Abstract
The methods currently available for genotype-specific diagnosis of T. cruzi infection still present relevant limitations, especially to identify mixed infection. In the present investigation, we have evaluated the performance of Chagas-Flow ATE-IgG2a test for early and late differential diagnosis of single and dual genotype-specific T. cruzi infections. Serum samples from Swiss mice at early and late stages of T. cruzi infection were assayed in parallel batches for genotype-specific diagnosis of single (TcI, TcVI or TcII) and dual (TcI+TcVI, TcVI+TcII or TcII+TcI) infections. The intrinsic reactivity to TcI, TcVI and TcII target antigens, including amastigote (AI/AVI/AII), trypomastigote-(TI/TVI/TII) and epimastigote (EI/EVI/EII), at specific reverse of serum dilutions (500 to 64,000), was employed to provide reliable decision-trees for “early” vs “late”, “single vs “dual” and “genotype-specific” serology. The results demonstrated that selective set of attributes “EII 500/EI 2,000/AII 500” were able to provide high-quality accuracy (81%) to segregate early and late stages of T. cruzi infection. The sets “TI 2,000/AI 1,000/EII 1,000” and “TI 8,000/AII 32,000” presented expressive scores to discriminate single from dual T. cruzi infections at early (85%) and late stages (84%), respectively. Moreover, the attributes “TI 4,000/TVI 500/TII 1,000”, “TI 16,000/EI 2,000/EII 2,000/AI 500/TVI 500” showed good performance for genotype-specific diagnosis at early stage of single (72%) and dual (80%) T. cruzi infections, respectively. In addition, the attributes “TI 4,000/AII 1,000/EVI 1,000”, “TI 64,000/AVI 500/AI 2,000/AII 1,000/EII 4,000” showed moderate performance for genotype-specific diagnosis at late stage of single (69%) and dual (76%) T. cruzi infections, respectively. The sets of decision-trees were assembled to construct a sequential algorithm with expressive accuracy (81%) for serological diagnosis of T. cruzi infection. These findings engender new perspectives for the application of Chagas-Flow ATE-IgG2a method for genotype-specific diagnosis in humans, with relevant contributions for epidemiological surveys as well as clinical and post-therapeutic monitoring of Chagas disease.
Author summary
Trypanosoma cruzi shows great genetic diversity, and was subdivided into six DTUs (Discrete Typing Units), named TcI-TcVI. This genetic and biological variability, coupled with natural reinfection of hosts, may play an important role in the clinical and epidemiological features of the disease. Furthermore, hosts infected with different T. cruzi genotypes demonstrated distinct therapeutic response. Thus, the development of new methods for genotype-specific diagnosis of T. cruzi infection is very important for clinical and epidemiological studies and for post-therapeutic monitoring of patients treated. Biochemical and molecular methods are used for the purpose, however, these techniques have methodological limitations. In addition, the standardized serological methods for genotype-specific diagnosis of Chagas disease present antigenic limitations and also do not evaluate the reactivity of serum samples from mixed infections. In order to overcome these challenges, our group developed the Chagas-Flow ATE-IgG2a technique with good performance for universal and genotype-specific diagnosis of single T. cruzi infection in the chronic phase. Based on our previous results, in the present investigation, we evaluated the applicability of Chagas-Flow ATE-IgG2a in the genotype-specific diagnosis at early and late stages for single and dual T. cruzi infections.
Introduction
Chagas disease, caused by the protozoan parasite Trypanosoma cruzi, is an endemic antropozoonosis in Latin America that affects about 6–7 million people worldwide [1]. The disease progresses from an acute phase, with high parasitemia and unspecific symptoms, to a lifelong chronic infection, with subpatent parasitemia and asymptomatic clinical status in most patients, or cardiovascular/gastrointestinal involvement in a minor proportion of infected individuals [2–4].
It has been proposed that besides the influence of genetic background of the infected subjects, the genetic variability of T. cruzi contribute to the clinical course of Chagas disease, defined by the particular tropism of T. cruzi genotypes to distinct host tissues [5, 6]. The current classification consensus proposes six genetic groups or Discrete Typing Units (DTUs) of T. cruzi stocks, referred as TcI to TcVI, based on different molecular markers and biological features [7]. In general, it has been considered that vertebrate and invertebrate hosts may present mixed infections with multiple T. cruzi genotypes that can lead to changes in the parasite biological properties and also the clinical course of Chagas disease [5, 8–19]. The resulting characteristics of single infections and mixed infections can also influence the outcome of specific etiological treatment [14, 15].
In this context, the identification of single and mixed T. cruzi infections upon the diagnosis is of extreme importance, as it can have direct impact in prognosis of disease morbidity and response to chemotherapy. Usually, the diagnosis of acute Chagas disease is carried out mostly by direct parasitological methods (fresh blood examination, thick smear and dry smear) while in the chronic phase is usually diagnosed by serological methods [20]. Nowadays, molecular methods have been widely used for genotype-specific diagnosis of single and mixed T. cruzi infection during acute and chronic infections [21]. In general, the molecular approaches require the use of several genetic markers to distinguish the T. cruzi genotype [22–24]. Moreover, in cases of mixed infections, the molecular tests may identify distinct T. cruzi genotypes, depending on: i) the biological sample tested (distinct tissue tropism); ii) the need for parasite isolation (clonal selection) and iii) the requirement of further expansion of parasites by in vitro or in vivo growth to obtain large amounts of genetic material (low sensitivity) [5, 6, 13, 17–19, 22, 25].
Aiming at developing a more reliable method for genotype-specific diagnosis of T. cruzi infections, with simple system of high performance, innovative serological methods have been proposed [26–30]. In general, most serological methods already described for genotypic-specific diagnosis of T. cruzi infection have employed polymorphic B-cell epitopes identified amongst distinct T. cruzi strains. Bhattacharyya et al. (2010, 2014 and 2015) [27, 29, 30], working with TSSA (trypomastigote small surface antigen), representative of the TcMUC III gene family, have demonstrated lineage-specific epitopes within TcII-VI with implications for sero-epidemiological studies [27, 29, 30]. Moreover, Mendes et al. (2013) [28] have described specific T. cruzi peptides, derived from RNA binding proteins, RNA polymerase III and GTPase activating protein, with high-precision ability to discriminate the experimental infections with distinct T. cruzi genotypes using ELISA or affinity-ELISA. Despite the great advance to overcome the limitations of molecular methods, some of these innovative methods have been test only with single infections and were not able to identify all T. cruzi genotypes. Recently, the Chagas-Flow ATE-IgG2a methodology, originally proposed by Alessio et al. (2014) [31] was optimized for genotype-specific diagnosis of single T. cruzi chronic infection, with high-quality performance to discriminate TcI and TcII genotypes [32].
In the present study, the performance of genotype-specific Chagas-Flow ATE-IgG2a [32] was further evaluated for early and late differential diagnosis of single and dual T. cruzi infections.
Methods
Ethics statement
All procedures involving experimental animals were carried out in compliance with the guidelines issued by National Council for Control of Animal Experimentation for ethical conduct in use of animals in research. The study protocol was approved by the Ethics Committee on Animal Experimentation of the Universidade Federal de Ouro Preto (Protocol approval numbers #2013/48 from December, 6th, 2013 for the experimental infection and collected blood).
Standard Trypanosoma cruzi genotypes stocks
Three standard T. cruzi strains, representative of the major genotypes associated with the domestic cycle of Chagas disease in Brazil, were selected for this study, including: Colombiana strain, “COL” (TcI) [33], CL strain (TcVI) [34] and Y strain (TcII) [35]. These T. cruzi stocks were used for the experimental infection and also as target antigens for Chagas-Flow ATE-IgG2a. All isolates were acquired from the T. cruzi cryobank at Grupo de Genômica Funcional e Proteômica de Leishmania spp e Trypanosoma cruzi, Instituto René Rachou (IRR-FIOCRUZ-Minas) and maintained by consecutive in vivo passages in Swiss mice to prepare inoculums for experimental infection and in vitro growth in tissue and axenic cultures to obtain target antigens for Chagas-Flow ATE-IgG2a.
Experimental infections with distinct T. cruzi genotypes
Female Swiss mice (28–30 days old) were obtained from the Animal Science Center at Universidade Federal de Ouro Preto (UFOP), MG, Brazil, and maintained in a temperature-controlled room with access to water and food ad libitum.
For single T. cruzi infections, animals were intraperitoneally inoculated with 50 blood trypomastigotes of each T. cruzi strains and allocated into three subgroups referred as “TcI/COL”, “TcVI/CL” or “TcII/Y”. For dual T. cruzi infections animals were intraperitoneally inoculated with 25 blood trypomastigotes of each T. cruzi strains and subdivided into three subgroups referred as “TcI/COL+TcVI/CL”, “TcVI/CL+TcII/Y” or “TcII/Y+TcI/COL”. All infections were confirmed with parasitemia using fresh blood examination performed either at day 7, 10 or 15 post-infection. A group of non-infected mice (NI, n = 10) were included as a “control”.
Blood samples were collected without anticoagulant from each mouse by ocular plexus puncture at distinct time points after infection, referred as “early stage” (30 days post-infection) and “late stage” (90 and 180 days post-infection). Serum samples were prepared from whole blood, inactivated at 56°C, 30 min and stored at -20°C until use. According to the animal mortality during the experimental timeline, the total number of animals included in each group was: i) “early stage” of single infection: “TcI/COL” (n = 16), “TcVI/CL” (n = 15) and “TcII/Y” (n = 19); ii) “late stage” of single infection: “TcI/COL” (n = 29), “TcVI/CL” (n = 29) and “TcII/Y” (n = 35); iii) “early stage” of dual infection:“TcI/COL+TcVI/CL” (n = 16), “TcVI/CL+TcII/Y” (n = 24), “TcII/Y+TcI/COL” (n = 20) and iv) “late stage” of dual infection:“TcI/COL+TcVI/CL” (n = 28), “TcVI/CL+TcII/Y” (n = 43), “TcII/Y+TcI/COL” (n = 36).
Preparation of target antigens for the Chagas-Flow ATE-IgG2a
The Amastigote/Trypomastigote/Epimastigote (ATE) target antigens were obtained as described previously by Alessio et al. (2014) [31]. Briefly, enriched live Trypomastigotes (“T”) and Amastigote (“A”) preparations of TcI, TcVI and TcII T. cruzi genotypes were harvested from supernatant of desynchronized in vitro tissue cultures after 4–6 days and 8–15 days post-inoculation, respectively. Epimastigote (“E”) forms of TcI, TcVI and TcII T. cruzi genotypes were obtained at log-phase growth in axenic culture and pre-fixed with FACS fix solution (10.0 g of paraformaldehyde, 10.2 g of sodium cacodylate and 6.65 g of sodium chloride/l, pH 7.2; Sigma Aldrich, St Louis, MO, USA).
