and IZUMO1. Our results predict that CD9 is interacting with both proteins, and can be followed up with additional experimental evidence to resolve the genetic interactions between these proteins. We saw evidence of positive selection in CRISP1, which can inform us about the rate of compensatory changes occurring between CD9 and CRISP1. We would predict that for every single amino-acid change that occurs in CD9, there are multiple amino acid changes that occur in the CRISP1 protein based on the selection analysis and also the slope of the correlation line (m =0.67). In contrast, IZUMO1 is not evolving under positive selection and has a shallower correlation slope when compared against CD9 (m =0.25). These tests remain exploratory in nature, and provide a simple analytical framework for investigating potentially interacting proteins. The well-characterized interacting abalone sperm–egg proteins, lysin and VERL, remain one of the best examples of how analytical data can be used to support the physical interaction of two proteins (Swanson and Vacquier, 1997; Clark et al., 2009).
Our likelihood analysis suggests that CD9 is not interacting with the sperm protein CRISP2 or the sperm proteins involved in zona pellucida-binding (PKDREJ and ZAN). CRISP2 is involved in the sperm–egg fusion process (Busso et al., 2007b; Cohen et al., 2008), and we show that it is evolving under positive selection and exhibits a correlation of evolutionary rates with CD9 by the weighted regression analysis. Our likelihood analysis may be picking up other statistical components in the data than the regression analysis uses, so these two results will not necessarily correspond. For example, the evolution of domains within CRISP2 and CD9 may be driving correlation that can be measured by one and not another method. It is also clear that some proteins may not be under long-term coevolution with another protein, so perhaps CRISP2 does not interact with another egg membrane protein. Further, some reproductive proteins may experience bouts of selection along specific lineages, but not within an entire clade, which may alter the signature of correlation that we measured.
It is worth noting that CD9 showed no evidence of correlation with the zona pellucida-binding sperm proteins. This suggests that no coevolution is occurring between CD9 and PKDREJ or ZAN, and more importantly, that sperm–egg fusion and zona pellucida-binding proteins may be evolving at different rates. Indeed, for mammalian systems, evidence suggests that the zona pellucida may play an important role in species-specificity because it is one of the first barriers the sperm needs to cross for fertilization to occur. In future studies, it would be of interest to compare the evolution rate of zona pellucida-binding proteins versus sperm–egg fusion proteins. By assessing evolutionary correlations, we can predict the interaction of candidate proteins for further functional characterization in mammalian fertilization studies.
METHODS
Sequencing and Identification of Sites Under Positive Selection
We gathered sequences for the coding exons of CD9, CRISP1, CRISP2, and IZUMO1 from a panel of 12 primate species, which included 5 species from the hominids, 2 Old World monkeys, and 5 New World monkeys (Fig. 2). We performed Sanger sequencing on genomic DNA received from the Coriell Cell Repositories for a bonobo (Pan paniscus, NG05253), a gorilla (Gorilla gorilla, NG05251), an orangutan (Pongo pygmaeus, NG12256), a pig-tailed macaque (Macaca nemestrina, NG08452), a red-chested mustached tamarin (Saguinus labiatus, NA05308), a spider monkey (Ateles geoffroyi, NG05352), a woolly monkey (Lagothrix lagotricha, AG05356), a patas monkey (Erythrocebus patas, AG06116), and a marmoset (Callithrix jacchus, NA07404). In addition, we obtained coding sequences from the publicallyavailable reference genomes of human (Homo sapiens, hg18), chimpanzee (Pan troglodytes, panTro2), and rhesus macaque (Macaca mulatta, rheMac2). PCR primers were designed to the human reference genome in conserved regions surrounding each exon, and are available upon request. Sequences were manually inspected in Seqman, aligned using the ClustalW program, and were deposited into GenBank (accession numbers KJ847058-KJ847091).
We tested each of these genes for evidence of positive selection using maximum likelihood methods implemented in the codeml program of the PAML 4.4 package (Yang, 2007). These methods compare neutral models of codon evolution to models of positive selection in which a subset of codons are allowed to have dN/dS > 1 (M7 versus M8 and M8a versus M8). The consensus species tree shown in Figure 2 was used for these analyses. We then performed likelihood ratio tests with a χ2 approximation to calculate P-values and to assess statistical significance. To determine if the dN/dS value was significantly different from 1, we compared a neutral model with a class of sites fixed at dN/dS = 1 (Model M8a) to a model of selection (Model 8). Codeml was run three times for each gene, with different starting dN/dS values to ensure convergence of model parameter estimates (no gene failed to converge). To identify individual codon sites subject to positive selection, we used the Bayes empirical Bayes approach from codeml’s model M8 (Yang, 2007). In addition, we tested for variation in the selective pressure across lineages by comparing a model that estimates one dN/dS ratio for all branches (fixed, Model 0) to a model that estimates one dN/ dS value for each branch (free-ratio model).
