Abstract
MicroRNAs have been reported to play an important role in diverse biological processes and progression of various cancers. MicroRNA-29a has been observed to be downregulated in human lung cancer tissues, but the function of microRNA-29a in lung cancer has not been well investigated. In this study, we demonstrated that the expression levels of microRNA-29a were significantly downregulated in 38 pairs of lung cancer tissues when compared to adjacent normal tissues. Overexpression of microRNA-29a inhibited the activity of cell proliferation and colony formation of lung cancer cells, H1299 and A549. Furthermore, microRNA-29a targeted NRAS proto-oncogene in lung cancer cells. In human clinical specimens, NRAS proto-oncogene was highly expressed in human lung cancer tissues compared to normal tissues. More interestingly, microRNA-29a also sensitizes lung cancer cells to cisplatin (CDDP[Please replace “CDDP” with its expansion in the abstract and also provide expansion for the same in its first occurrence in text, if appropriate.]) via its target, NRAS proto-oncogene. Thus, our results in this study demonstrated that microRNA-29a acted as a tumor suppressor microRNA, which indicated potential application of microRNAs for the treatment of human lung cancer in the future.
Keywords: miR-29a, NRAS, cisplatin, lung cancer, tumor suppressor
Introduction
Lung cancer is the first cause of cancer-related deaths in the world, which is characterized with lower survival and higher relapse rate after surgery.1-3 Non-small cell lung cancer (NSCLC) is the most frequently occurring category of lung cancer, accounting for approximately 80% to 85% of all cases.4 Tumor invasion and facile metastasis are the main factors responsible for failure of NSCLC treatment.5,6 Previous studies have shown that the tumorigenesis of lung cancer is involved in genetic and epigenetic alterations, including the activation of oncogenes and/or the suppression of tumor suppressor genes. Mounting evidence showed that noncoding microRNAs (miRNAs) play key roles in the progression of lung cancer. MicroRNAs stabilize and circulate in body fluids and therefore may serve as biomarkers for early diagnosis of lung cancer as well as for predicting prognosis of patients.7,8
MicroRNAs have been associated with various types of cancer, which involved in many important biological and pathological processes, including growth, differentiation, and tumorigenesis.9-11 By binding to the 3′-untranslated region (3′-UTR) of candidate messenger RNA (mRNA), miRNA suppresses protein synthesis through degradation of mRNA or repression of translation. As a tumor suppressor gene in several cancer types, miR-29a can affect tumor cell growth, migration, invasion, and apoptosis. Some genes have been identified as target genes of miR-29a, including c-MYC, CLDN1, CDC42, KDM5B, and TNFR1.12-16 However, the function and underlying mechanism of miR-29a in regulating tumorigenesis of lung cancer are still to be further investigated.
RAS sarcoma (Ras) genes have been reported as the most frequently activated oncogenes in various human cancer. NRAS proto-oncogene, as 1 of the 3 members of the RAS oncogene family, encodes small GTP enzymes (GTPases) that are functioned in cellular signal transduction. NRAS proto-oncogene is implicated and altered in various cancer and plays potential roles in regulation of cancer cell growth, migration, invasion, survival, and angiogenesis.17,18 Oncogenic NRAS promotes progression of tumorigenesis through activation of multiple downstream pathways, including mitogen-activated protein kinase 1 (MAPK)/extracellular signal-regulated kinase (ERK), phosphoinositide 3-kinase/AKT, and nuclear factor-kappa B (NF-κB) pathway.19-22 Thus, NRAS silencing may be an efficient therapeutic strategy in tumors.
It has been reported that miR-29a may have function in lung cancer via affecting various signaling pathways in tumorigenesis. In our study, we aimed to further identify the roles of miR-29a and its molecular as well as cellular mechanisms in the progression of lung cancer. Overexpression of miR-29a inhibited the activity of cell proliferation and colony formation of lung cancer cells by suppressing a key target NRAS. We further defined miR-29a-induced chemosensitivity of lung cancer cells to cisplatin through NRAS suppression. In this study, our data showed that miR-29a was significantly downregulated in human lung cancer tissues compared to the adjacent paired normal controls. Our results revealed a novel mechanism of miR-29a in lung cancer, and it possessed a potential to be used as a novel strategy to develop miR-29a-based therapeutics.