Live “A/T” and pre-fixed “E” were labeled with fluorescein isothiocyanate/FITC (Sigma Aldrich, St Louis, MO, USA) as described previously by Alessio et al. (2014) [31]. In summary, “A/T” and “E” suspensions (1×107/ml) were stained with FITC (100μg/mL for TcI/COL and 200μg/mL for TcVI/CL and TcII/Y) for 30 min at 37°C. After staining, “A/T” mix were re-incubated at 37°C for 60 min for differential quenching of FITC and “E” re-incubated at 4°C overnight as described previously by Alessio et al., (2014) [31]. Following, FITC-labeled ATE target antigens were mixed and monitored by flow cytometry to obtain a proportional suspension (33% “A”, 33% “T” and 33% “E”). Representative flow cytometric dot plot distribution of differential FITC labeling of target antigens are provided in the S1 Fig.
TcI/TcVI/TcII genotype-specific Chagas-Flow ATE-IgG2a
The Chagas-Flow ATE-IgG2a methodology was carried out as described previously by Alessio et al. (2017) [32] and the samples were tested in a blind study design for each genotype-specific platforms. Aliquots of 50μL of pre-diluted (1:500 to 1:64,000 final concentration) 0.22μm-filtered serum samples were incubated in the presence of 50μL of “A/T/E” target antigens at 37°C for 30 min using parallel batches of TcI, TcVI and TcII genotype-specific platforms in U-bottom 96-well plates. After incubation, “A/T/E” target antigens were washed twice with phosphate-buffered-saline supplemented with 10% fetal bovine serum (PBS-10%FBS). The “A/T/E” target antigens were re-incubated with 50μL of pre-diluted (1:3,000) biotin-conjugated anti-mouse IgG2a (BD Bioscience, San Jose, CA, USA) plus 20μL of pre-diluted (1:800) streptavidin phycoerytrin–SAPE (BD Bioscience, San Jose, CA, USA) at 37°C for 30 min. “A/T/E” target antigens were washed twice with PBS-10%FBS, fixed with 200μL of FACS fixing solution and store at 4°C for 30 min and up to 24 hours prior flow cytometric acquisition. Second step reagents control (anti-mouse IgG2a-biotin+SAPE) was included in all experiments to monitor unspecific bindings.
Flow cytometric measurements were performed on a FACSCan flow cytometer (BD Bioscience, San Diego, CA, USA) using the CellQuest software package for data acquisition and storage. Appropriate instrument settings were used on log scale (Forward Scatter-FSC = E00, Side Scatter-SSC = 427, threshold = 400; FL1 = 620 and FL2 = 500). A total of 10,000 events were acquired per each tested sample. The FlowJo software version 10.1 (TreeStar, San Diego, CA, USA) was used for data analysis. Serum reactivity to “A/T/E” target antigens of each T. cruzi genotype was determined as follows: target antigens were selected based on their differential FITC-label characteristic and the analysis of the percentage of positive fluorescent parasites (PPFP) calculated on FL2 (α-IgG2a-biotin/SAPE) one-dimensional histograms upon the setting of positivity limit (PPFP<2%) on the internal control. An overview of the Chagas-Flow ATE-IgG2a experimental procedure is provided in S1 Fig.
Statistical analysis and data mining for selecting sets of attributes
The R-project software, Version 3.0.1, was used to identify for each target antigen (“A”, “T” and “E”) the higher modular distance between the mean reactivity (PPFP) along the titration curves (1:500 a 1:64000) and identify the pairs of attributes (“target antigen and serum dilution”) for differential diagnosis “early” vs “late”, “single” vs dual and “genotype-specific” serology.
The GraphPad Prism software, Version 5.0 (San Diego, CA, USA) was used to perform the non-parametric Kruskal–Wallis test followed by the Dunns' post-test for multiple comparisons to select the “top” pairs of attributes (“target antigen and serum dilution”) with the most outstanding significant differences (p ≤ 0.05) of reactivity to target antigens amongst subgroups.
The MedCalc software package 7.3 (Ostend, Belgium) was used to obtain the Receiver Operating-Characteristic features and the Microsoft Excel 2010 employed to calculate the global median or global mean values as putative cut-off edges to define set of attributes (“target antigen and serum dilution/ cut-off”) to segregate the reactivity to target antigens amongst subgroups.
The WEKA software, version 3.6.11 (University of Waikato, New Zealand) was used to construct the decision trees based on the pre-selected set of attributes (target-antigen/serum dilution/cut-off) to create algorithms (root and branch) for differential diagnosis “early” vs “late”, “single” vs dual and “genotype-specific” serology. The global accuracy and the “leave-one-out-cross-validation” (LOOCV) values were employed as performance indices. The LOOCV, also referred as “rotation estimation” is a mathematical validation model for generalizing the results of a given statistical analysis to an independent data set. This strategy is usually applied in settings where the major aim is to estimate how accurate the predictive model will behave when applied in other studies.
The Microsoft Excel software was used for (i) radar charts assemblage and the (ii) reactivity board drawings. (i) The radar charts, also referred as “spider diagram” or “polar coordinate” is a graphical method to display multivariate data using a two-dimensional format with quantitative variables represented on axes starting from the same point, giving the plot a “flower-like” appearance, where the area of each spokes reflects the quantitative magnitude of each variable. (ii) The reactivity board summarizes the results of individual samples and gives an overall profile of the serological reactivity categorized by stage (early or late), type (single or dual) and genotype (TcI, TcVI and TcII). The GraphPad Prism package was used to create symbol&line graphs as well as box plots, and the Microsoft PowerPoint software was employed to draw the decision tree and table layouts.
Results
Panoramic overview of Chagas-Flow ATE-IgG2a reactivity during early and late experimental T. cruzi infections
The overall profile of Chagas-Flow ATE-IgG2a reactivity observed at early and late T. cruzi infections is presented in the Fig 1.
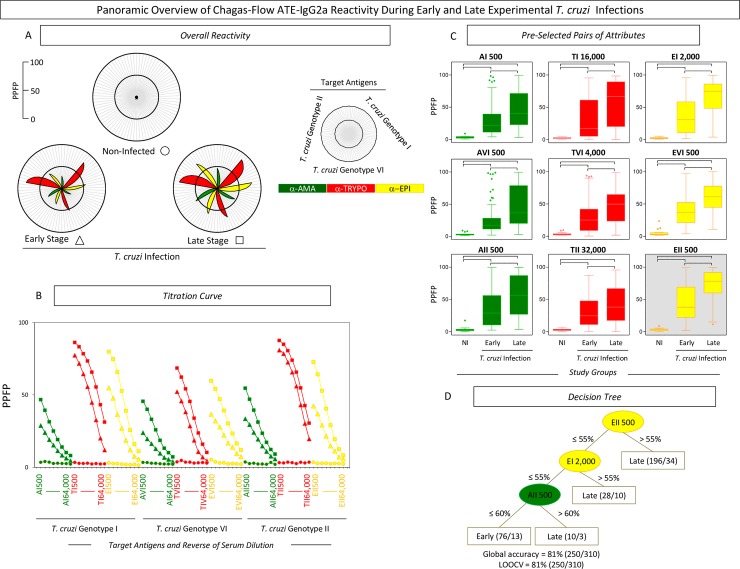
Fig 1. Chagas-Flow ATE-IgG2a performance to discriminate serological reactivity during early and late T. cruzi infections.
(A) Overall reactivity profile of sera samples from early (n = 110) and late (n = 200) T. cruzi infections and non-infected hosts (n = 10). The IgG2a reactivity was evaluated along the titration curve with each target-antigens amastigote (AMA = green), trypomastigote (TRYPO = red) and epimastigote (EPI = yellow) from TcI, TcVI and TcII genotypes of T. cruzi. The results are displayed in radar charts and expressed as mean of percentage of positive fluorescent parasites (PPFP). (B) Titration curves were assessed at eight serum dilutions (1:500 to 1:64,000) using target-antigens AI, AVI and AII; TI, TVI and TII along with EI, EVI and EI at early (triangle) and late (square) stages of T. cruzi infection as compared to non-infected hosts (circle). (C) The reactivity amongst groups was compared for pre-selected pairs of attributes, including: AI 500; AVI 500; AII 500; TI 16,000; TVI 4,000; TII 32,000; EI 2,000; EVI 500 and EII 500. The results are expressed as median PPFP values in box plot format with the outliers underscored by shaded dots. Comparative analyses were performed by the Kruskal-Wallis/Dunn’s post test for multi-group comparisons. Significant differences were considered at p<0.05 and highlighted by connecting lines. The light gray background underscores the pair of attributes (“target antigen and serum dilution”) with the higher performance to discriminate early and late T. cruzi infections. (D) Decision trees were constructed using sets of attributes (“target-antigen and serum dilution/cut-off”) to create algorithms (root and branches) to classify individual samples at early and late T. cruzi infections. Global accuracy and leave-one-out-cross-validation-LOOCV are provided in the Figure.
An overall profile of IgG2a reactivity to distinct target-antigen (“A”, “T” and “E”) of TcI, TcVI and TcII T. cruzi genotypes was first assessed along the titration curves using radar chart compilations of PPFP mean values. This panoramic snapshot provided evidences of differential reactivity during early and late T. cruzi infections as compared to non-infected hosts (Fig 1A and 1B). The results demonstrated persistent high reactivity to “T” at early and late stages of infection. Conversely, higher reactivity to “A” and “E” was evident during late infection (Fig 1A and 1B).
Comparative analysis, based on the modular distance between the mean reactivity (PPFP) along the titration curves and on box plot charts of selected target antigens, allowed the selection of pairs of attributes (“target antigen and serum dilution”) with the most promising performance to distinguish early and late T. cruzi infections (Fig 1C). The pairs of attributes “AI 500”; “TI 16,000”; “EI 2,000”; “AVI 500”; “TVI 4,000”; “EVI 500”; “AII 500”; “TII 32,000” and “EII 500” were pre-selected based on their ability to distinguish reactivity between early and late infections. The pair “EII 500” presented the “top” modular distance to discriminate the median reactivity at early and late infections (Fig 1C—gray background).
The ROC curve analyses were used to define specific cut-off edges for each pre-selected pairs of attributes and subsidize the construction of the most accurate decision tree to segregate the IgG2a reactivity to discriminate, at individual level, early and late infections.
Decision tree algorithm proposed the use of a sets of attributes (“target-antigen and serum dilution/cut-off”), including “EII 500/55%” (root), “EI 2,000/55%” (first branch) and “AII 500/60%” (second branch) to classify individual samples from early vs late infections, with elevated global accuracy (81.0%, LOOCV = 81.0%) (Fig 1D).
