Abstract
Objective
The cancer stem cell (CSC) paradigm hypothesizes that successful clinical eradication of CSCs may lead to durable remission for patients with ovarian cancer. Despite mounting evidence in support of ovarian CSCs, their phenotype and clinical relevance remain unclear. We and others have found high aldehyde dehydrogenase 1 (ALDHhigh) expression in a variety of normal and malignant stem cells, and sought to better characterize ALDHhigh cells in ovarian cancer.
Methods
We compared ALDHhigh to ALDHlow cells in two ovarian cancer models representing distinct subtypes: FNAR-C1 cells, derived from a spontaneous rat endometrioid carcinoma, and the human SKOV3 cell line (described as both serous and clear cell subtypes). We assessed these populations for stem cell features then analyzed expression by microarray and qPCR.
Results
ALDHhigh cells displayed CSC properties, including: smaller size, quiescence, regenerating the phenotypic diversity of the cell lines in vitro, lack of contact inhibition, nonadherent growth, multi-drug resistance, and in vivo tumorigenicity. Microarray and qPCR analysis of the expression of markers reported by others to enrich for ovarian CSCs revealed that ALDHhigh cells of both models showed downregulation of CD24, but inconsistent expression of CD44, KIT and CD133. However, the following drugable targets were consistently expressed in the ALDHhigh cells from both models: mTOR signaling, her-2/neu, CD47 and FGF18 / FGFR3.
Conclusions
Based on functional characterization, ALDHhigh ovarian cancer cells represent an ovarian CSC population. Differential gene expression identified drugable targets that have the potential for therapeutic efficacy against ovarian CSCs from multiple subtypes.
Keywords: ovarian cancer, cancer stem cells, aldehyde dehydrogenase 1, stem cell-targeted therapy
Introduction
Eighty percent of patients with advanced ovarian cancer show initial clinical responses to therapy, but almost all eventually relapse [1]. This transient clinical response is consistent with the cancer stem cell (CSC) hypothesis, which posits that the initial response would be attributed to the eradication of the bulk, differentiated cells [2, 3]. The persistence of a drug resistant subpopulation of cancer cells exhibiting a stem cell phenotype is further theorized to be responsible for relapse. A substantial body of recent laboratory evidence supports the existence of cell populations in ovarian cancer with stem cell features [4–8]. However, controversy exists regarding the phenotype of such so-called ovarian CSCs, as well as their clinical relevance [9–11].
Various “stem cell” markers have been proposed to enrich for ovarian CSCs, including CD44+, CD133+, CD24+, KIT+ (CD117) and aldehyde dehydrogenase 1 (ALDH1) [4–8, 12, 13], although conflicting data exist for each of these markers [9–11]. Work in our laboratory and others has identified ALDH1 as a marker of CSCs in many hematologic malignancies and solid tumors [14–21]. Furthermore, recent reports indicate that high ALDH1 expression (ALDHhigh) may serve as a superior ovarian CSC marker [12, 13]. Accordingly, we analyzed ALDHhigh ovarian cancer cells for CSC properties. Two ovarian cancer cell lines were tested: FNAR-C1 [22] and SKOV3 [28,29]. FNAR-C1 developed spontaneously in a female Lewis rat and displayed striking morphologic similarities to the human endometrial subtype of ovarian carcinoma, expressing estrogen receptor α, progesterone receptor, androgen receptor, her-2/neu, epithelial cell adhesion molecule, CA125, and nuclear b-catenin [22]. It can be carried as a cell line or passaged in the peritoneal cavity of immunocompetent female Lewis rats, a potential advantage over models requiring immunocompromised mice [22]. SKOV3 is a human ovarian cancer cell line, the subtype of which has been described as both clear cell and serous [28,29]. We find that ALDHhigh cells from both models display phenotypic, biologic, and functional stem cell properties. Expression analysis identified genes and pathways consistently expressed in ALDHhigh cells that may serve as therapeutic targets for the eradication of ovarian CSCs.
Materials and Methods
Cell Lines and Culture
FNAR-C1 rat ovarian cancer cells were derived as previously described [22]. The human ovarian cancer cell line SKOV3 and Taxol-resistant subclone were the kind gifts of Drs. Alexander Stoeck, Tian-Li Wang and Ie-Ming Shih [23]. Cells were maintained in either standard medium [DMEM (Life Technologies, Grand Island, NY) for FNAR-C1 or RPMI (Life Technologies, Grand Island, NY) for SKOV3 + 10% FBS (Sigma, St. Louis, MO)] or KnockOut Medium [KnockOut DMEM (Life Technologies, Grand Island, NY) + 10% KnockOut Serum Replacement (Life Technologies, Grand Island, NY)]. Both media were supplemented with 2 mM L-glutamine (Life Technologies, Grand Island, NY), 100 U/mL penicillin (Life Technologies, Grand Island, NY) and 100 µg/mL streptomycin (Life Technologies, Grand Island, NY). Taxol-resistant SKOV3 cells were maintained in 33.3 nM paclitaxel (Taxol; Sigma-Aldrich, St. Louis, MO). All experiments were conducted on cells passaged less than 30 times within our lab.
Cell Sorting and Flow Cytometry
Adherent cells were released from flasks with 0.05% trypsin-EDTA (Life Technologies, Grand Island, NY) and pipetting. Cells were stained with Aldefluor reagent (STEMCELL Technologies Inc., Vancouver, BC, Canada) to assess ALDH1 activity, using the manufacturer’s protocol then stained with 0.5–1 µg/mL propidium iodide (PI; Sigma-Aldrich, St. Louis, MO) for 5 minutes. FNAR-C1 cells grown in KnockOut medium showed sensitivity to the mechanical forces of cell sorting and therefore were sorted using a MoFlo cell sorter (Beckman Coulter, Miami, FL), which uses a lower pressure than the FACSAria II. Because of substantial death after sorting, these cells were allowed to recover for three to four days in KnockOut medium before use in the following experiments: doubling time, cell cycle analysis and drug resistance. All other cells were sorted using a FACSAria II cell sorter (BD Biosciences, San Jose, CA).
To measure cell size, unsorted cells were trypsinized then stained with Aldefluor and PI as described above. The relative forward scatter, or size, of gated ALDHhigh and ALDHlow cells was then compared. For cell cycle analysis, sorted cells were allowed to recover overnight after sorting in their corresponding medium. Cells were released from flasks as described above. No greater than 2.5x105 cells underwent methanol fixation (Fisher, Waltham, MA) followed by rehydration in PBS (Life Technologies, Grand Island, NY) and staining with 50 µg/ml PI and 10 µg/ml RNase (Roche, Indianapolis, IN). A minimum of 5000 singlet events were collected. To assay phenotypic diversity after culture, sorted cells were plated into vented T25 tissue culture flasks with either KnockOut or standard medium. After four days, cells were trypsinized and stained with Aldefluor and PI as described above. Cells were analyzed on a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA). All flow cytometry plots and calculations were generated using FlowJo 8 (Tree Star, Inc, Ashland, OR).