Materials and Methods
Cell Culture and Clinical Tissues
Human lung cancer cell lines, H1299 and A549, were cultured in RPMI 1640 medium, and HEK293 T cells were cultured in dulbecco's modified eagle medium (DMEM) medium. All cells were incubated at 37°C with 5% CO2. Paired human lung cancer specimens and matched normal samples were collected from patients undergoing a surgical procedure in the First Affiliated Hospital of Zhengzhou University, with the informed consent of the patients. Parts of tissue samples were immediately snap-frozen in liquid nitrogen, and parts were fixed in formalin for histological examination. All samples were histologically classified by a clinical pathologist. The experiment protocols have been approved by the ethics committees of Zhengzhou University.
Lentiviral Packaging and Stable Cell Line Establishment
To stably overexpress miR-29a in lung cancer cells, the lentiviral packaging kit was used (Thermo Fisher Scientific, Shanghai, China). Lentivirus carrying miR-29a or miR-NC was packaged. Following the manufacturer’s manual, lentivirus was packaged in HEK293 T cells. Cells were infected by lentivirus carrying miR-29a or miR-NC with the presence of polybrene (Sigma-Aldrich, St Louis, Missouri) and selected by puromycin (Sigma-Aldrich) for 10 days to obtain stable cell lines.
Oligonucleotides and Cell Transfection
Micro RNA-7 mimics and miR-NC were chemically synthesized by GenePharma. Cells at 50% to 70% confluence were transfected with miR-29a or miR-NC using Lipofectamine reagent (Invitrogen, Shanghai, China), according to the manufacturer’s instructions. Transfected cells were harvested at 24 or 48 hours after transfection.
RNA Extraction and Real-Time Polymerase Chain Reaction
RNA was extracted using TRIzol reagent (Takara, Dalian, China), and purified RNA was stored at −80°C prior to further analysis. Real-time reverse transcription polymerase chain reaction (RT-PCR) analysis for mature miR-29a was carried out using the RT Reagent Kit (Takara), according to the manufacturer’s instructions. Briefly, 500 ng total RNA was reversely transcribed into complementary DNA, and quantitative RT-PCR was performed using SYBR Green Master Mix (Takara) on a 7500HT system. The miR-29a expression in each group was determined relative to that of U6, and fold changes were calculated by relative quantification (2−▵▵Ct).
Cell Proliferation Assay
Cell Counting Kit-8 (Dojindo Laboratories, Japan) assay was used to determine cell viability. Cells were seeded in 96-well plates and cultured as described above for 48 hours after transfection. After 24, 48, 72, and 96 hours of incubation, CCK-8 was added into each well, followed by 1 to 2 hours of incubation. Absorbance at 450 nm was then determined. Experiments were carried out in triplicate.
Colony Formation Assay
Cells (N = 200) were placed in 12-well plates. The medium was replaced every 4 days. After 14 to 18 days, the cells were fixed with methanol and stained with 0.1% crystal violet (Sigma-Aldrich). Visible colonies were counted. For each treatment group, experiments were independently repeated 3 times.
Western Blot
Cells were treated as previously described, and cells were harvested after 24 hours and lysed in radioimmunoprecipitation assay buffer. After 15-minute centrifugation, protein concentrations were determined by the BCA(bicinchoninic acid)protein assay (BCA) method (Beyotime, Jiangsu, China) and then separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Subsequently, protein was electrically transferred onto a nitrocellulose membrane (Whatman, UK). The membrane was incubated with NRAS antibody (1:1000; Cell Signaling Technology, Shanghai, China) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:5000; Bioworld Technology, USA) at 4°C overnight.
Luciferase Reporter Assay
TargetScan software was used to predict miR-29a binding sites. 3′-Untranslated region of NRAS containing the putative miR-29a-binding site was amplified by PCR. To generate a construct containing the mutant miR-29a binding site, 2 nucleotides corresponding to the 5′-seeding region of the miR-29a binding site on the wild-type fragment were substituted. Its complementary sequence in the 3′-UTR of NRAS (UGGUGCU) was replaced by UGCAGGU. The PCR products were digested using HindIII and SacI, then inserted into pMIR-reporter, and validated by DNA sequencing. Constructs were transfected into H1299 cells in 24-well plates and cotransfected with miR-29a or miR-NC. Luciferase assays were performed after 24 hours using the dual luciferase reporter assay system (Promega, USA).
In Vitro Chemosensitivity Array
Cancer cells were seeded in a 96-well plate overnight. Freshly prepared cisplatin (Sigma-Aldrich) was added with the final concentration ranging from 1.25 to 80 μM. Forty-eight hours later, cell viability was assayed by CCK-8 kit.