Sets of attributes for differential diagnosis of single and dual at early and late T. cruzi infections
Similar approaches were employed to select the sets of attributes (“target-antigen and serum dilution/cut-off”) for differential diagnosis of single and dual at early and late T. cruzi infections (Figs 2 and 3).
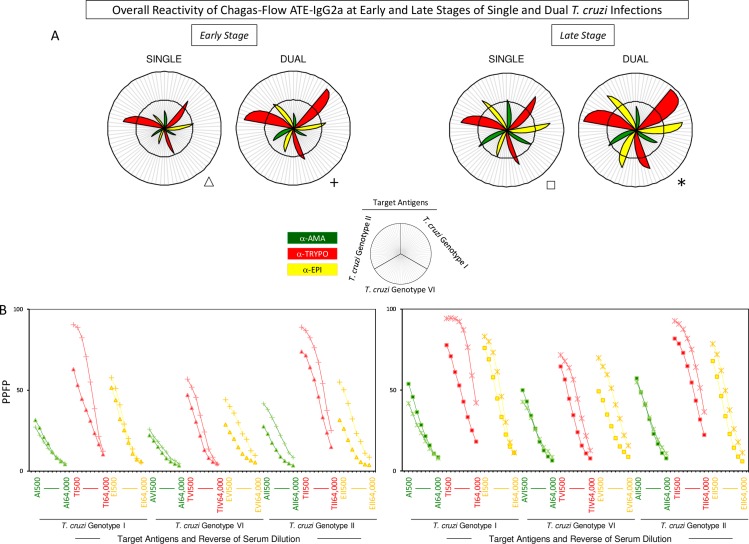
Fig 2. Overall Chagas-Flow ATE-IgG2a reactivity at early and late stages of single and dual T. cruzi infections.
(A) Panoramic reactivity profile of sera samples at early stage of single (Triangle, n = 50) and dual (Cross, n = 60) as well as at late stage of single (Square, n = 93) and dual (Asterisk, n = 107) T. cruzi infections. The IgG2a reactivity was evaluated along the titration curve with each target-antigens amastigote (AMA = green), trypomastigote (TRYPO = red) and epimastigote (EPI = yellow) from TcI, TcVI and TcII genotypes of T. cruzi. The results are shown in radar charts and expressed as mean of percentage of positive fluorescent parasites (PPFP). (B) Titration curves at eight serum dilutions (1:500 to 1:64,000) using target-antigens AI, AVI and AII; TI, TVI and TII and EI, EVI and EII were assessed at early and late stages of single and dual T. cruzi infections.
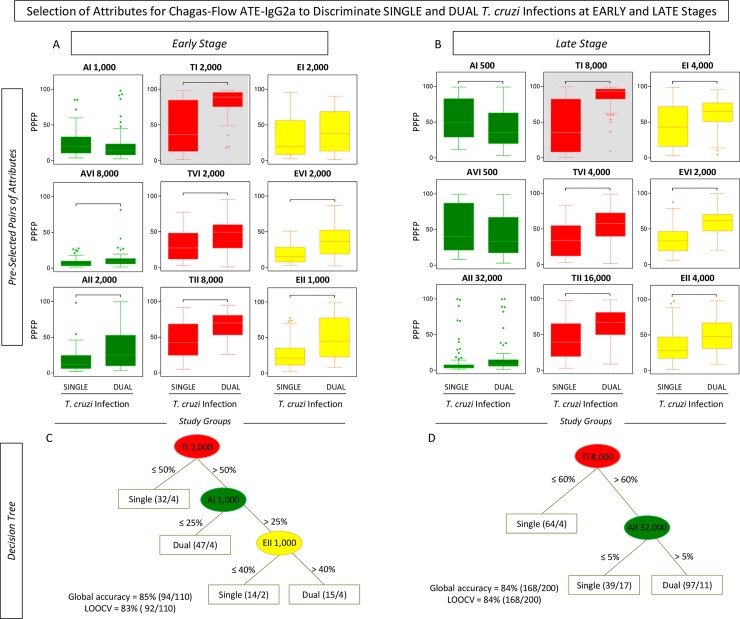
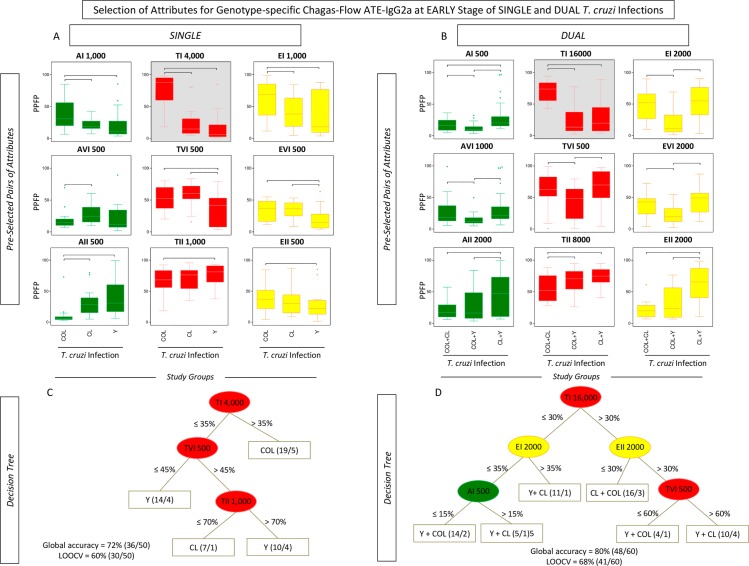
Fig 3. Selection of attributes for Chagas-Flow ATE-IgG2a to discriminate single and dual T. cruzi infections at early and late stages.
(A) Comparative analysis of IgG2a reactivity at early stage of single (n = 50) and dual (n = 60) T. cruzi infections, using pre-selected pairs of attributes, including: AI 1,000; AVI 8,000; AII 2,000; TI 2,000; TVI 2,000; TII 8,000; EI 2,000; EVI 2,000 and EII 1,000. (B) Comparative analysis of IgG2a reactivity at late stage of single (n = 93) and dual (n = 107) T. cruzi infections, using pre-selected pairs of attributes, including: AI 500; AVI 500; AII 32,000; TI 8,000; TVI 4,000; TII 16,000; EI 4,000; EVI 2,000 and EII 4,000. The results are expressed as median of PPFP values in box plot format with the outliers showed by shaded dots. Comparative analyses were performed by the Kruskal-Wallis/Dunn’s post test. Significant differences were considered at p<0.05 and highlighted by connecting lines. The light gray background highlights the pairs of attributes (“target antigen and serum dilution”) with the higher performance to discriminate single and dual T. cruzi infections at early and late stages. Decision trees were constructed using the sets of attributes (“target-antigen and serum dilution/cut-off”) to create algorithms (root and branches) to classify individual samples from single and dual T. cruzi infections at (C) early and (D) late stages. Global accuracy and leave-one-out-cross-validation-LOOCV are provided in the Figure.
The overall reactivity profile of dual infection demonstrated that regardless the stage, the mean reactivity to “T” was the highest. In general, reactivity to “A” and “E” were higher in late stage in both single and dual infections. Reactivity of late dual infection samples to “E” was even higher (Fig 2A and 2B).
Pairs of attributes (“target-antigen and serum dilution”) were selected to distinguish single and dual T. cruzi infections during early and late stages. In this sense, “TI 2,000”; “AVI 8,000”; “TVI 2,000”; “EVI 2,000”; “AII 2,000”; “TII 8,000” and “EII 1,000” were pre-selected to discriminate single from dual infections at early stage (Fig 3A). On the other hand, “AI 500”; “TI 8,000”; “EI 4,000”; “TVI 4,000”; “EVI 2,000”; “TII 16,000” and “EII 4,000” presented the highest ability to distinguish single and dual T. cruzi infections at late stage (Fig 3B).
Comparative analyses, based on the modular distance between the mean reactivity (PPFP) along the titration curves and on box plot charts of selected target antigens, indicated “TI 2,000” and “TI 8,000” as the top attributes to distinguish single and dual T. cruzi infections at early and late stages, respectively (Fig 3A and 3B—gray background).
The ROC curve analyses were used to calculated cut-off edges and define sets of attributes (“target-antigen and serum dilution/cut-off”) to construct decision tree algorithms (Fig 3C and 3D). The decision tree comprised of the following sets “TI 2,000/50%” (root), “AI 1,000/25%” (first branch) and “EII 1,000/40%” (second branch) segregated individual samples from single vs dual T. cruzi infections at early stage, with elevated global accuracy (85.0%, LOOCV = 84.0%) (Fig 3C). Moreover, the algorithm comprised of “TII 8,000/60%” (root) and “AII 32,000/5%” (branch) identified individual samples from single vs dual T. cruzi infections at late stage, also with elevated global accuracy (84.0%, LOOCV = 84.0%) (Fig 3D).
Performance of Chagas-Flow ATE-IgG2a for genotype-specific diagnosis of T. cruzi infections
Aiming at providing a feasible approach to employ the overall Chagas-Flow ATE-IgG2a methodology to genotype-specific diagnosis, the overall reactivity observed at early and late stages of single and dual infections with distinct T. cruzi genotypes was first evaluated by radar charts followed by the construction of decision trees (Figs 4, 5, 6 and 7).
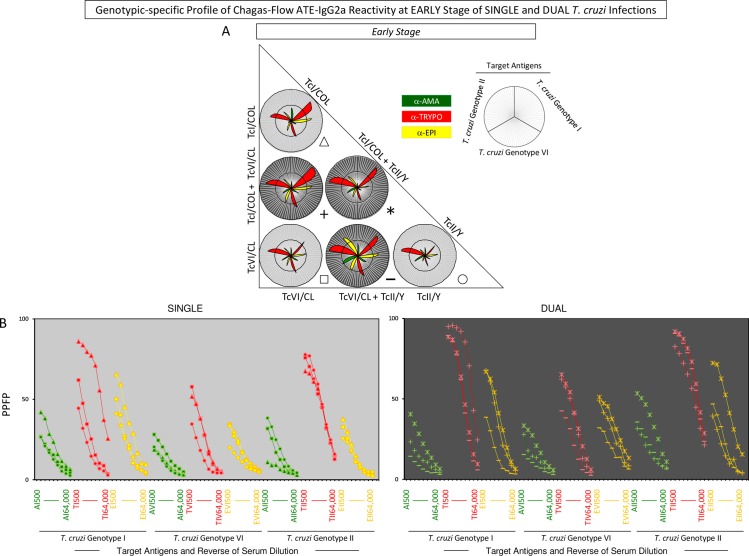
Fig 4. Genotype-specific profile of Chagas-Flow ATE-IgG2a reactivity at early stage of single and dual T. cruzi infections.