Photomicrographs
Sorted cells were plated into 6-well tissue culture plates. When cells were at the desired level of confluency, photomicrographs were generated using a Nikon Eclipse TE2000E with a 20x phase contrast objective, Nikon DS-Qi1Mc CCD camera, and Nikon NIS Elements 3 software (Nikon Instruments Inc., Melville, NY).
Doubling Time
Sorted cells were plated into vented T25 tissue culture flasks in the medium they were cultured in prior to sorting. At 24-hour intervals ranging from 24 to 120 hours, cells were released from flasks with trypsin and pipetting as described above. Aggregates formed by ALDHhigh FNAR-C1 cells were disrupted using Accumax and pipetting (EMD Millipore, Billerica, MA). Viable cells were identified by trypan blue exclusion (Life Technologies, Grand Island, NY), and counted using a hemacytometer (Hausser Scientific, Horsham, PA). Time points in exponential growth were used to calculate doubling times (Roth V. 2006 http://www.doubling-time.com/compute.php). R2 was calculated using Prism 6 for Mac (GraphPad Software, La Jolla, CA). Graphs were generated using Excel 2008 for Mac (Microsoft, Redmond, WA).
Microarray and Pathway Analysis
Cells were sorted into RNAprotect Cell Reagent (Qiagen, Valencia, CA). Total RNA was isolated with the RNeasy Mini Kit with QIAshredder columns (Qiagen, Valencia, CA). Quality assessment and microarray analysis was performed at The Sidney Kimmel Cancer Center Microarray Core Facility. RNA quality was determined with a NanoDrop ND-1000 spectrometer (Thermo Scientific, Waltham, MA) and a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA).
Rat samples were analyzed in triplicate using the Rat Gene Expression Microarray 4x44K v3 (Agilent Technologies, Santa Clara, CA). RNA spike-in controls (Agilent Technologies, Santa Clara, CA) were added, then samples were amplified and labeled using the Low RNA Input Fluorescent Linear Amplification Kit (Agilent Technologies, Santa Clara, CA) and purified with the RNeasy Mini Kit. 0.825 µg of each Cy3-labeled sample was used for hybridization according to manufacturers protocol. Microarrays were scanned using an Agilent G2565AA Scanner with Agilent Scan Control 7.0 software (Agilent Technologies, Santa Clara, CA). Data were extracted with Agilent Feature Extraction 9.5.3.1 software (Agilent Technologies, Santa Clara, CA).
Samples from human cell lines were analyzed in triplicate using the Human HT-12 v4 bead chip (Illumina, San Diego, CA). RNA was amplified and labeled using the Illumina Total Prep RNA Amplification Kit (Ambion, Austin, TX). 750 ng biotin-labeled cRNA was hybridized and stained with streptavidin-Cy3. Arrays were scanned with the iScan System (Illumina, San Diego, CA). Data were extracted with the Gene Expression Module in GenomeStudio Software (Illumina, San Diego, CA). (GEO accession numbers will be provided during review.)
GeneSpring GX software version 11 (Agilent Technologies, Inc, Santa Clara, CA) was used for data analysis, and differentially expressed genes were calculated based on moderated ttest and fold-change. Pathway analysis was performed using Ingenuity Pathway Analysis Tool (Ingenuity® Systems, www.ingenuity.com, Redwood City, CA).
PCR Validation of Microarray
PCR validation of microarray data was performed at The Sidney Kimmel Cancer Center Microarray Core Facility using custom 96-well TaqMan array plates (Life Technologies, Grand Island, NY). 20 ng of cDNA was added to each well along with TaqMan Universal PCR Master Mix (Life Technologies, Grand Island, NY) in a final volume of 20 µl. Each sample was assayed in triplicate. Data was collected using the ABI 7500 Real-Time PCR System (Life Technologies, Grand Island, NY) and analyzed with 7500 Fast System SDS software version 1.4 (Life Technologies, Grand Island, NY). The following endogenous controls were used: CXCL1 for FNAR-C1 and EEF1A1 for SKOV3.
In vivo Tumorigenicity
All animals were handled in accordance with the Johns Hopkins University Animal Care and Use guidelines (protocol numbers RA08M154 and RA11M116). Female Lewis rats received intraperitoneal injections with graded numbers of cells. As the FNAR-C1 cell line originated in a normal Lewis rat, the cells can be readily propagated in immunocompetent Lewis rats. Rats were monitored weekly and euthanized when becoming moribund or when tumors were detected by abdominal palpation. Two years after the injection date, remaining rats underwent necropsy. This time point was chosen as the endpoint for the study in order to maximize the time for our injected cells to form tumors yet minimize the chances of spontaneous tumors due to old age. All tumors underwent histological evaluation in a blinded manner to confirm tumor origin.
In Vitro Drug Resistance
Sorted cells were plated in sextuplicate at a density of 100 cells per well into 24-well tissue culture plates containing standard medium. Cells were exposed to Taxol and gemcitabine continuously and carboplatin for 72 hours. Data are presented as the percentage of wells that grew relative to no drug control (FNAR-C1: weeks 2–4, SKOV3: weeks 2–3). The number of biological replicates for each concentration is presented as “n” below. FNAR-C1 cells were exposed to the following concentrations of Taxol: 10 nM, 20 nM, 40 nM, and 60 nM (ALDHhigh: n=4; ALDHlow: n=5). Taxol-resistant SKOV3 cells were exposed to the following concentrations of Taxol: 100 nM (n=6), 200 nM (n=7), 400 nM (n=8), 600 nM (n=7), and 800 nM (n=3); carboplatin (Sigma-Aldrich, St. Louis, MO): 250 ng/ml (n=1), 500 ng/ml (n=5), 750 ng/ml (n=5), 1 µg/ml (n=5), and 1.25 µg/ml (n=4); and gemcitabine (Gemzar; Eli Lilly and Company, Indianapolis, IN): 2 nM (n=3), 3 nM (n=3), 4 nM (n=5), 5 nM (n=5), 6 nM (n=2), and 7 nM (n=2). Excel 2008 was used to generate graphs. The Kruskal-Wallis test was used to assess statistical significance.