Apoptosis Assay
Following the manufacturer’s manual, apoptosis assay kit (BD Pharmingen, Shanghai, China) was used in the samples, and the samples were incubated for 15 minutes at room temperature in the dark . Then the samples were analyzed by flow cytometry (FACS Canto II; BD Biosciences, Shanghai, China) within 1 hour. The data were analyzed using FlowJo software, version 10.4. Three experiments were performed in triplicate.
Caspase-3 Activity Assay
The activity of caspase-3 was determined using caspase-3 activity kit (Beyotime). Caspase-3 activity assay was performed on 96-well plates by incubating 10 μL protein of cell lysate per sample in 80 μL reaction buffer containing 10 μL caspase-3 substrate (Ac-DEVD-pNA; 2 mM) at 37°C for 2 hours according to the manufacturer’s protocol. The reaction was measured at optical density (OD) 405 nm for absorbance.
Statistical Analysis
All experiments were performed in triplicate, and data were analyzed with GraphPad Prism 5 (La Jolla, California). Correlation between miR-29a expression and NRAS in tissues was analyzed by Spearman rank test. Statistical evaluation for data analysis was determined by t test. P < .05 was considered as statistically significant.
Results
MicroRNA-29a Is Significantly Downregulated in Lung Cancer Tissues
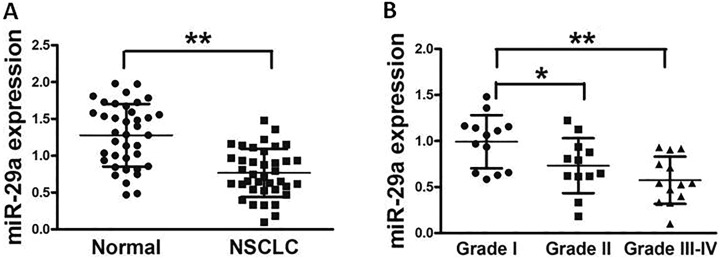
In this study, we assessed the miR-29a expression in 38 pairs of lung cancer tissues and normal tissues, and Figure 1A showed that the miR-29a expression in tumor tissues was significantly lower compared with those controls. In addition, samples were histologically classified by a clinical pathologist; miR-29a expression was lower in lung cancer tissues with World Health Organization (WHO) stage III and IV than those in stage I and stage II, indicating that miR-29a expression was significantly downregulated in late stages of lung cancer tissues (Figure 1B). Taken together, lower expression of miR-29a in patients with lung cancer may be used as a potential new biomarker, which could predict poor prognosis for lung cancer.
Figure 1.
MicroRNA-29a is significantly downregulated in lung cancer tissues. A, Relative miR-29a expression levels were analyzed by quantitative RT-PCR in 38 pairs of human lung cancer tissues and adjacent normal tissues. U6 RNA level was used as an internal control. B, All samples were histologically classified by a clinical pathologist. Relative expression levels of miR-29a in different stages of cancer tissues. Data represent mean ± SD of 3 replicates. *Significant difference at P < .05. **Significant difference at P < .01. MiR-29a denotes microRNA-29a; RT-PCR, reverse transcription polymerase chain reaction; SD, standard deviation.