(A) Overall reactivity profile of sera samples at early stage of single (TcI/COL, n = 16; TcVI/CL, n = 15 and TcII/Y, n = 19) and dual (TcI/COL+TcVI/CL, n = 16; TcVI/CL+TcII/Y, n = 24 and TcI/COL+TcII/Y, n = 20) T. cruzi infections with distinct genotypes. The IgG2a reactivity was evaluated along the titration curve with each target-antigens amastigote (AMA = green), trypomastigote (TRYPO = red) and epimastigote (EPI = yellow) from TcI, TcVI and TcII genotypes of T. cruzi. The results are shown on radar charts and expressed as mean of percentage of positive fluorescent parasites (PPFP). (B) Titration curves were assessed at eight serum dilutions (1:500 to 1:64,000) using target-antigens AI, AVI and AII; TI, TVI and TII along with EI, EVI and EI at early stage of single [TcI/COL (triangle), TcVI/CL (square), TcII/Y (circle)] and dual [TcI/COL+TcVI/CL (cross), TcVI/CL+TcII/Y (dash), TcI/COL+TcII/Y (asterisk)] T. cruzi infections.
Fig 5. Selection of attributes for genotype-specific Chagas-Flow ATE-IgG2a at early stage of single and dual T. cruzi infections.
(A) Comparative analysis of IgG2a reactivity at early stage of single infection with distinct T. cruzi genotypes (TcI/COL, n = 16; TcVI/CL, n = 15 and TcII/Y, n = 19) using pre-selected pairs of attributes, including: AI 1,000; AVI 500; AII 500; TI 4,000; TVI 500; TII 1,000; EI 1,000; EVI 500 and EII 500. (B) Comparative analysis of IgG2a reactivity at early stage of dual infection with distinct T. cruzi genotypes (TcI/COL+TcVI/CL, n = 16; TcVI/CL+TcII/Y, n = 24 and TcI/COL+TcII/Y, n = 20) using pre-selected pairs of attributes, including: AI 500; AVI 1,000; AII 2,000; TI 16,000; TVI 500; TII 8,000; EI 2,000; EVI 2,000 and EII 2,000. The results are expressed as median PPFP values in box plot format with the outliers showed by shaded dots. Comparative analyses were performed by the Kruskal-Wallis/Dunn’s post test. Significant differences were considered at p<0.05 and highlighted by connecting lines. The light gray background highlights the pairs of attributes (“target antigen and serum dilution”) with the higher performance for the genotype-specific diagnosis at early stage of single and dual T. cruzi infections. Decision trees were constructed using the sets of attributes (“target-antigen and serum dilution/cut-off”) to create algorithms (root and branches) to classify at early stage, individual samples from (C) single and (D) dual T. cruzi infections. Global accuracy and leave-one-out-cross-validation-LOOCV are provided in the Figure.
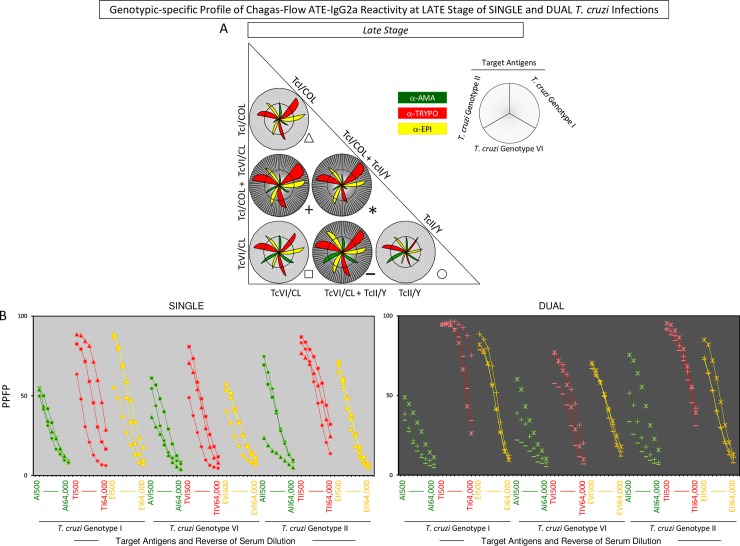
Fig 6. Genotype-specific profile of Chagas-Flow ATE-IgG2a reactivity at late stage of single and dual T. cruzi infections.
(A) Overall reactivity profile of sera samples at late stage of single (TcI/COL, n = 29; TcVI/CL, n = 29 and TcII/Y, n = 35) and dual (TcI/COL+TcVI/CL, n = 28; TcVI/CL+TcII/Y, n = 43 and TcI/COL+TcII/Y, n = 36) T. cruzi infections with distinct genotypes. The IgG2a reactivity was evaluated along the titration curve with each target-antigens amastigote (AMA = green), trypomastigote (TRYPO = red) and epimastigote (EPI = yellow) from TcI, TcVI and TcII genotypes of T. cruzi. The results are shown on radar charts and expressed as mean of percentage of positive fluorescent parasites (PPFP). (B) Titration curves were assessed at eight serum dilutions (1:500 to 1:64,000) using target-antigens AI, AVI and AII; TI, TVI and TII along with EI, EVI and EI at late stage of single [TcI/COL (triangle), TcVI/CL (square), TcII/Y (circle)] and dual [TcI/COL+TcVI/CL (cross), TcVI/CL+TcII/Y (dash), TcI/COL+TcII/Y (asterisk)] T. cruzi infections.
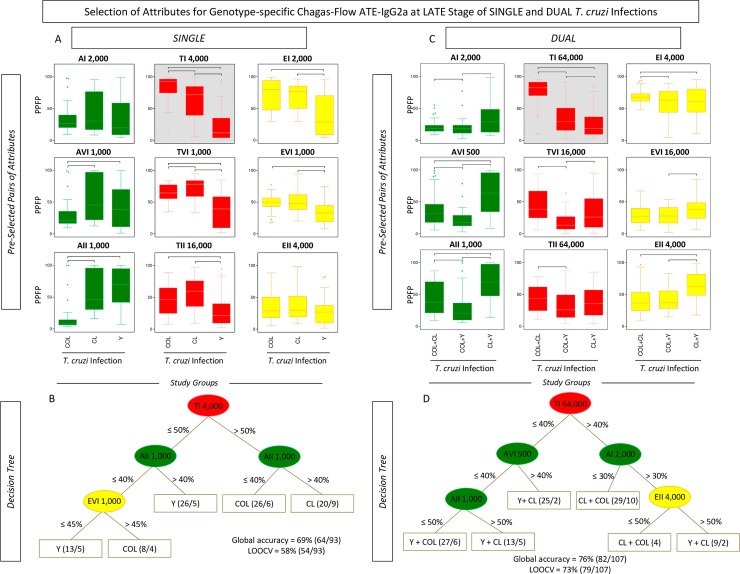
Fig 7. Selection of attributes for genotype-specific Chagas-Flow ATE-IgG2a at late stage of single and dual T. cruzi infections.
(A) Comparative analysis of IgG2a reactivity at late stage of single infection with distinct T. cruzi genotypes (TcI/COL, n = 29; TcVI/CL, n = 29 and TcII/Y, n = 35) using pre-selected pairs of attributes, including: AI 2,000; AVI 1,000; AII 1,000; TI 4,000; TVI 1,000; TII 16,000; EI 2,000; EVI 1,000 and EII 4,000. (B) Comparative analysis of IgG2a reactivity at late stage of dual infection with distinct T. cruzi genotypes (TcI/COL+TcVI/CL, n = 28; TcVI/CL+TcII/Y, n = 43 and TcI/COL+TcII/Y, n = 36) using pre-selected pairs of attributes, including: AI 2,000; AVI 500; AII 1,000; TI 64,000; TVI 16,000; TII 64,000; EI 4,000; EVI 16,000 and EII 4,000. The results are expressed as median PPFP values in box plot format with the outliers showed by shaded dots. Comparative analyses were performed by the Kruskal-Wallis/Dunn’s post test. Significant differences were considered at p<0.05 and highlighted by connecting lines. The light gray background highlights the pairs of attributes (“target antigen and serum dilution”) with the higher performance for the genotype-specific diagnosis at late stage of single and dual T. cruzi infections. Decision trees were constructed using the sets of attributes (“target-antigen and serum dilution/cut-off”) to create algorithms (root and branches) to classify at late stage, individual samples from (C) single and (D) dual T. cruzi infections. Global accuracy and leave-one-out-cross-validation-LOOCV are provided in the Figure.
A panoramic snapshot provided by the reactivity of sera samples during early and late stages of single and dual T. cruzi infections (Figs 4 and 6). The results demonstrated that the reactivity to “TII” was high regardless the stage (early or late) and type of infection (single or dual), re-enforcing the use of this attribute for universal diagnosis purposes. Additionally, higher reactivity to “TI” was usually associated with single infection with TcI/COL or dual infection. Conversely, the reactivity to “TI” was lower in TcII/Y infection. Moreover, specifically at late stage, the reactivity to “A” was lower in single or dual infections involving TcI/COL (Figs 4 and 6).
Line plots of mean reactivity profiles and box plot charts compiling the median PPFP values subsidize the pre-selection of pairs of attributes (“target-antigen and serum dilution”) with better performance for genotype-specific diagnosis at early stage of single and dual T. cruzi infections. The pair of attributes “AI 1,000”; “TI 4,000”; “EI 1,000”; “AVI 500”; “TVI 500”; “EVI 500”; “AII 500”; “TII 1,000” and “EII 500” were pre-selected based on their applicability of genotype-specific diagnosis at early stage of single T. cruzi infection (Fig 5A). The attributes “AI 500”; “TI 16,000”; “EI 2,000”; “AVI 1,000”; “TVI 500”; “EVI 2,000”; “AII 2,000”; “TII 8,000” and “EII 2,000” were pre-selected for genotype-specific diagnosis at early stage of dual T. cruzi infection (Fig 5B). The pairs of attributes with the most promising performance for genotypic-specific diagnosis at early stage of single and dual T. cruzi infections were “TI 4,000” and “TI 16,000”, respectively (Fig 5A and 5B).