Results
Aldefluor Defines Distinctive Populations
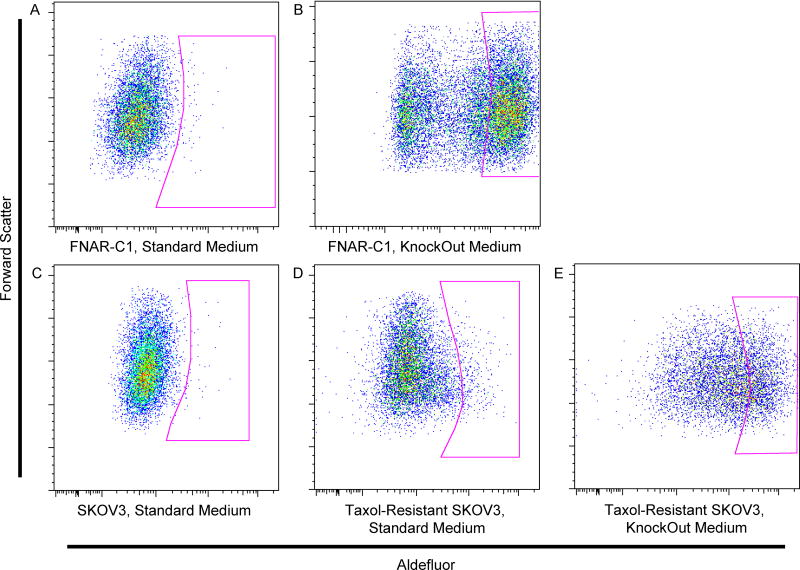
Initial experiments with the FNAR-C1 rat model of ovarian cancer showed a small ALDHhigh population, as measured with the Aldefluor reagent. However, continued culture in standard medium containing serum quickly resulted in the loss of ALDHhigh cells (Fig. 1A; mean = 2.05%, range = 0.23–9.3%). Culturing FNAR-C1 cells in Knockout medium, which is a proprietary, serum-free medium designed for the maintenance of embryonic stem cells in an undifferentiated state, maintained a large ALDHhigh population, representing approximately half of the cells (Fig. 1B; mean = 56.88%, range = 15.7 – 86.5%).
Figure 1. ALDH1 Activity of Ovarian Cancer Cell Lines.
Representative Aldefluor staining pattern of (A) FNAR-C1 cells grown in standard medium containing serum, (B) FNAR-C1 cells grown in KnockOut medium, (C) SKOV3 cells grown in standard medium, (D) Taxol-resistant SKOV3 cells grown in standard medium, and (E) Taxol-resistant SKOV3 cells grown in KnockOut medium. DEAB was used as a negative control for ALDH1 activity (shown in Figure S1).
Like FNAR-C1 cells, SKOV3 cells cultured in standard serum-containing medium showed few ALDHhigh cells (Fig. 1C; mean = 2.12%, range = 0.18 – 6.28%). In contrast to the FNAR-C1 cells, culture of SKOV3 cells in KnockOut medium did not substantially change the percentage of ALDHhigh cells (data not shown). However, when SKOV3 cells developed Taxolresistance, a distinct ALDHhigh population emerged in standard medium (Fig. 1D; mean = 3.25%, range = 1.18 – 12.1%). Furthermore, culturing Taxol-resistant SKOV3 cells in KnockOut medium expanded the ALDHhigh population (Fig. 1E; mean = 20.22, range = 10.0% to 42.2%).
ALDHhigh Cells Exhibit Stem Cell Morphologic and Growth Characteristics
Commonly accepted properties of stem cells include non-adherent growth and the absence of contact inhibition. FNAR-C1 cells grow adherently on plastic. Upon reaching confluency, ALDHlow cells showed substantial death, which resulted in large areas devoid of cells (data not shown). ALDHhigh FNAR-C1 cells failed to undergo growth arrest upon achieving confluency and instead formed large three-dimensional clusters of cells, which we refer to as nodules (Fig. S2A). In addition to forming nodules, a portion of ALDHhigh FNAR-C1 cells grew as nonadherent clusters, or spheroids (Fig. S2B). These spheroids formed regardless of the confluency of the culture and contained viable cells based on trypan blue exclusion. ALDHlow FNAR-C1 cultures produced no nodules, and supernatant from the cell cultures lacked viable cells (data not shown). ALDHhigh Taxol-resistant SKOV3 cells also displayed nonadherent growth through the formation of spheroids (Fig. S2C), which were not observed in ALDHlow SKOV3 cells (data not shown).
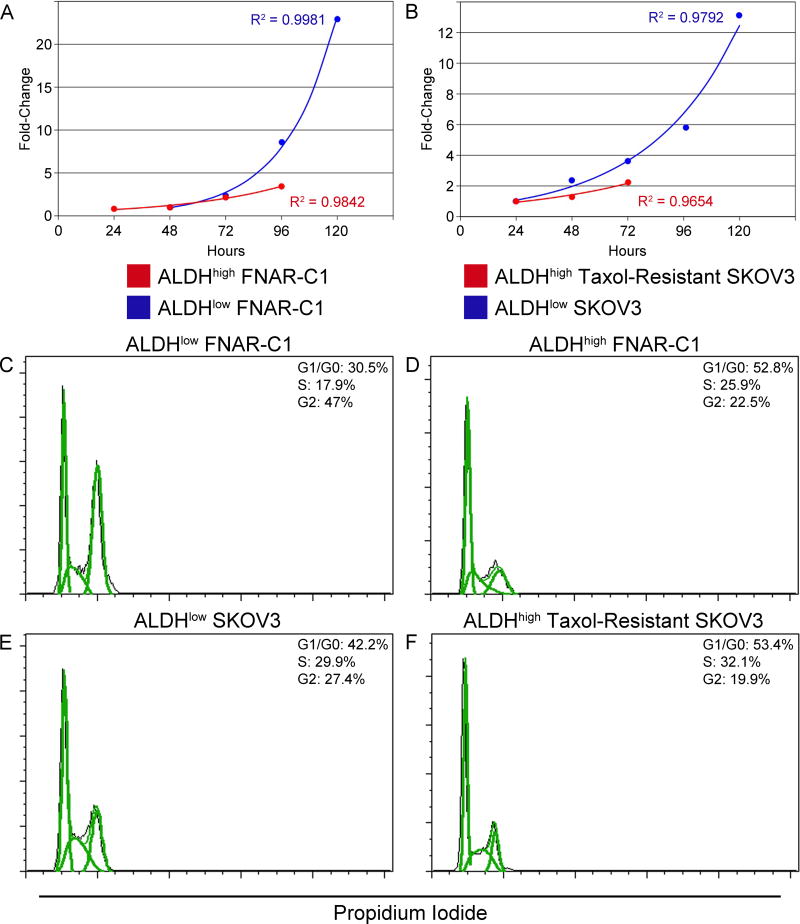
Stem cells are typically smaller in size than their differentiated counterparts; this is generally attributed to their relative quiescence. Accordingly, ALDHhigh cells from both FNARC1 and Taxol-resistant SKOV3 cells were smaller than their ALDHlow counterparts (Fig. S3). ALDHhigh cells had substantially longer doubling times than ALDHlow cells, revealing a slower growth rate (Fig. 2A, B; ALDHlow FNAR-C1: 15.58 hours, ALDHhigh FNAR-C1: 32.21 hours, ALDHlow SKOV3: 27.04 hours, ALDHhigh Taxol-resistant SKOV3: 39.62 hours). Cell cycle analysis showed that the slower growth rate was a result of a greater percentage of ALDHhigh cells in the G1/G0 phase of the cell cycle compared to ALDHlow cells (Fig. 2C–F). Furthermore, the decreased proliferation of ALDHhigh cells was associated with downregulation of genes important for progression from G1 to S phase relative to ALDHlow cells (Figs. S4 and S5). In the FNAR-C1 model, these genes included c-myc, neuregulin 1 and cyclin D1. ALDHhigh Taxolresistant SKOV3 cells exhibited downregulation of cyclin dependent kinase 6, transcription factor DP-1, and ABL1 with concurrent upregulation of cyclin-dependent kinase inhibitor 1A.