Correlation of Evolutionary Rates to Predict Interacting Proteins
A predicted signature of coevolution is the long-term correlation of evolutionary rates between two genes (Pazos et al., 1997; Goh and Cohen, 2002). If divergence of one protein selects for a compensatory change in its partner, then an increase in the evolutionary rate in one protein should be matched by an increase along the same lineage in its partner to maintain their joint interaction during divergence. This process will result in a correlation of evolutionary rates when comparing the corresponding branches of each protein’s phylogenetic tree. The similarity of the branch lengths can be statistically evaluated and used to infer coevolution (Fig. 4). We employed the method of Clark et al. (2009), in which the degree of correlation between two proteins is evaluated by the correlation of dN/dS values for corresponding branches of their phylogenetic trees. By dividing by the rate of synonymous substitutions, we can normalize for neutral divergence, and in effect remove correlation due to shared ancestry (Clark et al., 2009).
Figure 4.
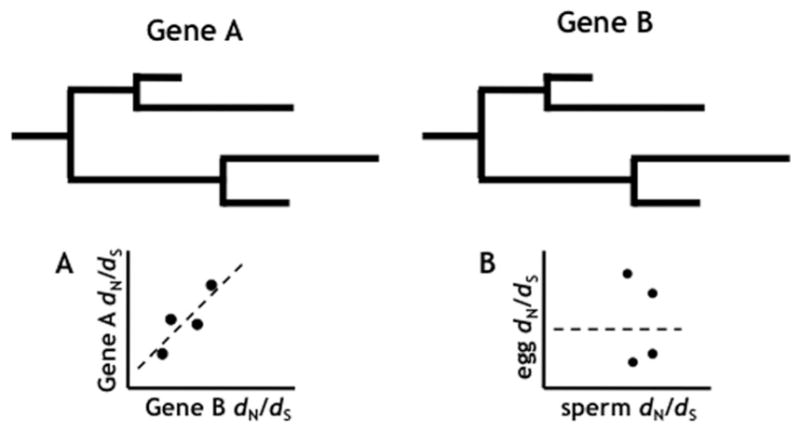
Detecting long-term coevolution using phylogenetics. Gene A and Gene B are potentially interacting proteins, and their hypothetical phylogenetic tree determined by multiple sequence alignment is shown. A: Correlated branch lengths for interacting proteins. B: Un-correlated branch lengths for non-interacting proteins.
We tested for a correlation of lineage-specific dN/dS values between each of our sperm–egg gene pairs (IZUMO1-CD9, CRISP1-CD9, and CRISP2-CD9) using two different methods: (1) linear regressions and (2) the likelihood models of Clark et al. (2009). These methods can have different powers to detect correlations between evolutionary rates because each uses a separate method for determining correlations. We estimated dN/dS values for each branch from the free-ratios model (model =1, NS sites =0) of codeml (Yang, 2007) from the multiple sequence alignments of IZUMO1, CRISP1, CRISP2, and CD9 from the 12 species. These dN/dS values were used as point estimates in a standard linear regression analysis. For branches with no synonymous substitutions, such that dN/dS = infinity, we rounded dN/dS up to the highest integer in the dataset. To limit the effects of branches with few substitutions, which may cause imprecise dN/dS estimates, we weighted the linear regressions by the substitution rate (total number of substitutions per codon) of each branch. We then used three likelihood models to jointly estimate the lineage-specific dN/dS values and the strength of correlation between them (Clark et al., 2009). We compared a correlated model, which constrains the dN/dS values to fall on a best-fit line of regression, with a null model, which sets the slope of this line to zero to represent an uncorrelated relationship. We also compared a free model, which allows dN/dS values to vary between pairs of proteins, with the correlated model. P-values were calculated from χ2 distributed likelihood ratio tests to assess the statistical significance between pairs of models. As controls, we tested for a correlation of dN/dS values between branches of CD9 and two other rapidly evolving male sperm proteins, PKDREJ and ZAN, which are thought to participate in sperm-zona pellucida binding rather than sperm–egg fusion (Gasper and Swanson, 2006; Hamm et al., 2007).
Supplementary Material
Acknowledgments
Grant sponsor: National Science Foundation Graduate Research Fellowship; Grant number: DEG-0718124; Grant sponsor: National Institute of Health; Grant numbers: HD042563, HD057974
This work was supported by a National Science Foundation Graduate Research Fellowship to K.G.C. under grant number DEG-0718124. W.J.S. was funded by National Institute of Health grant numbers HD042563 and HD057974.