The global mean or median values were used as specific cut-off edges to define sets of attributes (“target-antigen and serum dilution/cut-off”) to construct decision tree algorithms (Fig 5C and 5D). The sets “TI 4,000/35%” (root), “TVI 500/45%” (first branch) and “TII 1,000/70%” (second branch) were selected to compose the decision tree that classifies individual samples at early stage of single infection with a moderate global accuracy (72.0%, LOOCV = 60.0%) (Fig 5C). The algorithm composed by “TI 16,000/30%” (root), “EI 2,000/35%” and “EII 2,000/35%” (first branches) along with “AI 500/15%” and “TVI 500/60%” (second branches) classified individual samples at early stage of dual T. cruzi infection, also with a moderate global accuracy (80.0%, LOOCV = 68.0%) (Fig 5D).
Titration curves of mean reactivity profile and box plot charts displaying median PPFP values were used to pre-select the pairs of attributes (“target-antigen and serum dilution”) applicable to the genotype-specific diagnosis at late stage of single and dual T. cruzi infections (Fig 7A). The attributes “TI 4,000”; “EI 2,000”; “AVI 1,000”; “TVI 1,000”; “EVI 1,000”; “AII 1,000” and “TII 16,000” presented better performance for genotype-specific diagnosis at late stage of single T. cruzi infection (Fig 7A). Moreover, “AI 2,000”; “TI 64,000”; “EI 4,000”; “AVI 500”; “TVI 16,000”; “EVI 16,000”; “AII 1,000”; “TII 64,000” and “EII 4,000” were pre-selected for genotype-specific diagnosis at late stage of dual T. cruzi infection (Fig 7B). The “TI 4,000” and “TI 64,000” were elected as the “top” pairs of attributes for genotype-specific diagnosis at late stage of single and dual T. cruzi infections, respectively (Fig 7A and 7B).
The global mean or median values were used as specific cut-off edges to define sets of attributes (“target-antigen and serum dilution/cut-off”) to build decision trees for genotype-specific diagnosis at late stage of single and dual T. cruzi infections (Fig 7C and 7D). The “TI 4,000/50%” (root), “AII 1,000/40%” (first branches) and “EVI 1,000/45%” (second branch) to classify individual samples at late stage of single T. cruzi infection, with moderate global accuracy (69.0%, LOOCV = 58.0%) (Fig 7C). Besides, the algorithm proposed for genotype-specific diagnosis at late stage of dual infection include the attributes “TI 64,000/40%” (root), “AVI 500/40%” and “AI 2,000/30%” (first branches) along with “AII 1,000/50%” and “EII 4,000/50%” (second branches) to categorize the individual samples also with a moderate global accuracy (76.0%, LOOCV = 73.0%) (Fig 7D).
Overall performance of Chagas-Flow ATE-IgG2a for differential diagnosis of T. cruzi infection
The overall performance of Chagas-Flow ATE-IgG2a was estimated after gathering the reactivity profile of individual samples using the selected sets of attributes (Fig 8A).
Fig 8. Overall performance of Chagas-Flow ATE-IgG2a for differential diagnosis of experimental T. cruzi infection.
(A) Reactivity boards were employed to display the overall profile of Chagas-Flow ATE-IgG2a applied to the differential diagnosis of T. cruzi infection at early (left diagram) and late (right diagram), using the selected sets of attributes (“target-antigen and serum dilution/cut-off”) determined by the decision trees. (B) Sequential algorithms were applied to yield the “classified infection” status to each individual sample, referred as: “a” for early vs late; “b” for single vs dual infections at early stage; “c” for single vs dual infections at late stage; “d” for genotype-specific diagnosis at early stage of single infection; “e” for genotype-specific diagnosis at early stage of dual infection; “f” for genotype-specific diagnosis at late stage of single infection and “g” for genotype-specific diagnosis at late stage of dual infection. A color key was provided to identify early (yellow), late (brown), single (light gray), dual (dark gray), TcI/COL (red), TcVI/CL (green), TcII/Y (dark blue), TcI/COL+TcVI/CL (orange), TcVI/CL+TcII/Y (light blue) and TcI/COL+TcII/Y (pink). (C) Scatter graphs showed correspondence between each pair of “true” vs “classified” infection indicated by connecting lines and the percentage of agreement for “Stage & Type & Genotype”; “Type & Genotype” and “Type” of infection. (D) Discriminant analyses of combined Chagas-Flow ATE-IgG2a for genotype-specific diagnosis including T. cruzi single infection (COL, CL and Y) and dual infection (COL+CL, CL+Y and COL+Y). The global accuracy is provided in the Figure.
Following, the sequential algorithms (referred as “a”, “b”, “c”, “d”, “e”, “f” and “g”) were applied to yield the “classified infection” status to each individual sample (Fig 8B). Subsequently the results of each pair of “true infection” vs “classified infection” were compared to estimate the percentage of agreement (Fig 8C).
Data analysis showed that the “Stage & Type & Genotypes” were classified correctly in 163 samples (53%). Moreover, the “Type & Genotypes”, were classified correctly in 22 samples (7%), whereas the “Type” were classified correctly in 66 samples (21%) (Fig 8C). Together the results demonstrated that 251 out of 310 samples (81%) were classified correctly (Fig 8C). Our data demonstrated that 23 out of 310 samples were extreme outliers (7%), misclassified regarding to the stage, type and genotype, most from early infection (n = 22). The remaining 36 samples were misclassified according to the type and genotype, most of them form late infection (n = 30).
Detailed discriminant analysis of Chagas-Flow ATE-IgG2a applied to the genotype-specific diagnosis of T. cruzi infection, regardless the stage of infection is provided in the Fig 8D. In general, 71% of “true infection” with TcI/COL and 83% of “true infection” with TcII/Y were classified correctly, whereas only 27% of “true infection” with TcVI/CL received a correct classification status of single infection (Fig 8D). On the other hand, 73% of “true infection” with TcI/COL+TcVI/CL and 63% of “true infection” with TcVI/CL+TcII/Y were classified correctly, whereas only 39% of “true infection” with TcI/COL+TcII/Y received a correct classification status of dual infection (Fig 8D). Together, the overall performance of Chagas-Flow ATE-IgG2a for differential diagnosis of T. cruzi infection reached an elevated global accuracy (81%) (Fig 8D).
Discriminant analysis at early and late stages of infection is provided in the S2 Fig. In general, the performance of Chagas-Flow ATE-IgG2a applied to the genotype-specific diagnosis of T. cruzi infection was higher at late stage (85%) as compared to early (75%) stage of infection (S2 Fig). Generally, at early stage, the TcII/Y (90%) infection was better classified, whereas at late stage, the TcI/COL (80%), TcII/Y (80%) and TcVI/CL+TcII/Y (83%) infections received the higher percentage of agreement between “true vs classified” status (S2 Fig).
Discussion
Recently, an innovative technique was developed with applicability for universal and genotype-specific diagnosis of single experimental T. cruzi infection, in the chronic phase, named Chagas-Flow ATE-IgG2a [32], and originally proposed by Alessio et al. (2014) [31]. Chagas-Flow ATE-IgG2a is perform with distinct T. cruzi genotypes as target antigens, employing parasites strains from the three major genotypes (TcI, TcVI and TcII) associated with human Chagas disease in Latin America [21, 36, 37]. Our previous results demonstrated an excellent performance of Chagas-Flow ATE-IgG2a assay for universal diagnosis of experimental T. cruzi infection, with 100% sensitivity and specificity. Further, the method showed a good performance for genotype-specific diagnosis of single experimental T. cruzi infection during chronic phase, with accuracy of 94% to segregate TcI and TcII infections [32]. Thus, the present work aims to extend the early studies carried out by Alessio et al. (2017) [32], in order to improve the Chagas-Flow ATE-IgG2a for genotype-specific diagnosis of single and dual experimental T. cruzi infections, in early and late stages. The opportunity to investigate the antibody response at distinct stages of experimental T. cruzi infections intended to typify, in practical terms, the status of Chagas disease observed in children or young adults (early) as well as in middle age adults (late).
Endemic areas of Chagas disease allow individuals to undergo multiple re-infections that can result in different T. cruzi populations throughout the course of the infection [17, 38–42]. At mixed infections, distinct genotypes of T. cruzi may be found in blood and tissues, according to the phase or time of infection [13, 17, 19]. Indeed, Sales-Campos et al. (2014) [19] have demonstrated that during TcI/TcII mixed infections, TcII strains predominates in blood samples during both acute and chronic phases. On the other hand, in tissues samples, TcII strains were more prevalent during the acute phase, while TcI strains were more commonly identified during chronic phase of infection. Conversely, other studies have shown that TcI predominate over TcII strains in blood and tissue samples during acute and chronic infections [43]. In fact, the survival of one specific parasite population in blood can be associated with its capacity to escape from the host immune response [44, 45]. Thus, the standardization of the Chagas-Flow ATE-IgG2a methodology for the genotype-specific diagnosis at early and late stages for single and dual T. cruzi infection is extremely important to elucidate the genetic groups of T. cruzi which may result in distinct disease outcome.
Chagas-Flow ATE-IgG2a showed a good performance to discriminate early and late T. cruzi infections, with an elevated global accuracy (81%) (Fig 1). The correct knowledge regarding the stage of T. cruzi infection is essential since previous studies already demonstrated that distinct T. cruzi genotypes in host present particular behavior according to the time of infection [46–48]. Moreover, Toledo et al. (2003) [47] verified differences among stocks of T. cruzi in the response to treatment according to the genotype of the parasite and phase of infection. Stocks of genotype 39 (hybrids) were resistant to itraconazole treatment in acute phase and partially resistant in the chronic phase. On the other hand, stocks of genotype 32 (TcII) were partially susceptible to itraconazole treatment in acute phase and susceptible in the chronic phase. Therefore, Chagas-Flow ATE-IgG2a arises as a new tool that can distinguish early and late T. cruzi infections.
Our data demonstrated high reactivity of serum samples with the “T” antigen was high in early and late stages of T. cruzi infection. Conversely, low reactivity with the “A” antigen was observed during early stage which enhances in late infection (Fig 1). These results are consistent with the biological cycle of T. cruzi [49, 50]. Moreover, serum samples from T. cruzi infected mice showed high reactivity both in early and late stages with “TII” target antigen, demonstrating again to be a good antigen for universal diagnosis of T. cruzi infection, as observed previously by our group [32]. Additionally, high reactivity of the serum samples from mice in late stage compared to early stage was observed (Fig 1), suggesting an association of a better specificity with the establishment of the host immune response and therefore with the greater reactivity of antibodies during late infection [51–54].
Chagas-Flow ATE-IgG2a demonstrated an essential ability to discriminate single and dual T. cruzi infections with an elevated global accuracy of 85% in early stage and 84% in late stage (Figs 2 and 3). The capacity to differentiate between single and dual infections can provide better approaches for the appropriate treatment [14, 15]. A remarkable finding of the present work was the higher reactivity of sera samples from dual infection as compared to single infection, in both early and late stages (Figs 2 and 3). These findings reflect the fact that during dual infections the host immune system is exposed to a broader antigenic repertoire that leads to higher antibody response [51–54].