Figure 2. Proliferative Status of Cells Based on ALDH activity.
Growth curves of sorted: (A) FNAR-C1 cells and (B) SKOV3 cells. (C-F) Representative cell cycle analyses using propidium iodide: (C) ALDHlow FNAR-C1 cells, (D) ALDHhigh FNAR-C1 cells, (E) ALDHlow SKOV3 cells, and (F) ALDHhigh Taxol-resistant SKOV3 cells.
ALDHhigh Cells Are Capable of Regenerating the Phenotypic Diversity of the Cell Line and Are Tumorigenic In Vivo
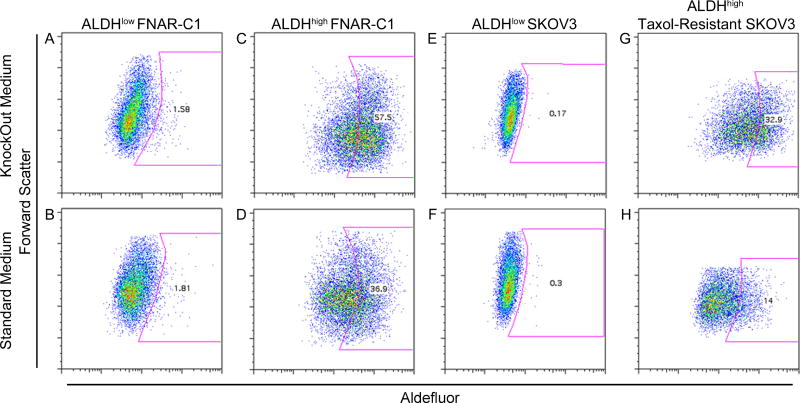
ALDHlow and ALDHhigh cells were isolated from both the FNAR-C1 and SKOV3 lines by cell sorting, then cultured in standard and KnockOut medium. ALDHlow cells isolated from both cell lines generated only ALDHlow cells in vitro, even when cultured in KnockOut medium that optimally supports the growth of ALDHhigh cells (Figs. 3A, B, E and F). Conversely, ALDHhigh cells remained ALDHhigh when cultured in KnockOut medium but rapidly generated ALDHlow cells in standard medium, indicating their ability to regenerate the phenotypic diversity of the cell line (Figs. 3C, D, G and H).M
Figure 3. Regeneration of Phenotypic Diversity.
Sorted ALDHlow FNAR-C1 cells (A, B), ALDHhigh FNAR-C1 cells (C, D), ALDHlow SKOV3 cells (E, F), and ALDHhigh Taxol-resistant SKOV3 cells (G, H) were plated in either KnockOut medium (A, C, E, G) or standard medium (B, D, F, H) for four days and then stained for Aldefluor using DEAB as a negative control (shown in Figure S6).
The gold standard assay for cancer stem cells is increased in vivo tumorigenicity. In order to test the tumorigenicity of ALDHhigh and ALDHlow cells, graded numbers of sorted FNAR-C1 cells were injected intraperitoneally into immunocompetent Lewis rats. The only rats to develop tumors were those receiving ALDHhigh cells. Three out of 16 rats receiving 105 ALDHhigh cells and one out of 12 rats receiving 103 ALDHhigh cells formed tumors (tumors became palpable at days 148, 177 and 222 for 105 cells and day 222 for 103 cells). None of the rats injected with ALDHlow cells developed tumors (105 cells, n=8; 103 cells, n=9). This provides in vivo support for the conclusion that ALDHhigh cells represent the tumorigenic population within FNAR-C1 cells.
ALDHhigh Cells Display Multi-Drug Resistance
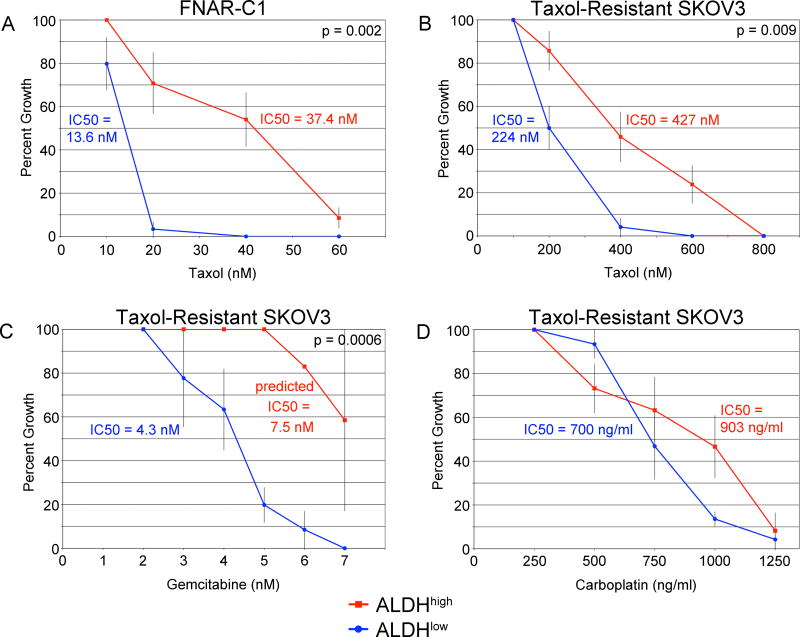
ALDHhigh FNAR-C1 cells showed a statistically significant increased resistance to Taxol (Fig. 4A). When examining the sensitivities of SKOV3 cells, it was inappropriate to compare Taxol-resistant cells to Taxol-sensitive cells, and so ALDHhigh Taxol-resistant SKOV3 cells grown in KnockOut medium were compared to ALDHlow Taxol-resistant SKOV3 cells grown in standard medium. ALDHhigh Taxol-resistant SKOV3 cells showed a statistically significant increased resistance to Taxol and gemcitabine (Fig. 4B, C) and a trend towards resistance to carboplatin (Fig. 4D). Therefore, ALDHhigh cells displayed resistance to chemotherapy drugs with different mechanisms of action and resistance.