Overexpression of MiR-29a Inhibits Cell Proliferation and Colony Formation of Lung Cancer Cells
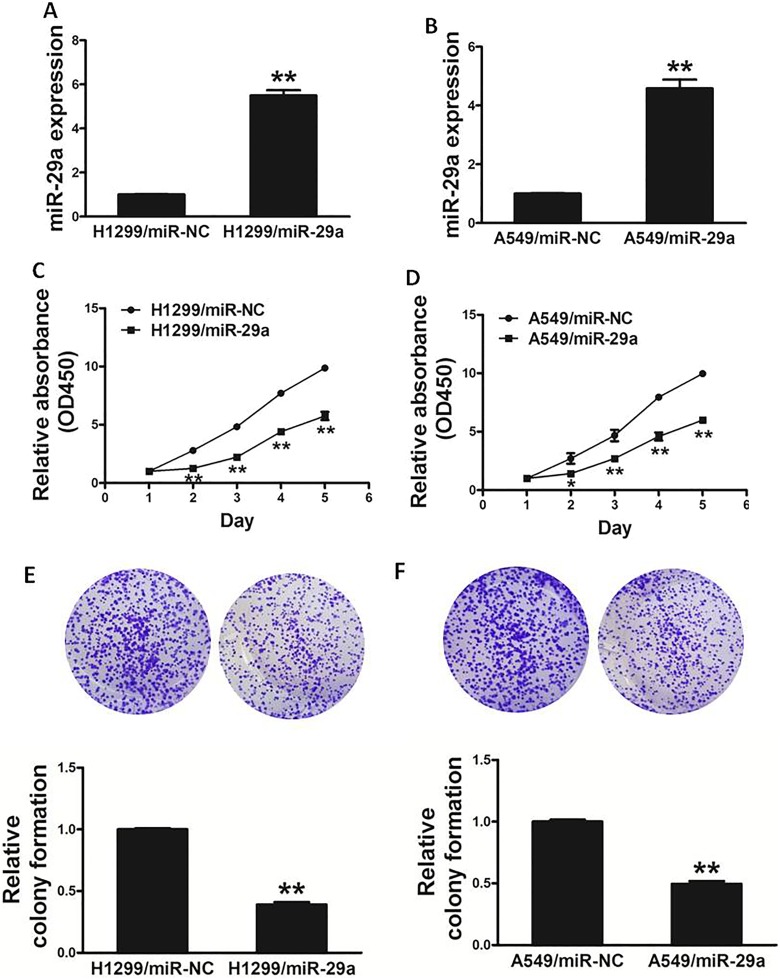
To explore the role of miR-29a in lung cancer cells, we established stable cell lines by infecting H1299 and A549 cells with lentiviral constructs harboring miR-29a or microRNA-negative control (miR-NC; Figure 2A and B). Cell viability assay indicated that the overexpression of miR-29a significantly reduced the rate of cell proliferation at 48 hours after cell seeding (Figure 2C and D). Moreover, we investigated the effects of miR-29a on colony formation in vitro. Forced expression of miR-29a also markedly decreased the activity of colony formation in cells (Figure 2E and F). Thus, these results showed that overexpression of miR-29a inhibits activity of cell proliferation and colony formation of lung cancer cells.
Figure 2.
Overexpression of miR-29a inhibits the ability of cell proliferation and colony formation in lung cancer cells. A and B, Relative expression levels of miR-29a in H1299/miR-29a, H1299/miR-NC, A549/miR-29a, and A549/miR-NC stable cell lines were confirmed by quantitative RT-PCR. C and D, Overexpression of miR-29a arrested cell proliferation in H1299 and A549 cells. E and F, MiR-29a overexpression reduced colony formation in H1299 and A549 cells. Data represent mean ± SD of 3 replicates. *Significant difference at P < .05. **Significant difference at P < .01. MiR-29a denotes microRNA-29a; mir-NC, microRNA-negative control; RT-PCR, reverse transcription polymerase chain reaction; SD, standard deviation.
NRAS Proto-Oncogene Is a Target of MiR-29a
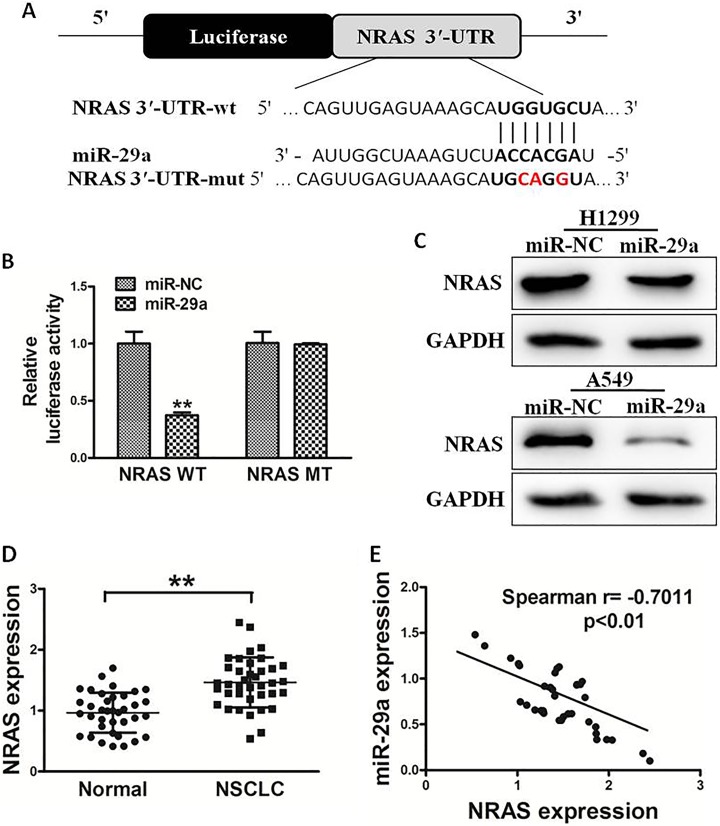
To analyze the molecular mechanism of miR-29a in lung cancer, TargetScan and miRanda were used to explore potential targets of miR-29a. Figure 3A shows that the 3′-UTR of NRAS contained the binding site for the seed region of miR-29a. To confirm whether NRAS was the direct target of miR-29a, human NRAS 3′-UTR, containing either wild-type or mutant miR-29a binding sequence, then was cloned downstream of the firefly luciferase reporter gene in the pMIR-reporter vector. H1299 cells were transfected with 2 reporter plasmids, plus miR-29a or miR-NC mimics. The luciferase activity of the vector containing the NRAS 3′-UTR wild type (WT) was significantly reduced by miR-29a, while NRAS 3′-UTR mutant type (MT) exhibited an insignificantly affected