This study is pioneer in the development of a serological method by flow cytometry able to distinguish single and dual T. cruzi infections during both early and late stages. Serological methods previous standardized for genotype-specific diagnosis evaluated particularly the reactivity of serum samples from single T. cruzi infection and not considering the phase of infection [27–30]. In general, the available serological approaches are not suitable to detect mixed infections in human samples from different countries [29]. One potential application of Chagas-Flow ATE is the putative genotypic characterization of Chagas disease in distinct geographic areas. Serodiscordant results are frequently observed when conventional serological methods are employed in endemic areas where TcI genotype is predominant [55]. Moreover, the existence of universal specific antigen in the Chagas-Flow ATE (TII antigen) could be very important to guarantee the sensitivity of Chagas disease diagnosis in areas with high variability of T. cruzi genotypes due to high number of immigrants from distinct endemic areas.
Chagas-Flow ATE-IgG2a showed a good performance for genotype-specific diagnosis of single and dual T. cruzi infections, both in early and late stages. During early stage, this methodology was able to differentiate single infection with a global accuracy of 72% and dual infection presenting a global accuracy of 80% (Fig 5). Regarding the late stage, this methodology was able to discriminate dual infection with a moderate global accuracy (76%) (Fig 7).
An interesting result highlights that in several algorithms of the decision trees, the root attribute was the “TI” target antigen (Figs 3, 5 and 7). The high reactivity of serum samples with “TI” can be associated with the predominant morphology of the parasite genotype (broad strains), and therefore with the better resistance to complement-mediated lyses, being a more immunogenic strain [56]. Also, single infection presented a similar profile both in early and late stages with mice serum samples infected with TcI/COL showing high reactivity with “TI” antigen and low reactivity with “AII’ antigen and mice serum samples infected with TcII/Y presenting high reactivity with “AII” antigen and low reactivity with “TI” and “EVI” antigens (Figs 5 and 7).
Some techniques of molecular biology are capable to detect mixed T. cruzi infections, such as: the ones used in the protocol of D´Ávilla et al. (2009) [22], which combines the analyses of the 24sα-LSU rDNA gene´s polymorphism [22, 57, 58], the polymorphism of mini-exon SL-IL intergenic spacer [59] as well as the profile of bands obtained after PCR-RFLP of the subunit II cytochrome oxidase gene [60]. Previous studies identified the mixed T. cruzi infections by kDNA PCR amplification and subsequent hybridization using DTU-specific probes (DTU-blot) [11, 40, 61, 62], using PCR-Real Time with Taq-Man probes (MTq-PCR) [63] and by the amplification of other T. cruzi genome targets, such as the cSC5D gene by PCR-RFL [64]. However, even using high complexity methodologies some of these approaches were not able to detect all the mixed infections present in hosts, as observed by the Chagas-Flow ATE-IgG2a.
The Chagas-Flow ATE-IgG2a identifies single and dual T. cruzi infections avoiding the need to isolate the parasite from the blood by low sensitivity techniques, such as hemoculture and xenodiagnosis, and in vitro cultures. Besides, molecular methods may lead to errors in the identification of genotypes that infect hosts, by selecting the populations. Indeed, previous studies demonstrated that parasite populations present in the blood hosts were distinct from those isolated by xenodiagnosis and cultured in culture media [11, 40, 62]. In addition, ORTIZ et al. (2015) [62] showed by kDNA PCR amplification and subsequent hybridization with DTU-specific probes, mixtures of TcI, TcII, TcV and TcVI DTUs in chronic chagasic host blood samples, although xenodiagnosis followed by the axenic cultures of the parasites identify mostly TcV.
The Chagas-Flow ATE-IgG2a seems to be an innovative serological method by flow cytometry with great potential for the genotype-specific diagnosis of single and dual T. cruzi infections. The analysis of the sequential algorithms of the decision trees demonstrated that this technique presented an accuracy of 81% as a complementary diagnostic test for the T. cruzi infection. Several advantages of Chagas-Flow ATE have been presented. The possible limitation of this method would be the fact that flow cytometers are not available in most laboratories in endemic areas. This putative limitation of using Chagas-Flow ATE in clinical studies can be overcome by the use of currently available portable flow cytometers or by establishing this method on reference laboratories. Moreover, as the Chagas-Flow ATE requires the use of live parasites (A and T antigens), the original proposal is to provide this innovative method at reference laboratory facilities. A perspective to overcome the use of live parasites, the second generation of the Chagas-Flow ATE, using purified or recombinant membrane antigens from distinct T. cruzi genotypes in a multiplex assay are under development. Additionally, it is important to mention that data generated from mouse model needs to be validate for human infections.
In conclusion, the Chagas-Flow ATE-IgG2a is a feasible tool to truly classify the serum samples infected with distinct T. cruzi genotypes, suggesting its applicability in the genotype-specific diagnosis of single and dual experimental T. cruzi infections. This methodology includes advantages such as high sensitivity and specificity, ability to differentiate serum samples during early and late stages, in single and dual infections and identify the specific genotype of T. cruzi. Moreover, the Chagas-Flow ATE can also be useful to establish clinical prognosis status of ongoing infection, since distinct T. cruzi genotypes has been associated with distinct severity of Chagas disease [21, 36, 42, 65]. An additional advantage of using the genotypic-specific Chagas-Flow ATE is its potential applicability for post-therapeutic monitoring, considering the distinct susceptibility of T. cruzi genotypes to etiological treatment [15, 47, 66]. In this context, the method would provide further information about post-therapeutic changes in the predominant T. cruzi DTU as compared to pre-treatment screening. The standardization of the Chagas-Flow ATE methodology for the genotype-specific diagnosis of human infection will be extremely important for further clinical purposes, epidemiological studies and post-therapeutic monitoring applications.
Supporting information
(A) Schematic flow chart overview of the experimental procedure Chagas-Flow ATE-IgG2a. Representative gating strategies to select the target antigens (amastigote-AMA = green, trypomastigote-TRYPO = red and epimastigote-EPI = yellow) and the histograms employed to quantify the percentage of positive fluorescent parasites (PPFP). (B) The compendium of the study population comprised of Swiss mice, categorized into subgroups referred as: early (yellow), late (brown), single (light gray), dual (dark gray), TcI/COL (red), TcVI/CL (green), TcII/Y (dark blue), TcI/COL+TcVI/CL (orange), TcVI/CL+TcII/Y (light blue) and TcI/COL+TcII/Y (pink). (C) Data mining approaches was achieved through the steps: Step 1 (algorithm for early & late stages); Step 2 (algorithms for single & dual infections); Step 3 (algorithm for genotypic-specific single & dual—early & late stages); Step 4 (assemblage sequential algorithm platform).
(TIF)
Discriminant analyses of combined Chagas-Flow ATE-IgG2a for genotype-specific diagnosis at (A) early and (B) late stages of T. cruzi single infection (COL, CL and Y) and dual infection (COL+CL, CL+Y and COL+Y). The global accuracy is provided in the Figure.
(TIF)
(TIF)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by European Community`s Seventh Framework Program No. 602773- Project KINDRED). Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenadoria de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Instituto René Rachou and Universidade Federal de Ouro Preto. GDA received PhD research fellowship form CAPES. OAM-F and AT-C received financial support from CNPq PQ Fellowship program. FFdA thanks PNPD/CAPES fellowship program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO- World Health Organization- Chagas disease (American trypanosomiasis) Fact sheet N° 340; 2017.