Figure 4. Relative Drug Resistance of ALDHhigh Cells.
Sorted cells were assayed for their ability to grow in increasing concentrations of drugs. Error bars show the standard error. Numbers of replicates are detailed in Materials and Methods. (A) FNAR-C1 cells cultured with Taxol. (B–D) Taxol-resistant SKOV3 cells cultured in: (B) Taxol, (C) gemcitabine, and (D) carboplatin.
Expression of ALDH1 family members in ALDHhigh Cells
In order to correlate the Aldefluor staining patterns in isolated cells with ALDH1 expression, the expression levels of ALDH1 family members were examined. In ALDHhigh FNAR-C1 cells, there was 86.522-fold upregulation (325.761-fold by qPCR) of ALDH1A2 relative to ALDHlow FNAR-C1 cells, but no differences in ALDH1A1 or ALDH1A3 were detected. ALDHhigh Taxol-resistant SKOV3 cells expressed ALDH1A1 at a 23.699-fold greater level (44.494-fold by qPCR) than ALDHlow SKOV3 cells. ALDH1A2 and ALDH1A3 were expressed at comparables levels in both SKOV3 cell populations.
Inconsistent Expression of Putative Ovarian Cancer Stem Cell Markers in ALDHhigh Cells
While CD24 has been proposed as an ovarian cancer stem cell marker, disagreements exist concerning whether ovarian CSCs express it to a greater or lesser degree than differentiated cells [6, 10]. ALDHhigh Taxol-resistant SKOV3 cells exhibited substantial downregulation of CD24 relative to ALDHlow SKOV3 cells (Table 1). ALDHhigh FNAR-C1 cells also showed downregulation of CD24 compared to ALDHlow cells, but to a lesser degree than in SKOV3 cells (Table 1). However, all populations expressed considerable levels of CD24, potentially limiting its usefulness as a marker for isolation.
Table 1. Differential Expression of Previously Reported Ovarian Cancer Stem Cell Markers.
Fold-change values from microarray and qPCR are presented
| Gene | FNAR-C1 Microarray |
FNAR-C1 PCR |
SKOV3 Microarray |
SKOV3 PCR |
|---|---|---|---|---|
| CD24 | −1.763 | −1.560 | −9.458 | −41.667 |
| CD44 | −21.481 | −22.727 | −1.188 | −1.003 |
| KIT | ** | 10.644 | −1.968 | −333.333 |
| CD133 (PROM1) |
** | ↓ | ** | ** |
FNAR-C1 data are presented as ALDHhigh FNAR-C1 cells versus ALDHlow FNAR-C1 cells. SKOV3 data are presented as ALDHhigh Taxol-resistant SKOV3 cells versus ALDHlow SKOV3 cells.
signifies that the gene was detectable in ALDHlow cells but not in ALDHhigh cells.
Signifies that the gene was not reliably detectable in either ALDHhigh or ALDHlow cells.
High expression of CD44 has been reported to enrich for ovarian CSCs [24]. However, ALDHhigh FNAR-C1 cells exhibited substantial downregulation of CD44 relative to ALDHlow FNAR-C1 cells (Table 1). Moreover, there was no difference in CD44 expression between ALDHhigh Taxol-resistant SKOV3 cells and ALDHlow SKOV3 cells, with both populations showing high levels of expression (Table 1).
Published reports of putative ovarian CSCs demonstrate stark variability in KIT expression, which is reflected in our findings [4, 7]. In FNAR-C1 cells, KIT expression was too low for differences to be reliably detected by microarray, however qPCR showed that KIT expression was largely confined to the ALDHhigh population (Table 1). ALDHhigh Taxol-resistant SKOV3 cells exhibited the opposite pattern with expression of KIT only reliably detected in ALDHlow SKOV3 cells (Table 1).
Putative ovarian CSCs have also been isolated based on CD133 positivity [5]. CD133 expression was too low to be reliably detected in FNAR-C1 cells using microarray analysis. By qPCR analysis, CD133 was undetectable in the ALDHhigh population but appeared to be expressed at very low levels in the ALDHlow fraction (Table 1). In ALDHhigh Taxol-resistant SKOV3 and ALDHlow SKOV3 cells, CD133 expression was too low for reliable detection by either microarray or qPCR (Table 1).
Potential Therapeutic Targets to Eliminate ALDHhigh Cells
In seeking potential therapeutic targets, we hypothesized that prognostic indicators in ovarian cancer would be differentially expressed in ovarian CSCs. However, steroid hormone receptors, gonadotropin receptors and CA125 were not consistently differentially expressed in the ALDHhigh cells of both models (Table S1). Similarly, ABC transporters and developmental pathways have been implicated in ovarian CSCs. Again, any differential expression in ALDHhigh cells was not consistent between the two cell lines (data not shown). As a result of this, a systematic survey of the microarray data was conducted to identify those genes and pathways expressed in the ALDHhigh cells of both models. This data was then examined for readily drugable targets. These genes and pathways are described as potential therapeutic targets that may be effective in eradicating ovarian CSCs.
Differential gene expression of ALDHhigh cells from both models indicates increased mTOR signaling (Figs. S7 and S8). The primary mechanism of increased mTOR signaling involved inhibition of the TSC complex, which inhibits mTOR activation through RHEB [25]. In FNAR-C1 cells, this inhibition was due to upregulation of TSC complex inhibitors (IRS1, PRKCH and RPS6KB2) and downregulation of a complex activator (PRKAG2). ALDHhigh Taxol-resistant SKOV3 cells primarily suppress the activity of the TSC complex via upregulation of complex inhibitors (PIK3R1 and RPS6KB2). ALDHhigh cells from both models also exhibited upregulation of the mTOR activator RRAGD (Table 2). Increased expression of many pathway components downstream of the TSC complex in ALDHhigh cells of both cell lines further supported activation of mTOR signaling in these cells.
Table 2. Differential Expression of Potential Therapeutic Targets.
Fold-change values from microarray and qPCR are presented
| Gene | FNAR-C1 Microarray |
FNAR-C1 PCR |
SKOV3 Microarray |
SKOV3 PCR |
|---|---|---|---|---|
| RRAGD | 3.467 | 2.309 | ||
| ERBB2 (her-2/neu) |
−1.040 | −1.147 | 1.320 | 1.308 |
| CD47 | 2.088 | 1.961 | 1.827 | 2.079 |
| FGF18 | 2.912 | 3.793 | 1.778 | 3.775 |
FNAR-C1 data are presented as ALDHhigh FNAR-C1 cells versus ALDHlow FNAR-C1 cells. SKOV3 data are presented as ALDHhigh Taxol-resistant SKOV3 cells versus ALDHlow SKOV3 cells. Empty cells signify that the gene was not tested with that assay.