luciferase activity (Figure 3B). Western blotting analysis was conducted to determine NRAS expression at protein level. We found that the NRAS expression was downregulated at the protein level in miR-29a-treated cells (Figure 3C). These results suggested that miR-29a directly targeted NRAS by binding to its 3′-UTRs in lung cancer cells. Furthermore, we measured the NRAS expression at the mRNA level in human lung cancer specimens and normal tissues. The results showed that the average expression level of NRAS was significantly higher in tumor tissues than that in normal tissues (Figure 3D). Then, we determine the correlation between NRAS levels and miR-29a expression levels in the same lung cancer tissues. As shown, Spearman rank correlation analysis showed that the expression levels of NRAS and miR-29a in cancer tissues were inversely correlated (Figure 3E, Spearman correlation r = −.7011). Thus, these results suggested that NRAS is a direct target of miR-29a.
Figure 3.
NRAS is a target of miR-29a. A, Sequence of the miR-29a binding site within the human NRAS 3′-UTR and a schematic diagram of the reporter construct showing the entire NRAS 3′-UTR sequence and the mutant NRAS 3′-UTR sequence. The mutant nucleotides of the NRAS 3′-UTR are labeled in red. B, Luciferase assay on H1299 cells which were cotransfected with miR-NC or miR-29a and a luciferase reporter containing the full length of NRAS 3′-UTR (WT) or a mutant type (MT) harboring 4 mutant nucleotides of the miR-29a binding site. Luciferase activities were measured 24 hours posttransfection. MicroRNA-29a markedly suppressed luciferase activity in NRAS 3′-UTR wild type (WT) reporter constructs. C, The immunoblotting showed that expression levels of NRAS were decreased in cells with miR-29a overexpression. D, The expression of NRAS in adjacent normal tissues and human lung cancer specimens was determined by quantitative RT-PCR analysis, and fold changes were obtained from the ratio of NRAS to GAPDH levels. E, Spearman rank correlation analysis showed that the expression levels of NRAS and miR-29a in lung cancer tissues were inversely correlated. Data represent mean ± SD of three replicates. **Significant difference at P < .01. MiR-29a denotes microRNA-29a; mir-NC, microRNA-negative control; NRAS, NRAS proto-oncogene; 3′-UTR, 3’-untranslated region; RT-PCR, reverse transcription polymerase chain reaction; SD, standard deviation.