- 2.Prata A. Classification of Chagas' infection in humans. Revista da Sociedade Brasileira de Medicina Tropical. 1990; 23: 109–113. [DOI] [PubMed] [Google Scholar]
- 3.Dias JCP. Natural history of Chagas’ disease. Arq Bras Cardiol. 1995; 65: 359–66. [PubMed] [Google Scholar]
- 4.Rassi AJR, Rassi A, Marin-Neto J. Chagas disease. Lancet. 2010; 375: 1388–1402. doi: 10.1016/S0140-6736(10)60061-X [DOI] [PubMed] [Google Scholar]
- 5.Macedo AM, Pena SDJ. Genetic variability of Trypanosoma cruzi: implications for the pathogenesis of Chagas disease. Parasitol. Today. 1998; 14: 119–123. [DOI] [PubMed] [Google Scholar]
- 6.Macedo AM, Machado CR, Oliveira RP, Pena SD. Trypanosoma cruzi: genetic structure of populations and relevance of genetic variability to the pathogenesis of chagas disease. Mem.Inst.Oswaldo Cruz. 2004; 99: 1–12. [DOI] [PubMed] [Google Scholar]
- 7.Zingales B, Andrade SG, Briones MR, Campbell DA, Chiari E, Fernandes O, Guhl F, Lages-Silva E, Macedo AM, Machado CR, Miles MA, Romanha AJ, Sturm NR, Tibayrenc M, Schijman AG. A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI to TcVI. Mem Inst Oswaldo Cruz. 2009; 104(7): 1051–1054. [DOI] [PubMed] [Google Scholar]
- 8.Deane MP, Souza MA, Pereira NM, Gonçalves AM, Momen H, Morel CM. Trypanosoma cruzi: inoculation schedules and reisolation methods select individual strains from doubly infected mice, as demonstrated by schyzodeme and zymodeme analyses. Memórias do Instituto Oswaldo Cruz. 1984; 79: 513–515. [DOI] [PubMed] [Google Scholar]
- 9.Pinto AS, Lana M, Bastrenta B, Barnabé C, Quesney V, Noel S, Tibayrenc M. Compared vectorial transmissibility of pure and biclonal infections of Trypanosoma cruzi in Triatoma infestans. Parasitology Research. 1998; 84: 348–353. [DOI] [PubMed] [Google Scholar]
- 10.Pinto AS, Lana M, Britto C, Bastrenta B, Tibayrenc M. Experimental Trypanosoma cruzi biclonal infection in Triatoma infestans: detection of distinct clonal genotypes using kinetoplast DNA probes. International Journal of Parasitology. 2000; 30: 843–848 [DOI] [PubMed] [Google Scholar]
- 11.Bosseno MF, Yacsik N, Vargas F, Breniére SF. Selection of T. cruzi clonal genotypes (clonet 20 and 39) isolated from Bolivian triatomines following subculture in liquid medium. Memórias do Instituto Oswaldo Cruz. 2000; 95: 601–607. [DOI] [PubMed] [Google Scholar]
- 12.Lana M, Pinto AS, Bastrenta B, Barnabé C, Noel S, Tibayrenc M. Trypanosoma cruzi: infectivity of clonal genotypes infections in acute and chronic phases in mice. Experimental Parasitology. 2000; 96: 61–66. doi: 10.1006/expr.2000.4552 [DOI] [PubMed] [Google Scholar]
- 13.Vago AR, Andrade LO, Leite AA, D'avilla RD, Macedo AM, Adad SJ, Tostes SJ, Moreira MC, Filho GB, Pena SD. Genetic characterization of Trypanosoma cruzi directly from tissues of patients with chronic Chagas disease: differential distribution of genetic types into diverse organs. Am J Pathol. 2000; 156(5): 1805–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martins HR, Toledo MJO, Veloso VM, et al. Trypanosoma cruzi: Impact of dual-clone infections on parasite biological properties in BALB/c mice. Exp Parasitol. 2006; 112: 237–46. doi: 10.1016/j.exppara.2005.11.006 [DOI] [PubMed] [Google Scholar]
- 15.Martins HR, Silva RM, Valadares HM, et al. Impact of dual infections on chemotherapeutic efficacy in BALB/c mice infected with major genotypes of Trypanosoma cruzi. Antimicrob Agents Chemother. 2007; 51: 3282–9. doi: 10.1128/AAC.01590-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Araújo CAC, Cabello PH, Jansen AM. Growth behaviour of two Trypanosoma cruzi strains in single and mixed infections: In vitro and in the intestinal tract of the blood-sucking bug, Triatoma brasiliensis. Acta Tropica. 2007; 101: 225–231. doi: 10.1016/j.actatropica.2007.02.004 [DOI] [PubMed] [Google Scholar]
- 17.Mantilla JC, Zafra GA, Macedo AM, González CI. Mixed infection of Trypanosoma cruzi I and II in a Colombian cardiomyopathic patient. Hum Pathol. 2010; 41(4): 610–3. doi: 10.1016/j.humpath.2009.11.005 [DOI] [PubMed] [Google Scholar]
- 18.Lo Presti MS, Esteves BH, Moya D, Banzán PC, Strauss M, Báez AL, Pizzi R, Quispe Ricalde MA, Valladares B, Rivarola HW, Paglini-Oliva PA. Circulating Trypanosoma cruzi populations differ from those found in the tissues of the same host during acute experimental infection. Acta Trop. 2014; 133: 98–109. doi: 10.1016/j.actatropica.2014.02.010 [DOI] [PubMed] [Google Scholar]
- 19.Sales-Campos H, Kappel HB , Andrade CP , Lima TP , Mattos ME JR , De Castilho A , Correia D , Giraldo LE , Lages-Silva E. A DTU-dependent blood parasitism and a DTU-independent tissue parasitism during mixed infection of Trypanosoma cruzi in immunosuppressed mice. Parasitol Res. 2014; 113(1): 375–85. doi: 10.1007/s00436-013-3665-z [DOI] [PubMed] [Google Scholar]
- 20.Sánchez NO, Sánchez VFJ, Lacunza CD, García BMF, Mora MC, Uncos AD, Basombrío MA. Serological evaluation of specific-antibody levels in patients treated for chronic Chagas' disease. Clin. Vaccine Immunol. 2008; 15: 297 doi: 10.1128/CVI.00106-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zingales B, Miles MA, Campbell DA, Tibayrenc M, Macedo AM, Teixeira MG, Schijman AG, Llewellyn MS, Lages-Silva E, Machado CR, Andrade SG, Sturm NR. The revised Trypanosoma cruzi subspecific nomenclature: Rationale, epidemiological relevance and research applications. Infect. Gene Evolut. 2012; 12: 240–253. [DOI] [PubMed] [Google Scholar]
- 22.D’ávilla DA, Macedo AD, Valadares HM, Gontijo ED, De Castro AM, Machado CR, Chiari E, Galvão LM. Probing Population Dynamics of Trypanosoma cruzi during Progression of the Chronic Phase in Chagasic Patients. J Clin Microbiol. 2009; 1718–1725. doi: 10.1128/JCM.01658-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis MD, Jonathan MA, Matthew YE, Hernán J, Carrasco MS, Llewellyn AN, Michael AM. Genotyping of Trypanosoma cruzi: Systematic Selection of Assays Allowing Rapid and Accurate Discrimination of All Known Lineages. Am J Trop Med Hyg. 2009; 81(6): 1041–1049. doi: 10.4269/ajtmh.2009.09-0305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliveira MT, Machado-de-Assis GF, Oliveira-Silva JC, Machado EM Da Silva GN, Veloso VM, Macedo AM, Martins HR, Lana M. Trypanosoma cruzi Discret Typing Units (TcII and TcVI) in samples of patients from two municipalities of the Jequitinhonha Valley, MG, Brazil, using two molecular typing strategies. Parasit Vectors. 2015; 8: 568 doi: 10.1186/s13071-015-1161-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgos JM, Diez M, Vigliano C, Bisio M, Risso M, Duffy T, Cura C, Brusses B, Favaloro L, Leguizamon MS, Lucero RH, Laguens R, Levin MJ, Favaloro R, Schijman AG. Molecular identification of Trypanosoma cruzi discrete typing units in end-stage chronic Chagas heart disease and reactivation after heart transplantation. Clin Infect Dis. 2010; 51(5): 485–95. doi: 10.1086/655680 [DOI] [PubMed] [Google Scholar]
- 26.Di Noia JM, Buscaglia CA, De Marchi CR, Almeida IC, Frasch AC. A Trypanosoma cruzi small surface molecule provides the first immunological evidence that Chagas´disease is due to a single parasite lineage. J Exp Med. 2002; 195(4): 401–413. doi: 10.1084/jem.20011433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhattacharyya T, Brooks J, Yeo M, Carrasco HJ, Lewis MD, Llewellyn MS, Miles MA. Analysis of molecular diversity of the Trypanosoma cruzi trypomastigote small surface antigen reveals novel epitopes, evidence of positive selection and potential implications for lineage-specific serology. Int J Parasitol. 2010; 40(8): 921–8. doi: 10.1016/j.ijpara.2010.01.002 [DOI] [PubMed] [Google Scholar]
- 28.Mendes TA, Reis-Cunha JL, de Almeida LR, Rodrigues LG, Lemos LD, dos Santos AR, da Câmara AC, Galvão LM, Bern C, Gilman RH, Fujiwara RT, Gazzinelli RT, Bartholomeu DC. Identification of strain-specific B-cell epitopes in Trypanosoma cruzi using genome-scale epitope prediction and high-throughput immunoscreening with peptide arrays. Plos Negl Trop Dis. 2013; 7(10): e2524 doi: 10.1371/journal.pntd.0002524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhattacharyya T, Falconar AK, Luquetti AO, Costales JA, Grijalva MJ, Lewis MD, Messenger LA, Tran TT, Ramirez JD, Guhl F, Carrasco HJ, Diosque P, Garcia L, Litvinov SV, Miles MA. Development of peptide-based lineage-specific serology for chronic Chagas disease: geographical and clinical distribution of epitope recognition. Plos Negl Trop Dis. 2014;8(5): e2892 doi: 10.1371/journal.pntd.0002892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhattacharyya T, Mills EA, Jansen AM, Miles MA. Prospects for T. cruzi lineage-specific serological surveillance of wild mammals. Acta Trop. 2015;151: 182–6. doi: 10.1016/j.actatropica.2015.06.017 [DOI] [PubMed] [Google Scholar]
- 31.Alessio GD, Côrtes DF, Machado-de-Assis GF, Sales-Júnior PA, Ferro EA, Antonelli LR, Teixera-Carvalho A, Martins-Filho OA, Lana M. Inovations in diagnosis and post-therapeutic monitoring of Chagas disease: Simultaneous flow cytometric detection of IgG1 antibodies anti-live amastigote, anti-live trypomastigote, and anti-fixed epimastigote forms of Trypanosoma cruzi. Journal of Immunological Methods. 2014; 413: 32–44. doi: 10.1016/j.jim.2014.07.005 [DOI] [PubMed] [Google Scholar]
- 32.Alessio GD, Araújo FF, Côrtes DF, Sales-Júnior PA, Lima DC, Gomes MS, Do Amaral LR, Xavier MAP, Teixeira-Carvalho A, Martins-Filho OA, Lana M. Performance of TcI/TcVI/TcII Chagas-Flow ATE-IgG2a for universal and genotype-specific serodiagnosis of Trypanosoma cruzi infection. Plos Negl Trop Dis. 2017; 11 (3): e0005444 doi: 10.1371/journal.pntd.0005444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Federeci EE, Albemann WH, Neva FA. Chronic and progressive myocarditis and myositis in C3H mice infected with Trypanosoma cruzi. Am J Trop Med Hyg. 1964; 13: 272–80. [DOI] [PubMed] [Google Scholar]
- 34.Brener Z, Chiari E. Morphological variations observed in different strains of Trypanosoma cruzi. Rev Inst Med Trop São Paulo. 1963; 5: 220–4. [PubMed] [Google Scholar]
- 35.Pereira da Silva LH, Nussenzweig. Sobre uma cepa de Trypanosoma cruzi altamente virulenta para o camundongo branco. Folia Clin Biol. 1953;20: 191–208. [Google Scholar]