Ovarian cancer specimens commonly express her-2/neu (ERBB2) [26]. FNAR-C1 cells did not display differential expression of her-2/neu (Table 2). However, strong expression was observed in both populations, with her-2/neu in the upper 40% of all genes when ranked by signal strength. Quantitative PCR confirmed this high level of expression with her-2/neu being detected less than 3 cycles after the endogenous control. SKOV3 cells expressed her-2/neu at a sufficiently high level as to suggest overexpression, with a slight, but consistent, upregulation of her-2/neu in ALDHhigh cells (Table 2). When ranked by signal strength, the microarray signal for ALDHhigh Taxol-resistant SKOV3 cells was in the top 3%.
CD47 prevents phagocytosis by macrophages and is highly expressed in normal and malignant stem cells [27–29]. ALDHhigh cells of both models expressed approximately 2-fold higher levels of CD47 than their ALDHlow counterparts as measured by microarray and qPCR (Table 2).
Both ALDHhigh populations also exhibited upregulation of fibroblast growth factor 18 versus their ALDHlow counterparts (FGF18; Table 2). FGF18 primarily signals through fibroblast growth factor receptor 3 (FGFR3), which was expressed in all populations tested. As measured by microarray, FNAR-C1 cells expressed FGFR3 at levels similar to her-2/neu, and expression in SKOV3 cells ranked in the upper 20% of measured genes.
Discussion
One of the best-studied hematopoietic stem cell markers is ALDH, specifically the ALDH1 family of enzymes also known as retinaldehyde dehydrogenases. Additionally, ALDH1 is a marker for stem cells from most other tissues, and its expression decreases as stem cells differentiate [30]. Putative CSCs from most malignancies also express ALDH1. Although the precise function of ALDH1 enzymes in stem cell biology is unclear, they act as general aldehyde scavengers and are a critical component of retinoic acid biosynthesis. We studied two different model systems representing two distinct ovarian cancer subtypes, a spontaneously occurring rat ovarian cancer of the endometrioid subtype and the human ovarian cancer cell line SKOV3 of the serous or clear cell subtype. We hypothesized that potential biologic similarities between putative ovarian CSCs from different subtypes and two species would suggest fundamental features of ovarian CSCs. Accordingly, we found that high ALDH1 activity isolated cells that exhibited stem cell characteristics. The ALDHhigh cells displayed an absence of contact inhibition, nonadherent growth, smaller size, quiescence, and multi-drug resistance. Moreover, the ability to regenerate the phenotypic diversity of the cell line in vitro and produce tumors in vivo was exclusive to the ALDHhigh subpopulation. These data suggest that ovarian CSCs in both models are likely contained within the ALDHhigh population.
CD44, KIT and CD133 have been reported to enrich for ovarian CSCs in both cell lines and patient material, but the data have been inconsistent. We also found them to be inconsistently expressed in the ALDHhigh cells of the two models. Both ALDHhigh populations did display decreased expression of CD24, although the difference in expression compared to the ALDHlow population may not be sufficient to permit isolation of the cells. A lack of consensus exists in the published literature concerning expression of these markers in ovarian CSCs with some studies finding higher expression of CD24, CD44, CD133 and KIT, but others finding reduced expression in the putative ovarian CSC population [4–13]. The inconsistencies in expression identified here and elsewhere in the literature may be due to biological differences between distinct ovarian cancer subtypes or intertumoral heterogeneity, but more work is needed to ascertain the relationship between these different markers.
These data confirm the utility of ALDH1 as an ovarian CSC marker, and raise questions about the broad applicability of other proposed ovarian CSC markers: CD44, KIT, and CD133. Few studies to this point have proposed targets for ovarian CSCs, perhaps in part because of inconsistent definitions for these cells. Several clinically targetable genes and pathways were highly expressed in ALDHhigh cells of both models, and thus may serve as ovarian CSC targets, including mTOR, her-2/neu, CD47, and FGF18 / FGFR3. Therapeutic interventions targeting the mTOR pathway or CD47 would likely preferentially target CSCs over their more differentiated counterparts. In contrast, targeting her-2/neu or FGF18 / FGFR3 would lack specificity, but would potentially affect CSCs in addition to more differentiated cell populations. Drugs targeting the mTOR pathway and her-2/neu are already in clinical use, and several FGFR3 inhibitors and antibodies targeting CD47 are beginning clinical trials. Future work will be necessary to identify the optimal targeted therapy and to validate these findings in primary human tumors.
Supplementary Material
DEAB controls of the representative Aldefluor staining patterns shown in Figure 1 from (A) FNAR-C1 cells grown in standard medium containing serum, (B) FNAR-C1 cells grown in KnockOut medium, (C) SKOV3 cells grown in standard medium, (D) Taxol-resistant SKOV3 cells grown in standard medium, and (E) Taxol-resistant SKOV3 cells grown in KnockOut medium.
Pictomicrographs showing nodules (A) and spheroid (B) in ALDHhigh FNAR-C1 cells and spheroid in ALDHhigh Taxol-resistant SKOV3 cells (C). For photography of ALDHhigh FNAR-C1 spheroids, the supernatant was moved to an empty well. All images used a 20x phase contrast objective.
Unsorted cells were stained for Aldefluor and gated on the population of interest using DEAB as a negative control. The forward scatter, or size, of these cells was then displayed: (A) FNAR-C1 cells and (B) SKOV3 cells.
Pathway analysis of microarray data. Data are presented as ALDHhigh FNAR-C1 cells relative to ALDHlow FNAR-C1 cells. Genes shaded in yellow were upregulated and genes shaded in blue were downregulated.
Pathway analysis of microarray data. Data are presented as ALDHhigh Taxol-resistant SKOV3 cells relative to ALDHlow SKOV3 cells. Genes shaded in yellow were upregulated and genes shaded in blue were downregulated.
DEAB controls for Figure 3. Sorted ALDHlow FNAR-C1 cells (A, B), ALDHhigh FNAR-C1 cells (C, D), ALDHlow SKOV3 cells (E, F), and ALDHhigh Taxol-resistant SKOV3 cells (G, H) were plated in either KnockOut medium (A, C, E, G) or standard medium (B, D, F, H) for four days and then stained for Aldefluor using DEAB as a negative control.
Pathway analysis of microarray data. Data are presented as ALDHhigh FNAR-C1 cells relative to ALDHlow FNAR-C1 cells. Genes shaded in yellow were upregulated and genes shaded in blue were downregulated. Confirmed by PCR: VEGFB: +7.209, PDGFC: +4.879, HMOX1: −5.155, VEGFA: −5.348.