Overexpression of MiR-29a Increases Chemosensitivity of Lung Cancer Cells to Cisplatin by Targeting NRAS
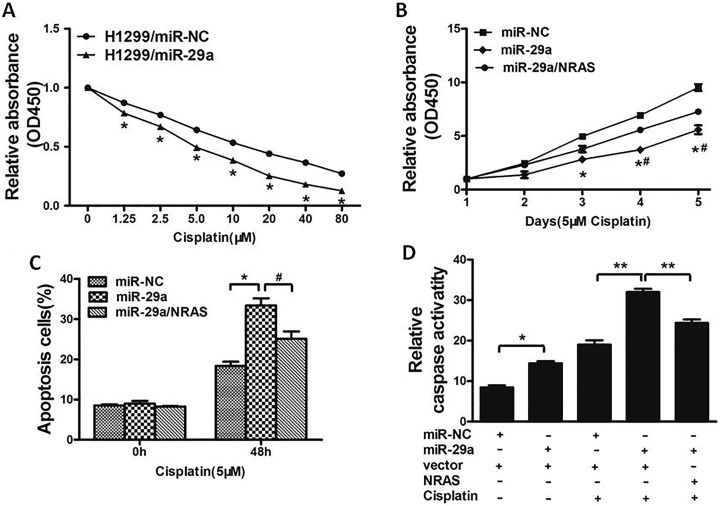
Resistance to cisplatin treatment is one of the major causes for the failure of clinical chemotherapy in treating various cancers including lung cancer. Therefore, it is urgent to discover new strategies to promote effectiveness of cisplatin. In this study, our results showed that miR-29a overexpressed in H1299 cells significantly promoted chemosensitivity to cisplatin (Figure 4A). Cell growth rate in the presence of cisplatin (5 μM) was determined by Cell Counting Kit-8 (CCK-8) assay at different time points, which showed that overexpression of NRAS reversed miR-29a-induced chemosensitivity to cisplatin (Figure 4B). To further investigate whether miR-29a and NRAS play a role in cell apoptosis in the presence of cisplatin, FACS analysis was performed. The combination of miR-29a and cisplatin significantly induced cell apoptosis, whereas forced expression of NRAS partially abolished the effect induced by miR-29a plus cisplatin treatment (Figure 4C). Moreover, we investigated that compared to miR-29a or cisplatin treatment alone, the activities of caspase-3, which acts as a key executor of cell apoptosis, were significantly upregulated upon treatment of miR-29a and cisplatin, whereas forced expression of NRAS attenuated the activation of caspase-3 (Figure 4D). These results indicated that miR-29a renders lung cancer cells more sensitive to cisplatin treatment and miR-29a and cisplatin combination promoted apoptotic effect through targeting NRAS in lung cancer cells.
Figure 4.
Overexpression of miR-29a increases chemosensitivity of lung cancer cells to cisplatin by inhibiting its target NRAS. A, H1299 cells stably expressing miR-NC or miR-29a were pretreated with cisplatin for indicated concentrations, then subjected to CCK-8 assay. B, H1299 cells stably expressing miR-NC, miR-29a, or miR-29a forced expression of NRAS were pretreated with 5 μM of cisplatin for indicated time points, then subjected to CCK-8 assay, apoptosis analyses by flow cytometry (C), and Caspase3 assay (D). Data represent mean ± SD of 3 replicates. */# P < .05. **P < .01. *Significant difference compared to control. #Significant difference compared to miR-29a forced expression of NRAS treatment. CCK-8 denotes Cell Counting Kit-8; miR-29a microRNA-29a; mir-NC, microRNA-negative control; NRAS, NRAS proto-oncogene; SD, standard deviation.
Discussion
Recent studies have demonstrated that miRNAs play important roles in carcinogenesis by various mechanisms, and certain miRNAs have been reported to be correlated with clinical characteristics and clinical outcomes. The role of some miRNAs in lung cancer and drug resistance has also been reported. For example, Zhao et al reported that miR-202 functions as a tumor suppressor by targeting signal transducer and activator of transcription 3 (STAT3) in non-small cell lung cancer.23 Shi et al reported that miR-218 contributes to epithelial–mesenchymal transition by targeting Slug/zinc finger E-box binding homeobox 2 (ZEB2) signaling and inhibits tumor metastasis in lung cancer.24 Chen et al reported that miRNA-145-3p inhibits NSCLC cell migration and invasion via the mechanistic target of rapamycin kinase (mTOR) signaling pathway by targeting pyruvate dehydrogenase kinase 1 (PDK1).25
Previous studies have reported that miR-29a is downregulated in several cancer types including lung cancer.12-16 Meanwhile, this study demonstrated that miR-29a targeted NRAS and inhibited proliferation and colony formation in lung cancer cells. Here, we found that miR-29a was downregulated in lung cancer tissues compared to normal controls, and the expression of miR-29a was inversely correlated with the histopathologic grade. More interesting, we further found overexpression of miR-29a-inhibited activity of cell proliferation and colony formation of lung cancer cells. Thus, we demonstrated that miR-29a regulated lung cancer growth, which provides more therapeutic strategies for lung cancer prevention and clinical treatment.