- 36.Lages-Silva E, Ramírez LE, Pedrosa AL, Crema E, Da Cunha GL, Junho PS, Macedo AM, Chiari E. Variability of kinetoplast DNA gene signatures of Trypanosoma cruzi II strains from patients with different clinical forms of Chagas' disease in Brazil. J Clin Microbiol. 2006;44: 2167–71. doi: 10.1128/JCM.02124-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramírez JD, Guhl F, Rendón LM, Rosas F, Marin-Neto JA, Morillo CA. Chagas cardiomyopathy manifestations and Trypanosoma cruzi genotypes circulating in chronic Chagasic patients. PLoS Negl Trop Dis. 2010;4(11): e899 doi: 10.1371/journal.pntd.0000899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bosseno MF, Telleria J, Vargas F, Yaksic N, Noireau F, Morin A, Brenière SF. Trypanosoma cruzi: study of the distribution of two widespread clonal genotypes in Bolivian Triatoma infestans vectors shows a high frequency of mixed infections. Exp.Parasitol. 1996; 83: 275–282. doi: 10.1006/expr.1996.0075 [DOI] [PubMed] [Google Scholar]
- 39.Brenière SF, Bosseno MF, Telleria J, Bastrenta B, Yacsik N, Noireau F, Alcazar JL, Barnabé C, Wincker P, Tibayrenc M. Diferent behavior of two T. cruzi major clones: transmission and circulation in young Bolivian patients. Experimental Parasitology. 1998; 89: 285–295. doi: 10.1006/expr.1998.4295 [DOI] [PubMed] [Google Scholar]
- 40.Solari A, Campilla YR, Ortiz S, Wallace A. Identification of Trypanosoma cruzi genotypes circulating in Chilean chagasic patients. Experimental Parasitology. 2001; 97: 226–233. doi: 10.1006/expr.2001.4607 [DOI] [PubMed] [Google Scholar]
- 41.Lehane MJ, MsangI AR, Whitaker CJ, Lehane SM. Grouping of trypanosome species in mixed infections in Glossina pallidipes. Parasitology. 2000; 120: 583–592. [DOI] [PubMed] [Google Scholar]
- 42.Monje-rumi MM, Brandán CP, Ragone PG, Tomasini N, Lauthier JJ, Alberti D’ Amato AM, Cimino RO, Orellana V, Basombrío MA, DIOSQUE P. Trypanosoma cruzi diversity in the Gran Chaco: mixed infections and differential host distribution of TcV and TcVI. Infect. Genet. Evol. 2015; 29: 53–9. doi: 10.1016/j.meegid.2014.11.001 [DOI] [PubMed] [Google Scholar]
- 43.Botero LA, Mejia AM, Triana O. Biological and genetic characterization of two Colombian clones of Trypanosoma cruzi groups I and II. 2007; Biomedica 27 (Suppl 1):64–4. [PubMed] [Google Scholar]
- 44.Krettli AU, Weisz-Carrington P, Nussenzweig RS. Membrane-bound antibodies to bloodstream Trypanosoma cruzi in mice: strain differences in susceptibility to complement-mediated lysis. Clin Exp Immunol. 1979; 37(3): 416–23. [PMC free article] [PubMed] [Google Scholar]
- 45.Wallace A, Sanchez G, Venegas J, Solari A. Lack of cross-reactivity of lytic antibodies with bloodstream forms of Trypanosoma cruzi zymodemes generated in a mouse experimental model. Exp Parasitol. 1995;80(2): 176–85. doi: 10.1006/expr.1995.1022 [DOI] [PubMed] [Google Scholar]
- 46.Andrade SG, Rassi A, Magalhães JB, Ferriolli FF, Luquetti AO. Specific chemotherapy of Chagas disease: a comparison between the response in patients and experimental animals inoculated with the same strains. Trans R Soc Trop Med Hyg. 1992; 86: 624–626. [DOI] [PubMed] [Google Scholar]
- 47.Toledo MJ, Bahia MT, Carneiro CM, Martins-Filho OA, Tibayrenc M, Barnabe C, Tafuri WL, Lana M. Chemotherapy with benznidazole and itraconazole for mice infected with different Trypanosoma cruzi clonal genotypes. Antimicrob Agents Chemother. 2003;47: 223–230. doi: 10.1128/AAC.47.1.223-230.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toledo MJ, Bahia MT, Veloso VM, Carneiro CM, Machado-Coelho GL, Alves CF, Martins HR, Cruz RE, Tafuri WL, Lana M. Effects of specific treatment on parasitological and histopathological parameters in mice infected with different Trypanosoma cruzi clonal genotypes. J Antimicrob Chemother. 2004; 53(6): 1045–53. doi: 10.1093/jac/dkh224 [DOI] [PubMed] [Google Scholar]
- 49.Brener Z. Biology of Trypanosoma cruzi. Annu. Rev. Microbiol. 1973; 27: 347–382. doi: 10.1146/annurev.mi.27.100173.002023 [DOI] [PubMed] [Google Scholar]
- 50.Dias JCP. Epidemiological surveillance of Chagas disease. Caderno de Saúde Pública. 2000; 16: 43–59. [PubMed] [Google Scholar]
- 51.Rowland EC, Mikhail KS , Cormick TS. Isotype determination of anti-Trypanosoma cruzi antibody in murine Chagas' disease. J Parasitol. 1992; 78(3): 557–61. [PubMed] [Google Scholar]
- 52.Brener Z, Gazzinelli RT. Immunological control of Trypanosoma cruzi infection and pathogenesis of Chagas’ disease. Int. Arch. Allergy Immunol. 1997; 114:103–110. doi: 10.1159/000237653 [DOI] [PubMed] [Google Scholar]
- 53.Cordeiro FD, Martins-Filho AO, Rocha MOC, Adad SJ, Correa Oliveira R, Romanha AJ. Anti-Trypanosoma cruzi immunoglobulin G1 can be a useful tool for diagnosis and prognosis of human Chagas disease. Clinical and Diagnostic Laboratory Immunology. 2001; 8:112–118. doi: 10.1128/CDLI.8.1.112-118.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Santos DM, Talvani A, Guedes PM, Machado-Coelho GL, De Lana M, Bahia MT. Trypanosma cruzi: Genetic diversity influences the profile of immunoglobulins during experimental infection. Exp. Parasitol. 2009; 121(1):8–14. doi: 10.1016/j.exppara.2008.09.012 [DOI] [PubMed] [Google Scholar]
- 55.Verani JR, Seitz A, Gilman RH, Lafuente C, Galdos-Cardenas G, Kawai V, De Lafuente E, Ferrufino L, Bowman NM, Pinedo-Cancino V, Levy MZ, Todd CW, Kirchhoff LV, Cabrera L, Verastegui M, Bern C. Geographic variation in the sensitivity of recombinant antigen-based rapid tests chronic Trypanosoma cruzi infection. Am J Trop Med Hyg. 2009;80(3): 410–415. [PubMed] [Google Scholar]
- 56.Krettli AU, Brener Z. Resistance against Trypanosoma cruzi associated to anti-living trypomastigote antibodies. Journal of Immunology. 1982; 128: 2009–2012. [PubMed] [Google Scholar]
- 57.Andrade SG, Campos RF, Steindel M, Guerreiro ML, Magalhães JB, Almeida MC, Reis JN, Santos VC, Valadares HM, Reis MG, Macedo AM. Biological, biochemical and molecular features of Trypanosoma cruzi strains isolated from patients infected through oral transmission during a 2005 outbreak in the state of Santa Catarina, Brazil: its correspondence with the new T. cruzi Taxonomy Consensus (2009). Mem Inst Oswaldo Cruz. 2011; 106(8): 948–56. [DOI] [PubMed] [Google Scholar]
- 58.Sá AR, Dias GB , Kimoto KY , Steindel M , Grisard EC , Toledo MJ , Gomes ML. Genotyping of Trypanosoma cruzi DTUs and Trypanosoma rangeli genetic groups in experimentally infected Rhodnius prolixus by PCR-RFLP. Acta Trop. 2016; 156: 115–21. doi: 10.1016/j.actatropica.2016.01.006 [DOI] [PubMed] [Google Scholar]
- 59.Burgos JM, Altchen J, Bisio M, Duffy T, Valadares HM, Seidenstein ME, Piccinali R, Freitas JM, Levin MJ, Macchi L, Macedo AM, Freilij H, Schijman AG. Direct molecular profiling of minicircle signatures and lineages of Trypanosoma cruzi bloodstream populations causing congenital Chagas disease. Int J Parasitol. 2007; 37(12): 1319–27. doi: 10.1016/j.ijpara.2007.04.015 [DOI] [PubMed] [Google Scholar]
- 60.Freitas JM, Augusto-Pinto L, Pimenta JR, Bastos-Rodrigues L, Gonçalves VF, Teixeira SMR, Chiari E, Junqueira ACV, Fernandes O, Macedo AM, Machado CR, Pena SDJ. Ancestral genomes, sex, and the population struture of Trypanosoma cruzi. PLOS Pathogens. 2006; 2: 226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Diez C, Lorenz V, Ortiz S, Gonzalez V, Racca A, Bontempi I, ManattinI S, Solari A, Marcipar I. Genotyping of Trypanosoma cruzi sublineage in human samples from a NorthEast Argentinaarea by hybridization with DNA probes and specific polymerase chain reaction (PCR). Am J Trop Med Hyg. 2010; 82(1): 67–73. doi: 10.4269/ajtmh.2010.09-0391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ortiz S, Zulantay I , Apt W , Saavedra M , Solari A. Transferability of Trypanosoma cruzi from mixed human host infection to Triatoma infestans and from insects to axenic culture. Parasitol Int. 2015; 64(1): 33–6. doi: 10.1016/j.parint.2014.09.005 [DOI] [PubMed] [Google Scholar]
- 63.Cura CI, Duffy T, Lucero RH, Bisio M, Péneau J, Jimenez-Coello M, et al. Multiplex real-time PCR assay using TaqMan probes for the identification of Trypanosoma cruzi DTUs in biological and clinical samples. Plos Negl Trop Dis. 2015; 9(5): 1371–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Consentino RO, Aguero F. A Simple Strain Typing Assay for Trypanosoma cruzi: Discrimination of Major Evolutionary Lineages from a Single Amplification Product. PLoS Negl Trop Dis. 2012; 6(7): 1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miles MA, Llewellyn MS, Lewis MD, Yeo M, Baleela R, Fitzpatrick S, Gaunt MW, Mauricio IL. The molecular epidemiology and phylogeography of Trypanosoma cruzi and parallel research on Leishmania: looking back and to the future. Parasitology. 2009;136(12): 1509–28. doi: 10.1017/S0031182009990977 [DOI] [PubMed] [Google Scholar]
- 66.Guedes PM, Veloso VM, Tafuri WL, Galvão LM, Carneiro CM, Lana M, Chiari E, Ataide SK, Bahia. The dog as model for chemotherapy of the Chagas´s disease. Acta Trop. 2002;84(1): 9–17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Schematic flow chart overview of the experimental procedure Chagas-Flow ATE-IgG2a. Representative gating strategies to select the target antigens (amastigote-AMA = green, trypomastigote-TRYPO = red and epimastigote-EPI = yellow) and the histograms employed to quantify the percentage of positive fluorescent parasites (PPFP). (B) The compendium of the study population comprised of Swiss mice, categorized into subgroups referred as: early (yellow), late (brown), single (light gray), dual (dark gray), TcI/COL (red), TcVI/CL (green), TcII/Y (dark blue), TcI/COL+TcVI/CL (orange), TcVI/CL+TcII/Y (light blue) and TcI/COL+TcII/Y (pink). (C) Data mining approaches was achieved through the steps: Step 1 (algorithm for early & late stages); Step 2 (algorithms for single & dual infections); Step 3 (algorithm for genotypic-specific single & dual—early & late stages); Step 4 (assemblage sequential algorithm platform).
(TIF)
Discriminant analyses of combined Chagas-Flow ATE-IgG2a for genotype-specific diagnosis at (A) early and (B) late stages of T. cruzi single infection (COL, CL and Y) and dual infection (COL+CL, CL+Y and COL+Y). The global accuracy is provided in the Figure.
(TIF)
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.