Pathway analysis of microarray data. Data are presented as ALDHhigh Taxol-resistant SKOV3 cells relative to ALDHlow SKOV3 cells. Genes shaded in yellow were upregulated and genes shaded in blue were downregulated. Confirmed by PCR: PPP2R2A: +1.534, HMOX1: −18.868.
Foldchange values from microarray and qPCR are presented. FNAR-C1 data are presented as ALDHhigh FNAR-C1 cells versus ALDHlow FNAR-C1 cells. SKOV3 data are presented as ALDHhigh Taxol-resistant SKOV3 cells versus ALDHlow SKOV3 cells. ↓signifies that the gene was detectable in ALDHlow cells but not in ALDHhigh cells. ** Signifies that the gene was not reliably detectable in either sample. Empty cells signify that the gene was not tested with that assay.
Highlights.
ALDHhigh ovarian cancer cells phenotypically resemble cancer stem cells (CSCs).
Based on gene expression ALDHhigh and ALDHlow cells are biologically distinct.
Other reported ovarian CSC markers do not consistently identify ALDHhigh cells.
Potential therapeutic targets include: mTOR signaling, Her-2/neu, CD47 and FGFR3.
Acknowledgments
This work was funded in part by the Department of Defense grant OC073136 and NIH grants P01 CA070970 and P30 CA006973. The authors wish to thank the UCLA Statistics Core, particularly Kevin McLaughlin and Holly LeClair, for their analysis of drug resistance data (supported by NIH/National Center for Advancing Translational Science (NCATS) UCLA CTSI Grant Number UL1TR000124).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
R.J.J. holds the patent for the stem cell marker Aldefluor and, under a licensing agreement between Aldagen and the Johns Hopkins University, is entitled to a share of royalties received by the University. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies. The remaining authors declare no competing financial interests.
References
- 1.Cannistra SA. Cancer of the Ovary. N Engl J Med. 1993;329(21):1550–9. doi: 10.1056/NEJM199311183292108. [DOI] [PubMed] [Google Scholar]
- 2.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 3.Ghiaur G, Gerber J, Jones RJ. Concise Review: Cancer Stem Cells and Minimal Residual Disease. Stem Cells. 2012;30(1):89–93. doi: 10.1002/stem.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bapat SA, Mali AM, Koppikar CB, Kurrey NK. Stem and Progenitor-Like Cells Contribute to the Aggressive Behavior of Human Epithelial Ovarian Cancer. Cancer Res. 2005 Apr 15;65(8):3025–9. doi: 10.1158/0008-5472.CAN-04-3931. 2005. [DOI] [PubMed] [Google Scholar]
- 5.Curley MD, Therrien VA, Cummings CL, Sergent PA, Koulouris CR, Friel AM, et al. CD133 Expression Defines a Tumor Initiating Cell Population in Primary Human Ovarian Cancer. Stem Cells. 2009 Dec;27(12):2875–83. doi: 10.1002/stem.236. 2009. [DOI] [PubMed] [Google Scholar]
- 6.Gao MQ, Choi YP, Kang S, Youn JH, Cho NH. CD24+ cells from hierarchically organized ovarian cancer are enriched in cancer stem cells. Oncogene. 2010 May 6;29:2672–80. doi: 10.1038/onc.2010.35. 2010. [DOI] [PubMed] [Google Scholar]
- 7.Luo L, Zeng J, Liang B, Zhao Z, Sun L, Cao D, et al. Ovarian cancer cells with the CD117 phenotype are highly tumorigenic and are related to chemotherapy outcome. Exp Mol Pathol. 2011 Oct;91(2):596–602. doi: 10.1016/j.yexmp.2011.06.005. 2011. [DOI] [PubMed] [Google Scholar]
- 8.Landen CN, Goodman B, Katre AA, Steg AD, Nick AM, Stone RL, et al. Targeting Aldehyde Dehydrogenase Cancer Stem Cells in Ovarian Cancer. Mol Cancer Ther. 2010 Dec 1;9(12):3186–99. doi: 10.1158/1535-7163.MCT-10-0563. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vathipadiekal V, Saxena D, Mok SC, Hauschka PV, Ozbun L, Birrer MJ. Identification of a Potential Ovarian Cancer Stem Cell Gene Expression Profile from Advanced Stage Papillary Serous Ovarian Cancer. PLoS ONE. 2012 Jan 17;7(1):e29079. doi: 10.1371/journal.pone.0029079. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi M, Jiao J, Lu W, Ye F, Ma D, Dong Q, et al. Identification of cancer stem cell-like cells from human epithelial ovarian carcinoma cell line. Cell Mol Life Sci. 2010 Nov;67(22):3915–25. doi: 10.1007/s00018-010-0420-9. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang B, Liu G, Xue F, Rosen DG, Xiao L, Wang X, et al. ALDH1 expression correlates with favorable prognosis in ovarian cancers. Mod Pathol. 2009 doi: 10.1038/modpathol.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silva IA, Bai S, McLean K, Yang K, Griffith K, Thomas D, et al. Aldehyde Dehydrogenase in Combination with CD133 Defines Angiogenic Ovarian Cancer Stem Cells That Portend Poor Patient Survival. Cancer Res. 2011 Jun 1;71(11):3991–4001. doi: 10.1158/0008-5472.CAN-10-3175. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abelson S, Shamai Y, Berger L, Shouval R, Skorecki K, Tzukerman M. Intratumoral Heterogeneity in the Self-Renewal and Tumorigenic Differentiation of Ovarian Cancer. Stem Cells. 2012 Mar;30(3):415–24. doi: 10.1002/stem.1029. 2012. [DOI] [PubMed] [Google Scholar]
- 14.Matsui W, Wang Q, Barber JP, Brennan S, Smith BD, Borrello I, et al. Clonogenic Multiple Myeloma Progenitors, Stem Cell Properties, and Drug Resistance. Cancer Res. 2008 Jan 1;68(1):190–7. doi: 10.1158/0008-5472.CAN-07-3096. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones RJ, Gocke CD, Kasamon YL, Miller CB, Perkins B, Barber JP, et al. Circulating clonotypic B cells in classic Hodgkin lymphoma. Blood. 2009 Jun 4;113(23):5920–6. doi: 10.1182/blood-2008-11-189688. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerber JM, Smith BD, Ngwang B, Zhang H, Vala MS, Morsberger L, et al. A clinically relevant population of leukemic CD34+CD38- cells in acute myeloid leukemia. Blood. 2012 Apr 12;119(15):3571–7. doi: 10.1182/blood-2011-06-364182. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma I, Allan AL. The Role of Human Aldehyde Dehydrogenase in Normal and Cancer Stem Cells. Stem Cell Rev and Rep. 2011;7(2):292–306. doi: 10.1007/s12015-010-9208-4. 2011/06/01. [DOI] [PubMed] [Google Scholar]