In this study, we found that miR-29a-targeted NRAS, a key downstream effector of the PI3K/AKT signaling pathway, has long been identified to play an important role in controlling cell growth. NRAS proto-oncogene was experimentally validated as the novel target of miR-29a. Firstly, luciferase-reporter assay showed that miR-29a directly recognized the 3′-UTR of NRAS. Secondly, the NRAS expression was significantly decreased in lung cancer cells stably expressing miR-29a. Thirdly, NRAS was upregulated in cancer tissues. These results show that miR-29a is a tumor suppressor through targeting NRAS. Other targets of miR-29a need further study in the future to clarify the molecular mechanism of lung cancer progression.
Recently, miRNAs have been proposed to play essential roles in the development of drug resistance.26-31 Resistance to chemotherapy is a complex process, which results from a lot of factors including individual differences in patients or genetic or epigenetic changes in tumors. In our study, we found that overexpression of miR-29a promoted the inhibition effects of CDDP. Flow cytometer assay demonstrated that lung cancer cells with miR-29a expression have higher CDDP sensitiveness. Therefore, miR-29a restoration approach may offer a new strategy to overcome chemoresistance to CDDP treatment in lung cancer.
Our study provided the first evidence that miR-29a suppressed lung cancer cell growth through inhibition of NRAS. Although we confirmed that miR-29a could inhibit the phenotype of lung cancer by targeting NRAS in this study, there might be many other targets of miR-29a, which could also affect the growth of lung cancer cells. Nonetheless, we showed that such effect was exerted through the suppression of NRAS. Taken together, these results suggest that miR-29a and NRAS may be potential new biomarkers for lung cancer, which would be interesting for further investigation in the future.
Abbreviations
- CCK-8
Cell Counting Kit-8
- mRNA
messenger RNA
- miRNA
microRNA
- miR-29a
microRNA-29a
- miR-NC
microRNA-negative control
- RT-PCR
reverse transcription polymerase chain reaction
- 3′-UTR
3′-untranslated region
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work is supported by grant from the National Natural Science Foundation of China (81672424).
References
- 1. Hayano T, Garg M, Yin D, et al. SOX7 is down-regulated in lung cancer. J Exp Clin Cancer Res. 2013;32:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Subramaniam S, Thakur RK, Yadav VK, Nanda R, Chowdhury S, Agrawal A. Lung cancer biomarkers: state of the art. J Carcinog. 2013;12:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global Cancer Statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. [DOI] [PubMed] [Google Scholar]
- 4. MacDonagh L, Gray SG, Finn SP, Cuffe S, O’Byrne KJ, Barr MP. The emerging role of microRNAs in resistance to lung cancer treatments. Cancer Treat Rev. 2015;41(2):160–169. [DOI] [PubMed] [Google Scholar]
- 5. Huang J, Song H, Liu B, Yu B, Wang R, Chen L. Expression of Notch-1 and its clinical significance in different histological subtypes of human lung adenocarcinoma. J Exp Clin Cancer Res. 2013;32:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Steeg PS. Metastasis suppressors alter the signal transduction of cancer cells. Nature Rev Cancer. 2003;3(1):55–63. [DOI] [PubMed] [Google Scholar]
- 7. Inamura K. Diagnostic and therapeutic potential of microRNAs in lung cancer. Cancers (Basel). 2017;9(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Inamura K, Ishikawa Y. MicroRNA in lung cancer: novel biomarkers and potential tools for treatment. J Clin Med. 2016;5(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. [DOI] [PubMed] [Google Scholar]
- 10. Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25(46):6188–6196. [DOI] [PubMed] [Google Scholar]
- 11. Inamura K. Major tumor suppressor and oncogenic non-coding RNAs: clinical relevance in lung cancer. Cells. 2017;6(2). Pii E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li J, Wan X, Qiang W, et al. MiR-29a suppresses prostate cell proliferation and induces apoptosis via KDM5B protein regulation. Int J Clin Exp Med. 2015;8(4):5329–5339. [PMC free article] [PubMed] [Google Scholar]