Abbreviation
- dN/dS
ratio of nonsynonymous substitutions per nonsynonymous site-to-synonymous substitutions per synonymous sites
Footnotes
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Busso D, Cohen DJ, Hayashi M, Kasahara M, Cuasnicu PS. Human testicular protein TPX1/CRISP-2: Localization in spermatozoa, fate after capacitation and relevance for gamete interaction. Mol Hum Reprod. 2005;11:299–305. doi: 10.1093/molehr/gah156. [DOI] [PubMed] [Google Scholar]
- Busso D, Cohen DJ, Maldera JA, Dematteis A, Cuasnicu PS. A novel function for CRISP1 in rodent fertilization: Involvement in sperm-zona pellucida interaction. Biol Reprod. 2007a;77:848–854. doi: 10.1095/biolreprod.107.061788. [DOI] [PubMed] [Google Scholar]
- Busso D, Goldweic NM, Hayashi M, Kasahara M, Cuasnicu PS. Evidence for the involvement of testicular protein CRISP2 in mouse sperm-egg fusion. Biol Reprod. 2007b;76:701–708. doi: 10.1095/biolreprod.106.056770. [DOI] [PubMed] [Google Scholar]
- Chen MS, Tung KS, Coonrod SA, Takahashi Y, Bigler D, Chang A, Yamashita Y, Kincade PW, Herr JC, White JM. Role of the integrin-associated protein CD9 in binding between sperm ADAM 2 and the egg integrin alpha6beta1: Implications for murine fertilization. Proc Natl Acad Sci USA. 1999;96:11830–11835. doi: 10.1073/pnas.96.21.11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NL, Gasper J, Sekino M, Springer SA, Aquadro CF, Swanson WJ. Coevolution of interacting fertilization proteins. PLoS Genet. 2009;5:e1000570. doi: 10.1371/journal.pgen.1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NL, Alani E, Aquadro CF. Evolutionary rate covariation reveals shared functionality and coexpression of genes. Genome Res. 2012;22:714–720. doi: 10.1101/gr.132647.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen DJ, Busso D, Da Ros V, Ellerman DA, Maldera JA, Goldweic N, Cuasnicu PS. Participation of cysteine-rich secretory proteins (CRISP) in mammalian sperm-egg interaction. Int J Dev Biol. 2008;52:737–742. doi: 10.1387/ijdb.072538dc. [DOI] [PubMed] [Google Scholar]
- Da Ros VG, Maldera JA, Willis WD, Cohen DJ, Goulding EH, Gelman DM, Rubinstein M, Eddy EM, Cuasnicu PS. Impaired sperm fertilizing ability in mice lacking Cysteine-RIch Secretory Protein 1 (CRISP1) Dev Biol. 2008;320:12–18. doi: 10.1016/j.ydbio.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellerman DA, Cohen DJ, Da Ros VG, Morgenfeld MM, Busso D, Cuasnicu PS. Sperm protein “DE” mediates gamete fusion through an evolutionarily conserved site of the CRISP family. Dev Biol. 2006;297:228–237. doi: 10.1016/j.ydbio.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Gasper J, Swanson WJ. Molecular population genetics of the gene encoding the human fertilization protein zonadhesin reveals rapid adaptive evolution. Am J Hum Genet. 2006;79:820–830. doi: 10.1086/508473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh CS, Cohen FE. Co-evolutionary analysis reveals insights into protein-protein interactions. J Mol Biol. 2002;324:177–192. doi: 10.1016/s0022-2836(02)01038-0. [DOI] [PubMed] [Google Scholar]
- Grayson P, Civetta A. Positive selection and the evolution of Izumo genes in mammals. Int J Evol Biol. 2012;2012:958164. doi: 10.1155/2012/958164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakes L, Lovell SC, Oliver SG, Robertson DL. Specificity in protein interactions and its relationship with sequence diversity and coevolution. Proc Natl Acad Sci USA. 2007;104:7999–8004. doi: 10.1073/pnas.0609962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm D, Mautz BS, Wolfner MF, Aquadro CF, Swanson WJ. Evidence of amino acid diversity-enhancing selection within humans and among primates at the candidate sperm-receptor gene PKDREJ. Am J Hum Genet. 2007;81:44–52. doi: 10.1086/518695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JD, Hibler DW, Fontenot GK, Hsu KT, Yurewicz EC, Sacco AG. Cloning and characterization of zona pellucida genes and cDNAs from a variety of mammalian species: The ZPA, ZPB and ZPC gene families. DNA Seq. 1994;4:361–393. doi: 10.3109/10425179409010186. [DOI] [PubMed] [Google Scholar]