- 18.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell. 2007 Nov 15;1:555–67. doi: 10.1016/j.stem.2007.08.014. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasper M, Schäfer A, Piontek G, Teufel J, Brockhoff G, Ringel F, et al. Aldehyde dehydrogenase 1 positive glioblastoma cells show brain tumor stem cell capacity. Neurooncol. 2010 Oct 1;12(10):1024–33. doi: 10.1093/neuonc/noq070. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang F, Qiu Q, Khanna A, Todd NW, Deepak J, Xing L, et al. Aldehyde Dehydrogenase 1 Is a Tumor Stem Cell-Associated Marker in Lung Cancer. Mol Cancer Res. 2009 Mar 1;7(3):330–8. doi: 10.1158/1541-7786.MCR-08-0393. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasheed ZA, Yang J, Wang Q, Kowalski J, Freed I, Murter C, et al. Prognostic Significance of Tumorigenic Cells With Mesenchymal Features in Pancreatic Adenocarcinoma. J Natl Cancer Inst. 2010 Mar 3;102(5):340–51. doi: 10.1093/jnci/djp535. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharrow AC, Ronnett BM, Thoburn CJ, Barber JP, II RLG, Armstrong DK, et al. Identification and characterization of a spontaneous ovarian carcinoma in Lewis rats. J Ovarian Res. 2010;3(9):9. doi: 10.1186/1757-2215-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu Y, Gaillard S, Phillip Jude M, Huang T-C, Pinto Sneha M, Tessarollo Nayara G, et al. Inhibition of Spleen Tyrosine Kinase Potentiates Paclitaxel-Induced Cytotoxicity in Ovarian Cancer Cells by Stabilizing Microtubules. Cancer Cell. 2015;28(1):82–96. doi: 10.1016/j.ccell.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang S, Balch C, Chan MW, Lai H-C, Matei D, Schilder JM, et al. Identification and Characterization of Ovarian Cancer-Initiating Cells from Primary Human Tumors. Cancer Res. 2008 Jun 1;68(11):4311–20. doi: 10.1158/0008-5472.CAN-08-0364. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol. 2003 Jun;5(6):578–81. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- 26.Lanitis E, Dangaj D, Hagemann IS, Song D-G, Best A, Sandaltzopoulos R, et al. Primary Human Ovarian Epithelial Cancer Cells Broadly Express HER2 at Immunologically- Detectable Levels. PLoS ONE. 2012;7(11):e49829. doi: 10.1371/journal.pone.0049829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaiswal S, Jamieson CHM, Pang WW, Park CY, Chao MP, Majeti R, et al. CD47 Is Upregulated on Circulating Hematopoietic Stem Cells and Leukemia Cells to Avoid Phagocytosis. 2009;138(2):271–85. doi: 10.1016/j.cell.2009.05.046. 07/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan KS, Espinosa I, Chao M, Wong D, Ailles L, Diehn M, et al. Identification, molecular characterization, clinical prognosis, and therapeutic targeting of human bladder tumorinitiating cells. Proceedings of the National Academy of Sciences. 2009 Aug 18;106(33):14016–21. doi: 10.1073/pnas.0906549106. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willingham SB, Volkmer J-P, Gentles AJ, Sahoo D, Dalerba P, Mitra SS, et al. The CD47- signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proceedings of the National Academy of Sciences. 2012 Apr 24;109(17):6662–7. doi: 10.1073/pnas.1121623109. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balber AE. Concise Review: Aldehyde Dehydrogenase Bright Stem and Progenitor Cell Populations from Normal Tissues: Characteristics, Activities, and Emerging Uses in Regenerative Medicine. Stem Cells. 2011;29(4):570–5. doi: 10.1002/stem.613. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DEAB controls of the representative Aldefluor staining patterns shown in Figure 1 from (A) FNAR-C1 cells grown in standard medium containing serum, (B) FNAR-C1 cells grown in KnockOut medium, (C) SKOV3 cells grown in standard medium, (D) Taxol-resistant SKOV3 cells grown in standard medium, and (E) Taxol-resistant SKOV3 cells grown in KnockOut medium.
Pictomicrographs showing nodules (A) and spheroid (B) in ALDHhigh FNAR-C1 cells and spheroid in ALDHhigh Taxol-resistant SKOV3 cells (C). For photography of ALDHhigh FNAR-C1 spheroids, the supernatant was moved to an empty well. All images used a 20x phase contrast objective.
Unsorted cells were stained for Aldefluor and gated on the population of interest using DEAB as a negative control. The forward scatter, or size, of these cells was then displayed: (A) FNAR-C1 cells and (B) SKOV3 cells.
Pathway analysis of microarray data. Data are presented as ALDHhigh FNAR-C1 cells relative to ALDHlow FNAR-C1 cells. Genes shaded in yellow were upregulated and genes shaded in blue were downregulated.
Pathway analysis of microarray data. Data are presented as ALDHhigh Taxol-resistant SKOV3 cells relative to ALDHlow SKOV3 cells. Genes shaded in yellow were upregulated and genes shaded in blue were downregulated.
DEAB controls for Figure 3. Sorted ALDHlow FNAR-C1 cells (A, B), ALDHhigh FNAR-C1 cells (C, D), ALDHlow SKOV3 cells (E, F), and ALDHhigh Taxol-resistant SKOV3 cells (G, H) were plated in either KnockOut medium (A, C, E, G) or standard medium (B, D, F, H) for four days and then stained for Aldefluor using DEAB as a negative control.
Pathway analysis of microarray data. Data are presented as ALDHhigh FNAR-C1 cells relative to ALDHlow FNAR-C1 cells. Genes shaded in yellow were upregulated and genes shaded in blue were downregulated. Confirmed by PCR: VEGFB: +7.209, PDGFC: +4.879, HMOX1: −5.155, VEGFA: −5.348.
Pathway analysis of microarray data. Data are presented as ALDHhigh Taxol-resistant SKOV3 cells relative to ALDHlow SKOV3 cells. Genes shaded in yellow were upregulated and genes shaded in blue were downregulated. Confirmed by PCR: PPP2R2A: +1.534, HMOX1: −18.868.
Foldchange values from microarray and qPCR are presented. FNAR-C1 data are presented as ALDHhigh FNAR-C1 cells versus ALDHlow FNAR-C1 cells. SKOV3 data are presented as ALDHhigh Taxol-resistant SKOV3 cells versus ALDHlow SKOV3 cells. ↓signifies that the gene was detectable in ALDHlow cells but not in ALDHhigh cells. ** Signifies that the gene was not reliably detectable in either sample. Empty cells signify that the gene was not tested with that assay.