- 13. Li Y, Wang Z, Li Y, Jing R. MicroRNA-29a functions as a potential tumor suppressor through directly targeting CDC42 in non-small cell lung cancer. Oncology Letters. 2017;13(5):3896–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mahati S, Xiao L, Yang Y, Mao R, Bao Y. . miR-29a suppresses growth and migration of hepatocellular carcinoma by regulating CLDN1. Biochem Biophys Res Commun. 2017;486(3):732–737. [DOI] [PubMed] [Google Scholar]
- 15. Saha MN, Abdi J, Yang Y, Chang H. MiRNA-29a as a tumor suppressor mediates PRIMA-1Met-induced anti-myeloma activity by targeting c-Myc. Oncotarget. 2016;7(6):7149–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao Y, Yang F, Li W, et al. miR-29a suppresses MCF-7 cell growth by downregulating tumor necrosis factor receptor 1. Tumour Biol. 2017;39(2):1010428317692264. [DOI] [PubMed] [Google Scholar]
- 17. Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296(5573):1655–1657. [DOI] [PubMed] [Google Scholar]
- 18. Shaw LM. Identification of insulin receptor substrate 1 (IRS-1) and IRS-2 as signaling intermediates in the alpha6beta4 integrin-dependent activation of phosphoinositide 3-OH kinase and promotion of invasion. Mol Cell Biol. 2001;21(15):5082–5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Birkenkamp KU, Geugien M, Schepers H, Westra J, Lemmink HH, Vellenga E. Constitutive NF-kappaB DNA-binding activity in AML is frequently mediated by a Ras/PI3-K/PKB-dependent pathway. Leukemia. 2004;18(1):103–112. [DOI] [PubMed] [Google Scholar]
- 20. Cole AL, Subbanagounder G, Mukhopadhyay S, Berliner JA, Vora DK. Oxidized phospholipid-induced endothelial cell/monocyte interaction is mediated by a cAMP-dependent R-Ras/PI3-kinase pathway. Arterioscler Thromb Vasc Biol. 2003;23(8):1384–1390. [DOI] [PubMed] [Google Scholar]
- 21. Posch C, Moslehi H, Feeney L, et al. Combined targeting of MEK and PI3K/mTOR effector pathways is necessary to effectively inhibit NRAS mutant melanoma in vitro and in vivo. Proc Natl Acad Sci U S A. 2013;110(10):4015–4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Santarpia L, Myers JN, Sherman SI, Trimarchi F, Clayman GL, El-Naggar AK. Genetic alterations in the RAS/RAF/mitogen-activated protein kinase and phosphatidylinositol 3-kinase/Akt signaling pathways in the follicular variant of papillary thyroid carcinoma. Cancer. 2010;116(12):2974–2983. [DOI] [PubMed] [Google Scholar]
- 23. Zhao Z, Lv B, Zhang L, Zhao N, Lv Y. miR-202 functions as a tumor suppressor in non-small cell lung cancer by targeting STAT3. Mol Med Rep. 2017;16(2):2281–2289. [DOI] [PubMed] [Google Scholar]
- 24. Shi ZM, Wang L, Shen H, et al. Downregulation of miR-218 contributes to epithelial-mesenchymal transition and tumor metastasis in lung cancer by targeting Slug/ZEB2 signaling. Oncogene. 2017;36(18):2577–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen GM, Zheng AJ, Cai J, Han P, Ji HB, Wang LL. microRNA-145-3p inhibits non-small cell lung cancer cell migration and invasion by targeting PDK1 via the mTOR signaling pathway. J Cell Biochem. 2018;119(1):885–895. [DOI] [PubMed] [Google Scholar]
- 26. Wang L, Shi ZM, Jiang CF, et al. MiR-143 acts as a tumor suppressor by targeting N-RAS and enhances temozolomide-induced apoptosis in glioma. Oncotarget. 2014;5(14):5416–5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hong L, Han Y, Zhang Y, et al. MicroRNA-21: a therapeutic target for reversing drug resistance in cancer. Expert Opin Ther Targets. 2013;17(9):1073–1080. [DOI] [PubMed] [Google Scholar]
- 28. Liu R, Liu X, Zheng Y, et al. MicroRNA-7 sensitizes non-small cell lung cancer cells to paclitaxel. Oncology Lett. 2014;8(5):2193–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rolfo C, Fanale D, Hong DS, et al. Impact of microRNAs in resistance to chemotherapy and novel targeted agents in non-small cell lung cancer. Curr Pharm Biotechnol. 2014;15(5):475–485. [DOI] [PubMed] [Google Scholar]
- 30. He J, Yu JJ, Xu Q, et al. Downregulation of ATG14 by EGR1-MIR152 sensitizes ovarian cancer cells to cisplatin-induced apoptosis by inhibiting cyto-protective autophagy. Autophagy. 2015;11(2):373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang L, Jiang CF, Li DM, et al. MicroRNA-497 inhibits tumor growth and increases chemosensitivity to 5-fluorouracil treatment by targeting KSR1. Oncotarget. 2016;7(3):2660–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]