- Inoue N, Ikawa M, Isotani A, Okabe M. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature. 2005;434:234–238. doi: 10.1038/nature03362. [DOI] [PubMed] [Google Scholar]
- Inoue N, Hamada D, Kamikubo H, Hirata K, Kataoka M, Yamamoto M, Ikawa M, Okabe M, Hagihara Y. Molecular dissection of IZUMO1, a sperm protein essential for sperm-egg fusion. Development. 2013;140:3221–3229. doi: 10.1242/dev.094854. [DOI] [PubMed] [Google Scholar]
- Kaji K, Oda S, Shikano T, Ohnuki T, Uematsu Y, Sakagami J, Tada N, Miyazaki S, Kudo A. The gamete fusion process is defective in eggs of Cd9-deficient mice. Nat Genet. 2000;24:279–282. doi: 10.1038/73502. [DOI] [PubMed] [Google Scholar]
- Le Naour F, Rubinstein E, Jasmin C, Prenant M, Boucheix C. Severely reduced female fertility in CD9-deficient mice. Science. 2000;287:319–321. doi: 10.1126/science.287.5451.319. [DOI] [PubMed] [Google Scholar]
- Miyado K, Yamada G, Yamada S, Hasuwa H, Nakamura Y, Ryu F, Suzuki K, Kosai K, Inoue K, Ogura A, Okabe M, Mekada E. Requirement of CD9 on the egg plasma membrane for fertilization. Science. 2000;287:321–324. doi: 10.1126/science.287.5451.321. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura H, Cho C, Branciforte DR, Myles DG, Primakoff P. Analysis of loss of adhesive function in sperm lacking cyritestin or fertilin beta. Dev Biol. 2001;233:204–213. doi: 10.1006/dbio.2001.0166. [DOI] [PubMed] [Google Scholar]
- Okabe M, Adachi T, Takada K, Oda H, Yagasaki M, Kohama Y, Mimura T. Capacitation-related changes in antigen distribution on mouse sperm heads and its relation to fertilization rate in vitro. J Reprod Immunol. 1987;11:91–100. doi: 10.1016/0165-0378(87)90014-3. [DOI] [PubMed] [Google Scholar]
- Pazos F, Valencia A. Similarity of phylogenetic trees as indicator of protein-protein interaction. Protein Eng. 2001;14:609–614. doi: 10.1093/protein/14.9.609. [DOI] [PubMed] [Google Scholar]
- Pazos F, Helmer-Citterich M, Ausiello G, Valencia A. Correlated mutations contain information about protein-protein interaction. J Mol Biol. 1997;271:511–523. doi: 10.1006/jmbi.1997.1198. [DOI] [PubMed] [Google Scholar]
- Rochwerger L, Cohen DJ, Cuasnicu PS. Mammalian sperm-egg fusion: The rat egg has complementary sites for a sperm protein that mediates gamete fusion. Dev Biol. 1992;153:83–90. doi: 10.1016/0012-1606(92)90093-v. [DOI] [PubMed] [Google Scholar]
- Rubinstein E, Benoit P, Billard M, Plaisance S, Prenant M, Uzan G, Boucheix C. Organization of the human CD9 gene. Genomics. 1993;16:132–138. doi: 10.1006/geno.1993.1150. [DOI] [PubMed] [Google Scholar]
- Rubinstein E, Ziyyat A, Prenant M, Wrobel E, Wolf JP, Levy S, Le Naour F, Boucheix C. Reduced fertility of female mice lacking CD81. Dev Biol. 2006;290:351–358. doi: 10.1016/j.ydbio.2005.11.031. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. The abalone egg vitelline envelope receptor for sperm lysin is a giant multivalent molecule. Proc Natl Acad Sci USA. 1997;94:6724–6729. doi: 10.1073/pnas.94.13.6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Nielsen R, Yang Q. Pervasive adaptive evolution in mammalian fertilization proteins. Mol Biol Evol. 2003;20:18–20. doi: 10.1093/oxfordjournals.molbev.a004233. [DOI] [PubMed] [Google Scholar]
- Wright MD, Tomlinson MG. The ins and outs of the transmembrane 4 superfamily. Immunol Today. 1994;15:588–594. doi: 10.1016/0167-5699(94)90222-4. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Swanson WJ, Vacquier VD. Maximum-likelihood analysis of molecular adaptation in abalone sperm lysin reveals variable selective pressures among lineages and sites. Mol Biol Evol. 2000;17:1446–1455. doi: 10.1093/oxfordjournals.molbev.a026245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.